Abstract
Background
The genetic risk factors for susceptibility to chronic obstructive pulmonary disease (COPD) are still largely unknown. Additional genetic variants are likely to be identified by genome-wide association studies in larger cohorts or specific subgroups.
Methods
Genome-wide association analysis in COPDGene (non-Hispanic whites and African-Americans) was combined with existing data from the ECLIPSE, NETT/NAS, and GenKOLS (Norway) studies. Analyses were performed both using all moderate-to-severe cases and the subset of severe cases. Top loci not previously described as genome-wide significant were genotyped in the ICGN study, and results combined in a joint meta-analysis.
Findings
Analysis of a total of 6,633 moderate-to-severe cases and 5,704 controls confirmed association at three known loci: CHRNA3/CHRNA5/IREB2, FAM13A, and HHIP (10−12 < P < 10−14), and also showed significant evidence of association at a novel locus near RIN3 (overall P, including ICGN = 5•4×10−9). In the severe COPD analysis (n=3,497), the effects at two of three previously described loci were significantly stronger; we also identified two additional loci previously reported to affect gene expression of MMP12 and TGFB2 (overall P = 2•6x10−9 and 8•3×10−9). RIN3 and TGFB2 expression levels were reduced in a set of Lung Tissue Research Consortium COPD lung tissue samples compared with controls.
Interpretation
In a genome-wide study of COPD, we confirmed associations at three known loci and found additional genome-wide significant associations with moderate-to-severe COPD near RIN3 and with severe COPD near MMP12 and TGFB2. Genetic variants, apart from alpha-1 antitrypsin deficiency, increase the risk of COPD. Our analysis of severe COPD suggests additional genetic variants may be identified by focusing on this subgroup.
Funding
National Heart, Lung, and Blood Institute; the COPD Foundation through contributions from AstraZeneca, Boehringer Ingelheim, Novartis, and Sepracor; GlaxoSmithKline; Centers for Medicare and Medicaid Services; Agency for Healthcare Research and Quality; US Department of Veterans Affairs.
Introduction
Chronic obstructive pulmonary disease (COPD), characterized by persistent and usually progressive airflow obstruction, is one of the leading causes of morbidity and mortality worldwide. While cigarette smoking is the major environmental risk factor, the burden of COPD is increasing1,2 despite many successful efforts at tobacco control, and the response to cigarette smoke is characterized by high inter-individual variability3. Genetic factors are a major contributor to this variability4–6, but the specific genetic loci responsible for this variation remain largely unknown7. Genome-wide association studies have successfully identified loci which are often novel for a range of complex diseases, including COPD, that have subsequently replicated8–17, but the majority of genetic susceptibility due to common variation remains unexplained18. Identifying genetic loci may lead to improved risk prediction and subtype identification19, and is arguably the most promising unbiased approach to understand disease mechanisms in humans and enable future specific and rational therapies20.
We recently completed genome-wide genotyping in COPDGene, a large, genetic epidemiology study of over 10,000 non-Hispanic White and African American cigarette smokers (both current and ex-smokers) with and without COPD21. We sought to determine whether a genome-wide association study (GWAS), combining the results from COPDGene with previous association studies7, would reveal new genetic susceptibility loci.
Genome-wide association analyses in COPD to date have included subjects with mild or moderately severe airflow limitation7,9,12,22. To our knowledge, a genome-wide association case-control study of severe COPD has not been previously reported. The severity of airflow limitation in COPD correlates with many other important disease characteristics, such as emphysema23, functional limitation24, and higher mortality25. In addition to potentially identifying novel signals unique to severe disease, a genome-wide association study of severe COPD may have improved power compared with a study of moderate-to-severe COPD due to decreased phenotypic heterogeneity and misclassification in severe COPD cases, as well as enrichment for subjects with the highest genetic risk profile26–30, despite the decreased sample size.
Methods
COPDGene (NCT00608764) is a large, multicenter study designed to investigate the genetic and epidemiologic characteristics of COPD and other smoking-related lung diseases21. COPDGene subjects were of self-described non-Hispanic white or African-American ancestry, and genotyped using the HumanOmniExpress (Illumina, San Diego, CA). Genotype imputation on the COPDGene cohorts was performed using MaCH and minimac31,32 using 1000 Genomes33 Phase I v3 European (EUR) and cosmopolitan reference panels for the non-Hispanic whites and African-Americans, respectively. Detailed descriptions of the ECLIPSE, NETT/NAS, and Norway (GenKOLS) cohorts, including genotyping quality control and imputation, have been previously published7,9,12,21,34–36.
In all cohorts, ‘moderate-to-severe’ cases had GOLD Grade 2-4 COPD (moderate, severe, and very severe COPD; post-bronchodilator FEV1 < 80% predicted with FEV1/FVC < 0•7); individuals with severe alpha-1 antitrypsin deficiency were excluded.
Controls had normal spirometry with a history of cigarette smoking. For the analysis of ‘severe’ COPD, cases were limited to those with GOLD 3 and 4 disease (severe and very severe, post-bronchodilator FEV1 < 50% predicted). Baseline characteristics of each of the genome-wide cohorts are shown in Table 1. Logistic regression was performed within each cohort and racial / ethnic group adjusting for age, pack-years of smoking, and ancestry-based principal components using plink (v1•07)37, as previously described7,12. Fixed-effects meta-analysis was performed using METAL (version 2010-08-01)38. Heterogeneity was reported as both I2,39 using the meta package in R (v2•3•0) (www.r-project.org) and P-values for Cochrane's Q. Markers were included for analysis if they passed genotyping or imputation quality control (as appropriate) in all genome-wide cohorts. Regional association plots were created using LocusZoom40, using the 1000 Genomes EUR reference data for linkage disequilibrium (LD) calculations.
Table 1.
Baseline characteristics.
| COPDGene | ECLIPSE | NETT/NAS | GenKOLS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NHW Case | NHW Control | AA Case | AA Control | Case | Control | Case | Control | Case | Control | |
| n | 2812 | 2534 | 821 | 1749 | 1764 | 178 | 373 | 435 | 863 | 808 |
| Age | 64•7 (8•2) | 59•5 (8•7) | 59•0 (8•2) | 52•8 (6•0) | 63•6 (7•1) | 57•5 (9•4) | 67•5 (5•8) | 69•8 (7•5) | 65•5 (10•0) | 55•6 (9•7) |
| Pack-years | 56•3 (28•0) | 37•8 (20•3) | 42•4 (23•0) | 36•4 (20•1) | 50•3 (27•4) | 32•1 (24•8) | 66•4 (30•7) | 40•7 (27•9) | 32•0 (18•5) | 19•7 (13•6) |
| FEV1, % predicted | 49•6 (18•0) | 96•8 (11) | 52•2 (17•8) | 98•4 (12•2) | 47•6 (15•6) | 107•8 (13•6) | 28•1 (7•4) | 100•0 (13•2) | 50•6 (17•4) | 94•9 (9•2) |
| Sex (% male) | 55•7 | 49•3 | 55•2 | 58•1 | 67 | 57•9 | 63•8 | 100 | 60•1 | 50•1 |
Values given as mean (SD) or percent, as appropriate. NHW: Non-hispanic white. AA: African-American.
Results yielding a P value threshold of < 5×10−7 at loci not previously described7 in the moderate-to-severe and severe COPD meta-analysis of COPDGene, ECLIPSE, NETT/NAS, and GenKOLS (Norway) were subsequently genotyped in 983 probands and 1876 siblings from the family-based International COPD Genetics Network study (ICGN)34. Association analysis in ICGN was performed using PBAT (v3•61), under an additive model, adjusting for age and pack-years of smoking. Results from the family-based ICGN study were combined with case-control results using a joint meta-analysis41 weighted by sample size, using the number of informative transmissions in ICGN and the effective number of cases in each cohort. A joint meta-analysis P-value of < 5×10−8 was considered significant.
Differences in odds ratios between severe cases versus controls and between all cases (moderate-to-severe) versus controls were assessed by permutation. Region-based conditional analyses were performed using logistic regression, adjusting for the most significant (lead) single nucleotide polymorphism (SNP) in each region using genotyped or dosage data as appropriate, and testing all SNPs within a 250kb window on either side of the lead SNP for association with affection status. To estimate the combined effect of genetic risk variants, we constructed a genetic score based on the cumulative number of risk alleles in a logistic regression in the COPDGene non-Hispanic whites including age, pack-years, and ancestry-based principal components.
Additional analyses using the meta-analysis results included gene-based testing using VEGAS42 and the literature mining using GRAIL43. Gene expression levels of TGFB2 and RIN3 were measured in lung tissue samples from 15 COPD patients – 8 with moderate (FEV1 < 80% predicted), and 7 with severe (FEV1 < 50% predicted) disease – and 15 control subjects with normal lung function, obtained from the NHLBI Lung Tissue Research Consortium (LTRC), as described previously44.
Role of the funding source
GlaxoSmithKline was involved in study design and data collection for the ECLIPSE, GenKOLS (Norway), and ICGN studies. No other study sponsors had a role in study design or data collection, and none of the study sponsors had a role in data analysis, data interpretation, or writing of the report. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Results
Results from the GWAS of moderate-to-severe COPD in the COPDGene non-Hispanic whites and African-Americans are shown in Tables S6 and S7. The analysis in the non-Hispanic whites confirmed three previously known (CHRNA3/5/IREB2, HHIP, and FAM13A) COPD susceptibility loci, but neither study alone identified novel loci achieving conventional genome-wide significance (P < 5•0×10−8, Table S7 and S8).
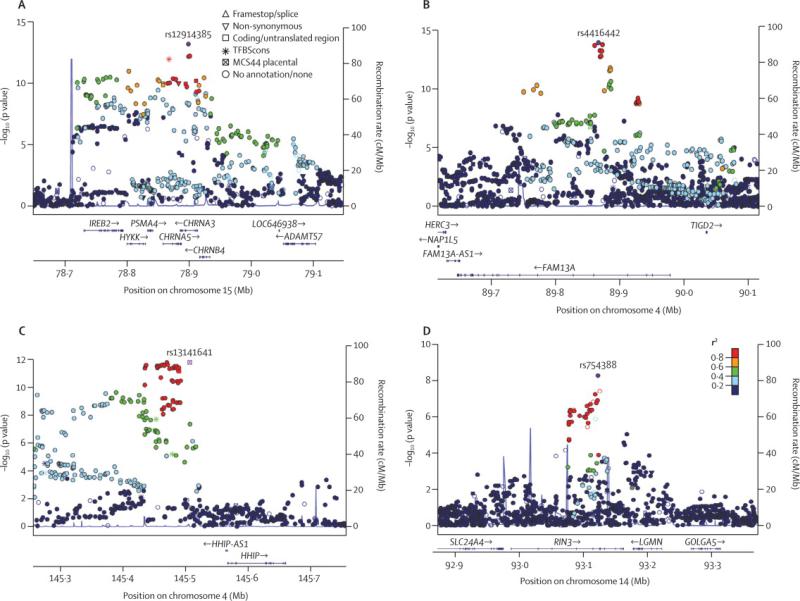
The combined GWAS of moderate-to-severe COPD included COPDGene non-Hispanic whites, COPDGene African-Americans, ECLIPSE, NETT/NAS, and GenKOLS (Norway), for a total of 6,633 cases and 5,704 controls. Both individual and overall quantile-quantile (Q-Q) plots showed no evidence of significant population stratification (individual study λGC all ≤ 1.04; overall λGC =1•03; λ 45GC1000 = 1•01, Figure S1). The top results at each of the loci with P < 10−7 are shown in Table 3 and Figure 1. The three most significant SNPs in this meta-analysis were either identical to, or in strong LD – r2 > 0•5 – with the top SNPs previously described at these three loci: 4q22 (FAM13A), 15q25 (CHRNA3/5/IREB2), and 4q31 (HHIP), confirming these previous association results 9,10,12.
Table 3.
Top results for the genome-wide association analysis of moderate-to-severe COPD versus smoking controls in COPDGene non-Hispanic white and African-American, ECLIPSE, NETT/NAS, and GenKOLS (Norway) studies.
| Locus | Nearest gene | SNP | Risk Allele | Frequency | Meta-analysis | ||||
|---|---|---|---|---|---|---|---|---|---|
| NHW | AA | OR (CI) | P | I2 | Q | ||||
| 4q22 | FAM13A | rs4416442 | C | 0•42 | 0•54 | 1•28 (1•2-1•36) | 1•12×10−14 | 0•23 | 0•27 |
| 15q25 | CHRNA3 | rs12914385 | T | 0•42 | 0•19 | 1•28 (1•2-1•36) | 6•38×10−14 | 0•26 | 0•25 |
| 4q31 | HHIP | rs13141641 | T | 0•59 | 0•89 | 1•27 (1•19-1•36) | 1•57×10−12 | 0•31 | 0•22 |
| 14q32 | RIN3 | rs754388 | C | 0•83 | 0•85 | 1•28 (1•18-1•39) | 5•25×10−9 | 0 | 0•59 |
Allele coding represents + strand, hg19. Allele frequency is given for the risk allele. Nhw = Non-Hispanic white; AA = African-American.
Figure 1.
Local association plots for significant loci for the analysis of moderate-to-severe COPD in COPDGene non-Hispanic whites and African-Americans, ECLIPSE, NETT/NAS, and GenKOLS (Norway). The x-axis is chromosomal position, and the y-axis shows the –log10 P-value. The most significant SNP at each locus is labeled in purple, with other SNPs colored by degree of linkage disequilibrium (r2).
We identified one novel additional locus with P < 5×10− at 14q32; the top SNP at this locus was rs754388 (nearest gene – RIN3), with a P-value of 5•25×10−9. We genotyped this SNP in the ICGN Study, and tested for association with COPD in ICGN using a family-based test. While the evidence of association at this SNP did not achieve statistical significance (one-sided P=0•20), the overall meta-analysis P-value (including ICGN) for rs754388 remained genome-wide significant (5•4×10−9). An analysis of the effect of this SNP on FEV1 as a quantative trait was not statistically significant.
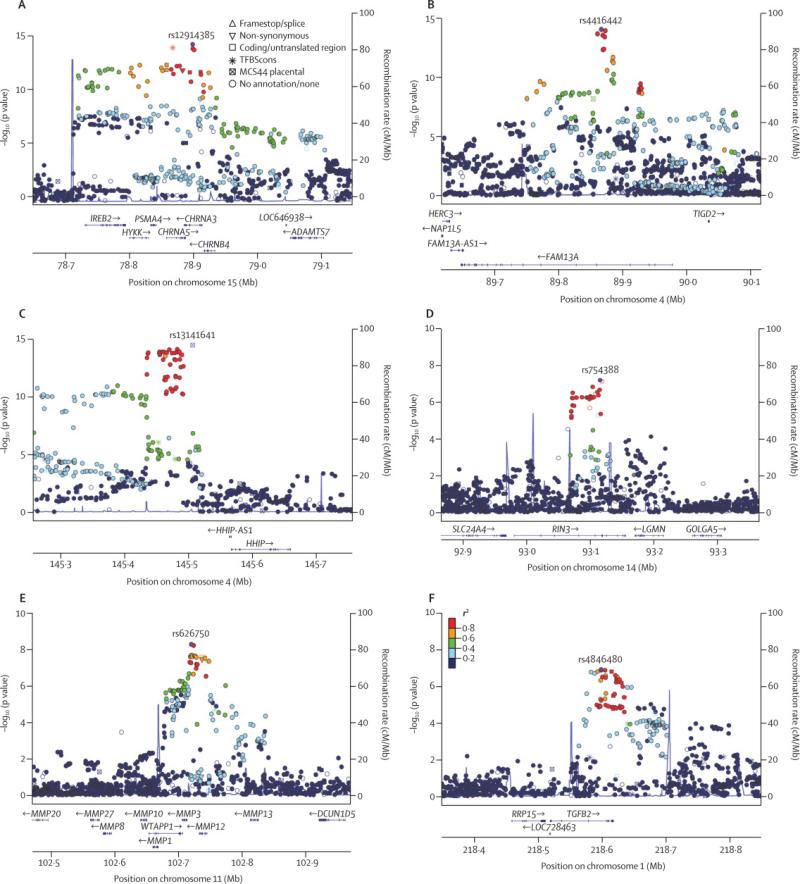
The analysis of severe COPD reduced the number of cases to 3,497, while the number of controls remained the same. Baseline characteristics of the severe subsets of COPDGene, ECLIPSE, and GenKOLS (Norway) cases are shown in Table 2 (characteristics of NETT subjects are included in Table 1, as all NETT cases are severe). Similarly to the analysis of moderate-to-severe cases, we found no evidence of inflation due to population stratification (individual λGC ≤ 1.04; overall λGC = 1•04; λGC1000 = 1•01, Figure S2) among severe cases and controls. We again confirmed the three previously described COPD loci: 4q22 (FAM13A), 15q25 (CHRNA3/5/IREB2), and 4q31 (HHIP) – as genome-wide significant for severe COPD (Table 4 and Figure 2). We noted effect estimates for these loci tended to be larger in severe COPD than in moderate to severe COPD cases; these differences were statistically significant at two markers (P < 0•01 for rs13141641 (15q25), and rs12914385, (HHIP)), and just above statistical significance for a third (P = 0•08 for rs4416442 (FAM13A)).
Table 2.
Baseline characteristics of severe COPD subsets (COPDGene, ECLIPSE, and GenKOLS; all NETT subjects have severe COPD and were included in the severe COPD analysis).
| COPDGene | ECLIPSE | GenKOLS | ||
|---|---|---|---|---|
| NHW | AA | |||
| n | 1390 | 352 | 999 | 383 |
| Age | 65•2 (7•8) | 60•6 (8•1) | 63•5 (7•0) | 66•7 (9•7) |
| Pack-years | 58•7 (28•4) | 43•9 (23•4) | 50•7 (26•3) | 33•0 (19•9) |
| FEV1, % predicted | 34•0 (9•9) | 34•8 (10•4) | 36•5 (8•6) | 34•4 (10•3) |
| Sex (% male) | 57•8 | 58 | 69•9 | 61•5 |
Values given as mean (SD) or percent, as appropriate. NHW: Non-hispanic white. AA: African-American.
Table 4.
Top results for the genome-wide association analysis of severe COPD versus smoking controls in COPDGene non-Hispanic white and African-American, ECLIPSE, NETT/NAS, and GenKOLS (Norway) studies.
| Locus | Nearest gene(s) | SNP | Risk Allele | Frequency | Meta-Analysis | ||||
|---|---|---|---|---|---|---|---|---|---|
| Nhw | Aa | OR (CI) | P | I2 | Q | ||||
| 15q25 | CHRNA3 | rs12914385 | T | 0•42 | 0•19 | 1•39 (1•29-1•51) | 2•70×10−16 | 0 | 0•76 |
| 4q31 | HHIP | rs13141641 | T | 0•59 | 0•89 | 1•39 (1•28-1•51) | 3•66×10−15 | 0 | 0•44 |
| 4q22 | FAM13A | rs4416442 | C | 0•42 | 0•54 | 1•36 (1•26-1•47) | 9•44×10−15 | 0 | 0•68 |
| 11q22 | MMP3/12 | rs626750 | G | 0•83 | 0•74 | 1•36 (1•23-1•51) | 5•35×10−9 | 0 | 0•62 |
| 14q32 | RIN3 | rs754388 | C | 0•83 | 0•85 | 1•33 (1•2-1•48) | 6•69×10−8 | 0 | 0•66 |
| 1q41 | TGFB2 | rs4846480 | A | 0•75 | 0•65 | 1•26 (1•16-1•37) | 1•25×10−7 | 0 | 0•99 |
Allele coding represents + strand, hg19. Allele frequency is given for the risk allele. Nhw = Non-Hispanic white; AA = African-American.
Figure 2.
Local association plots for significant loci for the analysis of severe COPD in COPDGene non-Hispanic whites and African-Americans, ECLIPSE, NETT/NAS, and GenKOLS (Norway). The x-axis is chromosomal position, and the y-axis shows the – log10 P-value. The most significant SNP at each locus is labeled in purple, with other SNPs colored by degree of linkage disequilibrium (r2).
We also identified two new genome-wide significant loci in the analysis of severe COPD versus controls. The first was at 11q22; the top-ranked SNP was rs626750 (nearest genes, MMP3 and MMP12). We found supportive evidence for association with severe COPD in ICGN (P = 0•06) and a genome-wide significant result in the joint meta-analysis (P = 2•6×10−9). This locus was previously reported in an analysis including subjects from NAS and NETT46. After excluding NETT/NAS subjects, the joint meta-analysis P-value remained significant (P = 7•0×10−9). The second locus was at 1q41, where the top-ranked SNP was rs4846480 (nearest gene, TGFB2). This locus was just below-genome wide significance in the genome-wide cohorts (1•3×10−). Including the results from ICGN (P = 0•007), brought the joint meta-analysis results to P = 8•3×10−9.
To determine whether gene expression levels of these two genes not previously described in association with COPD – RIN3 and TGFB2 – were different in lung tissue samples from COPD cases versus controls, we performed real-time quantitative reverse transcription PCR in 18 control samples and 15 COPD samples – 8 with moderate (FEV1 < 80% predicted), and 7 with severe (FEV1 < 50% predicted) disease) – from the NHLBI Lung Tissue Research Consortium. RIN3 expression was significantly lower in COPD cases versus controls (P = 0•003, Figure S3). Differences in TGFB2 expression were not significant when comparing all cases versus controls (P = 0•5), but were significant when the cases were limited to those with severe disease (P = 0•002, Figure S4).
While the definitions of cases and controls within each study – based on GOLD criteria – were similar, COPD is a highly heterogeneous disease, and differences exist between the studies 47. To explore these considerations, we used alternative methods for meta-analysis based on modified random-effects and binary effects model that may be more powerful in the presence of heterogeneity among studies48,49 (see Supplement). However, we were not able to identify new genome-wide significant results using these methods.
We next sought to determine whether there was evidence for secondary associations at each described locus. We performed analyses conditioning on the top (lead) SNP at each genome-wide significant locus reported in this analysis, examining all SNPs present in 250kb flanking regions around the top signal. We found evidence suggestive of secondary associations (P < 5×10−4) in the analysis of moderate-to-severe COPD at 15q25 (conditioning on rs12914385) for a SNP in strong LD (r2=0•92 in EUR) with the previously reported rs13180 in IREB2 (rs12903295, intronic in IREB2, P = 9•9 × 10−5). Suggestive evidence of a secondary association was also found near the 14q32 (RIN3) locus conditioning on rs754388 (rs11849228, P = 1•3×10−4). In severe COPD, evidence supporting a secondary association was found at 15q25 in another intronic SNP in CHRNA3 (rs3743073, P=3•3×10−4).
The number of loci identified as influencing risk to COPD to date is modest, despite the relatively large sample size of this study; these loci explain < 5% of the liability-scale variance. To explore whether additional true association signals of weaker effect – beyond the ability to detect in our current analysis – might be present, we examined the characteristics of the top results (P < 0•01) in a meta-analysis of three white cohorts (ECLIPSE, NETT/NAS, and GenKOLS) within the COPDGene non-Hispanic whites. We found the direction of effect in the first three cohorts was consistent with the direction of effect in COPDGene more often than expected by chance alone (P = 0•03). This result suggests additional signals of significance may be found in larger GWAS, and are consistent with a recent analysis of COPDGene data18. As with most GWAS studies, the effect sizes of these identified loci are relatively small; however one subject may carry multiple risk loci. Within the COPDGene non-Hispanic whites, each additional copy of a risk allele within a composite risk score resulted in an increase in odds for COPD of 1•24; this estimate was similar whether the model included only loci previously discovered in studies not including COPDGene (e.g. 15q25, HHIP, and FAM13A loci), or included the additional loci (RIN3, TGFB2, MMP3/12) described in this study.
To further explore additional signals not reaching genome-wide significance, we additionally performed a gene-based analyses – under the hypothesis that a given gene or genic region may harbor multiple susceptibility variants with p-values larger than the traditional GWAS significance level – with VEGAS, and a SNP and text-mining based analyses – identifying and prioritizing genes based on functional relationships identified using literature – with GRAIL. The top genes from the VEGAS analysis, using a Bonferroni correction for 17,640 genes, included previously implicated loci (FAM13A; CHRNB4, IREB2, CHRNA3/5, HYKK, and PSMA4 at 15q25); as well as RIN3 andAPOBR (Table S9). For the GRAIL analysis (Table S10), the top individual genes were OSM and OSMR; in contrast, genes at or near well-validated loci – HHIP, IREB2, andFAM13A – did not give significant P-values in the GRAIL analysis.
Discussion
In a large, genome-wide association meta-analysis of moderate to severe and severe COPD (and the first genome-wide association analysis to include African-Americans), we confirmed three previously described genome-wide significant loci, and identified three additional loci achieving genome-wide significance in moderate-to-severe and severe COPD. Our findings provide further evidence for a role of common genetic variants in contributing to COPD susceptibility (panel).
The association at 11q22 is located in a cluster of matrix metalloproteinases including MMP12 (matrix metalloproteinase 12, also known as macrophage metalloelastase or matrix metallopeptidase 12). MMP12 is produced by macrophages and degrades elastin, and has been extensively characterized in COPD both in mouse models50 and in human studies51,52. Several studies have described genetic associations with COPD or lung function for a SNP in the promoter region of MMP12, rs2276109 [-82A→G], where the minor allele leads to decreased promoter activity through less efficient binding of AP-146,53–55. In a combined analysis of a total of 7 cohorts, including subjects with both asthma and COPD, the minor allele (G) of rs2276109 was associated with improved lung function46. Of note, two of the COPD cohorts included in this study were enriched for severe disease. Similarly, in a study of 977 European cases and 876 controls, an association was identified for a haplotype including rs2276109 in MMP12 among severe cases (P = 0•0039) 54. SNP rs626750 is in strong LD with rs2276109 (r2 = 0•63). Our study thus confirms, with the same direction of effect, these previously described associations at genome-wide significance, and supports a role for MMP12 in severe COPD.
Meta-analyses across large population-based cohorts have previously reported an association at 1q41, near TGFB2, with FEV1 /FVC ratio56. However, the lead SNP for this association, rs993925, is not in strong LD (r2=0•027 in EUR) and lies over 250kb away from the SNPs reported here. Our top association is, however, in strong LD (r2=0•97 in EUR) with rs6684205, recently identified as an expression quantitative trait loci (eQTL) for TGFB2 in lung tissue57. The COPD risk allele has been associated with decreased expression, consistent with our findings of decreased TGFB2 expression in lung tissue from severe COPD cases versus controls. These lines of evidence strongly suggest effects of this locus on COPD susceptibility operate via changes in lung TGFB2 expression. While genetic variants in or near TGFB1 have been studied in association with COPD58–60, an association of variants near TGFB2 with COPD has not been previously described. Tgfb2 null mice have dilated conducting airways and collapsed terminal and respiratory bronchioles61, and loss-of-function mutations in TGFB2 have been associated with Loeys-Dietz syndrome, a disorder of connective tissue showing phenotypic overlap with Marfan syndrome, and has rarely been associated with emphysema62. TGF-β2 is also the predominant isoform present in airway tissue in severe asthma63,64; it is secreted in airway epithelial cells in response to injury or inflammatory cytokines (e.g. IL-13) and appears to play a major role in airway inflammation and remodeling 65–68.
The association at the RIN3 locus, while genome-wide significant in the overall analysis, was not significant in ICGN. This finding thus may represent a false positive, but it also may be due to the lower power of the family-based analysis. In support of the latter explanation, an alternative analysis using generalized estimating equations (which allows calculation of effect sizes) resulted in an odds ratio of 1•14 (95% confidence interval, 0•92-1•41), consistent with the estimates from our other cohorts. In addition, a lookup of a SNP in strong LD (rs17184313, r2 = 0•94 in EUR) in a recently published meta-analysis of COPD identified from population-based studies22 demonstrates nominal evidence of significance (P = 0•009), though the direction of association was not given. RIN3 is a Rab5 GTPase binding protein expressed in many tissues, including the lung, and is involved in transport from plasma membranes to early endosomes69,70. High levels of expression of RIN3 have been found on human mast cells71, a cell type that may be of interest in COPD72–74. Furthermore, we demonstrated, in a small number of lung tissue samples, that RIN3 expression differs between COPD cases versus controls. While RIN3 is the closest gene to the lead SNP, this locus is also approximately 1•7 megabases away from SERPINA1, the gene encoding alpha-1 antitrypsin, which could suggest an effect of distant rare variants75. For the loci reported in this study, and for most loci reported for GWAS, the role of candidate SNP(s) on a particular gene and on protein function cannot be deduced with certainty from linkage disequilibrium patterns and simple measures of gene expression, and requires further functional investigation including SNP-based functional studies76,77.
The 19q13 locus did not achieve genome-wide significant in this study, despite being identified in our prior meta-analysis in the ECLIPSE, NETT/NAS, Norway (GenKOLS), and the initial 1000 non-Hispanic White subjects from the COPDGene study7. In the current analysis of moderate-to-severe disease, rs7937 (nearest gene, RAB4B) was just below genome-wide significance (6•2×10−); however, the association was genome-wide significant (1•0×10−9) in a model adjusting only for principal components of genetic ancestry, and more significant when limited to non-Hispanic whites. A recent study in a Japanese population confirmed an association with smoking behavior with SNPs in this region78, and additional analyses of nicotine addiction and lung eQTLs suggest effects at this locus may be mediated through several different variants in CYP2A6 as well as EGLN2 79–81. Together, these data suggest effects of the 19q13 locus on COPD act through a mechanism involving cigarette smoking, and are complex, potentially in the presence of locus heterogeneity across populations.
Our gene-based analysis using VEGAS identified an association with APOBR, the apolipoprotein B receptor. Lipoproteins may have pathophysiologic importance in lung disease 82; differences in apolipoprotein B have been described in association with lung function and COPD83,84, and a recent study identified an association between SNPs near APOM, lung function, and emphysema85. Similarly, our GRAIL analysis suggested a role for oncostatin M and its receptor, which may be of interest in COPD and emphysema86,87. Additional studies will be needed to confirm these findings.
Racial differences in COPD may exist88,89. Thus, we also examined the case-control results only in the African-Americans. While underpowered, these results did not reveal any novel genome-wide significant loci (Table S8); furthermore, at the loci described in this work, there was no convincing evidence of heterogeneity (Tables 3 and 4) or differential effect sizes compared with those in non-Hispanic whites (Table S4 and S5). These data are consistent with a prior report finding little evidence that the relationship of smoking to lung function differed by genetic ancestry90, as well as genetic studies of other traits that have demonstrated overall similarities of loci shared between ethnically diverse groups91,92. While these results support our decision to combine the African-Americans and non-Hispanic whites to improve statistical power, our results should not be interpreted to imply including other ethnic groups is generally redundant; indeed, genetic studies in specific ethnic groups have led to discovery of novel loci93 and provided important information for identifying specific variants at individual loci91,94.
Our study does not address other genetic contributors to COPD susceptibility. We did not, for example, consider gene-gene interactions or gene-environment interaction. The genotyping and imputation in this study are not well-suited to address the role of rare variants, which may also be important in explaining COPD susceptibility95–97. Our definition of cases and controls was based only on the presence of moderate, or moderate-to-severe airflow obstruction, yet COPD is highly heterogeneous. Analysis of individual characteristics (e.g. emphysema) or of specific subtypes (e.g. severe disease, as we demonstrate here; radiographically defined subsets; or separate GOLD categories2,98) may provide greater insight into the development of this complex and heterogeneous disease27–30. Well-powered studies of lung function in the general population, as well as COPD ascertained through population-based studies, have not identified several of the loci reported here22,56,99–101. Additional studies will be helpful in determining whether heterogeneity in COPD definitions, including varying degrees of severity and case ascertainment, differential effects of genetic variants in disease versus lung function in the general population, or Type 1 or Type 2 error could account for these discrepancies. The number of loci achieving genome-wide significance described here for COPD is few compared to other complex diseases102,103, and the markers described here account for a very small fraction of the estimated heritability18. For unknown reasons, the number of discovered loci confirmed by GWAS for any given sample size can vary widely104,105.
However, despite differences in this ‘rate of return’, increasing sample size appears critical to discovering novel loci.
While the effect of an individual genetic variant may be small, discounting small effects in GWAS as unimportant would have ignored such critical effects as the insulin gene (INS) in diabetes and the HMG-CoA reductase gene (HMGCR) in cholesterol metabolism106,107; more dramatic perturbations of these causal genes – through experimental disruption76 or through identification of rare, more deleterious genetic variants108 – can highlight the importance of pathophysiology identified by GWAS. In addition, cumulative effects of these loci may be substantial. Although more accurate risk prediction estimates will require assessments in independent populations, the increased odds of 1·24 with each COPD GWAS risk allele in the COPDGene population suggest that harboring three risk alleles could nearly double odds of moderate to severe COPD. By comparison, the population-based BOLD study109 estimated an odds ratio for COPD per ten pack-years of smoking from1•16 to 1•28. These data, together with previous studies of familial aggregation and heritability of COPD, highlight the importance of genetic risk factors apart from alpha-1 antitrypsin deficiency in increasing risk of COPD.
Our work provides strong statistical support for association with moderate-to-severe or severe COPD susceptibility for three previously-described9–17 (CHRNA3/5/IREB2, HHIP, and FAM13A) and three additional (RIN3, MMP3/MMP12, TGFB2) loci. We provide evidence that additional GWAS in larger samples are likely to identify additional genetic determinants of COPD, and suggest using subsets of COPD (such as severe disease) may provide additional insight to genetic risk factors. Our work also suggests further studies to elucidate biological mechanisms77, which we hope will reveal new insights into COPD pathogenesis, and ultimately, treatment for this important disease.
Supplementary Material
Panel – Research in context.
Systematic Review
The genetic risk factors for COPD are still largely unknown. We searched PubMed with the search terms “genome-wide association” and “COPD” or “airflow”, as well as the genome-wide association study (GWAS) catalog (genome.gov/26525384). At the time of our search, the largest studies to date included approximately 3,500 cases. Evidence from GWAS in other diseases suggests larger sample sizes or analysis of specific subtypes could increase power and identify new genetic determinants of COPD.
Interpretation
Our study in moderate-to-severe and severe COPD confirms genome-wide associations near FAM13A, HHIP, and CHRNA3/CHRNA5/IREB2, and provides evidence in support of new associations near RIN3, MMP12 and TGFB2. GWAS continues to have potential to identify new genetic risk factors that could implicate novel disease mechanisms in COPD. Genetic variants, apart from alpha-1 antitrypsin deficiency, increase the risk of COPD; this burden may be higher in those with severe disease.
Acknowledgements
Funding:
This work was supported by NHLBI R01 HL084323, P01 HL083069, P01 HL105339 and R01 HL089856 (E.K.S.); K08 HL097029 and R01 HL113264 (M.H.C.), and R01 HL089897 (J.D.C.); the Alpha-1 Foundation (M.H.C.) and a VA Research Career Scientist award (D.S.). The COPDGene® study (NCT00608764) is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens and Sunovion. The National Emphysema Treatment Trial was supported by the NHLBI N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118 and N01HR76119, the Centers for Medicare and Medicaid Services and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/ERIC of the US Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). The Norway GenKOLS study (Genetics of Chronic Obstructive Lung Disease, GSK code RES11080), the ECLIPSE study (NCT00292552; GSK code SCO104960), and the ICGN study were funded by GlaxoSmithKline.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Appendix
Author Contributions
Study design: M.H.C., M.N.M., M.M., P.J.C., N.L., C.L., E.K.S., T.H.B. Data acquisition and quality control: M.H.C., X.Z., P.J.C., C.P.H, D.L.D., J.S.S., J. Z., A.A.L., D.S., R.C., R.G.B., E.A.R., B.J.M., J.E.H., T.M., H.F., J.B.H., R.T-S., D.A.L., P.B., A.G., J.D.C., E.K.S., T.H.B. Data Analysis: M.H.C., M.N.M, M.M., T.M., J.B.H., E.K.S., T.H.B. Critical revision of manuscript: all authors.
COPDGene® Investigators - Core Units
Administrative Core: James Crapo, MD (PI), Edwin Silverman, MD, PhD (PI), Barry Make, MD, Elizabeth Regan, MD, Sara Penchev, Rochelle Lantz, Lori Stepp, Sandra Melanson,
Genetic Analysis Core: Terri Beaty, PhD, Barbara Klanderman, PhD, Nan Laird, PhD, Christoph Lange, PhD, Michael Cho, MD, Stephanie Santorico, PhD, John Hokanson, MPH, PhD, Dawn DeMeo, MD, MPH, Nadia Hansel, MD, MPH, Craig Hersh, MD, MPH, Peter Castaldi, MD, MSc, Merry-Lynn McDonald, PhD, Jin Zhou, MD, PhD, Manuel Mattheisen, MD, Emily Wan, MD, Megan Hardin, MD, Jacqueline Hetmanski, MS, Margaret Parker, MS, Tanda Murray, MS
Imaging Core: David Lynch, MB, Joyce Schroeder, MD, John Newell, Jr., MD, John Reilly, MD, Harvey Coxson, PhD, Philip Judy, PhD, Eric Hoffman, PhD, George Washko, MD, Raul San Jose Estepar, PhD, James Ross, MSc, Mustafa Al Qaisi, MD, Jordan Zach, Alex Kluiber, Jered Sieren, Tanya Mann, Deanna Richert, Alexander McKenzie, Jaleh Akhavan, Douglas Stinson
PFT QA Core, National Jewish Health: Robert Jensen, PhD
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, PhD, Stacey Meyerer, Shivam Chandan, Samantha Bragan
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, PhD, Andre Williams, PhD, Carla Wilson, MS, Anna Forssen, MS, Amber Powell, Joe Piccoli
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, MPH, PhD, Marci Sontag, PhD, Jennifer Black-Shinn, MPH, Gregory Kinney, MPH, PhDc, Sharon Lutz, MPH, PhD
COPDGene® Investigators: Clinical Centers Ann Arbor VA: Jeffrey Curtis, MD, Ella Kazerooni, MD
Baylor College of Medicine, Houston, TX: Nicola Hanania, MD, MS, Philip Alapat, MD, Venkata Bandi, MD, Kalpalatha Guntupalli, MD, Elizabeth Guy, MD, Antara Mallampalli, MD, Charles Trinh, MD, Mustafa Atik, MD, Hasan Al-Azzawi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD
Brigham and Women's Hospital, Boston, MA: Dawn DeMeo, MD, MPH, Craig Hersh, MD, MPH, George Washko, MD, Francine Jacobson, MD, MPH, Hiroto Hatabu, MD, PhD, Peter Clarke, MD, Ritu Gill, MD, Andetta Hunsaker, MD, Beatrice Trotman-Dickenson, MBBS, Rachna Madan, MD
Columbia University, New York, NY: R. Graham Barr, MD, DrPH, Byron Thomashow, MD, John Austin, MD, Belinda D'souza, MD
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., MD, Lacey Washington, MD, H Page McAdams, MD
Reliant Medical Group, Worcester, MA: Richard Rosiello, MD, Timothy Bresnahan, MD, Joseph Bradley, MD, Sharon Kuong, MD, Steven Meller, MD, Suzanne Roland, MD
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, MD, MPH, Joseph Tashjian, MD
Johns Hopkins University, Baltimore, MD: Robert Wise, MD, Nadia Hansel, MD, MPH, Robert Brown, MD, Gregory Diette, MD, Karen Horton, MD
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Torrance, CA: Richard Casaburi, MD, PhD, Janos Porszasz, MD, PhD, Hans Fischer, MD, Matt Budoff, MD
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, MD, Charles Trinh, MD, Hirani Kamal, MD, Roham Darvishi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD
Minneapolis VA: Dennis Niewoehner, MD, Quentin Anderson, MD, Kathryn Rice, MD, Audrey Caine, MD
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, MD, MS, Gloria Westney, MD, MS, Eugene Berkowitz, MD, PhD
National Jewish Health, Denver, CO: Russell Bowler, MD, PhD, David Lynch, MB, Joyce Schroeder, MD, Valerie Hale, MD, John Armstrong, II, MD, Debra Dyer, MD, Jonathan Chung, MD, Christian Cox, MD
Temple University, Philadelphia, PA: Gerard Criner, MD, Victor Kim, MD, Nathaniel Marchetti, DO, Aditi Satti, MD, A. James Mamary, MD, Robert Steiner, MD, Chandra Dass, MD, Libby Cone, MD
University of Alabama, Birmingham, AL: William Bailey, MD, Mark Dransfield, MD, Michael Wells, MD, Surya Bhatt, MD, Hrudaya Nath, MD, Satinder Singh, MD
University of California, San Diego, CA: Joe Ramsdell, MD, Paul Friedman, MD
University of Iowa, Iowa City, IA: Alejandro Cornellas, MD, John Newell, Jr., MD, Edwin JR van Beek, MD, PhD
University of Michigan, Ann Arbor, MI: Fernando Martinez, MD, MeiLan Han, MD, Ella Kazerooni, MD
University of Minnesota, Minneapolis, MN: Christine Wendt, MD, Tadashi Allen, MD
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, MD, Joel Weissfeld, MD, MPH, Carl Fuhrman, MD, Jessica Bon, MD, Danielle Hooper, MD
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, MD, Sandra Adams, MD, Carlos Orozco, MD, Mario Ruiz, MD, Amy Mumbower, MD, Ariel Kruger, MD, Carlos Restrepo, MD, Michael Lane, MD Principal investigators and centers participating in ECLIPSE (NCT00292552, SCO104960) include: Bulgaria: Y. Ivanov, Pleven; K. Kostov, Sofia. Canada: J. Bourbeau, Montreal; M. Fitzgerald, Vancouver; P. Hernández, Halifax; K. Killian, Hamilton; R. Levy, Vancouver; F. Maltais, Montreal; D. O'Donnell, Kingston. Czech Republic: J. Krepelka, Praha. Denmark: J. Vestbo, Hvidovre. The Netherlands: E. Wouters, Horn. New Zealand: D. Quinn, Wellington. Norway: P. Bakke, Bergen, Slovenia: M. Kosnik, Golnik. Spain: A. Agusti, Jaume Sauleda, Palma de Mallorca. Ukraine: Y. Feschenko, Kiev; V. Gavrisyuk, Kiev; L. Yashina, Kiev. UK: L. Yashina, W. MacNee, Edinburgh; D. Singh, Manchester; J. Wedzicha, London. USA: A. Anzueto, San Antonio, TX; S. Braman, Providence. RI; R. Casaburi, Torrance CA; B. Celli, Boston, MA; G. Giessel, Richmond, VA; M.
Gotfried, Phoenix, AZ; G. Greenwald, Rancho Mirage, CA; N. Hanania, Houston, TX; D. Mahler, Lebanon, NH; B. Make, Denver, CO; S. Rennard, Omaha, NE; C. Rochester, New Haven, CT; P. Scanlon, Rochester, MN; D. Schuller, Omaha, NE; F. Sciurba, Pittsburgh, PA; A. Sharafkhaneh, Houston, TX; T. Siler, St Charles, MO; E. Silverman, Boston, MA; A. Wanner, Miami, FL; R. Wise, Baltimore, MD; R. ZuWallack, Hartford, CT.
Steering Committee: H. Coxson (Canada), C. Crim (GlaxoSmithKline, USA), L. Edwards (GlaxoSmithKline, USA), D. Lomas (UK), W. MacNee (UK), E. Silverman (USA), R. Tal Singer (Co-chair, GlaxoSmithKline, USA), J. Vestbo (Co-chair, Denmark), J. Yates (GlaxoSmithKline, USA).
Scientific Committee: A. Agusti (Spain), P. Calverley (UK), B. Celli (USA), C. Crim (GlaxoSmithKline, USA), B. Miller (GlaxoSmithKline, USA), W. MacNee (Chair, UK), S. Rennard (USA), R. Tal-Singer (GlaxoSmithKline, USA), E. Wouters (The Netherlands), J. Yates (GlaxoSmithKline, USA).
Co-investigators in the NETT Genetics Ancillary Study also include J. Benditt, G. Criner, M. DeCamp, P. Diaz, M. Ginsburg, L. Kaiser, M. Katz, M. Krasna, N. MacIntyre, R. McKenna, F. Martinez, Z. Mosenifar, J. Reilly, A. Ries, P. Scanlon, F. Sciurba and J. Utz.
International COPD Genetics Network (ICGN) investigators: Edwin K. Silverman, Brigham & Women's Hospital, Boston, MA, USA; David A. Lomas, Cambridge Institute for Medical Research, University of Cambridge, Cambridge, UK; Barry J. Make, National Jewish Medical and Research Center, Denver, CO, USA; Alvar Agusti and Jaume Sauleda, Hospital Universitari Son Dureta, Fundación Caubet-Cimera and Ciber Enfermedades Respiratorias, Spain; Peter M.A. Calverley, University of Liverpool, UK; Claudio F. Donner, Division of Pulmonary Disease, S. Maugeri Foundation, Veruno (NO), Italy; Robert D. Levy, University of British Columbia, Vancouver, Canada; Peter D. Paré, University of British Columbia, Vancouver, Canada; Stephen Rennard, Section of Pulmonary & Critical Care, University of Nebraska Medical Center, Omaha, NE, USA; Jørgen Vestbo, Department of Cardiology and Respiratory Medicine, Hvidovre Hospital, Copenhagen, Denmark; Emiel F.M. Wouters, University Hospital Maastricht, The Netherlands.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional data are available in the Supplement.
Conflict of Interest Statement: M.H.C. has received consultancy fees from Merck. C.P.H. has received lecture fees from Novartis and has been a consultant for CSL Behring. R.T.-S. is a current employee of GlaxoSmithKline. D.A.L has received grant support, honoraria, and consultancy fees from GlaxoSmithKline. He is the Chair of the GSK Respiratory Area Therapy Board. E.K.S. has received grant support from GlaxoSmithKline for studies of COPD genetics and honoraria and consulting fees from AstraZeneca, Merck, and GlaxoSmithKline. M.N.M., X.Z., M.M., P.J.C., J.S.S., J.Z., N.M.L., C.L., A.A.L., D.S., R.C., R.G.B., E.A.R., B.J.M., J.E.H., S.L., T.M., H.F., P.B., A.G., J.D.C., and T.B. report no conflicts of interest.
References
- 1.Minino M, Xu J, Kochanek J. Deaths: Preliminary Data for 2008. National Vital Statistics Reports. Hyattsville, MD: National Center for Vital Statistics. Natl Cent Vital Stat. 2010;59 [PubMed] [Google Scholar]
- 2.Vestbo J, Hurd SS, Agustí AG, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease: GOLD Executive Summary. Am J Respir Crit Care Med. 2013;187:347–65. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 3.Burrows B, Knudson RJ, Cline MG, Lebowitz MD. Quantitative relationships between cigarette smoking and ventilatory function. 1977;115:195–205. doi: 10.1164/arrd.1977.115.2.195. [DOI] [PubMed] [Google Scholar]
- 4.Ingebrigtsen T, Thomsen SF, Vestbo J, et al. Genetic influences on Chronic Obstructive Pulmonary Disease - a twin study. Respir Med. 2010;104:1890–5. doi: 10.1016/j.rmed.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 5.McCloskey SC, Patel BD, Hinchliffe SJ, Reid ED, Wareham NJ, Lomas DA. Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am J Respir Crit Care Med. 2001;164:1419–24. doi: 10.1164/ajrccm.164.8.2105002. [DOI] [PubMed] [Google Scholar]
- 6.Silverman EK, Chapman H a, Drazen JM, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–8. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 7.Cho MH, Castaldi PJ, Wan ES, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21:947–57. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindorff LA, Junkins HA, Manolio TA. [2009. 09.30];A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/26525384.
- 9.Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilk JB, Chen TH, Gottlieb DJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMeo DL, Mariani T, Bhattacharya S, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho MH, Boutaoui N, Klanderman BJ, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–2. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young RP, Hopkins RJ, Hay BA, Epton MJ, Black PN, Gamble GD. Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur Respir J. 2008;32:1158–64. doi: 10.1183/09031936.00093908. [DOI] [PubMed] [Google Scholar]
- 14.Hardin M, Zielinski J, Wan ES, et al. CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am J Respir Cell Mol Biol. 2012;47:203–8. doi: 10.1165/rcmb.2012-0011OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Durme YMTA, Eijgelsheim M, Joos GF, et al. Hedgehog-interacting protein is a COPD susceptibility gene: the Rotterdam Study. Eur Respir J. 2010;36:89–95. doi: 10.1183/09031936.00129509. [DOI] [PubMed] [Google Scholar]
- 16.Young RP, Whittington CF, Hopkins RJ, et al. Chromosome 4q31 locus in COPD is also associated with lung cancer. Eur Respir J. 2010;36:1375–82. doi: 10.1183/09031936.00033310. [DOI] [PubMed] [Google Scholar]
- 17.Young RP, Hopkins RJ, Hay BA, Whittington CF, Epton MJ, Gamble GD. FAM13A locus in COPD is independently associated with lung cancer - evidence of a molecular genetic link between COPD and lung cancer. Appl Clin Genet. 2011;4:1–10. doi: 10.2147/TACG.S15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou JJ, Cho MH, Castaldi PJ, Hersh CP, Silverman EK, Laird NM. Heritability of COPD and Related Phenotypes in Smokers. Am J Respir Crit Care Med. 2013 doi: 10.1164/rccm.201302-0263OC. doi:10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–58. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 20.Chakravarti A, Clark AG, Mootha VK. Distilling pathophysiology from complex disease genetics. Cell. 2013;155:21–6. doi: 10.1016/j.cell.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regan EA, Hokanson JE, Murphy JR, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilk JB, Shrine NRG, Loehr LR, et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med. 2012;186:622–32. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washko GR, Criner GJ, Mohsenifar Z, et al. Computed tomographic-based quantification of emphysema and correlation to pulmonary function and mechanics. COPD. 2008;5:177–86. doi: 10.1080/15412550802093025. [DOI] [PubMed] [Google Scholar]
- 24.Engstrom CP, Persson LO, Larsson S, Ryden A, Sullivan M. Functional status and well being in chronic obstructive pulmonary disease with regard to clinical parameters and smoking: a descriptive and comparative study. Thorax. 1996;51:825–30. doi: 10.1136/thx.51.8.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anthonisen NR, Wright EC, Hodgkin JE. Prognosis in chronic obstructive pulmonary disease. Am Rev Respir Dis. 1986;133:14–20. doi: 10.1164/arrd.1986.133.1.14. [DOI] [PubMed] [Google Scholar]
- 26.Risch N, Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science (80-) 1995;268:1584–9. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- 27.Holliday EG, Maguire JM, Evans T-J, et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012;44:1147–51. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung SA, Taylor KE, Graham RR, et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet. 2011;7:e1001323. doi: 10.1371/journal.pgen.1001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Traylor M, Bevan S, Rothwell PM, et al. Using Phenotypic Heterogeneity to Increase the Power of Genome-Wide Association Studies: Application to Age at Onset of Ischaemic Stroke Subphenotypes. Genet Epidemiol. 2013;37:495–503. doi: 10.1002/gepi.21729. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Closas M, Couch FJ, Lindstrom S, et al. Genome-wide association studies identify four ER negative–specific breast cancer risk loci. Nat Genet. 2013;45:392–8. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;834:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu G, Warren L, Aponte J, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176:167–73. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 35.Bell B, Rose CL, Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. 1972;3:5–17. [Google Scholar]
- 36.Fishman A, Martinez F, Naunheim K, et al. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348:2059–73. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 37.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 42.Liu JZ, Mcrae AF, Nyholt DR, et al. A Versatile Gene-Based Test for Genome-wide Association Studies. Am J Hum Genet. 2010;87:139–45. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raychaudhuri S, Plenge RM, Rossin EJ, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Qiu W, Sathirapongsasuti JF, et al. Gene expression analysis uncovers novel Hedgehog interacting protein (HHIP) effects in human bronchial epithelial cells. Genomics. 2013 doi: 10.1016/j.ygeno.2013.02.010. doi:10.1016/j.ygeno.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Bakker PIW, Ferreira M a R, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunninghake GM, Cho MH, Tesfaigzi Y, et al. MMP12, lung function, and COPD in high-risk populations. N Engl J Med. 2009;361:2599–608. doi: 10.1056/NEJMoa0904006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reilly JJ. COPD and declining FEV1--time to divide and conquer? 2008;359:1616–8. doi: 10.1056/NEJMe0807387. [DOI] [PubMed] [Google Scholar]
- 48.Han B, Eskin E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. Am J Hum Genet. 2011;88:586–98. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han B, Eskin E. Interpreting meta-analyses of genome-wide association studies. PLoS Genet. 2012;8:e1002555. doi: 10.1371/journal.pgen.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. 1997;277:2002–4. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 51.Woodruff PG, Koth LL, Yang YH, et al. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med. 2005;172:1383–92. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaudhuri R, McSharry C, Brady J, et al. Sputum matrix metalloproteinase-12 in patients with chronic obstructive pulmonary disease and asthma: relationship to disease severity. J Allergy Clin Immunol. 2012;129:655–663. e8. doi: 10.1016/j.jaci.2011.12.996. [DOI] [PubMed] [Google Scholar]
- 53.Joos L, He JQ, Shepherdson MB, et al. The role of matrix metalloproteinase polymorphisms in the rate of decline in lung function. 2002;11:569–76. doi: 10.1093/hmg/11.5.569. [DOI] [PubMed] [Google Scholar]
- 54.Haq I, Chappell S, Johnson SR, et al. Association of MMP-12 polymorphisms with severe and very severe COPD: a case control study of MMPs-1, 9 and 12 in a European population. BMC Med Genet. 2010;11:7. doi: 10.1186/1471-2350-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jormsjo S, Ye S, Moritz J, et al. Allele-specific regulation of matrix metalloproteinase-12 gene activity is associated with coronary artery luminal dimensions in diabetic patients with manifest coronary artery disease. Circ Res. 2000;86:998–1003. doi: 10.1161/01.res.86.9.998. [DOI] [PubMed] [Google Scholar]
- 56.Soler Artigas M, Loth DW, Wain LV, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–90. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hao K, Bossé Y, Nickle DC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho MH, Washko GR, Hoffmann TJ, et al. Cluster analysis in severe emphysema subjects using phenotype and genotype data: an exploratory investigation. Respir Res. 2010;11:30. doi: 10.1186/1465-9921-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Celedon JC, Lange C, Raby BA, et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) 2004;13:1649–56. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 60.Hersh CP, Demeo DL, Lazarus R, et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:977–84. doi: 10.1164/rccm.200509-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanford LP, Ormsby I, Gittenberger-de Groot AC, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–70. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boileau C, Guo D-C, Hanna N, et al. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat Genet. 2012;44:916–21. doi: 10.1038/ng.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu HW, Balzar S, Seedorf GJ, et al. Transforming growth factor-beta2 induces bronchial epithelial mucin expression in asthma. Am J Pathol. 2004;165:1097–106. doi: 10.1016/s0002-9440(10)63371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balzar S, Chu HW, Silkoff P, et al. Increased TGF-beta2 in severe asthma with eosinophilia. J Allergy Clin Immunol. 2005;115:110–7. doi: 10.1016/j.jaci.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 65.Jiang J, George SC. TGF-β2 reduces nitric oxide synthase mRNA through a ROCK-dependent pathway in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2011;301:L361–7. doi: 10.1152/ajplung.00464.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson HGR, Mih JD, Krasieva TB, Tromberg BJ, George SC. Epithelial-derived TGF-beta2 modulates basal and wound-healing subepithelial matrix homeostasis. Am J Physiol Lung Cell Mol Physiol. 2006;291:L1277–85. doi: 10.1152/ajplung.00057.2006. [DOI] [PubMed] [Google Scholar]
- 67.Bottoms SE, Howell JE, Reinhardt AK, Evans IC, McAnulty RJ. Tgf-Beta isoform specific regulation of airway inflammation and remodelling in a murine model of asthma. PLoS One. 2010;5:e9674. doi: 10.1371/journal.pone.0009674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wen F-Q, Kohyama T, Liu X, et al. Interleukin-4- and interleukin-13-enhanced transforming growth factor-beta2 production in cultured human bronchial epithelial cells is attenuated by interferon-gamma. Am J Respir Cell Mol Biol. 2002;26:484–90. doi: 10.1165/ajrcmb.26.4.4784. [DOI] [PubMed] [Google Scholar]
- 69.Kajiho H, Saito K, Tsujita K, et al. RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J Cell Sci. 2003;116:4159–68. doi: 10.1242/jcs.00718. [DOI] [PubMed] [Google Scholar]
- 70.Saito K, Murai J, Kajiho H, Kontani K, Kurosu H, Katada T. A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J Biol Chem. 2002;277:3412–8. doi: 10.1074/jbc.M106276200. [DOI] [PubMed] [Google Scholar]
- 71.Janson C, Kasahara N, Prendergast GC, Colicelli J. RIN3 is a negative regulator of mast cell responses to SCF. PLoS One. 2012;7:e49615. doi: 10.1371/journal.pone.0049615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ballarin A, Bazzan E, Zenteno RH, et al. Mast cell infiltration discriminates between histopathological phenotypes of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186:233–9. doi: 10.1164/rccm.201112-2142OC. [DOI] [PubMed] [Google Scholar]
- 73.Mortaz E, Folkerts G, Redegeld F. Mast cells and COPD. Pulm Pharmacol Ther. 2011;24:367–72. doi: 10.1016/j.pupt.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 74.Andersson CK, Mori M, Bjermer L, Löfdahl C-G, Erjefält JS. Alterations in lung mast cell populations in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;181:206–17. doi: 10.1164/rccm.200906-0932OC. [DOI] [PubMed] [Google Scholar]
- 75.Wang K, Dickson SP, Stolle CA, Krantz ID, Goldstein DB, Hakonarson H. Interpretation of association signals and identification of causal variants from genome-wide association studies. Am J Hum Genet. 2010;86:730–42. doi: 10.1016/j.ajhg.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou X, Baron RM, Hardin M, et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet. 2012;21:1325–35. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumasaka N, Aoki M, Okada Y, et al. Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PLoS One. 2012;7:e44507. doi: 10.1371/journal.pone.0044507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lamontagne M, Couture C, Postma DS, et al. Refining Susceptibility Loci of Chronic Obstructive Pulmonary Disease with Lung eqtls. PLoS One. 2013;8:e70220. doi: 10.1371/journal.pone.0070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bloom AJ, Harari O, Martinez M, et al. Use of a predictive model derived from in vivo endophenotype measurements to demonstrate associations with a complex locus, CYP2A6. Hum Mol Genet. 2012;21:3050–62. doi: 10.1093/hmg/dds114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bloom AJ, Baker TB, Chen L-S, et al. Variants in two adjacent genes, EGLN2 and CYP2A6, influence smoking behavior related to disease risk via different mechanisms. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt432. doi:10.1093/hmg/ddt432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gowdy KM, Fessler MB. Emerging roles for cholesterol and lipoproteins in lung disease. Pulm Pharmacol Ther. 2013;26:430–7. doi: 10.1016/j.pupt.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cirillo DJ. Lipids and Pulmonary Function in the Third National Health and Nutrition Examination Survey. Am J Epidemiol. 2002;155:842–8. doi: 10.1093/aje/155.9.842. [DOI] [PubMed] [Google Scholar]
- 84.Basili S, Ferroni P, Vieri M, et al. Lipoprotein(a) serum levels in patients affected by chronic obstructive pulmonary disease. Atherosclerosis. 1999;147:249–52. doi: 10.1016/s0021-9150(99)00192-6. [DOI] [PubMed] [Google Scholar]
- 85.Burkart KM, Manichaikul A, Wilk JB, et al. APOM and high-density lipoprotein are associated with lung function and percent emphysema. Eur Respir J. 2013 doi: 10.1183/09031936.00147612. doi:10.1183/09031936.00147612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morgan K, Marsters P, Morley S, et al. Oncostatin M induced alpha1-antitrypsin (AAT) gene expression in Hep G2 cells is mediated by a 3’ enhancer. Biochem J. 2002;365:555–60. doi: 10.1042/BJ20011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baines KJ, Simpson JL, Gibson PG. Innate immune responses are increased in chronic obstructive pulmonary disease. PLoS One. 2011;6:e18426. doi: 10.1371/journal.pone.0018426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dransfield MT, Bailey WC. COPD: racial disparities in susceptibility, treatment, and outcomes. Clin Chest Med. 2006;27:463–71. vii. doi: 10.1016/j.ccm.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 89.Foreman MG, Zhang L, Murphy J, et al. Early-onset chronic obstructive pulmonary disease is associated with female sex, maternal factors, and African American race in the COPDGene Study. Am J Respir Crit Care Med. 2011;184:414–20. doi: 10.1164/rccm.201011-1928OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Powell R, Davidson D, Divers J, et al. Genetic ancestry and the relationship of cigarette smoking to lung function and per cent emphysema in four race/ethnic groups: a cross-sectional study. Thorax. 2013;68:634–42. doi: 10.1136/thoraxjnl-2012-202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Coram MA, Duan Q, Hoffmann TJ, et al. Genome-wide Characterization of Shared and Distinct Genetic Components that Influence Blood Lipid Levels in Ethnically Diverse Human Populations. Am J Hum Genet. 2013 doi: 10.1016/j.ajhg.2013.04.025. doi:10.1016/j.ajhg.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waters KM, Stram DO, Hassanein MT, et al. Consistent association of type 2 diabetes risk variants found in europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:9. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–7. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 94.Wu Y, Waite LL, Jackson AU, et al. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet. 2013;9:e1003379. doi: 10.1371/journal.pgen.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho MH, Ciulla DM, Klanderman BJ, et al. Analysis of exonic elastin variants in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2009;40:751–5. doi: 10.1165/rcmb.2008-0340OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Silverman EK, Sandhaus RA. Clinical practice. Alpha1-antitrypsin deficiency. 2009;360:2749–57. doi: 10.1056/NEJMcp0900449. [DOI] [PubMed] [Google Scholar]
- 97.Kelleher CM, Silverman EK, Broekelmann T, et al. A functional mutation in the terminal exon of elastin in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2005;33:355–62. doi: 10.1165/rcmb.2005-0206OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han MK, Muellerova H, Curran-Everett D, et al. GOLD 2011 disease severity classification in COPDGene: a prospective cohort study. Lancet Respir Med. 2013;1:43–50. doi: 10.1016/S2213-2600(12)70044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hancock DB, Artigas MS, Gharib SA, et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Repapi E, Sayers I, Wain LV, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2009 doi: 10.1038/ng.501. doi:ng.501 [pii] 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2009 doi: 10.1038/ng.500. doi:ng.500 [pii] 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deloukas P, Kanoni S, Willenborg C, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–7. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buist AS, McBurnie MA, Vollmer WM, et al. International variation in the prevalence of COPD (the BOLD Study): a population-based prevalence study. 2007;370:741–50. doi: 10.1016/S0140-6736(07)61377-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.