Summary
As part of our screening for anti-HIV agents from marine invertebrates, the MeOH extract of Didemnum molle was tested and showed moderate in vitro anti-HIV activity. Bioassay-guided fractionation of a large scale extract allowed the identification of two new cyclopeptides, mollamides E and F (1 and 2), and one new tris-phenethyl urea, molleurea A (3). The absolute configurations were established using the advanced Marfey’s method. The three compounds were evaluated for anti-HIV activity in both an HIV integrase inhibition assay and a cytoprotective cell-based assay. Compound 2 was active in both assays with IC50 values of 39 and 78 µM, respectively. Compound 3 was only active in the cytoprotective cell-based assay with an IC50 value of 60 µM.
Human immunodeficiency virus (HIV) was first established as the causative agent of acquired immunodeficiency syndrome (AIDS) over 20 years ago.1, 2 In the past decade, clinical treatments of HIV-infected patients with inhibitors of reverse transcriptase and protease have led to significant improvements in reducing the viral load and the progression of AIDS. However, these treatments can become ineffective due to rapid mutations of the virus. HIV-1 integrase is an enzyme that is critical for integration of the HIV genome into the host genome.3 This process is unique to the virus and is absent in the host, and therefore presents an attractive target for the development of single and/or combination anti-HIV therapy.
As part of an International Cooperative Biodiversity Group (ICBG) involving a collaboration of institutions in Papua New Guinea and the United States, we have established a natural product-based antiviral drug discovery program specifically targeting HIV. Organisms of interest have included plants, marine invertebrates, and endophytic fungi. During an initial screen, a MeOH extract prepared from the ascidian Didemnum molle collected from Papua New Guinea was shown to inhibit HIV-1 replication at 100 µg/mL, while being devoid of cytoxicity at this concentration. Didemnum molle is known for producing cyclic hexa-, hepta-, and octa-peptides characterized by an alternating sequence of thiazole, thiazoline or oxazoline heterocycles and hydrophobic amino acids, many of which show remarkably high levels of cytotoxicity.4 Researchers have speculated that these peptides might be biosynthesized by symbiotic prochlorophytes. Recently, Schmidt et al. localized the genes responsible for the biosynthesis of patellamide to the symbiont Prochloron didemni.5 They also found that Prochloron spp. generate a diverse library of patellamides and related cyclic peptides, and they used this information to engineer the production of novel related peptides in E.coli.6
In the process of bioassay-guided fractionation of the extract of D. molle, we identified three compounds. Interestingly, two of the compounds (1 and 2) corresponded to peptides that were initially identified by genome sequencing of Prochloron spp. and their predicted products detected by LC-MS analysis of a single ascidian colony (1)7a or of E. coli heterologously expressing the corresponding biosynthetic pathway (2).7b However, this report marks the first time these compounds have been fully characterized as isolated natural products. These peptides were shown to be ribosomally produced with all residues presumably having the natural l configuration. Using the advanced Marfey’s method we found, however, that the thiazoline and phenylalanine residues in both 1 and 2 were present predominantly in a d configuration. Herein we report the isolation and structure elucidation of compounds 1–3, as well as their HIV-1 inhibitory activity.
Results and Discussion
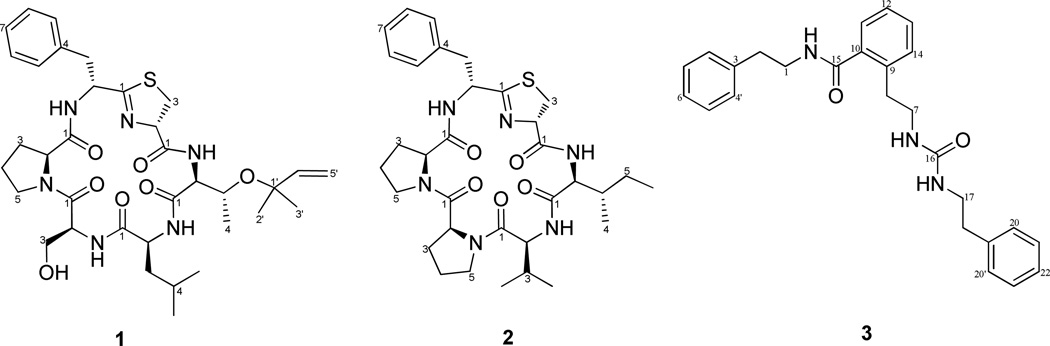
The molecular formula of C35H50N6O7S for compound 1 was provided by HRESIMS and supported by NMR data (Table 1). The 1H and 13C NMR spectra indicated peptidic metabolites due to the presence of four NH doublets (δH 6.62 – 9.28), six Hα multiplets (δH 3.97 – 5.40), and six putative amide carbonyl 13C signals (δC 168 – 175). Detailed analysis of the 1D and 2D NMR data of 1 established six amino acid residues: phenylalanine, proline, serine, leucine, threonine and cysteine present as a thiazoline (Tzn). In addition, a reversed-isoprene functionality attached to the Thr residue was apparent (Table 1).
Table 1.
NMR Data for 1 in DMSO-d6 (1H 500MHz, 13C 125MHz)
| position | δC | δH(J in Hz) | HMBC | ROESY |
|---|---|---|---|---|
| Pro | ||||
| 1 | 169.6, C | |||
| 2 | 60.0, CH | 4.67, d (7.0) | 1, 3, 4, 5, 1Ser | 3, 2Ser, 3Phe |
| 3 | 29.0, CH2 | 1.60, m | 1, 2, 4, 5 | 2, 4, 5 |
| 2.17, m | ||||
| 4 | 20.7, CH2 | 0.48, m | 2, 3 | 3, 5 |
| 1.49, m | ||||
| 5 | 45.7, CH2 | 2.93, br t (10.0) | 4, 3, 1Ser | 3, 4 |
| 3.22, m | ||||
| Ser | ||||
| 1 | 168.6, C | |||
| 2 | 55.2, CH | 4.08, m | 1, 3, 1Leu | 3, 2Pro, NH |
| 3 | 60.3, CH2 | 3.58, m | 1, 2 | 2, NH |
| NH | 9.07, br s | 3, 1Leu | 2, 3, 2Leu | |
| Leu | ||||
| 1 | 172.7, C | |||
| 2 | 50.3, CH | 4.34, dd (7.5) | 1, 3, 4, 1Thr-ether | 3, 5, 6, NH, NHSer |
| 3 | 42.8, CH2 | 1.44, m | 1, 2, 5, 6 | 2, 4, 5, 6 |
| 4 | 23.8, CH | 1.72, m | 2, 3, 5, 6 | 3, 5, 6 |
| 5 | 22.1, CH3 | 0.96, d (6.5) | 3, 4, 6 | 2, 3, 4 |
| 6 | 22.7, CH3 | 0.92, d (6.5) | 3, 4, 5 | 2, 3, 4 |
| NH | 6.62, d (7.5) | 1, 2, 1Thr-ether | 2, NHThr-ether | |
| Thr-ether | ||||
| 1 | 169.5, C | |||
| 2 | 60.7, CH | 3.97, dd (8.5, 2.5) | 1, 3, 4, 1Tzn | 4, NH |
| 3 | 66.7, CH | 4.05, m | 1′ | 2′, 3′, 4, NH |
| 4 | 20.9, CH3 | 1.17, d (6.0) | 2, 3 | 2, 3 |
| 1′ | 75.5, C | |||
| 2′ | 26.2, CH3 | 1.23, s | 1′, 3′, 4′ | 3, 4′, 5′, NH |
| 3′ | 26.2, CH3 | 1.23, s | l′, 2′, 4′ | 3, 4′, 5′, NH |
| 4′ | 143.7, CH | 5.85, dd (18.0, 11.0) | l′, 2′, 3′ | 2′, 3′, 5′ |
| 5′ | 113.6, CH2 | 5.05, d (11.0) | l′, 4′ | 2′, 3′4′ |
| 5.17, d (18.0) | ||||
| NH | 7.37, d (8.5) | 2, 3, 1Tzn | 2, 3, 2′, 3′, 2Tzn, NHLeu | |
| Tzn | ||||
| 1 | 170.3, C | |||
| 2 | 77.5, CH | 5.40, dd (11.0, 2.0) | 1, 1Phe | 3, NHThr-ether |
| 3 | 32.9, CH2 | 3.45, dd (11.0, 11.0) | 1, 2, 1Phe | 2 |
| 3.72, dd (11.0, 2.0) | ||||
| Phe | ||||
| 1 | 175.1, C | |||
| 2 | 51.6, CH | 4.97, m | 1, 3 | 3, 5/5′, NH |
| 3 | 36.8, CH2 | 3.17, m | 1, 2, 4, 5/5′ | 2, 5/5′, NH, 2Pro |
| 3.57, m | ||||
| 4 | 138.2, C | |||
| 5/5′ | 128.6, CH | 7.17, m | 3, 5′/5, 7 | 2, 3, NH |
| 6/6′ | 128.0, CH | 7.28, m | 4, 6′/6 | |
| 7 | 126.0, CH | 7.19, m | 5/5′ | |
| NH | 9.28, d (9.0) | 2, 3, 1Pro | 2, 3, 5/5′ |
The amino acid sequence of 1 was assigned from a combination of inter-residue ROESY and HMBC correlations (Table 1). The Hα of proline (δH 4.67) showed an HMBC correlation to the serine carbonyl carbon (δC 168.6) and ROESY correlations to both Hα of serine (δH 4.08) and Hβ of phenylalanine (δH 3.57). Hα (δH 4.08) and NH (δH 9.07) of serine both showed HMBC correlations to the leucine carbonyl carbon (δC 172.7). NH (δH 6.62) of leucine showed ROESY correlations to NH (δH 7.37) of threonine. Hα (δH 3.97) of threonine showed an HMBC correlation to the thiazoline carbonyl carbon (δC 170.3). Hα (δH 5.40) of thiazoline showed an HMBC correlation to the phenylalanine thioimide carbon (δC 175.1). Analysis of the MS-MS fragmentation of 1 identified several fragments consistent with the sequence as deduced by NMR (Fig. S17; Table S1). The sequence of residues for this peptide is consistent with the peptide mollamide E as predicted by genetic sequencing and confirmed by mass spectrometry.7a Hence we have used that name for 1.
The absolute configurations of the amino acid residues in 1 were established by acid hydrolysis (under argon) and derivatization with Marfey’s reagent8, 9 followed by comparative LC-MS analysis with derivatized standard d- andl- amino acids. It should be noted that the acid conditions used for hydrolysis simultaneously promote the cleavage of the reverse-prenylated ether group in 1. The analysis established the l-configuration for Cα of proline, serine, leucine and threonine. To determine the configuration of Cβ of prenyl-Thr, derivatization of standard l-Thr and l-allo-Thr was carried out and the resulting elution profiles compared against the corresponding residues from the peptide hydrolysate. Co-injection analysis with the two standards showed the peptide to contain exclusively l-Thr. Two peaks were observed for the phenylalanine residue corresponding to l-Phe and d-Phe at a ratio of 3:7, respectively (Fig. S16). It has been reported that the thiazoline based amino acid in cyclic peptides may undergo racemization during hydrolysis.10, 11 Given the 40% enantiomeric excess observered for d-Phe in the hydrolysate and absence of signs of stereoisomerism in the NMR spectra of the intact peptide, the natural product most likely contains exclusively d-Phe at this position. In order to determine the configuration of Cα of the thiazoline moiety, 1 was first hydrolyzed under argon. The hydrolysis product was derivatized with cystamine and then derivatized with Marfey’s reagent.d- and l-cysteine standards were processed in the same manner. The LC-MS analysis revealed that the product derived from 1 had the same retention time as d-cysteine standard (Fig. S15). Finally, the configuration of the Ser-Pro peptide bond was examined. A large difference in chemical shift between Cβ and Cγ of Pro (ΔδC=+8.3 ppm) and the presence of ROESY correlations between Hα of Ser and Hα of Pro are both consistent with a cis geometry for the Ser-Pro peptide bond.12
The HRESIMS spectrum of compound 2 suggested a molecular formula of C33H46N6O5S,which was supported by NMR data (Table 2). The 1H and 13C NMR spectra of 2 (Table 2) were very similar to those of 1 with minor differences such as the absence of the isoprene signal. Detailed analysis of the 1D and 2D NMR data of 2 established six amino acid residues: one phenylalanine, two prolines, one valine, one isoleucine and one cysteine present as a thiazoline. As with compound 1, the amino acid sequence of 2 was determined from a combination of inter-residue ROESY and HMBC correlations (Table 2) in comparison with MS-MS fragmentation (Fig. S18;Table S2), which was Phe, Pro, Pro, Val, Ile and Tzn. Similarly to 1, the absolute configurations for the amino acids were determined as d-Phe, l-Pro, l-Pro, l-Val, l-Ile and d-Tzn. Here again there was an enantiomeric excess of 20% favoring d-Phe over l-Phe (Fig. S16) but no evidence of multiple configurational isomers for 2 by NMR. Similar to 1, the Pro2-Pro1 peptide bond was determined to adopt a cis geometry based on the large chemical shift difference between Cβ and Cγ (ΔδC=+8.2 ppm) for Pro1 and the presence of ROESY correlations between Hα of Pro2 and Hα of Pro1. A trans peptide geometry was determined for Val-Pro2 based on the small chemical shift difference between Cβ and Cγ (ΔδC=+3.4 ppm) for Pro2 and the presence of ROESY correlations between Hα of Val and H2δ of Pro2.12 Compound 2 was named mollamide F.
Table 2.
NMR Data for 2 in DMSO-d6 (1H 500MHz, 13C 125MHz)
| position | δC | δH(J in Hz) | HMBC | ROESY |
|---|---|---|---|---|
| Pro1 | ||||
| 1 | 169.5, C | |||
| 2 | 59.8, CH | 4.56, d (7.0) | 1, 3, 4, 5, 1Pro2 | 3, 4, 2Pro2, NHPhe |
| 3 | 29.0, CH2 | 1.59, m | 1, 2, 4, 5 | 2, 4, 5 |
| 2.23, m | ||||
| 4 | 20.8, CH2 | 0.48, m | 2, 3 | 2, 3, 5 |
| 1.46, m | ||||
| 5 | 45.3, CH2 | 2.72, br t (10.0) | 3, 4, 1Pro2 | 3, 4 |
| 3.11, m | ||||
| Pro2 | ||||
| 1 | 169.4, C | |||
| 2 | 58.4, CH | 4.18, dd (7.3, 7.3) | 1, 3, 4, 5, 1Val | 3, 4, 2Pro1 |
| 3 | 28.1, CH2 | 1.60, m | 1, 2, 4, 5 | 2, 4, 5 |
| 2.27, m | ||||
| 4 | 24.7, CH2 | 1.79, m | 2, 3, 5 | 2, 3, 5 |
| 1.99, m | ||||
| 5 | 47.3, CH2 | 3.56, m | 2, 3, 4 | 3, 4, 2Val |
| 3.77, m | ||||
| Val | ||||
| 1 | 170.4, C | |||
| 2 | 54.6, CH | 4.35, dd (8.5, 8.5) | 1, 3, 4, 1Ile, | 3, 4, 5, NH, 5Pro2 |
| 3 | 30.5, CH | 2.00, m | 1, 2, 4, 5 | 2, 4, 5 |
| 4 | 18.2, CH3 | 0.93, d (6.5) | 2, 3, 5 | 2, 3, NH, 5Pro2 |
| 5 | 18.4, CH3 | 1.01, d (6.5) | 2, 3, 4 | 2, 3, NH, 5Pro2 |
| NH | 7.49, d (9.0) | 1, 2, 1ILe | 2, 3, 4, 2Ile, 3Ile, NHIle | |
| Ile | ||||
| 1 | 170.6, C | |||
| 2 | 59.6, CH | 4.01, dd (9.5, 9.5) | 1, 3, 4, 5, 1Tzn | 4, 5, 6, NH |
| 3 | 34.4, CH | 2.05, m | 1 | 4, 5, 6, NH, NHVal |
| 4 | 15.1, CH3 | 0.87, m | 2, 3, 5 | 2, 3, 5 |
| 5 | 24.9, CH2 | 1.14, m | 2, 3, 4, 6 | 2, 3, 4, NH |
| 1.52, m | ||||
| 6 | 10.2, CH3 | 0.85, m | 3, 5 | 2, 3 |
| NH | 7.77, d (9.0) | 2, 3, 1Tzn | 2, 3, 5, 2Tzn, NHVal | |
| Tzn | ||||
| 1 | 170.0, C | |||
| 2 | 77.3, CH | 5.30, dd (10.5, 2.5) | 1, 1Phe | 3, NHIle |
| 3 | 33.1, CH2 | 3.40, m | 1, 2, 1Phe | 2 |
| 3.71, dd (10.5, 4.0) | ||||
| Phe | ||||
| 1 | 173.3, C | |||
| 2 | 52.0, CH | 4.88, m | 1, 3 | 3, 5/5′, NH |
| 3 | 36.5, CH2 | 3.38, m | 1, 2, 4, 5/5′ | 2, 5/5′, NH |
| 3.51, m | ||||
| 4 | 138.8, C | |||
| 5/5′ | 129.2, CH | 7.19, m | 3, 7, ′5/5 | 2, 3, NH, 5Pro1 |
| 6/6′ | 128.0, CH | 7.25, m | 4, 6′/6 | 5Pro1 |
| 7 | 126.1, CH | 7.17, m | 5/5′ | |
| NH | 9.17, d (8.5) | 2, 3, 1pro1 | 2, 3, 5/5′, 2Pro1 |
The HRESIMS spectrum of compound 3 suggested a molecular formula of C26H29N3O2. The 1H and 13C NMR spectra exhibited signals corresponding to six methylene groups and three aromatic rings (Table 3). Examination of the 1D and 2D NMR data of 3 indicated unambiguously the presence of three phenethylamine fragments (A, B and C). Connections between these fragments were established through analysis of the HMBC spectrum. An HMBC correlation from the low field methylene residue of B (H-7, δH 3.52) to a carbonyl carbon (C-16, δC 158.1), and correlation from the low field methylene residue of C (H-17, δH 3.34) to the same carbonyl carbon indicated B and C were connected through a urea group. The methylene of A (H-1, δH 3.39) showed an HMBC correlation to a carbonyl carbon (C-15, δC 170.1), to which an aromatic proton of B (H-11, δH 6.96) was also correlated, suggesting A and B to be connected through a benzene-attached amide group. Thus the structure of 3, named molleurea A, was established. Compound 3 is similar to N, N’-diphenethylurea which is a common metabolite in tunicates, and has been reported to act as an antidepressant and to promote adipocyte differentiation.13, 14
Table 3.
NMR Data for 3 in C6D6 (1H 500MHz, 13C 125MHz)
| position | δC | δH(J in Hz) | HMBC |
|---|---|---|---|
| 1 | 41.1, CH2 | 3.39, m | 2, 3, 15 |
| 2 | 35.7, CH2 | 2.63, m | 1, 3, 4/4′, |
| 3 | 139.1, C | ||
| 4/4′ | 128.8, CH | 7.05, m | 2, 6, 4′/4 |
| 5/5′ | 128.6, CH | 7.13, m | 3, 5′/5 |
| 6 | 126.5, CH | 7.06, m | 4/4′ |
| 7 | 42.5, CH2 | 3.52, m | 8, 9, 16 |
| 8 | 33.6, CH2 | 2.73, m | 7, 9, 10, 14, |
| 9 | 138.5, C | ||
| 10 | 137.1, C | ||
| 11 | 126.6, CH | 6.96, m | 9, 13, 15 |
| 12 | 125.9, CH | 6.86, dd (7.2, 7.2) | 10, 14 |
| 13 | 129.9, CH | 7.03, m | 9 |
| 14 | 130.8, CH | 6.97, m | 10, 12 |
| 15 | 170.1, C | ||
| 16 | 158.1, C | ||
| 17 | 41.9, CH2 | 3.34, m | 16, 18, 19 |
| 18 | 36.8, CH2 | 2.60, m | 17, 19, 20, 20′ |
| 19 | 140.0, C | ||
| 20/20′ | 129.0, CH | 7.02, m | 18, 22, 20′/20 |
| 21/21′ | 128.4, CH | 7.11, m | 19, 21′/21 |
| 22 | 126.0, CH | 7.03, m | 20/20′ |
Compounds 1–3 were evaluated for anti-HIV activity in both an HIV integrase inhibition assay and a cytoprotective cell-based assay. In the cytoprotective assay, 2 and 3 showed HIV inhibition with IC50 values of 78 and 60 µM, respectively, whereas 1 did not show any inhibition at 78 µM. The cyclic peptides (1 and 2) are structurally similar to dolastatin 3 which was reported to have HIV integrase inhibition activity.15 We found 2 inhibited HIV-1 integrase with an IC50 value of 39 µM, whereas 1 and 3 showed no activity at 100 µg/mL. Based on our results, residues from the southern region of 2 as depicted appear to play a critical role in binding to HIV integrase.
There are only two other reports of S-(l-) thiazoline occurring in cyanobactin-like peptides.16, 17 The occurrence of epimerization in 1 and 2 was unexpected given the earlier study by Schmidt that detected 2 by sequencing and MS7 which used animals from the same collection site as this study. The absence of any apparent epimerase enzymes in the sequenced pathways suggests that epimerization of Phe and Tzn in the peptides reported here could result from thermodynamic relaxation under the constrained peptide geometry as allowed by the stereochemical lability of sites adjacent to the thioimide functionality.18 To test for formation of other stereoisomers under conditions favoring epimerization, intact 1 was incubated in 20% piperidine in DMF for 24 h at room temperature. The single ion recording for [M + H]+ of the reaction mixture showed no changes (Fig. S19) suggesting that no chromatographically distinguishable stereoisomers formed during the incubation. This result is consistent with the isolated material being at thermodynamic equilibrium with respect to the configuration of its most labile stereocenters.
Experimental Section
General Experimental Procedures
Optical rotations were measured on a Jasco DIP-370 polarimeter. UV spectra were acquired in spectroscopy grade MeOH using a Hewlett-Packard 8452A diode array spectrophotometer. IR spectra were recorded on a JASCO FT/IR-420 spectrophotometer. NMR data were collected using a Varian INOVA 500 (1H 500 MHz, 13C 125 MHz) NMR spectrometer with a 3 mm Nalorac MDBG probe with a z-axis gradient and utilized residual solvent signals for referencing (δH 2.50, δC 39.52 for DMSO-d6 and δH 7.16, δC 128.06 for C6D6). High-resolution ESIMS analyses were performed on a Bruker (Billerica, MA) APEXII FTICR mass spectrometer equipped with an actively shielded 9.4 T superconducting magnet (Magnex Scientific Ltd.), and an external Bruker APOLLO nanospray ESI source. LC-MS analyses were carried out using a Waters Micromass Q-TOF Micro integrated LC-MS system employing negative ion ESI mode with an ion source temperature of 100 °C, a desolvation temperature of 300 °C, and desolvation with nitrogen gas at a flow rate of 400 L/h. Analytical and semipreparative HPLC were accomplished utilizing a Beckman System Gold 126 solvent module equipped with a 168 PDA detector. All reagents were purchased and used without additional purification.
Biological Material
The Didemnum molle ascidian was collected by hand using SCUBA from New Britain, Papua New Guinea (S 5° 17.382’, E 150° 6.089’). A voucher specimen is maintained at University of Utah under accession number PNG07-2-050.
Extraction and Isolation
The frozen ascidian (480 g wet wt) was exhaustively extracted with MeOH to yield 8.4 g of extract. A portion of the extract (2.0 g) was separated on HP20SS resin using a gradient of H2O to IPA in 25% steps and a final wash of 100% MeOH to yield five fractions. The HIV active F2 (50/50 H2O/IPA) was further fractionated by a flash C18 column eluted with a gradient of MeOH/H2O to give five fractions (Fr2.1–2.5). The active Fr2.2 was chromatographed by HPLC using a Phenomenex Luna C18 column (250 × 10 mm) employing 60% CH3CN/40% H2O at 4 mL/min to yield compound 2 (1.4 mg, tR = 6.4 min) and compound 3 (1.0 mg, tR = 8.0 min). The active Fr2.3 was chromatographed by HPLC using a Phenomenex Luna C18 column (250 × 10 mm) employing 60% CH3CN/40% H2O at 4 mL/min to yield compound 1 (4.0 mg, tR = 9.2 min).
Mollamide E (1): colorless, amorphous powder; [α]20d–21 (c 0.1, MeOH); UV (MeOH) λmax(log ε) 208 (3.90), 254 (3.36) nm; IR (film) νmax3390, 3273, 2952, 2921, 2360, 2341, 1683, 1651, 1540, 1507, 1375, 1339, 1271, 1147, 1116, 1074, 1011, 954, 923, 865, 755, 700, 667 cm−1; 1H and 13C NMR, Table 1; HRESIMS m/z 699.35307 [M+H]+ (calcd for C35H51N6O7S, 699.35345; Δ −0.5 ppm).
Mollamide F (2): colorless, amorphous powder; [α]20 d −24 (c 0.1, MeOH); UV (MeOH) λmax(log ε) 208 (3.89), 256 (3.29) nm; IR (film) νmax 3384, 3264, 2965, 2932, 2877, 1652, 1624, 1539, 1507, 1455, 1387, 1313, 1275, 1209, 1106, 1031, 999, 921, 876, 751, 700, 667cm−1; 1H and 13C NMR, Table 2; HRESIMS m/z 639.33145 [M+H]+ (calcd for C33H47N6O5S, 639.33232; Δ −1.4 ppm).
Molleurea A (3): colorless, amorphous powder; UV (MeOH) λmax(log ε) 210 (3.74), 266 (2.68) nm; IR (film) νmax 3310, 3061, 3025, 2926, 2859, 1634, 1558, 1455, 1363, 1315, 1266, 1195, 828, 805, 748, 699 cm−1; 1H and 13C NMR, Table 3; HRESIMS m/z 416.23299 [M+H]+ (calcd for C26H30N3O2, 416.23326; Δ −0.6 ppm).
Acid Hydrolysis of Peptides. Compounds 1 and 2, 100 µg each, were separately dissolved in degassed 6 N HCl (600 µL) and heated in sealed glass vials (under argon) at 110 °C for 17 h. The solvent was removed in vacuo.
LC-MS Analysis of d/l-FDLA Derivatives.8, 9 The acid hydrolysates of 1 and 2 were dissolved in H2O (100 µL) separately, and 1 N NaHCO3 (20 µL) and 1% 1-fluoro-2,4-dinitrophenyl-5-l-leucinamide (l-FDLA solution in acetone, 100 µL) were added. The mixtures were then heated to 40 °C for 50 min. The solutions were cooled to room temperature, neutralized with 1 N HCl (20 µL) and then dried in vacuo. The residues were dissolved in 1:1 CH3CN: H2O and then analyzed by LC-MS. Amino acid standards were derivatized with l-FDLA in a similar manner. Analysis of the l-FDLA derivatives was performed on a Supelcosil LC-18 column (150 × 4.6 mm, 5 µm) employing a linear gradient of 25% CH3CN/75% 0.01 M formic acid to 70% CH3CN/30% 0.01 M formic acid at 0.5 mL/min over 45 min.
The LC-MS analysis of 1 established the presence of d-phenylalanine (tR = 37.11 min), (70%) [l-phenylalanine (tR = 31.60 min), (30%)], l-proline (tR = 24.29 min) [d-proline (tR = 27.60 min)], l-serine (tR = 20.38 min) [d-serine (tR = 21.16 min)], l-leucine (tR = 31.10 min) [d-leucine (tR = 39.15 min)], l-threonine (tR = 20.10 min) [d-threonine (tR = 24.88 min)].
Because l-Thr and l-allo-Thr could not be resolved clearly using this method, they were separated using a Luna C5 column (250 × 4.6 mm, 5 µm) with a mobile phase of 40 mM ammonium acetate (A), 70% CH3CN and 30% MeOH (B), from 5 – 40% B over 70 min at 1 mL/min. The derivative of 1 was co-injected with l-Thr and l-allo-Thr standards, which clearly revealed that the residue from 1 is l-Thr [l-Thr (tR = 56.00 min), l-allo-Thr (tR = 56.99 min)].
The LC-MS analysis of 2 established the presence of d-phenylalanine (tR = 37.11 min), (60%) [l-phenylalanine (tR = 31.60 min), (40%)], l-proline (tR = 24.29 min) [d-proline (tR = 27.60 min)], l-valine (tR = 28.37 min) [d-valine (tR = 35.33 min)], l-isoleucine (tR = 30.72 min) [d-isoleucine (tR = 38.71 min)].
Because l-Ile and l-allo-Ile could not be resolved clearly using this method, they were separated using an Agilent Eclipse Plus C18 column (150 × 4.6 mm, 3.5 µm) with a binary mobile phase of 40 mM ammonium acetate (A), CH3CN (B), with a gradient from 20 – 30% B over 30 min at 1 mL/min. The derivative of 2 was co-injected with l-Ile and l-allo-Ile standards, which clearly revealed that the residue from 2 is l-Ile [l-Ile (tR = 20.10 min), l-allo-Ile (tR = 19.78 min)].
The absolute configuration at the α-carbon of the thiazoline amino acids of 1 and 2 were determined to be S by adding cystamine after acid hydrolysis (under argon). 2-amino-3-((2-aminoethyl)disulfanyl)propanoic acid was detected by LC-MS. The mixtures were then derivatized with l-FDLA. l/d-cysteine standards were treated in a manner similar to the above. The analysis of the the l-FDLA derivatives was performed on a Supelcosil LC-18 column (150 × 4.6 mm, 5 µm) employing a linear gradient of 25% CH3CN/75% 0.01 M formic acid to 70% CH3CN/30% 0.01 M formic acid at 0.5 mL/min over 45 min: d-2-amino-3-((2-aminoethyl)disulfanyl)propanoic acid (tR = 34.49 min) [l-2-amino-3-((2-aminoethyl)disulfanyl)propanoic acid (tR = 34.06 min)].
HIV Integrase Assay
The HIV integrase inhibition assay used in this work was obtained as a kit purchased from XpressBio. This assay assesses compounds for their inhibition of wild-type HIV-1 integrase. Briefly, biotin linked HIV-1 LTR U5 DNA was applied to a streptavidin coated 96-well plate. Test compounds were then added along with a target substrate DNA and HIV integrase. The integrase then processes the HIV-1 LTR U5 and catalyzes covalent strand transfer of the target substrate DNA onto the HIV-1 LTR U5 DNA. An HRP-labeled antibody directed against the target substrate DNA was used to colorimetrically detect the modification. Sodium azide was used as a positive inhibitory control and yielded an IC50 of 46 mM.
Cytoprotective Cell-based Assay19
Briefly, 1A2 cells (a subclone of CEM-SS TART cells that are more prone to apoptosis upon HIV infection) were cultured in RPMI/20% FBS and plated in 96-well plates. The assay utilized controls that included cells without HIV, cells infected with HIV, and cells infected with HIV and treated with AZT at a final concentration of 50 µg/mL or test compounds. Uninfected control cells and HIV exposed cells were allowed to incubate for 96 h at 37 °C in 5% CO2. Cell viability was then assessed with a standard MTT assay.20 On each assay plate, each condition was performed in triplicate and p-values were determined between groups to gauge the performance of the assay. For a valid assay, HIV growth had to be less than 50% of controls; AZT rescue needed to be at least 50% above HIV killing and a test compound was considered active if it performed at 70% of the AZT value.
Supplementary Material
Acknowledgment
This work was supported by NIH support through the Fogarty International Center, ICBG 5U01T006671. Funding for the Varian INOVA 500 MHz NMR spectrometer was provided through NIH grant RR06262.
Footnotes
Supporting Information Available: 1H NMR, 13C NMR, HMBC and ROESY spectra for compounds 1–3. Ion chromatograms of the co-injection of l-FDLA derivatives of the hydrolysis product of 1 with l-Thr and l-allo-Thr; the co-injection of l-FDLA derivatives of the hydrolysis product of 2 with l-Ile and l-allo-Ile; selected ion chromatograms for l-FDLA derivatives of l- and D-Phe and the corresponding residues from 1 and 2 ; comparison between the hydrolysis product of 1 and 2 with l/d-cysteine standards in negative ion mode; MS-MS spectra and fragment assignments for 1 and 2; LC-MS chromatogram for reaction of 1 with 20% piperidine in DMF. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vézinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Broder S, Gallo RC. N. Engl. J. Med. 1984;311:1292–1297. doi: 10.1056/NEJM198411153112006. [DOI] [PubMed] [Google Scholar]
- 3.Craigie RJ. Biol. Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 4.(a) Arrault A, Witczak-Legrand A, Gonzalez P, Bontemps-Subielos N, Banaigs B. Tetrahedron Lett. 2002;43:4041–4044. [Google Scholar]; (b) Teruya T, Sasaki H, Suenaga K. Tetrahedron Lett. 2008;49:5297–5299. [Google Scholar]
- 5.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc. Natl. Acad. Sci. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat. Chem. Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 7.(a) Donia MS, Fricke WF, Ravel J, Schmidt EW. PLoS ONE. 2011;6:e17897. doi: 10.1371/journal.pone.0017897. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tianero MDB, Donia MS, Young TS, Schultz PG, Schmidt EW. J. Am. Chem. Soc. 2012;134:418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada K. Anal. Chem. 1997;69:3346–3352. [Google Scholar]
- 9.Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada K. Anal. Chem. 1997;69:5146–5151. [Google Scholar]
- 10.Yonetani K, Hirotsu Y, Shiba T. Bull. Chem. Soc. Jpn. 1975;48:3302–3305. [Google Scholar]
- 11.Szabo S, Szokan G, Khalafulla AM, Almas M, Kiss C, Rill A, Schon I. J. Pept. Sci. 2001;7:316–322. doi: 10.1002/psc.325. [DOI] [PubMed] [Google Scholar]
- 12.Randazzo A, Piaz FD, Orrù S, Debitus C, Roussakis C, Pucci P, Gomez-Paloma L. Eur. J. Org. Chem. 1998:2659–2665. [Google Scholar]
- 13.Iwai Y, Hirano A, Awaya J, Matsuo S, Omura S. J. Antibiot. 1978;31:375–376. doi: 10.7164/antibiotics.31.375. [DOI] [PubMed] [Google Scholar]
- 14.Choi SS, Cha B, Kagami I, Lee Y, Sasaki H, Suenaga K, Teruya T, Yonezawa T, Nagai K, Woo J. J. Antibiot. 2011;64:277–280. doi: 10.1038/ja.2010.168. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell SS, Faulkner DJ, Rubins K, Bushman FD. J. Nat. Prod. 2000;63:279–282. doi: 10.1021/np990353f. [DOI] [PubMed] [Google Scholar]
- 16.Zabriskie TM, Foster MP, Stout TJ, Clardy J, Ireland CM. J. Am. Chem. Soc. 1990;112:8080–8084. [Google Scholar]
- 17.Portmann C, Blom JF, Gademann K, Juttner F. J. Nat. Prod. 2008;71:1193–1196. doi: 10.1021/np800118g. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt EW. Personal communication. University of Utah, Salt Lake City, UT: Department of Medicinal Chemistry; 2012. Mar, [Google Scholar]
- 19.Kiser R, Makovsky S, Terpening SJ, Laing N, Clanton DJ. J. Virol. Methods. 1996;58:99–109. doi: 10.1016/0166-0934(95)01998-7. [DOI] [PubMed] [Google Scholar]
- 20.Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.