Abstract
Expression of the Growth Hormone (GH)-stimulated gene Socs2 (Suppressor of Cytokine Signaling 2) is mediated by the transcription activator STAT5 (Signal Transducer and Activator of Transcription 5) and the transcription repressor BCL6 (B-Cell Lymphoma 6). ChIP-Sequencing identified Cish (Cytokine-Inducible SH2-containing protein) and Bcl6 as having similar patterns of reciprocal occupancy by BCL6 and STAT5 in response to GH, though GH stimulates Cish and inhibits Bcl6 expression. The co-activator p300 occupied Socs2, Cish and Bcl6 promoters, and enhanced STAT5-mediated activation of Socs2 and Cish. In contrast, on Bcl6, p300 functioned as a repressor and inhibited in conjunction with STAT5 or BCL6. The co-repressor HDAC3 (Histone deacetylase 3) inhibited the Socs2, Cish and Bcl6 promoters in the presence of STAT5. Thus transcriptional outcomes on GH-regulated genes occupied by BCL6 and STAT5 are determined in a promoter-specific fashion by co-regulatory proteins which mediate the distinction between activating and repressive transcription factors.
Keywords: Socs2, Cish, Co-activator, Co-repressor, Adipocytes, ChIP-Seq
1. Introduction
The diverse actions of Growth Hormone (GH) are mediated by transcriptional programs which involve dynamic complexes containing activating and repressive transcription factors which bind DNA, and co-regulatory proteins which can modulate transcription through changes in chromatin (Kumar et al., 2004; Roeder, 2005; Rosenfeld et al., 2006). Well-studied examples of transcription factors for which transcription regulation involves coordinated activator and repressor molecules are nuclear receptors such as estrogen or thyroid hormone receptors (Lonard and O’Malley, 2007; O’Malley et al., 2012; Perissi et al., 2004): In the absence of ligand, transcription of their target genes is repressed by association with co-repressor proteins such as SMRT (Silencing Mediator of Retinoic acid and Thyroid hormone receptor) or NCoR (Nuclear receptor Co-Repressor) in complex with the DNA-bound receptor. Upon ligand binding, the co-repressor is released and a co-activator such as p300 or CBP (Cyclic AMP response element Binding Protein) is recruited, mediating hormone-induced gene activation (Kumar et al., 2004; Perissi et al., 2004).
It is well-recognized that GH signaling to the nucleus involves STAT (Signal Transducers and Activators of Transcription) proteins, a family of transcription factors that mediate diverse cellular events in response to a large number of cytokines and growth factors, and that STAT5 plays a prominent role in mediating activation of GH-regulated genes (Herrington et al., 2000; Litterst et al., 2005; Waters et al., 2006). In contrast to STAT5 and other activating transcription factors such as C/EBP beta and CREB (Cui et al., 2008, 2011), repression of transcription in response to GH has been poorly understood. STAT5b has recently been implicated in the GH-mediated repression of IGF1 binding protein 1 transcription by indirectly impairing the actions of the FoxO1 transcription factor at the IGF1 binding protein 1 promoter (Ono et al., 2007). More recently, the transcription factor B-Cell Lymphoma 6 (BCL6) was found to repress potently both basal and GH-stimulated expression of Socs2 (Chen et al., 2009; Chia and Rotwein, 2010; Meyer et al., 2009). BCL6 is a transcriptional repressor, most extensively studied in immune regulation where it is a key participant in B cell differentiation (Basso and Dalla-Favera, 2010; Jardin et al., 2007; Schebesta et al., 2002; Staudt et al., 1999). Non-immune functions of BCL6 include protecting testicular germ cells from apoptosis (Kojima et al., 2001) and repressing proliferation of pancreatic β cells (Glauser and Schlegel, 2009; Karnik et al., 2007). BCL6 serves as a male-specific transcription factor mediating GH-regulated sexual dimorphism of gene expression in the liver (Meyer et al., 2009; Zhang et al., 2012). BCL6 as well as STAT5 have been reported to play oncogenic roles in lymphomas and breast cancer (Basso and Dalla-Favera, 2010; Tran et al., 2010), and a role for GH in breast cancer and other malignancies has also been proposed (Perry et al., 2008). Further, BCL6 and STATs have been reported to function as a transcription repressor–activator pair. Such a mechanism is likely since DNA binding motifs for BCL6 and STAT5 are very similar (Chang et al., 1996; Dent et al., 2002; Mascle et al., 2003; Seyfert et al., 1996) and BCL6 and STAT5 have potential to occupy the same DNA regulatory sequences.
The gene for Suppressor of Cytokine Signaling (Socs) 2 is exquisitely sensitive to negative regulation by BCL6 (Chen et al., 2009; Meyer et al., 2009) as well as to activation by STAT5 (Vidal et al., 2007). BCL6 and STAT5 bind reciprocally to a GH-regulated sequence in the Socs2 promoter (Chen et al., 2009; Chia and Rotwein, 2010; Meyer et al., 2009): BCL6 occupies the Socs2 promoter in the absence of GH, and also represses Socs2 transcription (Chen et al., 2009). Upon GH treatment, occupancy of BCL6 decreases and STAT5 occupancy increases (Chen et al., 2009), resulting in potent activation of Socs2 expression. Based in part on their reciprocal occupancy, BCL6 and STAT5 have been postulated to function as reciprocal regulators of transcription for some GH-regulated genes (Chen et al., 2009; Conforto et al., 2012; Meyer et al., 2009), potentially contributing to GH responses.
In GH-regulated transcription complexes, negative regulation by BCL6 and positive regulation by STAT5 may also involve participation of co-regulators. BCL6 is well known to associate with co-repressors in immune cells, including histone deacetylases (HDACs), NCoR, SMRT (Bereshchenko et al., 2002; Dhordain et al., 1997, 1998; Huynh and Bardwell, 1998; Huynh et al., 2000), and the BCL6 co-repressor BCoR (Huynh et al., 2000). When functioning as a transcriptional activator, STAT5 interacts with co-activators such as p300, CBP and NCoA-1 (Nuclear receptor Co-Activator 1), which aid in driving STAT5-mediated transcription in response to cytokine stimulation (Chia and Rotwein, 2010; Kabotyanski et al., 2006; Litterst et al., 2003, 2005).
The reciprocal occupancy patterns and regulatory actions of BCL6 and STAT5 on Socs2 gene expression suggest that their reciprocal regulation is integral to their function in response to GH, which promotes linear growth, induces insulin resistance and alters lipid metabolism (Ahmed and Farquharson, 2010; Albertsson-Wikland and Rosberg, 1988; Barbour et al., 2005; Cohen and LeRoith, 2012; Ikeda et al., 1998). For insight into GH-regulated transcriptional mechanisms mediated by this repressor–activator pair, the present studies used genome-wide chromatin immunoprecipitation (ChIP) and deep sequencing (ChIP-Sequencing) to identify BCL6 target genes and focus on GH-regulated genes for which BCL6 and STAT5 show reciprocal occupancy patterns. The contrasting transcriptional outcomes of these genes in response to GH prompted further investigation revealing changes in chromatin state and varying contributions of co-regulators p300 and HDAC3 in modulating the balance between repression and activation of genes regulated by BCL6 and STAT5 in response to GH.
2. Materials and methods
2.1. Materials
Murine 3T3-F442A preadipocytes were provided by H. Green (Harvard University) and M. Sonenberg (Sloan-Kettering, NY). The 293T human kidney cell line was provided by M. Lazar (University of Pennsylvania) and O. MacDougald (University of Michigan). Recombinant human GH (lot # AFP8990A) for cell culture studies was purchased from the National Hormone and Pituitary Program (Torrance, CA). Recombinant human GH (Lot# N70315) for animal experiments was from Genentech (San Francisco, CA), courtesy of Dr. R.K. Menon (University of Michigan). Insulin, dexamethasone, isobutylmethylxanthine, formaldehyde, Bradford reagent, sodium orthovanadate and SYBR green were from Sigma (St. Louis, MO). Culture media, L-glutamine, and antibiotic-antimycotic were purchased from Invitrogen (Grand Island, NY). Fetal bovine serum (FBS) and calf serum (CS) were from Atlanta Biologicals (Lawrenceville, GA). Bovine serum albumin (BSA, fatty acid-free, cat. # 62150101) was purchased from Proliant (Ankeny, IA). Protease inhibitors leupeptin and aprotinin were purchased from Roche (Indianapolis, IN), and phenylmethylsulfonylfluoride (PMSF) from Mallinckrodt (St. Louis, MO).
2.2. Plasmids
For the Socs2, Cish, and Bcl6 promoter-luciferase constructs, a region from 750 bp upstream to 250 bp downstream from the BCL6/STAT5 site identified by ChIP-Seq of each gene was PCR amplified from mouse genomic DNA and cloned into the multiple cloning region of pGL3-Basic (Promega, Madison, WI). Constructs were confirmed by sequencing at the University of Michigan DNA Sequencing Core. Cloning primer sequences and restriction enzyme sites are listed in Appendix: Supplementary Table S2. The expression construct for mouse BCL6 in pCMV-SPORT6.1 was purchased from Open Biosystems (Lafayette, CO). The constitutively active rat STAT5b construct containing a point mutation of His for Asp at residue 642 (CA-STAT5b) was generously provided by P. Rotwein (Oregon Health Science University) (Woelfle and Rotwein, 2004). The human p300 and HDAC3 expression constructs were generously provided by R. Kwok (University of Michigan) (Cesena et al., 2007a).
2.3. Cell culture
3T3-F442A preadipocytes in 15 cm plates were differentiated into adipocytes as follows: Preadipocytes were grown to confluence in Dulbecco’s Modified Eagle Medium containing 1% L-glutamine, and 1% antibiotic-antimycotic (DMEM) and 8% calf serum. 2 days after confluence, cells were induced to differentiate by incubation in adipogenic medium (DMEM, 8% FBS, 0.5 mM isobutylmethylxanthine, 0.25 μM dexamethasone, 2 μg/ml insulin). After 48 h, the medium was replaced with DMEM containing 8% FBS and 1 μg/ml insulin. 48 h later, medium was changed to DMEM with 8% FBS, to maintain cells until use in experiments. Prior to experiments with GH treatment, confluent 3T3-F442A preadipocytes or adipocytes were deprived of serum for 16–18 h in DMEM containing 1% BSA instead of serum. Cells were then treated without or with human GH (500 ng/ml = 22 nM) for the times indicated, on day 7 following initiation of adipogenesis. 293T cells were cultured in DMEM with 8% calf serum.
2.4. In vivo studies
Tissues were obtained from mice treated chronically with GH or vehicle as follows: Mice used were control heterozygotes (+/−) in a colony crossed to generate targeted deficiency of BCL6 (Dent et al., 1997); for the responses measured, heterozygous mice were comparable to wild type controls (+/+). Mice were housed on a 12-h light, 12-h dark cycle (lights on at 0600 h), with food and water available ad libitum unless otherwise specified. Male mice were injected intraperitoneally with human GH (1.5 mg/kg BW) or with vehicle twice daily for 2.5 days, and were fasted for the final 16–18 h. Animals were euthanized between 1000 and 1200 h by CO2 inhalation. Liver tissue was rapidly removed, weighed, frozen on dry ice and stored at −80 °C. Animal protocols were approved by the University Committee on Use and Care of Animals at the University of Michigan.
2.5. ChIP-Sequencing (ChIP-Seq)
To generate ChIP DNA libraries for high throughput sequencing, 3T3-F442A adipocytes cultured in 15 cm culture plates were treated with GH (500 ng/ml) for 0 or 48 h (40 plates for each treatment condition). Lysates of cross-linked cells (input) were used for ChIP as described (Cui et al., 2005) using anti-BCL6 (N-3). ChIP DNA samples were pre-tested for quality by PCR using primers specific for the BCL6/STAT5 binding site in murine Socs2 (Vidal et al., 2007), and multiple ChIP DNA samples immunoprecipitated (IP) with the same antibody under the same GH treatment conditions were pooled to obtain a sufficient amount of DNA (>10 ng) for generating ChIP-Seq libraries as recommended by the Illumina ChIP-Seq Library Preparation Protocol. ChIP-Seq libraries were generated by ligation of adapter sequences and PCR amplification of chromatin fragments using the Illumina ChIP-Seq Sample Prep Kit following the manufacturer’s protocol. The quality of the ChIP-Seq libraries was validated by the University of Michigan DNA Sequencing Core using an Agilent Bioanalyzer and submitted for sequencing on the Illumina GAIIx sequencing platform. A DNA library was also prepared using input DNA from untreated adipocytes and sequenced in parallel as a background control. The ChIP-Seq experiment was independently performed twice.
Sequencing data obtained from the ChIP-Seq libraries were analyzed using the HPeak algorithm (http://www.sph.umich.edu/csg/qin/HPeak/), a hidden Markov model-based peak finding algorithm which allows for rigorous statistical inference (Qin et al., 2010). Distinct from other currently available algorithms, HPeak explicitly assumes probability distributions to model coverage profiles of DNA fragments. HPeak also provides the location of nearby genes and the genomic location of a peak. Occupancy profiles for Bcl6 on the mouse reference genome (mm9) were visualized using the UCSC Genome Browser (http://genome.ucsc.edu/). Data for replicate experiments were combined and peaks of occupancy with a peak height of 4 or greater, as determined by HPeak, were used in subsequent bioinformatic analyses.
To identify a subset of candidate peaks and related genes for experimental verification of BCL6 or STAT5 occupancy, ChIP-Seq peaks identified by HPeak (Qin et al., 2010) under each of the conditions (IP with antibody against BCL6, without or with GH for 48 h) were sorted by either peak height or P-value. The genes proximal to the top 200 ChIP-Seq peaks either upstream or downstream were prioritized according to previous studies reporting on them in conjunction with BCL6 or STAT5. Profiles of top peaks identified were selected using the ChIP-Seq data and the UCSC Genome Browser. Peak locations that showed signal across the various IP and treatment conditions were considered high priority for follow-up, as were locations that showed a single, distinct peak profile. Based on these multiple screens, a total of 35 ChIP-Seq peaks were selected for initial verification by standard ChIP-PCR of BCL6 and/or STAT5 binding in the presence or absence of GH. ChIP PCR primers were designed to flank the ChIP-Seq peaks identified by HPeak, with the final PCR product around 200 bp in length. Primers were designed using NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
2.6. Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously (Cui et al., 2005). Briefly, 3T3-F442A adipocytes or preadipocytes in 15 cm culture dishes were treated with 500 ng/ml GH for various times. Cells were washed twice in cold PBS and crosslinked in 1% formaldehye in PBS at room temperature for 15 min. For analysis of liver, frozen mouse liver (~200 mg) was thawed, minced in PBS and incubated in 1% formaldehyde in PBS for 15 min at room temperature before centrifugation for 2 min at 1500 rpm. Sonication was carried out using a Misonix S-3000 sonicator eighteen times for 15 sec, with a 1 min pause between cycles, to achieve chromatin fragments of approximately 200 bp. IP was carried out with 4 μg of antibodies against BCL6 (N-3, Santa Cruz), STAT5 (C-17, Santa Cruz), STAT5a (L-20, Santa Cruz), STAT5b (Millipore, Billerica, MA), phosphorylated STAT5 (pY694) (71-6900, Invitrogen), p300 (N-15, Santa Cruz), HDAC3 (H-99, Santa Cruz), acetylated histone H3 (Millipore), acetylated histone H4 (Millipore), and H3K4me3 (Abcam, Cambridge, MA), as indicated. Samples incubated with an equivalent amount of normal rabbit immunoglobulin (IgG, Cell Signaling, Danvers, MA) served as negative controls. 1% input was used to indicate the relative amount of each sample used for individual ChIP analysis.
ChIP samples were analyzed either by PCR followed by separation of products on 2% agarose gels and staining with ethidium bromide (Cui et al., 2005), or by quantitative real time PCR (qPCR). PCR primers for agarose gel analysis of the BCL6/STAT5 site in the Socs2 promoter have been described previously (Chen et al., 2009; Vidal et al., 2007). Primers for agarose gel analysis of the BCL6/STAT5 occupancy sites in the Cish and Bcl6 promoters identified by ChIP-Seq were designed using NCBI Primer-BLAST (Appendix: Supplementary Table S2). For quantitative ChIP PCR experiments, qPCR was carried out as described (Huo et al., 2006) using ChIP DNA as the PCR template. Signals for each IP condition were normalized to the signal for input ChIP DNA for each of the respective treatment conditions. Input DNA amplification signal was consistent between all experiments. Primer sequences for ChIP qPCR were designed using NCBI Primer-BLAST (Appendix: Supplementary Table S2).
2.7. Transcription assays
293T cells were transfected by calcium phosphate co-precipitation (Cui et al., 2005) with 200 ng luciferase reporter construct and 100 ng of plasmid for mouse BCL6, CA-STAT5b, p300, or HDAC3 per well of a 12 well culture plate, alone or in combination as indicated. Each transfection was performed in duplicate in each experiment. DNA transfection amounts were normalized across transfections using pcDNA3 (Invitrogen). After 24 h incubation at 37 °C, cells were washed twice in cold PBS, lysed in 250 μl of 1× passive lysis buffer (Promega) and analyzed using luciferase reagents from Promega Dual Luciferase Kit. Luciferase in lysates was analyzed on a Biotek Synergy 2 microplate reader (Biotek, Winooski, VT) and normalized for protein concentration (Bradford assay). Luciferase activity is expressed as RLU (relative luciferase units) and normalized to basal values for the respective promoter construct, which was set as 1.0.
2.8. Statistics
Values from ChIP-qPCR and luciferase measurements were analyzed statistically using Student’s t-test with GraphPad Prism (La Jolla, CA) software.
3. Results
3.1. Reciprocal occupancy of BCL6 and STAT5 is regulated by GH
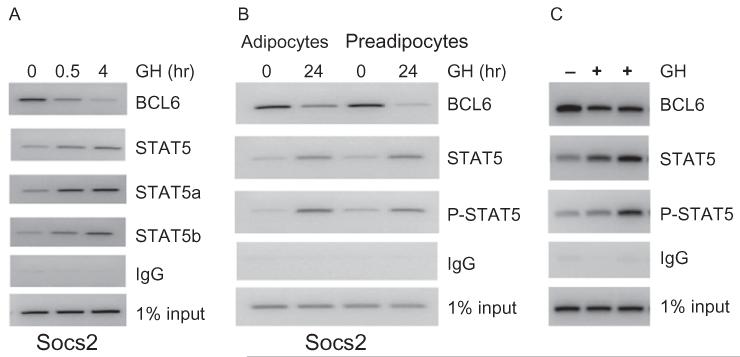
Reciprocal regulation of Socs2 by GH, mediated by the transcription repressor BCL6 and the activator STAT5, derives from observations that occupancy of BCL6 and STAT5 on Socs2 DNA is reciprocally regulated by GH. BCL6 occupancy on the STAT5 binding sequence in the Socs2 promoter is evident in the absence of GH, and decreases progressively 30 min and 4 h after GH treatment of 3T3-F442A preadipocytes (Fig. 1A). As the occupancy of BCL6 decreases, the occupancy of STAT5 increases progressively, as detected by ChIP with a pan-STAT5 antibody, as reported previously (Chen et al., 2009). The pattern of decreasing BCL6 occupancy and increasing STAT5 occupancy with GH treatment is consistently observed in both 3T3-F442A adipocytes and preadipocytes (Chen et al., 2009). The increase in STAT5 occupancy with GH treatment reflects increased occupancy of both endogenous STAT5a and STAT5b on Socs2 DNA in response to GH (Fig. 1A). The STAT5a and STAT5b isoforms share 91% amino acid identity (Grimley et al., 1999) and have both redundant and distinct functions (Adhikari et al., 2011; Hennighausen and Robinson, 2008; Teglund et al., 1998). Increased STAT5 occupancy on Socs2 is accompanied by a GH-induced increase in the occupancy of tyrosyl phosphorylated STAT5 (Fig. 1B). The reciprocal occupancy of endogenous BCL6 and STAT5 on Socs2 is detectable in vivo in liver of mice treated with GH (Fig. 1C; Meyer et al., 2009) and occupancy of endogenous BCL6 is also evident in mouse adipose tissue (not shown). For Socs2, functional consequences as well as occupancy of BCL6 and STAT5 are also reciprocal: BCL6 represses Socs2 promoter activation in response to GH, while STAT5 activates it (Chen et al., 2009; Laz et al., 2009; Vidal et al., 2007). Thus, the highly sensitive reciprocal regulation of BCL6 and STAT5 serves as a paradigm to probe mechanisms for functional interactions of a regulated transcriptional repressor–activator pair by GH.
Fig. 1.
BCL6 and STAT5 occupy Socs2 DNA reciprocally in response to GH. (A) 3T3-F442A preadipocytes were treated with GH for 0, 0.5 or 48 h and ChIP was carried out using antibodies against BCL6, STAT5, STAT5a or STAT5b. ChIP DNA was probed with primers specific to the STAT5 occupancy sequence on the Socs2 promoter. IgG served as a negative control and 1% input was used as an internal control, in this and subsequent figures. Data are representative of at least two experiments. (B) 3T3-F442A preadipocytes and adipocytes were treated with GH for 0 and 24 h and ChIP was carried out using antibodies against BCL6, STAT5, and phosphorylated STAT5 (P-STAT5). ChIP DNA was probed with primers specific to the STAT5 occupancy sequence on the Socs2 promoter. (C) Male littermate mice were treated ip with GH (1.5 mg/kg in 48 h) (+) or vehicle (−). Liver was used for ChIP analysis using antibodies against BCL6, STAT5 or P-STAT5 and probed for Socs2 as above.
3.2. Socs2, Cish and Bcl6 genes are reciprocally regulated by BCL6 and STAT5
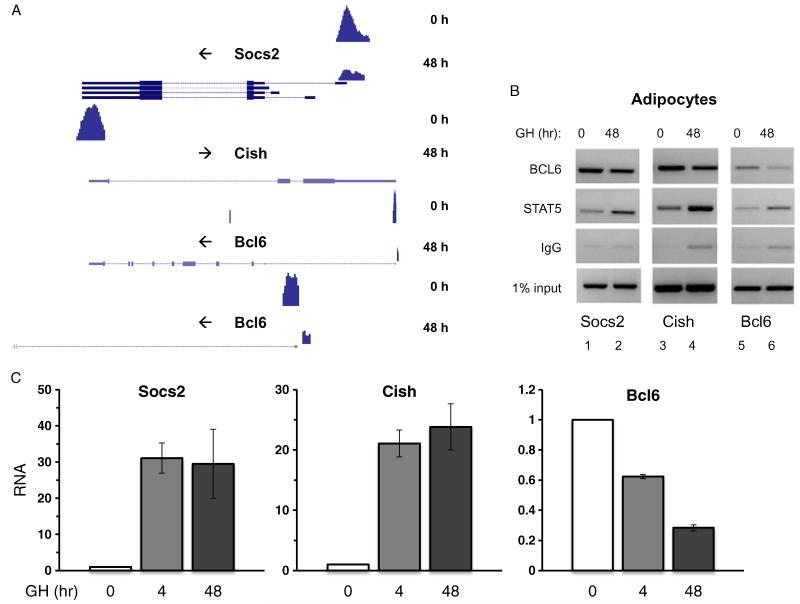
BCL6 occupancy throughout the genome was examined using high-throughput ChIP-Seq to identify candidate BCL6 target genes. GH-responsive 3T3-F442A adipocytes were treated with GH for 0 or 48 h, a time when Socs2 expression is strongly induced by GH (Chen et al., 2009; Huo et al., 2006). ChIP was carried out using antibodies against BCL6, and ChIP-Seq libraries were prepared from the resulting ChIP DNA, which were subjected to deep sequencing. Sequencing data were analyzed using the HPeak algorithm (Qin et al., 2010) which identified over 3000 potential binding sites for BCL6 across the genome (Lin G and Schwartz J, manuscript in preparation). Candidate peaks of BCL6 occupancy for follow-up were identified based on peak heights, peak significance, and peak profiles, as well as on the reported association of nearby genes with either BCL6 or STAT5; the gene or genes closest to a peak were predicted to be the most likely targets for transcriptional regulation. Based on these initial screens, 35 candidate BCL6 occupied loci (Appendix: Supplementary Table S1) were processed for initial verification by standard ChIP-PCR for BCL6 and STAT5, using primers specific for the genomic regions of ChIP-Seq signal for each of the 35 loci, on samples from 3T3-F442A adipocytes treated without or with GH. Among these, three showed reciprocal occupancy of BCL6 and STAT5 in response to GH. One was Socs2: Examination of the BCL6 ChIP-Seq peak profiles associated with the Socs2 locus showed a single BCL6 peak near the Socs2 transcription start site (TSS) (Fig. 2A, top panel); the peak height could be substantially reduced in cells treated with GH. The GH-regulated BCL6 occupancy on Socs2 detected by ChIP-Seq is consistent with prior ChIP observations of BCL6 occupancy on a functional STAT5 motif on Socs2, which overlaps with this ChIP-Seq locus (Chen et al., 2009; Chia and Rotwein, 2010; Meyer et al., 2009; Vidal et al., 2007).
Fig. 2.
BCL6 and STAT5 show reciprocal occupancy at transcription start sites of Socs2, Cish, and Bcl6. (A) Peaks of BCL6 occupancy determined by ChIP-Seq were identified by HPeak at the transcription start sites (TSS) of Socs2, Cish and Bcl6. Shown are the UCSC Genome Browser profiles for BCL6 occupancy in 3T3-F442A adipocytes treated with GH for 0 or 48 h. Y axis scales were set at 60 for Socs2, 35 for Cish, and 20 for Bcl6 as determined by maximum peak height in the absence of GH as calculated by HPeak. Top panel: Socs2. TSS: mouse chromosome (chr) 10: 94879491. Peak location 0 h GH: chr10: 94879276-94880050; 48 h GH: chr10: 94879326-94879975. Predicted BCL6/STAT consensus sites: chr10: 94879539-94879547 and chr10: 94879550-94879558. Second panel: Cish. TSS: chr9: 107199020. Peak location 0 h GH: chr9: 107198726-107199400; 48 h GH: chr9: 107198751-107199150. Predicted BCL6/STAT consensus sites: chr9: 107198991-107198999 and chr9: 107199002-107199010. Third panel: Bcl6. TSS: chr16: 23988698. Peak location 0 h GH: chr16: 23988401-23988850; 48 h GH: chr16: 23988776-23988900. Predicted BCL6/STAT consensus sites: chr16: 23988676-23988684 and chr16: 23988717-23988725. Bottom panel: Peak profile at Bcl6 promoter region with expanded scale to visualize peaks in detail. Arrows to the left of gene names indicate direction of transcription, from 5′ to 3′. Gene structure diagrammed at bottom of each profile. (B) 3T3-F442A adipocytes were treated with GH for 0 or 48 h and ChIP was carried out using antibodies against BCL6 and STAT5. ChIP DNA was probed with primers specific to the BCL6 occupancy sites corresponding to ChIP-Seq (peaks) on the Socs2 (lanes 1 and 2), Cish (lanes 3 and 4) and Bcl6 (lanes 5 and 6) genes. (C) 3T3-F442A adipocytes were treated with GH for 0, 4 or 48 h. RNA was prepared and analyzed by qPCR using primers for Socs2, Cish and Bcl6. Bars show mean + SE of triplicates in an experiment which was repeated at least 3 times.
In addition, two other genomic regions of BCL6 occupancy in ChIP-Seq were found to show clear reciprocal occupancy by BCL6 and STAT5 in response to GH: One region was identified as a ChIP-Seq peak of BCL6 signal near the TSS of the Cish (Cytokine-Inducible SH2-containing protein) gene (Fig. 2A, middle panel). The second region was a peak of BCL6 near the TSS of the Bcl6 gene itself (Fig. 2A, bottom panel). For Cish, a BCL6 peak was not detected after GH; for the Bcl6 locus, the peak height of BCL6 occupancy was reduced 60% after GH. Not only were the BCL6 peaks located in the proximal promoter region of each of these three genes (Fig. 2A), but they also corresponded with BCL6 or STAT motifs predicted by HPeak. Peaks tested for other candidate genes in the initial screen were primarily at genomic locations such as introns or intergenic regions (Appendix: Supplementary Table S1). Reciprocal occupancy of BCL6 and STAT5 was confirmed by standard ChIP in 3T3-F442A adipocytes treated with or without GH for 48 h. For both the Cish and the Bcl6 loci, occupancy of BCL6 was higher without GH and decreased after GH treatment (Fig. 2B). Conversely, STAT5 occupancy was lower in the absence of GH and higher with GH treatment (Fig. 2B) (Chen et al., 2009). The similarity in binding patterns among the three genes may be related to similarity in the BCL6 and STAT5 consensus DNA binding motifs (Baron et al., 1995; Hartatik et al., 2001; Ihle, 1996); for convenience, they are referred to here collectively as “BCL6/STAT5 regulatory region” in the proximal promoters of these genes (Fig. 5A).
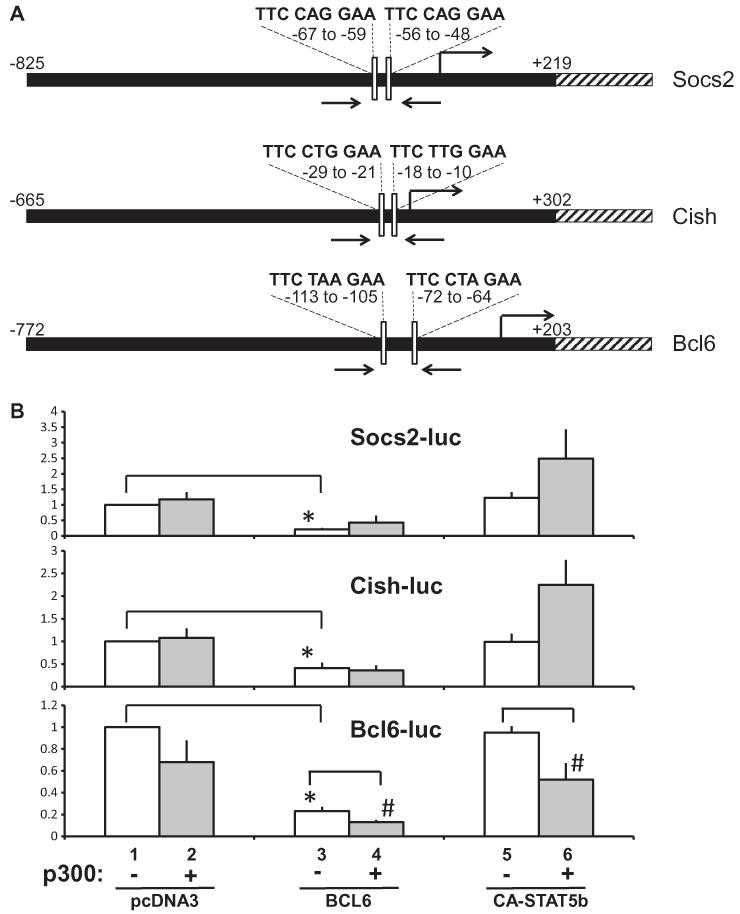
Fig. 5.
p300 represses transcription of the Bcl6 promoter. (A) Diagram of Socs2, Cish and Bcl6 genes showing relationship of genomic promoter sequences (black) driving luciferase (hatched) in reporter plasmids, predicted BCL6/STAT sites within occupied sequence (white bars), and location of ChIP primers (arrows). Predicted BCL6/STAT motifs are indicated above motif locations. Chromosomal locations are indicated in the legend of Fig. 2A. Diagrams are not to scale. (B) Plasmids for Socs2-luc (top), Cish-luc (middle) or Bcl6-luc (bottom) were transfected into 293T cells in the absence (bars 1–2) or presence of BCL6 (bars 3–4), or constitutively active STAT5b (CA-STAT5b) (bars 5–6). Plasmid for p300 (gray bars 2, 4, 6) or empty vector (pcDNA3, open bars 1, 3, 5) was co-expressed as indicated. Bars represent means + SE for 5 (Socs2, Cish) or 6 (Bcl6) independent experiments. Asterisks (*) indicate that BCL6 (bar 3) significantly (P < 0.05) represses expression compared to basal luciferase (bar 1). Hatch signs (#) show responses to p300 that are significantly (P < 0.05) different from the respective pair in the absence of p300.
Despite the similarity in BCL6 and STAT5 binding, the transcriptional responses to GH of the three genes do not coincide: GH stimulates the expression of Cish and Socs2, but GH potently inhibits the expression of Bcl6 mRNA (Fig. 2C). It is not surprising that Socs2 and Cish show similar binding and responses to GH, since the Cish gene product is a member of the SOCS family proteins (Croker et al., 2008; Kile et al., 2002): SOCS2 and CIS proteins suppress GH and cytokine signaling. Further, Cish is a STAT5 target gene (Matsumoto et al., 1997) which also binds BCL6 (Chia and Rotwein, 2010). On the other hand, for the Bcl6 gene, GH treatment and BCL6 itself repress transcription (auto-repression) (Chen et al., 2009; Mendez et al., 2008; Pasqualucci et al., 2003). STAT5 binding has been recognized to be cell type specific (Kang et al., 2013), and STAT5 has been reported either to activate or repress Bcl6 transcription depending on cell type (Diehl et al., 2008; Icardi et al., 2012; Karnik et al., 2007; Scheeren et al., 2005; Tran et al., 2010; Walker et al., 2007). The discrepancy between the shared reciprocal occupancy patterns of BCL6 and STAT5 in response to GH for Socs2, Cish and Bcl6, and their contrasting transcriptional outcomes in response to GH, where Bcl6 is inhibited while Socs2 and Cish are potently stimulated, was examined further to assess whether distinct mechanisms involving BCL6 and STAT5 might link transcription factor binding and transcriptional outcomes of the GH-regulated genes.
3.3. GH induces histone acetylation at the promoters of Socs2 and Cish, but not Bcl6
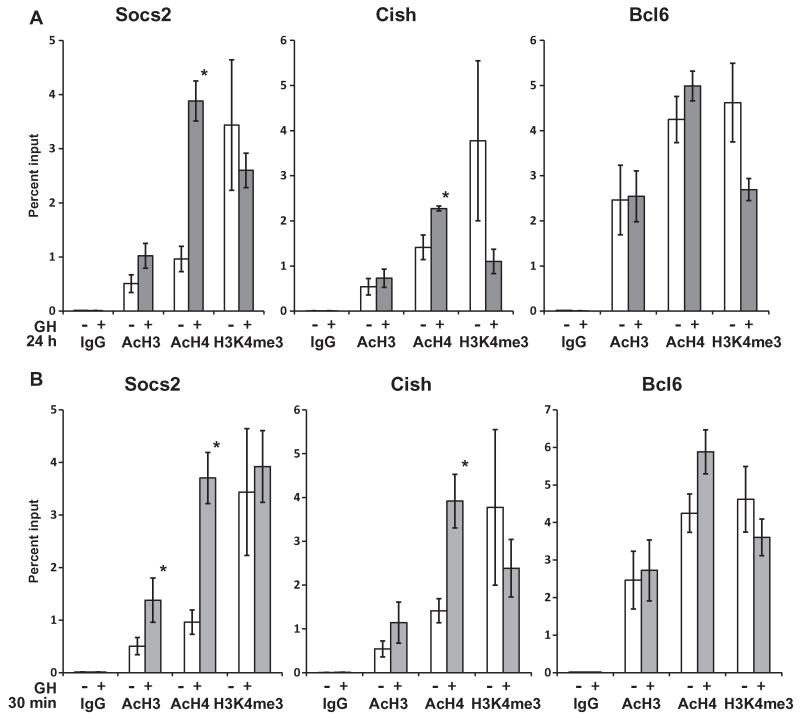
Transcription generally proceeds when chromatin is in an open conformation, which can be assessed by histone acetylation. To examine whether changes in the chromatin state of the respective BCL6/STAT5 occupied regions were different among Socs2, Cish and Bcl6, histone modifications were analyzed in the presence and absence of GH. Adipocytes were treated with GH for 24 h (Fig. 3A), as Socs2 and Cish expressions are strongly induced and Bcl6 expression is strongly inhibited with chronic GH (Fig. 2C) (Chen et al., 2009). ChIP was performed using antibodies against acetylated histone H3 (AcH3), acetylated histone H4 (AcH4), or trimethylated histone H3 lysine 4 (H3K4me3), general markers of open and accessible chromatin. Each of these histone marks was detectable on Socs2, Cish and Bcl6 in the absence of GH (Fig. 3), suggestive of active gene transcription (Berger, 2002; Grunstein, 1997; Ruthenburg et al., 2007; Strahl and Allis, 2000). On both Socs2 and Cish, levels of AcH4 increased significantly in response to GH (Fig. 3A), consistent with GH increasing active transcription of these genes. In contrast, AcH4 levels on Bcl6 barely changed with GH treatment. The significance of the higher basal levels of AcH3 and AcH4 on Bcl6 is not clear. Strong signal for H3K4me3 was detected with and without GH at the BCL6/STAT5 regulatory regions of Socs2, Cish, and Bcl6, consistent with accessible chromatin and active promoter regions for all three genes.
Fig. 3.
Enrichment of AcH3 and AcH4 increases in response to GH on Socs2 and Cish, but not Bcl6. 3T3-F442A adipocytes were treated without (open bars) or with GH (gray bars) for (A) 24 h or (B) 30 min. ChIP was performed using antibodies against AcH3, AcH4, or H3K4me3. QPCR was used to analyze enrichment of these activating histone marks at the BCL6/STAT5 sites on Socs2, Cish, and Bcl6 promoter sequences. Signal from ChIP DNA was normalized to input samples for respective treatment conditions. IgG was used as negative control. Each bar shows the mean ± SE from 4 independent experiments. Asterisks (*) indicate responses to GH that are statistically significant (P < 0.05).
Adipocytes were also treated with GH for 30 min (Fig. 3B), to examine changes in chromatin state during an earlier time of GH treatment. Timing of GH responses in adipocytes has long been of particular interest since acute (e.g. 30 min) and chronic (4 h or longer) metabolic responses to GH differ (Goodman, 1968), and pulsatile GH secretion patterns can govern its regulation of gene expression (Tannenbaum et al., 2001). Further, adipocyte gene expression is time-dependent, showing circadian rhythmicity (Wang and Lazar, 2008). Nevertheless, the histone modifications after 30 min GH (Fig. 3B) were generally comparable to those after 24 h, although AcH3 was significantly increased by GH on Socs2 at 30 min. On both Socs2 and Cish, levels of AcH4 increased significantly in response to GH, while AcH4 levels on Bcl6 barely changed with GH treatment. Strong signal for H3K4me3 was detected with and without GH at the BCL6/STAT5 regulatory regions of Socs2, Cish, and Bcl6. Overall, these findings suggest that stimulation of Socs2 and Cish by GH relative to its inhibition of Bcl6 may reflect differences among the genes in the state and changes in chromatin at the level of regulation of H4 acetylation.
3.4. Occupancy of p300 and HDAC3 does not distinguish Bcl6 from Socs2 and Cish
To examine whether co-regulatory proteins associated with BCL6 and/or STAT5 might distinguish Socs2 and Cish from Bcl6, recruitment of representative co-regulators, the co-activator p300 and the co-repressor HDAC3, to the three genes was compared by ChIP. p300 is a ubiquitous lysine acetyltransferase which in addition to acetylating histones, is known to associate with and acetylate numerous transcription factors (Gingras et al., 1999) including both BCL6 and STAT5 (Bereshchenko et al., 2002; Paulson et al., 1999). p300 is an effective co-activator in adipocytes (Erickson et al., 2001) and is regulated by GH (Cui et al., 2005, 2011). The role of p300 as a co-activator of STAT5 has been well described (Litterst et al., 2005; Paulson et al., 1999; Pfitzner et al., 1998). The acetylation status of STAT5, as modulated by p300 and HDACs, is a critical determinant of its activity (Icardi et al., 2012; Rascle et al., 2003). The HDACs have often been implicated in transcriptional repression (Fischle et al., 2001; Icardi et al., 2012; Narlikar et al., 2002), and multiple Class I and Class II HDACs associate with BCL6 (Jardin et al., 2007). HDAC3 is a Class I HDAC which plays a prominent role in regulation of genes mediating adipocyte metabolism and adipogenesis (Alenghat et al., 2008; Feng et al., 2011). Further, HDAC3 regulates a subset of BCL6 target genes involved in the innate immune response (Barish et al., 2010). Accordingly, p300 and HDAC3 were examined as representative co-activator and co-repressor respectively, to examine if they also participate in reciprocal regulation of transcription by BCL6 and STAT5.
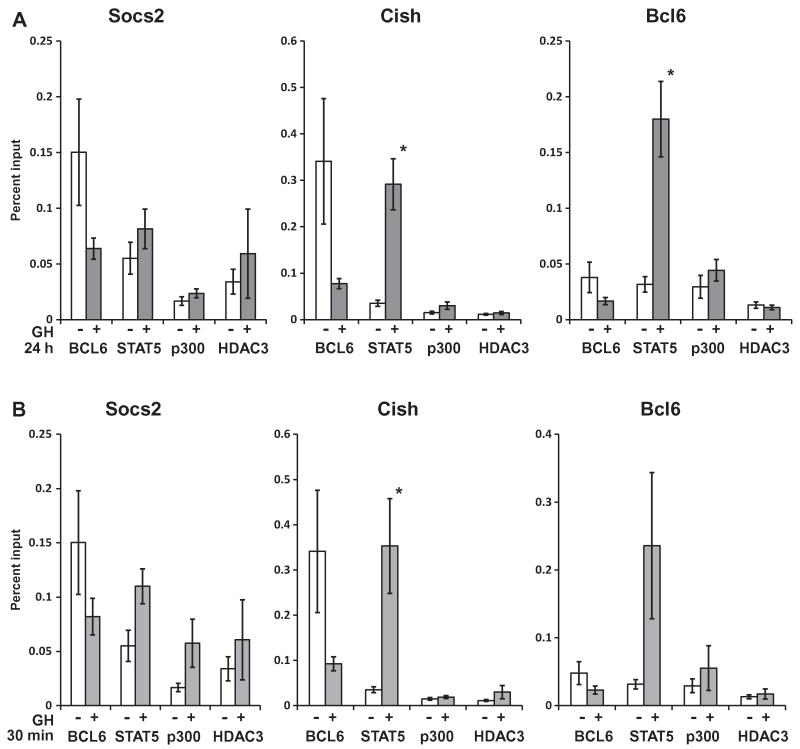
To compare the participation of p300 or HDAC3 in the regulation of Socs2, Cish and Bcl6, 3T3-F442A adipocytes were treated with GH for 24 h or 30 min (Fig. 4). ChIP was performed for p300 and for HDAC3, as well as for BCL6 and STAT5. Here, the ChIP samples were analyzed by qPCR for quantitative analysis. The ChIP-qPCR trends agree with the prior ChIP findings that for Socs2, Cish and Bcl6 genes, BCL6 occupancy is consistently higher in the absence of GH and decreases with GH treatment (Fig. 4). Conversely, occupancy of STAT5 is lower in the absence of GH, and increases upon GH treatment, consistent with the highly reproducible reciprocal changes in occupancy of BCL6 and STAT5 in response to GH determined previously by ChIP-PCR.
Fig. 4.
ChIP identifies p300 and HDAC3 on Socs2, Cish, and Bcl6. 3T3-F442A adipocytes were treated without (open bars) or with GH (gray bars) for (A) 24 h or (B) 30 min. ChIP was performed with antibodies against BCL6, STAT5, p300, or HDAC3. ChIP DNA was analyzed by qPCR with primers for the BCL6/STAT5 sites on Socs2, Cish, and Bcl6. Signals were normalized to input DNA from respective treatment conditions. Each bar shows the mean ± SE for 7 (BCL6 and STAT5) or 3 (p300 and HDAC3) independent experiments. Asterisks (*) indicate responses to GH that are statistically significant (P < 0.05).
In the same experiments, p300 occupancy was detected on both Socs2 and Cish in the basal state. Occupancy of p300 was also evident on Bcl6, suggesting that the absence of p300 does not distinguish Bcl6 from Socs2 and Cish. Occupancy of HDAC3 was detected on Socs2, Cish, and Bcl6 in the basal state. In general, the occupancy signals of HDAC3, as well as those of other HDACs tested (HDAC1, HDAC2, HDAC5, data not shown), and of p300, were lower than basal signals for BCL6 or STAT5 under the conditions of these experiments. Nevertheless, the detectable occupancy of HDAC3 on Socs2 and Cish as well as Bcl6 suggests that the presence vs absence of HDAC3 does not itself account for differences between repression and activation of these genes.
Although transcriptional responses to GH distinguish repression of Bcl6 from activation of Socs2 and Cish, the occupancy patterns of p300 or HDAC3 were changed minimally if at all by GH. Changes in p300 occupancy were not detected on any of the three genes after 24 h GH (Fig. 4A). Treatment with GH for 30 min increased the occupancy of the p300 on Socs2 (Fig. 4B), consistent with GH-stimulated Socs2 expression, though a comparable increase in p300 occupancy after GH was not detected on Cish, nor was a decrease of the co-activator observed on Bcl6 (Fig. 4B). Chronic exposure to GH did not alter the pattern: Thus, the differences in transcriptional responses to GH among Socs2, Cish and Bcl6 do not appear to reflect differences in occupancy of p300 in response to GH. The occupancy of HDAC3 also did not appear to be regulated by GH, since HDAC3 was not detectably altered by GH treatment when promoters of Socs2, Cish or Bcl6 were evaluated. Comparable HDAC3 occupancy with or without GH was observed after 24 h or 30 min GH (Fig. 4) for all three genes. Among other co-repressors that have been reported to associate with Bcl6, occupancy of NCoR, SMRT and mSin3A was detected in the basal state on Socs2 and Cish as well as on Bcl6. In initial trials, their patterns of occupancy were not different among the three genes, and were not reproducibly altered by GH (data not shown). These observations suggest that both the co-activator p300 and the co-repressor HDAC3 occupy the promoters of Socs2, Cish and Bcl6, but that differences in basal occupancy of p300 and HDAC3, or changes in their occupancy in response to GH, do not coincide with differences in their transcriptional responses.
3.5. Activation of Socs2 and Cish promoters by STAT5 is enhanced by p300
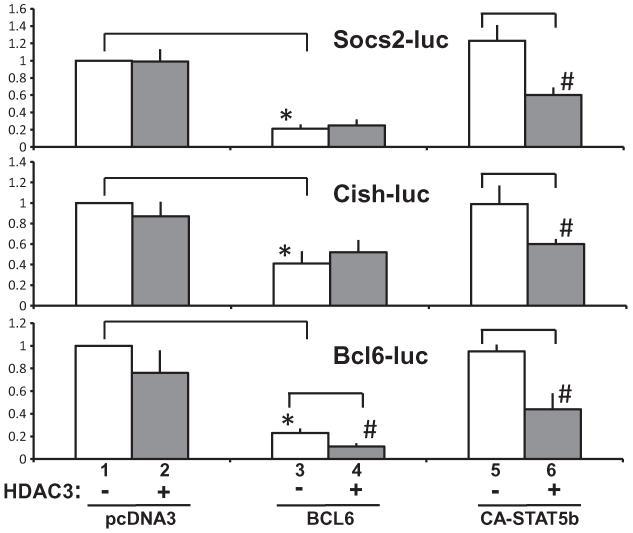
Since p300 and HDAC3 occupancy was detected at the BCL6/STAT5 regulatory regions of the Socs2, Cish, and Bcl6 genes, and since these co-regulators function in adipocytes and are regulated by GH, the functional contributions of p300 and HDAC3 on BCL6- or STAT5-mediated regulation of Socs2, Cish, and Bcl6 transcription were examined. BCL6/STAT5-occupied sequences for the Socs2, Cish, and Bcl6 promoters were fused upstream of the luciferase gene (Fig. 5A). The promoter constructs contained sequences spanning approximately 750 bp upstream and 250 bp downstream of the BCL6/STAT5 site identified by ChIP-Seq for each gene (Fig. 5A). To assess functional contributions of p300, activation of the three reporter constructs was evaluated in context of the expression of p300 alone, and p300 when co-expressed with BCL6 or with constitutively active STAT5b (CA-STAT5b). CA-STAT5b was used both to pinpoint the contributions of STAT5b on the regulation of the promoters of each of the three genes and as a representative of the downstream consequence of GH treatment, although overexpression of CA-STAT5b in these studies may not be directly reflective of physiological conditions in vivo.
For Socs2-luc (Fig. 5B, top), p300 alone did not alter Socs2-luc expression greatly under the conditions tested, compared to basal levels. BCL6 expression potently inhibited the Socs2 reporter gene as observed previously with a similar Socs2-luc construct (Chen et al., 2009). In the presence of BCL6, promoter activity was slightly greater with addition of p300 than with BCL6 alone, but Socs2-luc expression remained lower than basal Socs2-luc or with p300 alone, reinforcing the strong inhibition of the Socs2 promoter by BCL6. The addition of CA-STAT5b alone did not substantially elevate Socs2-luc above basal levels (Fig. 5B top). In support of CA-STAT5 as activator, the addition of p300 in the presence of CA-STAT5b doubled the activation of the promoter compared to CA-STAT5b alone or to basal Socs2-luc levels. A general trend toward activation was observed with p300 and STAT5 over STAT5 alone, though the increase was not significant. Regulation of Cish-luc (Fig. 5B, middle) by p300 was similar to the pattern for Socs2-luc: p300 alone did not appear to alter Cish-luc expression compared to basal levels. BCL6 alone potently and significantly inhibited Cish-luc expression demonstrating that BCL6 functions as a repressor for Cish. The inhibition by BCL6 was maintained despite co-expression of p300. CA-STAT5b alone did not activate the Cish-luc promoter above basal levels, though the expression of p300 in conjunction with CA-STAT5b doubled Cish-luc activation above levels with CA-STAT5b alone, with p300 alone, or with basal Cish-luc, consistent with the ability of p300 to enhance STAT5 activation of Socs2 and Cish.
3.6. p300 functions as a co-repressor on the Bcl6 promoter
Regulation of Bcl6-luc in the presence of p300 (Fig. 5B, bottom) contrasted with the changes observed for Socs2-luc and Cish-luc. The expression of p300 alone modestly reduced Bcl6-luc expression compared to basal levels under the conditions of these experiments. The expression of BCL6 alone strongly and significantly repressed Bcl6-luc promoter activation compared to basal conditions, as it did for Socs2 and Cish. However, for Bcl6-luc, the addition of p300 in combination with BCL6 protein repressed Bcl6-luc expression further, significantly below the repression exerted when BCL6 alone was expressed. Thus, p300 appears to exacerbate the repression by BCL6 on the Bcl6 promoter.
The Bcl6 promoter was also distinctly different from Socs2 and Cish promoters when p300 was co-expressed in combination with CA-STAT5b. Expression of CA-STAT5b alone did not alter Bcl6-luc expression relative to basal values (Fig. 5B, bottom), as observed for Socs2 and Cish promoters under the conditions of these experiments. Strikingly, however, p300 significantly inhibited Bcl6-luc expression in the presence of CA-STAT5b. Not only did p300 decrease Bcl6-luc, but this change was in sharp contrast to the apparent increases exerted by p300 with CA-STAT5b on Socs2 and Cish. These findings suggest that the Bcl6 promoter may be ‘primed’ for repression, such that p300 functions as a co-repressor and/or recruits other factors in the presence of STAT5 or BCL6 and renders STAT5b inhibitory on the Bcl6 promoter rather than activating. The relative changes elicited in the presence of p300 are consistent with the transcriptional outcomes for the three genes: increases for Socs2 and Cish and decreases with Bcl6.
3.7. HDAC3 serves as a co-repressor for STAT5 on Socs2 and Cish as well as Bcl6
HDAC3 alone was expressed under conditions in which it did not reduce basal promoter activation for Socs2 or Cish (Fig. 6, top and middle). In the context of the potent repression elicited by expression of BCL6 alone, the addition of HDAC3 did not repress expression of Socs2-luc or Cish-luc further. However, in the presence of CA-STAT5b which alone did not alter promoter activation, the addition of HDAC3 significantly inhibited both the Socs2 and Cish promoters, compared to either HDAC3 alone or CA-STAT5b alone. These observations are consistent with co-repression and/or recruitment of other factors by HDAC3 to STAT5b on Socs2 and Cish. Co-expression of HDAC3 significantly inhibited Bcl6-luc when expressed in combination with BCL6 protein (Fig. 6, bottom) or with STAT5.
Fig. 6.
HDAC3 co-represses STAT5b on Socs2, Cish and Bcl6. Plasmids for Socs2-luc (top), Cish-luc (middle) or Bcl6-luc (bottom) were transfected into 293T cells in the absence (bars 1–2) or presence of BCL6 (bars 3–4), or constitutively active STAT5b (CA-STAT5b) (bars 5–6). Plasmid for HDAC3 (gray bars 2, 4, 6) or empty vector (pcDNA3, open bars 1, 3, 5) was co-expressed as indicated. Bars represent means + SE for 5 (Socs2, Cish) or 6 (Bcl6) independent experiments. Asterisks (*) indicate that BCL6 significantly (P < 0.05) repressed expression compared to basal luciferase (bar 1). Hatch signs (#) show responses to HDAC3 that are significantly (P < 0.05) lower than respective pair in the absence of HDAC3.
Thus, the Bcl6 promoter is distinguished from Socs2 and Cish in being inhibited by HDAC3 in the presence of BCL6, and strikingly, in being inhibited by p300 in the presence of BCL6 or STAT5. Together these findings show that BCL6 is a potent inhibitor of three GH-regulated genes which share reciprocal regulation of BCL6 and STAT5 occupancy in response to GH. However, for Bcl6, a gene repressed rather than stimulated by GH, p300 is repressive rather than activating, and renders STAT5b inhibitory.
4. Discussion
4.1. Reciprocal occupancy of gene promoters by BCL6 and STAT5 mediates diverse transcriptional outcomes in response to GH
BCL6 is known as a transcription repressor, and STAT5 is named for its general role as a transcription activator (Basso and Dalla-Favera, 2010; Cesena et al., 2007b; Herrington et al., 2000; Schebesta et al., 2002; Staudt et al., 1999; Waters et al., 2006). This study demonstrates that BCL6 and STAT5 show reciprocal occupancy patterns in response to GH on multiple genes: BCL6 occupies the proximal promoters of GH target genes Socs2, Cish and Bcl6 in the absence of GH and decreases after GH treatment, while STAT5 occupancy is low in the absence of GH and increases after treatment. However, the similarity in their occupancy patterns does not lead to similar transcriptional outcomes, since Socs2 and Cish expressions are stimulated by GH while Bcl6 expression is inhibited. Differences in transcriptional response are specific for each promoter, and depend not only on the transcription factor occupancy but also on interactions with co-regulatory proteins.
Despite differences in transcriptional responses to GH, the occupied promoter sequences for Socs2, Cish and Bcl6 have several features in common. They contain the highly similar binding motifs shared by STAT5 and BCL6 and the site of BCL6/STAT5 occupancy is close to the TSS on each gene. ChIP-Seq showed that only 3% of all BCL6 peaks identified throughout the genome lie at the TSS, in contrast to approximately 80% in intronic or intergenic sequences; the remainder are located in exons or 3′UTR (Lin G and Schwartz J, manuscript in preparation). This is in general agreement with a report that most STAT sites reside outside of traditional promoters (Nelson et al., 2004).
The reciprocal relationship of BCL6 and STAT5 on DNA may reflect a variety of scenarios: The GH-induced decrease in BCL6 binding may be secondary to the potent inhibition of Bcl6 transcription by GH and resulting decrease in availability of BCL6 protein for binding. One can postulate that GH-activated STAT5 displaces BCL6 on a shared BCL6/STAT5 motif; STAT5 has recently been reported to outcompete STAT3 in occupying the Bcl6 promoter (Walker et al., 2013). In this regard, two predicted BCL6 and/or STAT5 binding sites are present within the BCL6/STAT5 occupancy regions identified by ChIP-Seq, for Socs2, for Cish and for Bcl6 (Fig. 5A). Thus, BCL6 and STAT5 may bind to different motifs within the occupied region. It is currently unclear which occupancy site or sites BCL6 and STAT5 occupy in these regulatory regions, and to what extent each of these sites is functional. Flanking sequences also impact the transcriptional outcome for the Bcl6 promoter (Ramachandrareddy et al., 2010; Walker et al., 2007). Although thousands of STAT sites were identified throughout the genome by ChIP-Seq in fibroblasts, the majority of STAT sites were reported to be ‘opportunistic’ while only 40% were associated with transcriptional responses (Zhu et al., 2012). Which among the abundant Bcl6 sites across the genome are functional remain to be determined.
BCL6 and STAT5 occupancy appear to be commonly detected on the Socs2, Cish, and Bcl6 promoters in cell types known to be responsive to STAT5 regulation, such as liver, adipocytes and immune cells (Appendix: Supplementary Table S3). Furthermore, the chromatin structure surrounding the Socs2, Cish, and Bcl6 promoters suggest that the promoters of these genes are potentially accessible to the transcriptional machinery in multiple cell types, based on association of these three genes with H3K9Ac, H3K27Ac, and possibly H3K4Me3 (Appendix: Supplementary Table S3). This suggests that reciprocal regulation of Socs2, Cish and Bcl6 by BCL6 and STAT5 in response to external stimuli may occur in multiple cells types, although the precise molecular mechanisms and co-regulatory molecules involved are likely to differ.
Post-translational modifications to BCL6 (Bereshchenko et al., 2002; Moriyama et al., 1997; Niu et al., 1998) and STAT5 (Cesena et al., 2007b; Herrington et al., 2000) such as phosphorylation and acetylation induced by signaling molecules activated by GH could also influence the association of co-activators or co-repressors with BCL6 and STAT5 and the recruitment of these complexes to specific genes (Icardi et al., 2012; Mendez et al., 2008). Recent studies have shown that Rac1 signaling induces a BCL6/STAT5 transcriptional switch on cell-cycle-associated target gene promoters (Barros et al., 2012). The reciprocal relationship between BCL6 and STAT5 occupancy may also contribute to regulation by other cytokines such as prolactin, a key factor involved in roles of BCL6 and STAT5 in normal breast development and oncogenic breast cancer growth (Tran et al., 2010). In light of the variety of factors which influence STAT5 and BCL6 binding, it remains to be determined whether or how other loci occupied by BCL6 have functional consequences, and the extent to which signaling pathways and cell type impact the binding at BCL6/STAT5 sites and the transcriptional outcomes.
4.2. Transcriptional responses to BCL6 and to STAT5 are modulated by co-regulatory proteins p300 and HDAC3
On the promoters of Socs2 and Cish, which are stimulated by GH, BCL6 serves as a repressor when it is expressed alone. A GH-induced decrease in Bcl6 mRNA and consequent loss of BCL6 repression of Socs2 and Cish are consistent with the increased expression of these two genes in response to GH. Though p300 often serves as a co-activator, the potent repression of Socs2 and Cish by BCL6 persists even when p300 is co-expressed. Interaction of p300 with BCL6 protein results in acetylation of BCL6 at a KKYK motif, leading to disruption of BCL6 interactions with co-repressive molecules and loss of BCL6 inhibition on some target genes (Bereshchenko et al., 2002). On Socs2, the slight increase in promoter activity observed with co-expression of p300 and BCL6 relative to BCL6 alone suggests that for the Socs2 promoter p300 may be sufficient to diminish the BCL6-induced repression slightly, consistent with loss of BCL6 repressive activity upon acetylation (Bereshchenko et al., 2002) in conjunction with p300. On the Cish promoter, p300 did not alter BCL6-induced repression when p300 was co-expressed with BCL6, suggesting promoter-specific differences in sensitivity to repressive and activating factors. Additionally, Socs2 was more responsive than Cish to GH-stimulated acetylation of H3 at 30 min GH, while the acetylation of H4 was equivalently stimulated by GH on Socs2 and Cish; in contrast, on Bcl6, histone acetylation was not regulated by GH.
For Socs2 and Cish, in contrast to the relative imperviousness of BCL6 to co-regulation by p300 when they were expressed together, STAT5 activation increased only when it was co-expressed with p300, but not alone. The addition of p300 with STAT5, relative to STAT5 alone, doubled the activation of the Socs2 and Cish promoters, a trend consistent with co-activation, though the increase was modest compared to the magnitude of p300 co-activation of some other genes (Chia and Rotwein, 2010; Pfitzner et al., 1998). These findings are in general agreement with in vivo observations in which, on Socs2 and Cish, p300 did not serve as an effective co-activator for STAT5b in response to GH (Chia and Rotwein, 2010; Chia et al., 2010); it was suggested that other undefined co-activators or acetylases serve this function.
Co-repressor proteins also occupy the Socs2 and Cish promoters. Deacetylase activity has been associated with both BCL6 and STAT5 (Bereshchenko et al., 2002; Dhordain et al., 1997; Icardi et al., 2012; Rascle et al., 2003). In fact, deacetylase activity is reported to be required for STAT5 to enable RNA Pol II recruitment (Rascle et al., 2003; Sebastián et al., 2008). For BCL6, its reported interactions with multiple HDAC proteins are important for the repression by BCL6 (Bereshchenko et al., 2002; Cotto et al., 2010; Dhordain et al., 1997). In particular, HDAC3 associates with BCL6 in repressing a subset of NF-κB target genes in bone marrow derived macrophages (Barish et al., 2010). The present results suggest that HDAC3 functions as a co-repressor for STAT5 but not for BCL6 on the Socs2 and Cish promoters. Multiple HDACs may be involved in regulating transcription of Socs2 and Cish by STAT5 at different steps of the transcriptional response.
4.3. The Bcl6 promoter is repressed by p300
Repression of the Bcl6 promoter by p300 clearly distinguishes Bcl6 from Socs2 and Cish promoters, whose activity is increased by p300 in the presence of STAT5. Co-expression of p300 with BCL6 inhibited the Bcl6 promoter significantly below the level with BCL6 expression alone, consistent with p300 serving as a co-repressor for BCL6 on this promoter. While p300 has acetylase activity, the repression by p300 is likely not related to acetylation of BCL6, as acetylation of BCL6 disrupts its interactions with co-repressors, resulting in a loss of BCL6-mediated repression (Bereshchenko et al., 2002). Other functions of p300 such as serving as a scaffold to recruit other interacting proteins may contribute to its repressive properties. It is likely that p300 exerts repression by interacting or coordinating with other nuclear factors that associate with BCL6, such as HDACs. Repression of Bcl6, Socs2 and Cish by HDAC3 is in agreement with well-accepted co-repressor roles of HDACs (Cunliffe, 2008; Fischle et al., 2001; Narlikar et al., 2002). These findings extend observations that HDAC3 occupies multiple immune-related genes within the BCL6 and NF-κB cistromes (Barish et al., 2010). Further, HDAC1 interacts with the POZ domain of BCL6, and HDAC inhibitors reduce BCL6-mediated repression (Dhordain et al., 1998). In repressing Bcl6, other co-repressor proteins likely work in conjunction with p300 and/or HDACs. Among these, NCoR and SMRT, which are reported to enhance the enzymatic function of HDAC3 (You et al., 2013), partner with HDACs in the repression of adipogenic genes (Alenghat et al., 2008), and the NCoR cistrome overlaps with the BCL6 cistrome in regulating atherogenic genes in bone marrow (Barish et al., 2012). Other co-repressor candidates include CtBP, a co-repressor in autorepression of Bcl6 (Mendez et al., 2008), and the BCL6 co-repressor BCoR (Huynh et al., 2000). A hybrid mechanism involving two independent repressive complexes associated with BCL6, one involving HDAC3 and p300, has recently been reported (Hatzi et al., 2013). Interactions among BCL6 and co-regulators are thought to play a role in the pathogenesis of conditions such as B-cell lymphoma (Cerchietti et al., 2010; Pasqualucci et al., 2011).
On the Bcl6 promoter, STAT5b is rendered repressive by both p300 and HDAC3. Based on deacetylase inhibitor studies, HDACs have been found to be required for recruitment of the basal transcription machinery to STAT5 target genes (Rascle et al., 2003; Sebastián et al., 2008); thus HDACs appear to participate in STAT5-mediated transcriptional activation. In its canonical role as a transcriptional co-repressor, as observed here, HDAC3 is likely to recruit additional co-repressors, many of which are known to interact with STAT5. Among these are FoxO1 and FoxO3 (Fernandez de Mattos et al., 2004; Ono et al., 2007), KRAB (Krebs et al., 2012) and Sin3a (Icardi et al., 2012), as well as NCoR/SMRT with HDAC3 (Fischle et al., 2002; Guenther et al., 2001). Such mechanisms involving co-repressors such as HDACs mediate STAT5 serving as a repressive transcription factor, for target genes in lymphocyte development (Heltemes-Harris and Farrar, 2012) and for genes such as IRF1 and other NF-κB regulated genes (Luo and Yu-Lee, 2000). STAT5 has also been shown to interact with the histone H3Lys27 methylase Ezh2, thus initiating the formation of repressive chromatin (Mandal et al., 2011). Similarly, this study documents that STAT5b is repressive when co-expressed with HDAC3 for Socs2, Cish and for Bcl6.
The role of p300 in STAT5-mediated repression of the Bcl6 promoter is non-traditional, since p300 has long been known to be recruited to STATs for transactivation (Paulson et al., 1999). The diverse roles of p300 as co-activator and co-repressor reflect the promoter-specific properties of co-regulators in determining transcriptional outcome (Luo and Yu-Lee, 2000). The ability of p300 to recruit or serve as a scaffold for other co-regulators is also a contributor in determining transcriptional outcome. For example, p300 recruits HDAC6 to the STAT5 regulated sequence in Spi2.1 to mediate negative regulation of GH-STAT5b-mediated transcription, which is dependent on activation of RhoA/ROCK (Ling and Lobie, 2004). Such versatility of co-regulator function on different promoters is reflected in recent demonstration that genes associated with NF-κB-driven inflammatory responses and tissue remodeling were highly enriched in the BCL6 and the overlapping NCoR/SMRT cistromes (Barish et al., 2012).
Taken together, the present findings demonstrate critical roles of a pair of transcription factors which exhibit reciprocal occupancy on multiple genes in response to GH, but which mediate contrasting transcriptional outcomes in response to GH dependent on the target gene. The occupancy of BCL6 appears to be a consistent determinant for repression of target genes, while occupancy of STAT5, which is often activating for GH-regulated genes, leads to repression for the Bcl6 promoter. The transcriptional outcomes associated with the similar occupancy pattern for BCL6 and STAT5 on the different target genes are in turn determined by co-regulatory proteins associated with BCL6 and/or STAT5. HDAC3 consistently mediated transcriptional repression in conjunction with STAT5 or BCL6 on all three of the genes studied. On the other hand, p300 enhanced activation of GH-stimulated Socs2 and Cish genes, but p300 mediated repression of Bcl6, which is inhibited by GH. Molecular interactions of p300 and HDAC3 with the transcription factors BCL6 and STAT5, or other associated regulatory proteins may be critical determinants in transcriptional outcome, and in response to signaling by GH and other regulators implicated in normal and oncogenic growth.
Supplementary Material
Acknowledgments
The authors thank Drs. Jindan Yu and Hong Cheng, and the University of Michigan Sequencing Core for their assistance in ChIP-Seq experiment design and sample preparation; and Jennifer Harley and Aaron Taylor for technical assistance.
This work was supported by grants to JS from NIH (DK46072), the American Diabetes Association (7-09-BS-168) and Center for Genetics in Health and Medicine at University of Michigan; and by NIH grant HG005119 to ZSQ. GL was supported by NIH T32 GM07315, Director’s and Loeb Fellowships from University of Michigan Cancer Center and by a Rackham Predoctoral Fellowship, University of Michigan. CRL was supported by a postdoctoral fellowship from the Center Q6 for Organogenesis (NIH T32 HD007505) at University of Michigan.
Footnotes
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.mce.2014.07.020.
References
- Adhikari A, Sen A, Brumbaugh RC, Schwartz J. Altered growth patterns of a mountain Ok population of Papua New Guinea over 25 years of change. Am. J. Hum. Biol. 2011;23:325–332. doi: 10.1002/ajhb.21134. [DOI] [PubMed] [Google Scholar]
- Ahmed SF, Farquharson C. The effect of GH and IGF1 on linear growth and skeletal development and their modulation by SOCS proteins. J. Endocrinol. 2010;206:249–259. doi: 10.1677/JOE-10-0045. [DOI] [PubMed] [Google Scholar]
- Albertsson-Wikland K, Rosberg S. Analyses of 24-hour growth hormone profiles in children: relation to growth. J. Clin. Endocrinol. Metab. 1988;67:493–500. doi: 10.1210/jcem-67-3-493. [DOI] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour LA, Mizanoor Rahman S, Gurevich I, Wayne Leitner J, Fischer SJ, Roper MD, et al. Increased P85α is a potent negative regulator of skeletal muscle insulin signaling and induces in vivo insulin resistance associated with growth hormone excess. J. Biol. Chem. 2005;280:37489–37494. doi: 10.1074/jbc.M506967200. [DOI] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri MS, Becerra D, Kim J, Tseng TW, et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 2012;15:554–562. doi: 10.1016/j.cmet.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron BW, Stanger RR, Hume E, Sadhu A, Mick R, Kerckaert JP, et al. BCL6 encodes a sequence-specific DNA-binding protein. Genes Chromosomes Cancer. 1995;13:221–224. doi: 10.1002/gcc.2870130314. [DOI] [PubMed] [Google Scholar]
- Barros P, Lam EW, Jordan P, Matos P. Rac1 signalling modulates a STAT5/BCL-6 transcriptional switch on cell-cycle-associated target gene promoters. Nucleic Acids Res. 2012;40:7776–7787. doi: 10.1093/nar/gks571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J. Clin. Invest. 2010;120:4569–4582. doi: 10.1172/JCI42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesena TI, Cardinaux JR, Kwok R, Schwartz J. CCAAT/enhancer-binding protein (C/EBP) beta is acetylated at multiple lysines: acetylation of C/EBP beta at lysine 39 modulates its ability to activate transcription. J. Biol. Chem. 2007a;282:956–967. doi: 10.1074/jbc.M511451200. [DOI] [PubMed] [Google Scholar]
- Cesena TI, Cui TX, Piwien-Pilipuk G, Kaplani JI, Calinescu AA, Huo JS, et al. Multiple mechanisms of growth hormone-regulated gene transcription. Mol. Genet. Metab. 2007b;90:126–133. doi: 10.1016/j.ymgme.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin G, Huo JS, Barney D, Wang Z, Livshiz T, et al. Computational and functional analysis of growth hormone (GH)-regulated genes identifies the transcriptional repressor B-cell lymphoma 6 (Bc16) as a participant in GH-regulated transcription. Endocrinology. 2009;150:3645–3654. doi: 10.1210/en.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DJ, Rotwein P. Defining the epigenetic actions of growth hormone: acute chromatin changes accompany GH-activated gene transcription. Mol. Endocrinol. 2010;24:2038–2049. doi: 10.1210/me.2010-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DJ, Varco-Merth B, Rotwein P. Dispersed chromosomal Stat5b-binding elements mediate growth hormone-activated insulin-like growth factor-I gene transcription. J. Biol. Chem. 2010;285:17636–17647. doi: 10.1074/jbc.M110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DH, LeRoith D. Obesity, type 2 diabetes, and cancer: the insulin and IGF connection. Endocr. Relat. Cancer. 2012;19:F27–F45. doi: 10.1530/ERC-11-0374. [DOI] [PubMed] [Google Scholar]
- Conforto TL, Zhang Y, Sherman J, Waxman DJ. Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Mol. Cell. Biol. 2012;32:4611–4627. doi: 10.1128/MCB.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto M, Cabanillas F, Tirado M, Garcia MV, Pacheco E. Epigenetic therapy of lymphoma using histone deacetylase inhibitors. Clin. Transl. Oncol. 2010;12:401–409. doi: 10.1007/s12094-010-0527-3. [DOI] [PubMed] [Google Scholar]
- Croker BA, Kiu H, Nicholson SE. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 2008;19:414–422. doi: 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui TX, Piwien-Pilipuk G, Huo JS, Kaplani J, Kwok R, Schwartz J. Endogenous CCAAT/enhancer binding protein β and p300 are both regulated by growth hormone to mediate transcriptional activation. Mol. Endocrinol. 2005;19:2175–2186. doi: 10.1210/me.2004-0502. [DOI] [PubMed] [Google Scholar]
- Cui TX, Kwok R, Schwartz J. Cooperative regulation of endogenous cAMP-response element binding protein and CCAAT/enhancer-binding protein beta in GH-stimulated c-fos expression. J. Endocrinol. 2008;196:89–100. doi: 10.1677/JOE-07-0169. [DOI] [PubMed] [Google Scholar]
- Cui TX, Lin G, LaPensee CR, Calinescu AA, Rathore M, Streeter C, et al. C/EBPβ mediates growth hormone-regulated expression of multiple target genes. Mol. Endocrinol. 2011;25:681–693. doi: 10.1210/me.2010-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT. Eloquent silence: developmental functions of class I histone deacetylases. Curr. Opin. Genet. Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Dent AL, Vasanwala FH, Toney LM. Regulation of gene expression by the proto-oncogene BCL-6. Crit. Rev. Oncol. Hematol. 2002;41:1–9. doi: 10.1016/s1040-8428(01)00164-0. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain P, Lin RJ, Quief S, Lantoine D, Kerckaert JP, Evans RM, et al. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl SA, Schmidlin H, Nagasawa M, van Haren SD, Kwakkenbos MJ, Yasuda E, et al. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J. Immunol. 2008;180:4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RL, Hemati N, Ross SE, MacDougald OA. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 2001;276:16348–16355. doi: 10.1074/jbc.m100128200. [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, et al. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol. Cell. Biol. 2004;24:10058–10071. doi: 10.1128/MCB.24.22.10058-10071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Kiermer V, Dequiedt F, Verdin E. The emerging role of class II histone deacetylases. Biochem. Cell Biol. 2001;79:337–348. [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Gingras S, Simard J, Groner B, Pfitzner E. p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res. 1999;27:2722–2729. doi: 10.1093/nar/27.13.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser DA, Schlegel W. The FoxO/Bcl-6/cyclin D2 pathway mediates metabolic and growth factor stimulation of proliferation in Min6 pancreatic β-cells. J. Recept. Signal Transduct. Res. 2009;29:293–298. doi: 10.3109/10799890903241824. [DOI] [PubMed] [Google Scholar]
- Goodman HM. Growth hormone and the metabolism of carbohydrate and lipid in adipose tissue. Ann. N. Y. Acad. Sci. 1968;148:419–440. doi: 10.1111/j.1749-6632.1968.tb20367.x. [DOI] [PubMed] [Google Scholar]
- Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartatik T, Okada S, Okabe S, Arima M, Hatano M, Tokuhisa T. Binding of BAZF and Bc16 to STAT6-binding DNA sequences. Biochem. Biophys. Res. Commun. 2001;284:26–32. doi: 10.1006/bbrc.2001.4931. [DOI] [PubMed] [Google Scholar]
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltemes-Harris LM, Farrar MA. The role of STAT5 in lymphocyte development and transformation. Curr. Opin. Immunol. 2012;24:146–152. doi: 10.1016/j.coi.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J, Smit LS, Schwartz J, Carter-Su C. The role of STAT proteins in growth hormone signaling. Oncogene. 2000;19:2585–2597. doi: 10.1038/sj.onc.1203526. [DOI] [PubMed] [Google Scholar]
- Huo JS, McEachin RC, Cui TX, Duggal NK, Hai T, States DJ, et al. Profiles of growth hormone (GH)-regulated genes reveal time-dependent responses and identify a mechanism for regulation of activating transcription factor 3 by GH. J. Biol. Chem. 2006;281:4132–4141. doi: 10.1074/jbc.M508492200. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Icardi L, De Bosscher K, Tavernier J. The HAT/HDAC interplay: multilevel control of STAT signaling. Cytokine Growth Factor Rev. 2012;23:283–291. doi: 10.1016/j.cytogfr.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Icardi L, Mori R, Gesellchen V, Eyckerman S, De Cauwer L, Verhelst J, et al. The Sin3a repressor complex is a master regulator of STAT transcriptional activity. Proc. Natl. Acad. Sci. USA. 2012;109:12058–12063. doi: 10.1073/pnas.1206458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Chang KT, Matsumoto Y, Furuhata Y, Nishihara M, Sasaki F, et al. Obesity and insulin resistance in human growth hormone transgenic rats. Endocrinology. 1998;139:3057–3063. doi: 10.1210/endo.139.7.6103. [DOI] [PubMed] [Google Scholar]
- Jardin F, Ruminy P, Bastard C, Tilly H. The BCL6 proto-oncogene: a leading role during germinal center development and lymphomagenesis. Pathol. Biol. (Paris) 2007;55:73–83. doi: 10.1016/j.patbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Kabotyanski EB, Huetter M, Xian W, Rijnkels M, Rosen JM. Integration of prolactin and glucocorticoid signaling at the beta-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers. Mol. Endocrinol. 2006;20:2355–2368. doi: 10.1210/me.2006-0160. [DOI] [PubMed] [Google Scholar]
- Kang K, Robinson GW, Hennighausen L. Comprehensive meta-analysis of Signal Transducers and Activators of Transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics. 2013;14:4. doi: 10.1186/1471-2164-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik SK, Chen H, McLean GW, Heit JJ, Gu X, Zhang AY, et al. Menin controls growth of pancreatic β-cells in pregnant mice and promotes gestational diabetes mellitus. Science. 2007;318:806–809. doi: 10.1126/science.1146812. [DOI] [PubMed] [Google Scholar]
- Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- Kojima S, Hatano M, Okada S, Fukuda T, Toyama Y, Yuasa S, et al. Testicular germ cell apoptosis in Bcl6-deficient mice. Development. 2001;128:57–65. doi: 10.1242/dev.128.1.57. [DOI] [PubMed] [Google Scholar]
- Krebs CJ, Schultz DC, Robins DM. The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration. Mol. Cell. Biol. 2012;32:3732–3742. doi: 10.1128/MCB.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Wang RA, Barnes CJ. Coregulators and chromatin remodeling in transcriptional control. Mol. Carcinog. 2004;41:221–230. doi: 10.1002/mc.20056. [DOI] [PubMed] [Google Scholar]
- Laz EV, Sugathan A, Waxman DJ. Dynamic in vivo binding of STAT5 to growth hormone-regulated genes in intact rat liver. Sex-specific binding at low- but not high-affinity STAT5 sites. Mol. Endocrinol. 2009;23:1242–1254. doi: 10.1210/me.2008-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L, Lobie PE. RhoA/ROCK activation by growth hormone abrogates p300/histone deacetylase 6 repression of Stat5-mediated transcription. J. Biol. Chem. 2004;279:32737–32750. doi: 10.1074/jbc.M400601200. [DOI] [PubMed] [Google Scholar]
- Litterst CM, Kliem S, Marilley D, Pfitzner E. NCoA-1/SRC-1 is an essential coactivator of STAT5 that binds to the FDL motif in the alpha-helical region of the STAT5 transactivation domain. J. Biol. Chem. 2003;278:45340–45351. doi: 10.1074/jbc.M303644200. [DOI] [PubMed] [Google Scholar]
- Litterst CM, Kliem S, Lodrini M, Pfitzner E. Coactivators in gene regulation by STAT5. Vitam. Horm. 2005;70:359–386. doi: 10.1016/S0083-6729(05)70012-1. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Luo G, Yu-Lee L. Stat5b inhibits NFκB-mediated signaling. Mol. Endocrinol. 2000;14:114–123. doi: 10.1210/mend.14.1.0399. [DOI] [PubMed] [Google Scholar]
- Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascle X, Albagli O, Lemercier C. Point mutations in BCL6 DNA-binding domain reveal distinct roles for the six zinc fingers. Biochem. Biophys. Res. Commun. 2003;300:391–396. doi: 10.1016/s0006-291x(02)02873-5. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, et al. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- Mendez LM, Polo JM, Yu JJ, Krupski M, Ding BB, Melnick A, et al. CtBP is an essential corepressor for BCL6 autoregulation. Mol. Cell. Biol. 2008;28:2175–2186. doi: 10.1128/MCB.01400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RD, Laz EV, Su T, Waxman DJ. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol. 2009;23:1914–1926. doi: 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama M, Yamochi T, Semba K, Akiyama T, Mori S. BCL-6 is phosphorylated at multiple sites in its serine- and proline-clustered region by mitogen-activated protein kinase (MAPK) in vivo. Oncogene. 1997;14:2465–2474. doi: 10.1038/sj.onc.1201084. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J. Biol. Chem. 2004;279:54724–54730. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley BW, Malovannaya A, Qin J. Minireview: nuclear receptor and coregulator proteomics – 2012 and beyond. Mol. Endocrinol. 2012;26:1646–1650. doi: 10.1210/me.2012-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Chia DJ, Merino-Martinez R, Flores-Morales A, Unterman TG, Rotwein P. Signal transducer and activator of transcription (Stat) 5b-mediated inhibition of insulin-like growth factor binding protein-1 gene transcription: a mechanism for repression of gene expression by growth hormone. Mol. Endocrinol. 2007;21:1443–1457. doi: 10.1210/me.2006-0543. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Perry JK, Mohankumar KM, Emerald BS, Mertani HC, Lobie PE. The contribution of growth hormone to mammary neoplasia. J. Mammary Gland Biol. Neoplasia. 2008;13:131–145. doi: 10.1007/s10911-008-9070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol. Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- Qin ZS, Yu J, Shen J, Maher CA, Hu M, Kalyana-Sundaram S, et al. HPeak: an HMM-based algorithm for defining read-enriched regions in ChIP-Seq data. BMC Bioinformatics. 2010;11:369. doi: 10.1186/1471-2105-11-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandrareddy H, Bouska A, Shen Y, Ming J, Rizzino A, Chan WC, et al. BCL6 promoter interacts with far upstream sequences with greatly enhanced activating histone modifications in germinal center B cells. Proc. Natl. Acad. Sci. USA. 2010;107:11930–11935. doi: 10.1073/pnas.1004962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol. Cell. Biol. 2003;23:4162–4173. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta M, Heavey B, Busslinger M. Transcriptional control of B-cell development. Curr. Opin. Immunol. 2002;14:216–223. doi: 10.1016/s0952-7915(02)00324-2. [DOI] [PubMed] [Google Scholar]
- Scheeren FA, Naspetti M, Diehl S, Schotte R, Nagasawa M, Wijnands E, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat. Immunol. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- Sebastián C, Serra M, Yeramian A, Serrat N, Lloberas J, Celada A. Deacetylase activity is required for STAT5-dependent GM-CSF functional activity in macrophages and differentiation to dendritic cells. J. Immunol. 2008;180:5898–5906. doi: 10.4049/jimmunol.180.9.5898. [DOI] [PubMed] [Google Scholar]
- Seyfert VL, Allman D, He Y, Staudt LM. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- Staudt LM, Dent AL, Shaffer AL, Yu X. Regulation of lymphocyte cell fate decisions and lymphomagenesis by BCL-6. Int. Rev. Immunol. 1999;18:381–403. doi: 10.3109/08830189909088490. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tannenbaum GS, Choi HK, Gurd W, Waxman DJ. Temporal relationship between the sexually dimorphic spontaneous GH secretory profiles and hepatic STAT5 activity. Endocrinology. 2001;142:4599–4606. doi: 10.1210/endo.142.11.8480. [DOI] [PubMed] [Google Scholar]
- Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A, et al. Prolactin inhibits Bcl6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res. 2010;70:1711–1721. doi: 10.1158/0008-5472.CAN-09-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal OM, Merino R, Rico-Bautista E, Fernandez-Perez L, Chia DJ, Woelfle J, et al. In vivo transcript profiling and phylogenetic analysis identifies suppressor of cytokine signaling 2 as a direct signal transducer and activator of transcription 5b target in liver. Mol. Endocrinol. 2007;21:293–311. doi: 10.1210/me.2006-0096. [DOI] [PubMed] [Google Scholar]
- Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- Walker SR, Nelson EA, Yeh JE, Pinello L, Yuan GC, Frank DA. STAT5 outcompetes STAT3 to regulate the expression of the oncogenic transcriptional modulator BCL6. Mol. Cell. Biol. 2013;33:2879–2890. doi: 10.1128/MCB.01620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol. Cell. Biol. 2008;28:2213–2220. doi: 10.1128/MCB.01608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MJ, Hoang HN, Fairlie DP, Pelekanos RA, Brown RJ. New insights into growth hormone action. J. Mol. Endocrinol. 2006;36:1–7. doi: 10.1677/jme.1.01933. [DOI] [PubMed] [Google Scholar]
- Woelfle J, Rotwein P. In vivo regulation of growth hormone-stimulated gene transcription by STAT5b. Am. J. Physiol. Endocrinol. Metab. 2004;286:E393–E401. doi: 10.1152/ajpendo.00389.2003. [DOI] [PubMed] [Google Scholar]
- You SH, Lim HW, Sun Z, Broache M, Won KJ, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 2013;20:182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Laz EV, Waxman DJ. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 2012;32:880–896. doi: 10.1128/MCB.06312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu BM, Kang K, Yu JH, Chen W, Smith HE, Lee D, et al. Genome-wide analyses reveal the extent of opportunistic STAT5 binding that does not yield transcriptional activation of neighboring genes. Nucleic Acids Res. 2012;40:4461–4472. doi: 10.1093/nar/gks056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.