Abstract
The protozoan parasite Cryptosporidium parvum causes severe enteric infection and diarrheal disease with substantial morbidity and mortality in untreated AIDS patients and children in developing or resource-limited countries. No fully effective treatment is available. Hypusination of eIF5A is an important post-translational modification essential for cell proliferation. This modification occurs in a two step process catalyzed by deoxyhypusine synthase (DHS) followed by deoxyhypusine hydroxylase. An ORF of 1086 bp was identified in the C. parvum (Cp) genome which encodes for a putative polypeptide of 362 amino acids. The recombinant CpDHS protein was purified to homogeneity and used to probe the enzyme’s mechanism, structure, and inhibition profile in a series of kinetic experiments. Sequence analysis and structural modeling of CpDHS were performed to probe differences with respect to the DHS of other species. Unlike Leishmania, Trypanosomes and Entamoeba, Cryptosporidium contains only a single gene for DHS. Phylogenetic analysis shows that CpDHS is more closely related to apicomplexan DHS than kinetoplastid DHS. Important residues that are essential for the functioning of the enzyme including NAD+ binding residues, spermidine binding residues and the active site lysine are conserved between CpDHS and human DHS. N1-guanyl-1.7-diaminoheptane (GC7), a potent inhibitor of DHS caused an effective inhibition of infection and growth of C. parvum in HCT-8 cells.
Keywords: Cryptosporidium parvum, Protozoan parasite, Deoxyhypsuine synthase, Hypusine pathway
1. Introduction
Cryptosporidium parvum is an opportunistic protozoan parasite responsible for enteric infection and severe diarrheal disease in various mammals, including humans [1]. The importance of C. parvum as a human pathogen became evident with the emergence of the AIDS epidemic and to date it remains a leading cause of death in untreated AIDS patients in developing or resource-limited countries [2,3]. Several major outbreaks of C. parvum infections associated with contaminated water supplies have been reported [4]. C. parvum has a multistage life cycle during which the merozoites develop within a specialized vacuole which has an intracellular but extra-cytoplasmic location within the host cell requiring drug candidates to cross both host and parasite membranes and presenting unique challenges for drug development. Cryptosporidium sporozoite antigens have been tested as vaccine candidates; however, a suitable vaccine is not yet available [7–9]. Nitazoxanide (NTZ), paromomycin, and azithromycin are the most commonly used drugs against cryptosporidiosis but they are only partially effective [5,6]. Nitazoxanide is effective in the immunocompetent but is ineffective in the immunocompromised patients [6].
Hypusine [Nε-(4-amino-2-hydroxybutyl) lysine] is formed by a post-translational modification of a lysine residue of the eukaryotic initiation factor 5A (eIF5A) [10,11]. Hypusine modification is important for cell proliferation and tumorigenesis [12,13]. The hypusine residue is also important in the binding of eIF5A to RNA and in its interaction with exportin 4, which was reported to facilitate the nucleo-cytoplasmic shuttle function of eIF5A [14–16]. Hypusine biosynthesis occurs in two steps [30]. First, deoxyhypusine synthase (DHS) synthesizes deoxyhypusine (Nε-(4-aminobutyl) lysine) by transferring the butyl amine moiety of spermidine to a specific lysine residue in NAD+-dependent reaction. Second, deoxyhypusine is hydroxylated by deoxyhypusine hydroxylase (DOHH) to form hypusine. eIF5A, DHS, and DOHH are highly conserved in all eukaryotes, indicating an important function of this modification [17,18].
Hypusine modification is essential in eukaryotic organisms as deletion of eIF5A or DHS in yeast or in mouse causes lethality [18–21]. However, a deletion mutant of DOHH is viable in yeast. DOHH is essential in higher eukaryotes [25,26]. Human DHS is a 41 kDa protein and forms a homo-tetramer of two identical dimers [25–28]. The crystal structure of human recombinant DHS shows that it has four active sites that bind four molecules of NAD+ and this binding site is present near the spermidine binding pocket [27]. Normally, in the complete reaction mixture containing DHS, cofactor NAD+, donor substrate spermidine, and acceptor substrate eIF5A, deoxyhypusine is formed in eIF5A [29]. However, in the absence of acceptor substrate, only half the reaction occurs [31].
DHS is present as a single copy gene in yeast and human but two copies of DHS, DHSL20 (DHS-like gene from chromosome 20) and DHS34 are present in the Leishmania parasite. DHS34 is a catalytically active enzyme form whereas DHSL20 is inactive as its lacks the active site lysine residue. The origin and significance of the two forms of DHS in the Leishmania parasite is unknown [21]. Trypanosoma brucei also encodes two deoxyhypusine synthase paralogs, one that is catalytically functional but grossly impaired, and the other is inactive. In T. brucei, both homologs are required for optimal enzyme activity [22]. In the human malaria parasites, P. falciparum and P. vivax a single copy of DHS is present and has been evaluated as a potential drug target [23]. Recent experiments show that down regulation by silencing the eIF5A, DHS and DOHH genes with short hairpin RNAs lead to impaired hypusine biosynthesis and growth retardation of the parasite [24].
In the present study we have characterized a functional DHS from C. parvum. The results indicate that C. parvum has a single DHS gene which, based on neighbor joining bootstrap analysis, has close similarity to other apicomplexan DHS sequences. The ability of several guanyldiamines to inhibit the enzyme, and infection and growth of C. parvum was examined.
2. Materials and methods
2.1. Chemicals
Radiolabeled spermidine trihydrochloride [1,8-3H] spermidine (16.6–32.2Ci/mmol) was purchased from PerkinElmer Life Sciences. All restriction enzymes and DNA-modifying enzymes were obtained from MBI Fermentas (Germany). N1-guanyl-1, 7-diaminoheptane (GC7), N1-guanyldiaminooctane (GC8), N1,N7-bisguanyl-l,7-diaminoheptane (GC7G), and N1, N8-bisguanyl-l.8-diaminooctane (GC8G) were synthesized as previously described [32]. Other materials used in this study were of analytical grade and were commercially available.
2.2. Parasite and culture conditions
C. parvum oocysts were obtained from G. & S. Pritchard (Bunch Grass Farm, Deary, ID 83823). Oocysts were passaged in 2–5 day old calves, collected, and purified on CsCl gradients as described [33,34]. Oocysts were surface sterilized using 10% (v/v) Clorox®, washed, and shipped in potassium dichromate. Prior to use, oocysts were washed with dH2O to rinse them free of potassium dichromate, and washed in 10% (v/v) Clorox® before being suspended in minimal essential medium (MEM) containing 10% horse serum [35,36].
2.3. Inhibition of C. parvum growth and development by amine analogs
Human adenocarcinoma cells (HCT-8, ATCC CCL-244) were grown to confluence in 12 well plates (3.8 cm2) containing MEM supplemented with 10% horse serum. C. parvum oocysts (3 × 104) were inoculated into HCT-8 containing plates and incubated for 1.5 h after which time the media was removed and replaced with fresh MEM + 10% horse serum and test compound (GC7, GC7G, diaminooctane, diaminononane, agmatine) and incubated for an additional 24 h. The media was removed and centrifuged (14,000 × g for 2 min) to obtain oocysts, which were stained using merifluor (Meridian Bioscience, OH) and counted using a plate reader with an excitation wavelength of 488 nm and emission wavelength of 518 nm (Spectromax, Molecular Devices, CA). The number of oocysts was determined by reference to a standard curve of fluorescence versus oocyst number. The ability of GC7 to prevent infection of HCT-8 cells was determined by pre-incubation of oocysts or sporozoites (3 × 104) with varying amounts of test compound for 30 min, followed by centrifugation at (14,000 × g for 2 min) and resuspension in MEM containing 10% horse serum. The GC7 pre-incubated oocysts or sporozoites were used to infect a confluent layer of HCT-8 cells and processed as described above.
2.4. Cloning of putative DHS genes from C. parvum and construction of CpDHS expression vector
Total RNA was isolated from C. parvum cells using RNAeasy kit (Qiagen, CA) and was treated with DNAse (Fermentas, MD) to remove gDNA contamination. Reverse transcription was performed using ThermoScript reverse transcription-PCR kit (Invitrogen, NY) in a 20 µl reaction containing 500 ng of purified RNA, according to the manufacturer’s instructions. The open reading frames of the C. parvum DHS were PCR-amplified using cDNA as a template and forward and reverse primers 5′-GGAAGATCTGATGCATTCTTTAGGGAATT-3′ containing Bg/II site and 5′CAAOTTCTATGTATCAAGAAAAGTAGAATAA-3′ containing HindΠI site, respectively (in which the restriction sites are underlined). PCR was performed using Jump Start REDTaq Ready Mix (Sigma, MO) and the program as follows: Initial denaturation at 95 °C for 5 min followed by 30 cycles at 95°C for 30 s, 58°C for 30 s and 72°C for 1 min and final extension at 72°C for 10 min. The PCR product was digested and inserted at the Bg/II and HindΠI sites of the pET30a (Novagen, Germany) vector. The recombinant plasmid CpDHS-pET30a was sequenced using both T7 promoter forward and reverse primers to confirm the accuracy of PCR amplification and correct insertion of the CpDHS open reading frames in the vector.
2.5. Sequence analysis
A comparative sequence analysis of DHS sequences from Cryptosporodium parvum Iowa II with its homologs from other eukaryotes was performed. The DHS homolog sequences were derived from Swissprot/UniprotKB [37], EupathDB [38] and GeneDB [39] databases. Multiple sequence alignment of these sequences was generated using CLUSTALW with default parameters. Phylogenetic analysis was performed using the neighbor joining tree generated using CLUSTALW [40]. MEGA v5 [41] was used both for visualization and analysis of the phylogenetic tree. The tree was annotated with bootstrap values (100 iterations).
2.6. Expression and purification of the recombinant CpDHS protein
The recombinant construct of CpDHS-pET30a was transformed into the BL21-DE3 strain of E. coli and protein expression was induced at 0.6 OD600nm with 0.5 mM isopropyl-1-D-galactopyranoside (IPTG) at 14 °C for 18 h. The cell lysate containing His-tagged CpDHS, was loaded onto pre-equilibrated Ni+2 nitrilotriacetic agarose resin (Qiagen). The recombinant CpDHS protein was eluted with increasing concentrations of imidazole. The purified protein (~95% purity), after removal of imidazole, was aliquoted and stored at −80°C in 50 mM Tris-Cl, pH 8.0, 300 mM NaCl buffer. The oligomerization state of CpDHS was determined by a glutaraldehyde cross-linking experiment. Briefly, 20 mM of glutaraldehyde was mixed with 6 µg of protein in a total volume of 20 µl. The mixture was incubated for 2 min, 5 min, 15 min, and 30 min and terminated by addition of 5 × SDS sample buffer and the cross- linked proteins were analyzed by SDS-PAGE. Recombinant human His-tagged DHS was used as a positive control.
2.7. Deoxyhypusine synthase assay
Deoxyhypusine synthase activity was assayed as described previously [42]. Briefly, a reaction mixture of 50 µl containing 0.2 M glycine-NaOH buffer pH 9.2, 1 mM dithiothreitol, 1 mM NAD+, 200 µg of bovine serum albumin, 2.5 µM [ 1,8-3H] spermidine, 5 µM eIF5A, and the indicated amount of recombinant DHS was incubated at 37 °C for 60 min and terminated by the addition of ice cold 10% trichloroacetic acid containing putrescine, spermidine, and spermine (1 mM each). The samples were then centrifuged at 10,000 × g for 5 min at 4°C. The precipitate obtained was repeatedly washed with 10% trichloroacetic acid containing polyamines to remove the [3H] spermidine non-covalently bound to the pellet. The [3H] deoxyhypusine formed was measured by dissolving the pellet in 100 µl of 0.1 N NaOH, and the radioactivity was measured in a scintillation counter. One unit of enzyme activity is defined as the amount of enzyme catalyzing the formation of one pmol/h of deoxyhypusine. Deoxyhypusine formation was also confirmed by fluorographic detection of radiolabeled eIF5A. Proteins in the reaction mixtures were separated by SDS-PAGE. The SDS gel was impregnated with Amplify (GE Healthcare), dried and exposed to X-ray film for one week at −80 °C and developed.
To test inhibitors, varying concentrations of GC7, GC8, GC8G and GC7G were added to the reaction mixture at time zero and incubated at 37 °C for 10 min prior to addition of eIF5A precursor.
2.8. NADH fluorescence measurement of DHS
Fluorescence data was generated as reported earlier [43]. Briefly, an assay mixture of 100µl containing 0.2 M glycine-buffer (pH 9.2), 200 µM NAD+, l00µM spermidine, and 2 µM CpDHS was prepared. For emission spectra of NADH fluorescence, excitation was at 340 nm and the measurement was recorded at 441 nm at different time points using Varian Cary Eclipse fluorescence spectrophotometer. Scan speed was 600 nm/min and slit widths were 5 nm for both excitation and emission. Background emission scan was collected on buffer containing CpDHS, and either spermidine or NAD+.
2.9. Model building and evaluation
A comparative structural model of C. parvum deoxyhypsuine synthase was built using Modeler v9 [44,45]. A sequence search using BLAST against the PDB database revealed a close relationship of CpDHS to the human DHS protein (P49366) with a sequence identity of 56% at reliable E-value of 2e-108. Hence, the C. parvum DHS sequence was modeled on the tertiary structure of the human protein template using Modeler v9 [44–46]. The models generated were energy minimized in GROMACS [47]. The stereochemical quality of the model was verified using PROCHECK in the PDB-SUM [48] web resource at EBI. Structural mapping of the active site residues was performed using Pymol [48]. 3-D structural models of the L. donovani and C. parvum enzymes were compared with the human template using DaliLite program [49]. The structural model was superposed on the template with a Z-score of 49.9 and with an RMSD of 2.4 Å respectively.
3. Results and discussion
3.1. Sequence and phylogenetic analysis
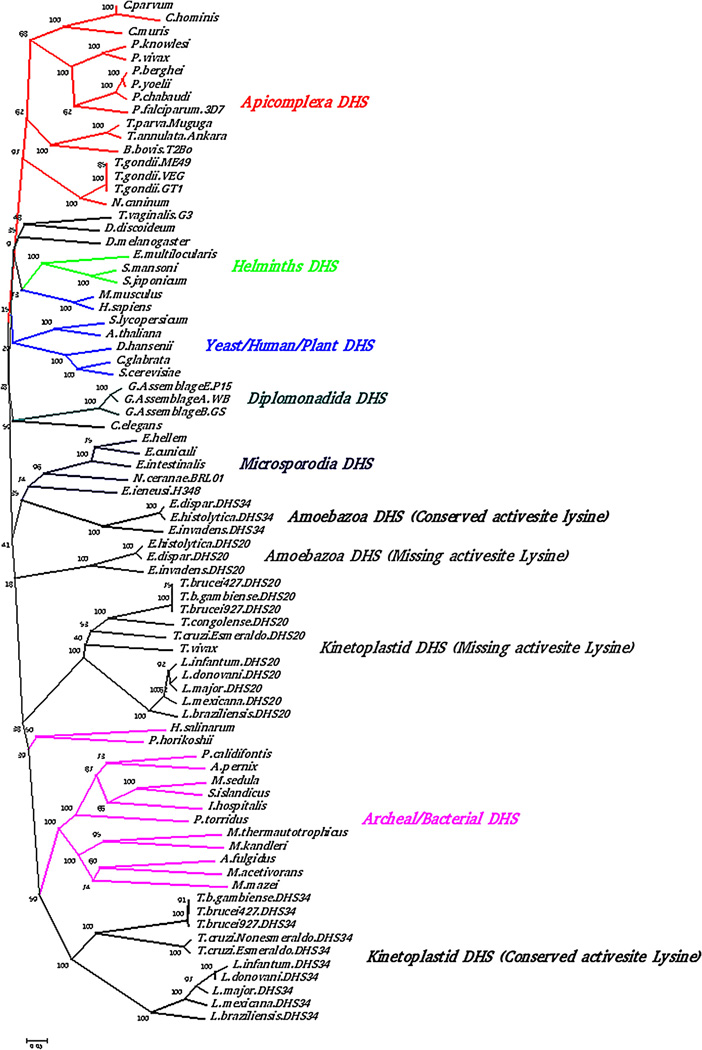
In common with Plasmodium vivax [53] and most other eukaryotic pathogens, human [54], yeast [55] and archea [56], a single DHS sequence was identified in the C. parvum genome database (EuPath.db.org). These groups are different from Leishmania, Trypanosoma [21] and Entamoeba genomes which have two DHS gene copies. DHS encoding genes are present on chromosomes 20 (DHS20) and 34 (DHS34) in L. major and L. infantum [21]. Multiple sequence alignment of the representative set of DHS homologs from kinetoplasts, Plasmodium falciparum and human with the C. parvum DHS sequence shows the conservation of important residues that are essential for the functioning of the enzyme which involves the cofactor (NAD+) binding residues, spermidine binding residues and the active site lysine (Supporting Information, Fig. S1). The C. parvum DHS shares 56% sequence identity with the human DHS homolog.
A phylogenetic tree was constructed using the putative DHS sequence from C. parvum along with its homologs from other eukaryotic pathogens, human, yeast, and archaea. The neighbor joining bootstrap tree suggests a close relationship of C. parvum DHS to other apicomplexan DHS sequences compared to the human protein (Fig. 1 ).
Fig. 1.
Phylogenetic analysis. Sequence based phytogeny of DHS sequences from C. parvum lowa II(CGD2_3930); C. muris (CMU_003030); E. multilocularis (EmW_000780100); S. japonicum (Sjp_0017580); S. mansoni (Smp_065120); E. hellem (EHEL_090880); N. ceranae (NCER_100851); G. Assemblage B (GL50581_338; GL50803_15535;GLP15_78;); T. gondii (TGGT1_070160); N. canium (NCLIV_050000); T. vaginalis (TVAG_359990); E. invadens (EIN_107550; EIN_017320); E. dispar (EDL161760; EDL198820); E. histolytica (EHI_098350; EHI_006030); T. brucei (Tb927.1.870; Tbg972.1.280; Tb427.10.2750; Tb927.10.2750; Tbg972.10.3430; Tb427.01.870); L. mexicana (LmxM.20.0250; LmxM.33.0330); T. cruzi (Tc00.1047053504119.29; Tc00.1047053506195.300); T. congolense (TcIL3000.1.360); P. yoelli (PY01546); P. berghei (PBANKA.103000); P. chabaudi (PCHAS_103080); P. knowlesi (PKH_133500); P. falciparum (PF14_0125); P. vivax (PVX_085825); T. parvum (TP02_0058); T. annulata (TA13570); B. bovis (BBOV_III010890); E. intestinalis (Ein09_0870); L. tarentolae (LtaP20.0250; LtaP34.0370); L. major(LmjF.20.0250; LmjF.34.0330); L. braziliensis (LbrM.20.4450; LbrM.20.0300); L.infantum (LinJ.20.0270; LinJ.34.0350); DHYS_PYRCJ (A3MVC9); DHYS_METS5 (A4YHK6); DHYS_IGNH4 (A8AA61); DHYS_HALS3 (B0R5L2); DHYS_SULIA (C3N5B4); DHYS_METTH (026230); DHYS1_ARCFU (028088); DHYS_PYRHO (050105); DHYS_HUMAN (P49366); DHYS_DEBHA (Q6BJH5); DHYS_CANGA (Q6FRN2); DHYS_PICTO (Q6KZL5); DHYS_SOLLC (Q9AXR0); DHYS_ENCCU (Q8SQN2); DHYS1_METAC (Q8TS38); DHYS_METKA (Q8TXD7); DHYS2_METMA (Q8Q051); DHYS_AERPE (Q9YE72); L. donovani (LdBPK_340350; LdBPK_200270) were derived using neighbor joining method. Bootstrap values are shown at the nodes.
3.2. Model building, evaluation and comparative structural analysis
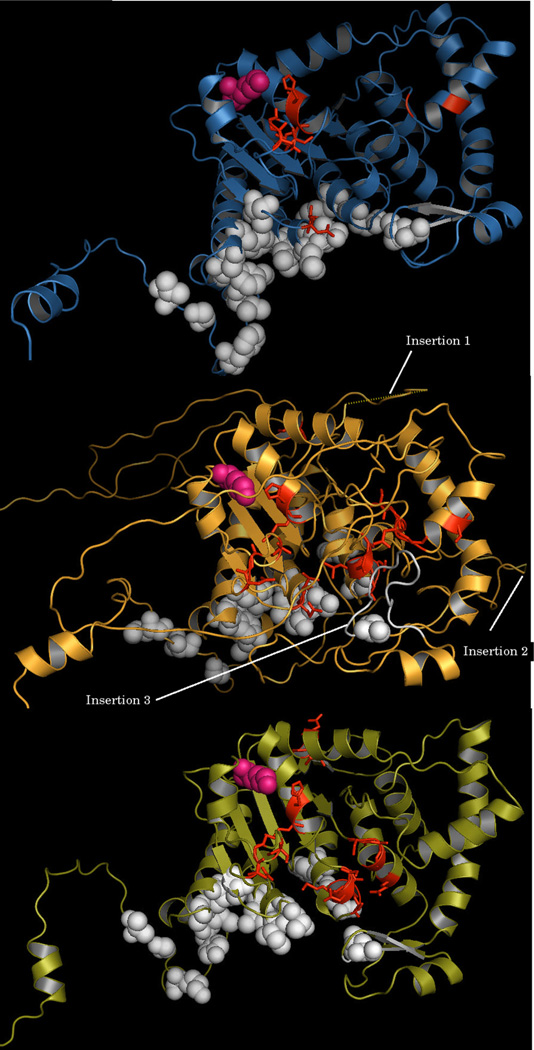
A C. parvum model was built using the human DHS crystal structure (PDB: 1DHS) [27,51] as the template. The dimer and tetramer interface residues were identified from the tetrameric structure of human DHS (PDB:1RQD) using the protein interaction calculator [52] (PIC Web interface) available at http://pic.mbu.iisc.ernet.in. The interface residues for the human multimer were mapped based on the residue–residue interactions predicted using the PIC server. Residue accessibility cut-off of ≤7% was used for calculating the interactions between the buried residues of the human DHS and hence, the interface residues were mapped onto the human structure (Fig. 2). The structurally equivalent residues are then mapped on to the C. parvum DHS structural model. All the interface residues are conserved or conservatively substituted in C. parvum enabling both dimer and tetramer formations similar to the human DHS. Characterization of a novel deoxyhypusine synthase (DHS34) from L. donovani revealed formation of only dimers [21]. Since, CpDHS and LdDHS are essential enzymes in these parasitic protozoa, the structural model of L. donovani DHS (DHS34) was built using the human structure (PDB: 1DHS) as the template as it shares 37% sequence identity with the human DHS. The model was built, energy minimized and the stereochemical quality was verified using a similar procedure described in Section 2. Structural comparison of all three structures reveals the presence of 3 loop insertions in L. donovani that are absent in the human and C. parvum structures (Fig. 2). Structural mapping of the equivalent interface residues as well as the loops in the structural models show the presence of one of the loops (Insertion 3) in L. donovani DHS near the interface residues. Thus, it is clear that the interface conservation helps in achieving a higher oligomeric state by CpDHS while a loop insertion in L. donovani DHS prevents the dimers from interacting further.
Fig. 2.
Structural comparison of the human DHS structure with C. parvum DHS Model and L. donovani DHS Model. Structural comparison of the human DHS structure (PDB: 1 DHS; Violet) with C. parvum DHS Model (cgd2_3930; Olive green) and L. donovani DHS Model (LdBPK_340350; Bright Orange). NAD+ binding residues are shown as red sticks. Catalytic lysine is shown as pink spheres. Residues at the tetramer interface in the human DHS structure (PDB:1RQD) and their equivalent residues in the C. parvum DHS and L. donovani DHS models are shown as white spheres. The loop insertions (Insertion 1 & Insertion 2) are shown in dotted lines in the L. donovani model. The 14 residue loop insertion (Insertion 3) in the L. donovani model and the equivalent interface residues in the C. parvum model and the human structure are shown in white. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.3. Over-expression and purification of recombinant CpDHS enzyme in E. coli
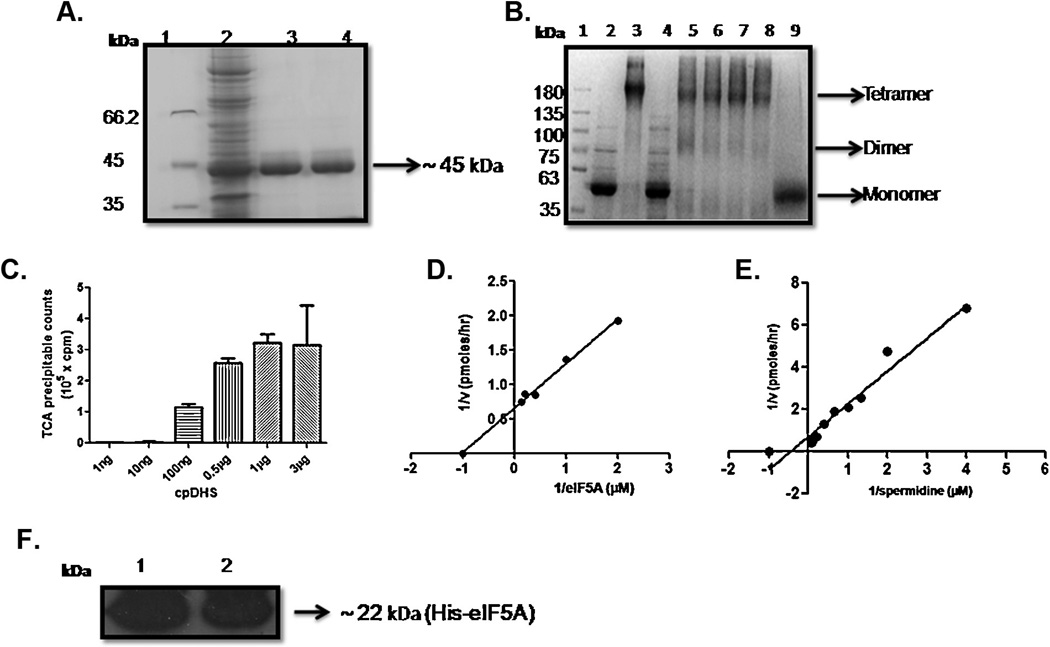
CpDHS-pET30a construct was transformed into E. coli BL21-DE3 and the recombinant enzyme was overexpressed. His-tagged protein with an estimated molecular mass of 45 kDa was induced by IPTG. The size matches well with the amino acid sequence of the ORF of CpDHS (40.8 kDa, 362 aa) with an added His-tag sequence (Fig. 3A). Purification of CpDHS by metal affinity chromatography yielded ~3 mg of purified protein per liter of bacterial culture.
Fig. 3.
Purification and biochemical characterization of recombinant CpDHS. (A) Purification of His-tagged CpDHS protein on Ni 2+Nitrolotriacetic acid affinity resin. Lane 1: Molecular weight marker; lane 2, flow through; lanes 3 and 4 eluted fractions showing purified protein. (B) Determination of oligomerization state of CpDHS by chemical crosslinking. Lane 1, Molecular weight marker; Lane 2, human His-DHS; lane 3, human His-DHS incubated with 20 mM glutaraldehyde; lane 4, His-CpDHS; lane 5–8, His-CpDHS incubated with 20 mM glutaraldehyde for 2,5,15 and 30 min respectively; lane 9, human His-DHS treated with 20% SDS and 20 mM glutaraldehyde. (C) Comparison of trichloroacetic acid precipitable counts obtained from enzyme assay mixture using different amount of recombinant CpDHS. Results are mean ± S.D. of triplicate samples. (D) Dependence of CpDHS reaction on eIF5A concentration. Varying concentrations of eIF5A (µM) with 1 mM NAD+, 2.5 µM (4 µCi) of [1,8-3H] spermidine and 1 µg of CpDHS were used. The Km and Vmax value were 0.9163 µM and 1.473 pmol/h, respectively. (E) Dependence of CpDHS reaction on spermidine concentration. Varying concentrations of spermidine (µM) with 1 mM NAD, 5µM eIF5Aand 1 µg of CpDHS were used. The Km and Vmax value were 12.29µM and 4.86 pmol/h, respectively. (F) Radiolabeling of eIF5A by in vitro CpDHS and human DHS reaction. Radiolabeling of eIF5A was confirmed by SDS-PAGE of the DHS reaction mixture. Fluorogram showing the position of eIF5A. Lane 1: recombinant CpDHS plus eIF5A, Lane 2: recombinant human DHS plus eIF5A.
Human DHS native enzyme is a homotetramer consisting of two dimers of 41-kDa subunits, while Leishmania DHS is a dimer. Therefore, we checked the oligomerization state of cpDHS by a crosslinking experiment. A band of ~180 kDa appears in reactions treated with glutaraldehyde (lanes 5, 6, 7, and 8) as compared to the untreated reaction (lane 4) (Fig. 3B).This correlates to the oligomerization state of human DHS (lane 3), indicating that cpDHS is also a tetramer.
3.4. Catalytic properties of CpDHS
The kinetic parameters of the enzyme CpDHS were determined using [1,8-3H] spermidine and human recombinant eIF5A as the substrates in vitro. [3H] Deoxyhypusine formation increased with increasing concentration of recombinant CpDHS (Fig. 3C) up to 1 µg. The Km value of CpDHS for eIF5A and spermidine was estimated to be 0.91 µM and 12.29 µM, respectively (Fig. 3D and E). The specificity of the enzyme for eIF5A was determined by fluorography. The appearance of a 22-kDa radiolabeled protein in lane1 confirmed deoxyhypusine formation on human eIF5A by CpDHS (Fig. 3F). The estimated Km of CpDHS for eIF5A was higher than that of the human (0.6 µM) [54] but lower than that of S. cerevisiae (1.01 µM) [57] whereas the Km value of CpDHS for spermidine (12.29 µM) was, higher than that of the human enzyme (7.2 µM) [54] but comparable to that of the S. cerevisiae enzyme (12.5 µM) [57].
3.5. Stochiometry of NAD+ binding to CpDHS
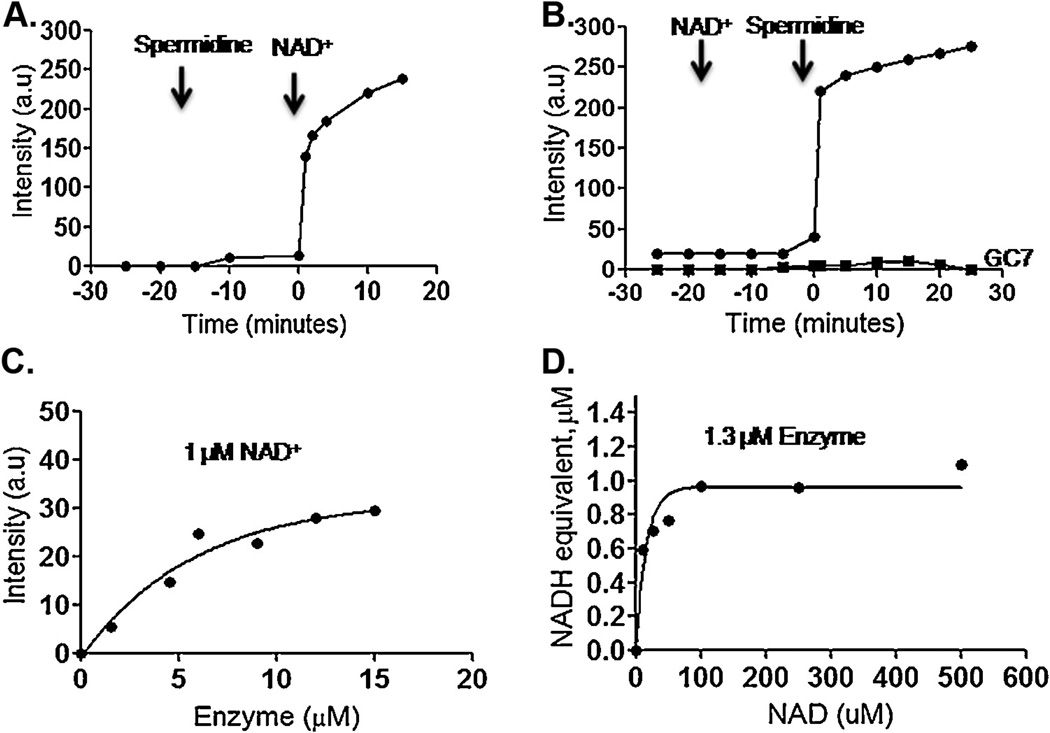
Free NADH emits a strong fluorescence at 457–465 nm when excited in the region of 340 nm, whereas, dehydrogenases bound NADH shows enhanced (up to 13-fold) fluorescence emission peak at 441 nm [50]. CpDHS-associated NADH formation was checked under various conditions in the absence of eIF5A precursor, as has been done with human DHS enzyme [43]. CpDHS with spermidine alone did not show any increase in intrinsic florescence at 441 nm with time. However, a rapid increase in fluorescence was observed upon addition of NAD+ to a mixture of the enzyme and spermidine, indicating that both spermidine and NAD+ are required for NADH fluorescence (Fig. 4A). Similar observation was made with addition of NAD+ prior to spermidine (Fig. 4B). This enhanced florescence was dependent on the enzyme concentration. External addition of NADH did not show any enhanced fluorescence at 441 nm (Data not shown). Addition of the competitive inhibitor (GC7) in the reaction completely abolished the NADH fluorescence.
Fig. 4.
Kinetics and stoichiometry of NADH bound to CpDHS. The final component of the reaction mixture was added in the fluorescence cell at r= 0 and emission at the fixed wavelength, 441 nm (excitation at 340 nm), was followed over the indicated time course. (A) NADH fluorescence with addition of 100 µM spermidine and 1 mM NAD+. (B) NADH fluorescence with addition of 1 mM NAD+ followed by 100 µM spermidine and its inhibition by 100 µM GC7. (C) Quantification of the AU of the NADH (1 µM) formed. 1.1 µM NAD+ and 100 µM spermidine were incubated with increasing the amount of DHS for 25 min. The fit of the experimental points to a hyperbola gave a maximum value of 31.92 (indicated by arrow). (D) DHS (1.3 µM) was incubated with 100 µM spermidine and increasing concentrations of NAD+.The arbitrary unit (A.U.) after the reaction was divided by 31.92 to give the equivalent NADH concentration in µM.
Stoichiometry of NADH molecules generated per monomer of the enzyme was determined in two steps. First, a very low concentration of NAD+ (1 µM) was incubated with high concentration of CpDHS (2.5–15 µM) and 100 µM of spermidine (Fig. 4C). Assuming that all the NAD+ present in the reaction gets reduced to NADH in the presence of excess spermidine and enzyme, the fluorescence corresponding to 1 µM of enzyme-bound NADH was estimated to be 31.2 A. U. (Fig. 4C). Secondly, low concentration of the CpDHS (1.3 µM) was incubated with 100 µM of spermidine and an increasing amount of NAD+ (50–500 µM). NADH equivalent (µM) reached the maximum of ~1 at high NAD+ concentration with 1.3 µM enzyme (Fig. 4D). This value (suggesting that 77% of proteins in recombinant CpDHS are active enzymes) approximates to the theoretical value of 1 derived from the crystal structure of human DHS.
3.6. Effect of guanyldiamines on the activity of CpDHS in vitro
Spermidine analogs were developed as inhibitors of human and rat DHS. Most potent compounds, are guanylated diamines with similar methylene chain length as in spermidine, and the order of inhibitory efficacy of these compounds for the mammalian enzyme is GC7 > GC8 > GC7G > GC8G [32]. The effect of four gunayl diamines on CpDHS activity was determined by the deoxyhypusine synthase assay as described under Section 2. Of the four compounds tested, GC7, which is the most effective inhibitor of human DHS, was also the most effective inhibitor of the C. parvum enzyme, causing inhibition of >90% at 1 µM and 99% at 10µM (Fig. 5).
Fig. 5.
Inhibition of CpDHS by various guanyldiamines. The reactions were carried out as described under methods using 1 µg of recombinant CpDHS in 50 µl of reaction volume with indicated concentrations of various guanyldiamine. Results are mean ± SD of triplicate samples. * p < 0.05; ** p < 0.01; *** p < 0.005 and ns indicates not significant (p>0.05).
3.7. Effect of diamines and N-guanyldiamines on C. parvum infection and growth
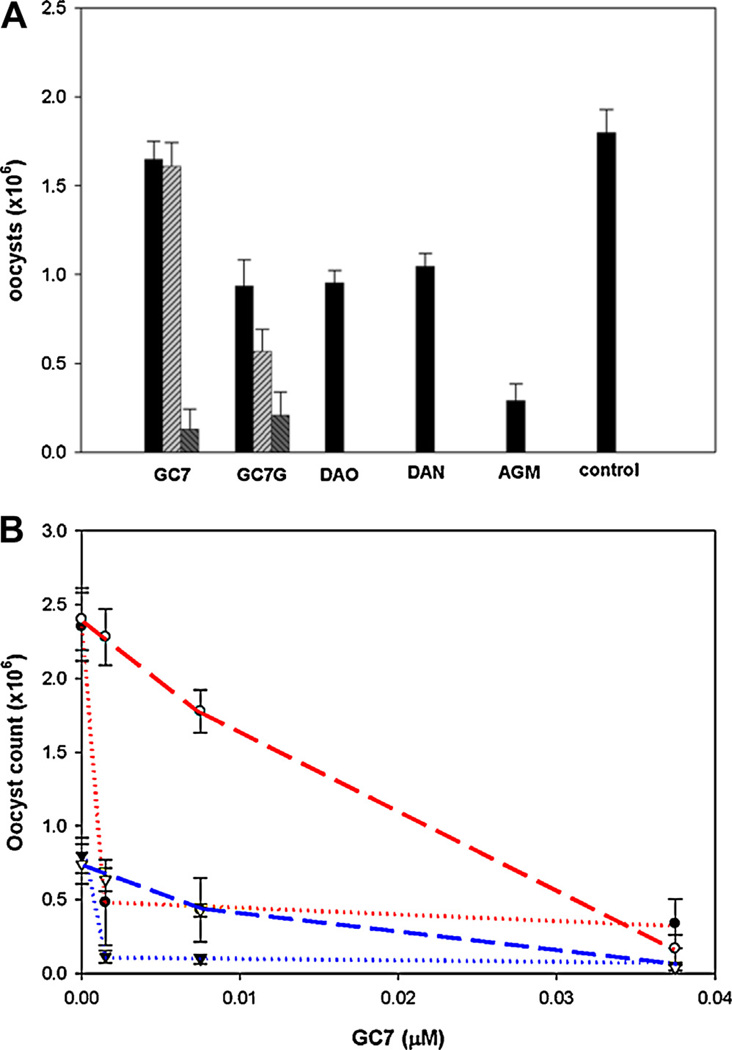
As CpDHS is quite sensitive to GC7, the ability of GC7 and GC7G to inhibit the infection, growth and development of a C. parvum in HCT-8 cells was determined using increasing concentrations (0.002–0.021 µM) of GC7, (0.45–4.5 µM) GC7G, diaminooctane (DAO) (24 µM), diaminononane (DAN) (36 µM) and agmatine (10 µM) (Fig. 6A). Both GC7 and GC7G demonstrated a dose-dependent inhibition, with GC7 being approx 200-fold more effective in reducing the parasitemia, showing approximately 90% inhibition at 0.021 µM whereas the same level of inhibition was achieved by GC7G at 4.50 µM (Fig. 6A). In contrast, the diamines, diaminooctane and diaminononane, were considerably less effective than GC7, requiring over 1000-fold higher concentration (Fig. 6A). Paromomycin, as has been reported earlier was only partially effective [5,6]. Paromomycin (150 µM) caused 58% inhibition of C. parvum oocyst production.
Fig. 6.
Inhibition of C. parvum infection and growth by GC7. (A) Inhibition of C. parvum oocyst production by DHS inhibitors C. parvum (3 × 104) oocysts were inoculated to a confluent layer of HCT-8 cells and incubated for 2 h after which time the media was replaced with fresh medium containing stated amount of test compound and incubated for 24 h; oocytes were harvested and counted as described under the Methods. C. parvum oocyst production in the presence of 0.02 µM ( ), 0.04 µM (
), 0.04 µM ( ), or 0.021 µM (
), or 0.021 µM ( ) GC7; 0.45 µM (
) GC7; 0.45 µM ( ), 0.90 µM (
), 0.90 µM ( ), or 4.5 µM (
), or 4.5 µM ( ) GC7G; 24µM diaminooctane (DAO); 36 µM diaminononane (DAN); 10 µM agmatine (AGM). Results are expressed as + SD of triplicate experiments compared to control oocysts lacking added compounds. Paromomycin (150 µM) caused 58% inhibition of C. parvum oocysts production. (B) Inhibition of C. parvum infection and growth by GC7. C. parvum oocyst production after 24 h incubation with HCT-8 cells. Oocysts (
) GC7G; 24µM diaminooctane (DAO); 36 µM diaminononane (DAN); 10 µM agmatine (AGM). Results are expressed as + SD of triplicate experiments compared to control oocysts lacking added compounds. Paromomycin (150 µM) caused 58% inhibition of C. parvum oocysts production. (B) Inhibition of C. parvum infection and growth by GC7. C. parvum oocyst production after 24 h incubation with HCT-8 cells. Oocysts ( ) or sporozoites (
) or sporozoites ( ) preincubated for 1.5 h with HCT-8 cells prior to addition of GC7; Oocysts (
) preincubated for 1.5 h with HCT-8 cells prior to addition of GC7; Oocysts ( ) or sporozoites (
) or sporozoites ( ) preincubated with GC7 for 30 min prior to infecting HCT-8 cells.
) preincubated with GC7 for 30 min prior to infecting HCT-8 cells.
In a second experiment the ability of GC7 to reduce the parasitemia was determined by pre-incubating 3 × 104 oocysts with 1.5 nM, 7.5 nM and 37.5 nM GC7 for 30 min prior to introduction to the HCT-8 monolayer cells (Fig. 6B). At 1.5 nM GC7, an 80% reduction in parasitemia was evident compared to controls lacking GC7, whereas only a 5% reduction in parasitemia was observed when GC7 was added 1.5 h post infection (Fig. 6B). These observations were supported using sporozoites, which exhibited an 86% reduction in parasitemia when pre-treated for 30 min with 1.5 nM GC7compared to control sporozoites lacking the inhibitor. Plates to which 1.5 nM GC7 was added 1.5 h post infection had a 27% reduction in growth (Fig. 6B). Exposure of oocysts or sporozoites to 1.5 nM GC7 for 30 min resulted in an 80% and 86% reduction in parasitemia, respectively, suggesting effective uptake of the inhibitors by the parasites during the pre-incubation.
The inhibitory effects of GC7 on the parasite infection and growth are quite remarkable. Although GC7 is a potent inhibitor of both parasite and human enzymes in in vitro assays, the parasite infection is effectively inhibited by GC7 at nM concentrations (1.5–37.5 nM) which would not affect deoxyhypusine synthesis in, and growth of host mammalian cells.
The present study, for the first time, reports the presence of a hypusine pathway in C. parvum. C. parvum enzyme is more closely related to the human enzyme than the kinetoplastid enzymes in sequence and structure. Although physical and catalytic properties and sensitivity to guanyldiamines in vitro are similar for human and C. parvum enzymes, our data suggest that C. parvum DHS is a druggable target, as GC7 effectively inhibits parasite infection and growth in cultured host cells (Fig. 6). In summary, our studies demonstrate that C. parvum has a functional hypusine pathway and has evolved differently from the kinetoplastids.
Supplementary Material
Acknowledgements
N.M. is a recipient of funding from the Council of Scientific and Industrial Research. V.S.G. and PT are both Kothari post-doctoral fellows supported by the University Grants Commission, India. RM is a J C Bose National Fellow.
Funding
The work is supported by a grant from the Council of Scientific and Industrial Research, India, (Grant No. 37(1328) /08/ EMR-II) to R.M. and in part by the Intramural Research Program of NIDCR, NIH, Bethesda Maryland, USA. Alison Quirch was recipient of a Pace University Faculty–Student Research Grant.
Abbreviations
- eIF5A
eukaryotic initiation factor 5A
- DHS
deoxyhypusine synthase
- DOHH
deoxyhypusine hydroxylase
- Cp
C. parvum
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.molbiopara.2014.05.005.
References
- 1.Fayer R. Cryptosporidium and cryptosporidiosis. CRC Press; 1997. [Google Scholar]
- 2.Shebl FM, Engels EA, Goedert JJ. Opportunistic intestinal infections and risk of colorectal cancer among people with AIDS. AIDS Res Hum Retroviruses. 2012;28:994–999. doi: 10.1089/aid.2011.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nissapatorn V, Sawangjaroen N. Parasitic infections in HIV infected individuals: diagnostic & therapeutic challenges. Indian J Med Res. 2011;134:878–897. doi: 10.4103/0971-5916.92633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza JC, Nichols G. Cryptosporidiosis surveillance and water-borne outbreaks in Europe. Euro Surveill. 2007;12:E13–E14. doi: 10.2807/esm.12.05.00711-en. [DOI] [PubMed] [Google Scholar]
- 5.Mead JR. Cryptosporidiosis and the challenges of chemotherapy. Drug Resist Update. 2002;5:47–57. doi: 10.1016/s1368-7646(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 6.Gargala G. Drug treatment and novel drug target against Cryptosporidium . Parasite. 2008;15:275–281. doi: 10.1051/parasite/2008153275. [DOI] [PubMed] [Google Scholar]
- 7.Manque PA, Tenjo F, Woehlbier U, Lara AM, Serrano MG, Xu P, et al. Identification and immunological characterization of three potential vaccinogens against Cryptosporidium species. Clin Vaccine Immunol. 2011;18:1796–1802. doi: 10.1128/CVI.05197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehigiator HN, Romagnoli P, Priest JW, Secor WE, Mead JR. Induction of murine immune responses by DNA encoding a 23-kDa antigen of Cryptosporidium parvum . Parasitol Res. 2007;101:943–950. doi: 10.1007/s00436-007-0565-0. [DOI] [PubMed] [Google Scholar]
- 9.He H, Zhao B, Liu L, Zhou K, Quin X, Zhang Q, et al. The humoral and cellular immune responses in mice induced by DNA vaccine expressing the sporozoite surface protein of Cryptosporidium parvum . DNA Cell Biol. 2004;23:335–339. doi: 10.1089/104454904323090967. [DOI] [PubMed] [Google Scholar]
- 10.Shiba T, Mizote H, Kaneko T, Nakajma T, Kakimoto Y, et al. Hypusine, a new amino acid occurring in bovine brain. Isolation and structural determination. Biochim Biophys Acta. 1971;244:523–531. doi: 10.1016/0304-4165(71)90069-9. [DOI] [PubMed] [Google Scholar]
- 11.Park MH, Wolff EC, Folk JE. Hypusine: its post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Bio-factors. 1993;4:95–104. [PubMed] [Google Scholar]
- 12.Caraglia M, Marra M, Giuberti G, D’ Alessandro AM, Budilon A, Prete del S, et al. The role of eukaryotic initiation factor 5A in the control of cell proliferation and apoptosis. Amino Acids. 2001;20:91–104. doi: 10.1007/s007260170050. [DOI] [PubMed] [Google Scholar]
- 13.Chen ZP, Chen KY. Marked elevation of hypusine formation activity on eukaryotic initiation factor 5A in v-HA-RAS transformed mouse NIH3T3 cells. Cancer Lett. 1997;115:235–241. doi: 10.1016/s0304-3835(97)04741-1. [DOI] [PubMed] [Google Scholar]
- 14.Xu A, Chen KY. Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J Biol Chem. 2001;276:2555–2561. doi: 10.1074/jbc.M008982200. [DOI] [PubMed] [Google Scholar]
- 15.Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, et al. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBOJ. 2000;19:4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosorius O, Reichart B, Kratzer F, Heger P, Dabauvalle MC, Hauber J, et al. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci. 1999;112(Pt 14):2369–2380. doi: 10.1242/jcs.112.14.2369. [DOI] [PubMed] [Google Scholar]
- 17.Bartig D, Klink SHF. The unique posttranslational modification leading to deoxyhypusine or hypusine is ageneral feature of the Archebacterial kingdom. Syst Appl Microbiol. 1990;13:112–116. [Google Scholar]
- 18.Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohl T, Klier H, Ammer H, Lottspeich F, Magdolen V. The HYP2 gene oisaccha-romyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. Mol Gen Genet. 1993;241:305–311. doi: 10.1007/BF00284682. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki K, Abid MR, Miyazaki M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae . FEBS Lett. 1996;384:151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- 21.Chawla B, Jhingran A, Singh S, Tyagi N, Park MH, Srinivasan N, et al. Identification and characterization of a novel deoxyhypusine synthase in Leishmania donovani . J Biol Chem. 2010;285:453–463. doi: 10.1074/jbc.M109.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen S, Jones DC, Wyllie S, Fairlamb AH, Phillips MA. Allosteric activation of trypanosomatid deoxyhypusine synthase by a catalytically dead paralog. J Biol Chem. 2013 May;288(21):15256–15267. doi: 10.1074/jbc.M113.461137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Njuguna JT, Nassar M, Hoerauf A, Kaiser AE. Cloning, expression and functional activity of deoxyhypusine synthase from Plasmodium vivax. BMC Microbiol. 2006;16(6):91. doi: 10.1186/1471-2180-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwentke A, Krepstakies M, Mueller AK, Hammerschmidt-Kamper C, Motaal BA, Bernhard T, et al. In vitro and in vivo silencing of plasmodial dhs and elf-5a genes in a putative, non-canonical RNAi-related pathway. BMC Microbiol. 2012 Jun;12:107. doi: 10.1186/1471-2180-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weir BA, Yaffe MP. Mmd1p, a novel, conserved protein essential for normal mitochondrial morphology and distribution in the fission yeast Schizosaccha-romyces pombe . Mol Biol Cell. 2004;15:1656–1665. doi: 10.1091/mbc.E03-06-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel PH, Costa-Mattioli M, Schulze KL, Bellen HJ. The drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J Cell Biol. 2009;185:1181–1194. doi: 10.1083/jcb.200904161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umland TC, Wolff EC, Park MH, Davies DR. A new crystal structure of deoxy-hypusine synthase reveals the configuration of the active enzyme and of an enzyme.NAD. Inhibitor ternary complex. J Biol Chem. 2004;279:28697–28705. doi: 10.1074/jbc.M404095200. [DOI] [PubMed] [Google Scholar]
- 28.Chawla B, Kumar RR, Tyagi N, Subramanian G, Srinivasan N, Park MH, et al. A unique modification of the eukaryotic initiation factor 5A shows the presence of the complete hypusine pathway in Leishmania donovani . PLoS ONE. 2012;7:33138. doi: 10.1371/journal.pone.0033138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen KY, Dou QP. NAD+ stimulated the spermidine-dependent hypusine formation on the 18 kDa protein in cytosolic lysates derived from NB-15 mouse neuroblastoma cells. FEBS Lett. 1988;229:325–328. doi: 10.1016/0014-5793(88)81149-9. [DOI] [PubMed] [Google Scholar]
- 30.Murphey RJ, Gerner EW. Hypusine formation in protein by a two-step process in cell lysates. J Biol Chem. 1987;262:15033–15036. [PubMed] [Google Scholar]
- 31.Abbruzzese A, Park MH, Folk JE. Deoxyhypusine hydroxylase from rat testis. Partial purification and characterization. J Biol Chem. 1986;261:3085–3089. [PubMed] [Google Scholar]
- 32.Jakus J, Wolff EC, Park MH, Folk JE. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono-guanylated diamines and polyamines. J Biol Chem. 1993;268:13151–13159. [PubMed] [Google Scholar]
- 33.Arrowood MJ, Sterling CR. Isolation of cryptosporidium oocysts and sporo-zoites using discontinuous sucrose and isopycnic Percoll gradients. J Parasitol. 1987;73:314–319. [PubMed] [Google Scholar]
- 34.Kilani RT, Sekla L. Purification of cryptosporidium oocysts and sporozoites by cesium chloride and Percoll gradients. Am J Trop Med Hyg. 1987;36:505–508. doi: 10.4269/ajtmh.1987.36.505. [DOI] [PubMed] [Google Scholar]
- 35.Upton SJ, Tilley M, Brillhart DB. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol Lett. 1994;118:233–236. doi: 10.1111/j.1574-6968.1994.tb06833.x. [DOI] [PubMed] [Google Scholar]
- 36.Upton SJ, Tilley M, Nesterenko MV, Brillhart DB. A simple and reliable method of producing in vitro infections of Cryptosporidium parvum (Apicomplexa) FEMS Microbiol Lett. 1994;118:45–49. doi: 10.1111/j.1574-6968.1994.tb06801.x. [DOI] [PubMed] [Google Scholar]
- 37.Magrane M, Consortium U. UniProt Knowledgebase: A Hub of Integrated Protein Data. Oxford: Database; 2011. [bar009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucl Acids Res. 2010;38:D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Logan-Klumpler FJ, De Silva N, Boehme U, Rogers MB, Velarde G, McQuillan JA, et al. GeneDB-an annotation database for pathogens. Nucl Acids Res. 2012;40:D98–D108. doi: 10.1093/nar/gkr1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff EC, Lee SB, Park MH. Assay of deoxyhypusine synthase activity. Methods Mol Biol. 2011;720:195–205. doi: 10.1007/978-1-61779-034-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff EC, Wolff J, Park MH. Deoxyhypusine synthase generates and uses bound NADH in a transient hydride transfer mechanism. J Biol Chem. 2000;275:9170–9177. doi: 10.1074/jbc.275.13.9170. [DOI] [PubMed] [Google Scholar]
- 44.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;9 doi: 10.1002/0471140864.ps0209s50. [Ch 2: Unit 2] [DOI] [PubMed] [Google Scholar]
- 45.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using modeller. Curr Protoc Bioinformatics. 2006;6 [Ch 5: Unit 5] [Google Scholar]
- 46.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van DerSpoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GRO-MACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 48.DeLano WL. The Pymol Molecular Graphics System. 2002 [Google Scholar]
- 49.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 50.David Dolphin OA, Rozanne Poulson. Pyridine nucleotide coenzymes: chemical, biochemical, and medical aspects, Part 1. New York: John Wiley & Sons; 1987. [Google Scholar]
- 51.Liao DI, Wolff EC, Park MH, Davies DR. Crystal structure of the NAD complex of human deoxyhypusine synthase: an enzyme with a ball-and-chain mechanism for blocking the active site. Structure. 1998;6:23–32. doi: 10.1016/s0969-2126(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 52.Tina KG, Bhadra R, Srinivasan N. PIC: protein interactions calculator. Nucl Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Njuguna JT, Nassar M, Hoerauf A, Kaiser AE. Cloning, expression and functional activity of deoxyhypusine synthase from Plasmodium vivax. BMC Microbiol. 2006;6:91. doi: 10.1186/1471-2180-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joe YA, Wolff EC, Park MH. Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins.J Biol Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- 55.Tao Y, Chen KY. Molecular cloning and functional expression of Neurospora deoxyhypusine synthase cDNA and identification of yeast deoxyhypusine synthase CDNA. J Biol Chem. 1995;270:23984–23987. doi: 10.1074/jbc.270.41.23984. [DOI] [PubMed] [Google Scholar]
- 56.Brochier C, Lopez-Garcia P, Moreira D. Horizontal gene transfer and archael origin of deoxyhypusine synthase homologous genes in bacteria. Gene. 2004;330:169–176. doi: 10.1016/j.gene.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 57.Kang KR, Wolff EC, Park MH, Folk JE, Chung SI. Identification of YHR068w in Saccharomyces cerevisiae chromosome VIII as a gene for deoxyhypusine synthase. Expression and characterization of the enzyme. J Biol Chem. 1995;270:18408–18412. doi: 10.1074/jbc.270.31.18408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.