Abstract
Purpose of review
Understanding the effects of in-utero exposures to environmental agents is of great importance as the resulting deregulation of biological processes can affect both fetal development and health outcomes that manifest later in life. Due to their established role in developmental processes and inherent stability ex vivo, microRNAs (miRNAs) have emerged as attractive candidates to explore the impact of such exposures during this critical window of susceptibility. In this review, we summarize the findings of studies assessing miRNAs as markers of in-utero environmental exposures and as candidates for the molecular basis through which these exposures exert their influence on children's health.
Recent findings
To date, miRNA expression profiles due to various in-utero environmental exposures, including xenochemicals, endogenous factors, and nutritional status, have been reported.
Summary
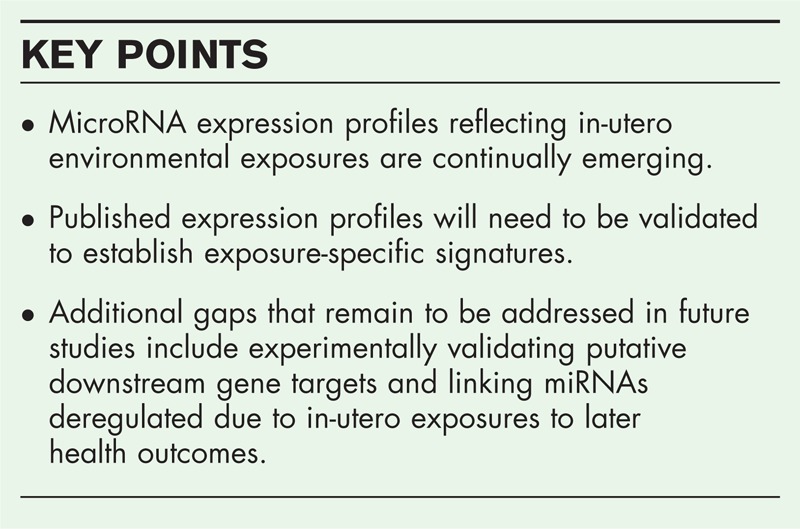
While the validity of the identified exposure-specific miRNA profiles remains to be established, the findings thus far do raise interesting questions worth addressing in future studies. Gaps that remain to be addressed include linking specific in-utero exposures to subsequent health outcomes based on established miRNA expression profiles and experimentally validating putative downstream targets of the deregulated miRNAs.
Keywords: endogenous exposures, in-utero exposures, miRNA, nutrition, xenochemicals
INTRODUCTION
The impact of environmental exposures on human health ranges from acute effects, such as skin irritation, to chronic long-term consequences, including cognitive and reproductive dysfunction. The life stage during which the exposure occurs has been deemed a critical determinant on the health consequences realized. The physiologic and metabolic characteristics of fetal development, including the immaturity of the blood–brain barrier and the reduced detoxification and elimination capacity of the internal organs such as liver and kidneys, mark the intrauterine experience as a period of heightened susceptibility [1]. In addition to impacting fetal development, environmental exposures can also modify the genome to program health outcomes experienced later in life. The molecular mechanism underlying this fetal programming, also referred to as the ‘developmental origins of health and disease (DOHAD)’ [2], has not been clearly delineated. However, mounting evidence suggests that epigenetics plays a vital role.
The epigenome refers to heritable, quasi-stable yet dynamic and environmentally responsive elements that regulate gene expression without changes in the DNA sequence itself [3]. Various epigenetic marks have been identified, including transcriptional regulation through DNA methylation and histone modification, as well as posttranscriptional regulation through microRNAs (miRNAs).
miRNAs are small noncoding RNAs, typically 21–23 nucleotides in length, formed from transcripts that take on a characteristic hairpin structure [4]. The process to generate miRNAs is initiated through the transcription of primary miRNA (pri-miRNA) by RNA polymerase II [5]. Processing by DROSHA, an RNAse III endonuclease, generates a 60–70-nt stem loop intermediate with a 3’ overhang [6,7]. This precursor miRNA (premRNA) is actively transported from the nucleus to the cytoplasm via Ran-GTP and Exportin-5, where it is further processed by an additional RNAse III endonuclease, DICER, into a double-stranded miRNA:miRNA∗ complex [8,9,10]. The mature single-stranded miRNA is loaded into the RNA-induced silencing complex (RISC), where it serves as a guide for targeting the RISC to mRNAs based on complementarity [11,12]. The degree of complementarity between a miRNA and its targets determines the mechanism of posttranscriptional regulation. Near perfect pairing results in cleavage of the target mRNA, whereas partial pairing, a phenomenon more commonly observed in animals, results in either mRNA decay or translational repression [13].
Box 1.
no caption available
To date, over 2000 unique mature human miRNAs have been annotated and catalogued in the publicly accessible database miRBase (www.miRbase.org), and up to one-third of all mRNA transcripts are believed to be regulated by miRNAs [14]. As targeting only requires partial complementarity, a single miRNA can bind multiple mRNA transcripts and each mRNA can be bound to multiple miRNAs. In this way, miRNAs are able to modulate entire gene networks, including biological processes such as development, stem cell differentiation, hematopoiesis, cardiac/muscle development, neurogenesis, insulin secretion, cholesterol metabolism, and immune response [15]. Significant modifications in the expression of miRNAs likely result in the deregulation of these processes, ultimately triggering disease. Children are a population of special interest in determining the impact of miRNA deregulation as the heightened susceptibility to early life environmental exposures may play a role in modulating miRNA expression levels. Additionally, the downstream effects owing to the resulting deregulation of miRNAs are likely more pronounced as many of the miRNA-regulated physiologic processes are most active early in the developmental process. Hence, this review focuses on miRNAs as markers of in-utero environmental exposures and as candidates for the molecular basis through which these exposures exert their influence on children's health.
microRNA AND CHILDREN'S HEALTH
A number of studies have implicated miRNAs in various pediatric health outcomes. As summarized in Table 1, these studies showcase the wide-ranging utility of miRNAs as markers of disease, in terms of diagnosis, prognosis, disease subtype classification, treatment monitoring, and agents of intervention. However, greater understanding is also required in identifying the upstream elements capable of disrupting the expression profile of miRNAs, including environmental factors, ultimately resulting in disease.
Table 1.
Studies assessing microRNAs in pediatric health outcomes
| Outcome | miRNA-related assessment | Impact on outcome | Reference |
| Acute lymphocytic leukemia (ALL) | miRNA expression signature profile | Diagnostic: case–control differences | Schotte et al. 2009 [16]; Zhang et al. 2009 [17]; Oliveira et al. 2012 [18] |
| miRNA expression signature profile | Disease subtype classification: T-ALL/MLL vs. ALL | Schotte et al. 2009 [16] | |
| miRNA expression signature profile | Disease subtype classification: T-ALL vs. B-ALL | Fulci et al. 2009 [19] | |
| miRNA expression signature profile | Treatment monitoring: response to prednisone | Zhang et al. 2009 [17] | |
| miRNA expression signature profile | Clinical prognosis | Zhang et al. 2009 [17] | |
| miRNA expression signature profile | Clinical prognosis | Kaddar et al. 2009 [20] | |
| Asthma | Presence of SNPs in HLA-G impacts miRNA binding to 3’UTR region | Cause: Case–control difference | Tan et al. 2007 [21] |
| Presence of SNPs in miRNA genetic sequence | Cause: Case–control differences | Su et al. 2011 [22] | |
| AntagomiR-targeted downregulation of miRNA | Intervention: reduces eosinic inflammation, mucus hypersecretion, TH2 cytokine production, airway hyperresponsiveness | Collison et al. 2011 [23] | |
| Autism | miRNA expression signature profile | Diagnostic: Case–control differences | Abu-Elneel et al. 2008 [24] |
| miRNA expression signature profile | Diagnostic: Case–control differences | Sarachana et al. 2010 [25] | |
| In-silico analysis | Etiology: miRNAs known to be deregulated in autism present in CNV loci | Vaishnavi et al. 2013 [26] | |
| Congenital heart disease (CHD) | Presence of SNP in miRNA coding region | Etiology: Case–control differences | Xu et al. 2009 [27] |
| In-silico analysis | Etiology: miRNAs present in CHD-related CNV loci | Xing et al. 2013 [28] | |
| miRNA expression signature profile | Diagnostic: Case–control differences | Zhu et al. 2013 [29] | |
| Cystic fibrosis | miRNA expression signature profile | Diagnostic: Case–control differences | Oglesby et al. 2010 [30] |
| In-silico/in-vitro analysis of CFTR 3’UTR binding targets | Etiology: miRNAs regulate CFTR gene expression | Megiorni et al. 2011 [31] | |
| In-silico/in-vitro analysis of miRNAs and binding targets involved in CFTR regulation | Treatment: restore function of mutated CFTR protein | Ramachandran et al. 2012 [32] | |
| miRNAs involved in CFTR regulation | Cause: miRNAs regulate CFTR gene expression | Oglesby et al. 2013 [33] | |
| In-vitro analysis: biogenesis and maturation of candidate miRNA | Cause: RNA binding proteins regulate maturation of miRNA upregulated in CF | Bhattacharyya et al. 2013 [34] | |
| Diabetes, Type 1 (T1D) | Candidate miRNA expression analysis | Clinical prognosis | Sebastiani et al. 2011 [35] |
| miRNA expression signature profile | Diagnostic: Case–control differences | Nielsen et al. 2012 [36] | |
| Candidate miRNA expression analysis | Diagnostic: Case–control differences | Salas-Pérez et al. 2013 [37] | |
| Neuroblastoma | miRNA expression signature profile | Disease subtype classification | Chen and Stallings, 2007 [38] |
| miRNA expression signature profile | Clinical prognosis | Bray et al. 2009 [39] | |
| Obesity | miRNA expression signature profile | Diagnostic: miRNA expression pattern associated with obesity markers | Prats-Puig et al. 2013 [40] |
3’ UTR, 3’ untranslated region; ALL, acute lymphoblastic leukemia; B-ALL, B-cell acute lymphoblastic leukemia; CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; CHD, chronic heart disease; CNV, copy number variation; HLA-G, human leukocyte antigen G; miRNA, microRNA; MLL, mixed lineage leukemia; SNP, single nucleotide polymorphism; T-ALL, T-cell acute lymphoblastic leukemia; TH2, Type 2 helper cells.
microRNA AND THE IN-UTERO ENVIRONMENT
Studies that have investigated the responsiveness of miRNAs to early life environmental exposures are summarized in Table 2. The assessed exposures fall into three general categories: xenochemicals, endogenous agents, and nutrition. The group of xenochemicals includes ethanol and tobacco smoke, established teratogens known to cross the placenta and induce fetal malformation. The remaining chemicals in this group, including the endocrine disruptor bisphenol A, have suspected but not yet verified detrimental effects on normal human fetal development. Studies revolving around endogenous factors, including parental stress and hypoxia, indicate that miRNAs are responsive not only to exogenously introduced chemical exposures, but also to the dysregulation of the internal environment. Finally, nutrition is highlighted as its own distinct category, since the importance of the nutritional state in fetal development and programming has been previously established, the best-documented of which links fetal malnutrition with later onset of metabolic diseases in studies conducted on individuals exposed to the Dutch famine [55]. The following sections highlight findings from representative exposures in each category.
Table 2.
Impact of in-utero environmental exposures on microRNA expression levels
| Environmental agent | miRNA | Target gene/pathway | Function | Species | Biospecimen/cell type | Reference |
| Xenochemical | ||||||
| BPA | miR-146a | TLR/cytokine signaling | Human | Placenta cell line | Avissar-Whiting et al. 2010 [41] | |
| Ethanol | miR-21 | Jag-1 | Notch ligand/proliferation of neuroepithelial cells | Mouse | Cerebral cortex-derived neurospheres | Sathyan et al. 2007 [42] |
| miR-335 | Ela Vl2 | Promotes neuronal maturation | ||||
| miR-9 | ||||||
| miR-153 | ||||||
| miR-10a | HOXa1 | Mouse | Brain | Wang et al. 2009 [43] | ||
| miR-10b | ||||||
| miR-9 | ||||||
| miR-145 | ||||||
| miR-153a | Pou4f1 | Regulation of transcription | Zebrafish | Embryo | Soares et al. 2012 [44] | |
| miR-30d | Pou4f1 | |||||
| miR-725 | ||||||
| let7k | ||||||
| miR-100 | ||||||
| miR-738 | ||||||
| miR-732 | ||||||
| Gold nanoparticles | let-7a | Cell proliferation | Mouse | Liver and lung | Balansky et al. 2013 [45] | |
| k-RAS activation | ||||||
| Apoptosis | ||||||
| miR-183 | Apoptosis | |||||
| Cell adhesion | ||||||
| Maternal smoking | miR-146a | BCL2L2/EDA | Pro-survival | Human | Placenta | Maccani et al. 2010 [46] |
| NF-κβ pathway | ||||||
| miR-16 | PLAG1/ SATB1 | Cell cycle/cell proliferation transcription factors | ||||
| miR-21 | TRAF6 | NF-κβ/TLR4 pathway | ||||
| miR-223 | Progenitor cell proliferation/granulocyte differentiation | Human | Cord blood | Herberth et al. 2013 [47▪] | ||
| PCBs | miR-762 | Wnt1 | Cardiovascular differentiation | Mouse | P19 cells embryonal carcinoma | Zhu et al. 2012 [48] |
| miR-29a | Gsk3β | Cardiovascular differentiation | ||||
| miR-324- | Nkx2.5 | Heart development | ||||
| 5p | ||||||
| PFOS | miR-19b-c | Cdk5 Smad1 Sox11b Pou5f1 | Brain and nervous system development | Zebrafish | Embryo | Zhang et al. 2011 [49] |
| Bax | Apoptosis | |||||
| Vsx1 | ||||||
| miR-19d | ||||||
| miR-181b-c | ||||||
| miR-735 | ||||||
| miR-739 | ||||||
| miR-297 | Ypel5 | Rat | Brain | Wang et al. 2012 [50] | ||
| miR-672 | Fgfr3 | |||||
| Syt1 | ||||||
| TCDD | mir-122 | Metabolism | Mouse | Thymus | Singh et al. 2012 [51] | |
| miR-181a | T-cell sensitivity and selection | |||||
| miR-23a | Fas | Apoptosis | ||||
| miR-18b | FasL | Apoptosis | ||||
| miR-31 | Cyp1a1 | Metabolism | ||||
| miR-182 | AhR | Metabolism | ||||
| Endogenous factor | ||||||
| Hypoxia | mR-520c-3p | Human | Placenta | Donker et al. 2012 [52▪] | ||
| Paternal stress | miR-322 | β-glycan | TGF superfamily | Mouse | Brain | Morgan et al. 2011 [53▪] |
| miR-574-3p | ||||||
| miR-873 | ||||||
| Nutrition | ||||||
| High-fat diet | miR-483* | Zswim3 | Mouse | Liver | Zhang et al. 2009 [54] | |
AhR, aryl hydrocarbon receptor; BAX, BCL2-associated X protein; BCL2L2, BCL2-like 2; BPA, bisphenol A; CDK5, cyclin-dependent kinase 5; CYP1A1, cytochrome P450, family 1, subfamily A, polypeptide 1; EDA, ectodysplasin A; ELAVL2, ELAV-like neuron-specific RNA binding protein 2; Fas, Fas cell surface death receptor; FasL, Fas ligand; GSK3B, glycogen synthase kinase 3 beta; HOXa1, homeobox A1; Jag-1, jagged 1; k-RAS, Kirsten rat sarcoma viral oncogene homolog; NFkB, nuclear factor kappa B; NKX2.5, NK2 homeobox 5; PCBs, polychlorinated biphenyls; PFOS, perfluorooctane sulfonate; PLAG1, pleiomorphic adenoma gene 1; Pou4f1, POU class 4 homeobox 1; Pou5f1, POU class 5 homeobox 1; SATB1, SATB homeobox 1; Smad1, SMAD family member 1; Sox11b, SRY-box containing gene 11b; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; TGF, transcription growth factor; TLR, Toll-like receptor; TLR4, Toll-like receptor 4; TRAF6, TNF receptor-associated factor 6; Vsx1, visual system homeobox 1; Wnt1, wingless-type MMTV integration site family, member 1; ZSWIM3, zinc finger, SWIM-type containing 3.
Tobacco smoke
Two human observational studies have been carried out to date assessing the role of miRNAs as potential mediators of the impact of tobacco smoke on in-utero development. Maccani et al.[46] compared the expression level of candidate miRNAs in 25 placentas obtained at the time of delivery from women with and without a history of smoking during pregnancy. Out of the four selected targets, a significant downregulation of miR-16, miR-21, and miR-146a was observed due to maternal cigarette smoking. Furthermore, miR-146a was significantly downregulated following exposure to both nicotine and benzopyrene in TCL-1 (immortalized human trophoblast cell line), a cell line derived from third trimester extravillous cells, whereas no impact due to either agent was observed on the remaining miRNAs.
The impact on miRNA expression levels due to tobacco smoke exposure during the gestational period was also addressed in a study conducted by Herberth et al.[47▪]. As this study was particularly concerned with the effect of tobacco smoke on immune-related responses, two candidate miRNAs, miR-155 and miR-223, previously implicated in Treg cell formation and function, were selected for analysis. In this prospective study of mother–child pairs, increasing levels of miR-223 were observed in both maternal and cord blood with increasing levels of cotinine in maternal urine.
Parental stress
In a study by Morgan and Bale [53▪], the impact of prenatal stress on brain development was assessed. For this purpose, the offspring of male mice that were prenatally exposed to maternal stressors were analyzed. Microarray analysis of brain-derived miRNAs revealed that the profile of F2 male offspring stemming from a stress-exposed lineage was more similar to the profile of nonexposed F2 female offspring than nonexposed F2 male offspring, suggesting dysmasculinization of the brain among the F2 stress-exposed males. Specifically, three miRNA species, miR-322, miR-574-3p, and miR-873, were observed to be downregulated among the F2 stress-exposed offspring, whereas the expression level of β-glycan, a predicted target common to all three miRNAs, was determined to be upregulated compared with F2 control offspring. Finally, following treatment with the aromatase inhibitor formestane, the brain miRNA profile of formestane-treated male mice clustered more closely to control female mice than control male mice, further suggesting that the miRNA environment is sensitive to hormonal dysregulation. The overall findings from this study, therefore, suggest that the changes incurred during in-utero development can be transmitted transgenerationally.
Nutrition
The effects of a maternal high-fat diet on the miRNA profile of the resulting offspring were assessed in a murine model by Zhang et al.[54]. Microarray analysis of liver-derived miRNAs from female offspring identified 10 miRNAs that were upregulated and 23 miRNAs that were downregulated among offspring exposed to a high-fat diet. Although all targets were not successfully validated, the downregulation of miR-483∗ was consistently observed as the greatest fold-change among high-fat diet-exposed offspring. The authors determined that the genetic location of miR-483∗ lies within the intron of Igf2, a gene known to be critical in regulating fetal growth, indicating that the transcription of both is likely regulated by the same promoter. Paradoxically, among high fat-exposed mice, the downregulation of miR-483∗ was observed alongside an upregulation of Igf2.
FUTURE PERSPECTIVE
Findings relaying the impact of in-utero exposures on miRNA expression levels thus far have raised several interesting questions that will need to be further addressed. Principal among these is determining whether the observed changes in miRNA expression levels reflect a causal pathway between exposure and outcome, identifying the downstream gene targets impacted by deregulated miRNAs, and delineating variability introduced because of methodological and biospecimen specifications.
Role of microRNAs in linking exposure to outcome
To address this question, it will first have to be established whether the observed changes in miRNA expression levels due to environmental exposures are the result of a direct effect or a surrogate indication of a different mechanism. This issue was highlighted in the study by Herberth et al., in which the expression level of miR-223 in blood was found to vary among different blood cell-types, leading the authors to question whether the observed tobacco-related increase in miR-223 levels is indeed a direct response to the exposure or a bystander effect of smoke-induced changes in blood cell-type composition [47▪]. Although the utility as a marker of exposure is not diminished in either case, the former would more directly associate miRNAs to the causal pathway linking exposures to putative deregulated physiological processes.
Given a substantiated impact of environmental exposures on miRNA expression levels and related physiological processes, more efforts into how these changes in expression profiles translate into health outcomes will also be warranted. To date, studies relating changes in miRNAs to various health outcomes and studies relating exposures to changes in miRNAs are often conducted separately. Few studies have linked the observed changes in miRNAs due to environmental exposures to known health effects. One such example includes the finding by Herberth et al.[47▪] that higher levels of miRNA-223 in cord blood were also correlated with decreasing levels of Treg cells in newborns, which in turn was shown to be associated with a significantly higher risk of developing atopic dermatitis by the age of 3. Such findings pave the way to further understand the etiologic processes underlying the exposure–outcome relationship and offer possibilities for remediation and intervention.
Prediction of microRNA targets
In order to identify potential physiological processes that are affected by the exposure-incurred changes in miRNA profiles, studies often employ various existing web-based programs (e.g., miRBase, Targetscan, PicTar) to identify putative mRNA targets of the aberrantly expressed miRNAs. Using slightly varying algorithms, these programs scan a library of mRNA 3’UTR regions to identify potential binding sites for the miRNAs of interest. However, as binding of miRNAs to their targets requires only partial complementarity, prediction of targets typically yields up to 20% false positives [56]. The substantial rate of false positives and the observed discrepancy in predicted targets across the various algorithms highlight the need to experimentally validate reported putative targets.
Methodological differences: next generation sequencing vs. microarray
Several high-throughput methodologies are currently in use to determine miRNA expression levels. Utilization of microarrays relies on nucleic-acid hybridization of labeled miRNA-derived cDNAs to complementary oligonucleotide probes immobilized on the array, with fluorescence intensity indicating the level of expression. This methodology is currently the most widely applied means of high-throughput determination of miRNA expression levels. However, innate properties of miRNAs limit the sensitivity and specificity achieved with this method. For example, miRNAs of low abundance likely fall below the limit of detection on the array. There is also considerable sequence homology among miRNAs, which probes on the array may not be sensitive enough to distinguish. Next generation sequencing is a methodology that is able to address several of these limitations. Additionally, unlike array-based methods, profiles generated using sequencing are not limited to previously identified miRNAs. However, the multiple steps involved in setting up the sequencing reaction offer various opportunities to introduce bias [57]. Furthermore, although the price is continuously dropping, the cost of sequencing at present is still higher than array-based methods.
Specimen issues: placenta, cord blood, and maternal plasma
As with any biomarker-related study, the biospecimen source of miRNAs greatly influences the interpretation of meaningful results. Placental tissue, umbilical cord blood, and maternal sera are common sources for biomarkers reflecting the in-utero experience in human observational studies. All three serve as easily obtainable, noninvasive sources of miRNAs. However, there are factors driving distinctions in the expression profile generated from these biospecimens that need to be considered. Both placenta and blood are composite tissues consisting of heterogeneous cell types. Therefore, minimizing variability in cell-type composition across samples needs to be accounted for prior to analysis to reduce the likelihood of introducing bias into the study. Cord blood does provide access to specific cell lineages that make this particular biospecimen an attractive source for studies focusing on the impact of environmental exposures on the differentiation potential of stem cell populations, such as neuronal precursors, and on the immune response of cytokines.
While the expression profile generated from placental tissue and cord blood reflects the exposure experience toward the end of pregnancy, maternal plasma offers a means to monitor dynamic changes in expression level throughout pregnancy, enabling the focus on specific gestational periods. However, the proportion of placental miRNA in circulation likely fails to account for the comprehensive expression profile of the placenta. Therefore, maternal plasma is best suited to develop screening markers, whereas cord blood and placental levels can also be analyzed to further etiologic understanding.
CONCLUSION
The implementation of miRNAs as indicators of environmental exposures relevant to children's health is still a nascent field, and the promise of their utility is just beginning to be realized. While mRNA transcript levels, the ultimate targets of miRNAs, can also be utilized for this purpose, one of the major attractions over their labile transcript counterparts is the inherent stability and robustness of miRNAs in bodily fluids under various conditions [58,59]. Furthermore, technological advances have made high-throughput assessment affordable, providing a means of feasible and sensitive detection.
The relevance of this marker in these types of studies is also established by the fact that miRNAs are known to be involved in processes with heightened activity during early development. Hence, beyond reflecting extent of exposure to an environmental agent, an observed deregulation of miRNA levels can also point to the etiologic mechanism, ultimately linking a given exposure to an outcome.
Already various exposure and outcome-related signatures have been identified. However, the lack of comparability in experimental conditions prevents deriving meaningful conclusions from the findings reported thus far. Even in studies focusing on the same exposure of interest, variability exists in the form of dosage, study population, biospecimen analyzed, detection methods utilized, and definitions for fold cut-offs. Hence, while technological advances yet to come will further facilitate the ability to assay and analyze expression profiles under various conditions, resulting in novel contributions to the literature, future studies should also focus on replicating existing expression profiles. Only by establishing reproducible expression profiles and validating putative gene targets can the ultimate goal of building a cohesive narrative tying environmental exposures to children's health outcomes be realized.
Acknowledgements
This work was supported by grants from the National Institutes of Health (R01 CA172460, R01HD067611/ R01ES022223-01A1, U01 ES019451) and Mount Sinai Children's Environmental Health Center Pilot Fund.
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.Chance GW, Harmsen E. Children are different: environmental contaminants and children's health. Can J Public Health 1998; 89 Suppl 1:S9–S13S10-5. [PubMed] [Google Scholar]
- 2.Barker D. The origins of the developmental origins theory. J Intern Med 2007; 261:412–417. [DOI] [PubMed] [Google Scholar]
- 3.Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb) 2010; 105:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001; 294:853–858. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004; 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003; 425:415–419. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Jeon K, Lee JT, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 2002; 21:4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohnsack MT, Czaplinski K, Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of premiRNAs. RNA 2004; 10:185–191 10.1261/rna.5167604.Most [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutvágner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001; 293:834–838. [DOI] [PubMed] [Google Scholar]
- 10.Ketting RF, Fischer SE, Bernstein E, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 2001; 15:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature 2000; 404:293–296. [DOI] [PubMed] [Google Scholar]
- 12.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005; 123:631–640. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A 2003; 100:9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr Protoc Bioinformatics 2010; 1–10Chapter 12 Unit 12. [DOI] [PubMed] [Google Scholar]
- 15.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci 2008; 65:545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schotte D, Chau FC, Sylvester G, et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia 2009; 23:313–322. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Luo XQ, Zhang P, et al. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One 2009; 4:e7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira JC, Scrideli CA, Brassesco MS, et al. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk Res 2012; 36:293–298. [DOI] [PubMed] [Google Scholar]
- 19.2009; Fulci V, Colombo T, Chiaretti S, et al. Characterization of B- and T -lineage acute lymphoblastic leukemia by integrated analysis of microRNA and mRNA expression profiles. 1082:1069–1082. [DOI] [PubMed] [Google Scholar]
- 20.Kaddar T, Chien WW, Bertrand Y, et al. Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leuk Res 2009; 33:1217–1223. [DOI] [PubMed] [Google Scholar]
- 21.Tan Z, Randall G, Fan J, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet 2007; 81:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su XW, Yang Y, Lv ML, et al. Association between single-nucleotide polymorphisms in premiRNAs and the risk of asthma in a Chinese population. DNA Cell Biol 2011; 30:919–923. [DOI] [PubMed] [Google Scholar]
- 23.Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol 2011; 128:160–167e4. [DOI] [PubMed] [Google Scholar]
- 24.Abu-Elneel K, Liu T, Gazzaniga FS, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 2008; 9:153–161. [DOI] [PubMed] [Google Scholar]
- 25.Sarachana T, Zhou R, Chen G, et al. Investigation of posttranscriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med 2010; 2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaishnavi V, Manikandan M, Tiwary BK, Munirajan AK. Insights on the functional impact of microRNAs present in autism-associated copy number variants. PLoS One 2013; 8:e56781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Hu Z, Gu H, et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum Mutat 2009; 30:1231–1236. [DOI] [PubMed] [Google Scholar]
- 28.Xing HJ, Li YJ, Ma QM, et al. Identification of microRNAs present in congenital heart disease associated copy number variants. Eur Rev Med Pharmacol Sci 2013; 17:2114–2120. [PubMed] [Google Scholar]
- 29.Zhu S, Cao L, Zhu J, et al. Identification of maternal serum microRNAs as novel noninvasive biomarkers for prenatal detection of fetal congenital heart defects. Clin Chim Acta 2013; 424:66–72. [DOI] [PubMed] [Google Scholar]
- 30.Oglesby IK, Bray IM, Chotirmall SH, et al. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol 2010; 184:1702–1709. [DOI] [PubMed] [Google Scholar]
- 31.Megiorni F, Cialfi S, Dominici C, et al. Synergistic posttranscriptional regulation of the cystic fibrosis transmembrane conductance regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS One 2011; 6:e26601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran S, Karp PH, Jiang P, et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci U S A 2012; 109:13362–13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oglesby IK, Chotirmall SH, McElvaney NG, Greene CM. Regulation of cystic fibrosis transmembrane conductance regulator by microRNA-145, -223, and -494 is altered in ΔF508 cystic fibrosis airway epithelium. J Immunol 2013; 190:3354–3362. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharyya S, Kumar P, Tsuchiya M, et al. Regulation of miR-155 biogenesis in cystic fibrosis lung epithelial cells: antagonistic role of two mRNA-destabilizing proteins, KSRP and TTP. Biochem Biophys Res Commun 2013; 433:484–488. [DOI] [PubMed] [Google Scholar]
- 35.Sebastiani G, Di U, Onlus M. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev 2011; 27:862–866 10.1002/dmrr [DOI] [PubMed] [Google Scholar]
- 36.Nielsen LB, Wang C, Sorensen K, et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res 2012; 2012:896362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salas-Pérez F, Codner E, Valencia E, et al. MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology 2013; 218:733–737. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res 2007; 67:976–983. [DOI] [PubMed] [Google Scholar]
- 39.Bray I, Bryan K, Prenter S, et al. Widespread dysregulation of miRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS One 2009; 4:e7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prats-Puig A, Ortega FJ, Mercader JM, et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab 2013; 98:E1655–E1760. [DOI] [PubMed] [Google Scholar]
- 41.2010; Avissar-Whiting M, Veiga KR, Uhl KM, et al. Reprod Toxicol. 29:401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci 2007; 27:8546–8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang LL, Zhang Z, Li Q, et al. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod 2009; 24:562–579. [DOI] [PubMed] [Google Scholar]
- 44.Soares AR, Pereira PM, Ferreira V, et al. Ethanol exposure induces upregulation of specific microRNAs in zebrafish embryos. Toxicol Sci 2012; 127:18–28. [DOI] [PubMed] [Google Scholar]
- 45.Balansky R, Longobardi M, Ganchev G, et al. Transplacental clastogenic and epigenetic effects of gold nanoparticles in mice. Mutat Res 2013; 751–752:42–48 10.1016/j.mrfmmm.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 46.Maccani MA, Avissar-Whiting M, Banister CE, et al. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics 2010; 5:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47▪.Herberth G, Bauer M, Gasch M, et al. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol 2013; 10.1016/j.jaci.2013.06.036 [DOI] [PubMed] [Google Scholar]; In this study, maternal cotinine levels were positively associated with miR-223 expression levels in cord blood. Additionally, miR-223 expression levels were negatively associated with Treg cell numbers, while low Treg cell counts were associated with an increased risk of atopic dermatitis. These findings are of interest as they suggest a potential etiologic mechanism linking an exposure to an outcome through the deregulation of miRNAs.
- 48.Zhu C, Yu ZB, Zhu JG, et al. Differential expression profile of microRNAs during differentiation of cardiomyocytes exposed to polychlorinated biphenyls. Int J Mol Sci 2012; 13:15955–15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Li Y, Zeng H, et al. MicroRNA expression changes during zebrafish development induced by perfluorooctane sulfonate. J Appl Toxicol 2011; 31:210–222. [DOI] [PubMed] [Google Scholar]
- 50.Wang F, Liu W, Ma J, et al. Prenatal and neonatal exposure to perfluorooctane sulfonic acid results in changes in miRNA expression profiles and synapse associated proteins in developing rat brains. Environ Sci Technol 2012; 46:6822–6829. [DOI] [PubMed] [Google Scholar]
- 51.Singh NP, Singh UP, Guan H, et al. Prenatal exposure to TCDD triggers significant modulation of microRNA expression profile in the thymus that affects consequent gene expression. PLoS One 2012; 7:e45054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52▪.Donker RB, Mouillet JF, Chu T, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod 2012; 18:417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study attempted to characterize the impact of hypoxia on expression levels in exosomal miRNA in addition to unfractionated miRNA. Although no differential exosomal packaging due to hypoxia was observed, the findings are still noteworthy as they address the potential impact of environmental exposures on the role of miRNAs in intercellular communication, in addition to their influence on intracellular activities.
- 53▪.Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci 2011; 31:11748–11755. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors of this study report an altered brain miRNA expression profile in the F2 generation of mice exposed to stress during pregnancy compared with the F2 generation of unexposed mice. These findings indicate that changes incurred in utero can be transmitted transgenerationally.
- 54.Zhang J, Zhang F, Didelot X, et al. Maternal high fat diet during pregnancy and lactation alters hepatic expression of insulin like growth factor-2 and key microRNAs in the adult offspring. BMC Genomics 2009; 10:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Reprod Toxicol 2005; 20:345–352. [DOI] [PubMed] [Google Scholar]
- 56.Gusev Y, Schmittgen TD, Lerner M, et al. Computational analysis of biological functions and pathways collectively targeted by co-expressed microRNAs in cancer. BMC Bioinformatics 2007; 8 Suppl 7:S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Planell-Saguer M, Rodicio MC. Analytical aspects of microRNA in diagnostics: a review. Anal Chim Acta 2011; 699:134–152. [DOI] [PubMed] [Google Scholar]
- 58.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18:997–1006. [DOI] [PubMed] [Google Scholar]
- 59.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010; 56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]


