Summary
To successfully establish infection Flaviviruses have to overcome the antiviral state induced by type I interferon (IFN-I). The nonstructural NS5 proteins of several flaviviruses antagonize IFN-I signaling. Here we show that yellow fever virus (YFV) inhibits IFN-I signaling through a unique mechanism that involves binding of YFV NS5 to the IFN-activated transcription factor STAT2 only in cells that have been stimulated with IFN-I. This NS5-STAT2 interaction requires IFN-I-induced tyrosine phosphorylation of STAT1 and the K63-linked polyubiquitination at a lysine in the N-terminal region of YFV NS5. We identified TRIM23 as the E3 ligase that interacts with and polyubiquitinates YFV NS5 to promote its binding to STAT2 and trigger IFN-I signaling inhibition. Our results demonstrate the importance of YFV NS5 in overcoming the antiviral action of IFN-I and offer a unique example of a viral protein that is activated by the same host pathway that it inhibits.
Introduction
There is an urgent need for therapeutics against flaviviruses. There were 5674 cases of WNV infection in the US in 2012 resulting in 286 deaths (Hadler et al., 2014), yet this number pales in comparison to the more than 200,000 cases of hemorrhagic disease and death caused annually by DENV. Even in the case of YFV, for which there is a highly efficacious vaccine, there are 200,000 reported cases annually with at least 30,000 deaths (Barrett and Monath, 2007). Despite the success of the live-attenuated YFV-17D vaccine, occurrences of vaccine-associated viscerotropic and neurotropic disease have brought its safety into question (Barrett et al., 2007; Biscayart et al., 2014; Breugelmans et al., 2013; Monath et al., 2005).
The flavivirus genome is a single-stranded, positive-sense RNA that encodes a polyprotein that is cleaved co- and post-translationally by viral and host proteases generating three structural proteins, C (core), prM-M (membrane protein and its precursor) and E (envelope) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) (Pierson and Diamond. 2007). The structural proteins form the virion particle whereas the NS proteins have roles in viral RNA replication, polyprotein processing and host immune evasion (Aguirre et al., 2012; Chambers et al., 1990a; Chambers et al., 1990b; Diamond, 2009; Rice et al., 1985). NS5 is the largest and most conserved of the flavivirus proteins. Its C-terminus encodes the viral RNA-dependent RNA polymerase (Lindenbach and Rice, 2003), whereas its N-terminus contains an S-adenosyl-methyltransferase (SAM) domain and is involved in methylation of the 5′ RNA cap structure of the viral RNA (Issur et al., 2009; Zhou et al., 2007a). The middle portion contains at least two nuclear localization sequences whose roles in viral replication are still uncharacterized (Brooks et al., 2002).
Type I IFN (IFN-I or IFN-α/β) induces an antiviral state in cells in order to curb viral replication and dissemination (Versteeg and Garcia-Sastre, 2010). Binding of IFN-I to its receptor (IFNAR) activates JAK1 and TYK2, which phosphorylate and activate STAT1 and STAT2, resulting in the formation of IFN-stimulated gene factor 3 (ISGF3), a transcription factor complex comprised of IRF9 and phosphorylated STAT1 and STAT2. ISGF3 translocates to the nucleus where it binds to IFN-I-stimulated response elements (ISREs) and promotes the transcription of IFN-stimulated genes (ISGs), which encode proteins with antiviral activities (Der et al., 1998; Horvath et al., 1996; Schoggins et al., 2011).
Several flaviviruses including DENV, WNV, Japanese encephalitis virus (JEV), Langat virus (LGTV) and tick-borne encephalitis virus (TBEV) encode IFN-I antagonists (Ashour et al., 2009; Best et al., 2005; Jones et al., 2005; Laurent-Rolle et al., 2010; Lin et al., 2006; Liu et al., 2005; Werme et al., 2008; Mazzon et al., 2009; Morrison et al., 2013). Our present work describes a mechanism of virus-mediated IFN-I signaling inhibition that is unique among all viruses described thus far. We show that IFN-I induces binding of YFV NS5 to STAT2 preventing its transcriptional activity by: 1) increasing K63-linked polyubiquitination at residue K6 of YFV NS5 thereby promoting the capacity of YFV NS5 to inhibit IFN-I signaling via STAT2 binding, and 2) promoting STAT1 tyrosine (Tyr) phosphorylation, which is required for NS5/STAT2 binding.
Results
YFV antagonizes IFN-I-stimulated gene expression
YFV and DENV share a similar host tropism; are spread by the same vector species; and can both cause hemorrhagic disease. Since DENV was known to antagonize IFN-I signaling (Ashour et al., 2009; Jones et al., 2005), we examined whether YFV (YFV-17D) would also have this ability. YFV infection caused a 5-fold reduction in IFN-I-mediated ISRE promoter activation (Figure 1A), but did not reduce IFN-II (IFN-γ)-stimulated GAS activity (Figure 1B). In addition, IFN-I but not IFN-II, signaling was reduced in YFV-infected Vero cells as demonstrated with an NDV-GFP bioassay (Park et al., 2003) (Figure 1C).
Figure 1. YFV inhibits IFN-I but not IFN-II signaling.
293T cells were transfected with (A) an IFN-I-inducible firefly-luciferase (f-luc) reporter plasmid (ISRE-luciferase) or (B) an IFN-II-inducible f-luc reporter (GAS-luciferase) along with a constitutively expressing Renilla luciferase (R-Luc) gene plasmid. Cells were infected with DENV-2 or YFV-17D viruses (MOI=5) for 24 hrs and treated with IFN-β or IFN-γ (1000 U/ml) for 16 hrs prior to assaying for luciferase activity. Fold induction of f-luc was normalized to R-luc. The bars represent the mean fold induction of 3 independent experiments compared to untreated, mock-infected controls. (C) Vero cells were mock infected or infected with YFV-17D virus (MOI=5). At 24 hrs post infection (hpi), cells were mock treated or treated with the indicated amounts of IFN-β or IFN-γ for 16 hrs then infected with NDV-GFP. NDV-GFP replication was monitored by fluorescence microscopy at 14 hpi. (D, E) Confocal microscopy images showing subcellular localization of the indicated protein in mock-infected or YFV-17D-infected (MOI=1) Vero cells before or after treatment with IFN-β for 30 minutes. (F) Intracellular fractionation of mock infected or YFV-17D-infected HeLa cells (MOI=10). The proteins in the cytoplasmic and nuclear fractions were detected by western blotting with the indicated antibodies. (G) Vero cells were mock-infected or infected with DENV-2 or YFV-17D viruses for 24 hours (MOI=10), then either mock-stimulated or stimulated with IFN-β for 8 hours. Cell extracts were analyzed by EMSA with ISRE elements derived from theOas1b gene. Statistical analyses were conducted using the unpaired t test in Prism 4 for Macintosh (GraphPad Software, USA). ns = no significance, where p>0.05; *** = p<0.0001.
To identify the step of the IFN-I signaling pathway that is targeted by YFV, the expression, phosphorylation and nuclear translocation of STAT1 and STAT2 were examined in YFV-infected cells. Immunofluorescence analysis (Figure 1D, 1E), as well as Western blots (Figure 1F), revealed that STAT1 and STAT2 levels were not reduced by YFV infection. If anything, STAT2 levels were increased (Figure 1F). Furthermore, IFN-I-induced phosphorylation and nuclear translocation of the STATs were unaffected (Figure 1D, 1E, 1F), suggesting that YFV interference is downstream of tyrosine phosphorylation and nuclear translocation of the STATs.
We next examined whether YFV inhibits IFN-I signaling at the level of DNA binding of ISGF3. For this purpose, electrophoretic mobility shift assays (EMSA) were carried out with lysates from IFN-I-treated YFV-infected Vero cells. ISGF3 failed to bind to the ISRE derived from the Oas1b promoter in extracts from IFN-I-treated, YFV-infected or DENV-2-infected Vero cells (Figure 1G). Antibodies against IRF7 did not disrupt DNA-protein complex formation whereas antibodies against STAT2 did, confirming the identity of the complex as ISGF3 (Figure 1G).
YFV NS5 inhibits IFN-I signaling
As the NS5 proteins of LGTV, JEV, DENV, and WNV had previously been identified as IFN antagonists (Ashour et al., 2009; Best et al., 2005; Laurent-Rolle et al., 2010; Lin et al., 2006; Mazzon et al., 2009; Morrison et al., 2013), we reasoned that YFV NS5 might also be an IFN-I signaling antagonist. Expression of YFV NS5 reduced IFN-I mediated ISRE promoter activation comparable to the positive controls, Nipah virus (NiV) V, DENV-2 NS5 and WNV NS5 (Figure 2A). Expression of DENV-2 Core did not inhibit CAT activity (Figure 2A).
Figure 2. YFV NS5 inhibits IFN-I signaling and interacts with STAT2 in IFN-I- and IFN-III-treated cells.
(A) 293T cells were co-transfected with a plasmid encoding an ISRE-54-CATGFP reporter and with a constitutively expressing firefly-luciferase (f-luc) plasmid plus an empty vector or viral protein encoding plasmids. At 24 hrs post transfection, cells were treated with IFN-β for 16 hrs prior to assaying for CAT activity. Induction of CAT activity was normalized to fluc activity. The lower panel of (A) is a western blot showing the relative expression of each transfected viral protein. (B) 293T cells were co-transfected with an IFN-I-inducible f-luc reporter plasmid (pISRE-luc) along with a constitutively expressing Renilla luciferase (R-luc) gene plasmid and empty vector or YFV-17D and YFV-Asibi NS5 expression plasmids. At 24 hrs post transfection, cells were treated with IFN-β for 16 hrs prior to assaying for luciferase activities. Fold induction of f-luc was normalized to R-luc. The bars represent the mean fold induction of 3 independent experiments compared to untreated empty vector controls. The lower panel of (B) is a western blot (WB) showing the relative expression of each transfected viral protein. (C) 293T cells were transfected with the indicated NS5 expression plasmids. At 24 hrs, cells were mock stimulated or stimulated with IFN-β for 45 minutes. Cells were lysed and immunoprecipitation was performed followed by WB with the indicated antibodies. (D) Same experiment as in (C) with cells mock-stimulated or stimulated with 1000 U/ml of IFN-β, IFN-γ or IFN-λ for 45 minutes prior to lysis. Abbreviations: WCE = whole cell extract; CAT = chloramphenicol acetyl transferase.
The NS5 proteins of the virulent YFV-Asibi strain and the YFV-17D vaccine strain differ by three amino acids (aa) (dos Santos et al., 1995; Hahn et al., 1987). We examined the ability of both YFV NS5 proteins to inhibit IFN-I signaling by investigating their impact on ISG54 promoter activation after stimulation with exogenous IFN-β. Both proteins behaved similarly in the ISG54 reporter assay (Figure 2B).
YFV NS5 interacts with STAT2 in the presence of IFN-I and IFN-III
YFV’s antagonism of IFN-I but not IFN-II signaling (Figure 1) suggested that the virus was targeting a factor, such as TYK2, STAT2 or IRF9, that is specific to the IFN-I pathway. Since DENV-2 NS5 targets STAT2, and DENV and YFV NS5 are ~60% identical at the aa level, we examined whether YFV NS5, like DENV-2 NS5, would interact with STAT2 (Ashour et al., 2009). Immunoprecipitation experiments in 293T cells expressing YFV-17D NS5, YFV-Asibi NS5, empty plasmid, or DENV-2 NS5 revealed STAT2 co-precipitation with DENV-2 NS5 but not YFV NS5 (Figure 2C). The DENV NS5 expression construct used in this study does not enable proteolytic processing of its N-terminus; therefore, only STAT2 binding, and not degradation, is observed (Ashour et al., 2009). When immunoprecipitation experiments were performed in cells stimulated with IFN-I, both YFV-17D NS5 and YFV-Asibi NS5 associated with STAT2 (Figure 2C). Similar results were observed in cells stimulated with IFN-III (IFN-λ), but not IFN-II (Figure 2D). Like IFN-I, IFN-III signals through JAK1/TYK2 and ISGF3, leading to an IFNI-like transcriptional profile (Ank et al., 2006; Zhou et al., 2007b). Since both the 17D and Asibi NS5 proteins behaved similarly in the reporter and binding assays (Figure 2B, 2C), we used YFV-Asibi NS5 for the remainder of the study.
IFN-I-induced modification of STAT2 is not required for STAT2 interaction with YFV NS5
YFV NS5 localizes to both the cytoplasm and nucleus but is predominantly nuclear (Figure 1F and Figure S1A). Since YFV NS5 binds STAT2 in cells treated with IFN-I, we examined whether IFN-I-mediated nuclear translocation of STAT2 was required for YFV NS5 to interact with STAT2. Cell fractionation assays revealed that YFV NS5 bound STAT2 in the cytoplasm and the nucleus of IFN-I-treated cells (Figure S1A). Thus IFN-I-induced nuclear translocation of STAT2 is not required for binding to YFV NS5.
We examined whether IFN-I-mediated modification of STAT2 was required for YFV NS5 to interact with STAT2. Co-immunoprecipitation assays were performed with lysates from U6A cells, which lack endogenous STAT2 (Leung et al., 1995), transfected with YFV NS5 along with wild-type STAT2 or a STAT2 mutant that was incapable of IFN-induced tyrosine phosphorylation (STAT2 Y690F) (Leung et al., 1995; Qureshi et al., 1996). YFV NS5 associated with both wild-type STAT2 and mutant STAT2 in IFN-I-treated cells indicating that IFN-I-mediated STAT2 tyrosine phosphorylation is not required for this interaction (Figure S1B). We also confirmed that STAT1 phosphorylation remained intact in U6A cells upon IFN stimulation (Figure S1C). Confocal immunofluorescence showed YFV NS5 co-localization with wild-type and mutant STAT2 only after IFN-I treatment (Figure 3A).
Figure 3. Neither IFN-I-mediated phosphorylation nor nuclear translocation of STAT2 are required for STAT2 interaction with YFV NS5.
(A) Confocal microscopy images showing subcellular localization of the indicated proteins in Vero cells before or after treatment with IFN-β for 45 minutes. (B) U6A cells were transfected with the indicated plasmids. At 24 hours, cells were mock stimulated or stimulated with IFN-β for 45 minutes. Cells were lysed and immunoprecipitation was performed against the HA epitope for 2 hrs using anti-HA beads. The beads were then washed three times with lysis buffer. 2fTGH cells were mock stimulated or stimulated with IFN-β then lysed and added to the NS5-bound anti-HA beads for 2 additional hrs. The protein was eluted from washed beads then used for western blot.
We next determined if IFN-I was inducing modifications of YFV NS5 and/or cellular proteins other than STAT2 to promote YFV NS5-STAT2 interaction. U6A cells were transfected with a construct expressing HA-tagged YFV NS5. After 24 hours, the cells were either mock-stimulated or treated with IFN-I. U6A cell lysates were subjected to immunoprecipitation using anti-HA beads to precipitate NS5. The beads were washed then incubated with lysates from mock-treated or IFN-I-treated 2fTGH cells, the STAT2-proficient U6A parental line. YFV NS5 interacted with STAT2 only when YFV NS5 had been precipitated from IFN-I-treated U6A cells, regardless of whether 2fTGH cells were IFN-I-treated or mock-treated (Figure 3B). In contrast, DENV-2 NS5 bound in all cases to STAT2. The difference in binding efficiencies of YFV NS5 and DENV NS5 to STAT2 is likely due to the fact that 1) the complexes are made in different lysates decreasing complex formation; 2) DENV NS5 may interact directly with STAT2 while YFV NS5 interaction may be indirect; 3) YFV NS5 may require both STAT1/2 to be complexed before its interaction with STAT2 whereas DENV NS5 does not. Thus, these data suggest that IFN-I signaling modifies YFV NS5 and/or a cellular interactor promoting NS5 binding to STAT2.
The N-terminus of YFV NS5 is required for IFN-I signaling antagonism
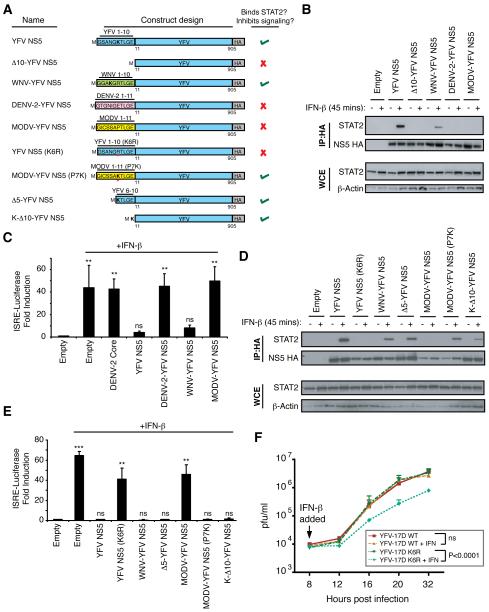
To map the domain of YFV NS5 required for IFN-I signaling inhibition, N-terminally truncated NS5 mutants were tested for STAT2 binding. Removal of the first 10 aa resulted in a loss of YFV NS5-STAT2 association (Figure S2). Chimeric proteins were constructed to ensure that mutant YFV NS5 was expressed in the context of a full-length NS5 molecule then tested for their ability to bind STAT2 and inhibit IFN-I signaling. Substitution of the first 10 aa of YFV NS5 with the first 10 or 11 residues of WNV, DENV-2 and Modoc virus (MODV) (Figure 4A) revealed that only WNV-YFV NS5 associated with STAT2 and inhibited IFN-I signaling (Figure 4B and 4C). Next we examined whether the first 10 aa of YFV NS5 were sufficient for binding STAT2 by substituting the first 10 aa of WNV NS5 for those of YFV NS5 (YFV-WNV NS5) (Figure S3). YFV-WNV NS5 did not bind STAT2, indicating that the first 10 aa of YFV NS5 were necessary but insufficient for this association.
Figure 4. A single lysine residue at position 6 of YFV NS5 is critical for IFN-I antagonist function.
(A) Diagram of the YFV NS5 expression constructs used in Figure 4. (B and D) 293T cells were transfected with the indicated plasmids. At 24 hours post transfection, cells were mock stimulated or stimulated with IFN-β for 45 minutes. Cells were lysed and immunoprecipitation was performed followed by western blot analysis with the indicated antibodies. (C and E) 293T cells were co-transfected with an IFN-β-inducible firefly-luciferase (fluc) reporter plasmid (pISRE-luc) along with a constitutively expressing R-luc plasmid plus empty vector or plasmids encoding the indicated viral proteins. At 24 hrs, cells were treated with IFN-β for 16 hrs prior to assaying for luciferase activities. Fold induction of f-luc activity was normalized to R-luc activity. The bars represent the mean fold induction of 3 independent experiments compared to the untreated, empty vector controls. (F) Vero cells were infected with either YFV-17D WT or YFV-17D K6R at an MOI of 10. At 8 hpi, the cells were either mock-treated or treated with 100 U/ml IFN-β. Virus was harvested at the indicated time points and viral titers were quantified by plaque assay on BHK-21 cells. Each point on the graph represents the mean of 4 independent experiments. Statistical analyses were conducted using the unpaired t test in Prism 4 (GraphPad Software, USA). ns = no significance, where p>0.05; *** = p<0.0001; ** = p<0.001.
A lysine residue within the first 10 amino acids of YFV NS5 is required for its IFN-I antagonist function
A comparison of the first 10 aa of the flaviviral NS5 proteins revealed a lysine (K) at position 6 of YFV NS5 and a K at position 4 (K4) of WNV NS5 that could account for the YFV NS5/STAT2 interaction. Importantly, the 10 aa of DENV and MODV did not have K residues. Mutation of K6 to an arginine, YFV NS5 K6R (Figure 4A) abolished the ability of YFV NS5 to associate with STAT2 after IFN-I stimulation (Figure 4D), and inhibit the ISRE promoter (Figure 4E). Substitution of the proline at position 7 of MODV-YFV NS5 with a K (MODV-YFV NS5 P7K) (Figure 4A) rescued MODV-YFV NS5’s capacity to bind STAT2 and inhibit ISRE promoter activity (Figure 4D, 4E).
The dissimilarity of the N-termini of YFV and MODV and the ability of MODV-YFV NS5 P7K to bind STAT2 suggested that the only residue of importance in the first 10 aa of YFV NS5 was lysine. We generated YFV NS5 constructs missing the first 5 aa (Δ5-YFV NS5) or the first 10 aa except K (K-Δ10-YFV NS5) (Figure 4A). Co-immunoprecipitation assays (Figure 4D) and reporter assays (Figure 4E) showed that these proteins were able to bind STAT2 and inhibit IFN signaling, emphasizing the requirement of the lysine residue in the N-terminal region of NS5. An alignment of the YFV genome sequences in the Virus Pathogen Resource (ViPR) revealed that K6 of YFV NS5 is conserved across strains (Figure S4A).
Introduction of the K6R mutation into YFV-17D (YFV-17D K6R) did not affect viral replication as compared to wild-type virus (YFV-17D WT) (Figure 4F). However, IFN-I treatment at 8 hours post infection, decrease YFV-17D K6R titers by one log relative to YFV-17D WT (Figure 4F). In addition, the upregulation of ISGs in YFV-17D K6R-infected cells was comparable to IFN-treated mock-infected cells (Figures S4B and S4C). These results indicate that K6 of YFV NS5 is critical for NS5-STAT2 interaction and inhibition of IFN-I-mediated antiviral activity.
IFN-I enhances K63-linked polyubiquitination of YFV NS5 and promotes YFV NS5-STAT2 interaction
Lysine residues are critical for the posttranslational modification of proteins through conjugation to acetyl (acetylation), ubiquitin (ubiquitination), small ubiquitin-like modifier (SUMO) (SUMOylation), Nedd8 (NEDDylation) and interferon-stimulated gene-15 (ISG15) (ISGylation) (Chen and Gerlier, 2006; Hemelaar et al., 2004). Thus, we investigated whether the K6 residue of YFV NS5 was posttranslationally modified and if that modification was required for STAT2 binding. 293T cells were cotransfected with empty vector, MODV-YFV NS5, YFV NS5, TRIM25, or TRIM19 along with UB, ISG15, or SUMO (Figure S5). TRIM25 serves as a positive control for ubiquitination and ISGylation (Gack et al., 2007) while TRIM19 serves as a positive control for SUMOylation (El Bougrini et al., 2011). Co-immunoprecipitation assays demonstrated that YFV NS5 was possibly ubiquitinated, but not ISGylated or SUMOylated (Figure S5). An increase in ubiquitination of YFV NS5 was also observed upon the addition of IFN (compare lanes 3 and 7 of Figure S5A). Immunoprecipitation experiments further demonstrated that a lysine in the first 10 aa of the protein was required for the interaction of YFV NS5 and STAT2 as well as the association of YFV NS5 with ubiquitin (Figure 5A).
Figure 5. K63-linked ubiquitination is required for binding of YFV NS5 to STAT2.
(A) 293T cells were transfected with various NS5 mutants. At 24 hrs post transfection, cells were mock stimulated or stimulated with IFN-β for 45 minutes. Cells were lysed and immunoprecipitation was performed followed by western blot analysis (WB) with the indicated antibodies. (B) Same as (A) but with more stringent conditions. (C) U6A cells were transfected with HA-tagged YFV NS5. At 24 hrs, cells were mock stimulated or stimulated with IFN-β. Cells were lysed and immunoprecipitation was performed against the HA epitope for 2 hrs using HA beads. Beads were washed three times with lysis buffer followed by incubation with reaction buffer or OTU. 2fTGH cells were mock stimulated or stimulated with 1000 U/ml IFN- β. Cells were lysed and were added to the HA beads for 2 hrs. Beads were then washed followed by WB with the indicated antibodies. (D) Diagram of the YFV NS5 constructs used in 5E and 5F. (E) Same experiment as in (A) with the indicated constructs. (F) 293T cells were co-transfected with an IFN-α/β-inducible firefly-luciferase (f-luc) reporter plasmid (pISRE-luc) along with a constitutively expressing Renilla luciferase (R-luc) gene plasmid plus empty vector or plasmids encoding the indicated viral proteins. At 24 hrs, cells were treated with IFN-β for 16 hrs prior to assaying for luciferase activities. Fold induction of f-luc activity was normalized to R-luc activity. The bars represent the mean fold induction of 3 independent experiments compared to the untreated empty vector controls. (G) 293T cells were transfected with FLAG-tagged wild-type YFV NS5 and various HA-tagged ubiquitin mutants. At 24 hours, cells were mock stimulated or stimulated with IFN-β for 45 minutes. Cells were lysed and immunoprecipitation was performed against the HA epitope followed by WB with the indicated antibodies. Statistical analyses were conducted using the unpaired t test in Prism 4 (GraphPad Software, USA). ns = no significance, where p>0.05; *** = p<0.0001.
Our results suggested direct ubiquitination of YFV NS5 K6 although K6 could also be required for NS5 to associate with unanchored polyubiquitin chains or an ubiquitinated protein. When immunoprecipitated YFV NS5 was washed in a high salt buffer, to disrupt NS5/STAT2 interaction, a reduction in STAT2 binding was evident but there was no change in ubiquitin binding to NS5 (Figure 5B). The strength of the association between ubiquitin and NS5 suggested covalent linkage of ubiquitin to NS5. We next assessed the effect of a deubiquitinating enzyme, OTU domain of Crimean-Congo hemorrhagic fever virus L protein, L(1-169) (Frias-Staheli et al., 2007) on the NS5-STAT2 interaction. Incubation of NS5 with OTU decreased the NS5-STAT2 interaction (Figure 5C), confirming that ubiquitination of NS5 at K6 is required to mediate the interaction with STAT2.
Given these results, we reasoned that covalently attaching an ubiquitin chain to the N-terminus of YFV NS5 K6R might allow this protein to behave like wild type NS5. We constructed clones with a linear four-ubiquitin chain fused to the N-terminus of wild-type or the K6R mutant. Since cellular hydrolases normally remove ubiquitin chains by cleaving the LRGG sequence of the ubiquitin C-terminus, we designed ubiquitin fusions lacking the GG portion of this sequence between ubiquitins and between ubiquitin and NS5 (Figure 5D). Co-immunoprecipitation assays (Figure 5E) and reporter gene assays (Figure 5F) showed that the YFV NS5 K6R regained the ability to bind STAT2 and inhibit IFN-I signaling when fused to linear ubiquitin. However, binding to STAT2 was still dependent on IFN-I treatment of cells. This may indicate that in addition to promoting YFV NS5 ubiquitination, IFN-I signaling is required for another modification that allows YFV NS5 to bind to STAT2. On the other hand, it might also be possible that YFV NS5 K6R coupled to linear ubiquitin is not competent for binding to STAT2, and that IFN-I treatment results in the formation of competent polyubiquitin chains that conjugate to the fused linear tetraubiquitin at the N-terminus of YFV NS5 K6R. In any case, the functional replacement of K6 in YFV NS5 by linear ubiquitin clearly indicates that ubiquitination mediates the role of K6 in promoting STAT2 binding.
Most polyubiquitinated proteins are modified through K48 or K63 ubiquitin linkage. We examined whether the NS5-STAT2 interaction was mediated by K63- or K48-linked polyubiquitin. Upon IFN-I treatment, wild-type and K48R but not K63R ubiquitin co-immunoprecipitated with both NS5 and STAT2 (Figure 5G) indicating that K63-linked ubiquitination of YFV NS5 is necessary for the interaction.
TRIM23 interacts with and promotes YFV NS5 ubiquitination
Since TRIM E3 ligases are important modulators of IFN signaling pathways, (McNab et al., 2011; Uchil et al., 2013; Versteeg et al., 2013), and IFN-I treatment increased ubiquitination of YFV NS5, we screened a panel of TRIM proteins from the different TRIM family subgroups (Groups I-IX) and examined their ability to increase ubiquitination of YFV NS5. Of the TRIMs examined, expression of TRIM23 was observed to increase ubiquitination of NS5 in IFN-I-stimulated cells (data not shown). We examined the cellular distribution of TRIM23 with wild-type or K6R NS5 using confocal immunofluorescence. Over-expressed TRIM23 localized in punctate perinuclear patterns, while overexpression of NS5 resulted in a diffuse pattern predominantly in the nucleus. However, co-expression of TRIM23 and YFV NS5 resulted in relocalization of NS5 to the punctate dot-like structures containing TRIM23 (Figure 6A). NS5 did not colocalize or co-immunoprecipitate with the other TRIMs tested (Figure S6A, S6B). In addition, NS5 precipitated endogenous TRIM23 (Figure 6B). Association of NS5 with TRIM23 was, in contrast to association of NS5 with STAT2, independent of IFN-I treatment (Figure 6B). Confocal immunofluorescence analysis and co-immunoprecipitation assays showed that TRIM23 associated with both wild-type and K6R NS5 (Figure 6A, 6B). We also show that overexpression of TRIM23 enhances ubiquitination of WT but not mutant NS5 (Figure 6C), and that the TRIM23 RING domain is required for its interaction with NS5 (Figure S7). In addition, silencing of TRIM23 in IFN-competent cells resulted in increased susceptibility of YFV-17D to IFN-I and increased induction of the ISG, MxA, in YFV-infected cells (Figure 6D). Furthermore, silencing TRIM23 reduced NS5 ubiquitination and binding to STAT2 (Figure 6E).
Figure 6. TRIM23 ubiquitinates YFV NS5.
(A) Confocal microscopy images showing subcellular localization of the indicated proteins in HeLa cells before or after treatment with IFN-β for 30 minutes. (B) 293T cells were transfected with YFV NS5. At 24 hours, cells were mock stimulated or stimulated with IFN-β then lysed followed by immunoprecipitation. Western blot analysis (WB) was done with the indicated antibodies. (C) Ubiquitination assay conducted in 293T cells with increasing amounts of TRIM23. Cells were transfected with FLAG-tagged wild-type YFV NS5 or YFV NS5 K6R with increasing amounts of HA-tagged TRIM23. Lysis and immunoprecipitation against the FLAG epitope was followed by western blot analysis with the indicated antibodies. (D) siRNA-expressing A549 cells were infected with YFV-17D WT at an MOI of 10. At 8 hpi, the cells were mock-treated or treated with 100 U/ml IFN-β. Virus was harvested at 24 hours and viral RNA, TRIM23 and MxA RNA quantified by qPCR. Viral titers were quantified by plaque assay on BHK-21 cells. (E) siRNA-expressing 293T cells were transfected with empty plasmid or YFV NS5 and mock treated or treated with IFN-I for 45 minutes. Lysis and immunoprecipitation were followed by WB with the indicated antibodies. Statistical analyses were conducted using the unpaired t test in Prism 4 (GraphPad Software, USA). ns = no significance, where p>0.05; *** = p<0.0001; ** = p<0.001.
STAT1 is necessary for IFN-I-induced YFV NS5-STAT2 interaction
It was intriguing that STAT1 was not precipitated along with STAT2 after YFV NS5 immunoprecipitation (Figure 2D). Co-immunoprecipitation experiments using less stringent wash conditions resulted in precipitation of NS5 with both STAT2 and STAT1 (Figure 7A). These results indicate that NS5 does not compete with STAT1 for STAT2 binding after IFN-I signaling. Rather, NS5 strongly interacts with STAT2 whereas binding to STAT1 likely occurs by means of IFN-I-induced STAT1/2 dimerization. Consistent with this, co-immunoprecipitation of STAT1 with DENV-2 NS5/STAT2 complexes was also induced by IFN-I treatment.
Figure 7. STAT1 is necessary for IFN-I induced YFV NS5-STAT2 interaction.
(A) 293T cells were transfected with the indicated constructs. Cells were mock stimulated or stimulated with IFN-β for 45 minutes at 24 hours post transfection. Cell lysates were immunoprecipitated followed by western blot analysis with the indicated antibodies. (B) Same experiment as in (A) done in U3A cells. In the second panel, U3A cells were complemented with wild-type STAT1 and the same experiment was repeated. (C) U3A cells were complemented with wild-type STAT1 or mutant STAT1 Y701F and the experimental procedure in (B) was repeated.
STAT degradation by the V proteins of some paramyxoviruses requires both STAT1 and STAT2 components, even though only one of the STAT proteins is targeted for degradation (Horvath, 2004). To explore whether the IFN-I-dependent NS5-STAT2 interaction required STAT1, we examined the NS5-STAT2 interaction using U3A cells, which are STAT1-deficient (McKendry et al., 1991; Muller et al., 1993). Immunoprecipitation assays showed requirement of STAT1 for YFV NS5 to interact with STAT2 since it did not bind STAT2 in STAT1-deficient cells. However, DENV-2 NS5 bound STAT2 in STAT1-deficient cells as previously described (Ashour et al., 2010). Complementation of U3A cells with a wild-type STAT1 construct rescued the ability of YFV NS5 to interact with STAT2 after IFN-I treatment indicating that STAT1 is required for NS5-STAT2 interaction (Figure 7B). Immunoblot analysis revealed that STAT2 was phosphorylated on Y690 upon IFN-I treatment in U3A cells, which is consistent with published reports (Figure 7C) (Improta et al., 1994). However, our results showed that YFV NS5 associated with STAT2 in response to IFN-I treatment when cells were reconstituted with wild-type but not mutant (Y701F) STAT1. Given that tyrosine phosphorylation induces STAT1/2 heterodimers to transform from a parallel to an antiparallel association with respect to their C- and N-termini (Lin et al., 2009), these results suggest that the NS5 binding domain is only exposed when STAT2 interacts with phosphorylated STAT1 resulting in a different STAT2 conformation. Thus, our results indicate that IFN-I promotes YFV NS5/STAT2 interaction by inducing STAT1 tyrosine phosphorylation. However, a modification in YFV NS5, i.e. ubiquitination is also required for STAT2 interaction. This modification is enhanced in the presence of IFN and this may explain why NS5 from untreated cells is unable to bind STAT2 compared to NS5 from IFN-treated cells.
Discussion
The experiments described in this paper demonstrate that YFV and its NS5 protein inhibit IFN-I signaling by binding to STAT2 and preventing ISGF3 engagement with ISREs. The results also show that IFN-I triggers YFV NS5 binding to STAT2. This is a unique example of a viral IFN-I signaling antagonist that is itself activated by IFN-I. In the absence of IFN-I treatment, NS5 is incapable of binding STAT2, but IFN-I-activated NS5 binds to both the phosphorylated and unphosphorylated forms of STAT2 indicating that the ability of IFN-I to promote NS5-STAT2 interaction is not due to IFN-I-mediated STAT2 modification. We have identified K63-linked polyubiquitination as one of the IFN-I activating modifications of NS5. In addition, IFN-I induced tyrosine phosphorylation of STAT1 is also required for NS5-STAT2 interaction, most likely by promoting conformational changes in STAT2 that trigger its association with ubiquitinated NS5.
YFV NS5 exhibits functional similarities to DENV-2 NS5, however DENV-2 NS5 binds STAT2 regardless of IFN treatment and targets it for proteasome-mediated degradation (Ashour et al., 2009). YFV NS5, on the other hand, binds STAT2 both in the cytoplasm and nucleus only after IFN treatment, and appears to inactivate ISGF3 within the nucleus. Mapping studies identified the first 10 residues of the YFV NS5 protein as essential for STAT2 interaction and IFN signaling inhibition. The first 10 aa of DENV-2 NS5 are critical for mediating proteasomal degradation of STAT2 although they are dispensable for STAT2 association (Ashour et al., 2009). Therefore, the extreme N-termini of both YFV NS5 and DENV-2 NS5 contain motifs required for their IFN antagonist function. Remarkably, the two N-termini initiate entirely different mechanisms that ultimately converge on STAT2 inhibition. A glycine residue and a threonine residue within the N-terminus of DENV NS5 are required for DENV NS5 to bind UBR4 (a potential E3 ligase) and mediate STAT2 degradation (Morrison et al., 2013). By contrast, other E3 ligases, such as TRIM23, polyubiquitinate K6 of YFV NS5, and this, together with STAT1 tyrosine phosphorylation is required for YFV NS5/STAT2 association and inhibition of ISGF3 engagement with the ISRE.
K6 of YFV NS5 is critical for NS5-STAT2 interaction and IFN-I signaling inhibition. When a K6R mutation was placed in the context of YFV-17D, the virus displayed an enhanced sensitivity to the antiviral action of IFN-I, indicating that K6 is important for viral replication in the context of an ongoing IFN-I response. As no replication defect was seen for the K6R mutant virus in mock-treated Vero cells, it appears that K6 may only be important for IFN antagonism. YFV-17D causes neurotropic and viscerotropic disease in immune-compromised individuals (Barrett et al., 2007; Biscayart et al., 2014; Breugelmans et al., 2013; Klein et al., 2010; Lindsey et al., 2008; Monath et al., 2005). Decreasing the ability of YFV-17D to antagonize IFN-I signaling may limit the replication of this vaccine in immunocompromised individuals thereby decreasing the incidence of vaccine-associated adverse events. A YFV vaccine that is more attenuated than, but equally or more immunogenic to, the current YFV-17D vaccine would be an attractive alternative for YF vaccination.
The data here show that K63-linked polyubiquitination of NS5 is required for STAT2 binding and IFN-I signaling inhibition. We have shown that a linear four-ubiquitin chain fused to the N-terminus of YFV NS5 K6R binds STAT2 upon stimulation with IFN-I. A possible explantion for these results is that the fused linear chain serves as a K63-linked ubiquitin chain mimic since the structures of K63-linked and linear ubiquitin are more similar to each other than they are to the structure of K48-linked ubiquitin (Tse et al., 2011). However we cannot rule out the possibility that the four-ubiquitin chain acts as a new scaffold for additional K63 polyubiquitination, which in turns activates NS5 binding to STAT2. It is of interest that the only requirement in the first 10 aa of NS5 to bind to STAT2 and inhibit IFN signaling was the presence of a K, as all these residues could be substituted by a K without loss of function. We postulate that binding of an E3 ligase (such as TRIM23) to YFV NS5 requires a domain outside of the first 10 residues, and this brings the required E2 protein into close contact with the N terminal of NS5 resulting in polyubiquitination irrespective of the precise K location within the N-terminus.
We have also identified TRIM23, an E3 ligase involved in innate immune signaling (Arimoto et al., 2010; Versteeg et al., 2013), as a YFV NS5 interacting protein. TRIM23 has been shown to promote IFN-β production by ubiquitinating NEMO (Arimoto et al., 2010). Taylor and colleagues recently demonstrated that TRIM79α restricts LGTV/TBEV replication by interacting with NS5 (Taylor et al., 2011). In contrast, we show that TRIM23 promotes YFV replication by polyubiquitinating YFV NS5 at its K6 residue, a modification that is required for YFV NS5-STAT2 binding. Of note, our experiments do not rule out the possible contribution of other E3 ligases in ubiquitinating YFV NS5, as there might be redundancy in their function (Morishima et al., 2008). Nevertheless, silencing of TRIM23 results in decreased ability of YFV to inhibit IFN-induction of MxA, and increased sensitivity of YFV to IFN-I, supporting a prominent role of TRIM23 in YFV-mediated inhibition of IFN signaling.
Despite the high level of amino acid conservation amongst flavivirus NS5 proteins, IFN antagonistic functions appear to be mediated by different mechanisms. This would suggest that IFN signaling inhibition evolved independently among flaviviruses, but that the NS5 protein acquired this function in all cases. It might be that the polyprotein-based strategy of viral protein expression that is common to all flaviviruses results in excess production of NS5, as only catalytic amounts of this protein are required for RNA synthesis, making it an ideal candidate for acquisition of additional functions.
Experimental Procedures
Cells and viruses
293T, Vero and HeLa cells were cultured in DMEM supplemented with 10% FCS. 2fTGH and 2fTGH derivatives were kindly provided by Dr. George Stark and were described previously (Leung et al., 1995; Pellegrini and Schindler, 1993). A previously described Newcastle disease virus expressing GFP (NDV-GFP) was grown in 10-day-old embryonated chicken eggs (Park et al., 2003). High titer stocks of DENV-2 16681 were obtained by passage in C6/36 cells (Diamond et al., 2000). High titer stocks of YFV-17D strain were obtained by passage in BHK-21 cells. The YFV-17D K6R mutant virus was made by PCR mutagenesis (primer sequences are available upon request) of a YFV-17D cDNA clone, pACNR-YF17D, kindly provided by Dr. Charles Rice (Bredenbeek et al., 2003; Lindenbach and Rice, 1999). The mutant virus and wild-type control virus were rescued by linearizing the cDNA clone with XhoI, and using the SP6 Cap-Scribe kit™ (Roche, Germany) to generate RNA. mRNA was transfected into BHK-21 cells using the Transit® mRNA transfection kit (Mirus Bio, USA), and virus was harvested 3 days later.
Plasmids
All flavivirus genes were cloned in the mammalian expression vector, pCAGGS. Primer sequences used in the generation of the constructs are available upon request. HA-NiV V plasmid was a gift from Dr. Megan Shaw. pCAGGS-Firefly luciferase and ISRE-54-CAT reporter plasmids were gifts from Dr. Luis Martinez-Sobrido. HA-ubiquitin expressing plasmids were gifts from by Dr. Gijs Versteeg.
Virus infections
Monolayers of cells were initially adsorbed with YFV-17D at the indicated multiplicity of infection (MOI) for 1 hr at room temperature. Unbound virus was removed, and cells were maintained in DMEM 10% FBS at 37°C. For the YFV-17D K6R versus YFV-17D growth curves, Vero cells were incubated with virus at an MOI of 10 for 1 hr at 37°C. Viral replication was measured by plaque assay on BHK-21 cells.
Transfections
Lipofectamine™ 2000 (Invitrogen, USA) was used for all transfections. 293T cells were transfected using a 1:1 ratio of plasmid DNA to Lipofectamine™ 2000. Vero cells were transfected using a 1:2 ratio. 2fTGH and 2fTGH derivatives were transfected using a 1:3 ratio.
Antibodies and Cytokines
The following antibodies were utilized in this study: TRIM23 monoclonal antibody (M01) (Abnova), rabbit polyclonal anti-STAT1 (BD Biosciences, USA), anti-STAT2 and anti-YFV E (Santa Cruz Biotechnology, USA), antiphospho-STAT1 (Tyr 701) (Cell Signaling Technology, USA), antiphospho-STAT2 (Tyr 689) (Upstate Biotechnology, USA), anti-HA, anti-FLAG, anti-GFP, anti-tubulin, and anti-actin (Sigma-Aldrich, USA), anti-ubiquitin (Enzo Life Sciences, USA), anti-PKR (Abcam), anti-MDA5 (Enzo AT113), anti-DENV-2 E (Hybridoma Facility of Icahn School of Medicine at Mount Sinai, New York, USA). Rabbit antibody against DENV-2 NS5 was generated in our lab and previously reported (Ashour et al., 2009). Anti-YFV NS5 antibody (YF17D NS5 C7) was kindly provided by Dr. Charles Rice (Chambers et al., 1990b). Universal IFN-I, human IFN-β, human IFN-λ, and human IFN-γ (PBL Interferon Source, USA) were used at 1000 U/ml unless otherwise specified.
Interferon Signaling Inhibition Assays and Antibody-based techniques
Reporter assay, NDV GFP assay, immunoprecipitation and immunoblot assay, immunoflourescence and subcellular fractionation assays are described in Supplemental Experimental Procedures.
Quantitative RT-PCR
Total RNA, isolated from cells using RNeasy (Qiagen), was subjected to DNase digestion with Turbo DNase (Ambion). Reverse transcription was performed with a cDNA reverse transcription kit (Applied Biosystems). qPCR was done in triplicates using SYBR green I master mix (Roche) in a Roche LightCycler 480. Relative mRNA values were calculated using ddCt method with 18S as an internal control and shown as fold change by normalizing to mock-control. The primers used are:
YFV NS5 Forward: GAGCTCATTGGGAGAGGAAG, Reverse: AGGCAAGCTGTTTCCTTGAT, All other primers used were published in (Rajsbaum et al., 2014).
siRNA Knockdown
TRIM23 siRNA-mediated knockdown experiments are described in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
These studies were partly supported by NIAID grant U54AI057158 (to AG-S) and by a National Institute of Health fellowship (to M.L.-R.). We thank Richard Cadagan and Osman Lizardo for technical assistance, and Dr. Adriana Forero for editorial help. Confocal laser scanning microscopy was performed at ISMMS-Microscopy Shared Resource facility.
Footnotes
Author Contributions J.M. and M.L.-R. performed all aspects of this study. R.R., J.M.L.M., G.P., A.P., J.A., L.M. and C.M. performed experiments. B.R.T. provided reagents and advice. M.L.-R, J.M. and A.G.-S. organized the study and prepared the manuscript. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS pathogens. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. Journal of virology. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. Journal of virology. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, et al. Mouse STAT2 restricts early dengue virus replication. Cell host & microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AD, Monath TP, Barban V, Niedrig M, Teuwen DE. 17D yellow fever vaccines: new insights. A report of a workshop held during the World Congress on medicine and health in the tropics, Marseille, France, Monday 12 September 2005. Vaccine. 2007;25:2758–2765. doi: 10.1016/j.vaccine.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. Journal of virology. 2005;79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscayart C, Carrega ME, Sagradini S, Gentile A, Stecher D, Orduna T, Bentancourt S, Jimenez SG, Flynn LP, Arce GP, et al. Yellow fever vaccine-associated adverse events following extensive immunization in Argentina. Vaccine. 2014;32:1266–1272. doi: 10.1016/j.vaccine.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Bredenbeek PJ, Kooi EA, Lindenbach B, Huijkman N, Rice CM, Spaan WJ. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. The Journal of general virology. 2003;84:1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Breugelmans JG, Lewis RF, Agbenu E, Veit O, Jackson D, Domingo C, Bothe M, Perea W, Niedrig M, Gessner BD, et al. Adverse events following yellow fever preventive vaccination campaigns in eight African countries from 2007 to 2010. Vaccine. 2013;31:1819–1829. doi: 10.1016/j.vaccine.2013.01.054. [DOI] [PubMed] [Google Scholar]
- Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem. 2002;277:36399–36407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990a;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, McCourt DW, Rice CM. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990b;177:159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- Chen M, Gerlier D. Viral hijacking of cellular ubiquitination pathways as an anti-innate immunity strategy. Viral immunology. 2006;19:349–362. doi: 10.1089/vim.2006.19.349. [DOI] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS. Mechanisms of evasion of the type I interferon antiviral response by flaviviruses. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2009;29:521–530. doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. Journal of virology. 2000;74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos CN, Post PR, Carvalho R, Ferreira II, Rice CM, Galler R. Complete nucleotide sequence of yellow fever virus vaccine strains 17DD and 17D-213. Virus Res. 1995;35:35–41. doi: 10.1016/0168-1702(94)00076-o. [DOI] [PubMed] [Google Scholar]
- El Bougrini J, Dianoux L, Chelbi-Alix MK. PML positively regulates interferon gamma signaling. Biochimie. 2011;93:389–398. doi: 10.1016/j.biochi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell host & microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Hadler JL, Patel D, Bradley K, Hughes JM, Blackmore C, Etkind P, Kan L, Getchell J, Blumenstock J, Engel J, et al. National capacity for surveillance, prevention, and control of West Nile virus and other arbovirus infections--United States, 2004 and 2012. MMWR Morbidity and mortality weekly report. 2014;63:281–284. [PMC free article] [PubMed] [Google Scholar]
- Hahn CS, Dalrymple JM, Strauss JH, Rice CM. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:2019–2023. doi: 10.1073/pnas.84.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J, Borodovsky A, Kessler BM, Reverter D, Cook J, Kolli N, Gan-Erdene T, Wilkinson KD, Gill G, Lima CD, et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Molecular and cellular biology. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CM. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur J Biochem. 2004;271:4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- Horvath CM, Stark GR, Kerr IM, Darnell JE., Jr. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Molecular and cellular biology. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Improta T, Schindler C, Horvath CM, Kerr IM, Stark GR, Darnell JE., Jr. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4776–4780. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. Journal of virology. 2005;79:5414–5420. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. Journal of virology. 2010;84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung S, Qureshi SA, Kerr IM, Darnell JE, Jr., Stark GR. Role of STAT2 in the alpha interferon signaling pathway. Molecular and cellular biology. 1995;15:1312–1317. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Buettner R, Yuan YC, Yip R, Horne D, Jove R, Vaidehi N. Molecular dynamics simulations of the conformational changes in signal transducers and activators of transcription, Stat1 and Stat3. Journal of molecular graphics & modelling. 2009;28:347–356. doi: 10.1016/j.jmgm.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. Journal of virology. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. Journal of virology. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- McKendry R, John J, Flavell D, Muller M, Kerr IM, Stark GR. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Current opinion in immunology. 2011;23:46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Monath TP, Cetron MS, McCarthy K, Nichols R, Archambault WT, Weld L, Bedford P. Yellow fever 17D vaccine safety and immunogenicity in the elderly. Hum Vaccin. 2005;1:207–214. doi: 10.4161/hv.1.5.2221. [DOI] [PubMed] [Google Scholar]
- Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Human molecular genetics. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, Garcia-Sastre A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS pathogens. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell JE, Jr., Stark GR, Kerr IM. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. Embo Journal. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. Journal of virology. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi SA, Leung S, Kerr IM, Stark GR, Darnell JE., Jr. Function of Stat2 protein in transcriptional activation by alpha interferon. Molecular and cellular biology. 1996;16:288–293. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, Martinez-Romero C, Patel JR, Morrison J, Pisanelli G, Miorin L, et al. Unanchored K48-Linked Polyubiquitin Synthesized by the E3-Ubiquitin Ligase TRIM6 Stimulates the Interferon-IKKepsilon Kinase-Mediated Antiviral Response. Immunity. 2014;40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RT, Lubick KJ, Robertson SJ, Broughton JP, Bloom ME, Bresnahan WA, Best SM. TRIM79alpha, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell host & microbe. 2011;10:185–196. doi: 10.1016/j.chom.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse MK, Hui SK, Yang Y, Yin ST, Hu HY, Zou B, Wong BC, Sze KH. Structural analysis of the UBA domain of X-linked inhibitor of apoptosis protein reveals different surfaces for ubiquitin-binding and self-association. PloS one. 2011;6:e28511. doi: 10.1371/journal.pone.0028511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J, Mothes W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. Journal of virology. 2013;87:257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, García-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werme K, Wigerius M, Johansson M. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell Microbiol. 2008;10:696–712. doi: 10.1111/j.1462-5822.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. Journal of virology. 2007a;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. Journal of virology. 2007b;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.