Abstract
Background
Genome-wide association studies (GWAS) have identified thousands of genetic variants that influence a variety of diseases and health-related quantitative traits. However, the causal variants underlying the majority of genetic associations remain unknown. The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Targeted Sequencing Study aims to follow up GWAS signals and identify novel associations of the allelic spectrum of identified variants with cardiovascular related traits.
Methods and Results
The study included 4,231 participants from three CHARGE cohorts: the Atherosclerosis Risk in Communities Study, the Cardiovascular Health Study, and the Framingham Heart Study. We used a case-cohort design in which we selected both a random sample of participants and participants with extreme phenotypes for each of 14 traits. We sequenced and analyzed 77 genomic loci, which had previously been associated with one or more of 14 phenotypes. A total of 52,736 variants were characterized by sequencing and passed our stringent quality control criteria. For common variants (minor allele frequency ≥1%), we performed unweighted regression analyses to obtain p-values for associations and weighted regression analyses to obtain effect estimates that accounted for the sampling design. For rare variants, we applied two approaches: collapsed aggregate statistics and joint analysis of variants using the Sequence Kernel Association Test.
Conclusions
We sequenced 77 genomic loci in participants from three cohorts. We established a set of filters to identify high-quality variants, and implemented statistical and bioinformatics strategies to analyze the sequence data, and identify potentially functional variants within GWAS loci.
Keywords: genetics, epidemiology, CHARGE, sampling, targeted sequencing
In the past few years, genome-wide association studies (GWAS) have successfully identified associations of common genetic variations with a variety of diseases and health-related quantitative traits.1 However, in most cases neither the gene underlying disease susceptibility nor the spectrum of candidate functional variants has been identified. Within a genomic locus identified by GWAS, detailed examination of all genetic variants is required to discover causal variant(s), to estimate their impact on disease susceptibility, and to identify their functional roles. The large number of low-frequency and rare variants that exist within any given GWAS locus vastly outnumber common variants and may contribute significantly to the genetic architecture of disease.2 With the advent of genome sequencing using next-generation technologies, targeted sequencing can be conducted at high-throughput to identify lower frequency variants within regions identified by GWAS associations. Targeted sequencing of protein-coding genes identified by GWAS has been demonstrated to identify a large excess burden of rare functional alleles in persons at extreme ends of quantitative traits such as level of circulating triglycerides.3 However, many GWAS signals have been located in introns or flanking regions of protein coding genes and are poorly correlated with functional variants in protein-coding genes, and at least 40% of GWAS signals are located in genomic regions uncorrelated with known missense variants,4 suggesting that most GWAS signals are regulatory in nature.2 Targeted sequencing of implicated genomic regions beyond exons may identify functional alleles involved in gene regulation. One emerging feature of GWAS is the existence of multiple apparently pleiotropic regions that underlie a number of different disease phenotypes, and targeted sequencing may aid in defining the genetic architecture of such regions.
The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium5 is a collaborative program of prospective population-based cohorts to leverage existing clinical, laboratory, and computational resources to identify susceptibility genes using genome-wide approaches such as GWAS for subclinical quantitative measures and clinical manifestations of cardiovascular, lung, and blood diseases and their risk factors. CHARGE cohorts have led or contributed to GWAS that have uncovered hundreds of loci for many dozens of heritable phenotypes. Clinical disease phenotypes studied by GWAS include atrial fibrillation, stroke and chronic obstructive pulmonary disease. Quantitative measures of subclinical cardiovascular measures that have been the focus of GWAS include electrocardiographic intervals, echocardiographic left ventricular internal diameter, and ultrasonographic carotic artery intimal medial thickness, and quantitative measures of CVD risk factors such as systolic blood pressure, body mass index and fasting insulin. Common pleiotropic regions appear to underlie genetic variation contributing to several of these measures, for example SNPs in 12q24.12 were associated with coronary heart disease, hypertension, anemia and retinal vein caliber.6
The CHARGE Targeted Sequencing Study aims to follow up GWAS signals to comprehensively localize the functional variants, to evaluate the contribution of rare variants to a wide array of cardiovascular related traits. A total of 77 genomic loci previously implicated by GWAS were selected and sequenced in participants from three CHARGE cohorts: the Atherosclerosis Risk in Communities Study (ARIC),7 the Cardiovascular Health Study (CHS),8 and the Framingham Heart Study (FHS).9 Here we summarize the study design and the bioinformatic and statistical analysis strategies used in the CHARGE Targeted Sequencing Study.
Methods
Study Design
The CHARGE Targeted Sequencing Study used a case-cohort study design in which a random sample was selected from all three cohorts at baseline. We planned for the Cohort Random Sample to include approximately 2,000 individuals, 1,000 participants from ARIC, 500 participants from CHS, and 500 participants from FHS, with proportions from each study reflecting relative cohort sizes with equal numbers of men and women. In addition to the Cohort Random Sample, approximately 200 participants (generally 100 from ARIC, 50 from CHS, and 50 from FHS) from each of 14 key phenotypes were selected for sequencing on the basis of either case status for discrete phenotypes or extreme values of quantitative traits. The phenotypes studied (Table 1) were atrial fibrillation, blood pressure, body mass index, bone mineral density, C-reactive protein, carotid intima-media thickness, echocardiography, electrocardiographic PR and QRS interval, fasting insulin, hematocrit, pulmonary function, retinal venule diameter, and stroke. Because individuals initially selected for the Cohort Random Sample or some Phenotype Groups could satisfy the criteria for one or more Phenotype Group’s extreme sampling, the achieved number with extreme values for each phenotype was often larger than the target number of 200. Detailed information regarding the criteria for the selection of study participants for each phenotype is provided in the Supplemental Materials.
Table 1.
Phenotype Groups and Sample Selection Strategies
| Phenotype Group | Strategy | Targeted Number of Extreme Participants* | ||
|---|---|---|---|---|
| FHS | CHS | ARIC | ||
| EKG PR interval | High residual† | 50 | 50 | 100 |
| EKG QRS interval | High | 50 | 50 | 100 |
| Stroke | Ischemic stroke | 50 | 70 | 80 |
| Blood Pressure | Low residual† | 25 | 25 | 54 |
| High residual† | 25 | 25 | 46 | |
| Body Mass Index | High residual† | 50 | 50 | 100 |
| Fasting Insulin | High | 50 | 50 | 100 |
| Bone mineral density by DEXA | Low z-score | 100 | 100 | - |
| Left ventricular diastolic diameter | High residual† | 100 | 100 | - |
| C-reactive protein | High residual† | 50 | 50 | 100 |
| Hematocrit | Low residual† | 50 | 50 | 100 |
| Retinal venule diameter | High residual† | - | 34 | 166 |
| Carotid wall thickness | High | 50 | 50 | 100 |
| Pulmonary: FEV1/FVC | Low | - | - | 200 |
| Atrial Fibrillation | Lone atrial | - | - | - |
| fibrillation‡ | ||||
These numbers represent the number of participants with extreme phenotypes targeted for selection by each Phenotype Group, but do not reflect the additional participants who may have met the criteria for an extreme phenotype but had already been selected by other Phenotype Groups or as part of the Cohort Random Sample
Extreme samples were selected by taking either the extreme high or low distribution based on age, sex, and phenotype specific variable adjusted residuals
200 cases with lone atrial fibrillation were selected from Massachusetts General Hospital
Participants in the CHARGE Targeted Sequencing Study had sufficient DNA for sequencing, self-reported ethnicity as non-Hispanic white, and availability of prior genotyping results. In addition, participants from ARIC and CHS had no evidence for relatedness to other individuals within the study. However, FHS participants in one Phenotype Group could be related to participants in another Phenotype Group, and could be related to members of the Cohort Random Sample. Institutional Review Boards at participating centers approved the study, and participants gave informed consent. The detailed description of each cohort is available in the Supplemental Materials.
Target selection
The 77 targeted regions selected for sequencing encompassed approximately 2 megabases (Mb) of the genome. Thirty-three of these regions had been shown to be associated with one of the investigated phenotypes by previous GWAS (Table 2a). The remaining 44 targeted regions had been shown to exhibit pleiotropy. For this work, we defined evidence of pleiotropy as a region or locus containing one to many genes having displayed strong associations (p < 5 × 10−8) with 2 or more traits in multiple genome-wide association studies (Table 2b).
Table 2.
| a: Primary Targets for the Phenotype Groups* | |||||||
|---|---|---|---|---|---|---|---|
| Phenotype Group | Target Name | Chr | Start Position† | Stop Position† | #SNPs | #Coding | #Novel‡ |
| Body Mass Index | TMEM18 | 2 | 586,432 | 677,539 | 2,180 | 66 | 1,629 |
| Atrial Fibrillation | PRRX1 | 1 | 168,853,417 | 168,975,265 | 198 | 105 | 165 |
| Atrial Fibrillation | CAV2 | 7 | 115,925,580 | 115,935,931 | 97 | 68 | 71 |
| Atrial Fibrillation | CAV1 | 7 | 115,950,975 | 115,988,574 | 135 | 103 | 98 |
| Atrial Fibrillation | ZFHX3 | 16 | 71,378,464 | 71,651,135 | 450 | 381 | 372 |
| Blood pressure | CACNB2 | 10 | 18,468,647 | 18,871,794 | 862 | 154 | 648 |
| Blood pressure | CYPI7AI | 10 | 104,579,177 | 104,619,322 | 660 | 52 | 532 |
| Blood pressure | PLEKHA7 | 11 | 16,764,687 | 16,993,639 | 797 | 145 | 613 |
| Blood pressure | ATP2B1 | 12 | 88,504,856 | 88,616,005 | 1,906 | 112 | 1,580 |
| C-reactive protein | LEPR | 1 | 65,830,704 | 65,880,672 | 1,069 | 125 | 829 |
| C-reactive protein | IL6R | 1 | 152,641,684 | 152,709,758 | 1,295 | 170 | 1,086 |
| C-reactive protein | GCKR | 2 | 27,567,127 | 27,603,561 | 674 | 129 | 557 |
| C-reactive protein | HNF1A | 12 | 119,899,944 | 119,925,301 | 338 | 45 | 273 |
| Fasting Insulin | 2q36.3 | 2 | 226,728,697 | 226,856,269 | 2,621 | 0 | 2,024 |
| Fasting Insulin | IRS1 | 2 | 227,236,803 | 227,388,200 | 1,913 | 151 | 1,522 |
| Fasting Insulin | IGF1 | 12 | 101,312,706 | 101,455,233 | 1,393 | 192 | 1,124 |
| Left ventricular diastolic diameter | PLN | 6 | 118,970,965 | 118,989,480 | 387 | 34 | 319 |
| EKG QRS interval | EXOG | 3 | 38,511,667 | 38,552,500 | 393 | 76 | 306 |
| EKG QRS interval | SCN10A | 3 | 38,694,789 | 38,811,605 | 3,209 | 204 | 2,434 |
| Retinal venule diameter | MEF2C | 5 | 87,819,438 | 88,215,292 | 802 | 17 | 666 |
| Hematocrit | TFR2 | 7 | 100,054,874 | 100,079,499 | 331 | 74 | 262 |
| Hematocrit | HK1 | 10 | 70,698,661 | 70,832,743 | 612 | 125 | 458 |
| Hematocrit | SH2B3 | 12 | 110,322,484 | 110,374,300 | 279 | 113 | 229 |
| Pulmonary: FEV1/FVC | HTR4 | 5 | 147,815,702 | 147,837,626 | 531 | 6 | 407 |
| Pulmonary: FEV1/FVC | ADAM19 | 5 | 156,830,995 | 156,936,446 | 2,630 | 207 | 2,046 |
| Bone mineral density by DEXA | WLS | 1 | 68,336,635 | 68,480,941 | 3,494 | 52 | 2,662 |
| Bone mineral density by DEXA | MEF2C | 5 | 88,335,639 | 88,457,219 | 2,375 | 0 | 1,969 |
| Bone mineral density by DEXA | JAG1 | 20 | 10,565,132 | 10,609,308 | 1,205 | 151 | 967 |
| Stroke | NINJ2 | 12 | 543,643 | 740,130 | 4,001 | 43 | 3,077 |
| EKG PR interval | SCN5A | 3 | 38,564,457 | 38,667,267 | 325 | 222 | 263 |
| EKG PR interval | SOX5 | 12 | 23,576,398 | 24,607,747 | 268 | 93 | 205 |
| EKG PR interval | C12ORF67 | 12 | 24,611,065 | 24,629,469 | 72 | 0 | 56 |
| Carotid wall thickness | SLC17A4 | 6 | 25,857,845 | 25,987,550 | 3,001 | 201 | 2,236 |
| b: Targets for Pleiotropic Loci* | ||||||
|---|---|---|---|---|---|---|
| Locus | Genes (5’->3’)† | Start Position‡ | Stop Position‡ | #SNPs | #Coding | #Novel§ |
| 6q23.3 | HBS1L, MYB | 135,322,113 | 135,582,124 | 1,894 | 282 | 1,486 |
| 7q22.3 | PIK3CG | 105,917,292 | 106,379,327 | 766 | 159 | 539 |
| 7q36.1 | PRKAG2 | 150,856,537 | 151,267,715 | 1,022 | 83 | 664 |
| 8p21.1 | SCARA5 | 27,783,553 | 27,906,232 | 631 | 108 | 436 |
| 11p11.2 | DGKZ, AMBRA1, ATG13, ARHGAP1, ZNF408, F2, CKAP5, LRP4, C11ORF49, DDB, ACP2, NR1H3, MADD, MYBPC3, SPI1, SLC39A13, PSMC3, RAPSN, CELF1, NDUFS3, MTCH2, AGBL2, FNBP4, NUP160 | 46,308,577 | 47,851,121 | 3,059 | 583 | 2,309 |
| 12q24.12–13 | ATXN2, BRAP, ACAD10, ALDH2, MAPKAPK5, ADAM1, TMEM116, NAA25, TRAFD1, C12ORF51, RPL6, PTPN11 | 110,374,301 | 111,436,622 | 3,818 | 989 | 3,130 |
| 13q34 | COL4A1, COL4A2 | 109,599,195 | 109,967,539 | 1,043 | 330 | 712 |
References for each target are provided in Supplemental Table 2a. Phenotype analyses might evaluate additional targets beyond their primary targets.
The chromosomal positions were based on the reference human genome (NCBI Build 36, 2006)
Novel was defined as not being found in 1000G Phase1
References for each locus are provided in Supplemental Table 2b. Phenotype analyses might evaluate additional targets beyond their primary targets.
The loci contain both gene and intergenic regions selected due to GWAS and eQTL associations, conserved regions and SNPs in LD (selection details in Supplemental Material).
The chromosomal positions were based on the reference human genome (NCBI Build 36, 2006).
Novel was defined as not being found in 1000G Phase1.
Library Preparation, Sequencing and Variant Calling
Detailed description of library preparation and sequencing could be found in the Supplemental Materials. In brief, the targeted regions were captured by a specific SOLiD™ platform-based multiplexed capture sequencing protocol developed at the Baylor College of Medicine Human Genome Sequencing Center (HGSC). The enriched libraries were then pooled to form an 8-sample pool for multiplexed sequencing. Each sequencing pool was subsequently sequenced on one quadrant of a SOLiD™ V4 slide using Life Technologies’ Barcode Fragment Sequencing Kits and methods.
The raw short reads were then aligned to the reference human genome (NCBI Genome Build 36, hg18) using BFAST,10 producing BAM files containing various mapping information. For samples requiring multiple sequencing ‘events’, multiple BAM files were merged to generate a single BAM file per sample. The current project was focused on single nucleotide polymorphisms (SNPs), whereas neither small indels nor large copy number variations were investigated. We applied SAMtools11 to each sample-level BAM and generated pileup format files containing a base-by-base summary of the reads overlapping each variant site and a variant call. This list of putative SNPs was post-processed to filter variants with apparent strand-bias, low allele fraction, low coverage, or low quality to produce a high-quality variant list.
Quality Control (QC)
Because data from sequencing experiments can have errors at multiple levels, such as variant calls and read mapping, we implemented a multilevel approach to identify sites with true variation for use in downstream association analyses. All QC procedures were carried out in the statistical platform R or Java, in combination with SAMtools.11
Preliminary QC Procedures in Sequencing Laboratory
The first level of quality control took place through laboratory procedures. After sequencing a sample to the target depth, we evaluated several QC metrics including alignment rate and uniqueness to validate that the sequencing performed as expected. Base and quality calling for the SOLiD data was performed on-instrument using standard vendor software and settings. To gauge the overall performance of the capture process, sample-level BAMs were also subjected to a capture analysis QC pipeline to obtain additional metrics such as the proportion of the aligned reads that mapped to the targeted region and the proportion of targeted bases at various coverage levels. Samples that met a minimum of 65% of the targeted bases at 20× or greater coverage were submitted for subsequent analysis and QC.
For each successfully sequenced sample, we confirmed sample identity and checked purity using the ERIS tool suite (https://github.com/dsexton2/ERIS) to compare sequence data to genotypes from available GWA SNP arrays. Using an ‘e-GenoTyping’ approach, we screened all sequence reads for exact matches to ‘probe’ sequences defined by the variant and position of interest, along with 11 bases of sequence flanking either side of the SNP site. In this process, we removed SNP array sites that were non-specific and over- or under-covered before comparing the read data to the variants for all samples in the project. Based on our previous empirical experience, we used thresholds of 90% self-concordance and next-best matches below 75% to identify samples that demonstrated minimal contamination and confirmed sample identity. We informatically unswapped any samples with clear evidence of mislabeling by attaching the appropriate sample names. Any samples that appeared to be either cryptically swapped or significantly contaminated were resequenced and rescreened for inclusion in the study.
Variant-level QC
Each cohort individually implemented an extensive QC pipeline for all of their own samples that passed the laboratory QC procedures. Our QC pipeline consisted of a series of variant-level filtering steps followed by QC on individual samples (summarized in Table 3). Before applying these steps, we first pre-filtered the raw data to remove any variants that mapped more than 100 base pairs from the requested target capture region. We further removed potentially low quality reads by filtering variants with a Phred-scaled base quality score12 (−10 log10 p, where p is the probability of calling error) less than 30, with less than two reads of the alternate alleles, and variants with a depth of coverage of less than 10 total reads.
Table 3.
SNP Quality Filters
| Stage | SNPs should satisfy all the following criteria | |
|---|---|---|
| Pre-Filter | Off-target distance | ≤100 bp |
| Phred score | ≥30 | |
| Depth of coverage | ≥10 | |
| Depth of alternate allele coverage | ≥2 | |
| Sample SNP | Allelic imbalance | Between 0.2 and 0.8 |
| Strand bias | At least one read on each strand | |
| Whole SNP | % Missing | < 20% |
| HWE exact test* | P≥ 1×10−5 | |
| # Alleles | ≤2 | |
| SNP cluster | ≤ 2 SNPs in 10bp | |
HWE: Hardy-Weinberg Equilibrium, calculated based on Cohort Random Sample only
At the “sample-SNP” filtering stage, we assessed each variant within each sample in terms of allelic imbalance and strand bias. Heterozygote genotypes were removed if their alternate to reference allele ratio was disproportionate, defined to be smaller than 0.2 or larger than 0.8 for one allele. We did not take into account of copy number variations (Supplemental Materials). For strand bias, we kept only variants with alternate allele reads obtained from both the positive and negative strands.
Finally, each variant was evaluated across all samples. We removed SNPs that had greater than 20% missingness, had more than 2 observed alleles, or were part of an overly dense SNP cluster (3 or more variants in a 10 bp window) since too many variants within a short genomic interval can indicate regional sequencing errors. Then, using only samples from the Cohort Random Sample, we filtered SNPs that deviated from the expectations of Hardy-Weinberg equilibrium (HWE, P<1×10−5) to identify excess heterozygosity that may have been induced by mismapped reads.
Sample-level QC
After variant-level QC was completed, each cohort conducted a quality assessment of the final sequence data based on a number of measures. Within each cohort, a sample was flagged as potentially poor quality if it fell beyond the lower or upper 2.5th percentile of any of 8 selected measures: mean mapping quality score across all variants; mean fold coverage; mean transition to transversion (Ti/Tv) ratios; mean heterozygote to homozygote ratio; mean nonsynonymous to synonymous ratio; number of singletons; number of doubletons; and percentage of sites with coverage greater than 20×. However, none of samples showed systematically low quality. We therefore kept all the sequenced samples, but recorded these quality metrics in a joint sample information file. Phenotype groups, however, could further examine these samples, and decide whether to remove some of them in their respective association analyses.
SNP Information and Functional Annotation
A SNP information file combining information across the three cohorts and all sequence data was produced after QC, including summaries and functional annotations for the SNPs. The summaries included the SNP position, reference and alternative alleles, sample size, genotype counts, allele counts, allele frequencies, average mapping quality, average SNP calling quality, lower 2.5 and upper 97.5 percentiles of read depths, genotype missing rate, and minimum p-value of the Hardy-Weinberg equilibrium test within the Cohort Random Sample. Functional annotations were produced using a combination of ANNOVAR,13 dbNSFP14 and custom internal tools. SNP positions referring to the RefSeq15 gene definition were annotated with ANNOVAR. Functional predictions for nonsynonymous mutations, including LRT,16 SIFT,17 PolyPhen-2,18 and MutationTaster,19 were annotated with dbNSFP. Other essential functional annotations included conservation scores, such as GERP++,20 allele frequencies observed in the 1000 Genomes Project,21 and various regulatory region annotations from the ENCODE Project,22 the ORegAnno database23 and the TRANSFAC database24 accessed through the UCSC Genome Browser.25 We recommended that phenotype working groups take into account various types of supporting evidence in the interpretation of association results.
Statistical Analysis
Common Variants
The CHARGE Analysis and Bioinformatics Committee recommended performing single marker analyses for each common variant within a target. Although individual Phenotype Groups implemented this threshold differently, common variants were loosely defined as those with allele frequency of ≥1%, which corresponded to variants where there were at least 50 individuals with one or two minor alleles across the entire study.
We performed two regression analyses – an unweighted analysis to obtain p-values for association and a weighted analysis to obtain effect estimates and estimated standard errors. The weighted analysis accounted for the sampling design by assigning different weights to extreme samples and to individuals from the Cohort Random Sample. Extreme samples were weighted by 1, whereas individuals of Cohort Random Sample were weighted by the inverse of their probability of inclusion in each cohort. More details of the sampling weight is described at Lumley T, Dupuis J, Rice KM, Barbalic M, Bis JC, Cupples LA, et al. http://stattech.wordpress.fos.auckland.ac.nz/files/2012/05/design-paper.pdf. For both analyses, we used data from all subjects. To produce p-values for association between each variant and the phenotype of interest, we used standard regression methods: linear regression or linear mixed effects models (FHS) for continuous phenotypes, logistic regression or generalized estimating equation models (FHS) for dichotomous outcomes, and Cox proportional hazards regression with robust variance or Cox proportional hazards regression (with clustering on pedigrees with robust variance in FHS)26,27 for survival outcomes. The different models used in FHS aimed to address relatedness in FHS subjects. Because these analyses were intended to follow up on GWAS loci, working groups typically used the same phenotype definition, adjustment variables, and additive genetic models (0/1/2 copies) as in the discovery GWAS analyses.
Results from each study (estimated regression coefficient (beta-hat) and estimated standard error) were then shared and combined, applying inverse-variance weighted fixed effects meta-analysis. P-values from this meta-analysis were reported. Because of our sampling scheme, we reported the corresponding meta-analytic estimate of effect (beta-hat) from the weighted analysis and p-values from the unweighted analysis. Each working group made their own decisions towards control of type I error. Some groups used an alpha cutoff according to their priori hypotheses and others used more than one cutoff, depending on the focus of their investigation. All the analyses were performed using R software (www.r-project.org/).
Rare Variants
Single-marker based association analysis generally has low power for rare variants. Therefore, a number of methods for rare variant tests have recently been developed. Basu and Pan28 performed an extensive comparison of many of the currently-available methods under different circumstances. For the CHARGE Targeted Sequencing Study, we recommended that working groups use analyses that either collapse variants in each genomic region using a burden test or jointly analyze associations with variants in each genomic region using the Sequence Kernel Association Test (SKAT).
Collapsing Tests
The primary recommendation for analyses that collapse variants in a genomic region into a single summary measure was to use the T1 count, defined as the number of variants with at least 1 rare allele among variants in the region with a study-wide MAF < 1%. A secondary recommendation was a Madsen-Browning type test, which aggregates all variants with MAF< 1% in a genomic region, weighting each variant by a function of its MAF.29 Although all variants in a region can be considered in the Madsen-Browning test statistic, we recommended restricting to rare variants with MAF < 1%. For these methods that collapse variants, the same regression analyses described above for common variants were used, with the aggregate collapsed regional burden replacing the usual genotype dosage.
Joint Analysis of Variants
The recommendation for jointly analyzing variants in a genomic region was a specific version of a general score test available as the Sequence Kernel Association Test (SKAT).30 The SKAT score test can be written as a weighted sum of squares of z-statistics from score tests in single-variant regression models. These single-variant tests were computed in each study and meta-analyzed using standard methods to give the SKAT statistic using weights based on combined allele frequencies across all studies. The reference distribution for the SKAT test requires the covariance matrix of the genetic variants, which was computed as a simple weighted average of the covariance matrices in the three cohorts. Each study implemented the SKAT analyses using custom R scripts that included a SKAT extension to account for familial relatedness.31 The scripts are provided in the CHARGE wiki website (http://depts.washington.edu/chargeco/wiki/CHARGE-S). Simulations confirmed that this approach agrees closely with the SKAT test performed on individual data, and that the power is higher than when the meta-analysis is performed on p-values (Lumley T, Brody J, Dupuis J, Cupples LA http://stattech.wordpress.fos.auckland.ac.nz/files/2012/11/skat-meta-paper.pdf).
Results
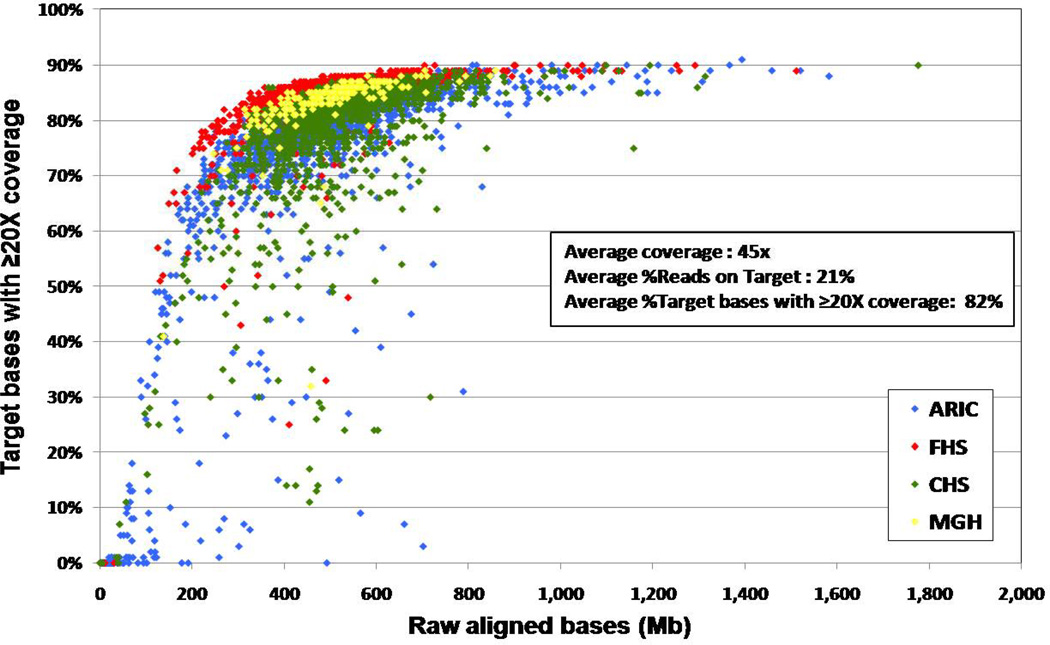
A total of 4,646 samples were target captured and sequenced for the project. After applying initial sequencing QC for sample identity, contamination and target coverage described above, 4,440 samples qualified for additional analysis, providing a 95.5% capture sequencing and QC success rate. Data produced from all these samples is summarized in Figure 1. Individual samples from the three cohorts (ARIC, CHS and FHS) plus one additional sample set (200 lone atrial fibrillation cases from Massachusetts General Hospital) are shown with the percent coverage of the target bases at 20× coverage in relation to the actual Mbs generated. Approximately 70–80% of short reads were successfully aligned to the reference genome (hg18) across the three studied cohorts. We found that 40–45% of short reads were mapped to the target regions. After removal of duplicate and low-quality reads, approximately 21% of total aligned reads were kept for downstream analyses. On average 82% of the targeted bases were covered at ≥ 20×, and the average coverage for each sample was ~45×. Nearly all of the targeted probe sets were successfully captured, and 95–96% of the targeted bases had ≥1 read for coverage. The number of targeted bases with a given depth of coverage closely followed a Poisson distribution, indicating uniform capture and sequencing of the targeted regions. After removing duplicate samples, a total of 4,231 unique individuals from the three cohorts were used for downstream analysis, including 2,003 from ARIC, 1,132 from CHS, and 1,096 from FHS. The Cohort Random Sample included 1,917 individuals and the remaining 2,314 individuals were distributed across the fourteen Phenotype Groups. Demographic characteristics of the investigated participants are presented in Table 4.
Figure 1.
Distribution of Targeted Sequence Coverage in 4,646 samples using SOLiD multiplexed capture sequencing. Each dot represents one sample. The x-axis represents the total depth of each sample (in terms of raw aligned bases), whereas the y-axis represents the proportion of targeted regions with more than 20× coverage. ARIC = Atherosclerosis Risk in Communities, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, MGH = Massachusetts General Hospital.
Table 4.
Characteristics of Study Participants
| Sample Size, N | Women, N (%) | Age*, mean (SD) | ||
|---|---|---|---|---|
| ARIC | ||||
| All | 2,003 | 978 (48.8%) | 54.8 (5.69) | |
| Cohort Random Sample | 946 | 469 (49.6%) | 54.5 (5.64) | |
| Phenotype Groups | 1057 | 509 (48.2%) | 55.1 (5.72) | |
| CHS | ||||
| All | 1,132 | 607 (53.6%) | 72.5 (5.46) | |
| Cohort Random Sample | 471 | 240 (51.0%) | 72.5 (5.40) | |
| Phenotype Groups | 661 | 367 (55.5%) | 72.5 (5.50) | |
| FHS | ||||
| All | 1,096 | 564 (51.5%) | 61.2 (11.83) | |
| Cohort Random Sample | 501 | 249 (49.7%) | 62.0 (9.35) | |
| Phenotype Groups | 595 | 315 (52.9%) | 60.5 (13.55) | |
ARIC = Atherosclerosis Risk in Communities, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study
Age in FHS was assessed at the time when DNA was drawn. Phenotype Groups might use the age at other exams.
A total of 52,736 variants were identified that passed QC among the three cohorts. This number included 30,912 variants in ARIC, 21,150 in CHS, and 21,267 in FHS. Across all samples, the average Ti/Tv ratio after SNP filtering was 2.44, in accordance with what would be expected given that the CHARGE targeted sequencing regions were a mixture of exonic, intronic and intergenic regions. A cross-validation with previous genotype data showed a concordance rate of 98.0% (Supplemental Materials). The summary statistics of SNPs found in each individual are shown in Supplemental Table 1.
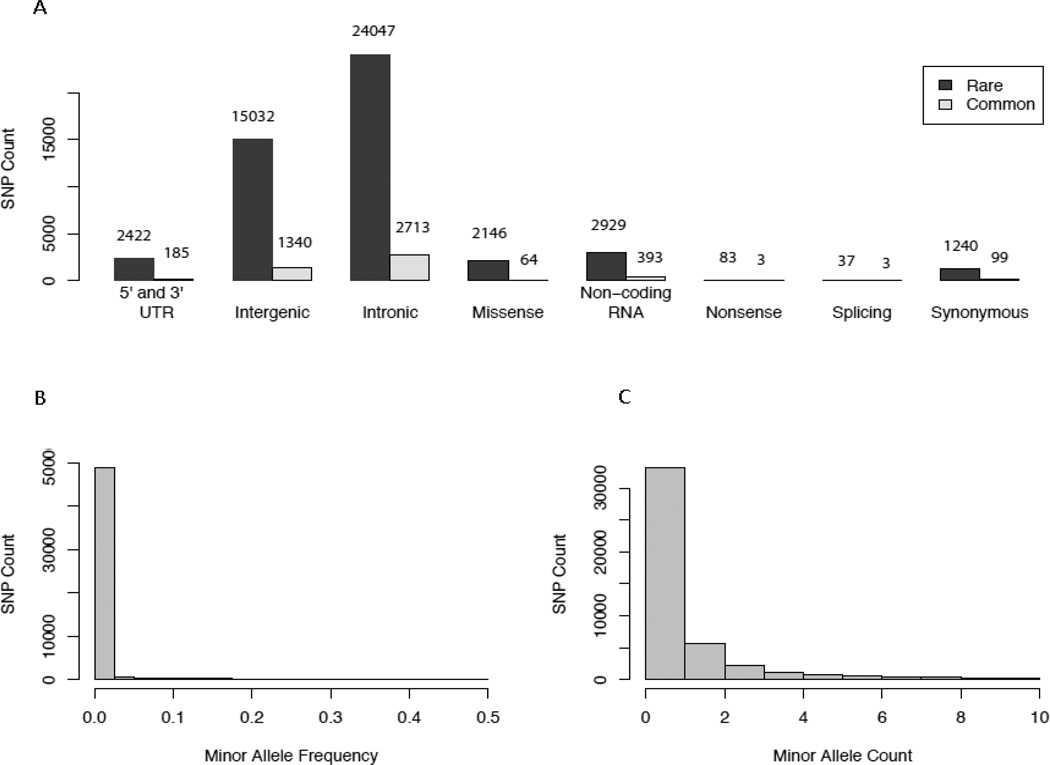
Figure 2 displays the distribution of functional classes and MAF combining filtered variants from all cohorts. The majority of variants were located within the intergenic (31.0%) or intronic regions (50.7%), and only 11.7% of variants were within known protein coding regions. A total of 4,800 (9.1%) were common variants (MAF ≥1%), and the remaining 47,936 were rare variants. Overall, most (93%) common variants were observed in multiple cohorts, while rare variants were more likely to be unique to a single cohort. Of the common variants, 98% have already been reported in phase 1 of the 1000 Genomes Project,21 whereas only 15% of rare variants have been reported. Among the 4800 common variants identified in this project, only 2501 (52.1%) of them were available in the HapMap CEU panel, which was used for genotype imputation and thus GWAS. In particular, we identified 70 damaging variants (missense, nonsense, or splicing variants), of which only half were available in the HapMap CEU panel. As an example, four gene regions were selected for sequencing because of prior associations with circulating C-reactive protein levels.32 We found 13 SNPs remained significant after adjusting for multiple testing, including one missense SNP rs2228145 within the IL6R locus (Supplemental Materials). The SNP was not studied in GWAS, but it was in linkage disequilibrium with the GWAS lead SNP (rs4129267) at the IL6R locus. Previous studies have found that rs2228145 was strongly associated with circulating concentrations of interleukin-6 soluble receptor,33,34 which is a pro-inflammatory cytokine regulating a variety of inflammatory responses.35,36 Our results suggest that rs2228145 might be the functional SNP explaining the association of the IL6R locus with C-reactive protein levels.
Figure 2.
Minor allele frequency distributions for variants passing QC (all three cohorts combined). (A) Distribution of functional classes in common/rare variants (B) Minor allele frequency spectrum (C) Frequencies of minor allele count <= 10
Discussion
The objective of the CHARGE Targeted Sequencing Study was to localize the GWA signals and to evaluate the contribution of rare variants to 14 phenotypes. We implemented a case-cohort study design, in which both a random sample of participants and participants with extreme trait values were selected from each of three participating cohorts. We also developed and implemented robust analysis strategies to analyze sequence data in relation to each individual phenotype. In addition, our sequencing project was able to accommodate different hypotheses proposed by Phenotype Groups regarding to the target selection. For some targets (e.g., ZFHX3 and SCN5A), only exonic regions were sequenced, and for some other targets (e.g., PLN and SCN10A), the entire gene region was sequenced. Some targeted regions were even outside of any known gene regions (e.g., 2q36.3 and MEF2C), demonstrating the flexibility of our target selection. The full data set has been registered with dbGaP and will be deposited soon.
Our study design provides a cost-effective way to evaluate genetic associations for multiple phenotypes. The same Cohort Random Sample was included in the analyses of all phenotypes, and thus sample sizes were larger than would be achieved with phenotype-specific analysis populations. In addition, analyses were typically conducted across all available samples from the Phenotype Groups. That is, extreme samples chosen by one phenotype working group were used by others, significantly increasing the overall sample size and allowing more rare variants to be observed in each analysis. Because the Phenotype Group sampling was based on trait values, we applied a weighting approach so that the distributions of all variables would be the same as in the full cohort.37 While testing can, in our circumstances, be performed without the sampling weights, they are needed for unbiased estimation of effects (Lumley T, Dupuis J, Rice KM, Barbalic M, Bis JC, Cupples LA, et al. http://stattech.wordpress.fos.auckland.ac.nz/files/2012/05/design-paper.pdf). Under plausible scenarios, for a single phenotype, our design’s use of the Cohort Random Sample is less powerful than sampling extreme values from both tails, but for studying multiple phenotypes the repeated use of the Cohort Random Sample provides greater power.38 An alternative sampling strategy that selected control subjects only from those participants without extreme values for any phenotype of interest, might offer larger power if a small number of phenotypes were studied. Given that a small proportion of samples in this study had familiar relatedness, we have very limited power to perform family cosegregation analysis of rare variants.
In summary, we sequenced and analyzed 77 genomic loci associated with various phenotypes as implicated in previous GWAS. A cost-effective case-cohort study design and robust analysis strategies were implemented to analyze sequence data.
Supplementary Material
Acknowledgments
Funding Sources: Support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Data for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium” was provided by Eric Boerwinkle on behalf of the Atherosclerosis Risk in Communities (ARIC) Study, L. Adrienne Cupples, principal investigator for the Framingham Heart Study, and Bruce Psaty, principal investigator for the Cardiovascular Health Study. The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN2682011000010C, HHSN2682011000011C, and HHSN2682011000012C), R01HL087641, R01HL59367 and R01HL086694. The authors thank the staff and participants of the ARIC study for their important contributions. The Framingham Heart Study is conducted and supported by the NHLBI in collaboration with Boston University (Contract No. N01-HC-25195), and its contract with Affymetrix, Inc., for genome-wide genotyping services (Contract No. N02-HL-6-4278), for quality control by Framingham Heart Study investigators using genotypes in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA) computing resources at Boston University Medical Campus. This CHS research was supported by NHLBI contracts N01-HC-85239, N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85084, N01-HC-85085, N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HHSN268201200036C and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. See also http://www.chs-nhlbi.org/pi.htm.
Footnotes
Conflict of Interest Disclosures: B.M.P. serves on the DSMB of a clinical trial of a device funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Medtronic. Other authors declare no commercial conflicts of interest.
References
- 1.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson AD, O'Donnell CJ. An open access database of genome-wide association results. BMC Med Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikram MK, Sim X, Jensen RA, Cotch MF, Hewitt AW, Ikram MA, et al. Four novel Loci (19q13, 6q24, 12q24, and 5q14) influence the microcirculation in vivo. PLoS Genet. 2010;6:e1001184. doi: 10.1371/journal.pgen.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 8.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 9.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 10.Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS ONE. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 18.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 20.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, et al. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith OL, Montgomery SB, Bernier B, Chu B, Kasaian K, Aerts S, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–D113. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ripatti S, Palmgren J. Estimation of multivariate frailty models using penalized partial likelihood. Biometrics. 2000;56:1016–1022. doi: 10.1111/j.0006-341x.2000.01016.x. [DOI] [PubMed] [Google Scholar]
- 27.Therneau TM, Grambsch PM, Pankratz VS. Penalized Survival Models and Frailty. J. Comp. Graph. Stat. 2003;12:156–175. [Google Scholar]
- 28.Basu S, Pan W. Comparison of statistical tests for disease association with rare variants. Genet Epidemiol. 2011;35:606–619. doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet Epidemiol. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, et al. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am J Hum Genet. 2007;80:716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens OW, Zhang Q, Qu P, Zhou Y, Chavan S, Tian E, et al. An intermediate-risk multiple myeloma subgroup is defined by sIL-6r: levels synergistically increase with incidence of SNP rs2228145 and 1q21 amplification. Blood. 2012;119:503–512. doi: 10.1182/blood-2011-07-367052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calabro P, Willerson JT, Yeh ET. Inflammatory cytokines stimulated C-reactive protein production by human coronary artery smooth muscle cells. Circulation. 2003;108:1930–1932. doi: 10.1161/01.CIR.0000096055.62724.C5. [DOI] [PubMed] [Google Scholar]
- 36.Naka T, Nishimoto N, Kishimoto T. The paradigm of IL-6: from basic science to medicine. Arthritis Res. 2002;4(Suppl 3):S233–S242. doi: 10.1186/ar565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvitz DG, Thompson DJ. A generalization of sampling without replacement from a finite universe. J Am Statist Assoc. 1952;47:663–685. [Google Scholar]
- 38.Abecasis GR, Cookson WO, Cardon LR. The power to detect linkage disequilibrium with quantitative traits in selected samples. Am J Hum Genet. 2001;68:1463–1474. doi: 10.1086/320590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.