Successful isolation of a highly active b6f complex including the PetP subunit allowed detailed analysis of the function of this recently discovered subunit. PetP appears to promote the stability and activity of the b6f complex and to influence linear electron transport.
Abstract
The cyanobacterial cytochrome b6f complex is central for the coordination of photosynthetic and respiratory electron transport and also for the balance between linear and cyclic electron transport. The development of a purification strategy for a highly active dimeric b6f complex from the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 enabled characterization of the structural and functional role of the small subunit PetP in this complex. Moreover, the efficient transformability of this strain allowed the generation of a ΔpetP mutant. Analysis on the whole-cell level by growth curves, photosystem II light saturation curves, and P700+ reduction kinetics indicate a strong decrease in the linear electron transport in the mutant strain versus the wild type, while the cyclic electron transport via photosystem I and cytochrome b6f is largely unaffected. This reduction in linear electron transport is accompanied by a strongly decreased stability and activity of the isolated ΔpetP complex in comparison with the dimeric wild-type complex, which binds two PetP subunits. The distinct behavior of linear and cyclic electron transport may suggest the presence of two distinguishable pools of cytochrome b6f complexes with different functions that might be correlated with supercomplex formation.
INTRODUCTION
Oxygenic photosynthesis, which is able to utilize the energy of electromagnetic radiation for the formation of carbon bonds, i.e., to transform and store it as chemical energy, is one of the most important achievements of evolution. Cyanobacteria, according to the endosymbiotic theory (Mereschkowsky, 1905; Margulis, 1975; Martin and Kowallik, 1999) the evolutional ancestors of chloroplasts, are the simplest model system performing oxygenic photosynthesis. In contrast to purple and green sulfur bacteria that perform anoxygenic photosynthesis with only one photosynthetic reaction center, cyanobacteria use, for the first time in evolution, two photosystems, i.e., photosystem II (PSII) and photosystem I (PSI). They are connected in series by the cytochrome b6f complex (b6f), the central component and switching point of photosynthetic electron transport. The detailed structural characterization of all three major photosynthetic membrane protein complexes from cyanobacteria, PSII (Zouni et al., 2001; Ferreira et al., 2004; Umena et al., 2011), PSI (Jordan et al., 2001), and b6f (Kurisu et al., 2003; Hasan et al., 2013), provided the basis for a functional understanding on the molecular level of each complex. By contrast, the coordinated functional interaction between these complexes is still widely unknown (Iwai et al., 2010).
Despite many, mainly functional, similarities between chloroplasts and cyanobacteria, there are fundamental differences especially in the morphology of their membrane systems: While thylakoids of chloroplasts consist of stacked grana enriched in PSII and light-harvesting complex II complexes and unstacked stroma thylakoids enriched in PSI and ATP synthase (Allen and Forsberg, 2001), cyanobacteria are lacking a stroma and grana compartmentalization. Instead, their membranes show PSII dimers organized in parallel rows (Olivea et al., 1997; Folea et al., 2008) with apparently randomly distributed PSI trimers in between (Westermann et al., 1999). However, for both thylakoid membrane systems of plants and cyanobacteria, the distribution of b6f is still unclear with most authors believing in an even distribution of this complex over the whole membrane (Hinshaw and Miller, 1993; Kirchhoff et al., 2000; Allen and Forsberg, 2001) or a state dependent distribution of b6f (Vallon et al., 1991). In cyanobacterial thylakoids, b6f is central to both photosynthetic and respiratory electron transport chain (Norling et al., 1997; Zak et al., 2001; Huang et al., 2002; Schultze et al., 2009), which are separated into chloroplasts and mitochondria in plants. Also, mitochondria contain a bc1 complex instead of b6f (Widger et al., 1984). Instead of separate organelles and membrane types, cyanobacterial thylakoids contain function-correlated discrete patches (Rexroth et al., 2011), which apparently can regulate electron transport by redistribution of the complexes involved. This is suggested by the respiratory complexes NADH:ubiquinone oxidoreductase 1 (NDH-1) and succinate dehydrogenase, which have been shown to form discrete patches when the plastoquinone (PQ) pool is predominantly oxidized (Liu et al., 2012). Due to its central role in the cyanobacterial electron transport network, a similar mechanism is likely for b6f, which links plastoquinone reducing membrane protein complexes (i.e., PSII and NADH dehydrogenase), with plastocyanin and cytochrome c oxidizing complexes (PSI and cytochrome oxidase). Also, b6f is involved in two cyclic electron transport pathways: The complex-internal “Q-cycle” and an “external” one, of which some components are still ambiguous (Yeremenko et al., 2005; Battchikova et al., 2011).
The structure at 2.7 Å of several dimeric cytochrome b6f complexes from cyanobacteria and green algae shows eight subunits per monomer (Kurisu et al., 2003; Stroebel et al., 2003; Baniulis et al., 2009; Hasan et al., 2013). The four large subunits cytochrome f (cyt f), cytochrome b6 (cyt b6), the Rieske iron-sulfur protein, and subunit IV (SU IV) are directly involved in electron transport, while the four small subunits PetG, PetL, PetM, and PetN are apparently involved in the structural stabilization of the complex. The more highly resolved crystal structures of the lumen-exposed water soluble parts of the complex, i.e., cyt f and the Rieske protein, suggest novel structural features for proton transport (Carrell et al., 1999) and regulation of the Rieske domain movement (Carrell et al., 1999; Bernat and Rögner, 2011; Kallas, 2012; Veit et al., 2012).
In addition to these “consensus” subunits, further components with weaker binding affinities and/or transient interactions with the complex could be identified: In green algae and higher plants, binding of the ferredoxin-NADP reductase (FNR) (Zhang et al., 2001) and of the subunit PetO, which is possibly involved in the regulation of state transitions (Hamel et al., 2000) has been shown. Also, we recently showed the presence of a potentially new b6f subunit, PetP, in b6f-preprations from both the mesophilic cyanobacterium Synechocystis PCC 6803 and the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 (Volkmer et al., 2007; Gendrullis et al., 2008; Nowaczyk et al., 2010). Most remarkably, this protein is highly conserved in cyanobacteria and red algae, but not present in green algae and plants (Volkmer et al., 2007).
Here, we show the isolation of a highly active dimeric b6f complex with quantitatively bound PetP subunit. Due to the stability of this complex and the transformability of its host, the thermophilic T. elongatus, the effect of PetP both on the isolated complex and on the cellular electron transport could be characterized in detail. Pronounced differences involved a distinct decrease in the linear electron transport rate and stability of the complex in the absence of PetP.
RESULTS
Isolation and Subunit Quantification of b6f from T. elongatus
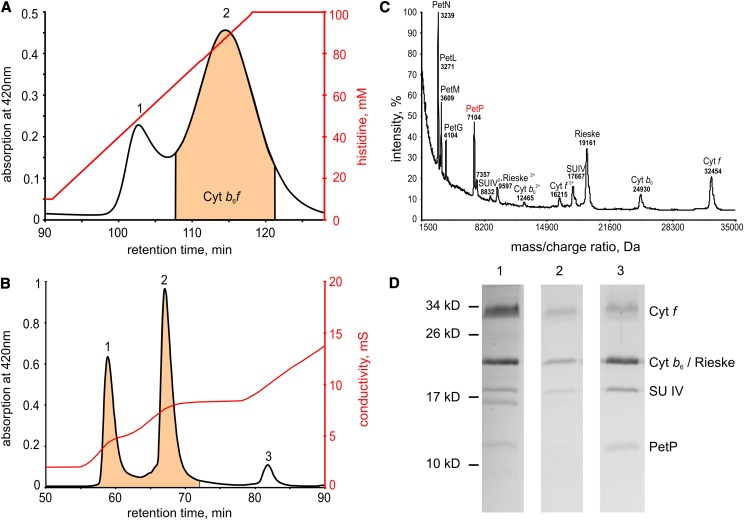
For the isolation of a functional dimeric b6f complex from T. elongatus, the use of a molecular tag system was crucial. Figure 1A shows the elution of b6f with C-terminal His-tag at the cyt f subunit from an immobilized metal ion affinity chromatography (IMAC) column. The tag shortened the biochemical separation considerably; also, the exposure time to unregulated proteases and reactive oxygen species could be minimized by this step. The b6f-containing fractions from this IMAC run were applied to an ion exchange column (Figure 1B), which separated fractions of monomeric from dimeric complexes and also yielded a third fraction containing coeluting PSI and NDH complex.
Figure 1.
Purification of Native b6f Complex from T. elongatus.
(A) Elution profile from a preparative IMAC column (first purification step): peak 1 with nonspecifically bound proteins, mainly NADH-dehydrogenase and PSI, peak 2 with enriched b6f. Chelating sepharose Fast Flow (GE Healthcare), 0.5 mL/min, 4°C, gradient: 10 to 100 mM histidine.
(B) Elution profile from preparative anion exchange chromatography (IEC) column as second purification step: peak 1 b6f-monomer and peak 2 b6f-dimer. Peak 3 with a mixture of nonspecifically bound proteins. UnoQ1 (Bio-Rad), 0.5 mL/min, 4°C; gradient 1: 0 to 4.2 mM NaCl, gradient 2: 4.2 to 8.2 mM NaCl, gradient 3: 8.2 to 20 mM NaCl.
(C) MALDI-TOF subunit analysis of isolated b6f (IEC fraction 2).
(D) SDS-PAGE gel after Coomassie staining. (1) b6f-peak after IMAC column (peak 2 of [A]), (2) b6f-monomer after IEC column (peak 1 of [B]), and (3) b6f-dimer after IEC column (peak 2 of [B]).
Subunit composition of the isolated monomeric and dimeric b6f complexes was analyzed by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) (Figure 1C; Supplemental Table 1) and SDS-PAGE (Figure 1D) with signals of the intact proteins in the mass spectrum being assigned on basis of the mass/charge ratio of the subunits in the b6f complex. By these combined methods, altogether nine subunits could be detected: four large (cyt f, cyt b6, the Rieske protein, and SU IV) and five small subunits (PetG, -L, -M, -N, and -P). The determined masses agree well with the theoretical masses based on protein sequence data (deviations <1%), if signals for single- and double-charged protein subunits and posttranslational modifications are considered (Whitelegge et al., 2002; de Vitry et al., 2004). Notably, a mass consistent with PetL was identified, although this subunit was not annotated in the genome of T. elongatus. Possibly this open reading frame was missed during genome annotation due to the small size of PetL (Nowaczyk et al., 2011). For this reason, we used the homolog of Synechocystis sp PCC 6803 as theoretical mass (for details, see Supplemental Table 1).
Subunit stoichiometry within T. elongatus cells was determined by absolute protein quantification based on SRM analysis with isotope-labeled standard peptides (Table 1). Although the accessibility of the protein during the proteolytic digest affects the quantification per cell, there is clear evidence for a 1:1:1 stoichiometry between Rieske, SU IV, and PetP. The estimated amount of FNR (PetH) per cell closely matches to the value determined for Synechocystis PCC 6803 (Moal and Lagoutte, 2012).
Table 1. Protein Copy Number of b6f Subunits per T. elongatus Cell According to SRM Analysis.
| Subunit | Copies per Cell ± sd |
|---|---|
| Rieske | 2,500 ± 300 |
| SU IV | 2,700 ± 200 |
| PetP | 3,300 ± 660 |
| FNR | 10,500 ± 3,000 |
Monomeric and dimeric b6f have also been carefully compared by a detailed subunit SDS-PAGE analysis (Figure 1D). While after the IMAC column (first purification step) all four major b6f subunits could be detected (lane 1), analysis after the ion exchange chromatography (IEC) showed a monomeric complex (lane 2), which in contrast to the dimeric complex (lane 3), had lost the Rieske protein, as confirmed by immunoblot analysis (Breyton et al., 1997; Cramer et al., 2005). Under long-term storage of the isolated dimeric complex, a loss of the Rieske subunit and a subsequent monomerization is observed (Supplemental Figure 1).
Similar to the Rieske, the 7.1-kD subunit PetP is present both after purification by IMAC and in the dimeric b6f after IEC, but apparently missing in the monomeric complex. By SDS-PAGE analysis, an apparent mass of 11 to 12 kD is observed both for the protein isolated from T. elongatus (Figure 1D) and that expressed heterologously in Escherichia coli (see below). Identity of the subunit was verified unambiguously by immunoblot and liquid chromatography-tandem mass spectrometry analysis, while the mass of 7104 ± 36 D was confirmed by MALDI-MS analysis of the intact protein (Figure 1C; Supplemental Table 1). A more detailed analysis by immunoblot and mass spectrometry (MS) confirmed a significantly reduced PetP content (<30%) in the b6f monomer compared with the dimer.
Localization of PetP Subunit in the Dimeric b6f Complex
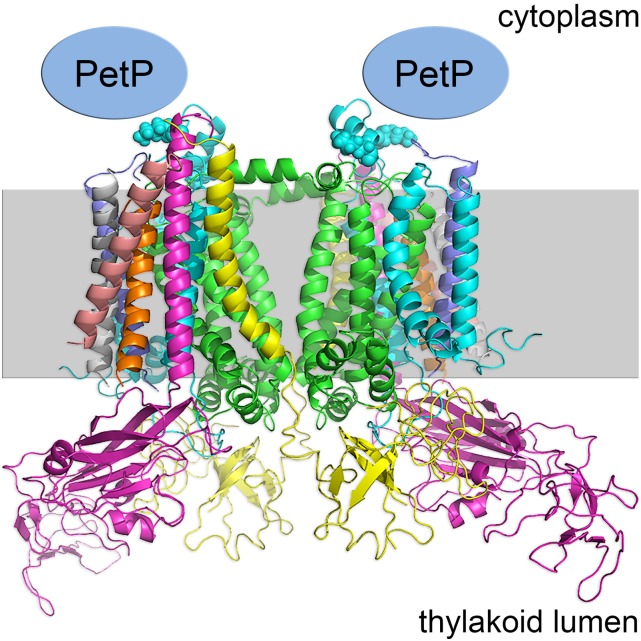
For the localization of PetP, we used a combination of cross-linking and high-resolution mass spectrometry. Subunits of the isolated b6f complex were cross-linked by incubation with the isotope-coded cross-linker BS3-H12/D12, and tryptic peptides were analyzed by high-resolution mass spectrometry. Data analysis revealed the presence of two cross-linked peptides between the N terminus of PetP and the N-terminal part (K7 and K20) of subunit IV (PetD) within the b6f complex (Supplemental Table 2). Both cross-links were identified based on MS2 spectra of the light and heavy variant (Supplemental Figure 2) of the cross-linker (H12/D12) using StavroX software (Götze et al., 2012). The independent identification of the cross-link by both the hydrogen and deuterium substituted linker served to suppress false positives and verify the identified cross-links. The 12-D shift is clearly visible in the MS and MS2 spectra (Supplemental Figure 2). This approach pinpoints PetP to the cytoplasmic side of the b6f complex (Figure 2)
Figure 2.
Localization of PetP within the b6f Structure.
The position of PetP is based on the cross-link between K7 and K20 of subunit IV (blue spheres) and the N terminus of PetP (unknown structure) as shown in the structural model of b6f from Nostoc sp PCC 7120 (PDB code: 4H44) (Hasan et al., 2013).
Effect of PetP on the Isolation of an Active Dimeric b6f Complex
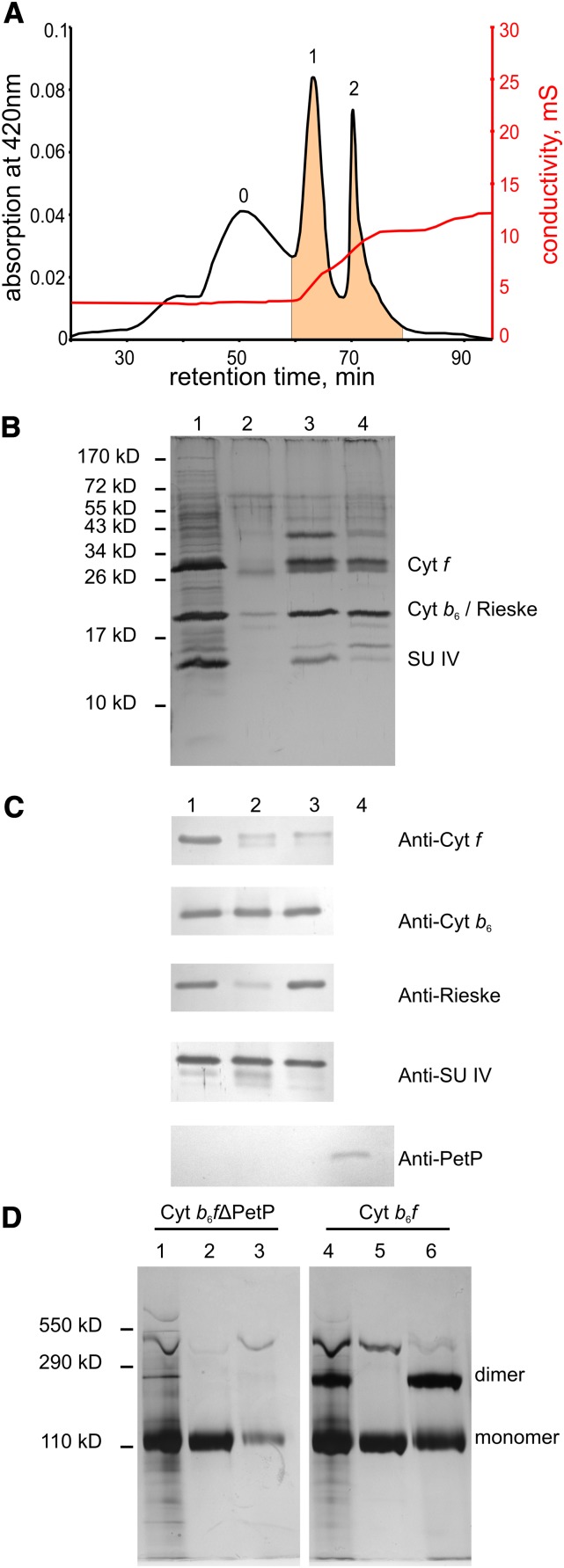
To analyze the physiological function of PetP, a ΔpetP mutant was generated in the genomic background of the tagged b6f variant with a His-tag on the cyt f subunit (for simplicity, the His-tagged variant is referred to as “WT” hereafter). Purification of the b6f-ΔPetP complex was done similar to that of the WT complex, i.e., by an IMAC column followed by an IEC column. Figure 3 shows the elution of b6f-ΔPetP from the IEC column, after prepurification by the IMAC column. In comparison with the WT complex (Figure 1B), the amount of monomeric b6f-ΔPetP (peak 1) clearly increased relative to the dimer (peak 2). Also, a new fraction consisting of unbound material (peak 0) appeared exclusively in the b6f-ΔpetP mutant. SDS-PAGE of the isolated complex after the IMAC column (Figure 3B, lane 1) showed, besides numerous weaker bands, three intense bands that could be identified by immunoblot and MS analysis as subunits cyt f, SU IV, cyt b6, and the Rieske protein with the latter two being superimposed in one band. Lane 2 showing IEC fraction 0 contained several b6f subunits that, however, appear to originate from a denatured or degraded complex as was apparent from a distinctly lower mass of the cyt f subunit. Similarly, IEC fraction 1 (b6f monomer, lane 3) and fraction 2 (dimer, lane 4) showed a double cyt f band, of which the lower was a degradation product as confirmed by immunoblot analysis (Figure 3C). Additionally, immunoblots of the other subunits indicated a degradation of SU IV and a massive loss of the Rieske protein especially in the b6f monomer, while cyt b6 remained approximately unchanged. Using a specific PetP antibody, Figure 3C also confirms the complete absence of PetP in the ∆petP mutant on the protein level with heterologously expressed PetP from T. elongatus being used as reference. Additionally, PetP deficiency in the mutant was demonstrated by liquid chromatography-mass spectrometry (LC-MS) analysis of both isolated complexes and whole-cell extracts.
Figure 3.
Purification of the b6f Complex from the b6f-ΔpetP Mutant.
(A) Elution profile from a preparative IEC column (second purification step): peak 0, with b6f degradation forms; peak 1, b6f-monomer; peak 2, b6f-dimer. UnoQ1 (Bio-Rad), 0.5 mL/min, 4°C; gradient 1: 0 to 4.2 mM NaCl, gradient 2: 4.2 to 8.2 mM NaCl, gradient 3: 8 0.2 to 20 mM NaCl.
(B) SDS-PAGE (silver-stained) of fractions from chromatography: (1) IMAC of b6f-ΔPetP, (2) peak 0 of IEC, (3) peak 1 of IEC, and (4) peak 2 of IEC.
(C) Immunoblot analysis of Cyt b6f-ΔPetP: (1) Cyt b6f-ΔPetP-peak of IMAC, (2) IEC fraction 1 (b6f monomer), (3) IEC fraction 2 (b6f dimer), and (4) PetP heterologously expressed in E. coli as reference (synthetic peptide-antibody; Davids Biotechologie).
(D) BN-PAGE (silver-stained) of isolated b6f-ΔPetP- and WT b6f-complex: (1) b6f-ΔPetP after IMAC, (2) IEC fraction 1 (b6f-ΔPetP monomer), (3) IEC fraction 2 (b6f-ΔPetP dimer), (4) b6f after IMAC, (5) IEC fraction 1 (b6f monomer), and (6) IEC fraction 2 (b6f dimer); for details, see text.
Figure 3D shows the blue-native gel electrophoresis (BN-PAGE) analysis of b6f complexes isolated from WT and the ΔpetP mutant. It is apparent that already after IMAC the b6f-ΔpetP complex was mainly a monomer with a very small amount of dimer remaining (lane 1). This is reflected by the IEC run (lane 2+3), which showed a strong monomer and a very weak dimer band (lane 3), in agreement with a small dimer peak in Figure 3A. Apparently, the originally dimeric b6f-ΔpetP complex easily dissociates during storage or under the conditions of native electrophoresis. By contrast, BN-PAGE of WT b6f (with His-tag) showed a distinct amount of dimer both after IMAC (lane 4) and IEC column (lane 6, dimer peak), with homogenous monomeric complexes (lanes 2 and 5, respectively).
In summary, the b6f dimer seems to be destabilized in the ΔpetP mutant, which is also indicated by an additional degradation peak in the IEC step.
The different stability of b6f complexes from WT and ΔpetP mutant is also clearly reflected by their electron transport activity: In contrast to WT complexes with activities of (110 ± 34) e−/s/cyt f after IMAC column and (256 ± 70) e−/s/cyt f for the dimeric IEC fraction, ΔPetP mutant complexes yielded only (18 ± 2) e−/s/cyt f after IMAC column and no activity after IEC.
Interaction of b6f with PetP
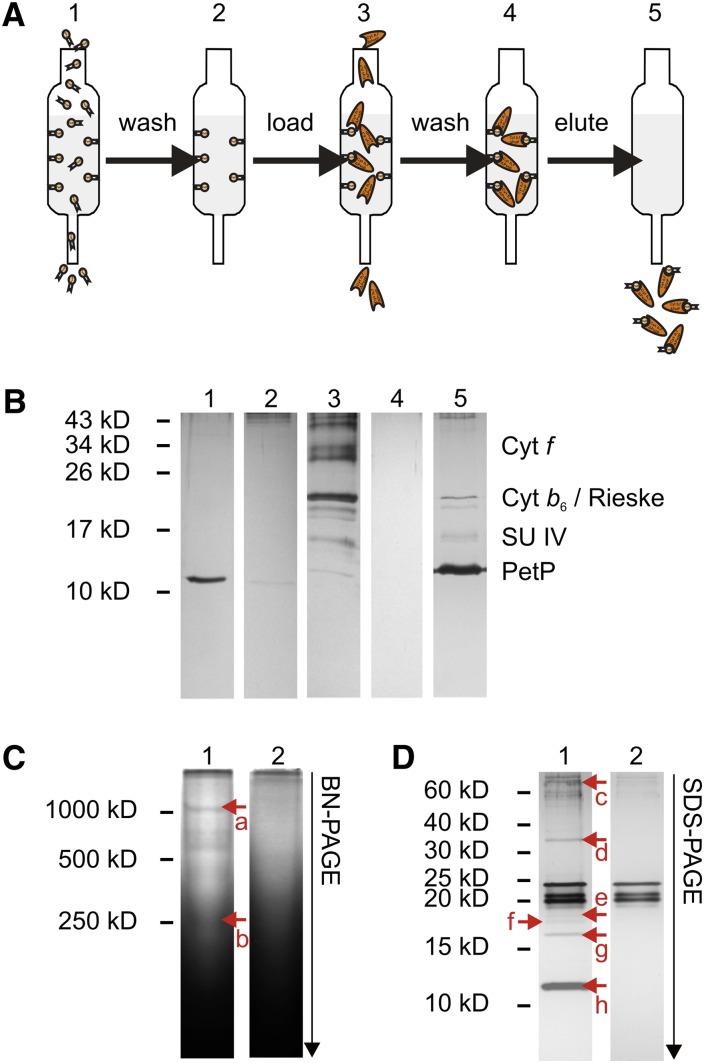
To test for direct interaction of the isolated partners in an in vitro system, PetP that had been heterologously expressed in E. coli was bound via a Strep-tag to a Strep-Tactin sepharose column and probed for interaction with a b6f-ΔPetP complex isolated from the corresponding mutant.
Figure 4A illustrates a pull-down assay to copurify potential interaction partners of PetP. When isolated b6f-ΔPetP complex was subjected to the pull-down assay, analysis of the various fractions by SDS-PAGE (Figure 4B) showed a coelution of b6f with PetP upon desthiobiotin treatment (lane 5), indicating specific binding of the complex via strep-tagged PetP to the column matrix. The identity of the protein subunits was confirmed by MS and immunoblotting analysis.
Figure 4.
Screening for Protein Interaction Partners of Heterologously Expressed PetP.
(A) Work flow of the assay: Strep-tagged PetP was bound as bait to the column, followed by washing (5 column volumes) and loading of the sample for interaction screening. After repeated washing (15 column volumes) PetP and potential interaction partners were specifically coeluted by desthiobiotin treatment.
(B) Silver-stained SDS-PAGE gel of the pull-down assay with isolated b6f-ΔPetP complex. (1) Heterologously expressed PetP, (2) wash fractions after PetP binding, (3) isolated b6f-ΔPetP, (4) wash fractions after b6f-ΔPetP-loading, and (5) elution by desthiobiotin.
(C) Silver-stained BN-PAGE gel of the pull-down assay with β-DDM treated membrane extract. 1, Membrane extract; 2, control. Proteins identified by LC-MS: (a) PsaA, PsaB, PsaD, PsaF, cyt f, cyt b6, Rieske, SU IV, PetP, NdhF3; (b) cyt f, cyt b6, Rieske, SU IV, and PetP.
(D) Silver-stained SDS-PAGE gel of the pull-down assay with β-DDM treated membrane extract. 1, Membrane extract; 2, control. Proteins identified by LC-MS: (c) NdhF3, (d) cyt f, (e) cyt b6, (f) Tll1812, (g) SU IV, and (h) PetP.
[See online article for color version of this figure.]
To exclude nonspecific binding of the complex to the column matrix, b6f-ΔPetP was also applied to a column matrix lacking immobilized PetP. The fact that neither SDS-PAGE nor LC-MS could identify any b6f subunit after various washing steps confirmed that the binding of b6f-ΔPetP to the column was mediated exclusively by PetP.
For the detection of new interaction partners, extracts of cytosolic and membrane proteins were applied to the column with immobilized PetP. BN-PAGE, SDS-PAGE, and LC-MS analysis were used to identify b6f subunits and potential interaction partners in the fractions eluted with desthiobiotin from the column. Table 2 summarizes proteins that have been identified by direct LC-MS analysis from the desthiobiotin eluates. In addition to b6f subunits, various PSI subunits as well as ferredoxin were identified specifically in the fractions eluted from the PetP column. In the blue-native gel, a weak band with ∼1000 kD mass was visualized (Figure 4C, a), which contained both PSI and b6f subunits. This indicates a direct interaction of b6f and PSI complexes as has been reported for green algae (Iwai et al., 2010). Also, SDS-PAGE (Figure 4D) and subsequent LC-MS indicate the binding of NdhF3 and protein Tll1812; the latter is a homolog of protein Sll2012 from Synechocystis sp PCC 6803, which is induced by CO2 limitation and has been proposed as constituent of a Na+-translocating NADH dehydrogenase (Wang et al., 2004; Daley et al., 2012).
Table 2. Potential Interaction Partners of PetP Isolated from Total Membrane Extracts on the Saturated PetP Column.
| Name | DBIDa | Immob. PetPb | Controlb,c |
|---|---|---|---|
| b6f, PetP | tsr0524 | 4.20 | – |
| Biotin carboxyl carrier protein (AccB) | tlr1295 | 1.98 | 1.62 |
| b6f, PetB | tlr0796 | 0.77 | – |
| b6f, cytochrome f (PetA) | tlr0960 | 0.68 | – |
| b6f, Rieske protein (PetC) | tlr0959 | 0.34 | – |
| PBS, allophycocyanin beta (ApcB) | tll0956 | 0.32 | 0.09 |
| b6f, subunit IV (petD) | tlr0797 | 0.29 | – |
| PBS, allophycocyanin alpha (apcA) | tll0957 | 0.27 | 0.11 |
| PBS, rod linker (CpcC) | tlr1959 | 0.13 | – |
| PBS, phycocyanin beta (CpcB) | tlr1957 | 0.13 | 0.20 |
| Ferredoxin (PetF) | tsl1009 | 0.12 | – |
| DnaK2 | tlr1733 | 0.10 | 0.08 |
| PBS, phycocyanin alpha (CpcA) | tlr1958 | 0.10 | 0.15 |
| PBS, small core linker (ApcC) | tsl0955 | 0.09 | – |
| Hypothetical protein | tsr1131 | 0.08 | – |
| PSI, subunit II (PsaD) | tll1724 | 0.06 | – |
| 60-kD chaperonin 1 (GroEL-1) | tll0185 | 0.05 | – |
| DNA binding protein HU | tll2267 | 0.05 | 0.07 |
| PSI, subunit VII (PsaC) | tsl1013 | 0.04 | – |
| PSI, subunit III (PSI-F) | tlr2411 | 0.04 | – |
Gene identifier according to CyanoBase (http://genome.microbedb.jp/CyanoBase).
Spectral abundance factors are used to represent the outcome of the label-free quantitation by LC-MS.
In the control experiment, an empty Strep-Tactin column instead of the PetP-saturated column was used.
Effect of PetP on Cell Growth
On the whole-cell level, the functional effect of PetP was analyzed via growth curves and electron transport kinetics of the ΔpetP mutant, WT, and petP-complementation mutant. Growth curves were recorded under different light conditions, revealing no significant change in the generation time at 80 μE m−2 s−1 between WT (10.4 ± 0.6) h, ΔpetP mutant (11.6 ± 1.1) h, and petP-complementation mutant (10.4 ± 0.9) h. By contrast, significant changes were observed at 120 μE m−2 s−1: While the generation time of WT and petP-complementation mutant remained constant (at 9.9 ± 1.1 and 10.1 ± 1.1 h, respectively), the ΔpetP mutant showed a distinctly increased generation time (of 15.9 ± 0.4 h). Also, in contrast to WT and petP-complementation mutant, the ΔpetP mutant showed a much slower growth rate, only 62% of WT, and a significantly lower maximum cell density, about OD 2.4 versus about OD 3.7 at 750 nm (Supplemental Figure 3).
Influence of PetP on Light Tolerance
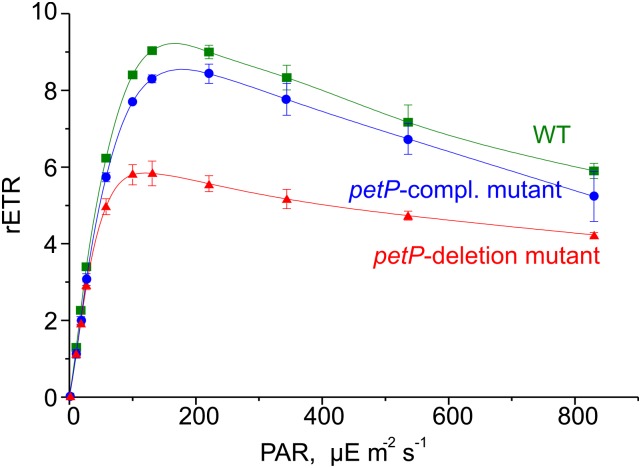
PSII light saturation curves of WT and mutant cells reveal differences in the relative electron transport rate, i.e., the ratio between absorbed light quanta and transported electrons, and in light adaptation mechanisms. When cells are exposed to gradually increasing light intensities, the electron transport increases in parallel up to its capacity limit. Beyond this point, increasing light intensity induces photoinhibition, i.e., a decrease of the relative electron transport rate. Figure 5 shows such light saturation curves for T. elongatus WT, ΔpetP mutant, and petP-complementation mutant. In contrast to WT with a maximum electron transport rate at ∼130 μE m−2 s−1, the ΔpetP mutant reaches its maximum already at 80 μE m−2 s−1, combined with a 30% lower electron transport rate. This is reverted in the petP-complementation mutant, which shows a similar light tolerance as WT cells, confirming the specific effect of PetP in preventing earlier photoinhibition. It is also in line with the observed slower growth rate of the ΔpetP mutant at 120 μE m−2 s−1 light (Supplemental Figure 3). At very high light intensities, i.e., >800 μE m−2 s−1, WT and ΔpetP mutant show similar electron transport rates. Under such conditions, the relative electron transport rates are generally reduced due to an overreduced PQ pool, and cyclic electron transport pathways strongly dominate over linear electron transport.
Figure 5.
Influence on PSII Light Saturation.
Light saturation measurements of T. elongatus WT (green, squares), the ΔpetP mutant (red, triangles), and the petP-complementation mutant (blue, circles) reveal the dependence of the relative electron transport rate (rETR), i.e., the ratio between absorbed light quanta and transported electrons, on the light intensity. PAR light intensity in μE m−2 s−1. Error bars indicate the sd of three independent measurements.
[See online article for color version of this figure.]
Effect of PetP on Electron Transport through b6f
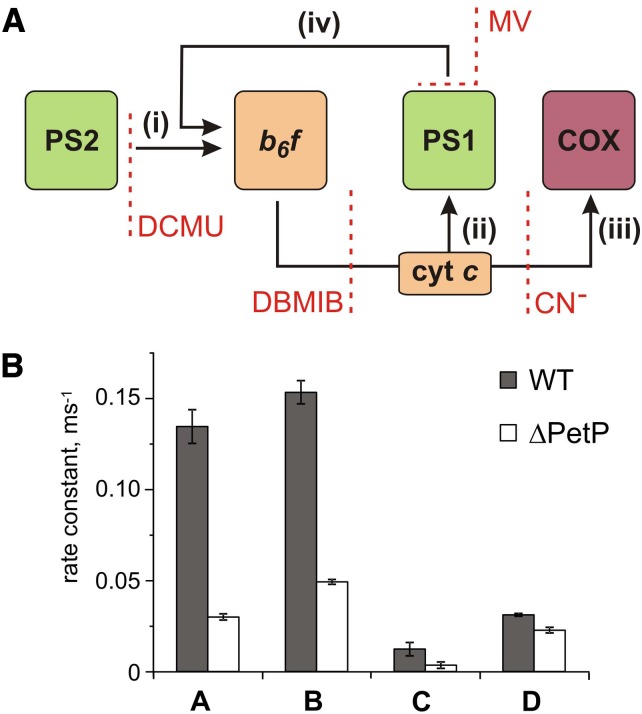
To quantify linear and cyclic photosynthetic electron transport as well as the respiratory electron transport toward cytochrome c oxidase (COX), P700 reduction kinetics after saturating light pulses were recorded in samples with and without inhibitors of specific electron transport routes (Figure 6A; Supplemental Figure 4). Such P700 reduction rates are shown in Table 3 for WT and the ΔpetP mutant. Based on these measurements, electron transport rates were determined for the different routes (Figure 6B; for details, see Methods). Apparently, the electron flow through b6f is considerably reduced in the ΔpetP mutant, with a decrease in comparison to WT down to 22% (linear electron transport from PSII; Figure 6A, i), 32% (flow toward PSI; Figure 6A, ii), and 28% (flow toward COX; Figure 6A, iii), respectively. As the ratio of the electron transport to PSI and COX stays constant despite the significant decrease in the transport capacity of b6f, distribution of electrons by the reduced cytochrome c is not affected by the absence of PetP. However, the over 3-fold decelerated linear electron transport in the mutant indicates distinctly slower electron transport through b6f in the absence of PetP resulting in delayed rereduction of PSI.
Figure 6.
P700+ Rereduction Kinetics.
(A) Model used for quantification of linear (i) and cyclic (iv) electron transport as well as the respiratory electron flow (C) based on P700 rereduction measurements. (i) Electron flow rate from PSII; (ii) electron flow rate to PSI; (iii) respiratory electron flow to COX; (iv) cyclic electron flow.
(B) Determined electron transport rates (i) to (iv) for T. elongatus WT (gray) and ΔpetP mutant (white) based on the kinetic measurements shown in Table 3. kD, ox and kD, red represent the rate for cyclic electron transport to an oxidized or reduced PQ pool, respectively.
[See online article for color version of this figure.]
Table 3. P700+ Rereduction Rates Obtained for the Wild Type and ΔpetP Mutant.
| Inhibitor | WT (ms−1)a | ΔpetP (ms−1)a |
|---|---|---|
| No inhibitor | 0.153 ± 0.004 | 0.049 ± 0.001 |
| Methyl viologen | 0.137 ± 0.007 | 0.039 ± 0.002 |
| DCMU | 0.019 ± 0.003 | 0.0192 ± 0.0003 |
| KCN+DCMU | 0.031 ± 0.001 | 0.022 ± 0.001 |
| DBMIB | 0.0014 ± 0.0003 | 0.0008 ± 0.0004 |
Mean and standard deviations based on at least three independent measurements.
Inhibition of the linear electron transport from PSII by DCMU decreases the P700 rereduction rate for WT and ΔpetP mutant to 12 and 39% of the rate observed in the untreated sample, respectively. This is due to the high percentage of linear relative to total electron transport. By contrast, there is only a minor change in the cyclic electron transport rate, which decreases in the mutant to 73 and 62% under conditions of oxidized or reduced PQ pool, respectively. This indicates a 25% contribution of cyclic electron flow to the P700 rereduction in the mutant and a 10% contribution in WT without any inhibitors.
In conclusion, the low electron transport rate in the ΔpetP mutant can be attributed mainly to reduced linear electron transport through b6f, which causes overreduction of the PQ pool.
DISCUSSION
Isolation of Stable Dimeric b6f Complexes Indicates Structural Constraints
Since the first isolation of an active b6f complex from spinach (Spinacia oleracea) chloroplasts (Hurt and Hauska, 1981), the purification strategy has been continuously improved and used for many photoautotrophic organisms ranging from cyanobacteria to green algae. Considerable progress resulted from the introduction of molecular genetic tag systems for the protein complex isolation, which reduced the purification time and with it the exposure time to detergents and other disintegrating components. Additionally, better column materials and milder detergents resulted in improved isolation of native, active b6f complexes that in turn enabled the elucidation of the b6f crystal structure from the green alga Chlamydomonas reinhardtii (Stroebel et al., 2003), the thermophilic cyanobacterium Mastigocladus laminosus (Kurisu et al., 2003), and the mesophilic cyanobacterium Nostoc sp PCC 7120 (Baniulis et al., 2009). Here, we report a method developed to isolate dimeric b6f complexes in high purity from T. elongatus. In contrast to previously reported preparations with active electron transport, the PetP subunit (Volkmer et al., 2007) was successfully coisolated with this b6f complex. The cause for the loss of PetP in the other preparations and also in all available crystal structures may be species dependent, method dependent, or a combination of both. One example of a possible method-specific effect is the treatment of thylakoid membranes with chaotropic salts for the removal of phycobilisomes (Zhang and Cramer, 2004), which most probably also removes transiently-bound components like PetP.
In contrast to other thermophilic systems (Kurisu et al., 2003), the genome of T. elongatus is completely sequenced (Nakamura et al., 2002) and the organism is easily accessible for molecular biology, which enables structural and functional studies of WT and mutant strains. Due to our gentle purification conditions, using a combination of IMAC (via a His-tag on cyt f) with anion exchange chromatography (IEC), the rather labile b6f-ΔpetP mutant complex also could be isolated and characterized in comparison with the WT complex. This yielded information on the stabilizing effect of PetP and the stoichiometry of bound PetP, two subunits per dimer, which could not be obtained on the full-cell level. Furthermore, the binding interface of PetP could be localized on the cytoplasmic side of b6f applying advanced cross-linking techniques to the isolated dimeric complex. The specific binding of isolated b6f-ΔpetP mutant complex to immobilized PetP could be demonstrated only by this biochemical approach. Finally, the stability and activity of this preparation now enables detailed characterization of numerous site-directed mutants on the level of isolated b6f complexes, which certainly will contribute to a better understanding of structure-function relationships on the molecular level.
Contribution of PetP to the Structure-Function Relationship in Isolated b6f
While oligomerization of autonomous monomeric complexes into dimers and trimers is a widespread principle for bioenergetic complexes of the photosynthetic electron transport chain, dimerization of the b6f complex is mandatory for its catalytic function (Breyton et al., 1997; Dietrich and Kühlbrandt, 1999). In contrast to PSI and PSII, which show high electron transport activities also as monomers (Takahashi et al., 2009; Watanabe et al., 2009; El-Mohsnawy et al., 2010), monomeric b6f is structurally impaired and functionally inactive (Breyton et al., 1997; Cramer et al., 2005).
The three-dimensional structure indicates why the dimerization of this complex is important for stability and functionality. As the hydrophobic and the hydrophilic domain of the Rieske protein are structurally and functionally linked with different monomers, this subunit connects the monomers in the dimer. Due to this, monomerization of the b6f complex must induce the, at least partial, loss of the Rieske subunit, which is combined with a loss of functionality (Breyton et al., 1997; Cramer et al., 2005). This is confirmed by our b6f preparation from T. elongatus, which is active only as a dimer and loses its activity proportional to its dissociation into monomers.
Remarkably, besides the small subunits PetG, -M, and -N, we observed an additional mass signal corresponding to subunit PetL, which had not been annotated in the genome of T. elongatus. While this may represent the missing link, a final assessment based on these data is not possible as the chemical background in the mass range of PetL contributes significantly to the signal.
Furthermore, a comparison of the b6f subunit composition from different organisms indicates a conservation of posttranslational processing of the subunits (Whitelegge et al., 2002; de Vitry et al., 2004). This includes modifications such as N-formylation, the cleavage of signal sequences and the covalent attachment of prosthetic groups. The electron transport activity of b6f isolated from WT T. elongatus, i.e., 256 e−/cyt f/s, is also in agreement with activities reported for other organisms (Table 4).
Table 4. Plastocyanin/Cytochrome c Reduction Activities of Isolated b6f Complexes.
| Organism | Reference | Activity ± sd [e−(cyt f/s] |
|---|---|---|
| T. elongatus | This work | 256 ± 70 |
| M. laminosus | Zhang and Cramer (2004) | 200-300 |
| Zhang et al. (2001) | 340 ± 50 | |
| Huang et al. (1999); Zhang et al. (2003) | 300-450 | |
| Nostoc sp PCC7120 | Baniulis et al. (2009) | 277 ± 14 |
| S. oleracea | Zhang et al. (2001) | 290 ± 60 |
| Dietrich and Kühlbrandt (1999) | 450 ± 60 | |
| C. reinhardtii | Pierre et al. (1995) | 270 ± 60 |
Evolutionary Implications for the Potential Physiological Role of PetP
PetP is a highly conserved cyanobacterial subunit apparently lacking homologs in green algae and plants. While this may reflect a loss of PetP during evolution, PetP homologs can surprisingly be found in red algae. Although sequence information available for red algae is very limited, a BLASTP analysis yielded seven PetP homologs, all from their plastid genomes. The peptide sequences of cyanobacterial and red algae PetPs are of similar length with sequence identities up to 40% (Figure 7). Also, a high number of N-terminal positive charges are conserved in the sequence of PetP, complementary to the negative charges in the N-terminal helix of cyt b6, the dominant domain of the b6f complex exposed to the cytoplasm. As cyanobacteria and red algae also both contain phycobilisomes for light harvesting (Enami et al., 1995), a functional relationship between PetP and phycobilisomes, possibly in the regulation of state transitions, should be considered and investigated in more detail. We previously reported the inability of Synechocystis petP deletion mutants to perform state transitions under varying light conditions; however, this effect may be indirect and could also be attributed to an overreduced PQ pool (Volkmer et al., 2007).
Figure 7.

Sequence Alignment of Various PetP Proteins.
Sequence alignment of various PetP proteins involving the following species. Cyanobacteria: T. elongatus (T. elongatus BP-1, pI: 4.8), Synechocystis (Synechocystis PCC 6803, pI: 8.0), Nostoc (Nostoc sp PCC 7120, pI: 8.0), A. marina (Acaryochloris marina MBIC 11017, pI: 6.2); red algae: Py. yezoensis (Pyropia yezoensis, pI: 9.1), Py. haitanensis (Pyropia haitanensis, pI: 9.7), Po. purpurea (Porphyra purpurea, pI: 9.8), Po. umbilicalis (Porphyra umbilicalis, pI: 9.4). Fully, highly, and relatively conserved residues are shaded dark green, green, and light green, respectively. Negatively and positively charged residues are highlighted with red and blue, respectively. The alignment was done with the program Clustal Omega (Sievers et al., 2011).
If PetP has a significant, possibly regulatory, role, the function of this subunit has to be taken over by other subunits in organisms lacking petP genes. A comparison with other deletion mutants shows that only the loss of subunit PetL in plants leads to a similar phenotype as the ∆PetP mutant in cyanobacteria. Both in C. reinhardtii and tobacco (Nicotiana tabacum), the reduced stability of b6f (Takahashi et al., 1996) lowers the amount of dimeric complex, Rieske protein (Schwenkert et al., 2007), and electron transport activity. Similarly to the cyanobacterial ∆PetP mutant, only the linear, but not the cyclic electron transport is affected.
Suggested Role of PetP in Cyanobacterial Electron Transport
The results presented here strongly support a model for the effect of PetP in cyanobacterial photosynthetic electron transport as shown in Figure 8. This model is based on the dramatic drop of linear electron transport and the essentially unaffected cyclic electron transport in the ΔpetP mutant. Evaluation of the isolated dimeric complexes of WT and mutant suggests that the impaired b6f complex of the mutant, which showed only 12% of the activity of WT complex, is the reason for an overreduced PQ pool leading to the drop in linear electron transport. By contrast, the rather unaffected cyclic electron transport of the mutants, based on the P700+ re-reduction kinetics, suggests the following models: (1) Each individual b6f complex lacking PetP is impaired in linear electron transport but still performs, although at a low level, cyclic electron transport. Or, (2) there are distinct b6f pools utilized exclusively for linear or cyclic electron transport. The b6f pool involved in cyclic transport, for instance, may be stabilized by structural interaction with PSI, NDH-1, and/or other constituents, while the linear electron transport pool is not. Such a supercomplex consisting of PSI, b6f, FNR, and other components involved in cyclic electron transport was reported for C. reinhardtii (Iwai et al., 2010) and is also suggested by our BN-PAGE analysis of PetP interaction partners. Additional hints of such a supercomplex in cyanobacteria are the coisolation of b6f and PSI from T. elongatus (IMAC column step, this report) and a similar copurification in the case of Synechocystis PCC 6803 (Bald et al., 1992), which could be avoided only by the use of a PSI-deficient mutant (Shen et al., 1993). PetP may play an active role in such a rearrangement or “sorting” of membrane complexes due to its ability to bind only transiently to the b6f complex. This could imply the transient binding of PetP to other partners as well, which may be involved in a regulatory cycle. We could identify NdhF3 and protein Tll1812, homologous to a potential NADH dehydrogenase subunit (Wang et al., 2004; Daley et al., 2012), as candidates for binding partners. Interestingly, this would also be in agreement with the formation of a supercomplex between b6f and NDH-1 with a potential role in cyclic (Ohkawa et al., 2000; Battchikova et al., 2011) and respiratory electron transport (Schmetterer, 1994). As recently observed for NDH-1 and succinate dehydrogenase, binding of PetP to b6f and NDH-1 could induce redistribution of these complexes, combined with changes in their electron transport activity (Liu et al., 2012). Our results further suggest that such changes may be triggered by stress conditions, such as photoinhibition, as the ΔPetP strain apparently lost the ability to tolerate external stresses like high light as evident from significant growth limitations and from the PSII light saturation curves.
Figure 8.
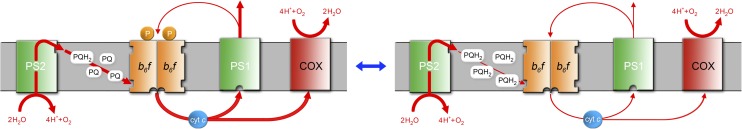
Model for the Suggested Role of the PetP Subunit in Cyanobacterial Photosynthetic Electron Transport.
On the left the situation in the WT is depicted. The linear electron transport involving water oxidation, PSII, the PQ pool, b6f, and PSI is significantly larger than the cyclic transport between b6f and PSI. When PetP is missing (right), the linear electron transport is strongly impaired, while the cyclic transport rate is still similar to the WT level. This impairment of linear electron transport leads to overreduction of the PQ pool. As determined on the cellular level, the model shows a stoichiometry of one PetP subunit per b6f monomer (which still has to be confirmed on the individual protein level and which may depend on the physiological state).
In summary, this report shows that PetP is an important b6f subunit both for structural integrity and functional efficiency. It has a major influence on the activity of linear electron transport and may fulfill a dynamic regulatory role on the cytosolic surface of the thylakoid membrane in contact with other complexes, especially for stress adaptation.
METHODS
Cultivation and Thylakoid Membrane Preparation
Thermosynechococcus elongatus was grown in a 25-liter foil photo bioreactor (Bioengineering) at 45°C in BG-11 medium, bubbled with CO2-enriched air (5%), and illuminated with white light of increasing intensity (50 to 250 μE m−2 s−1). Cells were harvested after 3 to 4 d at OD750 = 2 and concentrated to ∼2 liters using a hollow fiber system (Amicon DC10 LA). After disruption of the cells by a Parr bomb (2000 p.s.i.; according to Kuhl et al., 2000), thylakoid membranes were pelleted (Sorvall GSA rotor, 12,000 rpm, 15 min, 4°C) and resuspended in 200 mL buffer A (20 mM HEPES, pH 7.0, 20 mM CaCl2, and 10 mM MgCl2). After repeated centrifugation and resuspension in a small volume of buffer B (20 mM HEPES, pH 7.0, 20 mM CaCl2, 10 mM MgCl2, and 500 mM mannitol), the membranes were frozen in liquid nitrogen and stored at −80°C.
Construction of a ΔpetP Mutant
Genomic DNA isolated from T. elongatus was used as a template to amplify the up- and downstream sequences of the tsr0524 (petP) gene. For the upstream sequence, the following oligonucleotide primer pair was used: nt-up tsr0524 5′-AAGGAAAAGCGGCCGCATGCTGGGAAACTAGCACGTCCGG-3′ and ct-up tsr0524 5′-CCAATGCATTGGTTCTGCAGCAACTCCCAAACTTTGTATAATCC-3′. For the downstream sequence, the following oligonucleotide primer pair was used: nt-down tsr0524 5′-CCCAAGCTTCCTGCCCTGCCGCCTTTTTCACAGC-3′ and ct-down tsr0524 5′-CCGCTCGAGCCTATGCTTTCAGTTTTTCAGCC-3′. The PCR products were cloned into the pBluescript II KS(+) vector (Fermentas) containing a kanamycin resistance cassette. Cells of wild type and a petCA-His10 strain containing a chloramphenicol resistance cassette of T. elongatus were transformed by electroporation (Mühlenhoff and Chauvat, 1996; Iwai et al., 2004) with this vector to generate the ΔpetP mutant strains. Mutant cells were selected by gradually adding kanamycin to the medium up to a concentration of 200 μg/mL or both 16 µg/mL chloramphenicol and 200 μg/mL kanamycin for the double mutant. Single colonies were obtained by plating the mutant strain onto selective agar (200 μg/mL kanamycin and 8 μg/mL chloramphenicol). Segregation was checked by PCR using the following oligonucleotide primer pair: nt-SegCheck 5′-GAATCACCTCATAGCGATCGCCATTGTC-3′ and ct-SegCheck 5′-ATTGACCCAGCTCAGTTGCTGT-3′.
Isolation of His-Tagged b6f and b6f-ΔPetP Complexes
His-tagged b6f-complexes (10× His fused to the C terminus of the cyt f subunit) were isolated from T. elongatus cells that had been harvested in the exponential growth phase. Thylakoid membranes were solubilized in extraction buffer (20 mM HEPES, pH 7.0, 20 mM CaCl2, 10 mM MgCl2, 5 mM Na-ascorbate, 1 mM phenylmethylsulfonyl fluoride, 1 mM N-tosyl-l-lysine-chloromethyl ketone, 0.1 mg/mL DNase, 0.1 mg/mL RNase, 1% [w/v] n-dodecyl-β,d-maltoside [β-DM; Biomol]) at a chlorophyll concentration of 1 mg/mL. After incubation for 20 min, an identical volume of extraction buffer without β-DM was added, followed by centrifugation (Beckman Ti70, 47,000 rpm, 90 min, 4°C) and loading of the supernatant onto a chelating sepharose fast flow column (GE Healthcare) equilibrated with IMAC buffer (20 mM HEPES, pH 7.0, 20 mM CaCl2, 10 mM MgCl2, 500 mM mannitol, 5 mM Na-ascorbate, 0.03% [w/v] β-DM, and 300 mM NaCl). b6f was eluted by a linear gradient of 10 to 100 mM histidine. The pellet was resuspended in IEC buffer (16 mM HEPES, pH 7.0, 10 mM CaCl2, 5 mM MgCl2, 400 mM mannitol, 5 mM Na-ascorbate, and 0.032% [w/v] β-DM) and loaded onto a UNO Q1 column (Bio-Rad) that had been equilibrated with IEC buffer. b6f monomer and dimer were eluted with 1 M NaCl by a stepwise gradient (0 to 4.2 mM, 4.2 to 8.2 mM, 8.2 to 20 mM, and 20 to 50 mM), frozen in liquid nitrogen, and stored at −80°C.
Structural/functional integrity and purity of the complex was assayed by silver-stained (Blum et al., 1987) SDS-PAGE (Laemmli, 1970), immunoblotting (Towbin et al., 1992), and MS analysis. Activity of isolated complexes was assayed according to (Hurt and Hauska, 1981; Metzger et al., 1997) using 50 μM plastochinol as electron donor, 0.15 μM b6f complex and 20 μM cytochrome c as electron acceptor. The redox state of cytochrome c was monitored by spectrophotometry at 550 nm.
BN-PAGE
BN-PAGE with the Hoefer SE 600 system (18 × 16 × 0.15 cm3, 10 lanes) was performed as previously described (Schägger and von Jagow, 1991; Schwassmann et al., 2007); stacking gels with 3.5% acrylamide and separating gels with linear acrylamide gradients of 5 to 13% were used, and 250 μg solubilized protein was applied per lane.
Gels were stained with Coomassie R 250 or lanes of the blue-native gel were cut out and incubated in a solution of 1% (w/v) SDS and 1% (w/v) β-mercaptoethanol at 20°C. After 30 min, lanes were analyzed by Tricine-SDS-PAGE in the second dimension (Schägger et al., 1994).
MS Analysis and Protein Identification
For molecular mass determination of protein subunits, samples were analyzed on a Voyager-DE PRO (Applied Biosystems) MALDI-TOF mass spectrometer according to Poetsch et al., (2003) using sinapinic acid in 50% acetonitrile/0.1% trifluoroacetic acid as matrix for dried droplet target preparation.
LC-MS analysis for protein identification was performed according to Rexroth et al. (2012) with slight modifications. For reverse phase chromatography, a gradient of solvent A (95% water, 5% acetonitrile, and 0.1% formic acid) and solvent B (10% water, 85% acetonitrile, 5% isopropanol, and 0.1% formic acid) was used. MS analysis was done on a Thermo LTQ Orbitrap mass spectrometer, operated in a duty cycle consisting of one 400 to 2000 m/z FT-MS and four MS/MS LTQ scans.
Analysis of the liquid chromatography-tandem mass spectrometry data was performed using the Sequest algorithm (Eng et al., 1994) implemented in the Proteome Discoverer 1.3 software (Thermo Scientific) for peptide identification versus a database consisting of all T. elongatus proteins with a tolerance of 10 ppm for the precursor mass accuracy and 1 unit for the fragment mass accuracy. False discovery rates were estimated by the number of spectral matches to a decoy database (Käll et al., 2008). Acceptance criteria and filters were set to achieve a false positive rate of 5%. Spectral counting (Liu et al., 2004) and normalized spectral abundance factors (Zybailov et al., 2006; Usaite et al., 2008) were applied for relative quantification. The spectral abundance factors consider the length of a protein, which influences the number of spectral counts. Spectral abundance factors take into account that bigger proteins yield more peptides than small ones during proteolytic digest (Neilson et al., 2011).
Absolute Peptide Quantification
For absolute peptide quantification a triple quadrupole mass spectrometer (TSQ Vantage; Thermo Scientific) was operated in SRM mode (Gerber et al., 2003). Custom peptides were synthesized by Thermo Scientific for PetA, PetP, PetC, and PetD from T. elongatus with stable isotopes of lysine or arginine to induce mass shifts of 8 or 10 D per peptide, respectively. Supplemental Table 3 contains a list of the peptides and corresponding SRM transitions. These custom peptides were spiked into the tryptic digest right before the MS analysis. For the TSQ Vantage, an ion spray voltage of 1800 V was set, the temperature of the ion transferring tube was adjusted to 270°C, and the collision activated dissociation pressure was set to 1.5 mTorr. The S-lens voltage was experimentally defined, while the collision energy was determined by the skyline software (MacLean et al., 2010). The biological replicate was analyzed 3-fold.
SRM data have been deposited at the PeptideAtlas SRM Experiment Library and are accessible via the http://www.peptideatlas.org/PASS/PASS00282 (Farrah et al., 2012).
All SRM data were manually inspected for retention time and peak identification. The ratios between peak areas of light and heavy labeled peptides were calculated using the skyline software (MacLean et al., 2010). For the technical replicates, the median and sd were calculated.
Cross-Linking and Identification of Cross-Linked Peptides
Purified b6f complex (2.4 μM) was incubated with a 1:1 mixture of BS3-H12/D12 (Creative Molecules) in buffer (16 mM HEPES, pH 7.0, 10 mM CaCl2, 5 mM MgCl2, 4 mM Na-Ascorbat, and 0.03% β-DDM) with a final concentration of 5 mM of the cross-linker. After 30 min incubation on ice, the cross-linking reaction was terminated by addition of NH4HCO3 (100 mM). Tryptic digestion in solution and sample preparation was performed after protein precipitation by addition of cold acetone (Nowaczyk et al., 2011). Peptides were analyzed on an Orbitrap Elite mass spectrometer (Thermo Fischer Scientific) operated in a duty cycle, which consisted of one FT-MS scan (300 to 2000 m/z, resolution: 240,000) and 20 tandem mass spectrometry scans. Dynamic exclusion was set to 1 min.
Cross-linked peptides were identified with StavroX v. 3.1.19 (Götze et al., 2012) and a cytochrome b6f complex subunit-specific database. To sort out false-positive results, two runs per sample were made (H12-BS3 and D12-BS3) and the corresponding spectra were validated manually only in case of hits in both.
In Vivo Analysis: P700+ Oxidation-Reduction Kinetics/Light Saturation Curves
For measurement of P700 kinetics, T. elongatus wild type and mutants were grown in a continuous culture system (Photobioreactor FMT 150/400; Photon System Instruments) at 318.15 K in BG11 medium to an OD735 of 0.75 ensuring constant culture conditions. The half-life of P700+ reduction was determined by a Dual PAM-100 system (Walz). After 10 min dark incubation, cells were exposed to a 50-ms actinic light pulse (2000 μE) and a simultaneous 50-ms multiple turnover pulse to completely oxidize all P700 reaction centers. Subsequent measurement of the P700+ rereduction kinetics yielded electron transport rates toward PSI. Single exponential functions were used for fitting. To estimate linear and cyclic electron transport rates as well as the respiratory flow, 500 μM methyl viologen, 500 μM DCMU, and a combination of 2 mM KCN and 500 μM DCMU were used to block electron pathways as depicted in Figure 6A. Complete inhibition of the electron transport through the b6f complex was achieved by the addition of 50 μM DBMIB. For data interpretation, linear and cyclic electron transport routes for PQ reduction and cytochrome c oxidation by PSI and cytochrome c oxidase have been considered (Yu et al., 1993; Yeremenko et al., 2005). The following set of equations describes the inhibitor experiments (Figure 6A):
(1) no inhibitor :
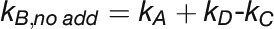
(2) MV :
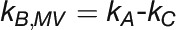
(3) DCMU :
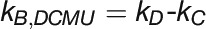
(4) KCN + DCMU :

Thus, individual rate constants can be obtained as
(5)
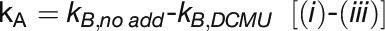 : Electron flow from PS2
: Electron flow from PS2(6)
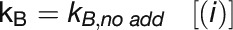 : Flow to PS1
: Flow to PS1(7)
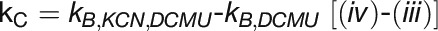 : respiratory flow to COX
: respiratory flow to COX(8)
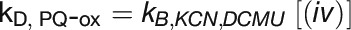 : Cyclic electron flow; oxidized PQ pool
: Cyclic electron flow; oxidized PQ pool(9)
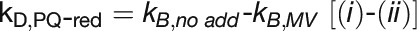 : cyclic electron flow; reduced PQ pool
: cyclic electron flow; reduced PQ pool
Light saturation curves of the photosynthetic electron transport rate were recorded at defined light intensities using a Dual PAM-100 system (Walz) according to Xu et al. (2008) and Imashimizu et al. (2011) with stepwise increase of the actinic light intensity from 0 to 830 μE m−2 s−1 with 30-s adaptation periods. Maximal fluorescence yields were obtained by saturating light pulses (duration 60 s, intensity 10,000 μE m−2 s−1) at the end of each 30-s period.
Heterologous Expression and Purification of PetP
Gene tsr0524 encoding the PetP subunit in T. elongatus was amplified from genomic DNA by PCR using the oligonucleotide primer pair petP BstBI for 5′-TGGTTCGAAAAAATCGAAGGGCGCATGGACGTGGGGCAAAAAGT-3′ and petP XhoI rev 5′-CGATCTCGAGTTAGCCAACGACTTCAACTTCATC-3′. The PCR fragment was subsequently inserted into pASK-IBA7 plus (IBA) between the BstBI and XhoI restriction sites. For overexpression, transformed Escherichia coli BL21 (DE3) was grown in Luria-Bertani medium supplemented with 200 μg/L anhydrotetracycline at 37°C. Cells were broken by French press treatment with four cycles at 10,000 p.s.i., and the supernatant was purified with Strep-Tactin sepharose resin (IBA) and a size exclusion column (Superdex75 column; GE Healthcare).
In Vitro Pull-Down Assay
The interaction study was done by conventional affinity chromatography. Heterologously Strep-tagged PetP, expressed in E. coli, was bound to a 5-mL gravity flow Strep-Tactin-sepharose column (IBA). Unbound PetP was removed by washing with five column volumes of buffer W (containing 0.03% [w/v] β-DM), followed by loading 0.7 μM b6fΔPetP onto the column and 30 min incubation at room temperature. After washing with 15 column volumes, samples were eluted with approximately seven column volumes of buffer E (IBA) containing 0.03% (w/v) β-DM. Eluted fractions with PetP- b6fΔPetP were analyzed by SDS-PAGE and LC-MS.
Accession Numbers
PetP protein sequences from this article can be found in the GenBank/EMBL databases under the following accession numbers: T. elongatus BP-1, BAC08076; Synechocystis PCC 6803, WP_010871443; Nostoc sp PCC 7120, WP_010998615; Acaryochloris marina MBIC11017, YP_001520284; Pyropia yezoensis, AGH27608; Pyropia haitanensis, YP_007947811; Porphyra purpurea, NP_053892; and Porphyra umbilicalis, AFC39951.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. 2D-BN/SDS Gel Analysis of Dimeric b6f Complex.
Supplemental Figure 2. MS2 Spectra and Peak Assignment of Cross-Linked PetP Peptides by the StavroX Software.
Supplemental Figure 3. Growth Curves of T. elongatus WT (Green), ΔpetP Mutant, and petP-Complementation Mutant at a Light Intensity of 120 μE m−2 s−1.
Supplemental Figure 4. P700 Rereduction Kinetics.
Supplemental Table 1. MALDI-TOF Analysis of Isolated b6f Protein Subunits.
Supplemental Table 2. Identified Cross-Linked PetP Peptides.
Supplemental Table 3. SRM Transitions Applied for Absolute Protein Quantification.
Acknowledgments
We acknowledge the financial support by the Deutsche Forschungsgemeinschaft (SFB 480, Project C1, MR), the Research School of the Ruhr-University Bochum (D.R. and K.U.C.), and the Greif-Stiftung (K.U.C.). We thank Navassard V. Karapetyan and David Fuente for helpful discussions of the P700 rereduction kinetics. We also appreciate the excellent technical assistance by R. Oworah-Nkruma.
AUTHOR CONTRIBUTIONS
S.R., D.R., and M.R. designed the research project. D.R. carried out most experiments. S.R. and N.P. performed the mass spectrometric analysis. K.U.C. and M.M.N. performed the cross-linking analysis. S.R. and S.V. contributed BLASTP analysis. D.R. and S.V. performed P700 rereduction measurements. S.V. performed screening for new interaction partners of PetP. S.R., D.R., S.V., and M.R. wrote the article.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- PQ
plastoquinone
- FNR
ferredoxin-NADP reductase
- IMAC
immobilized metal ion affinity chromatography
- MALDI-TOF
matrix-assisted laser desorption/ionization-time-of-flight
- IEC
ion exchange chromatography
- MS
mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
- BN-PAGE
blue-native gel electrophoresis
- β-DM
n-dodecyl-β,d-maltoside
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Allen J.F., Forsberg J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6: 317–326. [DOI] [PubMed] [Google Scholar]
- Bald, D., Kruip, J., Boekema, E.J., and Rögner, M. (1992). Structural investigations on cyt.b6f-complex from the cyanobacterium Synechocystis PCC 6803. Research in Photosynthesis. Proceedings of the IX. International Congress on Photosynthesis,Vol. I, N. Murata, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 629–632. [Google Scholar]
- Baniulis D., Yamashita E., Whitelegge J.P., Zatsman A.I., Hendrich M.P., Hasan S.S., Ryan C.M., Cramer W.A. (2009). Structure-function, stability, and chemical modification of the cyanobacterial cytochrome b6f complex from Nostoc sp. PCC 7120. J. Biol. Chem. 284: 9861–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battchikova N., Wei L., Du L., Bersanini L., Aro E.M., Ma W. (2011). Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 286: 36992–37001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat, G., and Rögner, M. (2011). Center of the cyanobacterial electron transport network: the cytochrome b6f complex. In Bioenergetic Processes of Cyanobacteria, G.A. Peschek, C. Obinger, and G. Renger, eds (Dordrecht, The Netherlands: Springer), pp. 573–606. [Google Scholar]
- Blum H., Beier H., Gross H. (1987). Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99. [Google Scholar]
- Breyton C., Tribet C., Olive J., Dubacq J.P., Popot J.L. (1997). Dimer to monomer conversion of the cytochrome b6 f complex. Causes and consequences. J. Biol. Chem. 272: 21892–21900. [DOI] [PubMed] [Google Scholar]
- Carrell C.J., Schlarb B.G., Bendall D.S., Howe C.J., Cramer W.A., Smith J.L. (1999). Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum. Biochemistry 38: 9590–9599. [DOI] [PubMed] [Google Scholar]
- Cramer W.A., Yan J., Zhang H., Kurisu G., Smith J.L. (2005). Structure of the cytochrome b6f complex: new prosthetic groups, Q-space, and the ‘hors d’oeuvres hypothesis’ for assembly of the complex. Photosynth. Res. 85: 133–143. [DOI] [PubMed] [Google Scholar]
- Daley S.M., Kappell A.D., Carrick M.J., Burnap R.L. (2012). Regulation of the cyanobacterial CO2-concentrating mechanism involves internal sensing of NADP+ and α-ketogutarate levels by transcription factor CcmR. PLoS ONE 7: e41286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry C., Desbois A., Redeker V., Zito F., Wollman F.A. (2004). Biochemical and spectroscopic characterization of the covalent binding of heme to cytochrome b6. Biochemistry 43: 3956–3968. [DOI] [PubMed] [Google Scholar]
- Dietrich J., Kühlbrandt W. (1999). Purification and two-dimensional crystallization of highly active cytochrome b(6)f complex from spinach. FEBS Lett. 463: 97–102. [DOI] [PubMed] [Google Scholar]
- El-Mohsnawy E., Kopczak M.J., Schlodder E., Nowaczyk M., Meyer H.E., Warscheid B., Karapetyan N.V., Rögner M. (2010). Structure and function of intact photosystem 1 monomers from the cyanobacterium Thermosynechococcus elongatus. Biochemistry 49: 4740–4751. [DOI] [PubMed] [Google Scholar]
- Enami I., Murayama H., Ohta H., Kamo M., Nakazato K., Shen J.R. (1995). Isolation and characterization of a photosystem II complex from the red alga Cyanidium caldarium: association of cytochrome c-550 and a 12 kDa protein with the complex. Biochim. Biophys. Acta 1232: 208–216. [DOI] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R., III (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Farrah T., et al. (2012). PASSEL: the PeptideAtlas SRMexperiment library. Proteomics 12: 1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838. [DOI] [PubMed] [Google Scholar]
- Folea I.M., Zhang P., Aro E.M., Boekema E.J. (2008). Domain organization of photosystem II in membranes of the cyanobacterium Synechocystis PCC6803 investigated by electron microscopy. FEBS Lett. 582: 1749–1754. [DOI] [PubMed] [Google Scholar]
- Gendrullis, M., Dyczmons, N., Gomolla, D., Gathmann, S., Bernat, G., Schneider, D., and Rögner, M. (2008). PetP, a new cytochrome b6f subunit and cytochrome bd oxidase – Two potential regulatory players of cyanobacterial electron transport? In Energy from the sun, J. Allen, E. Gantt, J. Goldbeck, and B. Osmond, eds (Dordrecht, The Netherlands: Springer Science + Media B.V.), pp. 585–590. [Google Scholar]
- Gerber S.A., Rush J., Stemman O., Kirschner M.W., Gygi S.P. (2003). Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA 100: 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götze M., Pettelkau J., Schaks S., Bosse K., Ihling C.H., Krauth F., Fritzsche R., Kühn U., Sinz A. (2012). StavroX—a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 23: 76–87. [DOI] [PubMed] [Google Scholar]
- Hamel P., Olive J., Pierre Y., Wollman F.A., de Vitry C. (2000). A new subunit of cytochrome b6f complex undergoes reversible phosphorylation upon state transition. J. Biol. Chem. 275: 17072–17079. [DOI] [PubMed] [Google Scholar]
- Hasan S.S., Yamashita E., Baniulis D., Cramer W.A. (2013). Quinone-dependent proton transfer pathways in the photosynthetic cytochrome b6f complex. Proc. Natl. Acad. Sci. USA 110: 4297–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J.E., Miller K.R. (1993). Mapping the lateral distribution of photosystem II and the cytochrome b6/f complex by direct immune labeling of the thylakoid membrane. J. Struct. Biol. 111: 1–8. [DOI] [PubMed] [Google Scholar]
- Huang, D., Zhang, H., Soriano, G.M., Dahms, T.E.S., Krahn, J.M., Smith, J.L., and Cramer, W.A.(1999). Characterization and christallation of a highly active cytochrome b6f complex from the thermophilic cyanobacterium Mastigocladus laminosus. In Photosynthesis: Mechanisms and Effects, G. Garab, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1577–1580. [Google Scholar]
- Huang F., Parmryd I., Nilsson F., Persson A.L., Pakrasi H.B., Andersson B., Norling B. (2002). Proteomics of Synechocystis sp. strain PCC 6803: identification of plasma membrane proteins. Mol. Cell. Proteomics 1: 956–966. [DOI] [PubMed] [Google Scholar]
- Hurt E., Hauska G. (1981). A cytochrome b6f complex of five polypeptides with plastoquinol-plastocyanin-oxidoreductase activity from spinach chloroplasts. Eur. J. Biochem. 117: 591–595. [DOI] [PubMed] [Google Scholar]
- Imashimizu M., Bernát G., Sunamura E., Broekmans M., Konno H., Isato K., Rögner M., Hisabori T. (2011). Regulation of F0F1-ATPase from Synechocystis sp. PCC 6803 by gamma and epsilon subunits is significant for light/dark adaptation. J. Biol. Chem. 286: 26595–26602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M., Katoh H., Katayama M., Ikeuchi M. (2004). Improved genetic transformation of the thermophilic cyanobacterium, Thermosynechococcus elongatus BP-1. Plant Cell Physiol. 45: 171–175. [DOI] [PubMed] [Google Scholar]
- Iwai M., Takizawa K., Tokutsu R., Okamuro A., Takahashi Y., Minagawa J. (2010). Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H.T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917. [DOI] [PubMed] [Google Scholar]
- Käll L., Storey J.D., MacCoss M.J., Noble W.S. (2008). Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7: 29–34. [DOI] [PubMed] [Google Scholar]
- Kallas, T. (2012). Cytochrome b6f complex at the heart of energy transduction and redox signaling. In Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation, J.J. Eaton-Rye, B.C. Tripathy, and T.D. Sharkey, eds (Dordrecht, The Netherlands: Springer), pp. 501–560. [Google Scholar]
- Kirchhoff H., Horstmann S., Weis E. (2000). Control of the photosynthetic electron transport by PQ diffusion microdomains in thylakoids of higher plants. Biochim. Biophys. Acta 1459: 148–168. [DOI] [PubMed] [Google Scholar]
- Kuhl H., Kruip J., Seidler A., Krieger-Liszkay A., Bunker M., Bald D., Scheidig A.J., Rögner M. (2000). Towards structural determination of the water-splitting enzyme. Purification, crystallization, and preliminary crystallographic studies of photosystem II from a thermophilic cyanobacterium. J. Biol. Chem. 275: 20652–20659. [DOI] [PubMed] [Google Scholar]
- Kurisu G., Zhang H., Smith J.L., Cramer W.A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302: 1009–1014. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Liu H., Sadygov R.G., Yates J.R., III (2004). A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76: 4193–4201. [DOI] [PubMed] [Google Scholar]
- Liu L.N., Bryan S.J., Huang F., Yu J., Nixon P.J., Rich P.R., Mullineaux C.W. (2012). Control of electron transport routes through redox-regulated redistribution of respiratory complexes. Proc. Natl. Acad. Sci. USA 109: 11431–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. (1975). Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp. Soc. Exp. Biol. 29: 21–38. [PubMed] [Google Scholar]
- Martin W., Kowallik K.V. (1999). Annotated English translation of Mereschkowsky's 1905 paper Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Eur. J. Phycol. 34: 287–295. [Google Scholar]
- Mereschkowsky C. (1905). Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Zent. Bl. 25: 593–604. [Google Scholar]
- Metzger S.U., Cramer W.A., Whitmarsh J. (1997). Critical analysis of the extinction coefficient of chloroplast cytochrome f. Biochim. Biophys. Acta 1319: 233–241. [DOI] [PubMed] [Google Scholar]
- Moal G., Lagoutte B. (2012). Photo-induced electron transfer from photosystem I to NADP(+): characterization and tentative simulation of the in vivo environment. Biochim. Biophys. Acta 1817: 1635–1645. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Chauvat F. (1996). Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol. Gen. Genet. 252: 93–100. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., et al. (2002). Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 9: 123–130. [DOI] [PubMed] [Google Scholar]
- Neilson K.A., Ali N.A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., Lee A., van Sluyter S.C., Haynes P.A. (2011). Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11: 535–553. [DOI] [PubMed] [Google Scholar]
- Norling B., Zarka A., Boussiba S. (1997). Isolation and characterization of plasma membranes from cyanobacteria. Physiol. Plant. 99: 495–504. [Google Scholar]
- Nowaczyk M.M., Sander J., Grasse N., Cormann K.U., Rexroth D., Bernát G., Rögner M. (2010). Dynamics of the cyanobacterial photosynthetic network: communication and modification of membrane protein complexes. Eur. J. Cell Biol. 89: 974–982. [DOI] [PubMed] [Google Scholar]
- Nowaczyk M.M., Wulfhorst H., Ryan C.M., Souda P., Zhang H., Cramer W.A., Whitelegge J.P. (2011). NdhP and NdhQ: two novel small subunits of the cyanobacterial NDH-1 complex. Biochemistry 50: 1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa H., Pakrasi H.B., Ogawa T. (2000). Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J. Biol. Chem. 275: 31630–31634. [DOI] [PubMed] [Google Scholar]
- Olivea J., Ajlanib G., Astierb C., Recouvreura M., Vernotte C. (1997). Ultrastructure and light adaptation of phycobilisome mutants of Synechocystis PCC 6803. Biochim. Biophys. Acta 1319: 275–282. [Google Scholar]
- Pierre Y., Breyton C., Kramer D., Popot J.L. (1995). Purification and characterization of the cytochrome b6 f complex from Chlamydomonas reinhardtii. J. Biol. Chem. 270: 29342–29349. [DOI] [PubMed] [Google Scholar]
- Poetsch A., Rexroth S., Heberle J., Link T.A., Dencher N.A., Seelert H. (2003). Characterisation of subunit III and its oligomer from spinach chloroplast ATP synthase. Biochim. Biophys. Acta 1618: 59–66. [DOI] [PubMed] [Google Scholar]
- Rexroth S., Mullineaux C.W., Ellinger D., Sendtko E., Rögner M., Koenig F. (2011). The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell 23: 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroth S., Poetsch A., Rögner M., Hamann A., Werner A., Osiewacz H.D., Schäfer E.R., Seelert H., Dencher N.A. (2012). Reactive oxygen species target specific tryptophan site in the mitochondrial ATP synthase. Biochim. Biophys. Acta 1817: 381–387. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199: 223–231. [DOI] [PubMed] [Google Scholar]
- Schägger H., Cramer W.A., von Jagow G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217: 220–230. [DOI] [PubMed] [Google Scholar]
- Schmetterer, G. (1994). Cyanobacterial respiration. In The Molecular Biology of Cyanobacteria, D.A. Bryant, ed (Dordrecht, The Netherlands: Kluwer), pp. 409–435. [Google Scholar]
- Schultze M., Forberich B., Rexroth S., Dyczmons N.G., Roegner M., Appel J. (2009). Localization of cytochrome b6f complexes implies an incomplete respiratory chain in cytoplasmic membranes of the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1787: 1479–1485. [DOI] [PubMed] [Google Scholar]
- Schwassmann H.J., Rexroth S., Seelert H., Dencher N.A. (2007). Metabolism controls dimerization of the chloroplast FoF1 ATP synthase in Chlamydomonas reinhardtii. FEBS Lett. 581: 1391–1396. [DOI] [PubMed] [Google Scholar]
- Schwenkert S., Legen J., Takami T., Shikanai T., Herrmann R.G., Meurer J. (2007). Role of the low-molecular-weight subunits PetL, PetG, and PetN in assembly, stability, and dimerization of the cytochrome b6f complex in tobacco. Plant Physiol. 144: 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G., Boussiba S., Vermaas W.F. (1993). Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5: 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D., Choquet Y., Popot J.L., Picot D. (2003). An atypical haem in the cytochrome b(6)f complex. Nature 426: 413–418. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Inoue-Kashino N., Ozawa S., Takahashi Y., Kashino Y., Satoh K. (2009). Photosystem II complex in vivo is a monomer. J. Biol. Chem. 284: 15598–15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Rahire M., Breyton C., Popot J.L., Joliot P., Rochaix J.D. (1996). The chloroplast ycf7 (petL) open reading frame of Chlamydomonas reinhardtii encodes a small functionally important subunit of the cytochrome b6f complex. EMBO J. 15: 3498–3506. [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. (1992). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. 1979. Biotechnology 24: 145–149. [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60. [DOI] [PubMed] [Google Scholar]
- Usaite R., Wohlschlegel J., Venable J.D., Park S.K., Nielsen J., Olsson L., Yates Iii J.R. (2008). Characterization of global yeast quantitative proteome data generated from the wild-type and glucose repression saccharomyces cerevisiae strains: the comparison of two quantitative methods. J. Proteome Res. 7: 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O., Bulte L., Dainese P., Olive J., Bassi R., Wollman F.A. (1991). Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions. Proc. Natl. Acad. Sci. USA 88: 8262–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit S., Takeda K., Tsunoyama Y., Rexroth D., Rögner M., Miki K. (2012). Structure of a thermophilic cyanobacterial b6f-type Rieske protein. Acta Crystallogr. D Biol. Crystallogr. 68: 1400–1408. [DOI] [PubMed] [Google Scholar]
- Volkmer T., Schneider D., Bernát G., Kirchhoff H., Wenk S.O., Rögner M. (2007). Ssr2998 of Synechocystis sp. PCC 6803 is involved in regulation of cyanobacterial electron transport and associated with the cytochrome b6f complex. J. Biol. Chem. 282: 3730–3737. [DOI] [PubMed] [Google Scholar]
- Wang H.L., Postier B.L., Burnap R.L. (2004). Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 279: 5739–5751. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Iwai M., Narikawa R., Ikeuchi M. (2009). Is the photosystem II complex a monomer or a dimer? Plant Cell Physiol. 50: 1674–1680. [DOI] [PubMed] [Google Scholar]
- Westermann M., Neuschaefer-Rube O., Mörschel E., Wehrmeyer W. (1999). Trimeric photosystem I complexes exist in vivo in thylakoid membranes of the Synechocystis strain BO9201 and differ in adsorption characteristics from monomeric photosystem I complexes. J. Plant Physiol. 155: 24–33. [Google Scholar]
- Whitelegge J.P., Zhang H., Aguilera R., Taylor R.M., Cramer W.A. (2002). Full subunit coverage liquid chromatography electrospray ionization mass spectrometry (LCMS+) of an oligomeric membrane protein: cytochrome b(6)f complex from spinach and the cyanobacterium Mastigocladus laminosus. Mol. Cell. Proteomics 1: 816–827. [DOI] [PubMed] [Google Scholar]
- Widger W.R., Cramer W.A., Herrmann R.G., Trebst A. (1984). Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6-f complex: position of the cytochrome b hemes in the membrane. Proc. Natl. Acad. Sci. USA 81: 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Bernát G., Singh A., Mi H., Rögner M., Pakrasi H.B., Ogawa T. (2008). Properties of mutants of Synechocystis sp. strain PCC 6803 lacking inorganic carbon sequestration systems. Plant Cell Physiol. 49: 1672–1677. [DOI] [PubMed] [Google Scholar]
- Yeremenko N., Jeanjean R., Prommeenate P., Krasikov V., Nixon P.J., Vermaas W.F., Havaux M., Matthijs H.C. (2005). Open reading frame ssr2016 is required for antimycin A-sensitive photosystem I-driven cyclic electron flow in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 46: 1433–1436. [DOI] [PubMed] [Google Scholar]
- Yu L., Zhao J., Muhlenhoff U., Bryant D.A., Golbeck J.H. (1993). PsaE is required for in vivo cyclic electron flow around photosystem I in the cyanobacterium Synechococcus sp. PCC 7002. Plant Physiol. 103: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak E., Norling B., Maitra R., Huang F., Andersson B., Pakrasi H.B. (2001). The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. USA 98: 13443–13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Cramer W.A. (2004). Purification and crystallization of the cytochrome b6f complex in oxygenic photosynthesis. Methods Mol. Biol. 274: 67–78. [DOI] [PubMed] [Google Scholar]
- Zhang H., Whitelegge J.P., Cramer W.A. (2001). Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J. Biol. Chem. 276: 38159–38165. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kurisu G., Smith J.L., Cramer W.A. (2003). A defined protein-detergent-lipid complex for crystallization of integral membrane proteins: The cytochrome b6f complex of oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 100: 5160–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouni A., Witt H.T., Kern J., Fromme P., Krauss N., Saenger W., Orth P. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409: 739–743. [DOI] [PubMed] [Google Scholar]
- Zybailov B., Mosley A.L., Sardiu M.E., Coleman M.K., Florens L., Washburn M.P. (2006). Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5: 2339–2347. [DOI] [PubMed] [Google Scholar]