Abstract
We present a method for identifying colitis in colon biopsies as an extension of our framework for the automated identification of tissues in histology images. Histology is a critical tool in both clinical and research applications, yet even mundane histological analysis, such as the screening of colon biopsies, must be carried out by highly-trained pathologists at a high cost per hour, indicating a niche for potential automation. To this end, we build upon our previous work by extending the histopathology vocabulary (a set of features based on visual cues used by pathologists) with new features driven by the colitis application. We use the multiple-instance learning framework to allow our pixel-level classifier to learn from image-level training labels. The new system achieves accuracy comparable to state-of-the-art biological image classifiers with fewer and more intuitive features.
Index Terms: histology, colitis, image classification
1. INTRODUCTION
Screening for colitis based on microscopic review of tissue sections taken by endoscopic biopsy occupies a prominent position in the overall practice of a general pathologist. As an example, in one tertiary pediatric hospital (the Children’s Hospital of Pittsburgh, CHP), screening for colitis occupies approximately 10–20% of pathologists’ time, making it both extremely important for patient care and one of the most common specimens in diagnostic pathology. Upper and lower gastrointestinal endoscopic biopsies are performed in many clinical scenarios including abdominal pain, vomiting, diarrhea, etc. In the majority of cases, a lower gastrointestinal endoscopy will be performed to exclude colitis.
Colitis is broadly defined as any pathological inflammatory state of the colon that can have multiple causes including infection, ischemia, immunological, and others. For example, inflammatory bowel disease (IBD; which includes ulcerative colitis and Crohns disease) is a major cause of colitis in children and adults and contributes heavily to the need for endoscopic biopsy both for diagnosis and subsequent follow-up after treatment. For instance, in the United States, the Centers for Disease Control and Prevention estimates that 1.4 million people have IBD, creating a health care cost of $1.7 billion [1]. Based on this estimation, the number of endoscopic biopsies of the colon for suspected IBD only (not including endoscopies done for other clinical scenarios) would be substantially greater since more individuals will undergo endoscopy biopsy than have actual disease. For instance, at CHP, approximately 1200 biopsies of the colon were done in 2011 out of approximately 3500–4000 endoscopies performed.
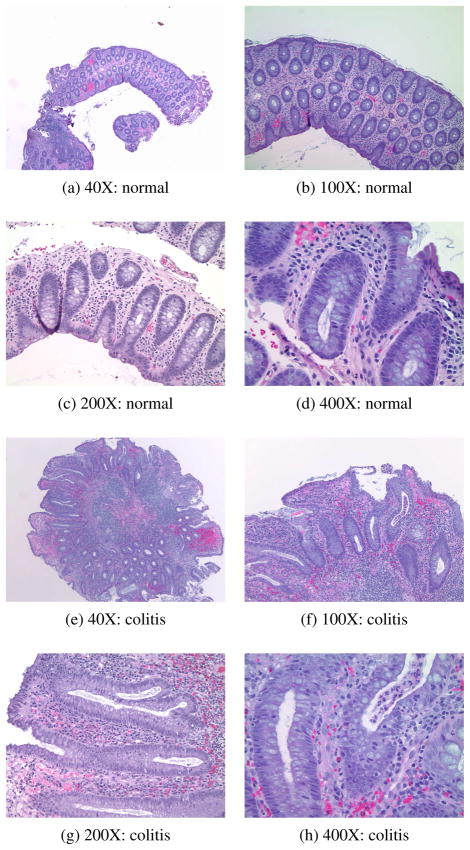
After standard tissue fixation and processing which include the generation of H&E-stained slides, a pathologist examines the colon tissue sections with a microscope and assesses for the presence or absence of colitis (see Figure 1 for examples of tissue sections from normal colon and colitis). Visual features used in this process include an increase in the number of mucosal inflammatory cells, involvement of the crypt epithelium by inflammatory cells, and overall architectural distortion. While histological examination of tissue specimens by a pathologist is standard practice in medicine and biomedical research, and many facets of the tissue processing and imaging can be/have been automated, the microscopic analysis of all tissue sections as described is still performed by eye and is time consuming and costly. Similar to systems available for screening of Pap smear cytological preparations, an automated image analysis tool that could screen images of tissue sections of frequently recurring specimens (such as endoscopic biopsies) for the presence or absence of a disease could prove valuable.
Fig. 1.
Example normal and colitis hematoxylin and eosin (H&E)-stained images at different magnification levels.
Related work on automated histology mainly includes systems for detecting or grading cancer [2–5]. Our lab, among others [6], has begun to develop a general framework for automated histology. Using our histopathology vocabulary (HV), we have had success with the identification and delineation of tissues in images of H&E-stained teratomas [7]. Here, we build upon this framework to develop a system for automated detection of colitis.
2. DOMAIN-BASED CLASSIFICATION FRAMEWORK
The problem at hand is a standard image-processing task: classification. We classify images into one of two diagnostic categories: normal or colitis. Here, we face a trade-off between universality and specificity: do we develop a general method that will work reasonably well in a number of application domains, or a specific method that will work extremely well in just this one application?
To address this trade-off, we adopt the following methodology: We aim to design a feature set understood by both pathologists and engineers based on the actual visual cues used by pathologists; we term this colitis histopathology vocabulary. We have used this method in [7] to design a general HV appropriate for many histopathology applications; here, we refine the HV to add colitis-specific features.
The method consists of the following steps:
Formulation of initial set of descriptions. The pathologist provides descriptions of the characteristics that best describe a given tissue and ranks them by their effectiveness in identifying the tissue.
Computational translation of key terms. From this set, the engineer distills the key terms and finds their computational synonyms, creating a computational vocabulary. For example, the pathologist’s term “long nuclei” can be translated into a computational term “nucleus eccentricity > 0.75”.
Computational translation of descriptions. Entire pathologist’s descriptions are similarly translated. For example, the pathologist’s description “small, oval-shaped nuclei” can be translated into two key terms as “mean nucleus eccentricity > 0.75” + “nucleus size < 0.2”.
Verification of translated descriptions. The pathologist then receives the descriptions translated using the computational vocabulary and tries to identify the tissue being described, emulating the overall classification system with translated descriptions as features and the pathologist as the classifier.
Refinement of insufficient terms. If the pathologist is unable to identify a tissue based on translated descriptions, or if a particular translation is not understandable, then that translation is refined and presented again to the pathologist for verification.
Histopathology vocabulary. If the pathologist is able to identify a tissue based on translated descriptions, then the discriminative power of the key terms is validated, and these terms are included as HV terms to create features.
Using this method we designed an initial HV vocabulary consisting of background/fiber color, cytoplasm color, clear areas (lumen), nuclei color, nuclei density, nuclei shape, nuclei orientation and nuclei organization. In the same work, we used pixel-level classification to identify and delineate tissues.
For the colitis problem, we identified inflammation, marked by an increase in the number and variety of cells present, as an important indicator of colitis. In our previous work, the nucleus density feature was based only on nucleus coverage (i.e. the local percentage of pixels inside nuclei), and neglected counting individual nuclei. To better describe inflammation, we include a more robust analysis of nuclei and a description of red blood cells, resulting in the following colitis HV set:
3. COLITIS CLASSIFICATION ALGORITHM
The colitis classification algorithm has two parts. First, we extract the colitis HV features for each pixel in the input image. We then classify all of its pixels, and assign the label colitis if the number of colitis pixels is above a threshold.
3.1. Feature Extraction
To compute the colitis HV features, we (1) locate the regions of nuclei, red blood cells, background tissue, and empty slide in each image; (2) use moment filters to count the nuclei and compute their size and eccentricity; and (3) gather local information at each pixel with an averaging filter.
We use color to assign each pixel to one of the four objects of interest. Since each of these objects has a distinct color under H&E staining, we use our prior knowledge to assign a set of color values to each object (shades of blue for nuclei, red for red blood cells, pink for background, and white for empty slide). For each image, we adjust these color values by running five iterations of k-means clustering. This step helps account for illumination and staining intensity variations. We then label pixels according to their nearest cluster (Euclidean distance in RGB space).
Moment Filters
After labeling each pixel of the image as nucleus, red blood cell, background tissue, or empty slide, we need to count the nuclei in the image and analyze their shape.
To do this, we use the pixel labels to create a nucleus mask, 1N, a binary version of the input image that takes a value of 1 wherever nuclei are present. Similarly, we create masks for red blood cells and background tissue, 1R and 1B, respectively. We extract local shape information from the nucleus mask using local moment filters in an approach similar to [8]. Briefly, we begin with a windowing filter, w, that is nonnegative, symmetric, and sums to 1. From this window, we define the order-p, q local moment filter
At location [m, n], the local moment transform of an image, I, is
| (1) |
We choose w to be Gaussian with a standard deviation of ⅓ the expected radius of a nucleus (in pixels).
We apply (1) to each image to obtain m00, m10, m01, m11, m20, and m02, convert them to central moments,
and form the inertia matrix , which describes the horizontal and vertical spread of the pixels within the support of w centered at the location [m, n], but calculated around the local center of mass [m + x̄, n + ȳ] rather than around [m, n]. From J, we calculate the local eccentricity
| (2) |
In a departure from [8], we then use the shape information computed by the moment filters to locate nucleus centers. We take as candidates each position where m00 reaches a local maximum, that is, every pixel for which m00 is larger than at any of its eight neighbors. We then remove candidates for which m00 is too small (less than 0.25) or for which the eccentricity is too high (greater than 0.1). Intuitively, this scheme tags pixels that are the centers of nucleus-sized blobs, rejecting those that are too oblong to be nuclei. An example comparing this scheme to simple maxima detection is shown in Figure 2.
Fig. 2.
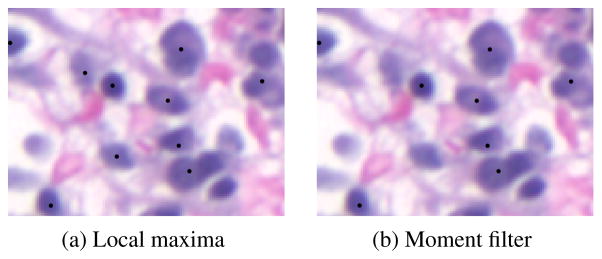
Example nucleus detections (marked with black dots) from simple maxima detection (a) and moment filter-based detection (b). The moment-filter approach rejects the two objects at the center-left because they are overly oblong.
Features
We use the nucleus center detections to form a new mask, 1C, which takes the value of 1 for those pixels that have been labeled as nucleus centers in the previous step. We proceed to use 1N and 1C and an averaging filter, ŵ (Gaussian with standard deviation of nine times the expected nucleus radius) to create the feature set we list in Table 1, where — and represent pointwise multiplication and division, respectively. These six features form a length-10 (because of two color features) feature vector for each pixel in the image.
Table 1.
Colitis HV feature set.
| Feature name | Expression |
|---|---|
| Nucleus density | w̄* 1C |
| Average nucleus size | [w̄* (m00 · 1C)]/[w̄ * 1C] |
| Average nucleus eccentricity | [w̄* (e · 1C)]/[w̄ * 1C] |
| Average nucleus color | [w̄ * (I · 1N)]/[w̄ * 1N ] |
| RBC coverage | w̄ * 1R |
| Average background color | [w̄ * (I · 1B)]/[w̄ * 1B] |
3.2. Pixel-level classification
After feature extraction, we classify pixels using an artificial neural network (MATLAB Neural Network Toolbox). The network has ten input nodes, no hidden layers, and one output node, with a bias connected only to the output node; activation functions are linear throughout. For training, we throw away all nontissue pixels, then label the remaining pixels according to the image they come from. We call this version of the system the pixel-level classifier (PLC).
Since our goal is to label images rather than pixels, we need a way to convert the pixel-level labels into into image-level labels. We accomplish this by selecting a threshold and labeling any image with a number of inflamed pixels greater than the threshold as colitis. The threshold is selected from the training data so as to give the least training error at the image level.
3.3. Multiple-instance learning
The PLC training we just described overlooks an important aspect of the colitis problem: even a small region in an image can contain features diagnostic of colitis. Thus, even in an colitis-labeled image, some pixels actually belong to normal tissue. To understand how to adapt our PLC training to this problem, we look at the classification task in the multiple-instance learning (MIL) framework [9].
Let X be a set of N digital images, . Each xi contains multiple instances (pixels) , where P and Q denote the height and width of each image, respectively. Each xi,j has a corresponding label, yi,j where yi,j ∈ {0, 1} (in our case, 0 denotes normal and 1 colitis), but only the overall label for each image, with yi ∈ {0, 1}, is known during training. For each image, the relationship between yi and is given by
In other words, a pathologist may label (diagnose) an image as colitis based on only a small region of the image.
The goal is to create an image-level classifier function d such that d(x*) = y* for any unseen image, x*, accomplished by learning a pixel-level classifier function p such that p(x*,*) = y*,* for any unseen pixel, x*,*, such that
We thus see that some of the training labels for the PLC training described earlier were incorrect: some colitis pixels should have been labeled as normal. To address this issue, we first use the basic PLC to identify the top 50% most inflamed pixels in each colitis image and create a new training set with only these pixels labeled as colitis, and retrain the classifier. We call the retrained classifier the PLC-MIL.
4. EXPERIMENTS AND RESULTS
We compared the performance of the PLC-MIL to two freely avail able biological image classifiers. The first is WND-CHARM [10], which extracts a large number generic image-level features, then classifies images with a nearest neighbor algorithm. We used the expanded color feature set. The second is the multiresolution classi fication (MRC) system [11], which decomposes the image into sub spaces via a multiresolution transform, extracts features and classi fies within each subspace separately, then combines these local de cisions via weighted voting. We used two levels of a wavelet packet decomposition and the expanded Haralick texture feature set.
Feature extraction for WND-CHARM required a powerful desk top computer (2× Intel Xeon E7540 2.00GHz and 64GB RAM), while the PLC and MRC features were computed on a laptop (In tel Core i7 2.67 GHz and 4 GB of RAM). For a single image, feature extraction required 30 seconds for the PLC, 5 seconds for the MRC, and 140 minutes for WND-CHRM. Each classifier was evaluated at each magnification level (40×, 100×, 200×, and 400×) with a leave one-out cross-validation over 20 normal and 20 colitis images.
Table 2 summarizes the results. They show that no classifier consistently better than the others at all scales. For all four classi fiers, accuracy is generally higher at lower magnifications and de creases as magnification increases. While the PLC classifier per forms well, the PLC-MIL classifier improves its performance at high magnification. Notably, the PLC-MIL performs comparably to both the MRC and WND-CHRM, with the advantage of simple, physio logically meaningful features and pixel-level labels (see Figure 3)
Table 2.
Comparison of classification accuracy (in %).
| Magnification | WND-CHRM | MRC | PLC | PLC-MIL |
|---|---|---|---|---|
| 40× | 100.0 | 95.0 | 97.5 | 97.5 |
| 100× | 97.5 | 90.0 | 90.0 | 87.5 |
| 200× | 87.5 | 85.0 | 77.5 | 90.0 |
| 400× | 77.5 | 85.0 | 80.0 | 85.0 |
Fig. 3.
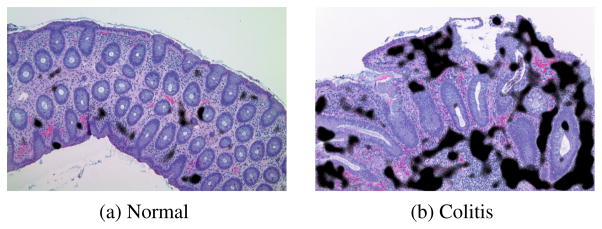
Example pixel-level classification results for PLC-MIL (100×). These results are physiologically reasonable: inflammation (black) is detected in regions of high nuclei density and not in crypts or in areas of hemorrhage (generally artifacts of the procedure).
5. CONCLUSIONS AND FUTURE WORK
We presented a framework and algorithm for classification of colon biopsy images. We build upon our previous work on automated histology and the HV and show that the colitis HV, especially when applied in a MIL framework, is well-suited to detecting colitis in images of colon biopsies. The PLC-MIL classifier compares favorably to generalized biomedical image classifiers, while using a small set of features, easily understood by physicians and fast to compute.
A near-future task is to, for any misclassified image, investigate which regions were mis-labeled. For those regions, we can see identify which aspect of the system (feature extraction or classification) failed, a task made easier because our features can be understood in simple terms. At a broader level, we plan to continue expanding the HV framework to create a truly general automated histology system.
Acknowledgments
The authors gratefully acknowledge support from the NIH through award EB009875, NSF through awards 1017278, 0750271, and the CMU CIT Award. Dr. Bhagavatula performed this research while at CMU.
References
- 1.Centers for Disease Control and Prevention. Inflammatory bowel disease (IBD) Nov, 2011. [Google Scholar]
- 2.Petushi S, Katsinis C, Haber MM, Garcia FU, Tozeren A. Large scale computations on histology images revels grade differentiating parameters for breast cancer. BMC Medical Imaging. 2006;6(14) doi: 10.1186/1471-2342-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle S, Monaco J, Feldman M, Tomaszewski J, Madabhushi A. An active learning based classification strategy for the minority class problem: application to histopathology annotation. BMC Bioinformatics. 2011 Oct;12(1):424. doi: 10.1186/1471-2105-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sieren J, Weydert J, Bell A, De Young B, Smith A, Thiesse J, Namati E, McLennan G. An automated segmentation approach for highlighting the histological complexity of human lung cancer. Ann Biomed Eng. 2010;38:3581–3591. doi: 10.1007/s10439-010-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sertel O, Kong J, Catalyurek UV, Lozanski G, Saltz JH, Gurcan MN. Histopathological image analysis using model-based intermediate representations and color texture: Follicular lymphoma grading. Journ Signal Proc Systems. 2009;55(1–3):169–183. [Google Scholar]
- 6.Meng T, Shyu M-L, Lin L. Multimodal information integration and fusion for histology image classification. International Journal of Multimedia Data Engineering and Management. 2011 Apr-Jun;2(2):54–70. [Google Scholar]
- 7.Bhagavatula R, Fickus MC, Kelly JW, Guo C, Ozolek JA, Castro CA, Kovačević J. Automatic identification and delineation of germ layer components in H&E stained images of teratomas derived from human and nonhuman primate embryonic stem cells. Proc. IEEE Int. Symp. Biomed. Imaging; Rotterdam, The Netherlands. Apr. 2010; pp. 1041–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sühling M, Arigovindan M, Hunziker P, Unser M. Multiresolution moment filters: Theory and applications. IEEE Trans Image Proc. 2004 Apr;13(4):484–495. doi: 10.1109/tip.2003.819859. [DOI] [PubMed] [Google Scholar]
- 9.Dietterich TD, Lathrop RH, Lozano-Perez T. Solving the Multiple-Instance problem with Axis-Parallel rectangles. Artif Intell. 1997;89:31–71. [Google Scholar]
- 10.Shamir L, Orlov N, Eckley DM, Macura T, Johnston J, Goldberg IG. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recogn Letters. 2008;29:1684–1693. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chebira A, Ozolek JA, Castro CA, Jenkinson WG, Gore M, Bhagavatula R, Khaimovich I, Ormon SE, Navara CS, Sukhwani M, Orwig KE, Ben-Yehudah A, Schatten G, Rohde GK, Kovačević J. Multiresolution identification of germ layer components in teratomas derived from human and nonhuman primate embryonic stem cells. Proc. IEEE Int. Symp. Biomed. Imaging; Paris, France. May 2008; pp. 979–982. [Google Scholar]