Abstract
NPF (formerly referred to as low-affinity NRT1) and ‘high-affinity’ NRT2 nitrate transporter genes are involved in nitrate uptake by the root, and transport and distribution of nitrate within the plant. The NPF gene family consists of 53 members in Arabidopsis thaliana, however only 11 of these have been functionally characterized. Although homologous genes have been identified in genomes of different plant species including some cereals, there is little information available for wheat (Triticum aestivum). Sixteen genes were identified in wheat homologous to characterized Arabidopsis low-affinity nitrate transporter NPF genes, suggesting a complex wheat NPF gene family. The regulation of wheat NFP genes by plant N-status indicated involvement of these transporters in substrate transport in relation to N-metabolism. The complex expression pattern in relation to tissue specificity, nitrate availability and senescence may be associated with the complex growth patterns of wheat depending on sink/source demands, as well as remobilization during grain filling.
Key words: Triticum aestivum, nitrate, transport, nutrition, senescence.
Introduction
Along with the initial uptake of nitrate by the root, the distribution of nitrate and N-containing metabolites between different organs and tissues is a complex process influenced by growth and development of a plant. For cereal crop production, one of the most critical stages is post-anthesis grain filling, involving both de novo uptake from the soil and remobilization of previously acquired N. In wheat and barley, up to 90% of the nitrogen is remobilized from the vegetative plant parts to the grain (Gregersen et al., 2008). The importance of nitrate transporter genes in determining overall efficiency is not well characterized.
Nitrate transporters have been best characterized in the model plant Arabidopsis. Plant nitrate transporters are divided into the NPF (former ‘low-affinity’ NRT1/PTR) and ‘high-affinity’ NRT2 gene families (Tsay et al., 2007; Léran et al., 2014). The Arabidopsis genome contains 53 NPF genes and 7 NRT2 genes (Tsay et al., 2007). The Arabidopsis NRT2 gene family is well characterized; most of the NRT2 genes are expressed in the root with the exception of AtNRT2.4 and AtNRT2.5, which are also expressed in leaves, and AtNRT2.7, which seems to be leaf-specific (Orsel et al., 2002). Mutant analysis of most NRT2 genes in Arabidopsis indicates involvement in nitrate transport (Cerezo et al., 2001; Wang et al., 2012). Gene expression of many NRT2 genes is regulated by nitrate availability and other factors (Zhuo et al., 1999; Orsel et al. 2002; Orsel et al., 2006).
Functional ‘high-affinity’ nitrate transport requires a second protein, Nar2, together forming a two-component nitrate uptake system (Zhou et al., 2000; Tong et al., 2006; Orsel et al., 2006).
As the NRT1/PTR gene family encompasses transporters for different molecules, including nitrate, peptides, amino acids, auxins, abscisic acid and glucosinolates (Tsay et al., 2007; Nour-Eldin et al., 2012; Krouk et al., 2010; Kanno et al., 2012; Chen et al., 2012), it is termed the NPF (‘NRT1-PTR-Family’). Phylogenetic analysis indicates eight subfamilies numbered NPF1–NPF8 (Léran et al., 2014). With the exception of AtNPF6.3, which is a dual-affinity (low and high) nitrate transporter (Liu et al., 1999), most of the NPF transporters characterized are ‘low affinity’. AtNPF6.3 is expressed predominantly in the root (Tsay et al., 2007) and is regulated by the N-status, but it is also expressed in guard cells (Guo et al., 2003) and influences stomatal function. AtNPF6.3 has bidirectional transport activities involved in root-to-shoot nitrate translocation (Léran et al., 2013). In addition to root dual-affinity nitrate uptake, AtNPF6.3 also functions as a nitrate sensor (Ho et al., 2009) with the switch between functions regulated by phosphorylation (Liu and Tsay, 2003; Ho et al., 2009). The nitrate signalling function of AtNPF6.3 is crucial for root architecture, influencing lateral root development (Forde, 2002). The control of lateral root development seems to be based on the dual nitrate/auxin transport activity, whereby high nitrate concentrations suppress auxin transport, which promotes lateral root development, and vice versa (Krouk et al., 2010). AtNPF4.6 is also involved in low-affinity root nitrate uptake. In contrast to AtNPF6.3, which has reduced expression under N-starvation and is inducible by nitrate resupply (Tsay et al., 1993), AtNPF4.6 is constitutively expressed.
Other Arabidopsis NPF transporters seem to be involved in the distribution of nitrate within the plant. For example, after uptake, the first step is loading nitrate into xylem vessels for root-to-shoot nitrate translocation, and AtNPF7.3 may fulfil this role (Lin et al., 2008). Furthermore NFP7.3 Arabidopsis mutants have reduced nitrate content in the xylem sap with reduced nitrate translocation to the shoots, supporting the role of NFP7.3 in root nitrate xylem loading (Lin et al., 2008). Two other NFP transporters, AtNPF7.2 and AtNPF2.9, may be involved in root-to-shoot nitrate translocation; in contrast to AtNPF7.3, AtNPF7.2 and AtNPF2.9 mutants showed enhanced root-to-shoot nitrate transport. The plasma membrane-located AtNPF7.2 is expressed mainly in the xylem parenchyma cells of roots, indicating a function in removing nitrate from the xylem back into the root cells (Li et al., 2010). Expression of AtNPF2.9 was detected in companion cells of the root phloem (Wang and Tsay, 2011). In Arabidopsis, two genes, At5G62680 and At3G47960, are phylogenetically closely related to AtNPF2.9. Functional analysis revealed that both transporters are capable of transporting glucosinolates; both gene products AtNPF2.10 and AtNPF2.11 have been shown to be plasma membrane located with cellular localization in adjacent mesophyll cells and leaf veins, respectively. Mutant analysis indicates a role in the transport of glucosinolates from source tissues to the grain (Nour-Eldin et al. 2012). The Arabidopsis NPF transporters AtNPF6.2 and AtNPF2.13 may have a role in leaf distribution of nitrate: AtNPF6.2 is mainly expressed in the leaf petioles and mutation of AtNPF6.2 reduces the nitrate content in the petiole and increases the nitrate content in the leaf lamina (Chiu et al., 2004). AtNPF2.13 is expressed in the phloem of older leaves and may be important for the source-to-sink remobilization of nitrate by allocation of nitrate from older to younger leaves (Fan et al., 2009). Recent data suggested that NPF1.1 and NPF1.2 are involved in xylem-to-phloem transfer to enable redistribution of nitrate into developing leaves, a critical step for optimal plant growth (Hsu and Tsay, 2013).
The plasma membrane-localized AtNPF2.12 is exclusively expressed in the vascular bundle of the siliques, suggesting involvement in nitrate transport to the developing seeds (Almagro et al., 2008).
Nitrogen remobilization during senescence in cereals is an important factor influencing grain quality and yields, however the extent to which NPF transporters determine the efficiency of these processes is unknown. In the rice (Oryza sativa) genome there are at least 80 genes belonging to the NPF family but few have been characterized. Modern wheat varieties have a high degree of nutrient use efficiency, and nitrogen mobilization from old leaves to new leaves and into the grain contribute to efficiency. This report identifies and characterizes the wheat NPF gene orthologues to the well characterized selected Arabidopsis NRT1 genes. Gene expression studies in relation to tissue specificity, N-status of wheat plants, as well as post-anthesis leaf senescence provides a first insight into the complex composition of wheat members of the NPF transporter gene family.
Materials and methods
Identification of wheat NFP genes in the wheat genome and phylogenetic analysis
Gene and mRNA sequences of the orthologous and phylogenetically closely related Brachypodium distachyon and rice sequences (Table 1) were used in a Blast analysis (Altschul et al., 1997; EMBL/NCBI; DFCI Gene index; Graingenes; CerealsDB; IWGSC survey sequence). The genomic exon-intron structures and chromosome localizations were identified by alignment of the derived sequences to the CerealsDB and the IWGSC survey sequence databases (the original CerealsDB sequence reads are archived in SRA of the European Nucleotide Archive (ENA) with Study ID ERP000319). For wheat sequence accessions where only incomplete coding regions have been identified, the complete coding sequence and the genomic structure were derived by genomic blast walking and assembling using the wheat genome CerealsDB (for NRT1 D-genome sequences see Supplementary Table S1) and IWGSC wheat genome survey sequence databases.
Table 1.
Symbol numbers and genome gene ID of selected Arabidopsis thaliana, Brachypodium distachyon and Oryza sativa (ssp japonica cv Nipponbare apart from OsNPF6.7 for which the ssp Indica annotation number was used) NFP genes
| Arabidopsis thaliana | Brachypodium distachyon | Oryza sativa | |||
|---|---|---|---|---|---|
| Symbol | TAIR Gene ID | Symbol | Brachypodium.org Gene ID | Symbol | MSU Gene ID |
| AtNPF6.3 | At1g12110 | BdNPF6.3 BdNPF6.4 BdNPF6.5 BdNPF6.6 |
Bradi3g16670 Bradi1g78330 Bradi3g33040 Bradi3g33030 |
OsNPF6.3 OsNPF6.5 OsNPF6.4 |
Loc_Os08g05910 Loc_Os10g40600 Loc_Os03g01290 |
| AtNPF4.6 | At1g69850 | BdNPF4.11 | Bradi1g37330 | OsNPF4.11 | Loc_Os06g38294 |
| AtNPF6.4 | At3g21670 | BdNPF6.7 | Bradi3g47010 | OsNPF6.7 OsNPF6.6 |
Os02g35830 Loc_Os04g39030 |
| AtNPF6.2 | At2g26690 | BdNPF6.2 | Bradi2g41060 | OsNPF6.2 | Loc_Os01g37590 |
| AtNPF7.3 | At1g32450 | BdNPF7.10 BdNPF7.11 |
Bradi3g52096 Bradi3g53380 |
OsNPF7.9 OsNPF7.10 OsNPF7.11 |
Loc_Os02g46460 Loc_Os06g21900 Loc_Os02g48570 |
| AtNPF2.12 | At1g27080 | N/A | N/A | N/A | N/A |
| AtNPF2.13 | At1g69870 | BdNPF2.6 | Bradi2g58470 | OsNPF2.5 | Loc_Os01g68510 |
| AtNPF7.2 | At4g21680 | N/A | N/A | N/A | N/A |
| AtNPF2.9A | At1g18880 | BdNPF2.4 | Bradi4g00530 | OsNPF2.2 OsNPF2.3 |
Loc_Os12g44100 Loc_Os12g44110 |
| AtNPF2.10 | At3g47960 | N/A | N/A | N/A | N/A |
| AtNPF2.11 | At5g62680 | N/A | N/A | N/A | N/A |
| AtNPF1.2 | At1g52190 | BdNPF1.2 | Bradi2g50580 | OsNPF1.2 | Loc_Os01g55610 |
Phylogenetic analysis was performed by multiple protein sequence alignment using ClustalX V. 2.1 (Larkin et al., 2007). MEGA 5.05 (Tamura et al., 2011) was used for calculation of phylogenetic trees [the neighbour-joining method (Saitou and Nei, 1987)]. Bootstrap values for the trees were calculated as a percentage of 1000 trials with a seed number for the random number generator of 1000 (Felsenstein, 1985). The evolutionary distances (expressed as number of amino acid differences per site) used the number of differences method (Nei and Kumar, 2000).
Plant material
The plant material for total RNA isolation was from hydroponically grown wheat (cv. Paragon), and from field-grown wheat (cv. Hereward) with 200kg N ha–1 (as ammonium nitrate) in triplicate repetition [Stackyard field (medium loam) at Rothamsted Research, Harpenden, UK, in 2006/7]. In field trials, the wheat varieties cv. Paragon and cv. Hereward exhibit similar characteristics in relation to N-uptake, N-remobilisation and grain N-utilization efficiencies (Barraclough et al., 2010; Barraclough et al., 2014). The spring wheat, cv. Paragon, requiring no vernalization, was used for all hydroponic experiments and the winter wheat cv. Hereward in field trials.
For the N-induction experiment, wheat was grown hydroponically in a modified Letcombe nutrient solution [1.5mM Ca(NO3)2, 5mM KNO3, 2mM NaNO3, 1mM MgSO4, 1mM KH2PO4, 25 μM FeEDTA, 160nM Cu(NO3)2, 9.2μM H3BO3, 3.6 µM MnCl2, 16nM Na2MoO4, 5 µM KCl and 770nM ZnCl2; Drew and Saker, 1984] under sufficient nitrate supply in a growth chamber and 16h/8h light/dark daily cycle. Nutrient solutions were exchanged three times a week. Two weeks after germination, plants (apart from the +N control plants) were N-starved for 1 week by replacement of nitrate salts with the corresponding chloride salt. After 1 week of N-starvation, N-induction was initiated by placing N-starved plants back into full nitrate nutrient solution (apart from –N control plants). Samples were taken at day 0, 3, 5 and 8 (+N) and at day 3, 5 and 8 (–N), and 30min, 1h, 2h, 4h, 8h and 24h after nitrate induction. Roots were washed, dried and frozen immediately in liquid nitrogen. Whole shoots were harvested, frozen immediately in liquid nitrogen, and stored at –80°C.
For field-grown wheat, the second leaf below the ear from 10 primary shoots per plot was harvested from anthesis, weekly until complete senescence. The leaves were frozen in liquid nitrogen and stored at –80°C.
The relative chlorophyll content was monitored in the middle part of at least 10 leaves using a Soil Plant Analysis Development (SPAD) meter (SPAD-502, Minolta, Japan) to monitor senescence. All plant materials were homogenized using a SPEX freezer mill (SPEX CertiPrep Ltd, UK) in liquid nitrogen, aliquoted into 2ml micro-tubes and stored at –80°C.
Plant total RNA isolation
Total RNA was isolated by a modified method based on Verwoerd et al. (1989) including additional phenol-chloroform-isoamyl alcohol extractions. Possible genomic DNA contamination was removed by RNase-free DNase treatment. The final air-dried pellet was dissolved in an appropriate volume of RNase-free water.
Nitrate content analysis
Ground, freeze-dried root and shoot samples (20–30mg sample–1) were extracted at 80°C in de-ionised water. After centrifugation and passage through a 0.2 µm filter, nitrate was measured on a Skalar Continuous Flow Analyser (Skalar SANPLUS System, Skalar, UK).
Reverse transcriptase – real-time PCR transcript analysis
Gene expression was analysed by quantitative and relative real-time PCR. First-strand cDNA synthesis was performed from 2 µg total RNA and dT-adapter primer (Invitrogen Superscript III; standard protocol, 2h synthesis time). Real-time PCR was performed using the Applied Biosystems 7500 Real Time PCR System and the SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma-Aldrich, UK). The 25 μl reactions contained 1 µl cDNA and 250nM of each primer. Partly degenerated primer combinations were used to cover gene expression of the genes from all three wheat genomes (Supplementary Table S2). The primer Tm and PCR conditions should allow binding of the primer as long as the wheat variety sequences analysed differed by one or two base sequence polymorphisms. Due to the sequence similarity of TaNPF2.4/2.5, a useful primer for real-time PCR could only be generated for the amplification of both isoform transcripts. Primer efficiency was analysed and only primer combinations were used with primer efficiencies between 85 and 115%. For greater accuracy, the mean primer efficiency was estimated using the linear phase of all individual reaction amplification curves (Ramakers et al., 2003) calculated by using the LinRegPCR package (Tuomi et al., 2010). By comparison of different constitutive normalization control wheat genes the Actin 3 gene showed the best performance and was used for the normalized relative quantification of expression. The normalized relative quantity (NRQ) of expression was calculated in relation to the CT values and the primer efficiency (E) of the target gene (X) and the normalizing reference gene (N) as Normalized Relative Expression (NRE) based on Rieu and Powers, 2009:
Statistically significant changes in relation to N-starvation/N-induction and time course of harvests were calculated by analysis of variance (ANOVA) of the log2-transformed NRE data (Gomez and Gomez, 1984).
For verification of transcripts, approximately 500-bp long cDNA-derived PCR fragments (Supplementary Table S3), genomic PCR fragments (for nonexpressed NRT1 genes), and corresponding real-time PCR fragments (data not shown) of the individual NRT1 genes were cloned into pGEM-Teasy (Promega, UK), sequenced (Eurofins, Germany), and submitted to EMBL (Supplementary Table S3). For each NPF gene analysed, quantitative real-time SYBR green PCR expression analysis in young roots and shoots was performed including a standard dilution series of each plasmid-PCR fragment in triplicate. Based on the molecular weight of plasmid and PCR fragments, the mRNA copy number per μl cDNA was calculated after actin normalization of CT values. The significance of differences in transcript copy number was analysed by ANOVA.
Results
Phylogenetic relationships of the putative wheat low-affinity nitrate transporters
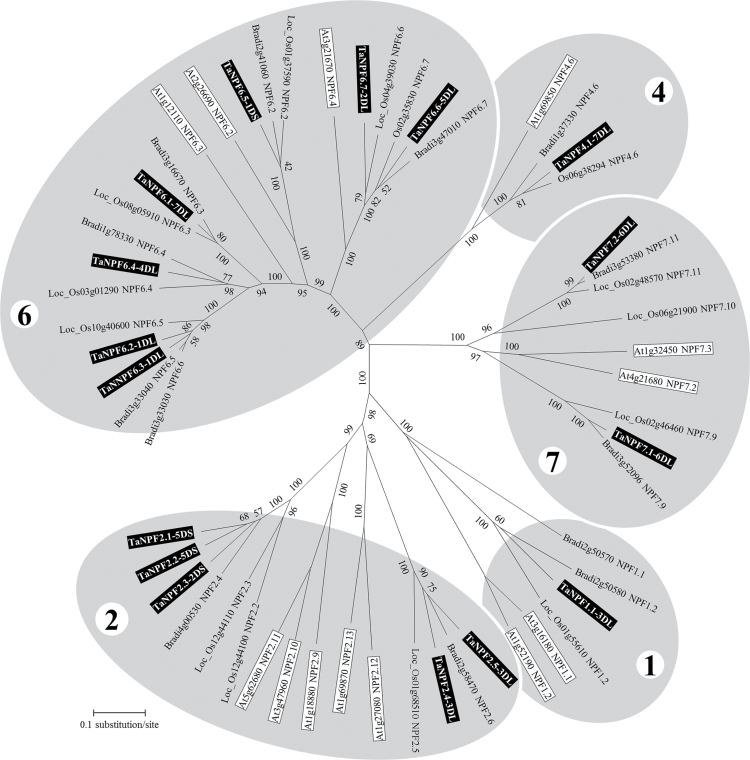
In Arabidopsis, 12 NPF transporter genes are well characterized (Table 1) with functional data for nitrate transport activity (for review, see Wang et al., 2012; Tsay et al., 2007; Léran et al., 2014). By sequence database BLAST analysis, orthologous full-length sequences and other phylogenetically closely related wheat NPF genes and their coding regions were identified (Table 2; Table S1 for all CerealsDB and IWGSC survey D-genome sequences). The symbol numbering used follows that suggested by Léran et al., 2014.
Table 2.
Wheat NPF and other wheat gene (analysed by gene expression) accession numbers of databases sequences
| Wheat gene symbol | IWGSC Wheat chromosome localization | Accession numbers |
|---|---|---|
| TaNPF1.1 | 3AL/B/DL | BQ806518;TaAffx.52263.1.S1_at;HF545002 |
| TaNPF2.1 | 5AS/BS/DS | AK334628;TC280584;HF544999 |
| TaNPF2.2 | 5AS/BS/DS | BJ244453;CV780655;BJ250261;HF545000 |
| TaNPF2.3 | 2AS/BS/DS | HF545001 |
| TaNPF2.4 | 3AL/B/DL | TC400476;HF544994 |
| TaNPF2.5 | 3AL/B/DL | HF544995 |
| TaNPF4.1 | 7AL/BL/DL | CA732431;CK207315;TC432669;HF544989 |
| TaNPF6.1 | 7AL/BL/DL | AK333802;HF544985 |
| TaNPF6.2 | 1AL/BL/DL | BJ279931;TC401315;GH722017;HF544986 |
| TaNPF6.3 | 1AL/BL/DL | BG907608;TC391493;HF544987 |
| TaNPF6.4 | 5AL/4BS/4DL | HF544988 |
| TaNPF6.5 | 1AS/BS/DS | AK330268;HF544991 |
| TaNPF6.6 | 5AL/BL/DL | AK332369;HF544990 |
| TaNPF6.7 | 2AL/BL/DL | HF545004 |
| TaNPF7.1 | 6AL/BL/DL | BJ279017;HX175323; TC380559;HF544992 |
| TaNPF7.2 | 6AL/BL/DL | TC334619;BU099863;TC433104;HF544993 |
| TaActin (3 like) | 5AL/BL/DL | Ta.28253.1.S1_at;TC441720 |
| TaSAG12 | 2AL/BL/DL | AB267407;TC232300 |
| TaRubiscoSSU | 5AL/BL/DL | Ta.27923.2.S1_x_a;TC263601 |
| TaGS1 | 6AL/BL/DL | DQ124209;DQ124210;DQ124211 |
| TaGS2 | 2AL/BL/DL | DQ124212;DQ124213;DQ124214 |
| TaGSe | 4AS/BS/DS | AY491970;AY491971 |
| TaGSr | 4AS/BS/DS | AY491968;AY491969 |
| TaGDH2 | 2AL/BL/DL | AK331666;TC266053 |
| TaNR1 | 6AS/DS/ BS? | TC236448;Ta.5633.1.S1_at;AL825459;AK333426 |
| TaNAM | 6AS/BS/DS | DQ869673;DQ869672;DQ869675 |
| TaNIR | 6DL/AL?/BL? | FJ527909;TC392193; HM989894 |
Phylogenetic analysis of the 16 identified wheat NPF D-genome proteins in comparison to the Arabidopsis, Brachypodium and rice orthologous and homologous proteins (gene ID Table 1) showed that they can be classified into the NPF subfamilies 1, 2, 4, 6 and 7 (Fig. 1 – group numbers for grey areas; Léran et al., 2014). As in Brachypodium, four wheat homologous proteins, TaNPF6.1, TaNPF6.2, TaNPF6.3, and TaNPF6.4, were identified as co-orthologous to Arabidopsis AtNPF6.3. The orthologous wheat NPF6.2 and NPF6.3 genes are both located on the long arm of chromosome 1D. Although TaNPF6.1, as with the Brachypodium and rice NPF6.3 proteins, has the shortest phylogenetic distance to Arabidopsis NPF6.3 compared to the other three co-orthologous proteins, a clear orthologous relationship was not verified, in agreement with Plett et al., 2010. In the Brachypodium genome, as in Arabidopsis, only one NPF6.4 orthologous gene exists. In wheat, two co-orthologous genes, TaNPF6.6 and TaNPF6.7, were identified in the D-genome, and they are closely related to the orthologous genes OsNPF6.6 and OsNPF6.7 in rice. Both wheat genes are located on different chromosomes (Fig. 1). As in rice and Brachypodium, only one gene orthologous to AtNPF6.2, TaNPF6.5, was identified in the wheat D-genome (Fig. 1).
Fig. 1.
Evolutionary radial relationships of the wheat NPF proteins to related NPF proteins from Arabidopsis thaliana, Oryza sativa, and Brachypodium distachyon (gene ID Table 1). Neighbour-joining evolutionary tree analyses (Saitou and Nei, 1987) were conducted in Mega5 (Tamura et al., 2011) from the multiple alignment (ClustalX V.2.1; Larkin et al., 2007) of the proteins of selected Triticum aestivum, A. thaliana, O. sativa, (ssp. japonica cv. Nipponbare, a part of OsNPF6.7 for which the ssp. Indica annotation number was used), and B. distachyon NPF genes. The TAIR genome, MSU, and Brachyposium.org IDs were used for the Arabidopsis (square frame, white highlighting), rice, and Brachypodium sequences, respectively, including the NPF numbering. For simplification only D-genome-located wheat protein sequences (square frame, black highlighting) were used. The chromosome number and chromosome arm location (short-S; long-L) follows the NPF numbering. Each group number (grey areas) represents the NPF subfamily number. Bootstrap values were calculated as a percentage of 1000 trials with a seed number for the random number generator of 1000 (Felsenstein, 1985). The evolutionary distances (number of amino acid differences per site) used the number of differences method (Nei and Kumar, 2000). The analysis involved 56 protein sequences. All positions containing gaps and missing data were eliminated. There was a total of 314 positions in the final dataset.
AtNPF4.6 of subfamily 4 is represented in wheat as well as in rice and Brachypodium by one orthologous gene, TaNPF4.1.
The Arabidopsis proteins AtNPF7.3 and AtNPF7.2 are phylogenetically closely related and located in subfamily 7 (Fig. 1). In wheat, two genes were identified which are phylogenetically closely related. Both wheat genes are orthologous to Brachypodium BradiNPF7.9 and BradiNPF7.11 and the three rice genes OsNPF7.9, OsNPF7.10, and OsNPF7.11 (Fig. 1). The phylogenetic distance of the TaNPF7.1 protein is closer to both Arabidopsis proteins than the TaNPF7.2 protein, but as already described in Plett et al. (2010), an orthologous relationship is not clear.
The Arabidopsis proteins for NPF2.12, NPF2.13, NPF2.9, NPF2.10, and NPF2.11 belong to the NPF subfamily 2 (Fig. 1). The wheat TaNPF2.4 and TaNPF2.5 proteins are phylogenetically related to AtNPF2.12 and AtNPF2.13 from Arabidopsis. Plett et al. (2010) suggested that Brachypodium BradiNPF2.6 and rice OsNPF2.5 are homologous to Arabidopsis AtNPF2.13, and that there may be no homologous/orthologous gene in either species related to Arabidopsis AtNPF2.12. Both TaNPF2.4 and TaNPF2.5 proteins from wheat are closely related to BradiNPF2.6, which suggests that there are two co-orthologous genes in wheat. The location of both genes on the long arm of chromosome 3D suggests gene duplication (Fig. 1).
Plett et al. (2010) did not include the Arabidopsis NPF genes for At1g18880 (AtNPF2.9), At3g47960 (AtNPF2.10) and At5g62680 (AtNPF2.11). Tsay et al. (2007) reported nitrate transporter activity for the AtNPF2.9 and AtNPF2.11 gene products. Further analysis by Nour-Eldin et al. (2012) indicated glucosinolate transporting capability for both AtNPF2.10 and AtNPF2.11. In the Brachypodium genome, only one gene, Bradi4g00530 (BradiNPF2.4), showed a close phylogenetic relationship with the Arabidopsis NPF2.9, NPF2.10, and NPF2.11 group. On chromosome 12 of the rice genome, there are two neighbouring genes, Os12g44100 and Os12g44110, whose gene products have the closest phylogenetic relationships. In the wheat D-genome there are three genes phylogenetically closely related to Arabidopsis NPF2.9, NPF2.10 and NPF2.11. Both TaNPF2.1 and TaNPF2.2 are located on the short arm of chromosome 5D, suggesting gene duplication, and TaeNPF2.3 is located on the short arm of chromosome 2D (Fig. 1).
The wheat NPF1.1 protein is similar to Brachypodium BradiNPF1.2 and rice OsNPF1.2, and all are orthologous to Arabidopsis NPF1.1 and NPF1.2 (Fig.1).
Quantitative gene expression of NPF in roots and shoots of wheat seedlings
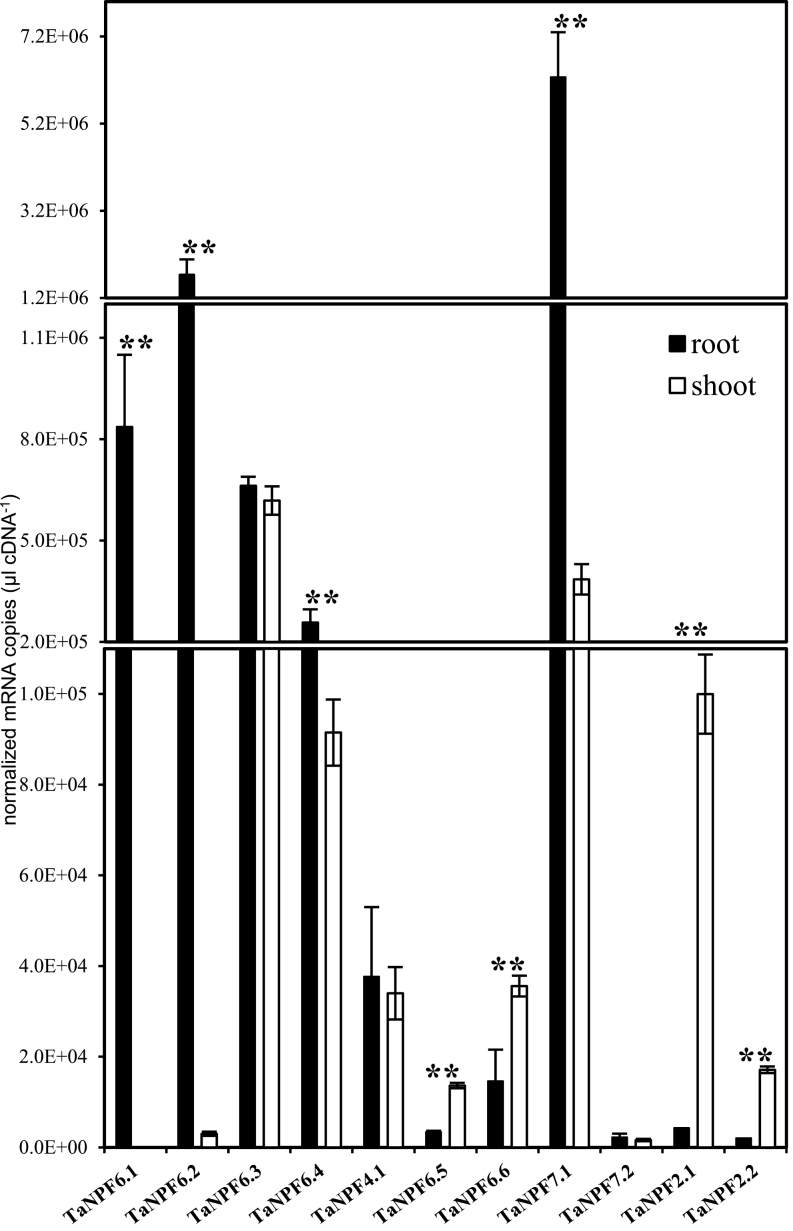
Profiles of the expression patterns in the root and shoot of young wheat plants 3 weeks after germination and grown hydroponically under sufficient nitrate nutrition (+N control plants of the N-starvation experiment) provide the first information on tissue specificity or dominance of the individual NPF transporters.
Expression of the four wheat genes NPF6.1 to 6.4, co-orthologous to AtNPF6.3, in the root and shoot of young wheat was distinctive. TaNPF6.1 and TaNPF6.2 transcripts were present with high abundance in the roots and very low abundance in the shoots (Fig. 2). The transcript abundance of TaNPF6.3 was very similar in the roots and in the shoots, and TaNPF6.4 transcript was significantly higher in the roots but also with reasonable transcript levels in shoots (Fig. 2).
Fig. 2.
Quantitative real-time PCR expression analysis of putative low-affinity nitrate transporters TaNPF6.1, TaNPF6.2, TaNPF6.3, TaNPF6.4, TaNPF4.1, TaNPF6.5, TaNPF6.6, TaNPF7.1, TaNPF7.2, TaNPF2.1, and TaNPF2.2 in roots and shoots of young hydroponically grown wheat plants. Each bar represents the mean ±SE of at least three biological replicates. **, significant differences between root and shoot (P < 0.01) of individual NPF genes.
As for TaNPF6.3 and TaNPF6.4, the transcript level of TaNPF4.1 was very similar in the roots and shoots (Fig. 2), but the overall transcript copy number was much lower.
TaNPF7.1 showed the highest mRNA copy number in roots amongst all NPF genes analysed (Fig. 2). Although there was a dominant high transcript level of TaNPF7.1 in roots, the shoot transcript level was still reasonably high, and similar to the shoot transcript abundances of TaNPF6.3, TaNPF6.4 and TaNPF2.1. The transcript level of TaNPF7.2 was very similar in roots and shoots, but was very low compared to TaNPF7.1 (Fig. 2).
TaNPF6.6, TaNPF6.5, TaNPF2.1 and TaNPF2.2 were highly expressed in the shoot (Fig. 2). Whilst the root transcript level of TaNPF6.6 was still around 50% of the shoot level, the root mRNA copy levels of TaNPF6.5, TaNPF2.1 and TaNPF2.2 were 25%, 12% and 4.3%, respectively, much lower compared to the shoot. Although in the wheat genome a third co-orthologous/homologous gene TaNPF2.3 is present, so far no clear transcript could be detected.
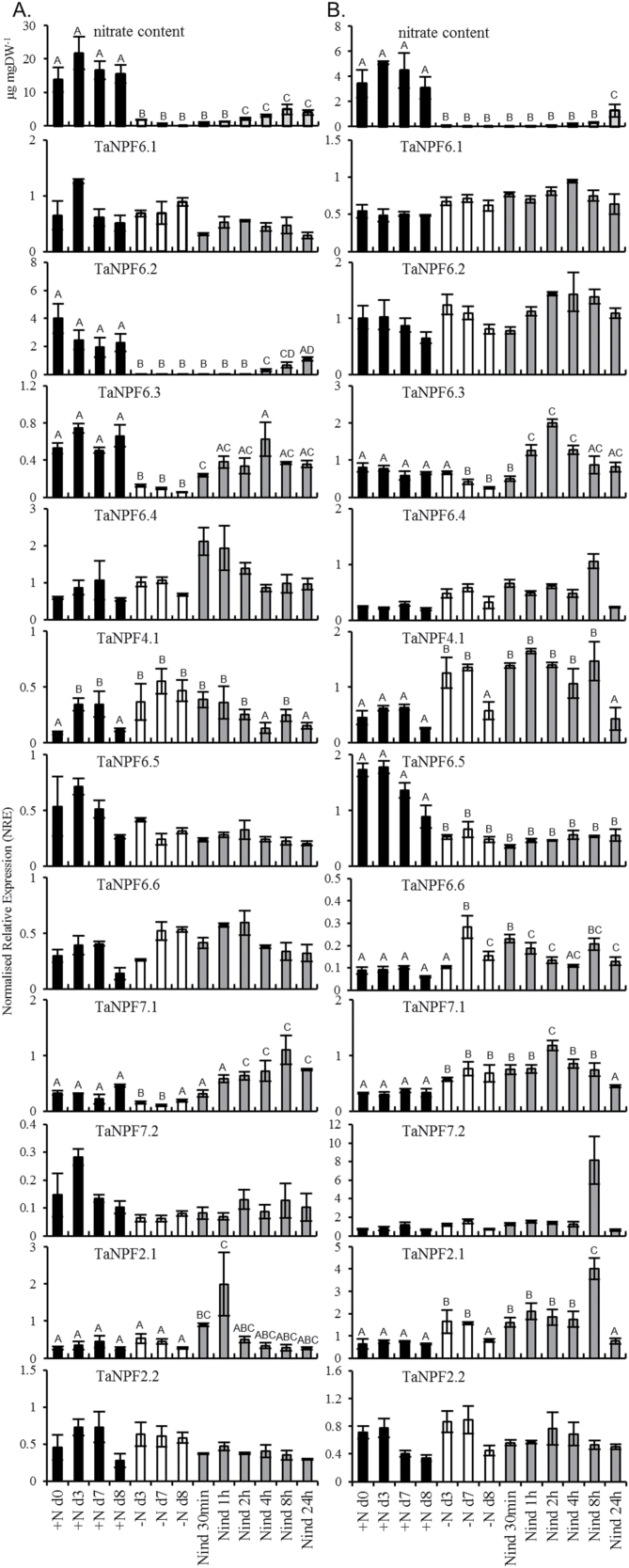
Influence of N-starvation and N-induction on the expression of NPF genes in roots and shoots
For some Arabidopsis NPF genes an influence of N-availability on gene expression has been reported (overview Wang et al., 2012). Three days’ nitrate starvation-deprivation of hydroponically grown wheat plants reduced the nitrate content in roots to 10% and in shoots to less than 1% compared to the controls, with further reduction to nearly zero in both with continuing starvation (Fig. 3). Nitrate resupply after 7 days of starvation resulted in a significant increase of nitrate in roots after 2h with a further increase until 24h, but without recovering to control content. In shoots a significant nitrate increase was only found after 24h of nitrate resupply (Fig. 3). The four wheat genes co-orthologous to AtNPF6.3 showed different patterns of expression in relation to the N-supply. In roots, TaNPF6.2 and TaNPF6.3 gene expression was clearly influenced by the N-status of the plants. The transcript of TaNPF6.2 was significantly reduced by N-starvation. After 3 days of N-starvation, only very low expression was detectable, and after nitrate resupply expression was induced very slowly, with a significant increase of expression 4h after nitrate induction and with a further increase until 24h after N-induction, reaching similar levels to control + nitrate culture (Fig. 3A). The expression of TaNPF6.3 was less down-regulated in roots by N-starvation compared to TaNPF6.2. With N-induction, the transcript level of TaNPF6.2 was restored after 1h to the average level found under sufficient N-supply (Fig. 3A). No influence, either by N-starvation or by N-induction, was detectable on expression of TaNPF6.1 and TaNPF6.4 genes in roots (Fig. 3A).
Fig. 3.
Influence of nitrate starvation and nitrate induction on the nitrate content and gene expression of wheat NPF genes in roots (A) and shoots (B) of young wheat plants. NRE, ‘Normalized Relative Expression’; black bars, + N treatment; white bars, – N-starvation; grey bars, nitrate induction. Each bar represents the mean ±SE of at least three biological replicates. Different letters on the top of the bars indicate significance of P < 0.05. Letters shared in common indicate no significant difference.
The expression of TaNPF7.1 was significantly reduced after 3 and 6 days of N-starvation. After nitrate resupply, the expression increased to the +N level within 30min with a further increase between 2h and 8h, significantly higher compared to the control culture, with a slight reduction after 24h (Fig. 3A).
The root expression of most of the other wheat NRT1 genes analysed was not influenced by nitrate starvation and nitrate induction (Fig 3A). TaNPF4.1 and TaNPF2.1 expression indicated non-N-supply related high variation with significant changes in the +N culture and/or N-starvation and N-induction (Fig. 3A).
There is little information about the influence of nitrate starvation and induction on the expression of NRT1 genes in vegetative shoot tissues. In Arabidopsis shoots, induction/up-regulation of gene expression was reported for AtNPF6.4 and AtNPF6.2 (Okamoto et al., 2003). In young wheat shoots the expression of several NPF genes is influenced by the N-supply. As in roots, TaNPF6.3 expression decreased in shoots with nitrate starvation, but much later compared to roots; however after 7 days of starvation the expression is reduced by around 50% compared to the initial/average level of the +N control culture (Fig. 3B). With nitrate resupply, TaNPF6.3 expression increased gradually to a maximum peak at 2h, which subsequently reduced gradually within 6h to the control +N level.
The expression of TaNPF4.1 was significantly up-regulated by nitrate starvation. With nitrate induction this higher expression level continued for almost 8h and decreased at 24h to the +N control level (Fig. 3B). With some minor differences a similar expression pattern was also found for TaNPF6.6, TaNPF7.1 and TaNPF2.1. In contrast to Arabidopsis, nitrate starvation in wheat resulted in a reduction of TaNPF6.5 expression. After 3 days of N-starvation, TaNPF6.5 expression decreased significantly to around 30% of the +N control level. This lower expression did not recover during nitrate resupply, but the +N control expression also decreased at day 8, suggesting a possible additional developmental influence.
Influence of post-anthesis senescence on the leaf expression of wheat NPF genes
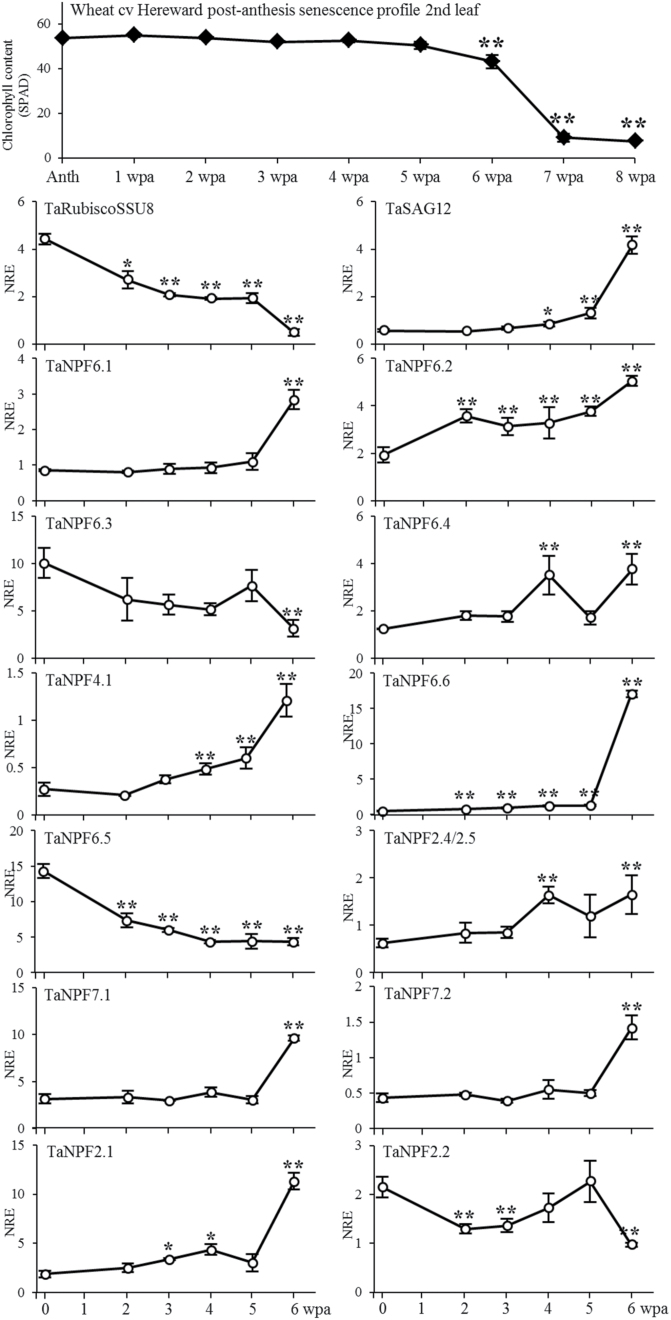
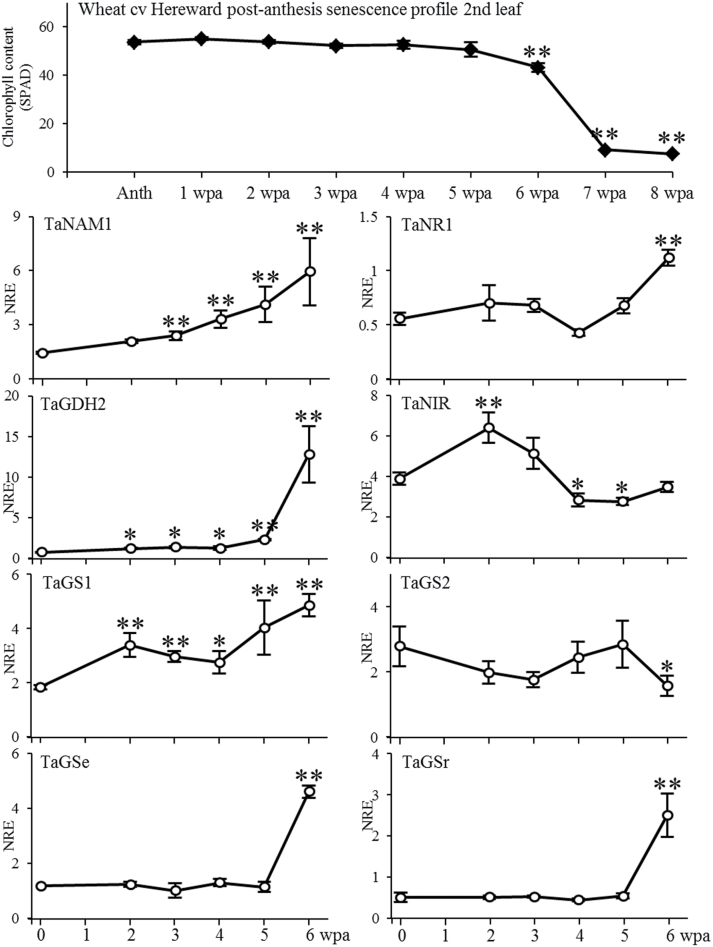
During wheat grain development the nitrogen in the vegetative tissues becomes an important N-reserve, as de novo root N uptake is insufficient for grain N demand and efficient re-translocation of N facilitated by shoot senescence is required (Foulkes et al., 2009). Here data is provided for post-anthesis senescence and selected gene expression profiles for a single year (2006/07) field experiment (Figs 4 and 5). This year was representative of typical UK growing conditions when comparing climatic conditions for this year (see supplementary figure S1) to other years. The relative chlorophyll/senescence profile of the SPAD analysis of the middle part of leaf 2 indicates at least 50% senescence at 6 weeks post-anthesis, with reduction to less than 10% after a further 7 days (Figs 4 and 5). As a marker of senescence, gene expression levels of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubiscoSSU) (Fig. 4), the expression profile of a wheat C1A cysteine protease (here called TaSAG12), orthologous to the Arabidopsis senescence-associated AtSAG12 involved in protein degradation (Noh and Amasino, 1999) (Fig. 4) and the wheat NAC transcription factor TaNAM-B1, known to be involved in regulating senescence and nutrient remobilization from canopy to developing grains (Fig.5; Uauy et al., 2006; Waters et al. 2009), were analysed. For RubiscoSSU, a reduction of relative expression compared to anthesis was already visible 2 weeks post-anthesis, remaining level for 3 weeks before decreasing to a low expression level 6 weeks post-anthesis (Fig. 4). TaSAG12 expression increased 4 weeks after anthesis, at which time the relative chlorophyll content in the middle of the leaf was still not reduced, but leaf tips showed signs of senescence (data not shown). With senescence, TaSAG12 expression increased eight-fold further (Fig. 4). An earlier increase of expression was found for TaNAM-B1 (Fig. 5). A significant increase of expression was detectable 3 weeks after anthesis with a continuous increase up to four times higher as compared to anthesis. The discrepancy between RubiscoSSU and TaSAG12/ TaNAM-B1 expression indicates that the process of leaf senescence and N-remobilization are independently regulated. Analysis of stay-green mutants has shown that in mutants with a defect in the chlorophyll catabolism pathway, soluble protein degradation during senescence may be close to normal, but light-harvesting and reaction centre thylakoid membrane proteins are much more stable (Thomas et al., 2002).
Fig. 4.
Expression analysis in relation to post-anthesis chlorophyll content/senescence in the second leaf of wheat. Real-time PCR expression analysis of ribulose bisphosphate carboxylase/oxygenase small subunit (RubiscoSSU on Chr5), senescence associated gene (SAG12), and putative low-affinity nitrate transporters NPF6.1, NPF6.2, NPF6.3, NPF6.4, NPF4.1, NPF6.5, NPF6.6, NPF7.1, NPF7.2, NPF2.4/2.5, NPF2.1, and NPF2.2. NRE, ‘Normalised Relative Expression’. Values are means ±SEM (n = 3). **, *, significantly different changes in time compared to anthesis (P < 0.01, P < 0.05, respectively). Anth, anthesis; wpa, weeks post-anthesis.
Fig. 5.
Expression analysis in relation to post-anthesis chlorophyll content/senescence in the second leaf of wheat. Real-time PCR expression analysis of NAC transcription factor (TaNAM), nitrate reductase 1 (TaNR1), nitrite reductase (TaNIR), glutamate dehydrogenase 2 (TaGDH2), glutamine synthetases TaGS1 (cytosolic), TaGS2 (plastidic), TaGse (cytosolic), and TaGSr (cytosolic). NRE, ‘Normalised Relative Expression’. Values are means ±SEM (n = 3). **, *, significantly different changes in time compared to anthesis (P < 0.01, P < 0.05, respectively). Anth, anthesis; wpa, weeks post-anthesis.
For the majority of the NPF genes analysed, there was a strong correlation of expression in relation to leaf senescence. The post-anthesis relative expression levels of TaNPF6.1, TaNPF6.6, TaNPF7.1, TaNPF7.2, and TaNPF2.1 were very similar with no change or only slightly increased transcript levels until 5 weeks post-anthesis, followed by a very strong increase at week 6, similarly to the TaSAG12 expression (Fig 4). The increased expression was in the range of 2-fold higher (TaNPF7.2) up to 35-fold higher (TaNPF6.6). The transcript levels of TaNPF6.2, TaNPF6.4, TaNPF4.1, and TaNPF2.4/2.5 increased much earlier, already at 3–4 weeks post-anthesis with a steady state higher level compared to anthesis, as found for TaNPF6.2, TaNPF6.6 and TaNPF2.4/2.5, or further increasing, as found for TaNPF4.1 (Fig. 4). For two NPF genes a post-anthesis reduction of the relative expression in leaf 2 was detected. The expression of TaNPF6.3 remained level until week 5, before it decreased by 50% at week 6 in comparison to anthesis (Fig. 4). The TaNPF2.2 transcript pattern was complex with a slight but significant reduction at 2 weeks post-anthesis, an increased expression at weeks 4 and 5, and a substantial decrease at 6 weeks post-anthesis (Fig. 4).
To put NPF expression in leaf 2 after anthesis in the context of nitrogen assimilation, gene expression patterns of nitrate reductase 1 (TaNR1), nitrite reductase (TaNIR), glutamate dehydrogenase 2 (TaGDH2), 3 cytosolic and 1 plastidic glutamine synthetases (TaGS1, TaG2e, TaGSr, and TaGS2, respectively) were analysed. A significant increase in relation to senescence was found for nitrate reductase TaNR1 transcript abundance at 6 weeks post-anthesis (Fig. 5). TaNIR gene expression increased slightly within 2 weeks and decreased back to the initial level with no further reduction until late senescence. Both cytosolic glutamine synthetase genes TaGse and TaGSr showed an expression pattern similar to some of the NPF genes with unchanged expression levels until week 5, and a drastic increase to a 3- to 5-fold higher level at 6 weeks post-anthesis (Fig. 5). The wheat glutamine synthetase TaGS1 expression was similar to TaNPF6.2 with increased post-anthesis expression at week 2, which stayed constant before a further increase at week 6 (Fig. 5). The glutamate dehydrogenase TaGDH2 gene expression showed a nearly identical pattern compared to the TaNPF6.6 gene, with a slow but significant steady increase in expression until week 5, which then drastically increased up to a 17-fold higher level (Fig. 5). The expression of TaGS2 (plastidic protein localization) followed an opposite pattern by decreasing at week 6, as would be expected with increasing senescence (Fig. 5).
Discussion
Expression studies of the wheat NPF genes identified in relation to tissue specificity, regulation by nitrate availability, and N-remobilisation in leaves during post-anthesis senescence indicate the functions of those genes.
Complex phylogeny of wheat NPF genes in the wheat genome
Analyses of other plant transporter gene families have identified similarities between phylogenetic relationships and function (Buchner et al., 2004; Takahashi et al., 2012). All of the Arabidopsis NPF transporters for which wheat homologues have been identified in this study are able to transport nitrate, as verified by oocyte assays (Tsay et al., 2007; Lin et al., 2008; Fan et al., 2009; Li et al., 2010). Given the background knowledge that the orthologous Arabidopsis genes are somehow involved in nitrate uptake as well as transport/distribution within the plant, the phylogenetically close relationships would favour similar functions/ substrate transport specificities for the orthologous wheat genes.
Plett et al. (2010) identified orthologous and paralogous cereal NPF genes in the genomes of rice, Sorghum, maize (Zea mays) and Brachypodium. Phylogenetic analysis of the identified wheat key orthologous and homologous NPF proteins, in comparison to Arabidopsis, rice and Brachypodium, identified clear single orthologues only for TaNPF4.1 and NPF6.5. For all other wheat NPF genes, orthologous genes to the related monocot species were identified, but as Plett et al. (2010) described, a clear analysis of orthologous relationships between the selected Arabidopsis NPF genes and the monocot species is not always possible. Plett et al. (2010) have shown that the NPF family structure in cereals seems to be much more complex than in Arabidopsis and other dicots. The differences found between wheat compared to rice and Brachypodium indicate a further complexity of the wheat NPF gene family and may be only partly explained by additional gene duplication and/or loss of genes during wheat evolution. The different gene composition for the NPF families in the different grass species may be explained by different evolutionary development influencing the genomes of the different grass species. The grass genomes differ in size, ploidy level and chromosome number. The hexaploid wheat genome (AABBDD; 2n=42) of ~16 000 Mbp size originated from two polyploidization events (Feldman et al., 1995). The diploid rice genome (2n=24) is, with ~430 Mbp, much smaller than the wheat genome; the Brachypodium genome (2n=10) with ~272 Mbp is nearly 60 times smaller than the wheat genome. In general, the gene order in the nuclear genomes of all grasses has been preserved; nevertheless genomic rearrangements, duplication and polyploidization events occurred during the evolution of the different grass species leading to differences in gene copy number. For example, in comparison to its progenitors, hexaploid wheat seems to have deleted many low-copy DNA sequences since the polyploidization event (Feldman et al., 1997). For rice, recent segmental duplications on Chromosomes 11 and 12 and massive ongoing individual gene duplications have been identified (Yu et al., 2005). For wheat, at least 10 duplicated regions, which represent 67.5% of the genome, were identified, and Salse et al. (2008) suggested an ancient duplication of the diploid wheat genomes before their hybridization into polyploid wheat.
Complex expression pattern of wheat NPF genes in different tissues and in relation to N-status of the plant
For most of the Arabidopsis NPF genes considered in this study, the expression pattern is closely related to their suggested function in relation to nitrate transport. With few exceptions, there is little available information on gene expression patterns of related cereal genes. The existence of co-orthologous genes in cereals and other plant species such as Lotus japonicus (Criscuolo et al., 2012), in comparison to the Arabidopsis single NPF gene loci, suggest split and/or individual additional functions of those co-orthologous/homologous NPF genes. Based on the existence of co-orthologous genes, the regulation of those genes would enable a much higher variability in relation to nitrate uptake as well as distribution within the plant. A good example can be seen for AtNPF6.3 in Arabidopsis. The four co-orthologous wheat NPF genes showed similar but also different patterns of expression in relation to tissue specificity and N-nutrition. The root dominance of TaNPF6.1 and TaNPF6.2 indicated a more specific function for nitrate uptake and/or translocation in the root. However, the expression patterns of the NPF6.2 and NPF6.3 genes in response to N-starvation and resupply suggest similar functions in relation to nitrate sensing, whereby the differences in the pattern in relation to the N-resupply response suggests an individual fine tuning of regulation. The regulation of TaNPF6.3 by the N-supply in the shoot has so far only been reported for the orthologous NPF gene of Brassica napus. In contrast to Arabidopsis, shoot gene expression of the orthologous Brassica gene was negatively correlated with shoot nitrate concentration, whereas it was positively correlated with root nitrate concentrations (Le Ny et al., 2013). This suggests an important specific function of TaNPF6.3 in regulation of nitrate distribution in wheat non-root tissues. This function is supplemented by a constitutive factor provided by the co-orthologous TaNPF6.4 protein.
The Lotus japonicus genome contains four genes co-orthologous to AtNPF6.3. Root expression analysis of two of these showed no induction by N-resupply after N-starvation, and the expression of one gene was even down-regulated (Criscuolo et al., 2012). Lauter et al. (1996) reported that the AtNPF6.3 co-orthologous tomato SolyNPF6.9 and SolyNPF6.10 genes had root-specific expression. Whilst SolyNPF6.10 root expression was nitrate inducible, the SolyNPF6.9 expression behaved constitutively (Lauter et al., 1996). Root expression analysis of two maize co-orthologous genes grown under low and adequate nitrate nutrition showed no response of either gene in relation to N-nutrition throughout the whole lifecycle (Garnett et al., 2013). This emphasizes the complexity and variation in gene regulation in relation to nutrient uptake in crop plants with complex genomes, in comparison to a simple genome model plant such as Arabidopsis.
Those differences are not only related to multi co-orthologous genes. TaNPF4.1, the single orthologous gene to AtNPF4.6, is expressed in roots and shoots to a similar level, suggesting involvement in substrate transport in roots and in shoots. The orthologous Arabidopsis NPF4.6 is described as a constitutive component of the low-affinity root nitrate uptake system (Okamoto et al., 2003). Huang et al. (1999) reported root-specific expression and an involvement in root nitrate uptake, although promoter-GUS studies of NPF4.6 indicated expression in the vascular tissue of the root and hypocotyl and also in the stem and leaves (Kanno et al., 2012). In contrast to AtNPF4.6, wheat TaNPF4.1 shoot expression is influenced by the N-status of the plant. Difference in the regulation of AtNPF4.6 orthologous genes was also reported for the Lotus japonicus orthologous gene. In contrast to AtNPF4.6 and TaNPF4.1, the Lotus japonicus orthologous NPF gene expression in roots is up-regulated by nitrate resupply after N-starvation (Criscuolo et al., 2012). These differences of orthologous gene expression in relation to tissue specificity and N-supply in different plant species suggest variation in the regulatory signalling pathways for those genes. This further suggests different functions for these orthologous NPFs in the different plant species in relation of substrate uptake and translocation in different tissues.
The demand for nitrate uptake by the root is driven by nitrate assimilation in the plant during development. Long-distance transport via the xylem, including uploading in the different parts of the plant, and also nitrate allocation between organs via the phloem, facilitates the distribution of nitrate and requires regulation. Several Arabidopsis NPF transporters are reported to be expressed in vascular tissues.
The expression pattern of the orthologous and phylogenetic close wheat NPF genes implicates similarities but also differences in relation to tissue specificity and N-nutrition compared to the related Arabidopsis NPF genes. The wheat TaNPF7.1 expression had root dominance with the highest root transcript level of all NPF genes analysed indicating importance for root nitrate transport. Further, the shoot transcript level of TaNPF7.1 was also still comparatively high (second highest) in relation to the low shoot expression described for Arabidopsis AtNPF7.3. In contrast to Arabidopsis NPF7.3, for which no regulation of shoot expression by the N-status was observed, the opposite regulation of TaNPF7.1 expression by N-starvation and nitrate resupply in the root and the shoot suggests a participation of TaNPF7.1, not only in root nitrate xylem loading, but also in nitrate translocation in the shoot regulated by the N-status. As there is a low transcript level in both roots and shoots for TaNPF7.2, a similar function to AtNPF7.2 in nitrate unloading from the xylem in the different tissues is possible.
Under stressed situations it has been observed that nitrate is reallocated to the root (Hernandez et al., 1997). Li et al. (2010) found that the nitrate reallocation process is regulated by the induction of NPF7.2 in Arabidopsis, which contributes essentially to Cd2+ stress tolerance. Furthermore it has been shown that, in contrast, AtNPF7.3 is down-regulated in roots by various stresses. Under these conditions, nitrate is retained in roots, which contributes to stress tolerance in a similar mechanism to that proposed for the NPF7.2 nitrate reallocation (Li et al., 2010; Chen et al., 2012). As there is no orthologous NPF7.2 in grasses (Plett et al., 2010), the up-regulation of the wheat TaNPF7.1 in shoots under N-starvation stress may suggest a similar function to AtNPF7.2 by reallocation of nitrate in the shoot to the vascular system and finally back to the root and reduction of expression in the root under N-starvation to retain high nitrate concentration.
Three wheat genes are co-orthologous to AtNPF2.9 but also closely related to the glucosinolate transporter AtNPF2.10 and AtNPF2.11. None of the wheat homologous NPF genes showed dominance for root expression. In contrast they are expressed more dominantly in the shoots with very low transcript levels in the roots. Similar expression pattern with shoot versus root dominance was also reported for the orthologous rice NPF2.2 gene (Ouyang et al., 2010). In Arabidopsis, the root-dominant AtNPF2.9 may be responsible for loading of nitrate into root phloem, with the apoplastic nitrate source for NPF2.9-mediated loading coming from efflux of vascular parenchyma cells or leakage of the xylem stream (Wang and Tsay, 2011). If the orthologous cereal genes of rice and wheat are also expressed in the phloem, the dominant shoot expression would favour a function more in loading and/or unloading of nitrate to or from the shoot/leaf phloem. The regulation of TaNPF2.1 expression by the N-status in the roots as well as in the shoots indicates that its function is controlled by the nitrate/nitrogen demand of the individual tissue.
Differences in expression patterns were also found for the wheat NPF6.5 and NPF6.6 genes. The orthologous Arabidopsis AtNPF6.2 is strictly leaf specific with expression in the petioles and with a unique function in nitrate homoeostasis regulation (Chiu et al., 2004). The wheat TaNPF6.5 showed a dominant shoot expression but with significant detectable expression in the root, indicating an additional root function. The down-regulation of expression by N-starvation in the shoot suggests a similar participation of TaNPF6.5 in regulation of nitrate homeostasis in non-root tissues. TaNPF6.6 showed higher transcript levels in the shoot and reasonable expression in the root. Unlike the Arabidopsis orthologous AtNPF6.4 gene, no up-regulation in the shoot or down-regulation in the root by nitrate resupply was observed for the orthologous TaNPF (Okamoto et al., 2003). Similarly no influence of N-provision on the root expression was reported for the two Lotus japonicus orthologous genes (Criscuolo et al., 2012). N-starvation resulted in a slow up-regulation of the wheat gene expression which went down slowly after nitrate resupply to the +N control level. Furthermore, Arabidopsis AtNPF6.4 gene expression was also suppressed in the root by nitrate application (Okamoto et al., 2003), which was not found for the wheat orthologues indicating a completely different regulation of TaNPF6.6 compared to the Arabidopsis AtNPF6.4 gene.
Correlation of NPF gene expression to leaf senescence
For wheat and other annual cereal crops, senescence is linked to seed reproduction (Noodén 1988a, b). This control of senescence by reproduction is not found in Arabidopsis. The life-span of single Arabidopsis leaves is independent of the development of reproductive structures (Noodén and Penney, 2001). This implicates a completely different demand for regulation of nitrate uptake and distribution/remobilization in wheat compared to Arabidopsis during senescence and grain development. Post-anthesis N-remobilisation and translocation of pre-anthesis stored N-assimilates to support grain growth is an important part of wheat development. Between 51 and 91% of the grain nitrogen comes from vegetative N remobilisation of organic N in the form of free amino acids and macromolecules (Van Sanford and MacKown, 1987; Palta and Fillery, 1995). In addition to post-anthesis N-remobilisation de novo N-uptake accounts for 5–40% of the total grain N in wheat, with high variation between wheat varieties (Kichey et al., 2007; Bogard et al., 2010). Several components of the assimilatory pathway have been implicated in the remobilization processes. Protein as well as gene expression studies indicated increased levels of wheat glutamine synthetase GS1 and GSr, as well as glutamate dehydrogenase, and decreased level of GS2, in wheat leaves with increasing senescence (Kichey et. al., 2005; Bernard et al., 2008). This could be confirmed by the analysis provided here (Fig. 5). Unusually there was an up-regulation of expression with senescence for the third cytosolic glutamine synthetase TaGSe gene, similarly to TaGSr. Additionally there was an up-regulation of the wheat nitrate reductase (TaNR) expression with senescence. During senescence-related N-remobilisation, the ammonia for glutamine synthesis can be provided by re-assimilation of ammonia released during protein hydrolysis, and by assimilation of nitrate via nitrate and nitrite reduction. In tobacco and tomato, nitrate reductase expression in leaves has been shown to be regulated by the nitrate content (Galangau et al., 1988). In wheat a good correlation was found for post-anthesis N-uptake and nitrate reductase activity (Kichey et al., 2007). The post-anthesis expression of NPF genes of different subfamilies in leaf 2 implicates a complex pattern of nitrate transport activity. The nitrate delivered by post-anthesis uptake needs to be taken up via the leaf vascular bundle and distributed within the leaf for further reduction and assimilation. The increased expression level of the wheat NPF genes with increasing senescence could be an indication of increased N-related substrate transport and accumulation. An increased level of nitrate would increase the demand for nitrate reduction requiring an increased expression of nitrate reductase. The expression level of the nitrite reductase at late senescence was the same as found at anthesis, and indicated continuing conversion of nitrite to ammonia. The senescence-correlated up-regulation of NPF genes in leaf 2 suggested two possible options: to enable direct transport of stored nitrate from the canopy tissue to the grain; or channelling of nitrate during senescence to nitrate/nitrite reduction to provide ammonium for glutamine synthetase. It is well established that N-assimilation to allow protein synthesis and leaf N remobilization occurs simultaneously. Therefore in addition to protein degradation during senescence, nitrate transport may be an important supportive part for the translocation process of N from senescing source tissues to provide sufficient N for grain filling influencing nitrogen use efficiency and yield. The NAM Nac-transcription factors are predicted to regulate genes that encode proteins that carry out physiological processes for nutrient remobilization and/or translocation to grain (Waters et al., 2009). The fact that some nitrate transporters, such as TaNPF4.1, showed a similar expression pattern compared to the wheat NAM Nac-transcription factor, indicated a highly regulated process.
Conclusion
The data presented for the selected wheat NPFs indicates the complexity of the phylogeny and expression of the 16 wheat NPF genes identified. The existence of multiple co-orthologous genes in wheat and other cereals as well as dicotyledonous crop plants, in comparison to Arabidopsis, suggests a higher complexity of pattern and regulation of N-related substrate uptake, translocation and redistribution compared to that described for Arabidopsis. The regulation of some of the analysed wheat NFP genes by N-supply suggested an involvement of those NFPs in substrate transport closely related to N-metabolism. An example is the existence of four orthologous NPF6.3 genes showing similarities and differences, in comparison the sole Arabidopsis NPF gene. The complexity of the expression pattern in relation to N-supply as well as N-remobilisation/leaf senescence suggests a requirement for a complex network of regulation of NPF gene expression in the process of N-uptake and distribution within the plant. This complexity described here for wheat and other important crop plants can be explained by the need to be able to adjust the nutrient uptake and distribution to a much more complex development including reproduction compared to the simple weed Arabidopsis. This demonstrates the limitation of simple model plants in generalising and adopting findings. Due to the broad range of substrates reported to be transported by NPF transporters, more detailed functional transport analysis and spatial tissue-specific expression patterns are required for a full understanding of the role of the NPF transporters in wheat.
Supplementary material
Supplementary data can be found at JXB online.
Supplementary Table S1. CerealsDB sequence IDs.
Supplementary Table S2. Oligonucleotide primer sequences used for SYBR Green real time RT-PCR expression analysis.
Supplementary Table S3. Oligonucleotide primer sequences used for cDNA-PCR fragment cloning and sequencing including database accession.
Supplementary Figure S1. Average rainfall and max.-min. air temperature at Rothamsted field trails between 1 September 2006 and 31 August 2007.
Acknowledgements
Rothamsted Research receives support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK as part of the 20:20 Wheat® project.
References
- Almagro A, Lin SH, Tsay YF. 2008. Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20, 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, Stephen F, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough PB, Howarth JR, Jones J, Lopez-Bellido R, Parmar S, Shepherd CE, Hawkesford MJ. 2010. Nitrogen efficiency of wheat: genotypic and environmental variation and prospects for improvement. European Journal of Agronomy 33, 1–11 [Google Scholar]
- Barraclough PB, Lopez-Bellido R, Hawkesford MJ. 2014. Genotypic variation in the uptake, partitioning and remobilisation of nitrogen during grain-filling in wheat. Field Crops Research 156, 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard SM, Blom Møller AL, Dionisio G, et al. 2008. Gene expression, cellular localization and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Molecular Biololgy 67, 89–105 [DOI] [PubMed] [Google Scholar]
- Bogard M, Allard V, Brancourt-Hulmel M, Heumez EL, Machet J-M, Jeuffroy M-H, Gate P, Martre P, Le Gouis J. 2010. Deviation from the grain protein concentration–grain yield negative relationship is highly correlated to post-anthesis N uptake in winter wheat. Journal of Experimental Botany 61, 4303–4312 [DOI] [PubMed] [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ. 2004. Plant sulphate transporters – coordination of uptake, intracellular and long distance transport. Journal of Experimental Botany 55, 785–1798 [DOI] [PubMed] [Google Scholar]
- Cerezo M, Tillard P, Filleur S, Muños S, Daniel-Vedele F, Gojon A. 2001. Major alterations of the regulation of root NO(3)(–) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiology 127, 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Lv XF, Li JY, Yi HY, Gong JM. 2012. Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiology 159, 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CC, Lin CS, Hsia AP, Su RC, Lin HL, Tsay YF. 2004. Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiology 45, 1139–1148 [DOI] [PubMed] [Google Scholar]
- Criscuolo G, Valkov VT, Parlati A, Alves LM, Chiurazzi M. 2012. Molecular characterization of the Lotus japonicus NRT1(PTR) and NRT2 families. Plant Cell and Environment 35, 1567–1581 [DOI] [PubMed] [Google Scholar]
- Drew MC, Saker LR. 1984. Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley. Evidence for non-allosteric regulation. Planta 160, 500–507 [DOI] [PubMed] [Google Scholar]
- Fan SC, Lin CS, Hsu PK, Lin SH, Tsay YF. 2009. The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Liu B, Segal G, Abbo S, Levy A, Vega J. 1997. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147, 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Lupton FGH, Miller TE. 1995. In: Smartt J, Simmonds NW, eds. Wheats. Evolution of crops. Ed 2 London: Longman Scientific, 184–192 [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- Forde BG. 2002. Local and long-range signalling pathways regulating plant responses to nitrate. Annual Reviews of Plant Biology. 5, 203–224 [DOI] [PubMed] [Google Scholar]
- Foulkes MJ, Hawkesford MJ, Barraclough PB, Holdsworth MJ, Kerr S, Kightley S, Shewry PR. 2009. Identifying traits to improve the nitrogen economy of wheat: Recent advances and future prospects. Field Crop Research 114, 329–342 [Google Scholar]
- Galangau F, Daniel-Vedele F, Moureaux T, Dorbe MF, Leydecker MT, Caboche M. 1988. Expression of leaf nitrate reductase genes from tomato and tobacco in relation to light-dark regimes and nitrate supply. Plant Physiology 88, 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett T, Conn V, Plett D, et al. 2013. The response of the maize nitrate transport system to nitrogen demand and supply across the lifecycle. New Phytologist 198, 82–94 [DOI] [PubMed] [Google Scholar]
- Gomez KA, Gomez AA. 1984. Statistical Procedures for Agricultural Research. 2nd ed. Chichester: John Wiley and Sons [Google Scholar]
- Gregersen PL, Holm PB, Krupinska K. 2008. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biology 10, 37–49 [DOI] [PubMed] [Google Scholar]
- Guo F-Q, Young J, Crawford NM. 2003. The Nitrate Transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez LE, Gárate A, Carpena-Ruiz R. 1997. Effects of cadmium on the uptake, distribution and assimilation of nitrate in Pisum sativum . Plant Soil 189, 97–106 [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hsu PK, Tsay YF. 2013. Two phloem nitrate transporters, nrt1.11 and nrt1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiology 163, 844–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF. 1999. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. 2012. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proceedings of the National Academy of Sciences, USA 109, 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichey L, Le Gouis J, Sangwan B, Hirel B, Dubois F. 2005. Changes in the cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase during flag leaf senescence in wheat (Triticum aestivum L.) Plant Cell Physiology 46, 964–974 [DOI] [PubMed] [Google Scholar]
- Kichey T, Hirel B, Heumez E. 2007. In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crops Research 102, 22–32 [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay YF. 2010. Nitrate signaling: adaptation to fluctuating environments. Current Opinion in Plant Biology 13, 266–273 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lauter FR, Ninnemann O, Bucher M, Riesmeie R JW, Frommer WB. 1996. Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proceedings of the National Academy of Sciences, USA 93, 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ny F, Leblanc A, Beauclair P, Deleu C, Le Deunff E. 2013. In low transpiring conditions, nitrate and water fluxes for growth of B. napus plantlets correlate with changes in BnNrt2.1 and BnNrt1.1 transporter expression. Plant Signaling & Behavior 8, e22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S, Brachet C, Tillard P, Gojon A, Lacombe B. 2013. The Arabidopsis NPF6.3/NRT1.1 is a bidirectional transporter involved in root-to-shoot nitrate translocation. Molecular Plant 6, 1984–1987 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, et al. 2014. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends in Plant Science 19, 5–9 [DOI] [PubMed] [Google Scholar]
- Li JY, Fu YL, Pike SM, et al. 2010. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, et al. 2008. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. 1999. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF. 2003. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO Journal 22, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular Evolution and Phylogenetics. New York: Oxford University Press [Google Scholar]
- Noh YS, Amasino RM. 1999. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Molecular Biology 41, 181–194 [DOI] [PubMed] [Google Scholar]
- Noodén LD. 1988a. The phenomena of senescence and aging. In: Noodén LD, Leopold AC, eds. Senescence and aging in plants. Academic Press, San Diego, 1–50 [Google Scholar]
- Noodén LD. 1988b. Whole plant senescence. In: Noodén LD, Leopold AC, eds. Senescence and aging in plants. Academic Press, San Diego, 392–439 [Google Scholar]
- Noodén LD, Penney JP. 2001. Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae). Journal of Experimental Botany 52, 2151–2159 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jørgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA. 2012. NRT/PTR transporters are essential for transclocation of glucosinolate defence compounds to seeds. Nature 488, 531–534 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Vidmar JJ, Glass AD. 2003. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiology 44, 304–317 [DOI] [PubMed] [Google Scholar]
- Orsel M, Filleur S, Fraisier V, Daniel-Vedele F. 2002. Nitrate transport in plants: which gene and which control? Journal of Experimental Botany 53, 825–833 [DOI] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ. 2006. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein–protein interaction. Plant Physiology 142, 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Cai Z, Kuaifei X, Wang Y, Duan J, Zhang M. 2010. Identification and analysis of eight peptide transporter homologs in rice. Plant Science 179, 374–382 [Google Scholar]
- Palta JA, Fillery IRP. 1995. N application increases pre-anthesis contribution of dry matter to grain yield in wheat grown on a duplex soil. Australian Journal of Agricultural Research 46, 507–518 [Google Scholar]
- Plett D, Toubia J, Garnett T, Tester M, Brent N, Kaiser B-N, Baumann U. 2010. Dichotomy in the NRT gene families of dicots and grass species. PLoS One 5, e15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter J, Lekanne-Deprez R, Moorman A. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–69 [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ. 2009. RT-PCR: Design, calculations, and statistics. Plant Cell 21, 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. 2008. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Buchner P, Yoshimoto N, Hawkesford MJ, Shiu S-H. 2012. Evolutionary relationships and functional diversity of plant sulphate transporters. Frontiers in Plant Science 2, 119. 10.3389/fpls.2011.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H, Ougham H, Canter P, Donnison I. 2002. What stay-green mutants tell us about nitrogen remobilization in leaf senescence. Journal of Experimental Botany 53, 801–808 [DOI] [PubMed] [Google Scholar]
- Tong Y, Zhou JJ, Li Z, Miller AJ. 2006. A two-component high-affinity nitrate uptake system in barley. Plant Journal 41, 442–450 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. 2007. Nitrate transporters and peptide transporters. FEBS Letters 581, 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. 1993. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713 [DOI] [PubMed] [Google Scholar]
- Tuomi JM, Voorbraak F, Jones DL, Ruijter JM. 2010. Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods 50, 313–322 [DOI] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. 2006. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sanford DA, MacKown CT. 1987. Cultivar differences in nitrogen remobilization during grain fill in soft red winter wheat. Crop Science 27, 295–300 [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. 1989. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Research 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Hsu P-K, Tsay YF. 2012. Uptake, allocation and signalling of nitrate. Trends in Plant Science 17, 458–467 [DOI] [PubMed] [Google Scholar]
- Wang YY, Tsay YF. 2011. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23, 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Uauy C, Dubcovsky J, Grusak MA. 2009. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. Journal of Experimental Botany 60, 4263–4274 [DOI] [PubMed] [Google Scholar]
- Yu J, et al. 2005. The genomes of Oryza sativa: A history of duplications. PLoS Biology 3, e38. 10.1371/journal.pbio.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass ADM. 1999. Regulation of a putative high-affinity nitrate transporter (At NRT2;1) in roots of Arabidopsis thaliana . Plant Journal 17, 563–568 [DOI] [PubMed] [Google Scholar]
- Zhou JJ, Fernandez E, Galvan A, Miller AJ. 2000. A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Letters 466, 225–227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.