Abstract
Although the polymorphisms of PTPN22 and the variants of CTLA-4 have been reported to be the susceptibility genes, which increased risk of latent autoimmune diabetes in adults (LADA), the results remained inconclusive. The aim of this meta-analysis was to evaluate the association between the polymorphisms of two genes and LADA. We performed a systematic review by identifying relevant studies and applied meta-analysis to pool gene effects. Data from ten studies published between 2001 and 2013 were pooled for two polymorphisms: rs2476601 in the PTPN22 gene and rs231775 in the CTLA-4 gene. Data extraction and assessments for risk of bias were independently performed by two reviewers. Fixed-effect model and random-effect model were used to pool the odds ratios; meanwhile, heterogeneity test, publication bias and sensitive analysis were explored. The minor T allele at rs2476601 and the minor G at rs231775 carried estimated relative risks (odds ratio) of 1.52 (95 % CI 1.29–1.79) and 1.39 (95 % CI 1.11–1.74), respectively. These alleles contributed to an absolute lowering of the risk of all LADA by 4.88 and 14.93 % when individuals do not carry these alleles. The estimated lambdas were 0.49 and 0.63, suggesting a codominant model of effects was most likely for two genes. In summary, our systematic review has demonstrated that PTPN22 rs2476601 and CTLA-4 rs231775 are potential risk factors for LADA. An updated meta-analysis is required when more studies are published to increase the power of these polymorphisms and LADA.
Electronic supplementary material
The online version of this article (doi:10.1007/s00592-014-0613-z) contains supplementary material, which is available to authorized users.
Keywords: PTPN22, CTLA-4, Polymorphism, LADA, Systematic review, Meta-analysis
Introduction
Latent autoimmune diabetes in adults (LADA) is commonly considered as a type of autoimmune diabetes that resembles type 1 diabetes (T1D); however, it masqueraded as type 2 diabetes (T2D) in the initial stage [1–3]. It is commonly recognized that LADA as the subgroup of adult phenotypic type 2 diabetes patients is positive for a GAD antibody [4]. Because of its clinical manifestation exhibits both presentation of two type diabetes, alternative terms have been used to describe this condition as type 1.5 diabetes [5]. There are 347 million people worldwide have diabetes, and LADA accounts for 2–12 % of all cases of diabetes [6].
The patients with LADA were present autoimmunity, immune-mediated β-cell dysfunction and damage as part of their disease process. The progression to insulin dependence in LADA patients is believed more rapidly than classic type 2 diabetes patients who were negative for islet autoantibodies that have been proved with no progressive damage in beta cell [7]. However, the pathogenesis of LADA is still unclear, and the criteria for diagnosing the condition vary between studies. Therefore, the prevalence of LADA patients varies from 2.8 to 22.3 % in different published studies [8], and 8–10 % of patients diagnosed with T2D are in fact misdiagnosed LADA case on average. So, efforts on establishing a targeted treatment strategy and exploring the early detection for primary prevention have come under the spotlight.
It has been clearly identified that there is a strong genetic component affects diabetes. Genome-wide association studies (GWAS) have had considerable success in identifying genetic contributions to T1D and T2D. Unfortunately, LADA is not arousing our attention, and the genetic studies of LADA are sorely lacking. However, some newly articles reported that the single nucleotide polymorphism (SNP) of some genes that associated with T1D and T2D is also showed relevancy with LADA [9]. The protein tyrosine phosphatase N22 gene (PTPN22), which localized on chromosome 1p13 [10] and constituted by 21 exons [11], encodes a lymphoid-specific phosphatase known as LYP. It is a powerful inhibitor of T cell activation [12], which is fundamental for T cell proliferation and maturation [13]. Mutation of PTPN22 gene may potentiate T cell activation and induce autoimmune diseases. Several studies showed that splice variants of PTPN22 rs2476601 may associated with type 1 diabetes [14] and other autoimmune diseases [15, 16]. The cytotoxic T-lymphocyte antigen-4 (CTLA-4) is a co-stimulatory molecular, which is located on chromosome 2 (2q33) [17]. It encodes a glycoprotein receptor of the immunoglobulin (Ig) family expressed on the surface of activated T cells [18], act as an important negative regulator of T cell activation, playing a protective role in autoimmunity [19]. A single nucleotide polymorphism of CTLA-4 rs231775 has been identified as potential risk factors contributing to the development of T1D [17].
Latent autoimmune diabetes in adults (LADA) has been considered as a subgroup of type 1 diabetes in the World Health Organization (WHO) classification. A number of studies have assessed the association between the polymorphism of PTPN22 rs2476601/CTLA-4 rs231775 and LADA in different population [ 20–23]. However, the individual study may not have enough statistical power to detect a true association, and some of the results are inconsistent. Our aim is to estimating strength, accuracy and feature of the association of polymorphism in PTPN22 rs2476601 with LADA, and the relationship between CTLA-4 rs231775 and LADA, performing a meta-analysis of the available literature.
Materials and methods
Literature search
Systematic computerized searches (up to May 2013) without language limitation were performing by using PubMed, Web of knowledge and Chinese National Knowledge Infrastructure (CNKI). A combination of keywords was applied as follows: [(gene or allele or polymorphism) and (PTPN22 or protein tyrosine phosphatase N22) and (CTLA-4 or cytotoxic T-lymphocyte antigen-4)], [(PTPN 1858 or rs2476601)], [(CTLA-4 +49A/G or rs231775)] and [(LADA or latent autoimmune diabetes in adults)]. Only published articles were considered and set no restriction on the source of controls. We browsed the title and abstract of all related manuscripts, manually examined reference lists for additional citations and obtained the full text of all potentially relevant articles. If there were more than one articles published by the same content, we choose the most complete and recent study.
Inclusion and exclusion criteria
Two reviewers (F.D. and W. K. L.) independently went through all titles and abstracts of the identified studies. Studies were selected if they met the following criteria: a case–control study that were written in English or Chinese; genotyped PTPN22 (PTPN22 1858 or rs2476601) or CTLA-4 (+49A/G or rs231775) polymorphisms and detailed data of each genotype; the outcome was latent autoimmune diabetes in adults (LADA); articles had to report the odds ratio and corresponding 95 % confidence interval or provided the sufficient information for estimation. Studies with insufficient data for pooling that with no frequencies of genotypes for each polymorphisms and outcomes were excluded.
Data extraction
For quality control, information was extracted from the studies independently by two investigators (F.D. and H. W. P.). If lack of genotype information, we will try to contact the corresponding author in order to obtain required data. If they did not provide data, those studies were excluded from our review. General characteristics (e.g., the ethnic, genotyping method and the number of male) of included studies were extracted. Any disagreement was resolved by consensus.
Risk of bias assessment
The quality of studies was also independently assessed by the same reviewer (F.D. and G. Y.) based on a risk of bias score for genetic association. This was modified on the basis of both traditional epidemiologic considerations and genetic issues, which were developed by Thakkinstian et al. [29]. The score was divided into five domains, including information bias, confounding bias, selective reporting of outcomes, population stratification and assessment of Hardy–Weinberg equilibrium (HWE) in the control group. Each item was classified with regard to “yes” or “no” or “unclear,” which represent low risk, high risk and insufficient information, respectively. Disagreement between the two reviewers was solved by a senior reviewer (C. X. J.).
Statistical analysis
We used the Comprehensive Meta-Analysis software (ver2.0) for all statistical analyses. The Hardy–Weinberg equilibrium (HWE) was examined in control groups by Fisher’s exact test. If the study was found not to be in HWE with P value less than 0.05, it was considered to be disequilibrium. We performed both per-allele and per-genotype approaches to estimate the strength of association between the polymorphism of genes and LADA risks.
Per-allele analysis
Suppose that D and d are risk and non-risk alleles, and DD, Dd and dd are minor homozygous, heterozygous and common homozygous genotype, respectively, for each polymorphism. The risk allele frequency in each group was estimated for each study by reported genotype data, and overall prevalence along with 95 % confidence intervals was estimated for each SNP. The Z-test was used to determine the statistical significance of the pooled OR, and its P value was used to determine whether the overall gene effect was significant (α = 0.05). Heterogeneity of odds ratios across studies was calculated by a Q test, and the degree of heterogeneity was quantified by I 2 test [30]. If the inspection result shows P > 0.10, a fixed-effect model was selected to pool the data, which can be considered as the evidence of homogeneity between studies. Otherwise, a random-effect model was used. In addition to this, the degree of heterogeneity was quantified using I 2 (I 2 < 25 %, no heterogeneity; 25 % < I 2 < 50 %, moderate heterogeneity; 50 % < I 2 < 75 %, large heterogeneity; and I 2 > 75 % extreme heterogeneity) [31]. We choose a random-effect model if I 2 was greater than 50 % [32]. If there is high heterogeneity exists, a set of subgroup meta-analysis were considering exploring the heterogeneity of current sources by ethnic group. The population-attributable risk (PAR) for risk allele was calculated based on results from discrete-time model [33, 34]. If the main effect of the genotype was statistically significant and with the appropriate effect model selection, further comparisons of OR1 and OR2 were explored.
Per-genotype analysis
We perform the model-free approach to estimate the genotype effect [35], two odds ratios: DD versus dd (OR1) and Dd versus dd (OR2) were estimated for each study. The model of genetic effect, measured by the parameter lambda (λ), which is defined as the ratio of logOR2 to logOR1, was then estimated using the model-free Bayesian approach. This parameter ranges from 0 to 1, which represents the heterozygote effect as a proportion of the homozygote variant effect and captures information about the genetic mode of action as follows: If λ = 0, a recessive (DD vs. Dd +dd) model is suggested; if λ = 1, a dominant model (DD + Dd vs. dd) is suggested; and if λ = 0.5, a codominant model (DD vs. dd; Dd vs.dd) is suggested. If λ > 1 or λ < 0, then a homozygous or heterosis model is likely, although this is rare. The two log odds ratios are modeled as either fixed or random effects, as described in the second statistical analysis enumerated above. Once the best genetic model is identified, this model is used to collapse the three genotypes into two groups and to pool the results again. For lambda, WinBugs 1.4.2 was used with vague prior to distributions for estimation of parameters (i.e., lambda and odds ratio). The models were run with a burn-in of 1,000 iterations, followed by 10,000 iterations for parameter estimates.
Publication bias was assessed using the cumulative forest plot and Egger’s regression intercept [36]. Cumulative forest plot can reflect the dynamic change trend of the research results and the potential impact of small samples on estimate effect size [37]. We did a sensitive analysis to estimate the stability of the meta-analysis with two statistical methods. We first omitted one study and observed the influence of the remaining results to the overall OR, and fail-safe number was also used to estimate the stability of the results.
Result
Characteristics of the studies
Thirteen relevant articles were identified after the primary literature search about PTPN22; seven articles were excluded after screening abstracts and full texts. Among these articles, some are described the irrelevant content to the LADA topic and others are lack of the detailed data we required. Finally, only six articles were left. There were 22 studies conform to the standard after the preliminary search about CTLA-4 gene. After extraction, a total of six case–control studies were selected according to the search criteria for LADA related to the polymorphism of CTLA-4. HWE was calculated for control groups in all articles; we found that one study was showed disequilibrium (Liu [25], P < 0.001).
Risk of bias assessment
As shown in the “Appendix,” the criteria for evaluating the quality of cases and controls were clearly described for all included studies. This work was conducted by two reviewers, and the disagreement was solved by consensus and discussion. The risk of bias was highest in the quality control for genotyping (unclear in 7 out of 10 studies, or 70 %), followed by not assessing HWE (4/10, 40 %) and confounding bias (3/10, 30 %).
Meta-analysis of PTPN22 rs2476601
There were six case–control studies described the association between PTPN22 rs2476601 polymorphism and LADA, which included 1,088 cases and 4,079 controls (Tables 1, 2). All except one study [25] did not observe HWE, and thus, this study was not included in further pooling. Results for these studies are summarized in Table 3. The pooled frequency of minor T allele was 16.9 % (95 % CI 9.7–24.0) in LADA group (I 2 = 92) and 9.9 % (95 % CI 7.6–12.7) in non-LADA group (I 2 = 91), which were both estimated by random model. The odds ratios (T vs. C) were not heterogeneous (χ 2 = 5.69, P = 0.34, I 2 = 12.14), with a pooled odds ratio of 1.52 (95 % CI 1.29–1.79). The overall gene effect estimated by fixed-effect model was significant (P < 0.001). This suggested that individuals carrying the minor T allele had 52 % increased risk of developing LADA than those carrying the major C allele (Table 4). Cumulative meta-analysis was performed for pooled odds ratio, which was used to estimate the publication bias (Fig. 1). From the shape of cumulative forest plot, we know that the point estimate of effect size is very stable transformation, after the process of article size in accordance with the order of accuracy gradually incorporated into the calculation model, which implied that there is no publication bias (seen in Fig. 2). The Egger test did not suggest any evidence of publication bias (SE = 1.79, P = 0.54). The sensitive analysis was performed by omitting one study at a time, which the method was used to make sure that no individual study was entirely responsible for the combined results. From the Table 5, we could found that none of the individual studies affect the final conclusion obviously about the gene. Classic fail-safe N value of PTPN22 rs2476601 was 21 (P = 0.00004, Z = 4.12) when α was set to 0.05, which suggest that 21 unpublished negative studies would have to be included to convert the combined P value to a nonsignificant value. The above results show that our results were statistically reliable.
Table 1.
Characteristics of the selected study
| Author | Year | Gene locus | Ethnic | Method | Male/number | Mean age | BMI | HWE | Confirmed standard | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | Case | Control (sources) | |||||||
| Kisand [20] | 2012 | rs2476601/rs231775 | Caucasian | RFLP/TaqMan | 25/65 | 91/229 | 54.5 | 45.9 | NA | NA | Yes | Initially diagnosis of T2DM but with positive antibody (ICA/IA2A/GADA), no insulin treatment for at least 6 months | Healthy blood donors and hospitalized with no diabetes | |
| Okruszko [24] | 2012 | rs2476601 | Caucasian | PCR | 42/80 | NA/151 | 45.4 | NA | NA | NA | Yes | WHO criterion | Medical staff and medical students with no family history or autoimmune disease | |
| Liu [25] | 2012 | rs2476601 | Asian | PCR | 129/229 | 72/210 | 51.8 | 47.2 | 22.0 ± 4.2 | 22.4 ± 2.9 | No | Age at onset > 35 year, positive at least one positive antibody (GAD-Ab/IA2-Ab), excluded other types of diabetes | Healthy volunteers with normal in OGTT test and no history of autoimmune disease or negative in antibody test | |
| Cervin [9] | 2008 | rs2476601 | Caucasian (Swedish) | MALDI-TOF–MS | 73/164 | 553/1000 | 52.4 | 70.1 | 25.9 ± 5.6 | 27.6 ± 4.4 | Yes | Age at onset > 35 year, GAD antibody positive | Without family history of diabetes or treatment of hypertension | |
|
Caucasian (Finnish) |
MALDI- TOF–MS |
83/113 | 83/113 | 54.6 | 53.7 | 26.8 ± 5.0 | 25.9 ± 3.7 | Yes | ||||||
| Petrone [21] | 2008 | rs2476601 | Caucasian | PCR | 131/250 | 278/545 | 50.3 | 30.0 | NA | 21.8 ± 2.2 | Yes | Initial diagnosis of T2DM, and with positive GADAs antibody, no insulin requirement and no ketosis disease duration between 6 months to 5 years | Normoglycemic subjects with no family history of autoimmune disease | |
| Pettersen [22] | 2010 | rs2476601/rs231775 | Caucasian | TaqMan | 68/126 | 740/1503 | NA | NA | NA | NA | Yes | Anti-GAD positive, no insulin treatment within 12 months | A questionnaire with answer “no” of the question: having diabetes? | |
| Jin [23] | 2011 | rs231775 | Asian | PCR | 79/135 | 297/476 | 49.6 | 48.2 | NA | NA | Yes | Immunology of Diabetes Society criterion | Non-diabetic individuals with no family history and no autoimmune disease | |
| Haller [26] | 2007 | rs231775 | Caucasian | RFLP | 24/61 | 101/252 | 64.6 | 45.5 | NA | NA | Yes | T2DM patients with at least on positive antibody (ICA/IA-2/GAD 65) | Younger: the blood donation, older: hospitalized for various reasons, without T1DM and T2DM | |
| Caputo [27] | 2005 | rs231775 | Caucasian | PCR | NA/63 | NA/168 | 51.4 | NA | NA | NA | Yes | WHO criterion | Healthy person with no family history and normally fasting blood glucose level | |
| Cosentino [28] | 2002 | rs231775 | Caucasian | PCR | 22/80 | 42/85 | 51 | 48 | NA | NA | Yes | Initially classified as T2DM, and with islet antibodies | Employees of the university with no family history of diabetes | |
BMI body mass index, RFLP restriction fragment length polymorphism analysis, MALDI-TOF–MS matrix-assisted laser desorption/ionization time of flight mass spectrometry, PCR polymerase chain reaction, NA not available
Table 2.
The risk of bias assessment
| Author | Ascertainment of LADA | Ascertainment of control | Quality control for genotyping | Population stratification | Confounding bias | Selective outcome report | HWE |
|---|---|---|---|---|---|---|---|
| Kisand [20] | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Okruszko [24] | Yes | Yes | Unclear | Yes | No | Yes | No |
| Liu [25] | Yes | Yes | Unclear | Yes | Yes | Yes | Yes |
| Cervin [9] | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Petrone [21] | Yes | Yes | Unclear | Yes | Yes | Yes | Yes |
| Pettersen [22] | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Jin [23] | Yes | Yes | Unclear | Yes | Yes | Yes | Yes |
| Haller [26] | Yes | Yes | Unclear | Yes | Yes | Yes | No |
| Caputo [27] | Yes | Yes | Unclear | Yes | No | Yes | No |
| Cosentino [28] | Yes | Yes | Unclear | Yes | Yes | Yes | No |
Table 3.
Genotype frequencies for PTPN22 rs2476601 and genotype effects of studies included in the meta-analysis
| Author | Total number | Case genotype | Control genotype | T allele prevalence | T versus C | TT versus CC | CT versus CC | HWE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | TT | CT | CC | TT | CT | CC | OR | 95 % CI | OR | 95 % CI | OR | 95 % CI | ||||||
| Kalle Kisand | 65 | 229 | 1 | 15 | 49 | 6 | 51 | 172 | 0.138 | 0.943 | 0.531 | 1.676 | 0.585 | 0.069 | 4.976 | 1.032 | 0.535 | 1.992 | P = 0.40 |
| Anna Okruszko | 80 | 151 | 9 | 20 | 51 | 1 | 36 | 114 | 0.126 | 2.164 | 1.315 | 3.561 | 20.118 | 2.483 | 163.008 | 1.242 | 0.656 | 2.352 | P = 0.47 |
| Camilla Cervin | 341 | 1,453 | 7 | 92 | 242 | 18 | 257 | 1,178 | 0.101 | 1.641 | 1.291 | 2.086 | 1.893 | 0.782 | 4.582 | 1.743 | 1.323 | 2.295 | P = 0.38 |
| Antonio Petrone | 250 | 545 | 1 | 31 | 218 | 2 | 47 | 496 | 0.047 | 1.440 | 0.917 | 2.261 | 1.138 | 0.103 | 12.612 | 1.501 | 0.928 | 2.427 | P = 0.33 |
| Elin Pettersen | 123 | 1,491 | 1 | 32 | 90 | 18 | 292 | 1,181 | 0.110 | 1.298 | 0.888 | 1.897 | 0.729 | 0.096 | 5.523 | 1.438 | 0.942 | 2.196 | P = 0.99 |
| Overall odds ratio | 1.518 | 1.286 | 1.792 | 1.861 | 0.940 | 3.684 | 1.523 | 1.260 | 1.840 | ||||||||||
HWE Hardy–Weinberg equilibrium
Table 4.
Genotype frequencies for CTLA-4 rs231775 and genotype effects of studies included in the meta-analysis
| Author | Total number | Case genotype | Control genotype | G allele prevalence | G versus A | GG versus AA | GA versus AA | HWE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | GG | AG | AA | GG | AG | AA | OR | 95 % CI | OR | 95 % CI | OR | 95 % CI | ||||||
| Kalle Kisand | 65 | 229 | 21 | 35 | 9 | 37 | 124 | 68 | 0.432 | 1.908 | 1.284 | 2.834 | 4.288 | 1.783 | 10.313 | 2.133 | 0.968 | 4.699 | P = 0.14 |
| Ping Jin | 135 | 476 | 51 | 73 | 11 | 167 | 239 | 70 | 0.602 | 1.218 | 0.920 | 1.615 | 1.943 | 0.957 | 3.948 | 1.944 | 0.977 | 3.866 | P = 0.34 |
| K.Haller | 61 | 252 | 20 | 33 | 8 | 40 | 135 | 77 | 0.427 | 2.003 | 1.339 | 2.996 | 4.812 | 1.948 | 11.891 | 2.353 | 1.035 | 5.350 | P = 0.16 |
| Mariela Caputo | 63 | 168 | 6 | 35 | 22 | 21 | 76 | 71 | 0.351 | 1.076 | 0.702 | 1.649 | 0.922 | 0.331 | 2.572 | 1.486 | 0.797 | 2.773 | P = 0.99 |
| Anna Cosentino | 80 | 85 | 4 | 55 | 21 | 5 | 40 | 40 | 0.294 | 1.559 | 0.986 | 2.463 | 1.524 | 0.369 | 6.285 | 2.619 | 1.344 | 5.103 | P = 0.30 |
| Elin Pettersen | 124 | 1,477 | 48 | 52 | 24 | 499 | 715 | 263 | 0.580 | 1.072 | 0.823 | 1.396 | 1.054 | 0.632 | 1.759 | 0.797 | 0.482 | 1.319 | P = 0.83 |
| Overall odds ratio | 1.391 | 1.114 | 1.738 | 1.961 | 1.098 | 3.502 | 1.682 | 1.119 | 2.527 | ||||||||||
HWE Hardy–Weinberg equilibrium
Fig. 1.
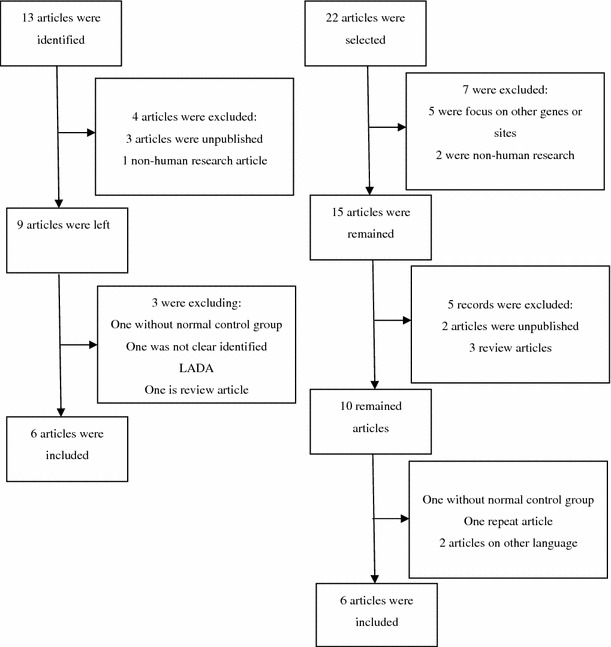
Flowchart for identify relevant studies for PTPN22 gene, CTLA-4 gene polymorphisms with LADA
Fig. 2.
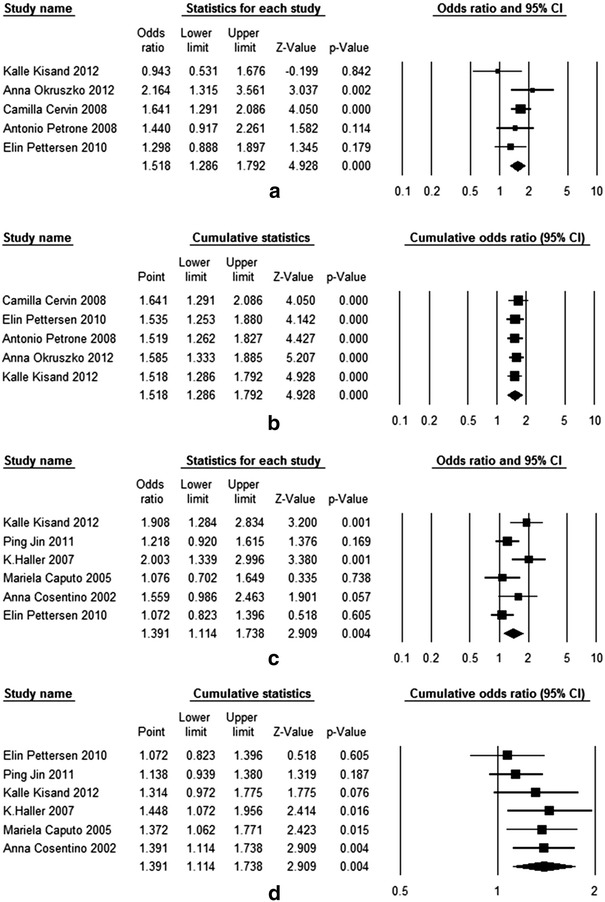
Forest plot and cumulative forest plot of PTPN22 and CTLA-4 genes with LADA. a Forest plot of the association between PTPN22 rs2476601 polymorphism and LADA risk (T vs. C), which was estimated by fixed-effect model. b Cumulative forest plot of PTPN22 gene (T vs. C). c. Forest plot of the association between CTLA-4 rs231775 polymorphism and LADA risk (G vs. A), which was calculated by random-effect model. d. Cumulative forest plot analysis of CTLA-4 gene (G vs. A). The size of each square is the proportion of percent weight of each study that contributed in the pooled odds ratio. The pooled odds ratios are indicated by the diamond. Horizontal bars represent the 95 % CI
Table 5.
The result of sensitive analysis
| Gene | Excluded study | Pooled OR | 95 % CI | P | I 2 (%) | P value for I 2 |
|---|---|---|---|---|---|---|
| PTPN22 rs2476601 | Okruszko [24] | 1.453 | 1.218–1.732 | <0.001 | 0.00 | 0.477 |
| Petrone [21] | 1.531 | 1.281–1.829 | <0.001 | 28.94 | 0.229 | |
| Cervin [9] | 1.414 | 1.124–1.778 | 0.003 | 18.58 | 0.296 | |
| Pettersen [22] | 1.575 | 1.310–1.894 | <0.001 | 18.05 | 0.300 | |
| Kisand [20] | 1.585 | 1.333–1.885 | <0.001 | 0.00 | 0.588 | |
| CTLA-4 rs231775 | Cosentino [28] | 1.314 | 1.132–1.526 | 0.001 | 63.688 | 0.026 |
| Pettersen [22] | 1.460 | 1.235–1.728 | <0.001 | 48.397 | 0.101 | |
| Haller [26] | 1.262 | 1.084–1.468 | 0.003 | 43.419 | 0.132 | |
| Kisand [20] | 1.268 | 1.089–1.476 | 0.002 | 49.531 | 0.094 | |
| Caputo [27] | 1.372 | 1.181–1.595 | <0.001 | 61.487 | 0.034 | |
| Jin [23] | 1.379 | 1.170–1.624 | <0.001 | 63.459 | 0.027 |
Genotype frequency and estimated OR for each study were shown in Table 3. The OR1 for TT versus CC was moderate heterogeneity (χ 2 = 7.08, P = 0.13, I 2 = 43.51), whereas the OR2 for CT versus CC was homogenous (χ 2 = 2.86, P = 0.72, I 2 = 0.00). They both calculated by fixed-effect model. The pooled OR1 and OR2 were 1.86 (95 % CI 0.94–3.68) and 1.52 (95 % CI 1.26–1.84), respectively, which suggested that individuals with TT and CT genotypes had 86 and 52 % higher risk of LADA than those carrying CC genotype. The λ = 0.49 (95 % CI 0.07–0.96) which suggested that a codominant effect was most likely, although one genotype effect did not reach statistical significant.
Meta-analysis of CTLA-4 rs231775
The associations between CTLA-4 rs231775 and LADA were investigated in six case–control studies, with 528 cases and 2,687 controls. The pooled frequency of minor G allele in LADA group was 49.2 % (95 % CI 35.2–63.3), along with high heterogeneity (I 2 = 95), and in non-LADA group was 44.9 % (95 % CI 36.3–53.9), which estimated by random model (I 2 = 97). The pooled OR was calculated by random-effect model due to the high heterogeneity (χ 2 = 11.50, P = 0.04, I 2 = 56.51). The odds ratio of G versus A is 1.39 (95 % CI 1.11–1.74) with statistical significance (P = 0.004), which indicated that individuals carrying G allele had 39 % increased risk of developing LADA than those carrying A allele. In cumulative plot, the summary OR was a bit different in the first two studies, whereas not much changed in a smooth curve with the increase of the literature. Egger’s regression test also suggest no publication bias (SE = 2.50, P = 0.18). In the sensitive analysis, after each removed a piece of literature have not seen a big difference in the OR values have changed. Classic fail-safe N value of CTLA-4 rs231775 is 24 (P = 0.00001, Z = 4.37) when α was set to 0.05, which suggest that 24 unpublished negative studies would have to be included to convert the combined P value to a nonsignificant value. This shows that our results are stable enough. When studies were divided according to the ethnic group, the result showed that there is a significant association in Caucasian. The pooled odds ratio is 1.45 (95 % CI 1.09–1.92), with a significantly statistical gene effect (P = 0.01) but a highly heterogeneity (I 2 = 63.46, P = 0.03). There is only one Chinese study that belongs to Asian population with no statistical significance (OR 1.22, 95 % CI 0.92–1.62, P = 0.17).
In heterogeneity test, there is a moderate heterogeneity across OR1 (χ 2 = 14.19, P = 0.01, I 2 = 64.77) and OR2 (χ 2 = 11.12, P = 0.05, I 2 = 55.04). The summary odds ratios for the GG and AG genotype were estimated by random model, they are 1.96 (95 % CI 1.10–3.50) and 1.68 (95 % CI 1.12–2.53), respectively. These point estimates can be interpreted as that person with the GG and GA genotypes had 96 and 68 % higher risks of developing LADA than persons with the AA genotype. The estimated λ = 0.63 (95 % CI 0.15–0.98) which suggested that a codominant effect was most likely.
Discussion
We performed a systematic review and meta-analysis to determine the effects of two gene polymorphisms (PTPN22 rs2476601 and CTLA-4 rs231775) on the LADA. The analyses included pooling data from five and six studies with a total sample size of 4,728 and 3,215 subjects. We were able to identify PTPN22 rs2476601 and CTLA-4 rs231775 polymorphisms as genetic markers that might increase the risk of LADA. Individuals who carried minor allele T in rs2476601 had 52 % increased risk of developing LADA relative to those carrying C allele, while individuals carrying the risk allele G in rs231775 may lead to an increasing risk of having LADA by 39 % compared with allele A. The results suggest association in Caucasians, that is, carriage of G in the CTLA-4 rs231775 increases 45 % relative to carriage of A allele. However, Asian populations showed an unrelated result. This difference may be due to the different genetic backgrounds and limited article.
The minor T risk allele of the PTPN22 rs2476601 polymorphism investigated is quite rare in non-LADA group, with frequency of 9.9 %. However, it is high in the LADA group, with frequency of 16.9 %. The PAR for the minor T was 4.88 %, which suggested that PTPN22 rs2476601 polymorphism probably serves as a marker for an absolute lowering of the risk of all LADA in Caucasians by 4.88 % when individuals do not carry T allele. The risk G allele in CTLA-4 rs231775 is common, with similar frequencies of 44.9 and 49.2 % in non-LADA group and LADA group, which might indicate an important effect at a population level. The PAR of CTLA-4 G allele was 14.93 %, which may provide a useful clinical estimation that might contribute an absolute lowering of the risk of all LADA by 14.93 % when individuals do not carry this allele.
Genotypic effects were also estimated for PTPN22 rs2476601 and CTLA-4 rs231775. For PTPN22 rs2476601, the estimated OR1 for TT versus CC and OR2 for CT versus CC were 1.86 and 1.52 in Caucasian, respectively, and estimated lambda was 0.49, suggesting a codominant mode of gene effect. However, the 95 % confident interval of lambda laid from 0.07 to 0.96, which suggested that the genetic mode could be recessive dominant and codominant. This pooling was based on small number of included studies, and thus, uncertainty of gene effects was still present. For CTLA-4 rs231775, the genotype effects of GG and GA versus AA were 0.96 and 0.68, respectively. The point estimated lambda was 0.63, suggesting a codominant mode of gene effect. The 95 % confident interval of lambda laid from 0.15 to 0.98, which suggested that the genetic mode could be recessive dominant and codominant.
Previous studies have shown that the SNPs of PTPN22 and CTLA-4 are associated with T1DM [38–43] and other autoimmune diseases [44–47], which have some meta-analysis to support it [48–51].Our research showed statistical evidence that the polymorphism of genes PTPN22 rs2476601 and CTLA-4 rs231775 is associated with LADA on the basis of population study, which could provide some clues on the research of fundamental to diabetes biology and uncover the major genetic factors involved in the pathogenesis of LADA.
LADA is a common subgroup of diabetes accounting for about 7 % of all diabetic patients in Europe (http://andis.ludc.med.lu.se). Multiple islet auto-antigens and autoantibodies could be detected before the development of autoimmune diabetes [52, 53], such as cytoplasmic islet cell autoantibodies (ICA) and glutamic acid decarboxylase autoantibody (GADA), which have been recognized as the most effective immune marker for LADA diagnosis [54, 55]. Huang Gan et al. also reported that combination testing of IAA with GADA and IA-2A could improve LADA diagnose rate by 2.39 % than GADA and IA-2A, which increased the evidence that autoimmunity to insulin may be central to disease pathogenesis [56]. It though should be noted that to date, no GWAS has been performed on LADA patients. Even though some newly articles reported that the single nucleotide polymorphism (SNP) of some genes that associated with T1D and T2D is also showed relevancy with LADA [9], the possible reason for inconsistence could be due to the diagnostic criteria for LADA or distinct interactions of genes and environment.
Heterogeneity is a potential factor affecting pooled results [57], which can be divided into genetic heterogeneity of effect and the genetic heterogeneity of the model. In our meta-analysis, a small heterogeneity was proved in the analysis of PTPN22 rs2476601 polymorphism with LADA in per-allele analysis; all studies included Caucasians, which may produce better consolidation effect. Beyond that, we excluded one study [25] which is out of Hardy–Weinberg equilibrium in the control group when we do the pooled odds ratio in order to make the results more precise. However, in the research of CTLA-4 rs231775 polymorphism with LADA, the result was suffered moderate heterogeneity influenced. We do a subgroup analysis according to ethnic population to explore the source of heterogeneity, and highly heterogeneity was observed in Caucasian groups. When we exclude the article by Elin [22] and Haller [26] during the sensitive analysis, the I 2 reduced and we conjecture that these two articles may increases the overall heterogeneity when we do the pooling. This analysis implies that different genetic backgrounds and small study sample size may be the source of heterogeneity. Analyses based on specific genetic models can produce misleading estimates of the odds ratios when an inappropriate model is assumed. The pooled genetic association was calculated by a genetic model-free approach, which does not assume that the underlying genetic model is known in advance but still makes use of the information available on all genotypes. We avoided multiple comparisons, which would lead to overly strong misjudge assumptions about the genetic model or of inefficient estimates, and offer a single method that could have been used in all of these examples giving a consistent presentation and to reduce heterogeneity.
There are still some limitations in our article. Firstly, we just conducted in English and Chinese literature retrieval, which may result in missing some related articles written by other languages. Secondly, the sources of control are not clearly and uniform that might lead to not enough estimation. Another potential disadvantage is that all the included studies were case–control study, which might overestimate the genetic association. To avoid such bias, the best way is to establish the population-based nested case–control study, although it is hard to implement. The last limitation is small sample size. There were only six studies included in two gene polymorphisms, which may lead to not powerful enough estimation. The small sample size study may have a low power and affecting the results in the process of pooled odds ratio. So a more precise association needs to be explored further with sufficient data. Thus, our results should be interpreted with caution until further verification of sequencing approaches plus larger and larger meta-analysis.
In conclusion, our meta-analysis suggests that both of the PTPN22 rs2476601 and the CTLA-4 rs231775 polymorphisms contribute to susceptibility to LADA. Future large, well-designed studies are warranted to examine the impact of PTPN22 and CTLA-4 on LADA risk. What’s more, a better understanding of the genetic basis is needed to more accurately place this disorder in the spectrum of diabetes phenotypes, which further research on genome-wide genotyped datasets, and more detailed genetic studies of LADA could help unravel the genetic etiology of LADA. In addition, a comprehensive interaction on gene–gene and gene–environment should also be evaluated in future analysis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported in part by National Natural Science Foundation of China (Grant Number 30901249; 81101267), the Guangdong Natural Science Foundation (Grant Numbers 10151063201000036; S2011010002526) and Project from Jinan university (Grant Number 21612426). We would like to thank Ammarin Thakkinstian from the section for Clinical Epidemiology and Biostatistics, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Thailand; John R. Thompson from the department of Health Sciences, University of Leicester, UK; Sasivimol Rattanasiri from the section for Clinical Epidemiology and Biostatistics, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Thailand; for providing the statistical computing algorithm support. Thanks also go to: Leif Groop from the Lund University Diabetes Centre, University Hospital Skåne, Sweden; Jasmina Kravić from the department of Clinical Sciences/Diabetes and Endocrinology, Lund University, Swede; Elin Pettersen from the department of Cancer Research and Molecular Medicine, Faculty of Medicine, The Norwegian University of Science and Technology, Norway; for providing the specific genotyping data.
Conflict of interest
The authors declare that there is no duality of interest associated with this manuscript.
Abbreviations
- LADA
Latent autoimmune diabetes in adults
- GWAS
Genome-wide association study
- SNP
Single nucleotide polymorphism
- BMI
Body mass index
- HWE
Hardy–Weinberg equilibrium
- PAR
Population-attributable risk
Appendix
Table 6.
Table 6.
Risk of bias assessment for genetic association studies of LADA of studies included in the meta-analysis
| Domain and item | Low risk of bias |
|---|---|
| Information bias | |
| Ascertainment of LADA | |
| Clearly described objective criteria of diagnosis of LADA | Yes |
| Not clearly described | No |
| Did not mention | Unclear |
| Ascertainment of controls | |
| Controls were non-LADA and without family history | Yes |
| Mentioned the sources of controls | Yes |
| Not described | No |
| Ascertainment of genotyping examination | |
| Genotyping done under “blind” conditions of case specimens and control specimens | Yes |
| Genotyping of cases and controls was performed together | Yes |
| Genotyping error rate <5 % | Yes |
| Quality control procedure (e.g., reanalysis of random specimens, by using different genotyping methods for analysis, analysis if replicate sample) | Yes |
| Unblind | No |
| Genotyping error rate >5 % | No |
| Did not mention what was done | Unclear |
| Confounding bias | |
| Population stratification | |
| No difference in ethnic origin between cases and controls | Yes |
| Use of controls who were not related to cases with clearly identification | Yes |
| Use of some controls who came from the same family | No |
| No report of what was done | Unclear |
| Other confounding bias | |
| Controls for confounding variables (e.g., age, gender, or BMI) in analysis | Yes |
| Not controlled for confounding variables | No |
| Not mentioned | Unclear |
| Selective reporting (for replication studies) | |
| Reported results of all polymorphisms mentioned in the objectives, no significant or not | Yes |
| Reported results of only significant polymorphisms | No |
| HWE | |
| HWE in the control group | Yes |
| HWD in the control group | No |
| HWE not checked or mentioned | No |
References
- 1.Tuomi T, Groop LC, Zimmet PZ, Rowley MJ, Knowles W, Mackay IR. Antibodies to glutamic acid decarboxylase reveal latent autoimmune diabetes mellitus in adults with a non-insulin-dependent onset of disease. Diabetes. 1993;42(2):359–362. doi: 10.2337/diab.42.2.359. [DOI] [PubMed] [Google Scholar]
- 2.Gale EA. Latent autoimmune diabetes in adults: a guide for the perplexed. Diabetologia. 2005;48(11):2195–2199. doi: 10.1007/s00125-005-1954-5. [DOI] [PubMed] [Google Scholar]
- 3.Rolandsson O, Palmer JP. Latent autoimmune diabetes in adults (LADA) is dead: long live autoimmune diabetes! Diabetologia. 2010;53(7):1250–1253. doi: 10.1007/s00125-010-1713-0. [DOI] [PubMed] [Google Scholar]
- 4.Zimmet PZ. The pathogenesis and prevention of diabetes in adults. Genes, autoimmunity, and demography. Diabetes Care. 1995;18(7):1050–1064. doi: 10.2337/diacare.18.7.1050. [DOI] [PubMed] [Google Scholar]
- 5.Schernthaner G, Hink S, Kopp HP, Muzyka B, Streit G, Kroiss A (2001) Progress in the characterization of slowly progressive autoimmune diabetes in adult patients (LADA or type 1.5 diabetes). Exp Clin Endocrinol Diabetes 109(Suppl 2):S94–S108. doi:10.1055/s-2001-18573 [DOI] [PubMed]
- 6.Stenstrom G, Gottsater A, Bakhtadze E, Berger B, Sundkvist G. Latent autoimmune diabetes in adults: definition, prevalence, beta-cell function, and treatment. Diabetes. 2005;54(Suppl 2):S68–S72. doi: 10.2337/diabetes.54.suppl_2.S68. [DOI] [PubMed] [Google Scholar]
- 7.Ekholm E, Gottsater A, Dahlin LB, Sundkvist G. No signs of progressive beta cell damage during 20 years of prospective follow-up of autoantibody-negative diabetes. Acta Diabetol. 2012;49(1):57–62. doi: 10.1007/s00592-011-0273-1. [DOI] [PubMed] [Google Scholar]
- 8.Turner R, Stratton I, Horton V, Manley S, Zimmet P, Mackay IR, Shattock M, Bottazzo GF, Holman R. UKPDS 25: autoantibodies to islet-cell cytoplasm and glutamic acid decarboxylase for prediction of insulin requirement in type 2 diabetes UK Prospective Diabetes Study Group. Lancet. 1997;350(9087):1288–1293. doi: 10.1016/S0140-6736(97)03062-6. [DOI] [PubMed] [Google Scholar]
- 9.Cervin C, Lyssenko V, Bakhtadze E, Lindholm E, Nilsson P, Tuomi T, Cilio CM, Groop L. Genetic similarities between latent autoimmune diabetes in adults, type 1 diabetes, and type 2 diabetes. Diabetes. 2008;57(5):1433–1437. doi: 10.2337/db07-0299. [DOI] [PubMed] [Google Scholar]
- 10.Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase Lyp. Blood. 1999;93(6):2013–2024. [PubMed] [Google Scholar]
- 11.Wang S, Dong H, Han J, Ho WT, Fu X, Zhao ZJ. Identification of a variant form of tyrosine phosphatase LYP. BMC Mol Biol. 2010;11:78. doi: 10.1186/1471-2199-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med. 1999;189(1):111–121. doi: 10.1084/jem.189.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannons JL, Schwartzberg PL. Fine-tuning lymphocyte regulation: what’s new with tyrosine kinases and phosphatases? Curr Opin Immunol. 2004;16(3):296–303. doi: 10.1016/j.coi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Dultz G, Matheis N, Dittmar M, Rohrig B, Bender K, Kahaly GJ. The protein tyrosine phosphatase non-receptor type 22 C1858T polymorphism is a joint susceptibility locus for immunthyroiditis and autoimmune diabetes. Thyroid Off J Am Thyroid Assoc. 2009;19(2):143–148. doi: 10.1089/thy.2008.0301. [DOI] [PubMed] [Google Scholar]
- 15.Dieude P, Teixeira VH, Pierlot C, Cornelis F, Petit-Teixeira E. Testing for linkage and association with rheumatoid arthritis a ptpn22 promoter polymorphism reported to be associated and linked with type 1 diabetes in the Caucasian population. Ann Rheum Dis. 2008;67(6):900–901. doi: 10.1136/ard.2007.077180. [DOI] [PubMed] [Google Scholar]
- 16.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MT, Chang M, Chang SY, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, Gregersen PK. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 18.Greene JL, Leytze GM, Emswiler J, Peach R, Bajorath J, Cosand W, Linsley PS. Covalent dimerization of CD28/CTLA-4 and oligomerization of CD80/CD86 regulate T cell costimulatory interactions. J Biol Chem. 1996;271(43):26762–26771. doi: 10.1074/jbc.271.43.26762. [DOI] [PubMed] [Google Scholar]
- 19.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, Thompson CB, Griesser H, Mak TW. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science (New York, NY) 1995;270(5238):985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 20.Kisand K, Uibo R. LADA and T1D in Estonian population—two different genetic risk profiles. Gene. 2012;497(2):285–291. doi: 10.1016/j.gene.2012.01.089. [DOI] [PubMed] [Google Scholar]
- 21.Petrone A, Suraci C, Capizzi M, Giaccari A, Bosi E, Tiberti C, Cossu E, Pozzilli P, Falorni A, Buzzetti R. The protein tyrosine phosphatase nonreceptor 22 (PTPN22) is associated with high GAD antibody titer in latent autoimmune diabetes in adults: non Insulin Requiring Autoimmune Diabetes (NIRAD) Study 3. Diabetes Care. 2008;31(3):534–538. doi: 10.2337/dc07-1457. [DOI] [PubMed] [Google Scholar]
- 22.Pettersen E, Skorpen F, Kvaloy K, Midthjell K, Grill V. Genetic heterogeneity in latent autoimmune diabetes is linked to various degrees of autoimmune activity: results from the Nord-Trondelag Health Study. Diabetes. 2010;59(1):302–310. doi: 10.2337/db09-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin P, Huang G, Lin J, Yang L, Xiang B, Zhou W, Zhou Z. High titre of antiglutamic acid decarboxylase autoantibody is a strong predictor of the development of thyroid autoimmunity in patients with type 1 diabetes and latent autoimmune diabetes in adults. Clin Endocrinol. 2011;74(5):587–592. doi: 10.1111/j.1365-2265.2011.03976.x. [DOI] [PubMed] [Google Scholar]
- 24.Okruszko A, Szepietowska B, Wawrusiewicz-Kurylonek N, Gorska M, Kretowski A, Szelachowska M. HLA-DR, HLA-DQB1 and PTPN22 gene polymorphism: association with age at onset for autoimmune diabetes. Arch Med Sci. 2012;8(5):874–878. doi: 10.5114/aoms.2012.31619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Liu J, Zheng TS, Li Q, Wang C, Pan XP, Lu H, Zhao YW. The -1123G > C variant of PTPN22 gene promoter is associated with latent autoimmune diabetes in adult Chinese Hans. Cell Biochem Biophys. 2012;62(2):273–279. doi: 10.1007/s12013-011-9291-4. [DOI] [PubMed] [Google Scholar]
- 26.Haller K, Kisand K, Pisarev H, Salur L, Laisk T, Nemvalts V, Uibo R. Insulin gene VNTR, CTLA-4 +49A/G and HLA-DQB1 alleles distinguish latent autoimmune diabetes in adults from type 1 diabetes and from type 2 diabetes group. Tissue Antigens. 2007;69(2):121–127. doi: 10.1111/j.1399-0039.2006.00745.x. [DOI] [PubMed] [Google Scholar]
- 27.Caputo M, Cerrone GE, Lopez AP, Villalba A, Krochik GA, Cedola FN, Targovnik HM, Frechtel GD. Cytotoxic T lymphocyte antigen 4 heterozygous codon 49 A/G dimorphism is associated to latent autoimmune diabetes in adults (LADA) Autoimmunity. 2005;38(4):277–281. doi: 10.1080/08916930500158203. [DOI] [PubMed] [Google Scholar]
- 28.Cosentino A, Gambelunghe G, Tortoioli C, Falorni A. CTLA-4 gene polymorphism contributes to the genetic risk for latent autoimmune diabetes in adults. Ann N Y Acad Sci. 2002;958:337–340. doi: 10.1111/j.1749-6632.2002.tb03000.x. [DOI] [PubMed] [Google Scholar]
- 29.Thakkinstian A, McKay GJ, McEvoy M, Chakravarthy U, Chakrabarti S, Silvestri G, Kaur I, Li X, Attia J. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2011;173(12):1365–1379. doi: 10.1093/aje/kwr025. [DOI] [PubMed] [Google Scholar]
- 30.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 31.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 33.Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA, Pandey JP, Newman LS, Magira E, Beznik-Cizman B, Monos D. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet. 2003;73(4):720–735. doi: 10.1086/378097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayden KM, Zandi PP, Lyketsos CG, Tschanz JT, Norton MC, Khachaturian AS, Pieper CF, Welsh-Bohmer KA, Breitner JC. Apolipoprotein E genotype and mortality: findings from the Cache County Study. J Am Geriatr Soc. 2005;53(6):935–942. doi: 10.1111/j.1532-5415.2005.53301.x. [DOI] [PubMed] [Google Scholar]
- 35.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34(6):1319–1328. doi: 10.1093/ije/dyi169. [DOI] [PubMed] [Google Scholar]
- 36.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clin res ed) 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egger M, Smith GD, Altman DG (2008) Systematic reviews in health care: meta-analysis in context, 2nd ed. doi:10.1002/9780470693926
- 38.Giza S, Goulas A, Gbandi E, Effraimidou S, Papadopoulou-Alataki E, Eboriadou M, Galli-Tsinopoulou A. The role of PTPN22 C1858T gene polymorphism in diabetes mellitus type 1: first evaluation in Greek children and adolescents. BioMed res int. 2013;2013:721604. doi: 10.1155/2013/721604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fichna M, Zurawek M, Januszkiewicz-Lewandowska D, Fichna P, Nowak J. PTPN22, PDCD1 and CYP27B1 polymorphisms and susceptibility to type 1 diabetes in Polish patients. Int J Immunogenet. 2010;37(5):367–372. doi: 10.1111/j.1744-313X.2010.00935.x. [DOI] [PubMed] [Google Scholar]
- 40.Lavrikova E, Nikitin AG, Seregin IuA, Zil’berman LI, Tsitlidze NM, Kuraeva TL, Peterkova VA, Dedov II, Nosikov VV. Association of the C1858T polymorphism of the PTPN22 gene with type 1 diabetes. Mol Biol. 2009;43(6):1040–1043. doi: 10.1134/S0026893309060089. [DOI] [PubMed] [Google Scholar]
- 41.Philip B, Isabel W. Association of cytotoxic T lymphocyte-associated antigen 4 gene single nucleotide polymorphism with type 1 diabetes mellitus in Madurai population of Southern India. Indian J Human Geneti. 2011;17(2):85–89. doi: 10.4103/0971-6866.86189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ei Wafai RJ, Chmaisse HN, Makki RF, Fakhoury H. Association of HLA class II alleles and CTLA-4 polymorphism with type 1 diabetes. Saudi J Kidney Dis Transpl. 2011;22(2):273–281. [PubMed] [Google Scholar]
- 43.Ahmedov G, Ahmedova L, Sedlakova P, Cinek O. Genetic association of type 1 diabetes in an Azerbaijanian population: the HLA-DQ, -DRB1*04, the insulin gene, and CTLA4. Pediatric Diabetes. 2006;7(2):88–93. doi: 10.1111/j.1399-543X.2006.00152.x. [DOI] [PubMed] [Google Scholar]
- 44.Alkhateeb A, Marzouka NA, Tashtoush R. Variants in PTPN22 and SMOC2 genes and the risk of thyroid disease in the Jordanian Arab population. Endocrine. 2013 doi: 10.1007/s12020-013-9908-z. [DOI] [PubMed] [Google Scholar]
- 45.Gianchecchi E, Palombi M, Fierabracci A. The putative role of the C1858T polymorphism of protein tyrosine phosphatase PTPN22 gene in autoimmunity. Autoimmun Rev. 2013;12(7):717–725. doi: 10.1016/j.autrev.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Spink C, Stege G, Tenbrock K, Harendza S. The CTLA-4 +49GG genotype is associated with susceptibility for nephrotic kidney diseases. Nephrol Dialysis Transpl. 2013;28(11):2800–2805. doi: 10.1093/ndt/gft381. [DOI] [PubMed] [Google Scholar]
- 47.AlFadhli S. Overexpression and secretion of the soluble CTLA-4 splice variant in various autoimmune diseases and in cases with overlapping autoimmunity. Genet Test Mol Biomark. 2013;17(4):336–341. doi: 10.1089/gtmb.2012.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L, Wei Y, Xun W, Han D. Meta-analysis of the correlation between PTPN22 gene polymorphisms and susceptibility to systemic lupus erythematosus. Asia-Pacific J Publ Health. 2013;25(4 Suppl):22S–29S. doi: 10.1177/1010539513496268. [DOI] [PubMed] [Google Scholar]
- 49.Xuan C, Lun LM, Zhao JX, Wang HW, Zhu BZ, Yu S, Liu Z, He GW. PTPN22 gene polymorphism (C1858T) is associated with susceptibility to type 1 diabetes: a meta-analysis of 19,495 cases and 25,341 controls. Ann Hum Genet. 2013;77(3):191–203. doi: 10.1111/ahg.12016. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z, Fei M, Fu D, Zhang L, Ma Y, Wang Y, Zhang F, Xia Q, Wang X. Association between cytotoxic T lymphocyte antigen-4 polymorphism and type 1 diabetes: a meta-analysis. Gene. 2013;516(2):263–270. doi: 10.1016/j.gene.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 51.Zhai JX, Zou LW, Zhang ZX, Fan WJ, Wang HY, Liu T, Ren Z, Dai RX, Ye D. CTLA-4 polymorphisms and systemic lupus erythematosus (SLE): a meta-analysis. Mol Biol Rep. 2013;40(9):5213–5223. doi: 10.1007/s11033-012-2125-7. [DOI] [PubMed] [Google Scholar]
- 52.Mayer A, Fabien N, Gutowski MC, Dubois V, Gebuhrer L, Bienvenu J, Orgiazzi J, Madec AM. Contrasting cellular and humoral autoimmunity associated with latent autoimmune diabetes in adults. Eur J Endocrinol. 2007;157(1):53–61. doi: 10.1530/EJE-07-0060. [DOI] [PubMed] [Google Scholar]
- 53.Roh MO, Jung CH, Kim BY, Mok JO, Kim CH. The prevalence and characteristics of latent autoimmune diabetes in adults (LADA) and its relation with chronic complications in a clinical department of a university hospital in Korea. Acta Diabetol. 2013;50(2):129–134. doi: 10.1007/s00592-010-0228-y. [DOI] [PubMed] [Google Scholar]
- 54.Groop L, Tuomi T, Rowley M, Zimmet P, Mackay IR. Latent autoimmune diabetes in adults (LADA)–more than a name. Diabetologia. 2006;49(9):1996–1998. doi: 10.1007/s00125-006-0345-x. [DOI] [PubMed] [Google Scholar]
- 55.Jin P, Huang G, Lin J, Luo S, Zhou Z. Epitope analysis of GAD65 autoantibodies in adult-onset type 1 diabetes and latent autoimmune diabetes in adults with thyroid autoimmunity. Acta Diabetol. 2011;48(2):149–155. doi: 10.1007/s00592-010-0250-0. [DOI] [PubMed] [Google Scholar]
- 56.Huang G, Wang X, Li Z, Li H, Li X, Zhou Z. Insulin autoantibody could help to screen latent autoimmune diabetes in adults in phenotypic type 2 diabetes mellitus in Chinese. Acta Diabetol. 2012;49(5):327–331. doi: 10.1007/s00592-010-0196-2. [DOI] [PubMed] [Google Scholar]
- 57.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


