Abstract
In multiple sclerosis (MS), an exchange of lymphocytes, in particular B cells, between the central nervous system (CNS) and periphery is believed to be required for the maintenance of active disease. Therapeutic monoclonal antibodies that prevent lymphocytes from crossing the blood-brain barrier (BBB) or induce near-complete peripheral B cell depletion rapidly mitigate MS disease activity. Using next-generation sequencing technology, we recently found that clonally related B cells exist in the cerebrospinal fluid (CSF) and peripheral blood (PB) of MS patients, establishing the existence of an immune axis across the BBB. However, it remains unclear which subpopulations of the highly diverse peripheral B cell compartment share antigen-specificity with intrathecal B cell repertoires, and whether their antigen stimulation occurs on both sides of the BBB. To address these questions, we combined flow cytometry sorting of PB B cell subsets with deep immune repertoire sequencing of CSF and PB B cells. Immunoglobulin (IgM and IgG) heavy chain variable (VH) region repertoires of five PB B cell subsets from MS patients (n=8) were compared with their CSF Ig-VH transcriptomes. In 6 of 8 patients, we identified peripheral CD27+IgD−memory B cells, CD27hiCD38hi plasma cells/plasmablasts, or CD27−IgD− B cells providing an immune connection to the CNS compartment. Pinpointing Ig class-switched B cells as key component of the immune axis thought to contribute to ongoing MS disease activity strengthens the rationale of current therapeutic strategies and may lead to more targeted approaches.
Introduction
Fuelled by recent advances in MS therapy using CD20-targeted B cell depletion(1-3), efforts to further understand features of disease-relevant (i.e. pathogenic) B cell autoimmunity have increased, both in MS patients and in animal models of MS. Clonal overlap between B cell receptor sequences in MS brain parenchyma, meningeal lymphoid follicles, and cerebrospinal fluid (CSF) indicates an immunological continuum inside the central nervous system (CNS)(4, 5). Furthermore, we have previously shown that antigen-driven B cell immune responses inside the CNS compartment are not sequestered from peripheral B cell responses(6), suggesting that disease-driving immune mechanisms against, as yet unknown, identical or highly similar antigenic epitopes operate in the CNS and periphery. Together these data establish that antigen-experienced B cells provide an immunological continuum across numerous different tissues where they may either directly or indirectly support CNS targeting autoimmunity. The ability to specifically identify and potentially target pathogenic B cells in autoimmune conditions like MS may provide an important basis for the development of immune therapies with improved efficacy and long-term safety. Based on previous studies, including our own, it is generally assumed that antigen-experienced B cell subsets provide a pathologically relevant link between the CNS and periphery. However, to date there is a complete lack of knowledge regarding which subsets of the highly diverse and complex peripheral B cell compartment support CNS-directed autoimmunity.
We applied multicolor flow cytometry sorting of peripheral blood (PB) B cell subsets in combination with next-generation Deep Ig heavy chain variable region (Ig-VH) Repertoire Sequencing (DIRS) of PB B cells and CSF lymphocytes to identify and characterize disease-associated peripheral B cell subsets. We found clusters of clonally related B cells involving CSF-derived Ig-VH and PB CD27−IgD− class-switched B cells, CD27+IgD− memory B cells, or CD27hiCD38hi plasmablasts/plasma cells. Our findings strongly suggest that Ig class-switched and/or post-germinal-center B cells provide an immunologically active connection between peripheral and CNS compartments and further support peripheral antigen-driven B cell activation as important contributor to CNS autoimmunity.
Results
Sequencing Output
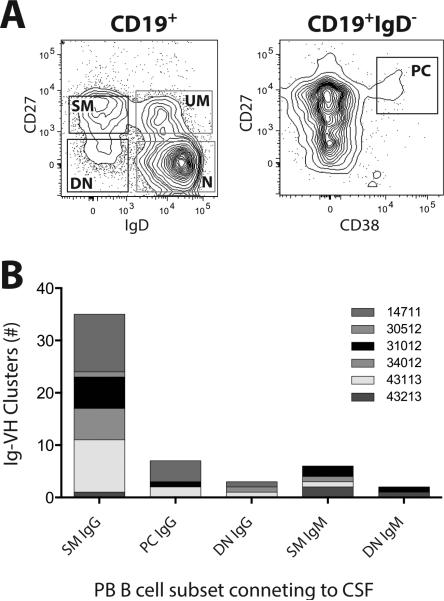
We examined the Ig repertoires in CSF and 5 different PB B cells subsets (Figure 1) of eight individual patients with clinically definitive MS (Table 1); 7 of 8 patients had >5 oligoclonal bands (OCB) in their CSF and an increased IgG-Index (Table 1); all patients were untreated and displayed MS-typical changes on MRI (Table 1). One patient (14711) had active disease at the time of lumbar puncture (Figure S1), all others either had clinically stable or quiescent relapsing-remitting MS or primary progressive MS without recent contrast enhancement on gadolinium enhanced MRI (Table 1). The distribution of sorted PB B cells used as input varied across subsets and patients (Table S1). CSF IgG-VH sequences were obtained from all patients; CSF IgM-VH were sequenced for 4 patients (31012, 34012, 43113, 43213). From PB of all patients, IgM-VH and IgG-VH transcripts were obtained from the following sorted subpopulations: Naïve (N) B cells, CD19+IgD+CD27− (IgM-VH); unswitched memory (UM) B cells, CD19+IgD+CD27+ (IgM-VH); switched memory (SM) B cells, CD19+IgD−CD27+ (IgM-VH/IgG-VH); double negative (DN) B cells, CD19+IgD−CD27− (IgM-VH/IgG-VH); plasmablasts/cells (PC), IgD−CD27hiCD38hi (IgM-VH/IgG-VH) (Figure 1).
Figure 1. Peripheral blood B cell subsets connecting to the CNS compartment.
A: Representative FACS plots (patient 34012) illustrating PB B cell subset identification and sorting strategy; plasmablasts/plasma cells (PC) are CD27hiCD38hi gated on CD19+IgD− cells; switched memory (SM), unswitched memory (UM), double negative (DN), and naïve (N) B cells were sorted based on the presence or absence of CD27 or IgD gated on CD19+ cells. Sorting gates are indicated in the FACS plots, see Figure S7 for single color histograms. PB B cell subsets with connections to the CSF are in black gates, those not connecting to the CSF are in gray gates. B: PB B cell clusters connecting to the CNS; each column represents the sum of Ig-VH clusters with contributions from CSF and the indicated PB B cell subset.
Table 1.
Patient characteristics.
| ID | Age | Sex | Dx | EDSS | MRI/Gd (weeks ago) | Last Relapse (weeks ago) | Tx | CSF OCB | CSF WBC | CSF IgG Index | CSF Volume (ml) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 14711 | 20 | m | RRMS | 3 | + (2 w) | 2 | none | >5 | 103 | 1.5 | 20 |
| 26712 | 54 | f | RRMS | 1.5 | − (6 w) | 8 | none | >5 | 1 | 1.3 | 9 |
| 29612 | 22 | m | RRMS | 1.5 | − (6 w) | 6 | none | neg. | 4 | 0.5 | 8 |
| 30512 | 34 | m | RRMS | 1.5 | − (4 w) | 12 | none | >5 | 7 | 1.2 | 9 |
| 31012 | 37 | f | RRMS | 1.5 | − (32 w) | 12 | none | >5 | 1 | 1.2 | 9 |
| 34012 | 43 | m | PPMS | 3 | + (20 w) | N/A | none | >5 | 4 | 1.4 | 9 |
| 43113 | 31 | f | PPMS | 6 | + (8 w) | N/A | none | >5 | 3 | 3.1 | 5 |
| 43213 | 34 | f | RRMS | 0 | + (5 w) | 8 | none | >5 | 10 | 1.1 | 14 |
ID, patient-specific identification number; Dx, diagnosis: RRMS, relapsing-remitting MS; PPMS, primary-progressive MS; EDSS, expanded disability status scale(55) at neurological exam closest to LP; MRI/Gd, presence (+) or absence (−) of gadolinium contrast-enhancing lesions on brain or spinal cord MRI, in parenthesis is the time in weeks between last MRI and LP; Tx, therapy at time of sample acquisition; CSF OCB, CSF restricted oligoclonal bands; CSF WBC, CSF leukocyte count per μl. CSF IgG-Index was calculated using the standard formula ([CSF IgG/CSF Albumin]/[Serum IgG/Serum Albumin]). CSF Volume, CSF volume used to obtain CSF lymphocytes for CSF IgM/G-VH DIRS.
Each sample was subjected to unbiased 5’-RACE and DIRS; separation into IgG-VH and IgM-VH expressing B cell subsets was achieved by using isotype-specific (i.e. IgM and IgG) reverse primers during Ig-VH 5’-RACE. Due to the generally low numbers of lymphocytes in CSF we chose to directly perform IgM-VH and IgG-VH 5’-RACE from each CSF cell-pellet without sorting of B cell subsets. We obtained a total of ~2.2 million sequencing reads, resulting in ~1.3 million usable IgM-VH or IgG-VH transcripts (Table S1). Sequencing reads were deposited at the NCBI Sequencing Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra/) under accession number SRP042205. Two sequence-clustering approaches were applied for Ig-VH analyses: 1) A distance metric approach (Hamming distance=1, (7)) was employed to group clonally-related Ig-VH with highly similar H-CDR3 amino acid sequence, identical H-CDR3 length, and usage of the same IGHV and IGHJ. Clonally related Ig-VH were used to identify and analyze bi-compartmental B cell clusters as previously described(6). 2) Ig-VH datasets of non-redundant Ig-VH reads were generated by considering sequences with identical H-CDR3 and usage of IGHV and IGHJ only once; non-redundant datasets were used to calculate IGHV usage as previously described (6, 8, 9) and to generate SHM profiles. An overview of the bioinformatics pipeline used for our studies is shown in Figure S2.
Among B cell subsets, numbers of redundant Ig-VH associated with each non-redundant Ig-VH sequence revealed overall mostly low counts for naïve B cells IgM-VH (N.IgM) and partially very high counts of sequences with identical H-CDR3, IGHV, and IGHJ usage in post-germinal center, Ig class-switched B cells, but also in CSF Ig-VH repertoires (Figure S3). Thus, to a reasonable degree, our sequencing approach approximated what is expected biologically: absent clonal expansion among naïve B cells, and extensive clonal activation in B cell subsets resulting from antigen-driven immune responses, such as SM B cells and plasmablasts/plasma cells. In addition, this finding supported previous reports of B cell activation in the CNS and CSF(10-14).
In the present study, we used PCR amplification of Ig-VH and nextgen immune repertoire sequencing (i.e. DIRS). In general, over-amplification or sequencing saturation of particularly prevalent Ig-VH transcripts could introduce skewing of data and affect conclusions drawn, particularly with regards to estimating clonal B cell stimulation. Furthermore, in nextgen sequencing, sequencing errors are a potential concern, particularly with respect to DIRS where read errors could be mistaken for evidence of somatic hypermutation(15, 16). In the following paragraphs we will discuss the potential impact of these issues on our results and interpretations where applicable.
Bi-compartmental B cell clusters
We identified a total of 46 bi-compartmental clusters of clonally related Ig-VH in 6 of 8 patients (14711 n=15 clusters; 30512 n=1; 31012 n=7; 34012 n=7; 43113 n=13; 43213 n=3) (Figures 1 and S4, Tables 2, S2, and S3) based on H-CDR3 aminoacid sequence similarity and usage of Ig germline segments, features that readily discern Ig-VH sequences of a common origin (17, 18). In all 6 patients with bi-compartmental B cell clusters, we identified CSF IgG-VH clusters connecting to PB IgG+ SM B cells (n=35 clusters); in all 4 patients from which CSF IgM-VH were sequenced we found clusters that were composed of CSF IgM-VH and IgM-expressing SM B cells from peripheral blood (n=5 clusters); in 3 patients 7 CSF IgG-VH clusters connected to PB IgG+ plasma cell (PC) clusters, 2 of which inter-connected with IgG+ SM B cells (Tables 2 and S2). We also identified a few clusters in which CD19+CD27−IgD− (DN) B cells connected to CSF B cells, or in which IgM+ SM B cells connected to CSF IgM-VH clusters. A single cluster that included CSF-IgG and both, IgG, and IgM expressing PB B cell subsets was identified in patient 31012 (Tables 2 and S2). No bi-compartmental clusters were found in a patient without OCB (29612) and in one other patient, the oldest patient in our cohort (26712, 54 year old female). It is noteworthy that, for these two patients, no CSF IgM-VH sequences were obtained; it is thus possible that a connection between IgM-expressing PB B cell subsets and the CSF did exist but was not identified for technical reasons. Notably, no connections were identified between CSF and naïve or UM B cells in PB.
Table 2.
B cell clusters connecting the periphery and CSF.
| CSF IgG | SM IgG | PC IgG | DN IgG | CSF IgM | SM IgM | DN IgM | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pat. | IGHV | H-CDR3 | IGHJ | # | SHM | # | SHM | # | SHM | # | SHM | # | SHM | # | SHM | # | SHM |
| 14711 | 1-18 | CARDSRGFADIW | 5 | 1124 | (13-16) | 48 | (14) | 50 | (14-16) | ||||||||
| 1-24 | CATGGISSSSDDWYNYFYGLDLW | 6 | 31 | (19-21) | 19 | (19) | |||||||||||
| 1-3 | CAGPVWAGSPNWKPWFGVDVW | 6 | 1766 | (8-19) | 34 | (16-18) | |||||||||||
| 1-8 | CARVVRYKITWYFDPW | 5 | 29 | (14-15) | 111 | (15) | |||||||||||
| 3-11 | CARDRFGVFDFW | 4 | 16 | (21) | 1 | (27) | |||||||||||
| 3-11 | CARDLRGRTRSIATVRGLLYDAFDIW | 3 | 9 | (14) | 33 | (16-18) | |||||||||||
| 3-23 | CAKDAGWYLYYYFDLW | 2 | 453 | (22-24) | 56 | (22) | |||||||||||
| 3-30/33rn | CARRPEGYAMDVW | 6 | 3 | (11) | 13 | (10) | |||||||||||
| 3-30/33rn | CAKDLVSDHYYYYGMDVW | 6 | 54 | (11-13) | 13 | (11) | |||||||||||
| 4-34 | CARGPKNKRFPMAPAEFFDYW | 4 | 15 | (21-22) | 7 | (27-28) | |||||||||||
| 4-34 | CARLGSWWLLTALRSTKGQYYYGMDVW | 6 | 210 | (11-24) | 35 | (22) | |||||||||||
| 4-4 | CARGGFSITWGGFDIW | 3 | 36 | (27-28) | 1 | (28) | |||||||||||
| 4-59/61 | CAKYDFWSGFDPW | 5 | 40 | (10) | 23 | (10) | |||||||||||
| 4-b | CARSYGDYVDPFFDYW | 4 | 64 | (18-34) | 689 | (6-19) | |||||||||||
| 6-1 | CARSGKPTGGGVLAWGPKKFVSSLYFDSW | 4 | 1146 | (26-32) | 8 | (26-29) | |||||||||||
| 30512 | 2-5 | CAHSKMATMGGELVFDYW | 4 | 68 | (3-4) | 7717 | (3-8) | ||||||||||
| 31012 | 1-24 | CATQSGMITASPLDYW | 4 | 1988 | (2-4) | 113 | (2-9) | ||||||||||
| 1-24 | CATGRKSGVVGAYFDYW | 4 | 35 | (0) | 48 | (1-2) | |||||||||||
| 3-21 | CATGGAAEHAYW | 4 | 116 | (36-37) | 19 | (36) | 2 | (30) | 11 | (27-32) | 6 | (29) | |||||
| 3-48 | CVRDRDIVTSDSW | 4 | 9 | (20) | 2 | (34) | |||||||||||
| 3-53/66 | CAKENIAALGNPLDFW | 4 | 68 | (17) | 1 | (18) | |||||||||||
| 3-9 | CVKDMEPYGDPLRPAEAFDFW | 3 | 279 | (23-25) | 4 | (26) | |||||||||||
| 4-34 | CVRGGPNTEYWPNFDSW | 4 | 1 | (26) | 33 | (23-25) | |||||||||||
| 34012 | 1-18 | CARTYFYGSENRQEYDWFDPW | 5 | 4892 | (15-22) | 1 | (16) | ||||||||||
| 3-21 | CARSTRTMHQGKSGMDVW | 6 | 45 | (23) | 4 | (24) | |||||||||||
| 3-21 | CARSTRSMHHRNSAMDVW | 6 | 83 | (23-26) | 2 | (25) | |||||||||||
| 3-30/33rn | CARNSFYCSSISCFYRPGSKRDYYHYGMDVW | 6 | 775 | (11-15) | 1 | (12) | |||||||||||
| 4-30-4/31 | CARGESSGSYVCFDCW | 4 | 498 | (17-21) | 6 | (19) | 13 | (19) | |||||||||
| 4-34 | CSRGALVATHVFDYW | 4 | 986 | (28-37) | 4 | (28-33) | |||||||||||
| 4-39 | CARRIILIGGAFDIW | 3 | 78 | (24) | 3 | (24) | |||||||||||
| 43113 | 3-7 | CARGTVFVVLLTSYFDYW | 4 | 4 | (29) | 1 | (28) | ||||||||||
| 3-7 | CARGRFFEFLLTSYFDYW | 4 | 30 | (29-30) | 32 | (28) | |||||||||||
| 4-39 | CARLQQWVEIW | 5 | 34 | (19-20) | 19 | (20-21) | |||||||||||
| 4-39 | CARGPVSLGTLDPW | 5 | 487 | (8-28) | 87 | (24-25) | |||||||||||
| 4-39 | CARRGRGWAPFDSW | 4 | 17 | (25) | 34 | (25) | |||||||||||
| 4-39 | CARGPVSRGDAPTPW | 5 | 4 | (23) | 1 | (24) | |||||||||||
| 4-39 | CATPLRDSSDYSTFDIW | 3 | 7 | (21) | 15 | (21-23) | |||||||||||
| 4-39 | CARHERDHTGFLNYYFDSW | 4 | 11 | (21) | 1 | (8) | |||||||||||
| 4-39 | CARRPQDFWSPYYTTYYFDSW | 4 | 360 | (7-25) | 1 | (7) | 32 | (18) | |||||||||
| 4-39 | CARLGASHYDSSGYYYYFDYW | 4 | 1 | (11) | 14 | (10) | |||||||||||
| 4-59/61 | CARATRFNMHWYPFVDLW | 2 | 56 | (20) | 8 | (18) | |||||||||||
| 4-b | CARPYFDGSGYYWDVLAFDVW | 3 | 10 | (7-8) | 9 | (7-9) | |||||||||||
| 6-1 | CARYTSGWFLDYW | 4 | 1 | (7) | 70 | (18-23) | |||||||||||
| 43213 | 4-59/61 | CAREYSRKEGGRWRVPKTGRYSYNSGMDVW | 6 | 6 | (29) | 16 | (35) | ||||||||||
| 6-1 | CARRTTLGFFDYW | 4 | 13 | (3) | 12 | (9) | |||||||||||
| 6-1 | CARYTSGWFLDSW | 5 | 2766 | (7-12) | 29 | (9-11) | 49 | ||||||||||
Clusters are grouped by patient; each row represents a bi-compartmental cluster. IGHV/IGHJ: closest related variable and joining germline segments utilized by Ig-VH sequences within each cluster. H-CDR3, representative H chain CDR3 aminoacid sequence per cluster, the N-terminal Cys (C) and C-terminal Trp (W) are shown for better visual orientation. Each B cell subset or CSF contributing to bi-compartmental clusters is represented in a column showing number of Ig-VH sequences (#) and range of SHM (SHM) along the IGHV sequence portion observed in CSF and PB B cell subsets (SM, switched memory B cells; PC, plasma cells/plasmablasts; DN, double negative B cells). Gray shades indicate clusters represented in Figure 2.
In linear regression analyses, the numbers of bi-compartmental IgG-VH clusters obtained did not correlate with CSF WBC (r2=0.39, P=0.1) or CSF volume [ranging from 5 to 20 ml] (r2=0.12, P=0.4) suggesting that detection of B cell clusters connecting the CSF and periphery does not depend on either factor (Table S4). Similarly, numbers of bi-compartmental IgG-VH were also independent from IgG-Index (r2=0.46, P=0.07), or total CSF IgG-VH reads (r2=0.12, P=0.4) (Table S4). Bi-compartmental clusters generally comprised less than 1% of PB B cell subset clusters (Figure S4 and Table S5). An exception were bi-compartmental clusters involving plasma cells/plasmablasts in patient 14711 comprising nearly 5% of Ig-VH clusters, which may be due to the overall low number of plasma cells/plasmablasts sorted form this patient, but may also suggest active egress of antibody-producing cells from the CSF (Table S1, Table S5).
Affinity-maturation in the CNS and periphery
We were interested to determine whether B cells belonging to bi-compartmental clusters undergo affinity-maturation in the periphery and/or the CNS compartment. Numerous IgM-VH and IgG-VH from either compartment contain multiple SHM along their IGHV-derived portion (Table 2) suggesting that they resulted from antigen-driven immune responses. From our data it is not possible to draw conclusions whether a B cell response originated in one or the other compartment; however, within clusters of related Ig-VH, we commonly identified a range of SHM (Figure 2, Table 2) in the periphery and/or CSF suggesting that B cells belonging to bi-compartmental clusters may have been exposed to antigen-stimulation in either compartment.
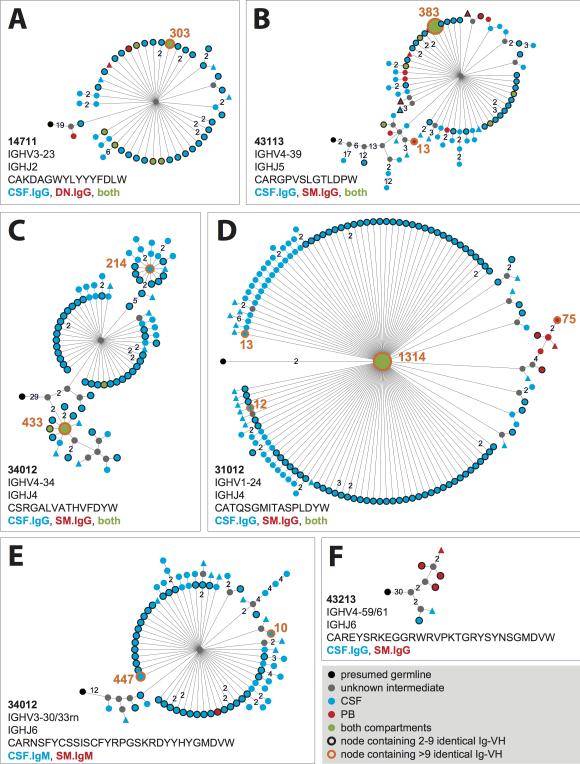
Figure 2. Affinity-maturation of B cells connecting CSF and PB.
Shown are representative lineages (A to F) of clonally related Ig-VH found in both compartments. Lineages were calculated using IgTree (see Methods) and visualized in Cytoscape using the proprietary Organic layout option. Numbers between nodes represent numbers of nucleotide mutations; connections without numbers represent a single nucleotide mutation. Round nodes without rim encompass a single Ig-VH sequence; round nodes with black rims contain 2 to 9 identical Ig-VH, those with orange rims contain >9 identical Ig-VH sequences (orange number next to respective nodes). Closest putative germline sequences represent the root of each lineage (black nodes). Gray nodes are lineage intermediates that were not identified as Ig-VH transcript and were calculated by IgTree. Shown are also IGHV and IGHJ used per lineage and representative H-CDR3. Alignments of representative Ig-VH are shown in Figure S5.
Sequencing errors cannot be fully differentiated from true SHM when sequencing natural Ig-VH repertoires where the mRNA templates are not known. In the analyses of our sequencing data we applied a reductionist approach to only use high-quality Ig-VH sequences for our conclusions regarding SHM occurring in CSF and/or periphery (see Methods and Figure S2). In 454 sequencing, the probability of sequencing errors due to single and multiple substitutions is less than 10% based on previous quantification(15). Since most nodes in our lineage analysis contain at least 2 identical in-frame sequencing reads, the probability of one single SHM due to substitution errors is less than 0.01 for a particular node containing 2 identical sequences (type I error rate less than 1%,(15)), with each additional identical sequencing read contained in a tree node, the probability of sequencing errors is reduced by a factor of 0.1; accordingly, for 3 identical sequences to represent a sequencing error, the probability is <0.001, for 4 sequences <0.0001, for 5 sequences <0.00001, etc. Thus, lineage relationships of Ig-VH generated by our sequencing approach and calculated using IgTree software (19, 20) support active antigen-driven immune responses to be involved in the shaping of bi-compartmental groups of related B cells; representative lineage trees are shown in Figure 2 and the respective Ig-VH sequence alignments are shown in Figure S5.
IGHV usage in Ig-VH repertoires in PB and CSF
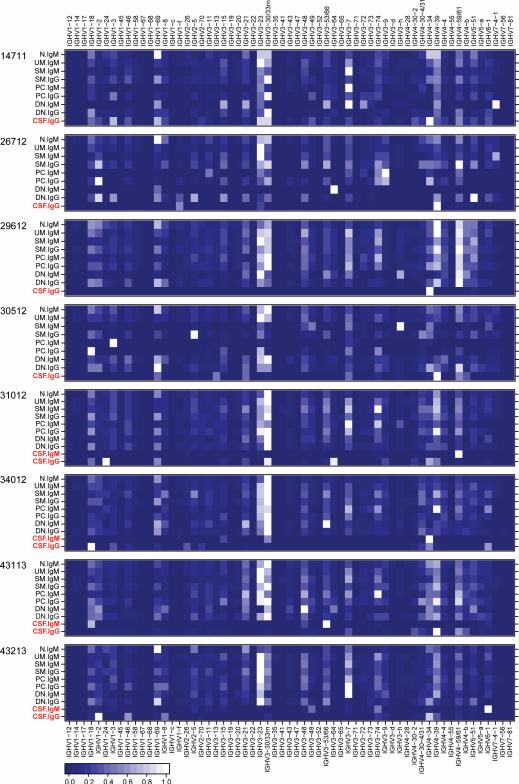
While our primary focus was to characterize B cells providing an immune axis between CNS and PB, our data also enabled Ig germline gene usage analyses in the overall CSF and PB B cell subsets Ig repertoires obtained by DIRS. We found diverse IGHV usage among the majority of PB B cell subsets (Figure 3, Table S6). Overall, IGHV usage in CSF was limited compared to PB B cells (Figure 3), while IGHV usage by different B cell subsets was generally similar within individual patients. Interestingly, patient 14711 who had active CNS inflammation with gadolinium contrast-enhancing cervical spinal cord lesions on MRI 2 weeks prior lumbar puncture (Table 1, Figure S1), displayed more diverse IGHV usage in CSF compared to all other patients (Figure S6). Previous studies, including our own, have shown over-representation of certain IGHV4 germline segments in MS CSF(6, 10, 21, 22); in the present study, IGHV4-39 was used by CSF IgG-VH in 7 of 8 patients, and IGHV4-59/61 by 6 of 8 patients’ CSF IgG-VH (Figure 3). Possibly artifactual limited IGHV usage was seen in PB B cells subsets for which low cell numbers were obtained by cell sorting, or when fewer non-redundant Ig-VH sequences were available; examples are 26712 DN IgM, 30512 PC IgM, 30512 PC IgG, etc. (Table S1, Figure 3).
Figure 3. IGHV usage in PB B cell subsets and CSF.
Shown are heatmaps representing relative usage of IGHV germline segments by IgM-VH or IgG-VH from the indicated PB B cell subsets or CSF for each patient studied (Table S6). To generate heatmaps, IGHV usage values were normalized to the most frequently used IGHV per sample and Ig isotype, which were set as 1. The resulting heatmaps show the most frequently used IGHV in white and the least frequently used IGHV in dark blue (see lower left corner for scale). Notably, IGHV usage is generally limited in CSF (red type) suggesting selective recruitment or survival of B cells in the CSF compartment. IGHV usage of CSF IgG-VH in patient 14711 is more balanced and similar peripheral B cell repertoires; this patient had signs of active disease 2 weeks prior to lumbar puncture (Table 1).
Biased IGHV usage representation could result from over-amplification of certain Ig-VH transcripts during PCR or saturation of the sequencing reaction by over-represented Ig-VH. For our studies we used 5’RACE of Ig-VH to eliminate potential biased introduced by IGHV specific-primers, and used non-redundant Ig-VH sequence counts to display IGHV usages (see Methods and Figure S2). We have previously shown similar IGHV usage in PBMC of unrelated individuals(8) using non-redundant Ig-VH datasets. In the present study, we again found similar IGHV distribution among most PB B cell subsets and in unrelated individuals (Figure 3), providing indirect evidence that over-amplification or sequencing saturation of certain Ig-VH rarely happens. Furthermore, we found low numbers of redundant sequences per non-redundant Ig-VH in naïve B cells, a population where clonal is absent (Figure S3 and Table S7).
SHM patterns in Ig-VH repertoires in PB and CSF
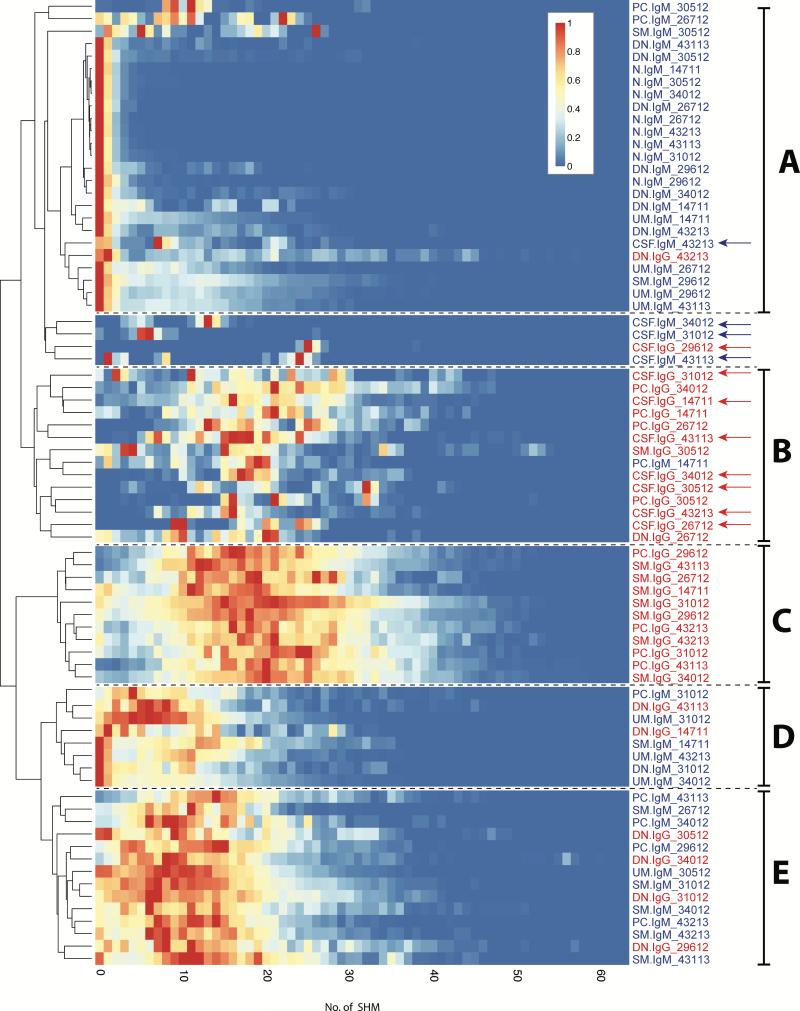
Our data was also conducive to understanding the effect of SHM on the IGHV portion of Ig-VH repertoires represented by each PB B cell subset and by CSF IgG-VH and IgM-VH (Figure 4, Table S8). We were particularly interested in SHM patterns of DN B cells which revealed an unexpected immune axis between CSF and PB in our study. As expected, naïve B cells displayed the lowest levels of SHM along their IgM IGHV (Figure 4 A), while IgG-expressing SM B cells and plasma cells displayed the highest level of SHM (Figure 4 B and C). SHM profiles of IgM-VH expressed by CD27−IgD− (DN) B cells were very similar to those seen in naïve B cells (Figure 4 A). IgG-expressing DN B cells clustered with IgM-expressing B cell subsets including UM, SM, and PC (Figure 4 E) overall suggesting lower levels of SHM having shaped the DN B cell repertoire. Within B cell subsets, IgG-VH had accumulated more SHM compared to IgM-VH (Figure 4). SHM profiles of CSF IgG-VH appeared most similar to IgG expressing SM and PC (Figure 4 B), while SHM profiles of CSF IgM-VH appeared more similar to IgM-expressing N, UM, and DN B cell subsets (Figure 4).
Figure 4. Dendogram and heatmaps of PB B cell and CSF Ig-VH SHM profiles.
Shown are heatmaps of Ig-VH SHM profiles; each row represents a PB B cell subset or CSF as indicated by row titles on the right. IgM are in blue type, IgG are in red type; CSF samples are indicated by arrows. Within each subset or CSF sample, SHM profiles were scaled to the highest peak set at 1.0 and colors assigned such that a count of 0 non-redundant sequences with a certain umber of SHM resulted in blue color and the highest number of sequences with an indicated number of SHM was labeled red; the coloring scale is shown in the upper right. PB B cells subsets or CSF samples clustering together based on SHM profile-similarity are indicated by letters on the right (see also Results). A: Group of subsets characterized by predominantly low SHM, including all naïve B cells and nearly all IgM+ DN B cells. B and C: Groups of subsets characterized by extensive SHM, and containing all IgG-expressing SM B cells and plasma cells, and the majority of CSF samples. D and E: Subsets characterized by SHM lower than B and C, but higher than A; these groups include mainly IgM-expressing subsets and IgG-expressing DN B cells. The data used to generate these heatmaps is shown in Table S8.
Overall, the SHM patterns displayed in Figure 4 support the validity of the B cell subset sorting approach based on CD19, CD27, CD38, and IgD, together with IgM and IgG specific PCR amplification of VH. Furthermore, the observed SHM profiles, i.e. very low levels of SHM in naïve B cells and the most extensive SHM in IgG-expressing SM B cells and antibody-producing plasma cells/plasmablasts (Figure 4), further suggests that our sequencing approach yielded biologically correct data.
Discussion
A central goal of MS research is to identify disease-relevant B cells among the vastly diverse peripheral B lymphocyte compartment. We recently demonstrated that an exchange of immunologically active clusters of related B cells occurs between the CNS and PB compartments(6). Here, we describe the identification of specific peripheral blood B cell subsets that are clonally related to CSF B cells, and could act as important contributors to disease-associated B cell repertoires in MS patients. Overall, our analyses afforded an unprecedented view of Ig germline gene usage and levels of affinity-maturation shaping Ig repertoires associated with different B cells subsets in PB and the CSF.
A key question is at which stage during maturation B cells enter the CNS compartment. Influx of naïve B cells would support fully functional secondary lymphoid tissue-like function of the CNS that can operate independent from the periphery. Influx of post-germinal center (GC) B cells, such as memory B cells and plasmablasts/plasma cells, on the other hand might suggest initial antigen-training in the periphery and further affinity maturation and B cell maturation inside the CNS. Overall, our findings suggest that Ig class-switched B cells provide an antigen-experienced immune axis connecting the periphery and the CNS in MS. B cells linking both compartments undergo SHM in PB and CSF, lending strong support 1) to the presence of potentially disease-driving antigens in the periphery, and 2) to the capability of the intrathecal tissues to support active B cell receptor affinity maturation.
The antigens targeted by B cells participating in the immune axis between periphery and CNS remain unknown. It was previously shown, that clonally expanded intrathecal plasma cells produce antibodies that bind to as yet unknown myelin or myelin-associated antigens in demyelinating lesions of MS(13), and soluble IgG binding to myelin-oligodendrocyte protein were identified in MS CSF(23, 24). Accordingly, bi-compartmental B cell clusters are assumed to participate in potentially CNS-reactive B cell responses. However, clonal CSF IgG have also been shown to react with EBV antigens (25) at least raising the possibility that bi-compartmental B cell clusters target viral or microbial epitopes and possibly cross-react with CNS epitopes via molecular mimicry, as was shown for T cells in MS(26) . Considering that in humans a multitude of viruses are constantly replicating(27) which could stimulate both, CNS-specific, and non-specific B cell responses via bystander activation, significant additional work is necessary to understand the involvement of bi-compartmental B cell clusters in MS disease activity.
In our study, class-switched memory B cells were found most frequently to provide an immune axis between PB and CSF. Memory B cells are strongly CD20-positive and are efficient antigen-presenting cells (28, 29); memory B cells reactive against myelin have been shown to be present in MS patients’ blood(30). CD20-targeting, B cell-depleting monoclonal antibodies (rituximab, ocrelizumab) are highly effective in reducing or eliminating MS disease activity(1-3), and in depleting B cells from CSF(31). Our finding that memory B cells participate in an exchange across the BBB further strengthens the rationale of using anti-CD20 mediated B cell depletion to interrupt disease-promoting peripheral B cell autoimmunity. Natalizumab, a monoclonal anti-VLA4 antibody that also effectively reduces MS disease-activity, inhibits transmigration of lymphocytes from PB into the CNS compartment and reduces CSF B cell numbers (32). Two studies described decreased intrathecal production of clonal Ig with natalizumab therapy(33, 34), an effect that could be due to limiting transmigration of B cells involved in OCB production. However, natalizumab was also shown to reduce CSF CD4+ and CD8+ T cells and CD19+ B cells in MS patients(32). Accordingly, intrathecal OCB production may require influx of B cells, or T cells providing signals for B cell differentiation into antibody-expressing plasma cells or plasmablasts, or a combination of both.
Intrathecal plasma cells and B cells are known to be responsible for local production of clonal IgG(35, 36). We found a few instances in three patients of PB plasma cells forming bi-compartmental clusters with CSF IgG-VH, which is in agreement with our recent finding that antibodies identical to OCB can theoretically also be produced in the periphery(37). However, it seems unlikely that a sustained supply of clonally restricted peripheral plasma cells entering the CNS compartment is responsible for intrathecal antibody production, as this would require specific recruitment of select plasma cell clones into the CNS. More likely, plasma cells that participate in bi-compartmental clusters are progeny of antigen-experienced memory B cells that became established in both, the CNS, and periphery. An important question that remains, is whether the initial antigen exposure of naïve B cells that ultimately mature to bi-compartmental memory B cells and OCB-producing plasma cells occurs in the periphery or CNS. Mature naïve B cells that migrate across the BBB could provide input into presumed GC in lymphoid follicle-like structures in the CNS (38, 39); consistent with this concept, IgD+CD27− naïve B cells have been found in the CSF of MS patients(40) and evidence for B cell differentiation were found in MS CNS(41). In further support of the presence of naïve B cells in CSF we found CSF IgM-VH harboring very few mutations suggesting an intrathecal presence of IgM-expressing B cells that have not yet undergone affinity-maturation. However, the absence of IGHV usage diversity among CSF IgM-VH in the 4 patients from which CSF IgM-VH were obtained argues against an ongoing unrestricted exchange of naïve B cells in MS.
We found diverse IGHV usage in patient 14711 CSF, a possible example for a more balanced exchange of B cells during active MS, as previously reported in another MS patient with active disease(6). We also found the largest number of bi-compartmental Ig-VH clusters in this patient, but not all IGHV found in this patient's CSF are also represented in clonally related B cell clusters spanning periphery and CSF. While speculative, it is possible that the observed clusters represent the pathologically relevant ones participating in active inflammation, while B cells expressing IGHV not represented by bi-compartmental clusters are pathologically irrelevant. In all other patients’ CSFs we found limited IGHV usage suggesting selective survival of B cells in the CNS compartment during disease sates that appear quiescent by clinical or MRI measures.
Interestingly, in 4 of 8 patients studied here we found Ig class-switched DN B cells that were clonally related to intrathecal Ig repertoires. As previously described by others(42, 43), SHM profiles of DN IgG-VH are frequently shifted towards fewer somatic mutations when compared to IgG+ SM B cells, which may support DN cells as progenitors of SM B cells. However, based on a high-throughput immune repertoire sequencing study comparing Ig-VH of SM and DN B cells, CD27− and CD27+ may also be developmentally related in either direction, i.e. with DN B cells being precursors of SM B cells, and vice versa(44). Peripheral blood DN B cells are expanded in patients with active systemic lupus erythematosus (SLE); these cells have been proposed to represent a class of memory B cells that fail to undergo productive GC maturation(42). In fact, B cells accumulating SHM outside of GCs likely also escape the scrutiny of peripheral B cell tolerance and may harbor increased autoimmune potential(45). Our observation in this cohort of MS patients, that SHM profiles of IgM-expressing DN B cells compare more closely to naïve B cells than non-class switched memory B cells, suggests that they are direct progeny of naïve B cells and undergo class-switch recombination (CSR) outside of GCs or exit GCs prematurely. Interestingly, DN B cells proliferate more readily upon TLR-mediated activation than with BCR-cross-linking, suggesting that they function in an antigen-independent manner(42). Further work will be necessary to understand the relevance of DN B cells to MS; however, our findings suggest that DN B cells may be a relevant part of the immune axis linking the periphery with the CNS.
The identification of PB B cell subsets participating in bi-compartmental immune response would not have been possible without high-throughput next-generation sequencing techniques that are far-more comprehensive than Sanger-sequencing based approaches. Nonetheless, there are several limitations of our approach that should be recognized:
We are unable to directly measure clonal expansion. However, the number of redundant Ig-VH associated with non-redundant reads appears to approximate biologically expected findings. With the technology used here it is difficult to determine whether a given sequence appears more frequently due to a large quantity of its mRNA being recovered from a single cell or multiple cells expressing that same sequence, or due to disproportional PCR amplification or sequencing of certain Ig-VH transcripts. Higher number of reads per non-redundant Ig-VH could also reflect low sequence diversity within these populations and thus sequencing oversaturation. New technologies incorporating nucleotide tags (46, 47) or high-throughput single cell sequencing technologies (48) may in the future permit better estimates of numbers of mRNA molecules obtained from single cells, thereby permitting more accurate measures.
Both, identifying clonal overlaps, and appropriate representation of IGHV usage depends on the sampling and sequencing depth of the interrogated compartments, and the likelihood of detecting clonally related Ig-VH between CSF B cells and PB B cell subsets is expected to correlate with the level of clonal expansion in each subset. Clonal expansion is a characteristic of activated memory B cells or plasma cells, theoretically facilitating identification of B cell clusters by DIRS. Conversely, absent or low-level clonal expansion (i.e. homeostatic proliferation (49)) found in the naïve B cell compartment likely impedes detection of PB naïve B cells related to CSF Ig-VH, and greater numbers of cells sampled and increased sequencing depth may, or may not, reveal additional clusters.
Sequencing errors introduced by 454 sequencing could result in artifactual clonal families of Ig-VH sequences(16). In our experiments we studied natural Ig-VH repertoires without knowledge of template sequences. It is thus impossible for us to determine exactly which mutations are sequencing errors and which result from SHM. We applied a reductionist approach to select Ig-VH sequences for lineage analyses that incorporate likely true SHM, rather than sequencing errors. SHM profiles resulting from PB B cell subsets reveal biologically expected information (i.e. low SHM in naïve B cells, extensive SHM in IgG-expressing memory B cells and plasma cells), and therefore suggest that, overall, sequencing errors do not exert extensive influence on the representation of SHM.
Despite these limitations, our data in summary reveal a highly dynamic and interconnected B cell repertoire shaping the PB and CNS immune landscape in MS. Our study represents an important initial snapshot of processes that are likely to come into better resolution as additional observations – especially those accompanying the early phases of disease development, during clinical attacks, and with transition to the late progressive phase of MS – are revealed. Indirectly, our findings suggest, that exchange of disease-associated B cell subsets between the CNS and periphery in MS is a highly selective process. This process may be supported by B cells that have undergone class-switch recombination and/or affinity maturation, in the periphery and/or CNS compartment. Although the target antigens and exact mechanisms driving the establishment of intrathecal B cell repertoires remain unknown, the ability to identify antigen-experienced B cells participating in MS immune pathology, as described here, should help to resolve these key issues. In particular identifying the specificity of bi-compartmental B cells will ultimately link B cell exchange between periphery and CNS to MS immune pathology.
Materials and Methods
Patient samples
We studied 8 patients with clinically definitive MS (Table 1) based on the latest diagnostic criteria for MS(50). Patients were selected based on availability of paired CSF and sufficient peripheral blood (PB). Based on a prior study(6), we expected to identify bi-compartmental clusters in approximately 80% of patients. No sample size calculations were performed prior to our experiments; data analyses were not performed blinded as no comparisons between different groups of patients were performed. Peripheral blood (48 ml) was obtained via standard venipuncture; 5-20 ml of CSF were collected during standard lumbar puncture (LP) during the same visit. To avoid potential CSF contamination with PB, the first 2 ml of CSF were discarded. Immediately after LP, the entire volumes of CSFs were centrifuged at 400xg for 15 minutes and cell pellets lysed in RLT buffer (Qiagen RNeasy kit). Peripheral blood mononuclear cells (PBMC) were isolated using a Ficoll gradient, red blood cells lysed and PBMC washed in phosphate buffered saline (PBS) containing 1% BSA. These studies were approved by the institutional review board of the University of California, San Francisco and informed consent was obtained from patients before CSF/PB collection.
Cell staining and sorting
After FcR blocking (mouse serum; Jackson ImmunoResearch), PBMC were incubated for 20 minutes on ice in dark with the following antibodies: CD19 (APC Cy7), IgD (PE Cy7), CD27 (Qdot605), CD38 (PerCP Cy5.5), CD3 (Pacific blue). Then, cells were washed and resuspended in PBS containing 1% BSA; DAPI (4',6-diamidino-2-phenylindole) was added to discriminate dead cells. B cell subsets were identified using the expression of following surface markers and collected on a FACS Aria (BD Biosciences): Naïve (N): CD19+IgD+CD27−, unswitched memory (UM): CD19+IgD+CD27+, switched memory (SM): CD19+IgD−CD27+, double negative (DN): CD19+IgD−CD27−. Plasmablasts/cells (PC) were gated on IgD− cells and selected based on high expression of CD27 and CD38 (IgD−CD27hiCD38hi) (Figure 1). Gating controls (single color histograms) for CD19, CD20, CD27, CD38, and IgD are shown in Figure S7. Dead cells and T cells were gated out using DAPI and CD3 expression respectively. B cell subsets were sorted and immediately lyzed in RLT buffer (Qiagen RNeasy kit) and stored at −80°C.
Unbiased Ig mRNA amplification and Ig repertoire sequencing
Sequencing work flow was performed as previously described(8). In brief, Total RNA was isolated from CSF (miRNeasy mini kit, Qiagen) and PB B cells subsets (RNeasy micro kit, Qiagen). RNA quality was assessed using an Agilent Bioanalyzer. Total isolated RNA was reverse transcribed (SMARTerTM RACE, Clontech), and approximately 27% of each cDNA reaction was used for IgM-VH and IgG-VH amplification via PCR using SMARTerTM RACE 10X Universal primer mix (Clontech) and IgG or IgM isotype specific 3’ primers (IgG, 5’-GGG AAG ACS GAT GGG CCC TTG GTG G-3’; IgM, 5’- GAT GGA GTC GGG AAG GAA GTC CTG TGC GAG-3’) for 31 cycles following the manufacturer's recommendation. These primer sequences were attached to Lib-L specific adaptor (454 sequencing, Roche) and barcode sequences to amplify IgG-VH and IgM-VH libraries. Barcoded IgM-VH (~715 bp) and IgG-VH (~640 bp) transcript libraries were purified using AMPure XP (Beckman Coulter Inc.), quantified using PicoGreen (Life Technologies) and normalized to 1×109 molecules/μl. Sequencing library pools of uniquely barcoded B cell subsets from each subject were created by pooling the normalized IgM-VH and IgG-VH amplified libraries at relative proportions to obtain 1X (DN, PC); 2X (N) and 4X (SM, UM) sequencing depth respectively. The final multiplexed library pools were subjected to emulsion PCR and unidirectional sequencing using the GS FLX Titanium Lib-L chemistry (454 Sequencing, Roche).
Sequence analysis
Figure S2 illustrates and summarizes the bioinformatics pipeline used for Ig-VH sequence analysis. For all reads, IGHV and IGHJ gene segment usage, CDR1-3 aminoacid sequence and number of somatic hypermutation (SHM) events were determined using VDJFasta as previously described(9). In general, Ig-VH sequences were considered related based on the fact that IGHV and IGHJ usage are stable features of clonally related B cells, that H-CDR3 sequence is identical or highly similar between clonally related B cell, and that the H-CDR3 sequence length is virtually invariable within groups of clonally related B cells. For calculations of IGHV usage frequency non-redundant Ig-VH sequence counts were considered; i.e. sequences with identical H-CDR3, IGHV, and IGHJ were counted only once. Numbers of non-redundant IgG/M-VH are shown in Table S1; per IGHV germline segment usage is displayed as percentage of all Ig-VH sequences obtained per sample. To generate SHM profiles, the most frequent SHM count encountered in a group of non-redundant Ig-VH was used. This approach ensured correction for potential over-amplification of more abundant Ig-VH mRNAs.
To identify bi-compartmental clusters containing IgM-VH or IgG-VH from CSF and PB B cell subsets clonally related Ig-VH were grouped based on H-CDR3 similarity and usage of identical Ig germline segments. This approach combined Ig-VH containing identical H-CDR3 aminoacid sequences and Ig-VH with H-CDR3 differing by a single aminoacid compared to all other H-CDR3 in a cluster based on a Hamming distance =1(6, 7). For lineage analysis, only Ig-VH sequences for which in-frame H-CDR3 were identified, and that spanned at least from the 5’ end of H-CDR1 to the 3’ end of H-CDR3 with a contiguous reading frame were considered; all out-of-frame sequences were thus automatically filtered based on the presence of uncommonly occurring insertions and deletions (indels). Putative germline sequences were obtained using SoDA (34) and were used as tree-root nodes (black) in our lineage tree calculations.
We utilized IgTree(19, 20) (kindly provided by Dr. Ramit Mehr, Bar-Ilan University, Ramat-Gan, Israel) to calculate B cell lineages contained within each IgG-VH cluster. This software has been successfully used with DIRS data(20). Automated multiple alignments of cluster-derived Ig-VH sequences and the corresponding germline sequence were performed using ClustalW 2.1(51). Lineage trees were displayed in Cytoscape (Version 2.8.3(52)) using the proprietary “organic” layout which permits a more compact depiction of lineages; tree nodes were colored according to their origin (CSF, PB cell subsets) and isotype (IgM, IgG). Putative germline nodes are labeled black, lineage intermediates not found by DIRS were calculated by IgTree and are labeled gray. The size of tree nodes is proportional to the number of identical sequences contained within each node. Ig-VH amino acid alignments in were generated using the MAFFT algorithm(53) within Jalview(54) software.
Statistics
Linear regression analyses and ANOVA were performed in GraphPad Prism 6.0. Comparisons of numbers of reads in each non-redundant Ig-VH sequence group between naïve PB B cells and all other B cell subsets and CSF per patient (Table S7) were performed using Kruskal-Wallis test (ANOVA with Dunn's multiple comparisons)(GraphPad Prism 6.0).
Supplementary Material
One-Sentence Summary.
Immunoglobulin class-switched B cell subsets including memory B cells and antibody-producing B cells connect the peripheral and CNS immune compartments in MS and suggest active peripheral immunity to be involved in MS immune pathogenesis.
Acknowledgements
The authors are deeply grateful to their patients who have agreed to participate in this research study. We thank Ramit Mehr, Bar-Ilan University, Ramat-Gan Israel for providing IgTree. We thank Shobha Potluri and Hong Wan (both Rinat-Pfizer) for helpful discussions and suggestions.
Funding: These studies were supported by grants from the NMSS (RG-4868 to HCvB), the NIH/NINDS (K02NS072288 to HCvB), and Pfizer Inc. HCvB was also supported by an endowment from the Rachleff Family Foundation.
Abbreviations
- MS
multiple sclerosis
- PB
peripheral blood
- CSF
cerebrospinal fluid
- Ig-VH
immunoglobulin heavy chain variable region
- CNS
central nervous system
- BBB
blood-brain barrier
- DIRS
deep Ig heavy chain variable region repertoire sequencing
- OCB
oligoclonal bands
- N
naïve B cells
- UM
unswitched memory B cells
- SM
switched memory
- DN
double negative B cells
- PC
plasmablasts/cells
- GC
germinal center
Footnotes
Author contributions: A.P. performed experiments, analyzed and interpreted data, and wrote the paper; L.A. performed bioinformatics analyses, interpreted data and wrote the paper; T.C.K. performed experiments, analyzed and interpreted data, and wrote the paper; M.S. performed bioinformatics analyses, interpreted the data, and wrote the paper; S.W. performed bioinformatics and statistical analyses and wrote the paper; S.J.P. performed bioinformatics analyses, and wrote the paper; P.D.S. performed experiments and wrote the paper; D.T. performed experiments; L.Z.Z. performed experiments; M.D. collected patient samples and provided clinical information; A.A. collected patient samples and provided clinical information; S.L.H. interpreted the data and wrote the paper; H.C.v.B. planned all experiments, analyzed data, and wrote the paper.
Competing interests: No competing interests.
Data and materials availability: Sequencing reads were deposited at the NCBI Sequencing Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra/) under accession number SRP042205.
References
- 1.Bar-Or A, Calabresi PA, Arnold D, Markowitz C, Shafer S, Kasper LH, Waubant E, Gazda S, Fox RJ, Panzara M, Sarkar N, Agarwal S, Smith CH. Rituximab in relapsing-remitting multiple sclerosis: a 72-week, open-label, phase I trial. Annals of neurology. 2008;63:395–400. doi: 10.1002/ana.21363. [DOI] [PubMed] [Google Scholar]
- 2.Hauser SL, Waubant E, Arnold DL, Vollmer T, Antel J, Fox RJ, Bar-Or A, Panzara M, Sarkar N, Agarwal S, Langer-Gould A, Smith CH, Group HT. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358:676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 3.Kappos L, Li D, Calabresi PA, O'Connor P, Bar-Or A, Barkhof F, Yin M, Leppert D, Glanzman R, Tinbergen J, Hauser SL. Ocrelizumab in relapsingremitting multiple sclerosis: a phase 2, randomised, placebo-controlled, multicentre trial. Lancet. 2011;378:1779–1787. doi: 10.1016/S0140-6736(11)61649-8. [DOI] [PubMed] [Google Scholar]
- 4.Lovato L, Willis SN, Rodig SJ, Caron T, Almendinger SE, Howell OW, Reynolds R, O'Connor KC, Hafler DA. Related B cell clones populate the meninges and parenchyma of patients with multiple sclerosis. Brain : a journal of neurology. 2011;134:534–541. doi: 10.1093/brain/awq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obermeier B, Lovato L, Mentele R, Bruck W, Forne I, Imhof A, Lottspeich F, Turk KW, Willis SN, Wekerle H, Hohlfeld R, Hafler DA, O'Connor KC, Dornmair K. Related B cell clones that populate the CSF and CNS of patients with multiple sclerosis produce CSF immunoglobulin. Journal of neuroimmunology. 2011;233:245–248. doi: 10.1016/j.jneuroim.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Büdingen HC, Kuo TC, Sirota M, van Belle CJ, Apeltsin L, Glanville J, Cree BA, Gourraud PA, Schwartzburg A, Huerta G, Telman D, Sundar PD, Casey T, Cox DR, Hauser SL. B cell exchange across the blood-brain barrier in multiple sclerosis. The Journal of clinical investigation. 2012;122:4533–4543. doi: 10.1172/JCI63842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamming RW. Error Detecting and Error Correcting Codes. Bell System Technical Journal. 1950;29:147–160. [Google Scholar]
- 8.Glanville J, Kuo TC, von Büdingen HC, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, Cox DR, Rajpal A, Pons J. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, Cox D, Rajpal A, Pons J. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baranzini SE, Jeong MC, Butunoi C, Murray RS, Bernard CC, Oksenberg JR. B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. J Immunol. 1999;163:5133–5144. [PubMed] [Google Scholar]
- 11.Colombo M, Dono M, Gazzola P, Roncella S, Valetto A, Chiorazzi N, Mancardi GL, Ferrarini M. Accumulation of clonally related B lymphocytes in the cerebrospinal fluid of multiple sclerosis patients. J Immunol. 2000;164:2782–2789. doi: 10.4049/jimmunol.164.5.2782. [DOI] [PubMed] [Google Scholar]
- 12.Owens GP, Bennett JL, Lassmann H, O'Connor KC, Ritchie AM, Shearer A, Lam C, Yu X, Birlea M, Dupree C, Williamson RA, Hafler DA, Burgoon MP, Gilden D. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid. Annals of neurology. 2009;65:639–649. doi: 10.1002/ana.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Büdingen HC, Harrer MD, Kuenzle S, Meier M, Goebels N. Clonally expanded plasma cells in the cerebrospinal fluid of MS patients produce myelin-specific antibodies. Eur J Immunol. 2008;38:2014–2023. doi: 10.1002/eji.200737784. [DOI] [PubMed] [Google Scholar]
- 14.Winges KM, Gilden DH, Bennett JL, Yu X, Ritchie AM, Owens GP. Analysis of multiple sclerosis cerebrospinal fluid reveals a continuum of clonally related antibody-secreting cells that are predominantly plasma blasts. J Neuroimmunol. 2007;192:226–234. doi: 10.1016/j.jneuroim.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Prabakaran P, Streaker E, Chen W, Dimitrov DS. 454 antibody sequencing - error characterization and correction. BMC research notes. 2011;4:404. doi: 10.1186/1756-0500-4-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, O'Dell S, Ofek G, Pancera M, Wu X, Zhang B, Zhang Z, Program NCS, Mullikin JC, Simek M, Burton DR, Koff WC, Shapiro L, Mascola JR, Kwong PD. Somatic Populations of PGT135-137 HIV-1-Neutralizing Antibodies Identified by 454 Pyrosequencing and Bioinformatics. Frontiers in microbiology. 2012;3:315. doi: 10.3389/fmicb.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Fire AZ. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barak M, Zuckerman NS, Edelman H, Unger R, Mehr R. IgTree: creating Immunoglobulin variable region gene lineage trees. J Immunol Methods. 2008;338:67–74. doi: 10.1016/j.jim.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Michaeli M, Barak M, Hazanov L, Noga H, Mehr R. Automated analysis of immunoglobulin genes from high-throughput sequencing: life without a template. Journal of clinical bioinformatics. 2013;3:15. doi: 10.1186/2043-9113-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens GP, Kraus H, Burgoon MP, Smith-Jensen T, Devlin ME, Gilden DH. Restricted use of VH4 germline segments in an acute multiple sclerosis brain. Annals of neurology. 1998;43:236–243. doi: 10.1002/ana.410430214. [DOI] [PubMed] [Google Scholar]
- 22.Owens GP, Winges KM, Ritchie AM, Edwards S, Burgoon MP, Lehnhoff L, Nielsen K, Corboy J, Gilden DH, Bennett JL. VH4 gene segments dominate the intrathecal humoral immune response in multiple sclerosis. J Immunol. 2007;179:6343–6351. doi: 10.4049/jimmunol.179.9.6343. [DOI] [PubMed] [Google Scholar]
- 23.Markovic M, Trajkovic V, Drulovic J, Mesaros S, Stojsavljevic N, Dujmovic I, Mostarica Stojkovic M. Antibodies against myelin oligodendrocyte glycoprotein in the cerebrospinal fluid of multiple sclerosis patients. Journal of the neurological sciences. 2003;211:67–73. doi: 10.1016/s0022-510x(03)00066-2. [DOI] [PubMed] [Google Scholar]
- 24.Xiao BG, Linington C, Link H. Antibodies to myelin-oligodendrocyte glycoprotein in cerebrospinal fluid from patients with multiple sclerosis and controls. J Neuroimmunol. 1991;31:91–96. doi: 10.1016/0165-5728(91)90014-x. [DOI] [PubMed] [Google Scholar]
- 25.Cepok S, Zhou D, Srivastava R, Nessler S, Stei S, Bussow K, Sommer N, Hemmer B. Identification of Epstein-Barr virus proteins as putative targets of the immune response in multiple sclerosis. The Journal of clinical investigation. 2005;115:1352–1360. doi: 10.1172/JCI23661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, Weill D, Bernstein D, Valantine HA, Quake SR. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bar-Or A, Oliveira EM, Anderson DE, Krieger JI, Duddy M, O'Connor KC, Hafler DA. Immunological memory: contribution of memory B cells expressing costimulatory molecules in the resting state. J Immunol. 2001;167:5669–5677. doi: 10.4049/jimmunol.167.10.5669. [DOI] [PubMed] [Google Scholar]
- 29.Duddy M, Niino M, Adatia F, Hebert S, Freedman M, Atkins H, Kim HJ, Bar-Or A. Distinct effector cytokine profiles of memory and naive human B cell subsets and implication in multiple sclerosis. J Immunol. 2007;178:6092–6099. doi: 10.4049/jimmunol.178.10.6092. [DOI] [PubMed] [Google Scholar]
- 30.Harp CT, Ireland S, Davis LS, Remington G, Cassidy B, Cravens PD, Stuve O, Lovett-Racke AE, Eagar TN, Greenberg BM, Racke MK, Cowell LG, Karandikar NJ, Frohman EM, Monson NL. Memory B cells from a subset of treatment-naive relapsing-remitting multiple sclerosis patients elicit CD4(+) T-cell proliferation and IFN-gamma production in response to myelin basic protein and myelin oligodendrocyte glycoprotein. European journal of immunology. 2010;40:2942–2956. doi: 10.1002/eji.201040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cross AH, Stark JL, Lauber J, Ramsbottom MJ, Lyons JA. Rituximab reduces B cells and T cells in cerebrospinal fluid of multiple sclerosis patients. J Neuroimmunol. 2006;180:63–70. doi: 10.1016/j.jneuroim.2006.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stuve O, Marra CM, Jerome KR, Cook L, Cravens PD, Cepok S, Frohman EM, Phillips JT, Arendt G, Hemmer B, Monson NL, Racke MK. Immune surveillance in multiple sclerosis patients treated with natalizumab. Annals of neurology. 2006;59:743–747. doi: 10.1002/ana.20858. [DOI] [PubMed] [Google Scholar]
- 33.Harrer A, Tumani H, Niendorf S, Lauda F, Geis C, Weishaupt A, Kleinschnitz C, Rauer S, Kuhle J, Stangel M, Weber F, Uhr M, Linnebank M, Wildemann B, Jarius S, Guger M, Ayzenberg I, Chan A, Zettl U, Wiendl H, Pilz G, Hitzl W, Weber JR, Kraus J. Cerebrospinal fluid parameters of B cell-related activity in patients with active disease during natalizumab therapy. Multiple sclerosis (Houndmills, Basingstoke, England) 2013;19:1209–1212. doi: 10.1177/1352458512463483. [DOI] [PubMed] [Google Scholar]
- 34.Villar LM, Garcia-Sanchez MI, Costa-Frossard L, Espino M, Roldan E, Paramo D, Lucas M, Izquierdo G, Alvarez-Cermeno JC. Immunological markers of optimal response to natalizumab in multiple sclerosis. Archives of neurology. 2012;69:191–197. doi: 10.1001/archneurol.2011.971. [DOI] [PubMed] [Google Scholar]
- 35.Obermeier B, Mentele R, Malotka J, Kellermann J, Kumpfel T, Wekerle H, Lottspeich F, Hohlfeld R, Dornmair K. Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat Med. 2008;14:688–693. doi: 10.1038/nm1714. [DOI] [PubMed] [Google Scholar]
- 36.von Büdingen HC, Gulati M, Kuenzle S, Fischer K, Rupprecht TA, Goebels N. Clonally expanded plasma cells in the cerebrospinal fluid of patients with central nervous system autoimmune demyelination produce “oligoclonal bands”. J Neuroimmunol. 2010;218:134–139. doi: 10.1016/j.jneuroim.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Bankoti J, Apeltsin L, Hauser SL, Allen S, Albertolle ME, Witkowska HE, von Budingen HC. In multiple sclerosis, oligoclonal bands connect to peripheral B-cell responses. Annals of neurology. 2014;75:266–276. doi: 10.1002/ana.24088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magliozzi R, Columba-Cabezas S, Serafini B, Aloisi F. Intracerebral expression of CXCL13 and BAFF is accompanied by formation of lymphoid follicle-like structures in the meninges of mice with relapsing experimental autoimmune encephalomyelitis. Journal of neuroimmunology. 2004;148:11–23. doi: 10.1016/j.jneuroim.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 39.Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Aloisi F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14:164–174. doi: 10.1111/j.1750-3639.2004.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas J, Bekeredjian-Ding I, Milkova M, Balint B, Schwarz A, Korporal M, Jarius S, Fritz B, Lorenz HM, Wildemann B. B cells undergo unique compartmentalized redistribution in multiple sclerosis. J Autoimmun. 2011;37:289–299. doi: 10.1016/j.jaut.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Corcione A, Casazza S, Ferretti E, Giunti D, Zappia E, Pistorio A, Gambini C, Mancardi GL, Uccelli A, Pistoia V. Recapitulation of B cell differentiation in the central nervous system of patients with multiple sclerosis. Proc Natl Acad Sci U S A. 2004;101:11064–11069. doi: 10.1073/pnas.0402455101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, Lee EH, Milner EC, Sanz I. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J Immunol. 2007;178:6624–6633. doi: 10.4049/jimmunol.178.10.6624. [DOI] [PubMed] [Google Scholar]
- 43.Fecteau JF, Cote G, Neron S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J Immunol. 2006;177:3728–3736. doi: 10.4049/jimmunol.177.6.3728. [DOI] [PubMed] [Google Scholar]
- 44.Wu YC, Kipling D, Dunn-Walters DK. The relationship between CD27 negative and positive B cell populations in human peripheral blood. Frontiers in immunology. 2011;2:81. doi: 10.3389/fimmu.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- 46.Morrissy S, Zhao Y, Delaney A, Asano J, Dhalla N, Li I, McDonald H, Pandoh P, Prabhu AL, Tam A, Hirst M, Marra M. Digital gene expression by tag sequencing on the illumina genome analyzer. Current protocols in human genetics / editorial board, Jonathan L. Haines ... [et al.] 2010 doi: 10.1002/0471142905.hg1111s65. Chapter 11, Unit 11 11 11-36. [DOI] [PubMed] [Google Scholar]
- 47.Shiroguchi K, Jia TZ, Sims PA, Xie XS. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci U S A. 2012;109:1347–1352. doi: 10.1073/pnas.1118018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eastburn DJ, Sciambi A, Abate AR. Ultrahigh-throughput Mammalian single-cell reverse-transcriptase polymerase chain reaction in microfluidic drops. Analytical chemistry. 2013;85:8016–8021. doi: 10.1021/ac402057q. [DOI] [PubMed] [Google Scholar]
- 49.van Zelm MC, Szczepanski T, van der Burg M, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J Exp Med. 2007;204:645–655. doi: 10.1084/jem.20060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, Lublin FD, Montalban X, O'Connor P, Sandberg-Wollheim M, Thompson AJ, Waubant E, Weinshenker B, Wolinsky JS. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Annals of neurology. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 54.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.