Abstract
Traditionally, the ketone body β-hydroxybutyrate (βOHB) has been looked upon as a carrier of energy from liver to peripheral tissues during fasting or exercise. However, βOHB also signals via extracellular receptors and acts as an endogenous inhibitor of histone deacetylases (HDACs). These recent findings support a model in which βOHB functions to link the environment, in this case the diet, and gene expression via chromatin modifications. Here, we review the regulation and functions of ketone bodies, the relationship between ketone bodies and calorie restriction, and the implications of HDAC inhibition by the ketone body βOHB in the modulation of metabolism, and diseases of aging.
Keywords: acetylation, HDAC, calorie restriction, longevity, epigenetics
Metabolites in aging pathways
The past two decades have witnessed an explosion of knowledge of the genetic and metabolic factors that affect aging and lifespan. Calorie restriction (CR) remains the surest path to increased longevity and resilience to diseases of aging across many organisms, from yeast to monkeys and perhaps humans [1]. Many of the beneficial effects of CR appear to be due to modification of specific nutrient-responsive pathways, such as the insulin/insulin-like growth factor (IGF-1) pathway, the target of rapamycin (TOR) signaling pathway, and the NAD+-dependent deacetylases sirtuins. For example, genetic modulation of any one step in the IGF-1 signaling pathway, from ligand to receptor, to downstream kinase cascades and target transcription factors, enhances lifespan in worms and mice [2]. Rapamycin, the first small molecule found to extend lifespan in mammals, works by inhibiting the nutrient-responsive TOR pathway [3]. Finally, the mitochondrial NAD+-dependent protein deacetylase SIRT3 is required for at least one of the benefits of CR in mice, prevention of age-related hearing loss [4].
Intriguingly, the ketone body β-hydroxybutyrate (βOHB) might also be a metabolic intermediary of the benefits of CR and fasting. Long viewed as a simple carrier of energy from the liver to peripheral tissues during prolonged fasting or exercise, βOHB also possesses signaling activities, perhaps most intriguingly as an endogenous inhibitor of HDACs [5]. Thus, it joins a small but growing list of metabolic intermediaries that affect gene expression via chromatin modifications [6]. These intermediaries may be key links between variations in the cellular environment and the epigenetic changes associated with increased healthspan and lifespan. Environmental factors, such as nutrition, dramatically alter cellular metabolism, and many also alter the epigenetic regulation of gene expression. Overall, energy balance controls the NAD/NADH ratio, which affects the activity of sirtuins [7]. Lipid-burning states, such as fasting, increase both acetyl-CoA production and levels of histone acetylation [5]. Intake of threonine affects the levels of the methyl donor S-adenosylmethionine, which in turn promotes histone methylation [8]. As discussed below, the activity of HDACshas already been linked to the regulation of lifespan (Box 1) and to diseases of aging, such as diabetes and cancer.
BOX 1. HDACs in longevity and aging: lessons from model organisms.
The association of class I HDACs with the regulation of lifespan in model organisms is most intriguing. Deletion of Rpd3, the yeast and fly homolog of mammalian class I HDACs (e.g., HDACs 1 and 2), extends replicative lifespan in yeast by 40–50% [121]. Rpd3 deletion enhances ribosomal DNA (rDNA) silencing [121], similar to the mechanism by which overexpression of the sirtuin Sir2 enhances yeast replicative longevity [122]. However, co-deletion of Hda1, the yeast homolog of class II HDACs that partially overlaps Rpd3 function, actually increases yeast mortality—one example of a “Goldilocks” zone of HDAC function [121]. Another possible mechanism of increased longevity of yeast Rpd3 mutants is through increased autophagy, which is regulated by histone acetylation of specific genes [123].
In Drosophila, flies heterozygous for a null or hypomorphic Rpd3 allele show a 30–40% extension of lifespan, with no further increase with CR [124]. Both CR and reduced Rpd3 activity increase expression of Sir2 [124]. Conversely, mutations in Sir2 block lifespan extension by either CR or Rpd3 mutations [125]. Altogether, this indicates that in Drosophila, CR, Rpd3, and Sir2, all function in the same longevity pathway. Notably, while these modest reductions in Rpd3 activity enhance lifespan, strong hypomorphic alleles are embryonic lethal [126]. The small molecule HDAC inhibitors trichostatin A and butyrate also extend lifespan in Drosophila, perhaps via increased expression of heat shock proteins hsp22 and hsp70 [127]. Feeding 4-phenylbutyrate throughout adulthood increases lifespan in Drosophila although high doses are toxic. Interestingly, it also increases stress resistance and climbing ability, and works even when given later in adult life [128]. Valproic acid, another HDAC inhibitor, extends lifespan in Caenorhabditis elegans, although again high doses are toxic [129].
HDAC knockouts in mammals highlight their importance in longevity and age-related diseases. Although HDAC1 knockout is embryonic lethal [130], similar to fly Rpd3 knockout, HDAC2 knockout mice are viable but 25% smaller than normal, with impaired IGF-1 signaling and reduced tumor formation when crossed with oncogenic adenomatous polyposis coli (APC) gene knockout mouse models [131]. Conditional knockouts in mouse embryonic fibroblasts and embryonic stem cells demonstrated roles for HDACs 1 and 2 in hematopoiesis [132] and stem cell differentiation [133]. Lifespan has not been rigorously reported for class I HDAC mutant mice, nor for HDAC inhibitor treatment in mammals. By analogy with yeast and fly studies a positive effect might require careful calibration of gene dosage or function, or inhibitor concentration.
Here we review the metabolism, regulation, and functions of ketone bodies, and how the newly discovered activity of βOHB as an endogenous HDAC inhibitor opens a broad new vista into its potential roles in regulation of lifespan and diseases of aging.
Metabolism, regulation and function of ketone bodies
Ketone bodies are small lipid-derived molecules that serve as a circulating energy source for tissues in times of fasting or prolonged exercise. Fatty acids in adipose tissue contain over 80% of the human body’s stored energy [9]. During fasting, muscle and liver stores of glycogen are depleted first. Then, fatty acids are mobilized from adipocytes and transported to the liver for conversion to ketone bodies. Ketone bodies are then distributed via the circulation to metabolically active tissues, such as muscle or brain, where they are converted to acetyl-CoA and used as a glucose-sparing energy source [9]. In humans, basal serum levels of βOHB are in the low micromolar range, but begin to rise to a few hundred micromolar after 12–16 hours of fasting, reaching 1–2 mM after 2 days of fasting [10, 11], and 6–8 mM with prolonged starvation [12]. Similar 1–2 mM levels of βOHB can be reached after 90 minutes of intense exercise [13]. Consistent levels above 2 mM are also reached with a ketogenic diet that is almost devoid of carbohydrates [14]. Children produce and utilize βOHB more efficiently than adults, a capability crucial in the days immediately after birth when the brain depends on ketone bodies as an energy source, and serum levels can reach 2–3 mM [12]. At the other end of life, the elderly generate ketone bodies after a fast or ketogenic meal to the same extent as younger adults [15, 16].
Ketone body production and utilization
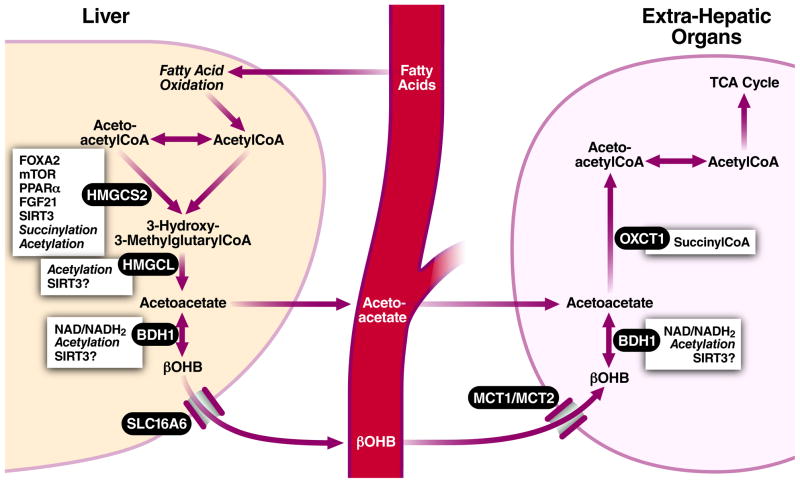
Most ketone body production occurs in the liver [9], although smaller amounts may be produced in other tissues through aberrant expression of ketogenic enzymes [17, 18] or reversal of the ketolysis pathway [19, 20]. In hepatic ketogenesis (Figure 1), fatty acids are first metabolized to acetyl-CoA via mitochondrial β-oxidation. Mitochondrial HMG-CoA synthase (HMGCS2, EC 2.3.3.10) condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA, from which acetoacetate is liberated by HMG-CoA lyase (HMGCL, EC 4.1.3.4) (Figure 1). Acetoacetate is the common precursor of the two other circulating ketone bodies, acetone and βOHB. Most acetoacetate is further metabolized by β-hydroxybutyrate dehydrogenase (BDH1, EC 1.1.1.30) to βOHB. βOHB is the most abundant circulating ketone body and is less likely to spontaneously degrade into acetone than acetoacetate. Once taken up by a target tissue, βOHB is converted back into acetoacetate by the same enzyme, but from there, the pathway of ketone body utilization diverges from the synthetic pathway. Succinyl-CoA donates its CoA to acetoacetate to form acetoacetyl-CoA, a reaction catalyzed in most tissues by succinyl-CoA:3-ketoacid coenzyme A transferase (OXCT1, also known as SCOT, EC 2.8.3.5). This reaction bypasses the essentially irreversible reaction catalyzed by HMG-CoA synthase. The differing enzymatic routes of synthesis and utilization prevent a futile cycle of βOHB synthesis and utilization in the liver since OXCT1 is not expressed in the liver [21]. Acetoacetyl-CoA can then be converted to two acetyl-CoA and fed into the TCA cycle for oxidation and ATP production [22].
Figure 1. Outline of ketone body metabolism and regulation.
The key irreversible step in ketogenesis is synthesis of 3-hydroxy-3-methylglutaryl-CoA by HMGCS2. Conversely, the rate limiting step in ketolysis is conversion of acetoacetate to acetoacetyl-CoA by OXCT1. HMGCS2 transcription is heavily regulated by FOXA2, mTOR, PPARα, and FGF21. HMGCS2 activity is post-translationally regulated by succinylation and acetylation/SIRT3 deacetylation. Other enzymes are regulated by cofactor availability (e.g., NAD/NADH2 ratio for BDH1). All enzymes involved in ketogenesis are acetylated and contain SIRT3 deacetylation targets, but the functional significance of this is unclear other than for HMGCS2. Although ketone bodies are thought to diffuse across most plasma membranes, the transporter SLC16A6 may be required for liver export, while several monocarboxylic acid transporters assist with transport across the blood-brain barrier.
Transcriptional and post-translational regulation of βOHB metabolism
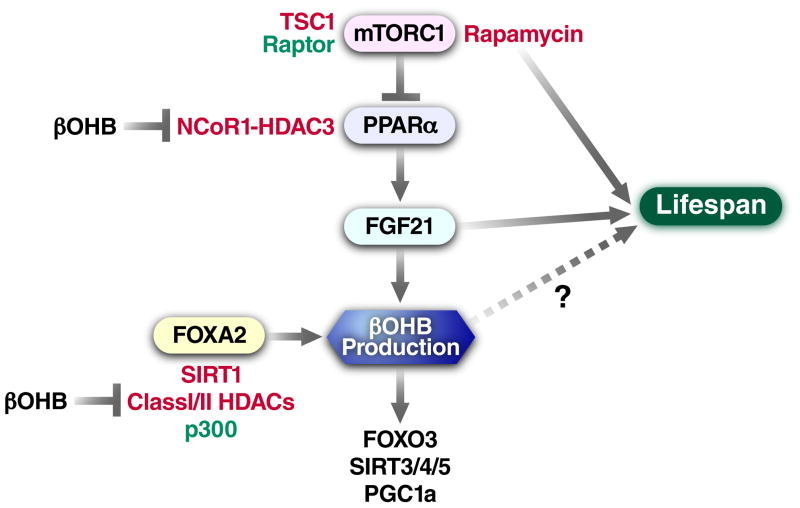
The rate-limiting step of ketone body synthesis is the condensation of acetyl-CoA and acetoacetyl-CoA into HMG-CoA by mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2) [23]. HMGCS2, and therefore production of ketone bodies, is transcriptionally regulated at least two nutrient-responsive pathways (Figure 2). The first involves the forkhead box transcription factor FOXA2, which binds to the HMGCS2 promoter and activates transcription [24]. FOXA2 itself is regulated by dueling hormonal signals: insulin signaling leads to inactivation of FOXA2 via phosphorylation and nuclear export [25], while glucagon activates FOXA2 via p300 acetylation [26]. FOXA2 deacetylation is controlled by yet another nutrient-responsive enzyme, SIRT1, working in cooperation with class I or II HDACs [26]. The second pathway of HMGCS2 transcriptional regulation involves mTORC1, peroxisome proliferator-activated receptor alpha (PPARα), and finally fibroblast growth factor (FGF21) [23, 27–29]. Both PPARα and its target gene FGF21 are dramatically up-regulated in liver after fasting or by ketogenic diet, and mice lacking either one have reduced levels of ketogenesis [27, 28]. The mTORC1 complex suppresses PPARα, thus inhibition of mTORC1 is required for the induction of PPARα [29], and in turn PPARα is required to induce FGF21 [27].
Figure 2. Intersection of longevity pathways and regulation of βOHB production.
βOHB production is controlled by at least two nutrient-responsible pathways that are implicated in longevity and may be subject to regulation by βOHB via HDAC inhibition. Rapamycin and down-regulation of the mTOR pathway promote ketogenesis; rapamycin and FGF21 enhance mammalian longevity. FOXA2 also enhances ketogenesis, and its activation is regulated by both class III (sirtuins) and class I/II HDACs.
The activity of HMGCS2 is also post-translationally regulated by succinylation and acetylation. HMGCS2 is deacetylated and activated by the primary mitochondrial deacetylase SIRT3 [30]. SIRT3 regulates many pathways involved in fasting metabolism, and mice lacking SIRT3 have reduced levels of βOHB upon fasting [30]. Interestingly, all of the enzymes involved in the generation of ketone bodies from lipids are acetylated, many of them heavily, and contain at least one site for SIRT3 deacetylation [31, 32]. Similar to acetylation, succinylation of HMGCS2 reduces its activity [33]. The mechanism that drives succinylation is not known, but some degree of dependence on succinyl-CoA levels is suggested by the fact that both liver succinyl-CoA abundance and succinylation of HMGCS2 are reduced after treatment of rats with glucagon [33, 34]. Lysine succinylation is removed from other proteins by the mitochondrial desuccinylase SIRT5, which regulates a variety of mitochondrial pathways involved in fasting metabolism [35], though it is not yet known if HMGCS2 is a target of SIRT5 desuccinylation. By contrast, the interconversion of acetoacetate and βOHB by BDH1 appears to be readily reversible and is regulated primarily by the ratios of substrates and cofactors (NAD/NADH2) [22]. BDH1 contains several SIRT3-regulated acetylation sites, though their functional significance is not yet known [31, 32]. OXCT1 activity may be inhibited by tyrosine nitration [36], but little else is known of its regulation.
βOHB transport, utilization and conservation
βOHB transport is relatively less well understood than its synthesis and utilization. A small, polar molecule, βOHB is readily soluble in water and blood [9]. Several monocarboxylic acid transporters, including MCT1 and MCT2, carry it across the blood-brain barrier [37]. Of interest, up-regulation of MCT1 in particular is associated with high utilization of ketone bodies in the neonatal period and on a ketogenic diet [38]. Recently, the monocarboxylate transporter SLC16A6 was identified as the key transporter for exporting βOHB from the liver, in model organisms; zebrafish lacking Slc16a6a develop fatty liver during fasting mainly due to the diversion of acetyl-CoA to lipid synthesis rather than ketone bodies [39].
Interestingly, the use of βOHB as a fasting energy source is evolutionarily ancient. Many species of bacteria synthesize polymers of βOHB to store energy [12], a rather interesting reaction that is utilized in the production of biopolymers as a plastic substitute [40]. A complete “suite” of ancestral βOHB biosynthetic enzymes, from HMG-CoA synthase through βOHB dehydrogenase, emerged early in eukarya and is present even in plants. This deep conservation likely reflects important roles in cholesterol biosynthesis, as these cytoplasmic enzymes are not known to participate in ketogenesis in vivo. Specialization for ketone body metabolism, along with mitochondria- and tissue-localization, emerged more recently and gradually. Mitochondrial HMGCS2 was the latest enzyme involved in ketone body metabolism to diverge from its cytoplasmic counterpart, and is conserved throughout amniota (including birds and humans) [41].
Signaling functions of βOHB
Although βOHB has long been known to be a circulating source of energy in the fasting state, its signaling functions have been recognized much more recently. Besides its predictable effects on cellular energy balance and metabolites, βOHB acts through at least two cell surface receptors and as an endogenous inhibitor of HDACs.
βOHB receptors
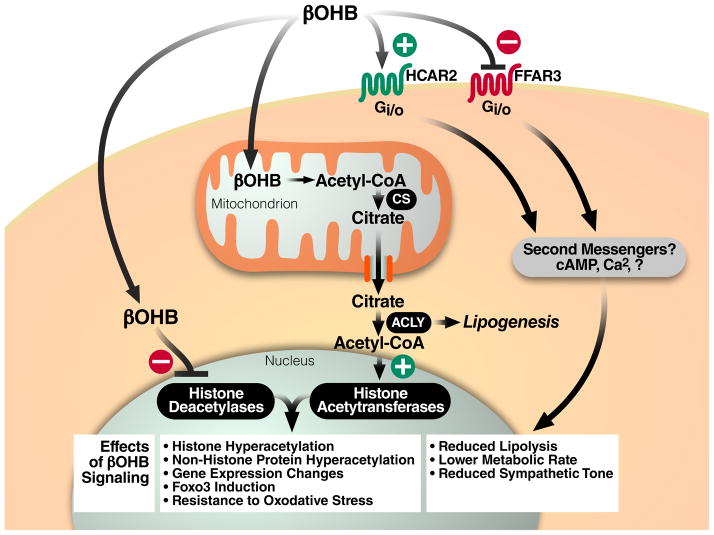
βOHB is a ligand for at least two G-protein coupled receptors (GPCRs) that bind short-chain fatty acids. HCAR2 (also known as PUMA-G or Gpr109), a Gi/o-coupled GPCR, first identified as a nicotinic acid receptor [42], was recently shown to bind and get activated by βOHB [43]. HCAR2 activation by βOHB reduces lipolysis in adipocytes, perhaps as a feedback mechanism to regulate availability of the fatty acid precursors of ketone body metabolism [43, 44]. βOHB also binds and antagonizes free fatty acid receptor 3 (FFAR3, also known as GPR41), another Gi/o-protein coupled receptor that is present in sympathetic ganglions, thereby suppressing sympathetic activity and, in turn, overall metabolic rate in mice [45]. Thus, through its actions on HCAR2 and FFAR3, βOHB may reduce lipolysis, reduce sympathetic tone, and lower metabolic rate (Figure 3) [43–45]. These receptors are part of a growing family of GPCRs, many with fatty acid ligands that have important roles in metabolism and metabolic disease [46, 47].
Figure 3. Cellular signaling mediated by βOHB.
βOHB is a ligand for at least two cell-surface G-protein-coupled receptors that modulate lipolysis, sympathetic tone, and metabolic rate. In addition, βOHB alters protein acetylation through at least two mechanisms: increasing the cellular pool of acetyl-CoA that is a substrate for histone acetyltransferases, and directly inhibiting the activity of class I histone deacetylases. Abbreviations: CS, citrate synthase. ACLY, ATP-citrate lyase.
βOHB binds to and inhibits class I HDACs
It was recently discovered that βOHB inhibits class I HDACs [48], a family of proteins that suppress gene expression by deacetylating lysine residues on histone and non-histone proteins (reviewed in [49–51]); histone hyperacetylation is generally associated with activation of gene expression. Although histones were the first known targets, many non-histone proteins are also subject to HDAC-mediated deacetylation including p53, c-Myc, MyoD, and others [52]. HDACs belong in four separate classes: Class I HDACs (e.g., HDACs 1, 2, 3 and 8) are short primarily nuclear proteins that consist of mainly the deacetylase domain and are usually found in large regulatory multi-protein complexes; Class IIa HDACs (e.g., HDAC4, 5, 7 and 9) are larger proteins with extensive regulatory domains in their N-terminus, that shuttle between the nucleus and cytoplasm; Class IIb HDACs (e.g., HDAC6 and 10) are primarily cytoplasmic proteins containing tandem deacetylase domains; Class III HDACs, the sirtuins, are a structurally distinct group of NAD-dependent deacylases; and Class IV, a single, poorly-understood representative (HDAC11) [49].
The short-chain fatty acid butyrate, differing structurally from βOHB by only a hydroxyl group, was the first HDAC inhibitor to be identified [53]. Since then, at least four major structurally distinct classes of HDAC inhibitors have been discovered [54]. Crystal structures of the human class I HDAC8 bound to several hydroxamic acid inhibitors [55, 56], as well as modeling of other inhibitors, show that a carbonic or hydroxamic acid group of an inhibitor is commonly bound to the catalytic zinc at the bottom of the hydrophobic active site channel of the HDAC [57]. The kinetics of butyrate suggest that it is a competitive inhibitor [58], supporting the notion that both the carboxylic acid groups of butyrate and βOHB chelate zinc in a similar manner, with the beta-hydroxyl group of βOHB sufficiently inconspicuous to fit within the deep hydrophobic channel. Notably, butyrate, which is aliphatic beyond the carbonic acid moiety, is more potent than βOHB; and acetoacetate, with a second carbonyl group instead of βOHB’s hydroxyl group, is much less potent than βOHB [58].
Exciting new data shows that βOHB inhibits HDACs 1, 3 and 4 (class I and IIa) in vitro with an IC50 of 2–5 mM. Treating cultured cells with βOHB induces dose-dependent histone hyperacetylation, particularly at histone H3 lysines 9 and 14. Interestingly, fasting also induces prominent histone hyperacetylation in many mouse tissues and especially the kidney. Along these lines, treating mice with βOHB via osmotic pump causes kidney histone hyperacetylation that is associated with specific changes in gene expression, including induction of forkhead box O3 (Foxo3a), the mammalian ortholog of the stress-responsive transcriptional factor DAF16 that regulates lifespan in worms [2]. Induction of Foxo3a appears to be a direct effect of HDAC inhibition, as HDAC1 and HDAC2 are found on its promoter, knockdown of both relieves the HDAC mediated Foxo3a repression, and βOHB causes hyperacetylation of histones at the Foxo3a promoter that results in increased Foxo3a expression [5].
βOHB indirectly promotes protein hyperacetylation
βOHB may also promote protein hyperacetylation more indirectly, by increasing the intracellular pools of acetyl-CoA (Figure 3). Metabolism of βOHB into acetyl-CoA should raise intracellular acetyl-CoA levels, providing additional substrate for both enzymatic and non-enzymatic protein acetylation thus driving the reaction equilibria towards acetylation. For example, CR, fasting and high-fat diets, all states associated with increased lipid utilization and therefore high acetyl-CoA production, cause increased mitochondrial protein acetylation, even though the HDACs that are inhibited by βOHB are not known to enter the mitochondria [35]. Mitochondrial acetylcarnitine is known to be a source of acetyl-CoA for histone acetylation [59]. Export of acetyl-CoA from the mitochondria is an active process primarily mediated by citrate synthase and ATP citrate lyase [60]. ATP citrate lyase is a key enzyme in fatty acid biosynthesis, but its role in facilitating acetyl-CoA export from mitochondria is also required for the increase in histone acetylation that occurs with growth factor stimulation [60]. An alternative pathway for acetyl-CoA export from mitochondria is via the enzymes carnitine acetyltransferase (CAT) and carnitine/acylcarnitine translocase [61]. Indeed, a muscle-specific knockout of CAT in mice compromises glucose tolerance and decreases metabolic flexibility [61], demonstrating the importance of intracellular acetyl-CoA transport to overall metabolic health.
Ketone bodies, fasting metabolism and the ketogenic diet
Energy-restricted metabolic states, such as CR or intermittent fasting (every other day), extend lifespan in animals [1]. All such states in vertebrates are necessarily associated with elevations in ketone bodies, whether consistent and modest as in CR or periodic and substantial as in intermittent fasting (see above). Surprisingly, the health benefits of intermittent fasting do not require overall reduced caloric intake. Mice fed every-other-day have increased longevity [62], and mice fed only during 8 hours at night are resistant to diet-induced obesity [63], both without altering overall calorie intake. With our growing understanding of the non-energy functions of βOHB, βOHB might be an intermediary of some of the benefits of energy-restricted states. As described below, many of the data on the metabolic effects of ketone bodies come from studies of ketogenic diets, particularly in rodents. Ketogenic diets in rodents are not a restricted-energy state, but phenocopy some of the biochemical characteristics of fasting, including several that are associated with longevity. In particular, ketogenic diets are associated with low insulin levels [64–68], reduced IGF signaling [69–71] and Foxo3 induction [5], AMP-activated protein kinase (AMPK) activation [71, 72], mTOR repression [71], and induction of antioxidant genes [5] (Table 1).
Table 1.
Comparison of longevity pathways regulated by ketogenic diets and CR
| Ketogenic Diet | Calorie Restriction | References | ||
|---|---|---|---|---|
| Glucose content of diet | ⬇⬇ | ⚊ | [140] | |
| Energy content of diet | ⚊ | ⬇ | [140] | |
| βOHB production | ⬆⬆ | ⬆ | [140] | |
| Insulin levels | ⬇⬇ | ⬇ | [2, 64–68] | |
| IGF signaling | ⬇ | ⬇ | [2, 69–71] | |
| AMPK activity | ⬆ | ⬆ | [2, 71, 72] | |
| mTOR activity | ⬇ | ⬇ | [2, 71] | |
| βOHB | Foxo3 | ⬆ | ⬆ | [5] |
| Protein acetylation | ⬆ | ⬆ | [5, 141] | |
| Stress resistance | ⬆ | ⬆ | [5, 84, 98–113] | |
| Longevity | ? | ⬆ | [1] |
Impact of ketogenic diet on energy homeostasis
Apart from inducing metabolic changes characteristics of fasting, ketogenic diets represent a high-fat, high-energy state and are also in some ways similar to non-ketogenic high-fat or Western diets. A ketogenic diet increases fasting leptin [67] and consistently causes hyperglycemia and insulin resistance, although basal insulin levels are lower [64–68]. It also promotes liver endoplasmic reticulum (ER) stress (based on the expression levels of genes involved in unfolded protein response), a phenotype associated with increased gluconeogenesis and insulin resistance in diabetic mice [68]. Similar to high-fat diets, there is strong induction of hepatic genes involved in fatty acid oxidation, including acyl coA dehydrogenases and trifunctional enzyme components [68, 72, 73].
Isolated studies have found ketogenic diets to be obesogenic in mice [65, 67], though the majority of studies have not [68, 69, 72–75]. Even among the same strain (C57BL/6), mice on a ketogenic diet have been reported to both gain weight [67] and lose weight [72], relative to controls, despite ingesting equal calories. This discrepancy may be due to differences in diet composition or genetic background. For example, in certain rodent genetic backgrounds, ketone bodies can suppress appetite through central effects in the hypothalamus (reviewed in [76]). The details of diet formulation are important as well. Two studies which found ketogenic diets to be obesogenic both used diets containing >20% calories from protein, similar to typical control diets [65, 67]. Meanwhile, the most popular ketogenic diet (Bioserv F3666), often found to be non-obesogenic, contains a very low 5% of calories from protein [68, 72–75] as well as low methionine content (0.22% vs. >0.4% for a typical control diet) [77]. Both protein restriction and methionine restriction extend lifespan in rodents [78, 79], and methionine restriction also reduces obesity from a high-fat diet [80].
However, the ketogenic diet is unusual in that it simultaneously activates “fasting” pathways despite being a high-energy state. For example, a high-fat, non-ketogenic diet induces not only fatty acid oxidation enzymes but also enzymes involved in fatty acid synthesis. Knockdown in liver of the fatty acid synthesis enzyme stearoyl-CoA desaturase-1 (SCD1), ameliorates the development of metabolic syndrome in SIRT3 knockout mice fed a high-fat diet [81]. In contrast, ketogenic diets suppress fatty acid synthesis enzymes including SCD1 [72, 73]. Instead, PPARα, a regulator of ketogenic genes, is strongly induced [73], as is one of its crucial downstream targets, FGF21 [68, 73, 74], resulting in increased transcription of ketogenic enzymes, such as HMGCS2 [68, 72, 73]. A ketogenic diet also increases expression of peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1α), a master regulator of mitochondrial function [74, 75], in mouse liver and brown adipose tissue, which may explain how the ketogenic diet promotes mitochondrial biogenesis in a mouse myopathy model [82]. Finally, one study also reported increased expression of SIRT1 [75], a mammalian homolog of the yeast Sir2 deacetylase that is increased during calorie restriction and regulates a variety of aging-related pathways (reviewed in [83]).
Mouse knockout models have shown that these “fasting” adaptations are driven by the actions of specific nutrient-regulated genes, including leptin, PPARα, and FGF21. For example, leptin-deficient ob/ob mice have a defective response to the ketogenic diet: they have elevated hepatic PPARα at baseline but do not increase hepatic FGF21 [73]. PPARα knockout mice on a ketogenic diet show reduced ketonemia, as well as fatty liver and lipemia, and suppressed hepatic fatty acid oxidation and ketogenic gene expression [27]. Similarly, FGF21 knockout mice on a ketogenic diet have less ketosis, higher levels of insulin and leptin, and more weight gain, along with reduced PGC1α and lipolytic gene expression, in adipocytes [28]. Interestingly, some of these genes respond differently to fasting and a ketogenic diet, for example hepatic FGF21 is induced by fasting but not by ketogenic diet in ob/ob mice [73], while PPARα-knockout mice induce FGF21 on a ketogenic diet but not fasting [27].
Ketogenic diet and longevity
Ketogenic diets alter other nutrient-sensitive pathways that are implicated in longevity. For example, a ketogenic diet is associated with high activity of AMPK in mouse muscle and liver [71, 72] and inhibition of the mTOR pathway including reduced phosphorylation of S6 kinase in rat liver and hippocampus [71], although the latter could be due in part to the low protein content of the ketogenic diet used. A ketogenic diet also lowers the serum ratio of IGF to insulin-like growth factor-binding protein 3 (IGFBP3) in mice [69, 70], that has been associated with metabolic syndrome and cancer, and reduces pAkt in rat liver and in mouse prostate tumor xenografts [70, 71]. Altogether, these crucial differences between a ketogenic and non-ketogenic high-fat diet may explain the beneficial metabolic effects of a ketogenic diet and have potentially important implications for longevity.
Benefits of ketogenic diet
Both obese mice and obese humans show improved metabolic measures when placed on a ketogenic diet. In contrast to the effects of a ketogenic diet on lean mice, obese or diabetic mice show improved glucose tolerance and insulin sensitivity [69, 73, 84]. A ketogenic diet can even reverse diabetic nephropathy in mice [84]. Nevertheless, ample caution must be exercised when extending laboratory rodent findings to humans. Ketogenic diets in adult humans should only be used under physician supervision, or in the context of a clinical trial. Still, such trials to date have had suggestive results; intermittent severe energy restriction, presumably ketogenic, is more effective than daily energy restriction at improving insulin sensitivity and promoting weight loss [85] and a recent clinical trial of a ketogenic diet found that obese, diabetic humans lost more weight with greater improvement in glucose control than a lower-calorie low-fat diet [86], a result echoing the findings of a recent meta-analysis [87].
Ketone bodies are neuroprotective and cytoprotective
Fasting has been used as an anticonvulsive therapy since ancient times, while the ketogenic diet has been in clinical use for over a century. It continues to be an effective therapy, particularly for certain childhood epilepsies that are resistant to anticonvulsant medications [88]. Ketogenic diets are clinically beneficial in mouse models of several common human neurodegenerative diseases, with promising early data from limited human clinical trials. In the 3xTgAD mouse model of Alzheimer’s disease, βOHB delivered via the diet in the form of a synthetic ester suppresses beta-amyloid pathology and improves learning and memory [89]. Two small clinical trials of C8 medium chain triglycerides improved cognitive function in patients with mild-moderate Alzheimer’s disease [90, 91]. βOHB infusion ameliorates the phenotype of drug-induced Parkinsonism in mice [92], and a preliminary uncontrolled study of ketogenic diet in humans with Parkinson’s disease showed improvement in disease severity scale [93]. Ketogenic diets improve motor performance and motor neuron number in a mouse model of amyotrophic lateral sclerosis [94] and are neuroprotective in mouse models of chronic epilepsy [95]. They also improve motor function and clinical measures in mouse models of genetic and drug-induced Huntington’s disease [96], a disease where aberrant histone hypoacetylation may be crucial to pathogenesis [97].
Neuroprotective actions of βOHB
βOHB appears to have broadly neuroprotective effects in these and other neurodegenerative disease models, but its mechanism of action has not been established. In vitro, βOHB protects cultured neurons from MPP (a chemical used to induce Parkinsonism in mice) and β-amyloid (Aβ42) induced toxicity (the peptide that accumulates in Alzheimer’s amyloid plaques) [98]. βOHB infusions or ketogenic diets protect against neuronal death in several animal models of brain injury. In rat models of traumatic brain injury, ketogenic diets reduce neuronal apoptosis, reduce brain edema, and improve sensorimotor and cognitive outcomes [99, 100]. Similarly, in rat models of stroke by either cerebral artery occlusion [101] or cardiac arrest [102], a ketogenic diet reduces neuronal loss and infarct size. In vitro, βOHB reduces apoptosis after hypoxia in rat hippocampal neuron cultures [103] and protects hippocampal cultures from a variety of insults including hypoglycemia, hypoxia and N-methyl-D-aspartate-induced excitotoxicity [104]. Both βOHB and acetoacetate also inhibit uptake of glutamate into synaptic vesicles by competing with chloride, an allosteric activator of vesicular glutamate transporters [105]. Acetoacetate is 10-fold more potent in this effect and reduces both glutamate release and seizure severity in a mouse model of epilepsy [105]. In vitro, βOHB enhances survival of cultured cortical neurons from exposure to hydrogen peroxide, both under no glucose (high cell death) and normal glucose (low cell death) conditions [84].
Resistance to cellular stressors by βOHB
One hallmark of longevity-enhancing pathways in model organisms is the induction of resistance to multiple forms of cellular stress [106]. In the roundworm Caenorhabditis elegans, for example, genetic mutations that confer enhanced longevity activate an array of cytoprotective pathways that are required for longevity extension [76]. Although a longevity effect for βOHB has not been established, the resistance provided to multiple cellular stressors by βOHB is reminiscent of longevity factors, and is not restricted to neurons. βOHB in Ringer’s solution reduces lung injury after hemorrhage and fluid resuscitation in rat models [107–110], as does βOHB alone [111, 112]. βOHB also reduces myocardial damage in a rat model of cardiac ischemia [113]. Administration of βOHB via osmotic pump enhances resistance of mouse kidney to oxidative stress from paraquat, with reduced accumulation of lipid peroxides and protein carbonylation. This resistance to oxidative stress may be due to induction of antioxidant genes, including Foxo3, metallothionein 2 (Mt2), superoxide dismutase 1 (Sod1) and catalase. Mt2 and Foxo3 in particular are both up-regulated by βOHB via its effect on HDAC inhibition and promoter hyperacetylation [5]. The full suite of genes regulated by βOHB via HDAC inhibition is not yet known, but HDAC inhibition provides a possible mechanism by which multiple stress-response pathways could be activated by βOHB.
HDACs in longevity and diseases of aging
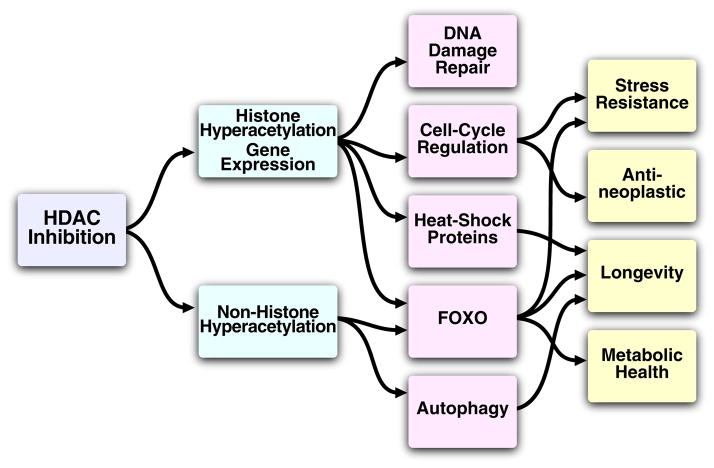
As discussed above, inhibition of HDACs by βOHB indicates that βOHB has specific regulatory effects in addition to its metabolic effects, particularly on HDACs and histone acetylation. Interestingly, reduced HDAC activity, either by genetic or pharmacologic means, also has beneficial metabolic and cytoprotective effects similar to βOHB. Moreover, HDACs regulate a variety of pathways implicated in longevity, including autophagy and IGF signaling (Figure 4), and modulation of HDAC activity regulates lifespan in model organisms (Box 1).
Figure 4. HDAC regulation of longevity pathways.
HDACs deacetylate both histone and non-histone proteins, regulating gene transcription and the post-translational function of proteins. HDACs regulate a variety of pathways implicated in longevity and age-related disease, and modulation of HDAC activity regulates lifespan in model organisms.
HDACs have a key role in regulating metabolic disease, and loss or inhibition of class I HDAC function appears to phenocopy some of the benefits of a ketogenic diet. HDAC3 regulates expression of gluconeogenic genes [114] and HDAC3 knockout mice have lower fasting glucose and insulin [115–117]. In fact, chronic treatment with butyrate keeps mice essentially metabolically normal on a high-fat diet, with lower glucose and insulin, better glucose tolerance, prevention of weight gain, and improved respiratory efficiency; butyrate also provides some of these benefits even to mice already obese from being fed a high-fat diet [118]. One mechanism for this may be up-regulation of PGC1α in liver, brown adipose tissue and muscle by butyrate [118], as seen in ketogenic diets.
Also reminiscent of βOHB, HDAC inhibitors are cytoprotective in animal models of tissue injury. Butyrate improves overall survival in a rat sepsis model [119], as well as reducing lung injury after lipopolysaccharide infusion in mice [120].
There is a growing literature on the importance of epigenetic regulation in learning and memory, and specifically mouse models of dementia. In the severely affected CK-p25 mouse (an inducible mouse model for p25 accumulation, the cleaved isoform of cyclin-dependent kinase 5 activator 1) of neurodegeneration, environmental enrichment that improves learning and memory is associated with increased histone H3 and H4 acetylation in the cortex and hippocampus [134]. Treatment with the HDAC inhibitor butyrate also improves learning and memory [134]. Age-related impairments in learning and memory in wild-type mice are associated with alterations in histone acetylation [135], and treatment with HDAC inhibitors improves memory performance in both young and aged mice [135, 136]. HDAC2 appears to be the crucial mediator of these effects, as overexpression of HDAC2, but not HDAC1, impairs learning and memory in wild-type mice [136]. Conversely, HDAC2 knockout mice show improved memory formation, which is not further improved by HDAC inhibitors [136]. HDAC2 expression is increased in the brains of two mouse dementia models as well as the brains of humans with Alzheimer’s disease, and knockdown of HDAC2 improves memory in the CK-p25 dementia mouse model [137].
Although HDAC2 seems to have a memory-impairing role, HDAC1 – another class I HDAC – has been reported to be neuroprotective [138]. HDAC1 activity is important for the repair of double-stranded DNA breaks in neurons, and its own deacetylase activity is enhanced by SIRT1 via deacetylation [139]. This provides a fascinating example of how multiple metabolically-responsive pathways (e.g. SIRT1 and βOHB) might intersect to provide overlapping epigenetic regulation: while βOHB inhibition of HDAC2 may be broadly beneficial, the potentially detrimental inhibition of HDAC1 by βOHB is offset by SIRT1 activation. Such cross-talk may be common, where fasting-activated sirtuins provide tissue- or subcellular-specific fine-tuning of the broad effects of βOHB. Alternatively, different target specificities of class I and class III HDACs could provide ample opportunity for coordinated regulation or targets, even perhaps via different lysines on the same protein. It remains to be determined if inhibition of HDACs by βOHB has similar effects on learning and memory in rodent models as chemical HDAC inhibition or genetic manipulation, and precisely how HDAC inhibition by βOHB intersects with other fasting-related mechanisms of epigenetic regulation.
Concluding Remarks and Future Perspectives
Ketone bodies are emerging as crucial regulators of metabolic health and longevity, via their ability to regulate HDAC activity and thereby epigenetic gene regulation. Ketogenic diets provide a partial phenocopy of CR, through their effects on insulin, IGF, Foxo3, fatty acid metabolism, AMPK and mTOR. The finding that βOHB is an inhibitor of HDACs, together with the coincidence of biological effects of ketone bodies and HDAC inhibition, suggests the fascinating possibility that βOHB could be an endogenous avenue to attain some of the benefits of lifespan extension seen with HDAC inhibition in model organisms. It will be of great interest to define the molecular targets of HDAC inhibition by βOHB in specific tissues and metabolic states, investigate whether βOHB regulates HDAC-targeted pathways like autophagy, and determine if these effects culminate in enhancement of longevity by βOHB in mammals.
Box 2. Outstanding questions.
What are the molecular targets of HDAC inhibition by βOHB in specific tissues and metabolic states?
Does βOHB regulate HDAC-targeted pathways like autophagy?
Do pathways downstream of βOHB actions increase longevity in mammals?
Are mammalian sirtuins downstream targets of βOHB and HDACs?
If fasting or ketogenic conditions promote inhibition of class I HDACs (via βOHB) but activation of class III HDACs (sirtuins), how are these potentially opposing activities coordinated?
Is there an acetylation code that may trigger the metabolic reprogramming of dietary restriction?
Highlights.
Ketone bodies have signaling functions as well as being a mobile source of cellular energy
Ketone bodies inhibit histone deacetylases and control gene transcription
Histone deacetylase function is implicated in the regulation of aging
Ketone bodies may link environmental cues such as diet to the regulation of aging
Acknowledgments
We thank John Carroll for artistic assistance. EV is supported by funds from NIH/NIDDK and the Gladstone Institutes. JCN is supported by funds from the Larry L. Hillblom Foundation, the John A. Hartford Foundation, the Glenn Foundation for Medical Research, and an NIH/NIA T32 training grant.
GLOSSARY
- β-hydroxybutyrate (βOHB)
is a molecule that can be used as an energy source by the brain when blood glucose is low. It is one of three metabolically related molecules known collectively as ketone bodies but is itself technically a carboxylic acid. It can also be used for the synthesis of biodegradable plastics, such as poly(3-hydroxybutyrate)
- Calorie restriction (CR)
is defined as reduced calorie intake. CR without malnutrition slows down the aging process, resulting in an increased lifespan in a variety of species, including yeast, flies, and rodents
- Ketone bodies
refers to three distinct molecule, acetone, acetoacetic acid, and βOHB, by-products of fatty acids oxidation in the liver under fasting conditions
- Ketogenic diet
a diet high in fat, low in carbohydrate and with adequate but often variable amounts of protein. Ketogenic diet has been used extensively to treat epilepsy in children. Due to its low amount of carbohydrates, the body switches to fatty acid oxidation as energy source that also results in the formation of ketone bodies. Elevated levels of ketone bodies in the blood, a state known as ketosis, has been shown to lead to a reduction in the frequency of epileptic seizures
- Histone deacetylases (HDACs)
refers to a class of enzyme that removes acetyl groups form lysine residues residing on histones as well as non-histone proteins often resulting in transcriptional repression
- Histone deacetylase inhibitors (HDIs)
a group of compounds that inhibit the action of histone deacetylases. Some common HDAC inhibitors are valproic acid, sodium butyrate and trichostatin A. HDIs are being investigated as possible treatments for cancers and inflammatory diseases
- Rapamycin
is an immunosuppressant drug and inhibitor of mTOR, the first compound found to extend lifespan in healthy mammals
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fontana L. Excessive adiposity, calorie restriction, and aging. Jama. 2006;295:1577–1578. doi: 10.1001/jama.295.13.1577. [DOI] [PubMed] [Google Scholar]
- 2.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 3.Bjedov I, Partridge L. A longer and healthier life with TOR down-regulation: genetics and drugs. Biochem Soc Trans. 2011;39:460–465. doi: 10.1042/BST0390460. [DOI] [PubMed] [Google Scholar]
- 4.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimazu T, et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katada S, et al. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Stein LR, Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab. 2012;23:420–428. doi: 10.1016/j.tem.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg JM, et al. Biochemistry. W.H. Freeman; 2012. [Google Scholar]
- 10.Cahill GF, Jr, et al. Hormone-fuel interrelationships during fasting. J Clin Invest. 1966;45:1751–1769. doi: 10.1172/JCI105481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson AM, Williamson DH. Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiol Rev. 1980;60:143–187. doi: 10.1152/physrev.1980.60.1.143. [DOI] [PubMed] [Google Scholar]
- 12.Cahill GF., Jr Fuel metabolism in starvation. Annu Rev Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 13.Koeslag JH, et al. Post-exercise ketosis. J Physiol. 1980;301:79–90. doi: 10.1113/jphysiol.1980.sp013190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim do Y, Rho JM. The ketogenic diet and epilepsy. Curr Opin Clin Nutr Metab Care. 2008;11:113–120. doi: 10.1097/MCO.0b013e3282f44c06. [DOI] [PubMed] [Google Scholar]
- 15.London ED, et al. Effects of fasting on ketone body concentrations in healthy men of different ages. J Gerontol. 1986;41:599–604. doi: 10.1093/geronj/41.5.599. [DOI] [PubMed] [Google Scholar]
- 16.Freemantle E, et al. Metabolic response to a ketogenic breakfast in the healthy elderly. J Nutr Health Aging. 2009;13:293–298. doi: 10.1007/s12603-009-0026-9. [DOI] [PubMed] [Google Scholar]
- 17.Thumelin S, et al. Developmental changes in mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene expression in rat liver, intestine and kidney. Biochem J. 1993;292 (Pt 2):493–496. doi: 10.1042/bj2920493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, et al. Proteomics analysis reveals diabetic kidney as a ketogenic organ in type 2 diabetes. Am J Physiol Endocrinol Metab. 2011;300:E287–295. doi: 10.1152/ajpendo.00308.2010. [DOI] [PubMed] [Google Scholar]
- 19.Fink G, et al. Pseudoketogenesis in the perfused rat heart. J Biol Chem. 1988;263:18036–18042. [PubMed] [Google Scholar]
- 20.Weidemann MJ, Krebs HA. The fuel of respiration of rat kidney cortex. Biochem J. 1969;112:149–166. doi: 10.1042/bj1120149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukao T, et al. Enzymes of ketone body utilization in human tissues: protein and messenger RNA levels of succinyl-coenzyme A (CoA):3-ketoacid CoA transferase and mitochondrial and cytosolic acetoacetyl-CoA thiolases. Pediatric research. 1997;42:498–502. doi: 10.1203/00006450-199710000-00013. [DOI] [PubMed] [Google Scholar]
- 22.Fukao T, et al. Pathways and control of ketone body metabolism: on the fringe of lipid biochemistry. Prostaglandins Leukot Essent Fatty Acids. 2004;70:243–251. doi: 10.1016/j.plefa.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Hegardt FG. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem J. 1999;338 (Pt 3):569–582. [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfrum C, et al. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–1032. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- 25.Wolfrum C, et al. Insulin regulates the activity of forkhead transcription factor Hnf-3beta/Foxa-2 by Akt-mediated phosphorylation and nuclear/cytosolic localization. Proc Natl Acad Sci U S A. 2003;100:11624–11629. doi: 10.1073/pnas.1931483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Meyenn F, et al. Glucagon-induced acetylation of Foxa2 regulates hepatic lipid metabolism. Cell Metab. 2013;17:436–447. doi: 10.1016/j.cmet.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Badman MK, et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Badman MK, et al. Fibroblast growth factor 21-deficient mice demonstrate impaired adaptation to ketosis. Endocrinology. 2009;150:4931–4940. doi: 10.1210/en.2009-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengupta S, et al. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 30.Shimazu T, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebert AS, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rardin MJ, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quant PA, et al. Glucagon activates mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase in vivo by decreasing the extent of succinylation of the enzyme. Eur J Biochem. 1990;187:169–174. doi: 10.1111/j.1432-1033.1990.tb15291.x. [DOI] [PubMed] [Google Scholar]
- 34.Quant PA, et al. Treatment of rats with glucagon or mannoheptulose increases mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase activity and decreases succinyl-CoA content in liver. Biochem J. 1989;262:159–164. doi: 10.1042/bj2620159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He W, et al. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab. 2012;23:467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Turko IV, et al. Diabetes-associated nitration of tyrosine and inactivation of succinyl-CoA:3-oxoacid CoA-transferase. Am J Physiol Heart Circ Physiol. 2001;281:H2289–2294. doi: 10.1152/ajpheart.2001.281.6.H2289. [DOI] [PubMed] [Google Scholar]
- 37.Pellerin L, et al. Cellular and subcellular distribution of monocarboxylate transporters in cultured brain cells and in the adult brain. J Neurosci Res. 2005;79:55–64. doi: 10.1002/jnr.20307. [DOI] [PubMed] [Google Scholar]
- 38.Balietti M, et al. Ketogenic diets: an historical antiepileptic therapy with promising potentialities for the aging brain. Ageing Res Rev. 2010;9:273–279. doi: 10.1016/j.arr.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Hugo SE, et al. A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 2012;26:282–293. doi: 10.1101/gad.180968.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hankermeyer CR, Tjeerdema RS. Polyhydroxybutyrate: plastic made and degraded by microorganisms. Rev Environ Contam Toxicol. 1999;159:1–24. doi: 10.1007/978-1-4612-1496-0_1. [DOI] [PubMed] [Google Scholar]
- 41.Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tunaru S, et al. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9:352–355. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- 43.Taggart AK, et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem. 2005;280:26649–26652. doi: 10.1074/jbc.C500213200. [DOI] [PubMed] [Google Scholar]
- 44.Offermanns S, et al. International Union of Basic and Clinical Pharmacology. LXXXII: Nomenclature and Classification of Hydroxy-carboxylic Acid Receptors (GPR81, GPR109A, and GPR109B) Pharmacol Rev. 2011;63:269–290. doi: 10.1124/pr.110.003301. [DOI] [PubMed] [Google Scholar]
- 45.Kimura I, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc Natl Acad Sci U S A. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blad CC, et al. G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat Rev Drug Discov. 2012;11:603–619. doi: 10.1038/nrd3777. [DOI] [PubMed] [Google Scholar]
- 47.Layden BT, et al. Short chain fatty acids and their receptors: new metabolic targets. Transl Res. 2013;161:131–140. doi: 10.1016/j.trsl.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Gregoretti IV, et al. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mihaylova MM, Shaw RJ. Metabolic reprogramming by class I and II histone deacetylases. Trends Endocrinol Metab. 2013;24:48–57. doi: 10.1016/j.tem.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.New M, et al. HDAC inhibitor-based therapies: can we interpret the code? Mol Oncol. 2012;6:637–656. doi: 10.1016/j.molonc.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glozak MA, et al. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Cousens LS, et al. Different accessibilities in chromatin to histone acetylase. J Biol Chem. 1979;254:1716–1723. [PubMed] [Google Scholar]
- 54.Bolden JE, et al. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 55.Vannini A, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somoza JR, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 57.Wang DF, et al. Toward selective histone deacetylase inhibitor design: homology modeling, docking studies, and molecular dynamics simulations of human class I histone deacetylases. J Med Chem. 2005;48:6936–6947. doi: 10.1021/jm0505011. [DOI] [PubMed] [Google Scholar]
- 58.Sekhavat A, et al. Competitive inhibition of histone deacetylase activity by trichostatin A and butyrate. Biochem Cell Biol. 2007;85:751–758. doi: 10.1139/o07-145. [DOI] [PubMed] [Google Scholar]
- 59.Madiraju P, et al. Mitochondrial acetylcarnitine provides acetyl groups for nuclear histone acetylation. Epigenetics: official journal of the DNA Methylation Society. 2009;4:399–403. doi: 10.4161/epi.4.6.9767. [DOI] [PubMed] [Google Scholar]
- 60.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muoio DM, et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012;15:764–777. doi: 10.1016/j.cmet.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anson RM, et al. The diet restriction paradigm: a brief review of the effects of every-other-day feeding. Age. 2005;27:17–25. doi: 10.1007/s11357-005-3286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bainbridge HW. The reduced sensitivity to insulin of rats and mice fed on a carbohydrate-free, excess-fat diet. J Physiol. 1925;60:293–300. doi: 10.1113/jphysiol.1925.sp002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burcelin R, et al. Heterogeneous metabolic adaptation of C57BL/6J mice to high-fat diet. Am J Physiol Endocrinol Metab. 2002;282:E834–842. doi: 10.1152/ajpendo.00332.2001. [DOI] [PubMed] [Google Scholar]
- 66.Kinzig KP, et al. Insulin sensitivity and glucose tolerance are altered by maintenance on a ketogenic diet. Endocrinology. 2010;151:3105–3114. doi: 10.1210/en.2010-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borghjid S, Feinman RD. Response of C57Bl/6 mice to a carbohydrate-free diet. Nutr Metab (Lond) 2012;9:69. doi: 10.1186/1743-7075-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garbow JR, et al. Hepatic steatosis, inflammation, and ER stress in mice maintained long term on a very low-carbohydrate ketogenic diet. Am J Physiol Gastrointest Liver Physiol. 2011;300:G956–967. doi: 10.1152/ajpgi.00539.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freedland SJ, et al. Carbohydrate restriction, prostate cancer growth, and the insulin-like growth factor axis. Prostate. 2008;68:11–19. doi: 10.1002/pros.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mavropoulos JC, et al. The effects of varying dietary carbohydrate and fat content on survival in a murine LNCaP prostate cancer xenograft model. Cancer Prev Res (Phila) 2009;2:557–565. doi: 10.1158/1940-6207.CAPR-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McDaniel SS, et al. The ketogenic diet inhibits the mammalian target of rapamycin (mTOR) pathway. Epilepsia. 2011;52:e7–11. doi: 10.1111/j.1528-1167.2011.02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kennedy AR, et al. A high-fat, ketogenic diet induces a unique metabolic state in mice. Am J Physiol Endocrinol Metab. 2007;292:E1724–1739. doi: 10.1152/ajpendo.00717.2006. [DOI] [PubMed] [Google Scholar]
- 73.Badman MK, et al. A very low carbohydrate ketogenic diet improves glucose tolerance in ob/ob mice independently of weight loss. Am J Physiol Endocrinol Metab. 2009;297:E1197–1204. doi: 10.1152/ajpendo.00357.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jornayvaz FR, et al. A high-fat, ketogenic diet causes hepatic insulin resistance in mice, despite increasing energy expenditure and preventing weight gain. Am J Physiol Endocrinol Metab. 2010;299:E808–815. doi: 10.1152/ajpendo.00361.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Srivastava S, et al. A ketogenic diet increases brown adipose tissue mitochondrial proteins and UCP1 levels in mice. IUBMB Life. 2013;65:58–66. doi: 10.1002/iub.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Laeger T, et al. Role of beta-hydroxybutyric acid in the central regulation of energy balance. Appetite. 2010;54:450–455. doi: 10.1016/j.appet.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 77.Schugar RC, Crawford PA. Low-carbohydrate ketogenic diets, glucose homeostasis, and nonalcoholic fatty liver disease. Curr Opin Clin Nutr Metab Care. 2012;15:374–380. doi: 10.1097/MCO.0b013e3283547157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Orentreich N, et al. Low methionine ingestion by rats extends life span. J Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 79.Pamplona R, Barja G. Mitochondrial oxidative stress, aging and caloric restriction: the protein and methionine connection. Biochim Biophys Acta. 2006;1757:496–508. doi: 10.1016/j.bbabio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 80.Ables GP, et al. Methionine-restricted C57BL/6J mice are resistant to diet-induced obesity and insulin resistance but have low bone density. PLoS One. 2012;7:e51357. doi: 10.1371/journal.pone.0051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirschey MD, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahola-Erkkila S, et al. Ketogenic diet slows down mitochondrial myopathy progression in mice. Hum Mol Genet. 2010;19:1974–1984. doi: 10.1093/hmg/ddq076. [DOI] [PubMed] [Google Scholar]
- 83.Houtkooper RH, et al. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poplawski MM, et al. Reversal of diabetic nephropathy by a ketogenic diet. PLoS One. 2011;6:e18604. doi: 10.1371/journal.pone.0018604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harvie M, et al. The effect of intermittent energy and carbohydrate restriction v. daily energy restriction on weight loss and metabolic disease risk markers in overweight women. The British journal of nutrition. 2013:1–14. doi: 10.1017/S0007114513000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Westman EC, et al. The effect of a low-carbohydrate, ketogenic diet versus a low-glycemic index diet on glycemic control in type 2 diabetes mellitus. Nutr Metab (Lond) 2008;5:36. doi: 10.1186/1743-7075-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nordmann AJ, et al. Effects of low-carbohydrate vs low-fat diets on weight loss and cardiovascular risk factors: a meta-analysis of randomized controlled trials. Arch Intern Med. 2006;166:285–293. doi: 10.1001/archinte.166.3.285. [DOI] [PubMed] [Google Scholar]
- 88.Hartman AL, Vining EP. Clinical aspects of the ketogenic diet. Epilepsia. 2007;48:31–42. doi: 10.1111/j.1528-1167.2007.00914.x. [DOI] [PubMed] [Google Scholar]
- 89.Kashiwaya Y, et al. A ketone ester diet exhibits anxiolytic and cognition-sparing properties, and lessens amyloid and tau pathologies in a mouse model of Alzheimer’s disease. Neurobiol Aging. 2013;34:1530–1539. doi: 10.1016/j.neurobiolaging.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henderson ST, et al. Study of the ketogenic agent AC-1202 in mild to moderate Alzheimer’s disease: a randomized, double-blind, placebo-controlled, multicenter trial. Nutr Metab (Lond) 2009;6:31. doi: 10.1186/1743-7075-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reger MA, et al. Effects of beta-hydroxybutyrate on cognition in memory-impaired adults. Neurobiol Aging. 2004;25:311–314. doi: 10.1016/S0197-4580(03)00087-3. [DOI] [PubMed] [Google Scholar]
- 92.Tieu K, et al. D-beta-hydroxybutyrate rescues mitochondrial respiration and mitigates features of Parkinson disease. J Clin Invest. 2003;112:892–901. doi: 10.1172/JCI18797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vanitallie TB, et al. Treatment of Parkinson disease with diet-induced hyperketonemia: a feasibility study. Neurology. 2005;64:728–730. doi: 10.1212/01.WNL.0000152046.11390.45. [DOI] [PubMed] [Google Scholar]
- 94.Zhao Z, et al. A ketogenic diet as a potential novel therapeutic intervention in amyotrophic lateral sclerosis. BMC Neurosci. 2006;7:29. doi: 10.1186/1471-2202-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noh HS, et al. Neuroprotective effects of the ketogenic diet. Epilepsia. 2008;49(Suppl 8):120–123. doi: 10.1111/j.1528-1167.2008.01855.x. [DOI] [PubMed] [Google Scholar]
- 96.Lim S, et al. D-beta-hydroxybutyrate is protective in mouse models of Huntington’s disease. PLoS One. 2011;6:e24620. doi: 10.1371/journal.pone.0024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sadri-Vakili G, Cha JH. Mechanisms of disease: Histone modifications in Huntington’s disease. Nat Clin Pract Neurol. 2006;2:330–338. doi: 10.1038/ncpneuro0199. [DOI] [PubMed] [Google Scholar]
- 98.Kashiwaya Y, et al. D-beta-hydroxybutyrate protects neurons in models of Alzheimer’s and Parkinson’s disease. Proc Natl Acad Sci U S A. 2000;97:5440–5444. doi: 10.1073/pnas.97.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu ZG, et al. Ketogenic diet reduces cytochrome c release and cellular apoptosis following traumatic brain injury in juvenile rats. Ann Clin Lab Sci. 2009;39:76–83. [PubMed] [Google Scholar]
- 100.Appelberg KS, et al. The effects of a ketogenic diet on behavioral outcome after controlled cortical impact injury in the juvenile and adult rat. J Neurotrauma. 2009;26:497–506. doi: 10.1089/neu.2008.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Puchowicz MA, et al. Neuroprotection in diet-induced ketotic rat brain after focal ischemia. J Cereb Blood Flow Metab. 2008;28:1907–1916. doi: 10.1038/jcbfm.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tai KK, et al. Ketogenic diet prevents cardiac arrest-induced cerebral ischemic neurodegeneration. J Neural Transm. 2008;115:1011–1017. doi: 10.1007/s00702-008-0050-7. [DOI] [PubMed] [Google Scholar]
- 103.Masuda R, et al. D-beta-hydroxybutyrate is neuroprotective against hypoxia in serum-free hippocampal primary cultures. J Neurosci Res. 2005;80:501–509. doi: 10.1002/jnr.20464. [DOI] [PubMed] [Google Scholar]
- 104.Samoilova M, et al. Chronic in vitro ketosis is neuroprotective but not anticonvulsant. J Neurochem. 2010;113:826–835. doi: 10.1111/j.1471-4159.2010.06645.x. [DOI] [PubMed] [Google Scholar]
- 105.Juge N, et al. Metabolic control of vesicular glutamate transport and release. Neuron. 2010;68:99–112. doi: 10.1016/j.neuron.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller RA. Cell stress and aging: new emphasis on multiplex resistance mechanisms. The journals of gerontology Series A, Biological sciences and medical sciences. 2009;64:179–182. doi: 10.1093/gerona/gln072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alam HB, et al. Resuscitation-induced pulmonary apoptosis and intracellular adhesion molecule-1 expression in rats are attenuated by the use of Ketone Ringer’s solution. J Am Coll Surg. 2001;193:255–263. doi: 10.1016/s1072-7515(01)01004-3. [DOI] [PubMed] [Google Scholar]
- 108.Koustova E, et al. Ketone and pyruvate Ringer’s solutions decrease pulmonary apoptosis in a rat model of severe hemorrhagic shock and resuscitation. Surgery. 2003;134:267–274. doi: 10.1067/msy.2003.245. [DOI] [PubMed] [Google Scholar]
- 109.Ayuste EC, et al. Hepatic and pulmonary apoptosis after hemorrhagic shock in swine can be reduced through modifications of conventional Ringer’s solution. J Trauma. 2006;60:52–63. doi: 10.1097/01.ta.0000200156.05397.0b. [DOI] [PubMed] [Google Scholar]
- 110.Jaskille A, et al. Hepatic apoptosis after hemorrhagic shock in rats can be reduced through modifications of conventional Ringer’s solution. J Am Coll Surg. 2006;202:25–35. doi: 10.1016/j.jamcollsurg.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 111.Klein AH, et al. Small-volume d-beta-hydroxybutyrate solution infusion increases survivability of lethal hemorrhagic shock in rats. Shock. 2010;34:565–572. doi: 10.1097/SHK.0b013e3181e15063. [DOI] [PubMed] [Google Scholar]
- 112.Mulier KE, et al. Treatment with beta-hydroxybutyrate and melatonin is associated with improved survival in a porcine model of hemorrhagic shock. Resuscitation. 2012;83:253–258. doi: 10.1016/j.resuscitation.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 113.Zou Z, et al. dl-3-Hydroxybutyrate administration prevents myocardial damage after coronary occlusion in rat hearts. Am J Physiol Heart Circ Physiol. 2002;283:H1968–1974. doi: 10.1152/ajpheart.00250.2002. [DOI] [PubMed] [Google Scholar]
- 114.Mihaylova MM, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Knutson SK, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. Embo J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fajas L, et al. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell. 2002;3:903–910. doi: 10.1016/s1534-5807(02)00360-x. [DOI] [PubMed] [Google Scholar]
- 117.Bhaskara S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gao Z, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang LT, et al. Sodium butyrate prevents lethality of severe sepsis in rats. Shock. 2007;27:672–677. doi: 10.1097/SHK.0b013e31802e3f4c. [DOI] [PubMed] [Google Scholar]
- 120.Ni YF, et al. Histone deacetylase inhibitor, butyrate, attenuates lipopolysaccharide-induced acute lung injury in mice. Respir Res. 2010;11:33. doi: 10.1186/1465-9921-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim S, et al. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yi C, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 124.Rogina B, et al. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- 125.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mannervik M, Levine M. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc Natl Acad Sci U S A. 1999;96:6797–6801. doi: 10.1073/pnas.96.12.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhao Y, et al. Lifespan extension and elevated hsp gene expression in Drosophila caused by histone deacetylase inhibitors. J Exp Biol. 2005;208:697–705. doi: 10.1242/jeb.01439. [DOI] [PubMed] [Google Scholar]
- 128.Kang HL, et al. Life extension in Drosophila by feeding a drug. Proc Natl Acad Sci U S A. 2002;99:838–843. doi: 10.1073/pnas.022631999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Evason K, et al. Valproic acid extends Caenorhabditis elegans lifespan. Aging Cell. 2008;7:305–317. doi: 10.1111/j.1474-9726.2008.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. Embo J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zimmermann S, et al. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]
- 132.Wilting RH, et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. Embo J. 2010;29:2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dovey OM, et al. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci U S A. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fischer A, et al. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 135.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 136.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Graff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim D, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dobbin MM, et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci. 2013;16:1008–1015. doi: 10.1038/nn.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kossoff EH, Hartman AL. Ketogenic diets: new advances for metabolism-based therapies. Curr Opin Neurol. 2012;25:173–178. doi: 10.1097/WCO.0b013e3283515e4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schwer B, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]