This work determines that the ARMADILLO-REPEAT KINESIN1 (ARK1) of Arabidopsis accumulates at the plus ends of growing microtubules and promotes microtubule catastrophe. By inducing such events, ARK1 enables the recycling of tubulin subunits, which is important for maintaining rapid microtubule polymerization, a process that is critical for maintaining unidirectional tip growth in root hairs.
Abstract
Microtubule dynamics are critically important for plant cell development. Here, we show that Arabidopsis thaliana ARMADILLO-REPEAT KINESIN1 (ARK1) plays a key role in root hair tip growth by promoting microtubule catastrophe events. This destabilizing activity appears to maintain adequate free tubulin concentrations in order to permit rapid microtubule growth, which in turn is correlated with uniform tip growth. Microtubules in ark1-1 root hairs exhibited reduced catastrophe frequency and slower growth velocities, both of which were restored by low concentrations of the microtubule-destabilizing drug oryzalin. An ARK1-GFP (green fluorescent protein) fusion protein expressed under its endogenous promoter localized to growing microtubule plus ends and rescued the ark1-1 root hair phenotype. Transient overexpression of ARK1-RFP (red fluorescent protein) increased microtubule catastrophe frequency. ARK1-fusion protein constructs lacking the N-terminal motor domain still labeled microtubules, suggesting the existence of a second microtubule binding domain at the C terminus of ARK1. ARK1-GFP was broadly expressed in seedlings, but mutant phenotypes were restricted to root hairs, indicating that ARK1’s function is redundant in cells other than those forming root hairs.
INTRODUCTION
Precise coordination of microtubule dynamics and organization is necessary for normal plant cell development, growth, and shape. Microtubules contribute to root hair morphogenesis by establishing and maintaining the polar growth process (Sieberer et al., 2002). Root hairs are tubular extensions of epidermal cells that protrude perpendicularly from the root and grow through a highly polarized process called tip growth in which the plasma membrane and cell wall components are asymmetrically secreted at the root hair apex (Carol and Dolan, 2002). Microtubule depolymerization or hyperstabilization by treatment with the drugs oryzalin and taxol, respectively, can lead to wavy and branched root hairs, stressing the importance of microtubules in focusing tip growth (Bibikova et al., 1999). Root hairs of Arabidopsis thaliana possess two types of microtubule populations: cortical microtubules found at the cortex and endoplasmic microtubules located within the transvacuolar strands and cytoplasmic-rich zone near the root hair apex (Sieberer and Timmers, 2009). Live-cell imaging of the green fluorescent protein fused to a microtubule binding domain (GFP-MBD) reporter protein in Arabidopsis plants showed that cortical microtubules are present throughout all developmental stages of the root hair, while endoplasmic microtubules are only present in elongating hairs (Van Bruaene et al., 2004).
Microtubules are dynamic polymers made up of heterodimeric tubulin subunits that self-assemble into a polar cylindrical polymer with a less dynamic minus end and a more dynamic plus end. The stochastic addition or removal of tubulin subunits from the microtubule ends causes the microtubule to either grow/polymerize or shrink/depolymerize, respectively (Desai and Mitchison, 1997). Microtubule dynamics and organization are achieved through the activity of microtubule-associated proteins (MAPs) such as the Arabidopsis MICROTUBULE ORGANIZATION1 protein that promotes microtubule dynamics by aiding in microtubule polymerization and depolymerization (Kawamura and Wasteneys, 2008). In eukaryotes, the highly conserved microtubule-based motor proteins called kinesins can, in addition to transporting cargo along microtubules, also modulate microtubule dynamics and organization. Thus far for Arabidopsis, in which 61 putative kinesins have been identified, the majority of those characterized have been shown to organize interphase and mitotic microtubule arrays through bundling, cross-linking, and translocation of other microtubules. A smaller proportion of Arabidopsis kinesins are related to organelle and vesicle motility along microtubules (Reddy and Day, 2011; Zhu and Dixit, 2012). To date, Kinesin-13A is the only kinesin shown to be a catastrophe factor. It acts specifically to promote microtubule depolymerization in the secondary cell wall pits of developing xylem cells (Oda and Fukuda, 2013). The depolymerizing function of Kinesin-13A is similar to that of its homologs in other organisms (Desai et al., 1999; Oda and Fukuda, 2013). Kinesins can also organize microtubules in tip-growing cells such as caulonemal cells of the moss Physcomitrella patens. The KINID1a and KINID1b kinesins in these cells are responsible for the bundling of microtubule plus ends. Loss of both these kinesins leads to reduced microtubule bundling and aberrant tip growth (Hiwatashi et al., 2014)
The ARMADILLO-REPEAT KINESIN1 (ARK1), originally named MORPHOGENESIS OF ROOT HAIR2 (MRH2) (Jones et al., 2006), is thought to participate in microtubule dynamics. Because ARK1 is an uncharacterized kinesin belonging to an ungrouped plant-specific family of kinesins, its function cannot be predicted based on phylogeny (Reddy and Day, 2011; Zhu and Dixit, 2012). However, with previous evidence showing that ark1 mutants have root hairs with increased endoplasmic microtubule abundance and wavy/branched morphologies, it has been suggested that ARK1 specifically promotes microtubule depolymerization in root hairs (Sakai et al., 2008; Yoo et al., 2008).
A single ARK1 polypeptide has an N-terminal catalytic motor domain that interacts with microtubules through the binding and hydrolysis of ATP (Sakai et al., 2008). Much like most conventional kinesins, ARK1 is predicted to function as a homodimer, as evidenced by its internal coiled-coil domain. The C terminus of ARK1 is composed of three tandem Armadillo (ARM) repeat domains that give rise to the kinesin’s name (Sakai et al., 2008). An ARM repeat forms three α-helices and, when in tandem, forms a superhelix that allows for its interaction with other proteins. ARM repeats have been shown to function in essential cellular processes related to signaling, the cytoskeleton, and protein-protein interactions in a variety of eukaryotes (Tewari et al., 2010), but their function in ARK1 is still unclear. Interestingly, previous studies using in vitro analyses have shown that the ARM domain of ARK1 is able to bind to actin (Yang et al., 2007) as well as the Never in Mitosis, Gene A (NIMA)-related kinase, NEK6 (Sakai et al., 2008). ARK1 is also thought to play a pivotal role in the tightly coordinated tip growth signaling pathway as a potential linker among the cytoskeletal, endomembrane, and GTPase signaling components (Yoo et al., 2008; Yoo and Blancaflor, 2013).
Although previous studies have confirmed ARK1’s ability to bind microtubules through fluorescent protein tagging (Yoo et al., 2008) and in vitro microtubule binding (Yang et al., 2007), its function in modulating microtubule dynamics and/or organization has not been confirmed. In this study, we use genetic, chemical, and overexpression analyses to show that ARK1 is a kinesin with plus-end tracking properties that promotes microtubule depolymerization.
RESULTS
Microtubule Catastrophe Frequency and Growth Rates Are Reduced in ark1-1 Mutants
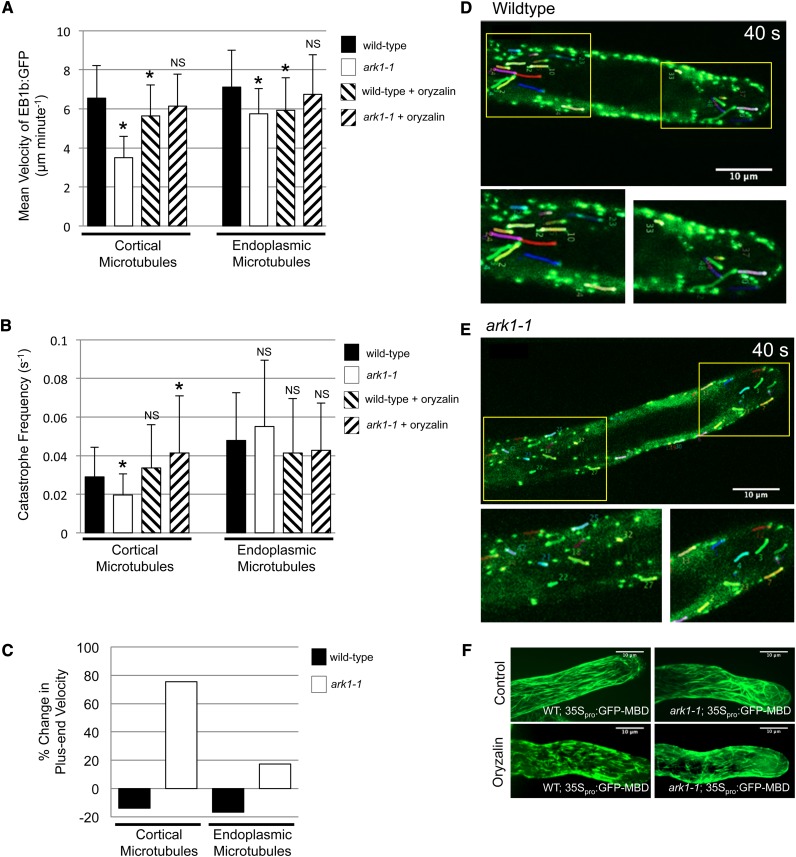
Based on the fact that microtubules are more abundant in the T-DNA insertional mutant ark1-1 (SALK_035063) (Sakai et al., 2008), we hypothesized that microtubule dynamics would be altered. In order to test this hypothesis, we quantified microtubule plus-end growth rates as well as the frequency at which microtubules undergo catastrophe by tracking the microtubule plus-end tracking protein END BINDING 1b (EB1b). This reporter was tagged with green fluorescent protein (GFP) and expressed using the 35S cauliflower mosaic virus promoter (35Spro:EB1b-GFP); thus, we were able to visualize microtubule plus-ends. EB1b-GFP movement was measured in wild-type and ark1-1 elongating root hairs by spinning-disc confocal microscopy. Both endoplasmic and cortical microtubule mean growth velocities were significantly reduced in ark1-1 (cortical microtubule [CMT], 3.5 ± 1.1 μm/min; endoplasmic microtubule [EMT], 5.7 ± 1.3 μm/min) relative to the wild type (CMT, 6.5 ± 1.7 μm/min; EMT, 7.1 ± 1.9 μm/min; two-sample unequal variance t test: CMT, P < 10−77; EMT, P < 10−05) (Figures 1A, 1D, and 1E, Table 1; Supplemental Figure 1 and Supplemental Movies 1 and 2). The frequency of cortical microtubule catastrophe was significantly reduced by 31% in ark1-1 (0.020 ± 0.011 events/second) relative to wild-type root hairs (0.029 ± 0.015 events/second; two-sample unequal variance t test: P < 10−09) (Figure 1B, Table 1). In contrast, we found no significant difference in catastrophe frequency between wild-type (0.048 ± 0.025 events/second) and ark1-1 endoplasmic microtubules (0.055 ± 0.034 events/second) (Figure 1B, Table 1) despite a greater abundance of endoplasmic microtubules in ark1-1 root hairs (Supplemental Figures 2A to 2C).
Figure 1.
Cortical Microtubule Plus-End Velocities and Catastrophe Frequencies Are Reduced in ark1-1 Root Hairs but Can Be Rescued by Oryzalin Treatment.
(A) Plus-end velocities of EB1b-GFP of CMTs and EMTs were measured in wild-type and ark1-1 elongating root hairs upon imaging with spinning-disc confocal microscopy. The mean ark1-1 microtubule plus-end velocities (CMT, 3.5 ± 1.1 μm/min, n = 111; EMT, 5.7 ± 1.3 μm/min, n = 85) are significantly reduced relative to wild-type microtubules (CMT, 6.5 ± 1.7 μm/min, n = 209; EMT, 7.1 ± 1.9, n = 149). Exposure to 100 nM oryzalin rescued the microtubule plus-end velocities of ark1-1 root hairs (CMT, 6.1 ± 1.6 μm/min, n = 47; EMT, 6.7 ± 2.0 μm/min, n = 106) so that there was no significant difference from oryzalin-free wild-type root hairs (CMT, 6.5 ± 1.7 μm/min, n = 209; EMT, 7.1 ± 1.9 μm/min, n = 149). Oryzalin treatment significantly reduced wild-type velocities (CMT, 5.6 ± 1.6 μm/min, n = 134; EMT, 5.9 ± 1.7 μm/min, n = 76). A minimum of five roots (for untreated) and three roots (for oryzalin-treated) (five root hairs per root) from each genetic background were imaged.
(B) Catastrophe frequencies of cortical and endoplasmic microtubules were measured in the same elongating root hairs as in (A). Cortical microtubule catastrophe frequencies in ark1-1 (0.020 ± 0.011 events/second) are significantly reduced relative to wild-type microtubules (0.029 ± 0.015 events/second). There is no significant difference for endoplasmic microtubules (wild type, 0.048 ± 0.025 events/second; ark1-1, 0.055 ± 0.034 events/second). Exposure to 100 nM oryzalin increased the catastrophe frequency of cortical microtubules in ark1-1 root hairs (0.041 ± 0.03 events/second) relative to untreated wild-type root hairs (0.029 ± 0.015 events/second). The same microtubules in (A) were used to calculate the catastrophe frequency.
(C) Exposure to 100 nM oryzalin reduced microtubule plus-end velocities in wild-type root hairs but increased velocities in ark1-1 root hairs.
(D) and (E) Confocal micrographs of EB1b-GFP in wild-type (D) and ark1-1 (E) root hairs showing both cortical and endoplasmic microtubule plus ends. The colored lines represent the EB1b-GFP trajectories after 40 s with the wild type showing faster EB1b-GFP movement than ark1-1. Images represent the medial plane of the root hair. Magnified images of the trajectories are seen below and the yellow boxes indicate the area where the magnified images were selected. For the full montage, refer to Supplemental Figure 1. Images are representations of Supplemental Movies 1 and 2 online. Bars =10 μm.
(F) Spinning-disc laser confocal micrographs of root hairs expressing the 35Spro:GFP-MBD microtubule marker in the wild type and ark1-1. No differences were noticeable between the microtubules of wild-type and ark1-1 root hairs treated with 100 nM oryzalin. Images are merged Z-projections of the entire root hair stack.
Data and bars are represented as means ± sd, respectively. Asterisk shows a significant reduction, while NS indicates no significant difference in velocity relative to untreated wild-type root hairs using a two-sample t test with unequal variance (α = 0.01).
Table 1. Plus-End Growth Velocities and Catastrophe Frequencies of the Wild Type and ark1-1 in Untreated and 100 nM Oryzalin-Treated Root Hairs.
| CMTs | EMTs | |||
|---|---|---|---|---|
| Wild Type | ark1-1 | Wild Type | ark1-1 | |
| Plus-end velocity (μm min−1) | 6.54 ± 1.66 | 3.39 ± 1.09a | 7.11 ± 1.90 | 5.74 ± 1.29b |
| With oryzalin | 5.63 ± 1.58a | 6.14 ± 1.64a | 5.93 ± 1.66b | 6.74 ± 2.03d |
| Catastrophe frequency (events s−1) | 0.029 ± 0.015 | 0.020 ± 0.0118a | 0.048 ± 0.024 | 0.055 ± 0.034d |
| With oryzalin | 0.034 ± 0.022c | 0.041 ± 0.030a | 0.041 ± 0.028d | 0.043 ± 0.024d |
The decreased frequency of microtubule catastrophe in ark1-1 mutants is consistent with the increased microtubule polymer mass and the occurrence of microtubule bundling. The slower rate of microtubule polymerization, however, is more difficult to explain because decreased polymerization rates should theoretically reduce the polymer mass. Given that microtubule polymerization rates are proportional to free tubulin concentrations, we considered the possibility that the reduced polymerization rates in ark1-1 are the consequence of a reduced free tubulin concentration, which is likely to result from the reduced catastrophe frequency. We hypothesized that increasing the availability of free tubulin should restore normal microtubule growth rates in ark1-1. To achieve this, we applied low concentrations of the microtubule destabilizing drug oryzalin and measured microtubule growth and catastrophe rates using the EB1b-GFP reporter.
Exposure to 100 nM oryzalin increased the ark1-1 plus-end growth rates (as measured by EB1b-GFP) such that they were equivalent to the oryzalin-free wild-type growth rate in both cortical and endoplasmic microtubule populations (Figures 1A and 1C, Table 1). Microtubules in wild-type root hairs, in contrast, showed reduced plus-end growth rates when exposed to 100 nM oryzalin (Figures 1A and 1C, Table 1), which is consistent with previously published data (Nakamura et al., 2004).
The cortical microtubule catastrophe frequency in 100 nM oryzalin-treated ark1-1 root hairs was increased relative to that of untreated wild-type and ark1-1 cells. The dynamics of endoplasmic microtubules in oryzalin-treated ark1-1 root hairs and untreated wild-type root hairs were not significantly different (Figure 1B, Table 1).
Despite a clear difference in plus-end growth velocities of oryzalin-treated wild-type and ark1-1 microtubules as assessed with EB1b-GFP, both genotypes exposed to the same oryzalin concentrations had fragmented microtubules that looked indistinguishable from each other when using the 35Spro:GFP-MBD (MBD is the microtubule binding domain of MAP4) marker (Figure 1F). This observation possibly resulted from increased resistance to microtubule destabilizing drugs and bundling typically associated with the 35Spro:GFP-MBD marker (Marc et al., 1998; Lechner et al., 2012).
Low Concentrations of Oryzalin Partially Rescue the ark1-1 Root Hair Morphology Phenotype
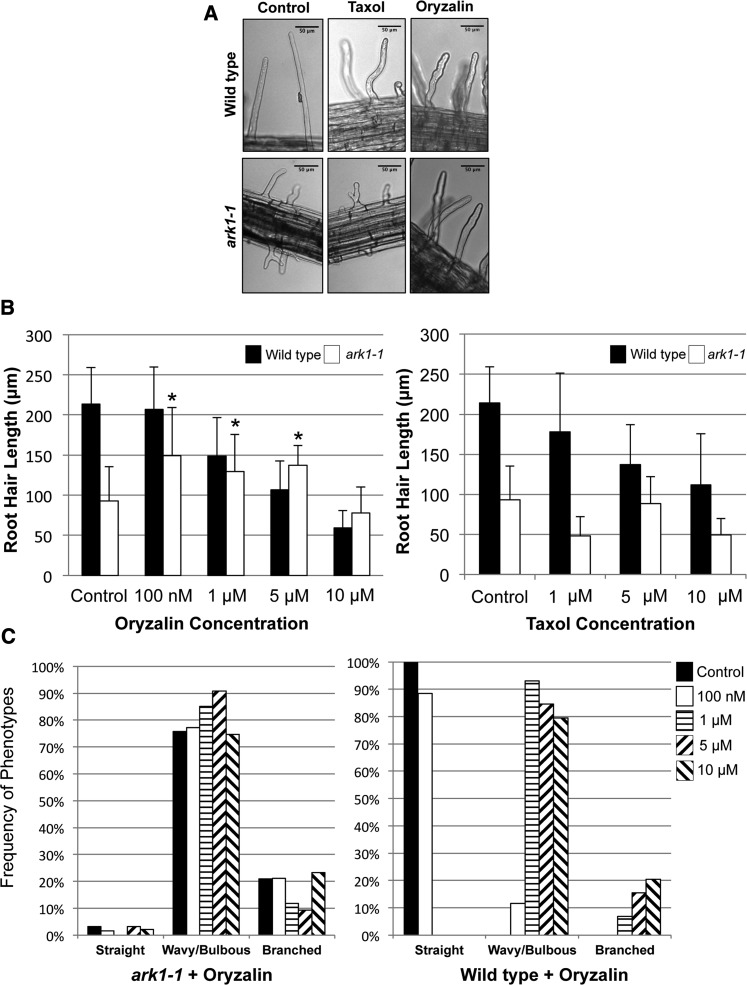
Based on the restoration of wild-type microtubule growth rates in the ark1-1 mutant by the oryzalin treatments, we hypothesized that these treatments would ameliorate the ark1-1 root hair morphological defects. It was previously shown that wavy and branched root hair phenotypes were indicative of a less severe and more severe loss of root hair polarity, respectively, and that the severity of polarity loss was positively correlated with increasing oryzalin and taxol concentrations (Bibikova et al., 1999). Exposure to 100 nM, 1 μM, and 5 μM oryzalin partially rescued the ark1-1 root hair phenotype by significantly increasing the root hair length relative to untreated ark1-1 root hairs (Figures 2A and 2B). Moreover, the frequency of the most severe root hair phenotype (branched) appeared to decrease with the less severe phenotype (wavy and bulbous) increasing in frequency at 1 and 5 μM oryzalin concentrations (Figures 2A and 2C). In the wild type, these same oryzalin concentrations decreased root hair length and led to wavy and branched root hairs that resembled the ark1-1 phenotype (Figures 2A and 2C). Oryzalin (10 μM) was unable to partially rescue the ark1-1 phenotype, suggesting a saturation effect of oryzalin on microtubules.
Figure 2.
Oryzalin Partially Rescues the ark1-1 Root Hair Morphology.
(A) Bright-field images of wild-type and ark1-1 root hairs treated with either 5 μM taxol or oryzalin. The root hair phenotype of ark1-1 is partially rescued with oryzalin but not with taxol. Bars = 50 μm.
(B) Mean length of wild-type and ark1-1 root hairs treated with various concentrations of taxol and oryzalin. ark1-1 exposed to 100 nM, 1 μM, and 5 μM oryzalin partially rescues the root hair length phenotype. Data and bars represent mean and sd, respectively. Asterisks represent a significant increase in mean length relative to untreated ark1-1 root hairs. A minimum of 25 root hairs were measured for each genotype and treatment.
(C) Frequency distribution histograms comparing root hair morphology types in the wild type and ark1-1 grown on varying oryzalin concentrations. Oryzalin (1 and 5 μM) partially mitigates the ark1-1 root hair defect by shifting the distribution of the most severe branched phenotype to a milder wavy/bulbous phenotype. As a control, wild-type root hairs show an increase loss of root hair polarity upon exposure to increasing oryzalin concentrations. A minimum of 10 roots and 150 root hairs were measured for each treatment.
In contrast to the oryzalin treatments, taxol, which promotes microtubule polymerization, further impaired root hair elongation in both wild-type and ark1-1 root hairs (Figures 2A and 2B) and increased root hair waving/branching (Figure 2A).
ARK1 Accumulates on Plus Ends of Growing Microtubules
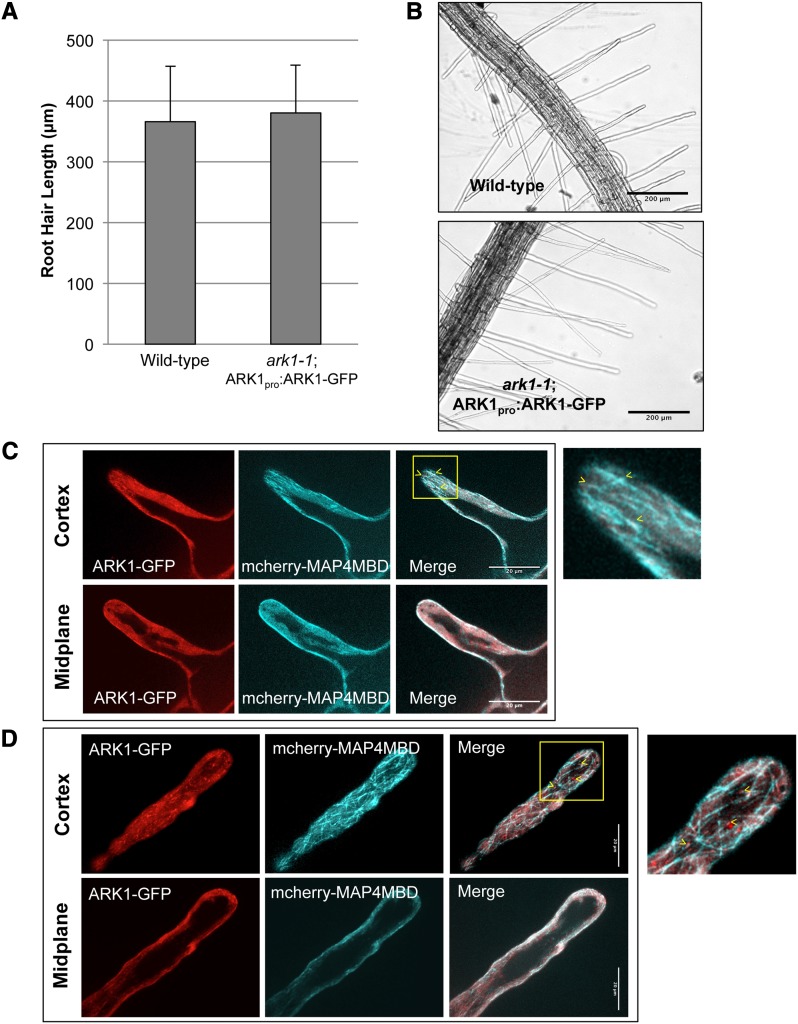
In order to see where ARK1 is localized on the microtubule, we made an ARK1pro:ARK1-GFP construct using the entire ARK1 genomic sequence including the putative 5′-end promoter region (888 bp upstream of the ARK1 start codon). ark1-1 plants expressing 35Spro:mCherry-MAP4MBD were transformed with the ARK1pro:ARK-GFP construct and a T3 homozygous line for the ARK1-GFP transgene was used for further analysis. ARK1pro:ARK1-GFP expression rescued the ark1-1 root hair phenotype (Figures 3A and 3B), indicating that the GFP-tagged ARK1 protein was fully functional.
Figure 3.
ARK-GFP Rescues the ark1-1 Root Hair Phenotype and Localizes to Microtubules in Elongating Root Hairs.
(A) Root hair lengths of ark1-1; ARK1pro:ARK1-GFP and the wild type are not significantly different. Data and bars represent mean and sd, respectively. A minimum of 110 root hairs were measured.
(B) Differential interference contrast images of wild-type and ark1-1; ARK1pro:ARK1-GFP root hairs. Bars = 200 μm.
(C) Laser confocal micrographs of ark1-1; ARK1pro:ARK1-GFP elongating root hairs. ARK1-GFP (indicated in red) localizes to microtubules (mCherry-MAP4MBD indicated in cyan). The cortex and the midplane of the root hairs are both shown. Arrowheads point to ARK1-GFP localization on microtubules. The right of the figure shows the magnified representation of the yellow-boxed area in the merged image. Images are representations of Supplemental Movie 3. Bar = 20 μm.
(D) Laser confocal micrographs of ark1-1; ARK1pro:ARK1-GFP fully grown root hairs. ARK1-GFP (indicated in red) is still expressed in fully grown root hairs and localizes to microtubules (indicated in cyan). The cortex and the midplane of the root hairs are both shown. Arrowheads point to ARK1-GFP localization on microtubules. The right of the figure shows the magnified representation of the yellow-boxed area in the merged image. Images are representations of Supplemental Movie 4. Bar = 20 μm.
ARK1-GFP was associated with microtubules and expressed in elongating and fully grown root hairs (Figures 3C and 3D; see Supplemental Movies 3 and 4 for z-stacks of ARK1-GFP in root hairs), although characterization of ARK1-GFP on microtubules was difficult to analyze in root hairs due to high cytoplasmic fluorescence of ARK1-GFP. However, we were able to observe ARK1-GFP expression in other cell types, including root atrichoblasts as well as epidermal cells of the hypocotyl, petiole, cotyledons, and root tip, which is consistent with previous ARK1 gene expression analysis (Figure 4A) (Yang et al., 2007; Sakai et al., 2008).
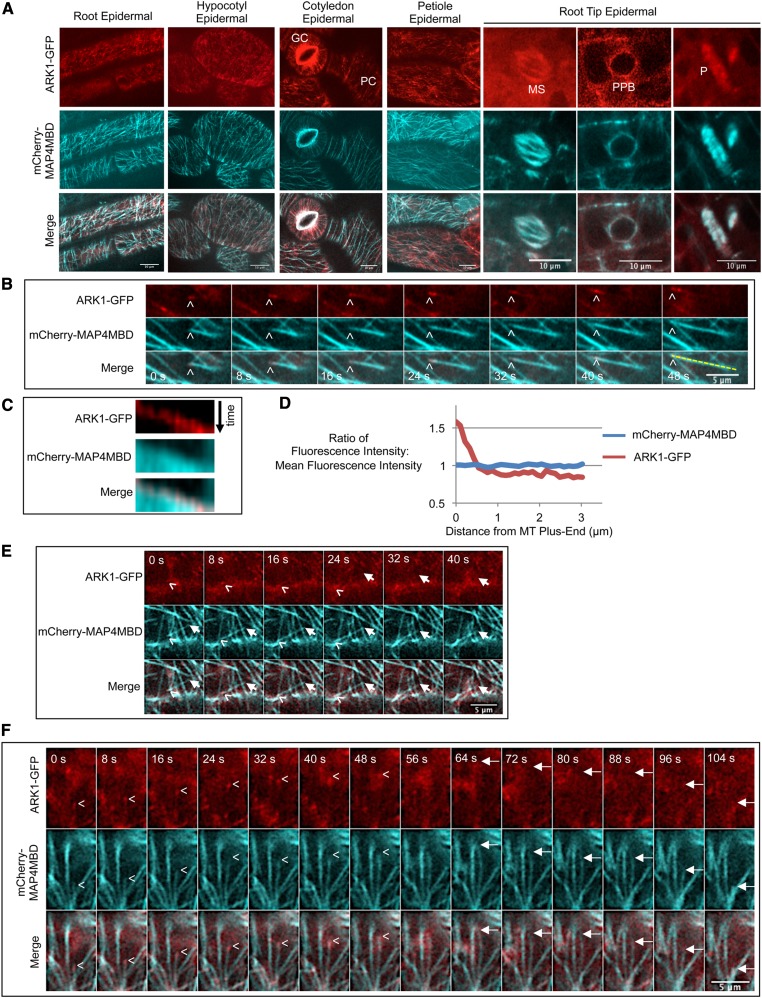
Figure 4.
ARK1-GFP Is Expressed in Non-Root Hair Cells and Accumulates on Growing Microtubule Plus Ends.
(A) ARK1-GFP is expressed in epidermal cells of the root, hypocotyl, cotyledon, petiole, and root tip. ARK1-GFP was coexpressed in ark1-1 35Spro:mCherry-MAP4MBD plants. GC, guard cell; PC, pavement cell; MS, mitotic spindle; PPB, preprophase band; P, phragmoplast. Bars = 10 μm.
(B) A time-lapse montage showing ARK1-GFP accumulating on microtubule plus ends (mCherry-MAP4MBD) in cotyledon epidermal cells. Arrowheads indicate the growing microtubule plus end. Time for each frame is indicated. Bar = 5 μm.
(C) A kymograph of the yellow line scan in the last panel of (B) showing ARK1-GFP moving along growing microtubules.
(D) Graph showing that ARK1-GFP accumulates at the plus end of microtubules but still remains bound to the sidewall of microtubules. Lines represent the ratio between fluorescence intensity of one point to the mean fluorescence intensity of one line scan. The ratio was highest within 1 μm of the microtubule plus end but decreased further from the plus end. As a control, the same ratio was calculated for microtubules with mCherry-MAP4MBD. Forty-six line scans were used to measure ARK1-GFP, and 26 line scans were used to measure mCherry-MAP4MBD.
(E) A time-lapse montage showing that ARK1-GFP localizes specifically to plus ends but not minus ends. Arrowheads indicate a growing microtubule plus end. Arrows label the microtubule minus end that is depolymerizing. Bars = 5 μm.
(F) A time-lapse montage showing that ARK1-GFP does not localize to shrinking microtubule plus ends. The microtubule plus end grows (indicated by the arrowhead) and eventually depolymerizes (indicated by arrow). ARK1-GFP disappears upon microtubule depolymerization. Bars = 5 μm.
In epidermal cells of cotyledons, we observed ARK1-GFP plus-end tracking on growing microtubules (Figure 4B; Supplemental Movie 5). Time-lapse imaging and kymographic analysis revealed that ARK1-GFP accumulates at and tracks along growing microtubule plus ends (Figures 4B and 4C; Supplemental Movie 6). ARK1-GFP was also observed along the microtubule sidewalls, although the fluorescence was not as strong (Figures 4B and 4D). Quantifying the ratio of local fluorescence intensity to the mean fluorescence intensity obtained from ARK1-GFP line scans confirmed these observations. As a control, the local-to-mean fluorescence intensity ratio for the mCherry-MBD fluorescence was ∼1:1 at each point along the length of microtubules, indicative of even distribution of the fluorescent protein (Figure 4D). In contrast, ARK1-GFP showed the highest intensity ratio closest to the microtubule plus end, with the ratio sharply declining at points farther away from the tip (Figure 4D). We did not detect ARK1-GFP on the depolymerizing minus ends (Figure 4E; Supplemental Movie 7) or depolymerizing plus ends (Figure 4F; Supplemental Movie 8) of microtubules, indicating that ARK1 is predominantly found at the plus end of growing microtubules.
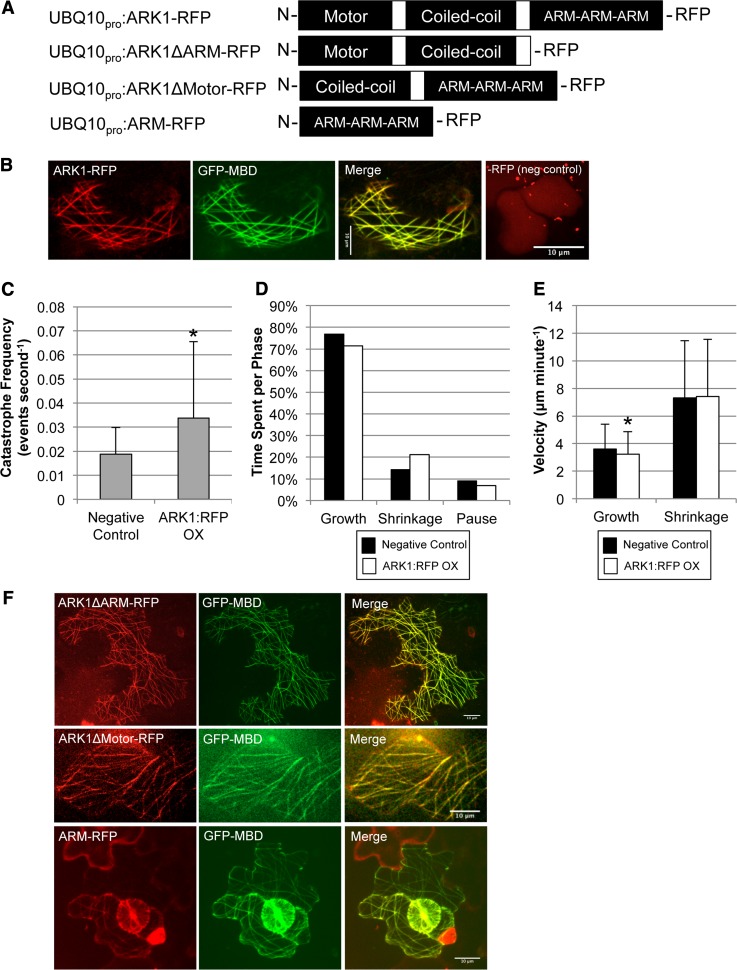
ARK1 Overexpression Increases Microtubule Catastrophe Frequency and the Amount of Time Spent in Shrinkage Phase
Based on the decreased microtubule catastrophe frequencies in ark1-1, we hypothesized that ARK1 overexpression would increase the incidence of microtubule catastrophe. In order to test this, we made an ARK1-RFP (red fluorescent protein) translational reporter construct driven by a UBIQUITIN10 promoter (Figure 5A) and transiently expressed it in ark1-1 Arabidopsis cotyledons from lines previously stably transformed to express the GFP-MBD microtubule marker under the 35S promoter. This experimental system, using the FAST technique involving Agrobacterium tumefaciens-mediated transformation of germinating seedlings (Li et al., 2009), made it possible to compare microtubule dynamics in cells overexpressing ARK1-RFP with nontransformed cells within the same cotyledon, which acts as internal negative controls. In transformed cells, ARK1-RFP appeared to label the entire length of microtubules and remained bound to both growing and shrinking microtubules (Figure 5B), indicative of the high levels of ARK1-RFP expression under the UBQ10 promoter. The cells expressing ARK1-RFP had a significantly increased catastrophe frequency (0.034 ± 0.032 events/second) relative to cells not expressing ARK1-RFP (0.019 ± 0.011 events/second; two-sample unequal variance t test: P < 0.02807) (Figure 5C). ARK1-RFP microtubules spent a greater amount of time in a shrinking phase (ARK1-RFP, 21.2%; negative control, 14.2%) (Figure 5D), yet there was no significant difference in the microtubule shrinkage velocity between cells expressing ARK1-RFP (7.4 ± 4.1 μm/min) and cells not expressing the construct (7.3 ± 4.1 μm/min) (Figure 5E). In contrast, the growth velocity of ARK1-RFP overexpressing cells (3.2 ± 1.6 μm/min) was reduced significantly (by ∼20%) relative to cells not expressing ARK1-RFP (3.6 ± 1.8 μm/min; two-sample equal variance t test: P < 2.6 × 10−5) (Figure 5E).
Figure 5.
ARK1-RFP Overexpression Leads to Increased Microtubule Catastrophe Frequencies.
The ARM domain is able to bind to microtubules. Data and bars represent the mean and sd, respectively. Asterisk represents a significant difference relative to the negative control (i.e., cells not expressing ARK1-RFP) (α = 0.05).
(A) Diagram of all the UBIQUITIN10 promoter translational fusion constructs used for transient expression in Arabidopsis cotyledons. RFP was tagged on the C terminus of all proteins.
(B) Confocal laser micrographs of the UBQ10pro:ARK1-RFP overexpression constructs in ark1-1; 35Spro:GFP-MBD plants. Negative control shows UBQ10pro:RFP expression. Bar = 10 μm.
(C) Cells transiently overexpressing ARK1-RFP have a significant increase in microtubule catastrophe frequency (0.034 ± 0.032 events/second) relative to cells not expressing ARK1-RFP (0.019 ± 0.011 events/second). For ARK1-RFP overexpression and the negative control, 26 and 42 microtubules were measured over a time period of 1700 and 2500 s, respectively.
(D) Distribution of time spent in each phase (growth, shrinkage, and pause) for cells overexpressing and not overexpressing ARK1-RFP in 35Spro:GFP-MBD plants. Microtubules in cells overexpressing ARK1-RFP spent a greater amount of time shrinking as a result of increased catastrophe frequency shown in (C). For ARK1-RFP overexpression and the negative control, 86 and 82 microtubules were visualized over a time period of 8000 and 5800 s, respectively.
(E) The mean microtubule growth and shrinkage velocities in cells overexpressing and not overexpressing ARK1-RFP. There is a significant reduction in growth velocities in ARK1-RFP overexpression cells (3.2 ± 0.1.6 μm/min) relative to the negative control (3.6 ± 1.8 μm/min). There is no significant difference in shrinkage velocities between the two treatments (ARK1-RFP, 7.4 ± 4.1 μm/min; negative control, 7.3 ± 4.2 μm/min). For each treatment, a minimum of 700 growth events and 150 shrinkage events were measured.
(F) Confocal laser micrographs of the various UBQ10pro overexpression constructs seen in (A) expressed in ark1-1; 35Spro:GFP-MBD plants. Microtubule labeling still occurs in constructs lacking the motor or ARM domain. Bars = 10 μm.
Stable transgenic lines expressing the UBQ10pro:ARK1-RFP construct could not be recovered. Some transgenic lines were identified according to antibiotic resistance, but, possibly due to posttranscriptional silencing, these lines neither displayed RFP fluorescence nor did they rescue the ark1-1 root hair phenotype. This suggests that overexpression of the ARK1 kinesin is detrimental to the cell and results in embryo lethality.
The N-Terminal Microtubule Binding Domain and the C-Terminal Armadillo-Repeat Domain Both Play a Role in ARK1 Microtubule Localization
To determine the function of the different ARK1 domains, we performed domain deletion analysis by engineering three RFP-fusion constructs driven by the UBIQUITIN10 promoter: a construct missing the C-terminal Armadillo-repeat domain (ARK1ΔARM-RFP), a construct missing the N-terminal motor domain (ARK1ΔMOTOR-RFP), and a construct with just the Armadillo-repeat domain (ARM-RFP) (Figure 5A). ARK1ΔARM-RFP showed strong microtubule labeling, confirming that the motor domain can bind microtubules (Figure 5F). Surprisingly, the ARK1ΔMOTOR-RFP (which includes the coiled-coil domain) (Figure 5F) also labeled microtubules, suggesting that the coiled-coil and/or the C-terminal ARM domain can bind microtubules independently of the motor domain. The ARM-RFP construct also labeled microtubules (Figure 5F), indicating that the ARM repeats comprise a second microtubule binding domain and that this domain is able to associate with microtubules even in monomeric form (as a result of deleting the coiled-coil domain).
ARK1 Function Is Redundant in Cell Types Other Than Elongating Root Hairs
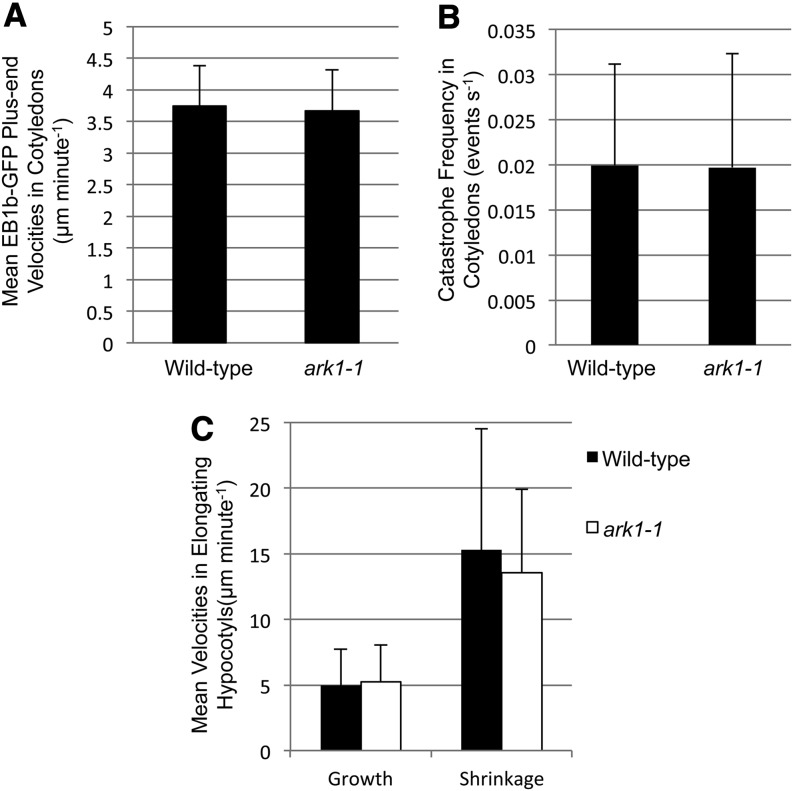
The expression ARK1-GFP in cell types other than root hairs under its endogenous promoter prompted us to explore ARK1 localization on other microtubule populations and to identify potential mutant phenotypes in non-root hair cell types. In root tips of ark1-1; ARK1pro:ARK1-GFP plants, ARK1-GFP labeled preprophase bands, mitotic spindles, and phragmoplasts (Figure 4A). Despite ARK1’s association with microtubules in mitotic and dividing cells, we did not detect any phenotypes related to cell size or cell plate positioning in ark1-1 mutants (Supplemental Figure 3). Moreover, cotyledon pavement cells, atrichoblasts, and hypocotyl epidermal cells in ark1-1 mutants showed no morphological differences from wild-type equivalents as previously shown (Yang et al., 2007; Sakai et al., 2008). No differences in microtubule dynamic parameters were detected in these cells (Figures 6A to 6C).
Figure 6.
ark1-1 Microtubule Phenotype Is Restricted to Root Hair-Forming Cells.
(A) Microtubule plus-end growth velocities are not significantly different in the pavement cells of wild-type (3.7 ± 0.6 μm/min, n = 409) and ark1-1 (3.7 ± 0.6 μm/min, n = 153) cotyledons. Four wild-type and ark1-1 plants each were imaged.
(B) Microtubule catastrophe frequencies in ark1-1 (0.020 ± 0.013 events/second) and wild-type cotyledon pavement cells (0.020 ± 0.011 events/second) are not significantly different. Catastrophe frequencies of microtubules were measured in the same cells as in (A).
(C) Microtubule growth and shrinkage velocities in wild-type (growth, 5.0 ± 2.8 μm/min, n = 371; shrinkage, 15.3 ± 9.2 μm/min, n = 37) and ark1-1 (growth, 5.3 ± 2.8 μm/min, n = 286; shrinkage, 13.6 ± 6.3 μm/min, n = 34) elongating hypocotyls are not significantly different from each other. Two (wild-type) and seven (ark1-1) plants were imaged.
Data and bars are represented as mean ± sd, respectively.
DISCUSSION
ARK1 has not been grouped into any of the 14 conventional kinesin families, which means that its function could not easily be predicted based on sequence homology. Using translational reporters, overexpression and loss-of-function analysis, we determined that ARK1 is a microtubule plus-end tracking catastrophe factor. The exact mechanism by which it promotes catastrophe remains to be determined. Specifically, it is unknown if the motor/ATP catalytic domain is used for kinesin movement along the microtubule and/or for physically removing tubulin heterodimers from the protofilament.
ARK1’s activity is potentially similar to that of the yeast (Saccharomyces cerevisiae) kinesin, Kip3p. A member of the Kinesin-8 family, Kip3p, was previously shown to be a plus-end tracking protein as well as a microtubule catastrophe factor (Gupta et al., 2006). Much like ark1-1 mutants, kip3p mutants have increased polymer mass (i.e., longer microtubules) (Cottingham and Hoyt, 1997) and reduced catastrophe frequency (Gupta et al., 2006). Movement of Kip3p along microtubules is ATP dependent (Gupta et al., 2006; Varga et al., 2006) and highly processive, which allows the kinesin to maintain plus-end association when depolymerization occurs. ATP hydrolysis in the motor domain was also shown to physically remove tubulin from the plus end of the microtubule protofilament (Gupta et al., 2006; Varga et al., 2006). In vitro experiments using purified ARK1 and tubulin are currently being pursued to determine ARK1’s exact mechanism of action. Nevertheless, we can conclude that ARK1 promotes catastrophe specifically at the plus ends of microtubules in vivo.
Although previous reports have supported the idea that ARK1 acts as a catastrophe factor (Yoo et al., 2008; Sakai et al., 2008), one study has hypothesized that ARK1 promotes microtubule stability and orientation (Yang et al., 2007). This hypothesis was based on confocal micrographs showing randomly oriented, fragmented microtubules in the root hairs of the cae1-1/mrh2-3 mutant allele of ark1, which, through a G-to-A substitution, has a premature stop codon and reduced transcript. Moreover, it was reported that upon taxol exposure, the mrh2-3 root hair morphological defects were partially rescued (Yang et al., 2007). The confocal images of microtubules in the Yang et al. (2007) study, however, were from root hairs that appeared to have completed growth. In wild-type fully elongated root hairs, endoplasmic microtubules are absent and cortical microtubules generally appear fragmented and sparse (Van Bruaene et al., 2004). The mrh2-3 phenotype rescue by taxol reported by Yang et al. (2007), however, remains puzzling and is clearly in contrast to our finding that taxol enhances the ark1-1 root hair phenotype.
The ability of the C-terminal ARM domain to independently associate with microtubules suggests that this domain plays a role in localizing ARK1 to microtubules. This interaction could potentially aid in ARK1’s ability to remain at the growing plus ends. The idea that a kinesin has two independent microtubule binding domains is not entirely new. The Arabidopsis ATK5, for example, possesses two microtubule binding domains: the C-terminal motor domain and an additional N-terminal domain that enables microtubule plus-end tracking (Ambrose et al., 2005). Members of the kinesin-8 family (S. cerevisiae Kip3p and human Kif18A) also contain secondary microtubule binding domains that allow these kinesins to remain bound to microtubules, resulting in high processivity and plus-end accumulations (Stumpff et al., 2011; Su et al., 2011; Mayr et al., 2011; Weaver et al., 2011). The second microtubule binding domain of Kif18A also functions independently of ATP (Mayr et al., 2011; Weaver et al., 2011) much like the Armadillo-repeats, which do not have an ATP catalytic site (Tewari et al., 2010).
Whether the ARM domain of ARK1 enables direct or indirect binding to microtubules remains to be determined. A previous study demonstrated that this domain, while showing some affinity for actin filaments in vitro, had little or no affinity for microtubules (Yang et al., 2007). One possibility is that the ARM domain interacts with microtubules via the NIMA-related kinase, NEK6. This is based on previous evidence that showed NEK6’s ability to bind to the ARM domain (Sakai et al., 2008) and NEK6-GFP’s ability to localize to microtubules (Motose et al., 2008, 2011).
Based on its widespread expression, ARK1 appears to function in cell types other than root hair-forming ones, with its expression pattern overlapping with that of its homologs, ARK2 and ARK3. ARK2 is thought to modulate microtubule dynamics in root epidermal cells, evident by its T-DNA insertional mutants having a root twisting phenotype (Sakai et al., 2008) typically associated with aberrant microtubule organization and dynamics (Nakajima et al., 2004; Nakamura et al., 2004; Sedbrook et al., 2004). ARK3/KinUa accumulates at preprophase bands and is cell cycle regulated during embryogenesis and stomatal development (Malcos and Cyr, 2011). Although ARK1-GFP localized to preprophase bands as well as mitotic spindles and phragmoplasts, it is unlikely to be cell cycle regulated. ARK3 was previously found to contain a D-Box motif, a regulatory sequence found in cyclins and cell cycle-regulated proteins, which is absent in ARK1 (Malcos and Cyr, 2011). Because ARK3 knockout mutants could not be recovered (either through homozygous T-DNA insertional mutants or ARK3 RNA interference), ARK3 is thought to be essential for plant growth and development through its function in mitotic and cytokinetic microtubule arrays (Malcos and Cyr, 2011). We did not see any ark1-1 phenotype related to disrupted cell cycle (i.e., abnormal cell plate placement and changes in cell shape/size), suggesting the presence of ARK3 is sufficient and necessary for cells to complete cell division while ARK1 is not.
By promoting periodic microtubule catastrophe, ARK1 can facilitate microtubule turnover, an important feature in array remodeling. In addition, our findings suggest that ARK1’s microtubule destabilizing activity ultimately serves to maintain sufficient levels of free tubulin, which is required to sustain rapid microtubule polymerization. Interestingly, the data obtained in the current and some previous studies suggest that rapid microtubule polymerization, rather than polymer level, is the single parameter of microtubule dynamics most critical for maintenance of polar tip growth in root hairs. Previously, it was shown that drugs and mutations that either overstabilize or destabilize microtubules can have the same consequence on cell shape (reviewed in Wasteneys and Ambrose, 2009). Taxol and oryzalin treatments, which increase and decrease microtubule polymer mass, respectively, both cause crooked and branched root hairs to develop (Bibikova et al., 1999). Crooked and branched root hairs are also found in ark1-1 and mor1-1 mutants, which have increased and decreased microtubule polymer mass, respectively (Whittington et al., 2001; Sakai et al., 2008). Despite these extremes in polymer mass, the common feature is that microtubule growth rates are consistently reduced in mor1-1 mutants at restrictive temperature (Kawamura and Wasteneys, 2008), in ark1-1 (this study) and in wild-type cells treated with oryzalin (this study; Nakamura et al., 2004). Importantly, the application of low concentrations of oryzalin to ark1-1 both increased microtubule polymerization rates and reduced root hair branching. With this evidence, we conclude that microtubule polymerization rates must exceed a threshold value (∼6 µm per min or 100 nm per second at 21°C) in order to maintain uniform tip growth. This supports the concept that microtubule dynamics need to be within a very narrow “Goldilocks zone” for maintenance of microtubule array organization and cellular development (Wasteneys and Ambrose, 2009).
Our results reveal that ark1-1 root hairs accumulate bundles of endoplasmic microtubules despite the fact that these microtubules have reduced growth velocities and catastrophe frequencies not significantly different from endoplasmic microtubules in wild-type root hairs. This apparently paradoxical result can be explained by the fact that the endoplasmic microtubules originate in the cortex such that their accumulation may be independent of their dynamics. By resisting catastrophe, the cortical microtubules in the apex of ark1-1 mutant root hairs eventually become detached from the cortex and, via the reverse-fountain cytoplasmic streaming (Sieberer and Emons, 2000), are swept back into the endoplasm where they accumulate in large bundles. It has also been postulated that the continuously changing positions and orientation of the endoplasmic microtubules are correlated with the highly dynamic cytoplasmic streaming found in the endoplasm of root hairs (Van Bruaene et al., 2004). This may have caused us to overestimate endoplasmic microtubule catastrophe frequencies despite our attempts to ensure we were imaging the entire endoplasmic region with frequent acquisition time points for an extended time period.
From our mutant, drug, and overexpression analysis, it is clear that the correct modulation of microtubule dynamics encompasses a broad range of cellular factors, including free tubulin concentration and the expression and activation of different microtubule-associated proteins. Although it was expected that ARK1-RFP overexpression would result in an increase in catastrophe frequency, we were surprised to observe a reduced microtubule growth rate. It is possible that overaccumulation of ARK1-RFP, particularly at the microtubule plus end, inhibits the binding of other microtubule-associated proteins or affects its own function. In Arabidopsis, several MAPs have been reported to interact with microtubule plus ends to orchestrate dynamics (e.g., EB1 [Chan et al., 2003], SKU6/SPIRAL1 [Nakajima et al., 2004; Sedbrook et al., 2004], ATK5 [Ambrose et al., 2005], and CLASP1 [Ambrose et al., 2007]), the activity of which might be altered by ARK1-RFP overexpression. In vitro analysis has previously shown that increasing concentrations of Kip3p and other MAPs can decrease Kip3p’s velocity along microtubules as a result of macromolecular crowding (Leduc et al., 2012). Given that the plus-end microtubule-associated proteins interact at the nanoscale level, an appropriate ratio of these proteins is required for precise regulation of microtubule dynamics. Based on our study, complete removal of ARK1 from microtubule plus ends or its overexpression is detrimental to proper microtubule dynamics.
METHODS
Plant Material and Culture
Arabidopsis thaliana wild-type (Columbia-0 ecotype) and ark1-1 plants with the 35Spro:GFP-MBD transgene were used, as previously described (Sakai et al., 2008). The 35Spro:EB1b-GFP reporter line (Mathur et al., 2003) and the 35Spro:mCherry-MAP4MBD construct (Gutierrez et al., 2009) were kindly provided by Jaideep Mathur (University of Guelph, Canada) and David Erhardt (Carnegie Institute of Science, Stanford, CA), respectively.
All seeds were sterilized in 70% ethanol, rinsed three times with double distilled water, and plated onto Petri dishes with Hoagland media (1.2% Bacto-agar [BD Diagnostics], no sucrose). Plates with seeds were stored in the dark at 4°C for 2 to 3 d and transferred to a 21°C growth cabinet (24 h light) where they were grown vertically until imaging. For the drug studies, seeds were initially vertically grown on Hoagland media supplemented with DMSO (Fisher Scientific) and then transferred to Hoagland media containing various concentrations of taxol (Sigma-Aldrich) or oryzalin (Sigma-Aldrich) for 2 d prior to imaging.
ARK1 Construct Design and Cloning Strategies
Gateway cloning technology (Invitrogen) was used for the ARK1 genomic and coding sequences. To generate the ARK1pro:ARK1-GFP construct, the ARK1 genomic sequence (between 888 bp upstream of the ATG/start codon and the TGA/stop codon) was amplified from the F28P10 BAC (from the ABRC, Ohio State University) using the full-length ARK1 genomic sequence primer set (Supplemental Table 1). A second PCR with the attB-adapter primers was performed according to the manufacturer’s protocol (see Invitrogen for attB-adapter sequence). For the amplification of the ARK1 coding sequences, different primers were used to amplify the cDNA templates for the ARK1, ARK1ΔARM, ARK1ΔMotor, and ARM constructs (see Supplemental Table 1 for primer list). A second PCR with the attB-adapter primers was performed according to the manufacturer’s protocol (Invitrogen). The mRNA extraction and cDNA synthesis protocol were performed as described (Galway et al., 2011). Following the BP reaction with the various attB-PCR products and the pDONR221 vector (Invitrogen), an LR reaction was performed with the pMDC107 vector (Curtis and Grossniklaus, 2003) for the ARK1 genomic sequence and the pUBC-RFP DEST vector (Grefen et al., 2010) for the various ARK1 coding sequences.
Generation of Transgenic Plant Materials
For stable transgenic lines, the ARK1pro:ARK1-GFP construct was first transformed into Agrobacterium tumefaciens (GV3101 strain) and then transformed into an Arabidopsis ark1-1 35Spro:mCherry-MAP4MBD line using the floral dip method (Clough and Bent, 1998). T3 lines homozygous for the ARK1pro:ARK1-GFP transgene were segregated and used for further experiments. Transient expression in cotyledons of the four different UBQpro-driven ARK1 fragment-RFP constructs was performed in ark1-1; 35Spro:GFP-MBD plants using the FAST technique (Li et al., 2009). ark1-1 (SALK_035063) plants were crossed into the 35Spro:EB1b-GFP reporter line (Mathur et al., 2003), and homozygous F3 ark1-1 and wild-type plants were segregated for imaging.
Live-Cell Imaging
For observing root hair morphology, whole seedlings were mounted on slides with cover slips and bright-field images of root hairs were then collected with a 20× (air) objective lens on a Leica DMR light microscope with a Q-CAM digital camera (Leica).
Live imaging of the various microtubule reporter proteins, the ARK1pro-driven ARK1-GFP, and the UBQ10pro-driven ARK1-RFP was done using a Perkin-Elmer Ultraview VoX Spinning Disc Confocal system (Perkin-Elmer) mounted on a Leica DMI6000 B inverted microscope and equipped with a Hamamatsu 9100-02 electron multiplier CCD camera (Hamamatsu). An argon 488-nm laser line with a complementary GFP (525/36) emission band-pass filter or a 561-nm laser with a complementary RFP (595/50) emission band-pass filter was used. Images were acquired with a 63× (water) objective lens every 8 s for 3 to 5 min with 0.3- to 0.5-μm optical z-slices. For imaging of microtubule dynamics during drug treatments, seedlings grown on Hoagland medium with 100 nM oryzalin were mounted in 100 nM oryzalin. The imaging temperature of the samples was maintained at 21°C using a Bionomic Controller BC-110 with a HEC-400 Heat Exchanger, a Bionomic Controller BC-100 (20-20 Technology) temperature-controlled stage, and an objective heater (Bioptechs).
Image and Data Analysis
All images were processed and analyzed using ImageJ (http://rsbweb.nih.gov/ij/). For root hair length analysis, line selections were superimposed on root hairs and then measured. Measuring the microtubule velocities of the EB1b-GFP and GFP-MBD microtubule marker lines were done using the Manual Tracking plug-in (http://rsbweb.nih.gov/ij/plugins/track/track.html). EB1b-GFP particles were considered endoplasmic when images from the medial confocal plane of the root hair were analyzed in the endoplasmic region of the root hair. Cortical EB1b-GFP particles were measured and analyzed from cortical plane of the root hairs or along the cortex/plasma membrane in the medial confocal plane of the root hair. For determining the catastrophe frequency of EB1b-GFP, the inverse of the duration of time spent tracking one EB1b-GFP comet was taken. This is based on the assumption that the disappearance of the EB1b-GFP signal meant the microtubule was undergoing catastrophe since EB1b-GFP has highest affinity for the growing plus end of microtubules. The same method for determining catastrophe frequency was used with the GFP-MBD marker in the ARK1 overexpression analysis. For determining the amount of time spent in each phase, the amount of time spent in one phase (i.e., growth, shrinkage, or pause) was divided by the total measured time of a microtubule lifespan (using the GFP-MBD marker). The mean, sd, F-tests, and t tests were calculated using Excel (Microsoft).
We also sought to measure potential changes in shrinkage velocities and rescue frequencies by imaging the microtubule markers 35Spro:GFP-MBD and UBQ1pro:mRFP-TUB6 in root hairs. However, we were unable to accurately observe and quantify plus-end dynamics of single microtubules with these markers in root hairs because of increased microtubule bundling, dense microtubule populations, or high background fluorescence.
For quantifying the ratio of fluorescence intensity to mean fluorescence intensity, the equations below were used. The procedure involved taking 3-μm line scans on microtubule plus ends from ARK1-GFP images and then measuring the fluorescence intensities/gray values (xn) every 0.108 μm along the line scan using the Plot Profile function in ImageJ. The fluorescence intensities were then averaged 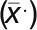 by the total number of points (N) measured in the line scan (Equation 1). Each fluorescence intensity (xn) value was then divided by the mean fluorescence intensity
by the total number of points (N) measured in the line scan (Equation 1). Each fluorescence intensity (xn) value was then divided by the mean fluorescence intensity 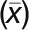 to get the desired ratio (Rn) at each point along the 3-μm line scan (Equation 2). All ratios are reported as mean values. The same measurements were done with the mCherry-MAP4 MBD images as a control.
to get the desired ratio (Rn) at each point along the 3-μm line scan (Equation 2). All ratios are reported as mean values. The same measurements were done with the mCherry-MAP4 MBD images as a control.
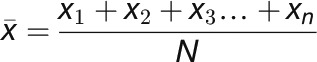 |
(1) |
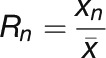 |
(2) |
Quantification of Endoplasmic and Cortical Microtubules
Confocal images of wild-type and ark1-1; 35Spro:GFP-MBD root hairs were analyzed in Image J according to Sakai et al. (2008).
Propidium Iodide Staining and Imaging of Wild-Type and ark1-1 Root Tips
Labeling of the cell wall in the root tips was done by incubating whole wild-type and ark1-1 seedlings in 10 μg/mL propidium iodide (Calbiochem) for 1 min, rinsing them with water, and mounting them on slides and cover slips. Laser confocal images of the stained root tips were done on a Zeiss AxioImager M1 microscope with a Zeiss PASCAL Excite two-channel LSM 780 system (Carl Zeiss). A helium-neon 543-nm laser line and a 560-nm emission long-pass filter were used. Images were acquired in 2-μm optical z-slices with a 63× (oil) objective lens.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or EMBL/GenBank databases under the following Arabidopsis Genome Initiative identifier: At3g54870 (ARK1/MRH2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. EB1b-GFP Velocity Is Slower in ark1-1 Root Hairs Than in Wild-Type Root Hairs.
Supplemental Figure 2. Endoplasmic Microtubules Are More Abundant in ark1-1 Than in Wild-Type Root Hairs.
Supplemental Figure 3. Cell and Tissue Patterns Are Not Affected in ark1-1 Root Tips.
Supplemental Table 1. Primer Sets Used for Cloning the Various ARK1 Constructs.
Supplemental Movie 1. EB1b-GFP Trajectories in Wild-Type Elongating Root Hairs Show That Microtubules Undergo Rapid Elongation and Frequent Catastrophe.
Supplemental Movie 2. EB1b-GFP Trajectories in ark1-1 Elongating Root Hairs Show That Microtubule Growth Rates and Catastrophe Frequencies Are Reduced.
Supplemental Movie 3. ARK1-GFP Localizes to Microtubules in Elongating Root Hairs.
Supplemental Movie 4. ARK1-GFP Localizes to Microtubules in Fully Grown Root Hairs.
Supplemental Movie 5. ARK1-GFP Is Concentrated at Microtubule Plus Ends in Cotyledon Pavement and Guard Cells.
Supplemental Movie 6. High-Magnification Time-Lapse Images Demonstrate the ARK1-GFP Plus-End Tracking.
Supplemental Movie 7. Time-Lapse Images of a Treadmilling Microtubule Demonstrates that ARK1-GFP Is Distributed Specifically to the Growing Microtubule Plus Ends and Not the Minus Ends.
Supplemental Movie 8. ARK1-GFP Is Not Associated with Shrinking Microtubule Plus Ends.
Supplementary Material
Acknowledgments
This research was funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant (298264-09) to G.O.W., an NSERC postgraduate scholarship to R.C.E., and the Canada Foundation for Innovation. We thank Jaideep Mathur (University of Guelph, Canada) and David Ehrhardt (Carnegie Institute of Science, Stanford, CA) for reporter lines, Chris Ambrose (University of British Columbia [UBC]) for technical insight and careful reading of the article, Lacey Samuels (UBC) for use of her Leica light microscope, and the UBC Bioimaging Facility for confocal microscope access and assistance.
AUTHOR CONTRIBUTIONS
R.C.E. performed experiments. G.O.W. and R.C.E. designed the research, analyzed the data, and wrote the article.
Glossary
- MAP
microtubule-associated protein
- CMT
cortical microtubule
- EMT
endoplasmic microtubule
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ambrose J.C., Li W., Marcus A., Ma H., Cyr R. (2005). A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 16: 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose J.C., Shoji T., Kotzer A.M., Pighin J.A., Wasteneys G.O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova T.N., Blancaflor E.B., Gilroy S. (1999). Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 17: 657–665. [DOI] [PubMed] [Google Scholar]
- Carol R.J., Dolan L. (2002). Building a hair: tip growth in Arabidopsis thaliana root hairs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G.M., Doonan J.H., Lloyd C.W. (2003). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5: 967–971. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cottingham F.R., Hoyt M.A. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138: 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13: 83–117. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96: 69–78. [DOI] [PubMed] [Google Scholar]
- Galway M.E., Eng R.C., Schiefelbein J.W., Wasteneys G.O. (2011). Root hair-specific disruption of cellulose and xyloglucan in AtCSLD3 mutants, and factors affecting the post-rupture resumption of mutant root hair growth. Planta 233: 985–999. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Gupta M.L., Jr, Carvalho P., Roof D.M., Pellman D. (2006). Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8: 913–923. [DOI] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y., Sato Y., Doonan J.H. (2014). Kinesins have a dual function in organizing microtubules during both tip growth and cytokinesis in Physcomitrella patens. Plant Cell 26: 1256–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Raymond M.J., Smirnoff N. (2006). Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 45: 83–100. [DOI] [PubMed] [Google Scholar]
- Kawamura E., Wasteneys G.O. (2008). MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J. Cell Sci. 121: 4114–4123. [DOI] [PubMed] [Google Scholar]
- Lechner B., Rashbrooke M.C., Collings D.A., Eng R.C., Kawamura E., Whittington A.T., Wasteneys G.O. (2012). The N-terminal TOG domain of Arabidopsis MOR1 modulates affinity for microtubule polymers. J. Cell Sci. 125: 4812–4821. [DOI] [PubMed] [Google Scholar]
- Leduc C., Padberg-Gehle K., Varga V., Helbing D., Diez S., Howard J. (2012). Molecular crowding creates traffic jams of kinesin motors on microtubules. Proc. Natl. Acad. Sci. USA 109: 6100–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.F., Park E., von Arnim A.G., Nebenführ A. (2009). The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcos J.L., Cyr R.J. (2011). An ungrouped plant kinesin accumulates at the preprophase band in a cell cycle-dependent manner. Cytoskeleton (Hoboken) 68: 247–258. [DOI] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao Th., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Kernebeck B., Srinivas B.P., Hülskamp M. (2003). A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr. Biol. 13: 1991–1997. [DOI] [PubMed] [Google Scholar]
- Mayr M.I., Storch M., Howard J., Mayer T.U. (2011). A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PLoS ONE 6: e27471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motose H., Tominaga R., Wada T., Sugiyama M., Watanabe Y. (2008). A NIMA-related protein kinase suppresses ectopic outgrowth of epidermal cells through its kinase activity and the association with microtubules. Plant J. 54: 829–844. [DOI] [PubMed] [Google Scholar]
- Motose H., Hamada T., Yoshimoto K., Murata T., Hasebe M., Watanabe Y., Hashimoto T., Sakai T., Takahashi T. (2011). NIMA-related kinases 6, 4, and 5 interact with each other to regulate microtubule organization during epidermal cell expansion in Arabidopsis thaliana. Plant J. 67: 993–1005. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Furutani I., Tachimoto H., Matsubara H., Hashimoto T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Naoi K., Shoji T., Hashimoto T. (2004). Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol. 45: 1330–1334. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2013). Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns. Plant Cell 25: 4439–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A.S.N., and Day, I.S.(2011). Microtubule motor proteins in the eukaryotic green lineage: Functions and regulation. In The Plant Cytoskeleon, Advances in Plant Biology, B. Liu, ed (New York: Springer), pp. 119–141. [Google Scholar]
- Sakai T., et al. (2008). Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 53: 157–171. [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Ehrhardt D.W., Fisher S.E., Scheible W.R., Somerville C.R. (2004). The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer, B., and Timmers, A. (2009). Microtubules in plant root hairs and their role in cell polarity and tip growth in root hairs. In Plant Cell Monograph, Vol. 12, A.M. Emons and T. Ketelaar, eds (Berlin: Springer), pp. 233–248. [Google Scholar]
- Sieberer B., Emons A.M.C. (2000). Cytoarchitecture and pattern of cytoplasmic streaming in root hairs of Medicago truncatula during development and deformation by nodulation factors. Protoplasma 214: 118–127. [Google Scholar]
- Sieberer B.J., Timmers A.C., Lhuissier F.G., Emons A.M. (2002). Endoplasmic microtubules configure the subapical cytoplasm and are required for fast growth of Medicago truncatula root hairs. Plant Physiol. 130: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., Du Y., English C.A., Maliga Z., Wagenbach M., Asbury C.L., Wordeman L., Ohi R. (2011). A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol. Cell 43: 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Qiu W., Gupta M.L., Jr, Pereira-Leal J.B., Reck-Peterson S.L., Pellman D. (2011). Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol. Cell 43: 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R., Bailes E., Bunting K.A., Coates J.C. (2010). Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 20: 470–481. [DOI] [PubMed] [Google Scholar]
- Van Bruaene N., Joss G., Van Oostveldt P. (2004). Reorganization and in vivo dynamics of microtubules during Arabidopsis root hair development. Plant Physiol. 136: 3905–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A.A., Tanaka T.U., Howard J. (2006). Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8: 957–962. [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Ambrose J.C. (2009). Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 19: 62–71. [DOI] [PubMed] [Google Scholar]
- Weaver L.N., Ems-McClung S.C., Stout J.R., LeBlanc C., Shaw S.L., Gardner M.K., Walczak C.E. (2011). Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr. Biol. 21: 1500–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington A.T., Vugrek O., Wei K.J., Hasenbein N.G., Sugimoto K., Rashbrooke M.C., Wasteneys G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613. [DOI] [PubMed] [Google Scholar]
- Yang G., Gao P., Zhang H., Huang S., Zheng Z.L. (2007). A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS ONE 2: e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.M., Blancaflor E.B. (2013). Overlapping and divergent signaling pathways for ARK1 and AGD1 in the control of root hair polarity in Arabidopsis thaliana. Front. Plant Sci. 4: 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.M., Wen J., Motes C.M., Sparks J.A., Blancaflor E.B. (2008). A class I ADP-ribosylation factor GTPase-activating protein is critical for maintaining directional root hair growth in Arabidopsis. Plant Physiol. 147: 1659–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Dixit R. (2012). Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 249: 887–899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.