In photosynthetic organisms, D-ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the major enzyme assimilating atmospheric CO2 into the biosphere1. Due to the wasteful oxygenase activity and slow turnover of Rubisco, the enzyme is among the most important targets for improving the photosynthetic efficiency of vascular plants2,3. It is also anticipated that introducing the CO2-concentrating mechanism (CCM) from cyanobacteria into plants could enhance crop yield4-6. However, the complex nature of Rubisco’s assembly has made manipulation of the enzyme extremely challenging, and attempts to replace it in plants with the enzymes from cyanobacteria and red algae have not been successful7,8. Here we report two transplastomic tobacco lines with functional Rubisco from the cyanobacterium Synechococcus elongatus PCC7942 (Se7942). We knocked out the native tobacco gene encoding the large subunit of Rubisco by inserting the large and small subunit genes of the Se7942 enzyme, in combination with either the corresponding Se7942 assembly chaperone, RbcX, or an internal carboxysomal protein, CcmM35, which incorporates 3 small subunit-like domains9,10. Se7942 Rubisco and CcmM35 formed macromolecular complexes within the chloroplast stroma, mirroring an early step in the biogenesis of cyanobacterial β-carboxysomes11,12. Both transformed lines were photosynthetically competent, supporting autotrophic growth, and their respective forms of Rubisco had higher rates of CO2 fixation per unit of enzyme than the tobacco control. These transplastomic tobacco lines represent an important step towards improved photosynthesis in plants and will be valuable hosts for future addition of the remaining components of the cyanobacterial CCM, such as inorganic carbon transporters and the β-carboxysome shell proteins4-6.
Rubisco catalyses the incorporation of CO2 into biological compounds in photosynthetic organisms1. During photorespiration, Rubisco also reacts wastefully with oxygen, leading to the release of previously fixed CO2, NH3 and energy13. Furthermore, catalysis by Rubisco is slow and very large amounts (up to 50% of leaf soluble protein, 25% leaf N) are needed to support adequate photosynthetic rates. Some variation in the catalytic properties of Rubisco from diverse sources is apparent. Harnessing this variation has the potential to confer superior photosynthetic characteristics to specific crops and environments14. C4 plants, cyanobacteria and hornworts have evolved forms of CO2-concentrating mechanisms (CCM) that allow them to utilize faster forms of Rubisco that have lower CO2 affinity, whereas C3 plants, which lack a CCM, are constrained to express forms of Rubisco with higher CO2 affinity but a relatively low rate of turnover2. In plants, Rubisco is a L8S8 hexadecamer consisting of eight small subunits (SSU) and eight large subunits (LSU). Although the SSU genes are located in the nucleus, the LSU is encoded by the chloroplast genome, which has complicated previous attempts to engineer improvements in higher plant Rubisco2,15.
Introduction of a CCM has been proposed as a means to improve the performance of Rubisco in C3 plant chloroplasts4-6,16. In cyanobacteria and several autotrophic prokaryotes, Rubisco and carbonic anhydrase are enclosed within polyhedral microcompartments known as carboxysomes, which maintain elevated CO2 levels in the vicinity of Rubisco, which both increases carbon fixation and suppresses photorespiration4,6. However, when a tobacco transplastomic line was created in which the LSU gene, rbcL, from the cyanobacterium Synechococcus PCC6301 replaced the native tobacco rbcL, the cyanobacterial LSU did not form a functional complex with the native tobacco SSU8. Although a simpler L2 homodimer Rubisco from Rhodospirillum rubrum was able to assemble inside tobacco chloroplasts17, red algal Rubisco subunits failed to produce functional L8S8 complexes within chloroplasts7. In order to test whether cyanobacterial LSU and SSU can assemble into a functional enzyme within higher plant chloroplasts, we generated two transplastomic tobacco lines, named SeLSX and SeLSM35, using the biolistic delivery system18, to express the two Rubisco subunits from Se7942 along with either RbcX or CcmM35, respectively. In each chloroplast transformant, three genes were co-transcribed from the tobacco rbcL promoter. Each downstream gene was preceded by an intercistronic expression element (IEE) and a Shine-Dalgarno sequence (SD) and equipped with a terminator to facilitate processing into translatable monocistronic transcripts19,20 (Fig. 1a).
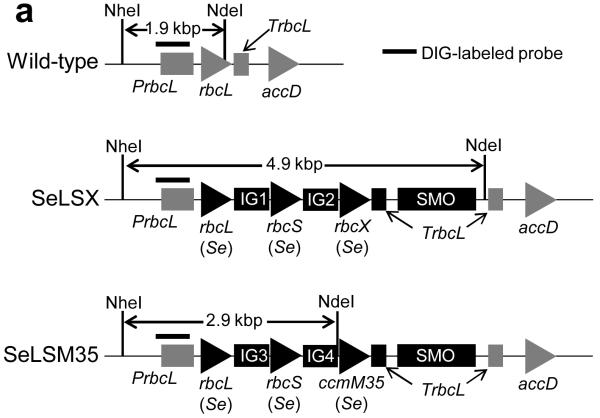
Figure 1. Replacement of the tobacco chloroplast rbcL with cyanobacterial genes.
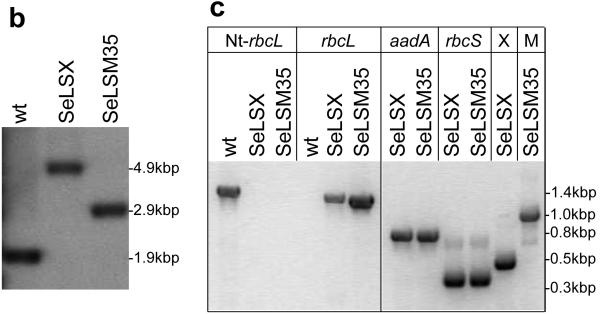
a, Gene arrangements of the rbcL locus in the wild-type, SeLSX and SeLSM35 tobacco lines. Endogenous chloroplast DNA elements are shown in grey and the newly introduced segments in black. The intergenic regions IG1, IG2, IG3 and IG4 include TpetD(At)-IEE-SD, TpsbA(At)-IEE-SD, Trps16(At)-IEE-SD and TpsbA(At)-IEE-SD18 respectively. The selectable marker operon (SMO) includes LoxP-PpsbA-aadA-Trps16-LoxP. The probe recognizes the rbcL promoter (PrbcL) region. The NheI and NdeI sites used in the DNA blot along with the lengths of the expected DNA fragments detected by the probe are indicated. b, DNA blot analysis of wild-type, SeLSX and SeLSM35 lines digested with NdeI and NheI. c, Analyses of RT-PCR products of 6 genes. Nt-rbcL is the only tobacco gene; all other genes are the transgenes introduced into the tobacco chloroplast genome. x = rbcX, M = ccmM35.
The two vectors we constructed were designed to replace the tobacco rbcL gene with the foreign DNA. To determine whether all chloroplasts in each plant contained the transgenic locus rather than endogenous tobacco rbcL, we examined blots of total leaf DNA digested with restriction enzymes that would produce fragment length polymorphism between the wild-type and transgenic loci (Fig. 1b). We found that shoots arising after two rounds on selective medium were homoplasmic for the transgene locus, lacking the fragment corresponding to the wild-type chloroplast genome (Fig. 1b). In order to verify these observations, we performed reverse transcription and PCR (RT-PCR) and observed no cDNA derived from the native rbcL transcript, while cDNAs produced from aadA, the selectable marker gene, and the cyanobacterial genes were detected (Fig. 1c).
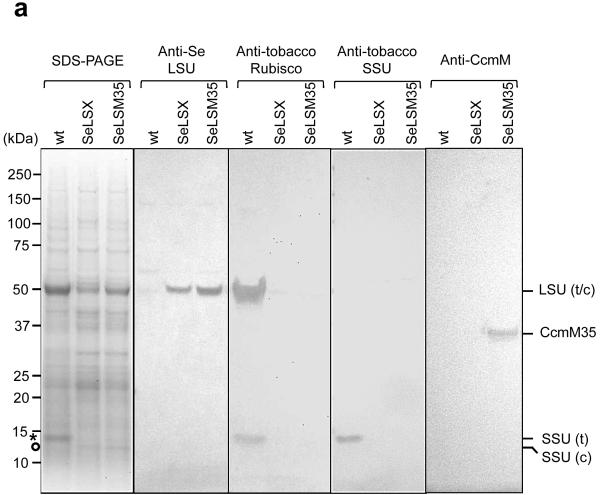
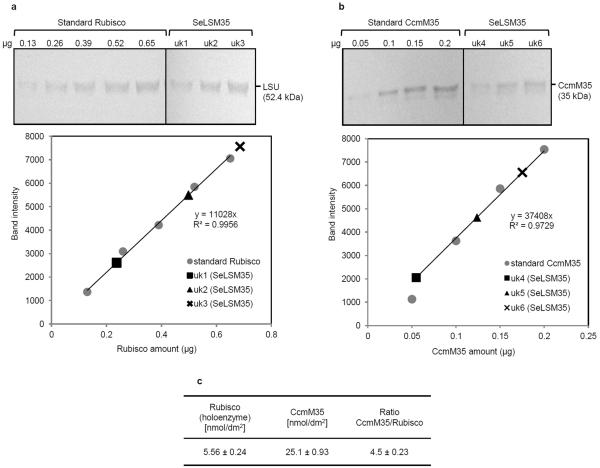
To observe the expression of the cyanobacterial proteins, we extracted total leaf proteins and examined them by SDS-PAGE and immunoblots. In Coomassie-stained gels, we detected protein bands at the predicted molecular masses of ~52 kDa for the LSU and ~13 kDa for the SSU of the cyanobacterial Rubisco in SeLSX and SeLSM35 samples, while wild-type tobacco exhibited a protein of the expected and distinct SSU mass of ~15 kDa (Fig. 2a). Immunoblots probed with antibodies specific for either the cyanobacterial LSU, tobacco Rubisco, tobacco SSU or cyanobacterial CcmM35 verified the presence of cyanobacterial proteins in the two transformants and tobacco Rubisco only in the wild-type plant (Fig. 2a). Although no engineering of tobacco SSU genes was performed in the transgenic lines, tobacco SSU protein was undetectable, as expected, since its stability is known to be severely affected in the absence of a compatible LSU8,17. The absence of the tobacco SSU in the transformants also indicated that it could not form a stable complex with the cyanobacterial LSU. The estimated stoichiometry of CcmM35 per Rubisco holoenzyme in SeLSM35 transformant is about 4.5, which is consistent with the values reported for cyanobacteria (Extended Data Fig. 1)21.
Figure 2. Cyanobacterial proteins in chloroplasts.
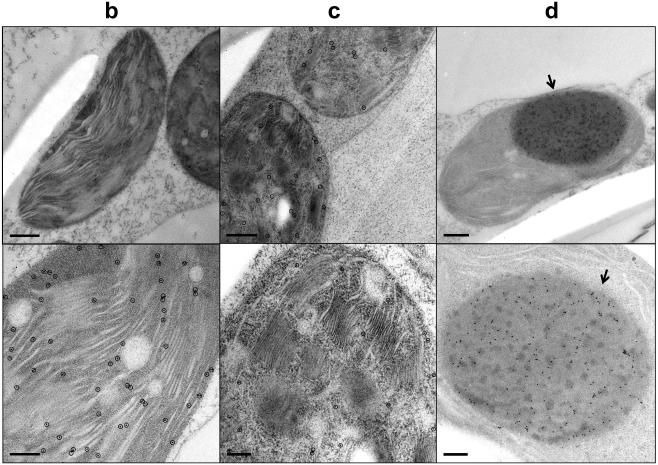
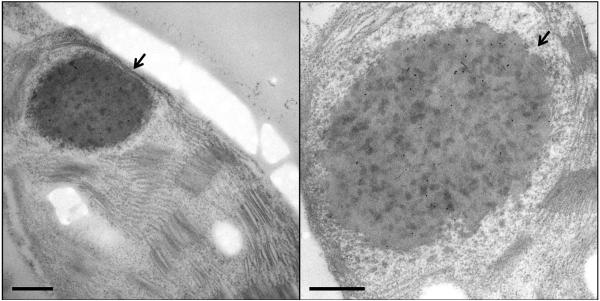
a, Coomassie-stained gel and immunoblot of 14 μg of total leaf protein from wild-type (wt), SeLSX and SeLSM35 tobacco lines. Immunoblots were probed with the antibodies indicated. Molecular weights (MW; kDa) of standard proteins are shown. (*) MW of tobacco SSU; (°) MW of cyanobacterial SSU. b-d, Electron micrographs of leaf sections showing Rubisco’s localization in the stroma of mesophyll chloroplasts of (b) wild-type, (c) SeLSX and (d) SeLSM35 tobacco lines. Leaf tissues were prepared by high pressure freeze fixation (HPF) in combination with immunogold labeling using an anti-tobacco Rubisco antibody (b) or an anti-cyanobacterial Rubisco antibody (c,d) and a secondary antibody conjugated with 10 nm gold particles, which are indicated with either black circles or arrows. Scale bars = 500 nm (for the top panels in b,d), 200 nm (for c and the bottom panels in b,d).
In order to observe the configuration of the cyanobacterial Rubisco in the two transgenic lines, we examined the plant material by transmission electron microscopy (TEM) in combination with immunogold labeling. Although the enzyme was localized to the chloroplast stroma in both transgenic lines, we observed markedly different patterns of molecular organization. In leaves of the SeLSX line, the cyanobacterial Rubisco showed a diffuse localization similar to endogenous Rubisco in wild-type tobacco (Fig. 2b,c). In contrast, in the SeLSM35 line, in which the Rubisco is co-expressed with CcmM35, the proteins were aggregated into a giant complex in each chloroplast (Fig. 2d and Extended Data Fig. 2). In Se7942, CcmM35 is translated from an internal ribosome entry site of the ccmM transcript, which also produces the full length protein, CcmM58, with an additional N-terminal domain22. Previous estimation of protein ratios suggested that Rubisco in Se PCC7942 likely exists as L8S5 units cross-linked by the SSU-like domains of CcmM35 resulting in their paracrystalline arrangement in the lumen of β-carboxysomes21. The cyanobacterial mutant lacking CcmM58 produces large electron-dense bodies of 300-500 nm with rectangular cross-section composed of Rubisco and CcmM3522. However, the structures formed inside chloroplasts are generally rounded in appearance without apparent internal order. This discrepancy probably arises from different ratios of Rubisco and CcmM35 or additional carboxysomal components potentially present in the cyanobacterial bodies. Remarkably, the structures observed in chloroplasts are highly similar in appearance to procarboxysomes recently identified as an important early stage in the carboxysome assembly11 and will potentially facilitate future attempts to assemble β-carboxysomes in chloroplasts through expression of other essential components.
The specificity of the carboxylase activity of cyanobacterial Rubisco relative to its competing oxygenase activity (specificity factor) is known to be lower than that in higher plants, making it more sensitive to the inhibitory effects of oxygen than tobacco Rubisco2. SeLSX and SeLSM35 plants did not survive on soil at the normal atmospheric CO2 level of ~400 ppm, but were able to grow in CO2 enriched (9000 ppm) air at a rate slower than the wild-type plant. Both transgenic plants have normal appearance (Fig. 3). Previous efforts to engineer tobacco Rubisco demonstrated that the growth rate and photosynthetic properties of transplastomic plants are generally consistent with the expression levels and catalytic properties of the recombinant Rubisco2,17. We believe it is also the case in our transplastomic plants. Our preliminary analyses to quantify the Rubisco content using the CABP (2-carboxy-D-arabinitol-1,5-bisphosphate) binding method indicate that the Rubisco concentrations in the two chloroplast transformants are approximately 12-18 % of that in the wild-type plant (Extended Data Table 1)23. In addition, the lower levels of total soluble proteins and chlorophyll concentrations likely contribute to the observed slow growth of the two chloroplast transformants (Extended Data Table 1).
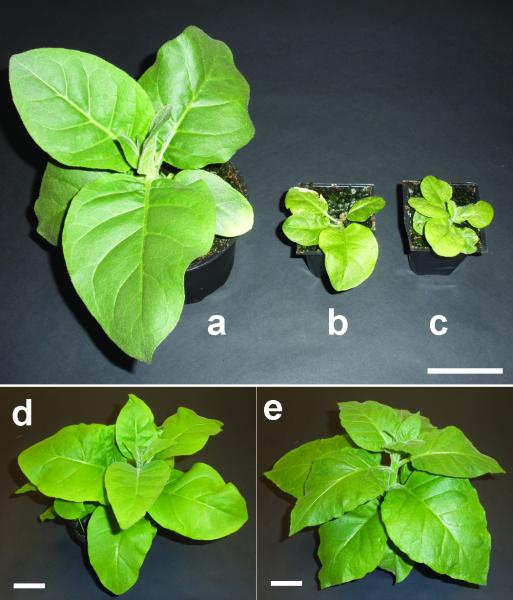
Figure 3. Phenotype of the wild-type and transplastomic tobacco lines.
Plant were grown at atmospheric CO2 level about 9000 ppm. Pictures showing 6-week old (a) wild-type (b) SeLSX, and (c) SeLSM35; and 10-week old (d) SeLSX and (e) SeLSM35 tobacco lines grown in the same conditions. Scale bars = 5 cm.
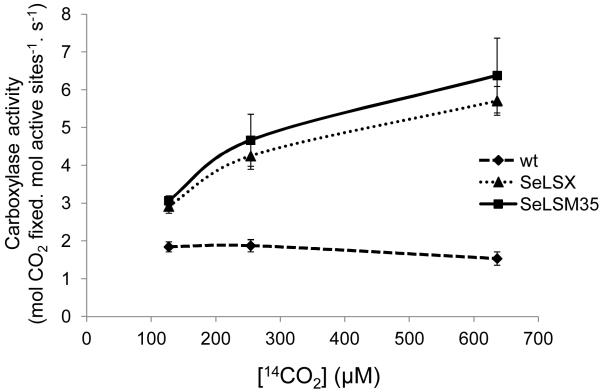
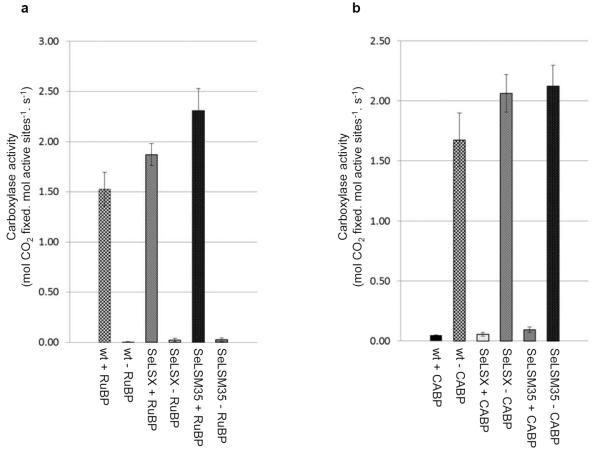
The fact that both transgenic lines could grow autotrophically indicated that active cyanobacterial Rubisco has assembled. We measured the carboxylase activities of the cyanobacterial Rubisco in the leaf homogenates at room temperature using ribulose bisphosphate (RuBP) and several concentrations of radiolabeled sodium bicarbonate (NaH14CO3). The assays were performed in the presence of 10 mM, 20 mM and 50 mM NaH14CO3, which at pH 8.0 would generate dissolved CO2 concentrations ([CO2]) of approximately 125 μM, 250 μM and 640 μM, respectively. The carboxylase activity of Rubisco in the tobacco control did not increase upon increasing [CO2], confirming that the native enzyme was already saturated at 125 μM [CO2] (Fig. 4). In contrast, cyanobacterial Rubisco displayed greater carboxylase activity at higher [CO2] values, with a rate of catalysis which exceeded that of the tobacco enzyme at each [CO2]. Our measured kinetic values are consistent with the reported rate and Michaelis constants for CO2 (~3 s−1 &10.7 μM for tobacco and ~12 s−1 & 200 μM for the enzyme in Synechococcus PCC6301, respectively)2,24. We confirmed that the carboxylase activities detected in our samples were specific to Rubisco, since they were entirely dependent on the presence of RuBP and were inhibited by CABP25 (Extended Data Fig. 3). The high carboxylase activities detected in the transformants are consistent with the absence of interference by tobacco SSU in the assembly of bona fide cyanobacterial Rubisco in the chloroplasts. Furthermore, both transgenic lines exhibited high Rubisco activities despite differences in its intra-organellar organization.
Figure 4. Carboxylase activities at different [14CO2].
CO2 fixation by crude leaf homogenates from tobacco lines expressing cyanobacterial Rubisco (SeLSX and SeLSM35) and wild-type tobacco (wt). The rates of carboxylase activity (mol CO2 fixed. mol active sites−1.s−1) at each point of the curves are the means ± standard deviation of the 2,4 & 6 min data obtained in two independent assays at different [CO2] (125 μM; 250 μM; 640 μM).
We included RbcX in one of our chloroplast transformation vectors because it has been shown to enhance the assembly of the LSU core complex before formation of the final hexadecameric complex9. However, Se7942 lacking RbcX suffered no defect in growth rate or Rubisco activity26. As line SeLSM35 lacks RbcX but has active Rubisco, evidently Se-RbcX is not essential for the assembly of functional cyanobacterial Rubisco in chloroplasts. CcmM35, through its SSU-like domains, might assist in the assembly of cyanobacterial Rubisco in SeLSM35 in the absence of RbcX.
The transgenic plants described here are absolutely dependent on the cyanobacterial Rubisco for carbon fixation. If the oxygenation reaction of cyanobacterial Rubisco can be suppressed and the local [CO2] in the vicinity of the enzyme can be raised by further engineering, CO2 assimilation may be enhanced, and the necessity to divert so much fixed nitrogen into this enzyme may be diminished. Recently, we demonstrated that the shell proteins of β-carboxysomes could form structures similar to empty microcompartments in the chloroplast stroma27. Introduction of the carboxysome shell proteins, the required internal proteins, and appropriate transporters into transgenic plants containing cyanobacterial Rubisco is predicted to result in significantly enhanced photosynthetic performance in vascular plants5,6. This report, demonstrating that cyanobacterial Rubisco can assemble into active enzyme in a C3 plant and support autotrophic photosynthesis, is an important step towards the introduction of a complete and functional CCM into the chloroplasts of vascular plants.
Methods
Construction of the transformation vectors
The Se-rbcL and Se-rbcS genes with codons optimized for chloroplast translation system were designed by Muhammad Waqar Hameed and synthesized by Bioneer. Extended Data Table 2 contains the primers ordered from Integrated DNA Technologies and used in this work. The amplifications of DNA molecules were carried out with Phusion High-Fidelity DNA polymerase (Thermo Scientific). The restriction enzymes and T4 DNA ligase were also purchased from Thermo Scientific.
The two tobacco chloroplast genomic loci (F1 and F2) immediately flanking the rbcL gene (bp56620-57599 and bp59034-60033 of NCBI Reference Sequence: NC_001879.2) were amplified from the DNA extracted from tobacco plants using the primer pairs F1for-F1rev and F2for-F2rev respectively and cloned into pCR8/GW/TOPO TA vector (Life Technologies) adding PstI and MluI restriction sites at the 5’ and 3’ end of F2, respectively. The Se-rbcL gene was amplified from pGEM-Teasy-Se-rbcL with F1OLrbcLfor and 4RErbcLrev primers adding an overlap to the 3’ end of F1 at the 5’ end of Se-rbcL and four restriction sites, MauBI, NotI, PstI and MluI, at the 3’ end of Se-rbcL. This amplified Se-rbcL gene was designed to replace the tobacco rbcL in frame and allow the synthetic expansion of the operon. F1for2 and F1rev primers were used to amplify F1 from its pCR8 vector and the resulting product was then joined with the Se-rbcL amplicon by the overlap extension PCR procedure. The F1-Se-rbcL segment was then digested with ApaI and MluI restriction enzymes and ligated into pGEM-Teasy-Se-rbcL template treated with the same two enzymes to obtain the pGEM-F1-rbcL vector. F2 was digested out of its pCR8 vector with PstI and MluI enzymes and ligated into the similarly disgested pGEM-F1-rbcL to yield the pGEM-F1-rbcL-F2 vector. The selectable marker operon (SMO) containing LoxP-PpsbA-aadA-Trps16-LoxP was amplified from a previously reported chloroplast transformation vector, pTetCBglC28, with SMOfor and SMOrev primers, digested with PstI and ligated in forward orientation to the PstI-digested pGEM-F1-rbcL-F2 vector to obtain the pGEM-F1-rbcL-SMO-F2 vector. The rbcL terminator (TrbcL) was amplified from the tobacco DNA with TrbcLfor and TrbcLrev primers, digested with MauBI and Bsp120I enzymes and ligated between the MauBI and NotI sites of the pGEM-F1-rbcL-SMO-F2 vector to obtain the pCT-rbcL vector, which is ready to replace the tobacco rbcL with Se-rbcL and the SMO by the chloroplast transformation procedure. The Se-rbcL operon driven by the native rbcL promoter in pCT-rbcL was then expanded at the MauBI site with Se-rbcS, Se-rbcX and Se-ccmM35 as follows.
Three terminators from the Arabidopsis thaliana (At) chloroplast genome, TpetD(At), TpsbA(At) and Trps16(At), were amplified with their respective primer pairs, TpetDAtfor-TpetDAtrev, TpsbAAtfor-TpsbAAtrev and Trps16Atfor-Trps16Atrev, adding an overlap to the intercistronic expression element (IEE) at the 3’ end and two restriction sites, MluI and MauBI at the 5’ end of each terminator. Each terminator was extended at the 3’ end by IEE-SD or IEE-SD18 fragment with primers IEESDrev or IEESD18rev-SD18rev2 respectively, resulting in the four intergenic regions, IG1, IG2, IG3, and IG4 in Fig. 1a. The Se-rbcX and Se-ccmM35 genes were amplified from the genomic DNA extracted from Se7942 using the primer pairs rbcXfor-rbcXrev and M35for-M35rev respectively, adding an overlap to the IEE-SD fragment at the 5’ end and an MluI site at the 3’ end of each gene. Similarly, Se-rbcS was amplified from pGEM-Teasy-Se-rbcL using the primer pair rbcSfor-rbcSrev. Then, IG1-rbcS, IG2-rbcX, IG3-rbcS and IG4-ccmM35 fragments were similarly generated by joining each intergenic fragment with the corresponding gene using the overlap extension PCR procedure. The MluI-digested IG2-rbcX and IG4-ccmM35 modules were each inserted into the MauBI site of the pCT-rbcL to obtain pCT-rbcL-rbcX and pCT-rbcL-ccmM35, respectively. Then the MluI-digested IG1-rbcS and IG3-rbcS modules were each inserted into the MauBI site of pCT-rbcL-rbcX and pCT-rbcL-ccmM35 to obtain pCT-LSX and pCT-LSM vectors, respectively, which were used in the following chloroplast transformation procedure to replace the native rbcL gene with the cyanobacterial genes.
Generation of transplastomic tobacco plants
We used the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, Inc.) and a tissue-culture based selection method18. Two-week-old tobacco (Nicotiana tabacum cv. Samsun) seedlings germinated in sterile MS agar medium were bombarded with 0.6 μm gold particles carrying the appropriate chloroplast transformation vector. Two days later, the leaves were cut in half and put on RMOP agar plates containing 500 mg/l of spectinomycin and incubated for 4-6 weeks at 23 °C with 14 hours of light per day. The shoots arising from this medium were cut into small pieces of about 5 mm2 and subjected to the second round of regeneration in the same RMOP medium for about 4-6 weeks. The shoots from the second selection round were then transferred to MS agar medium containing 500 mg/l of spectinomycin for rooting and then to soil for growth in a greenhouse chamber with elevated atmospheric CO2.
DNA blot analyses of the rbcL locus of the chloroplast genome
We synthesized the digoxigenin(DIG)-sUTP-labeled DNA probe (bp56907-57411 of NCBI Reference Sequence: NC_001879.2) with PCR DIG Probe Synthesis Kit by Roche and SBprbfor-SBprbrev primer pair. The total DNA from leaf tissues were extracted with a standard CTAB-based procedure. The leaf tissues frozen in liquid nitrogen were finely ground in Eppendort tubes in 600 μl of 2x CTAB buffer (2% hexadecyltrimethyl ammonium bromide, 1.4 M sodium chloride, 20 mM EDTA, 100 mM Tris pH 8.0, 0.2% b-mercaptoethanol) and incubated at 65 °C for 1 hour. The DNA was extracted with 600 μl of chloroform containing 4% isopropanol. The DNA present in the upper layer transferred to a clean tube was precipitated with 0.8 volume of isopropanol at −70 °C for 1 hour and pelleted with a microcentrifuge. The DNA pellet was washed with 200 μl of 70% ethanol and air-dried before it was dissolved in 100 μl of double distilled water. After the DNA samples’ quality and concentration were determined by a NanoDrop method, 1 μg of each DNA sample was digested by NdeI and NheI restriction enzymes, and the digested fragments were separated on a 1% agarose gel. The DNA pieces in the gel were depurinated, denatured and then transferred and cross-linked to a Nylon membrane according to the manufacturer’s protocols. The DNA samples on the membrane blot were hybridized with the DIG-labeled probe, which was then detected with Anti-Digoxigenin-alkaline phosphatase antibody using CDP-star chemiluminescent substrate (Roche) according to the manufacturer’s specifications.
Analyses of the transcripts by RT-PCR
Total RNA was extracted from each leaf tissue sample with a standard TRIzol procedure. The leaf tissues frozen with liquid nitrogen were ground in 800 μl of trizol and incubated at 22 °C for 5 min. After the insoluble pieces were removed by centrifugation, 160 μl of chloroform was added to the supernatant, mixed vigorously for 15 seconds and incubated at 22 °C for 3 min. The two aqueous phases were separated in a centrifuged at 4 °C for 15 min and the upper layer transferred to a new tube was mixed with 500 μl of isopropanol. The sample was incubated at 22 °C for 10 min and centrifuged at 4 °C for 10 min. The pellet was resuspended in 800 μl of 75% ethanol and centrifuged again at 4 °C for 10 min. The pellet was air-dried and re-suspended in 50 μl of molecular biology grade water. The RNA samples were treated with DNase using Ambion DNA-free kit (Life Technologies) and the cDNA for each gene was generated with its corresponding reverse primer using Sensiscript® Reverse Transcription kit (Qiagen) according to the manufacturer’s protocols. The cDNA samples were amplified with the PCR master mix (Bioline) and analyzed in a 1% agarose gel.
SDS page, immunoblot and determination of CcmM35/Rubisco content
The crude leaf homogenates used in the carboxylase activity measurements were separated by SDS-PAGE using 4 - 20% polyacrylamide gradient gels (Thermo Scientific, UK). For each sample, the same amount of protein, as determined by Bradford assay, was loaded onto the gel. After electrophoresis, the resolved proteins were transferred to a nitrocellulose membrane (Hybond-C Extra from GE Healthcare Life Sciences) using a Western blot apparatus. The nitrocellulose membranes were immunoblotted using one of four primary polyclonal antibodies raised against: cyanobacterial (Se PCC6301) Rubisco; tobacco Rubisco; the small subunit of tobacco Rubisco; and CcmM from Se PCC7942. The primary polyclonal antibody to detect CcmM35 was generated in rabbit with His-tagged CcmM58 protein purified from E. coli (Cambridge Research Biochemicals, UK) and used at a dilution of 1:500 in the immunoblots and from 1:500 to 1:2,000 for immunogold labeling, and was highly specific for CcmM (Fig. 2a). The primary antibodies were visualized by means of a secondary, goat anti-rabbit, peroxidase-conjugated, antibody (Sigma).
The absolute and relative content of Synechococcus Rubisco and CcmM35 in SeLSM35 leaves were determined using immunoblots with antibodies against CcmM and cyanobacterial Rubisco. The amounts of Rubisco and CcmM35 present in crude leaf homogenates were estimated by comparison with authentic protein standards (purified CcmM35 and cyanobacterial Rubisco). Amounts of CcmM35 and cyanobacterial Rubisco (nmols dm−2) were the mean ± standard deviation for duplicate determinations. The band intensities were obtained using ImageJ software (NIH, USA) and the standard curves using Microsoft Excel.
Purification of cyanobacterial Rubisco and CcmM35 proteins
Synechococcus Rubisco was expressed in E.coli BL21 (DE3) cells using the vector pAn92 as previously described29. This material was harvested by centrifugation and resuspended in buffer containing 0.1M Bicine-NaOH pH 8.0, 20 mM MgCl2, 50 mM NaHCO3, 100 mM PMSF and bacterial protease inhibitor cocktail (Sigma). All steps in the purification were conducted at 0°C. The harvested cells were sonicated and cell debris removed by centrifugation (17,400xg, 20 min, 4°C). PEG-4000 and MgCl2 were added to the supernatant, giving final concentrations of 20 % (w/v) and 20 mM, respectively. After 30 min at 0°C, the precipitated Rubisco was sedimented by centrifugation (17,400xg, 20 min, 4°C) and the pellet resuspended in 25 mM triethanolamine (pH 7.8, HCl), 5 mM MgCl2, 0.5 mM EDTA, 1 mM ε-aminocaproic acid, 1 mM benzamidine, 12.5 % (v/v) glycerol, 2 mM DTT and 5 mM NaHCO3. This material was subjected to anion-exchange chromatography using a 5 mL HiTrap Q column (GE-Healthcare) pre-equilibrated with the same buffer. Rubisco was eluted with a 0 – 600 mM NaCl gradient in the same buffer. Fractions containing the most Rubisco activity (as judged by RuBP-dependent 14CO2 assimilation) were further purified and desalted by size-exclusion chromatography using a 20 cm × 2.6 cm dia. column of Sephacryl S-200 HR (GE-Healthcare) pre-equilibrated and developed with (50 mM Bicine-NaOH pH 8, 20 mM MgCl2, 0.2 mM EDTA, 2 mM DTT). The resulting protein peak was concentrated by ultrafiltration using 20 ml capacity /150 kDa cut off centrifugal concentrators (Thermo Pierce). The PCR-amplified ccmM35 gene from Se PCC7942 was cloned into pCR8/GW/TOPO TA vector (Life Technologies) and subsequently transferred to the Gateway pDEST17 E. coli expression vector (Life Technologies), which utilizes the T7 promoter to express the inserted gene and incorporates a 6xHis tag at the N-terminus of the translated protein. The expression vector was transformed into Rosetta (DE3) competent cells, and the protein expression was induced with 0.5 mM IPTG at OD600 of 0.5. The cells in 0.5 L LB culture were harvested after 4 hours of growth at 37 °C and 250 rpm. The cells were resuspended in about 10 ml of ice cold 50 mM sodium phosphate, 300 mM sodum chloride, 20 mM imidazole at pH 8.0 and broken with sonication. The cell debris were removed by centrifugation and the supernatant was mixed with 2 ml of Ni-NTA resin, which was then washed with 15 ml of the cell suspension buffer in a gravity-flow column and the bound protein was eluted with the buffer containing 200 mM imidazole. The purity of CcmM35 was assessed with SDS-PAGE, and its concentration was determined by the Bradford method.
Cryo-preparation of leaf material and transmission electron microscopy
Leaf material was cryo-fixed at a rate of 20,000 Kelvins /sec using a high pressure freezer unit (Leica Microsystems EM HPM100). The second step of freeze substitution of cryo-fixed samples was performed in an EM AFS unit (Leica Microsystems) at −85°C for 48 hours in 0.5% uranyl acetate in dry acetone. The samples were then infiltrated at low temperature in Lowicryl HM20 resin (Polysciences) and polymerized with a UV lamp.
For the immunogold labelling, gold grids carrying ultrathin sections (60 – 90 nm) of leaf tissue embedded in HM20 were treated using different rabbit primary antibodies against: cyanobacterial Rubisco from Se PCC6301; tobacco Rubisco; and CcmM35 (produced by Cambridge Research Biochemicals). A secondary goat polyclonal antibody to rabbit IgG conjugated with 10 nm gold particles (Abcam, UK) was used for the labeling.
Images were obtained using a transmission electron microscope (Jeol 2011 F) operating at 200kV, equipped with a Gatan Ultrascan CCD camera and a Gatan Dual Vision CCD camera.
Plant material and growing conditions
Both transgenic and wild-type Nicotiana tabacum var. Samsun NN were grown in the same controlled environment chamber. 16 hours of fluorescent light (43%) and 8 hours dark, at 24°C during the day and 22°C during the night. The humidity level was set at 70% during the day and 80% during the night. The atmospheric CO2 level was kept constant at 9000 ppm (air containing 0.9% v/v CO2).
Quantification of protein,Rubisco, and chlorophyll
Total soluble protein in the leaf homogenates was determined by the standard Bradford method. Rubisco active site concentration in the crude homogenate was determined using the [14C]-CABP binding assay23 or by quantifying LSU band intensity by immunoblotting. Each approach gave very similar results. Chlorophyll concentration was determined from crude homogenates by spectrophotometry30.
Carboxylase activity measurements
Leaf discs (1 cm2) were cut and promptly homogenized using an ice-cold pestle and mortar, in the presence of 500 μL of ice-cold extraction buffer (50 mM EPPS-NaOH pH8.0; 10 mM MgCl2; 1 mM EDTA; 1 mM EGTA; 50 mM 2-mercaptoethanol; 20 mM DTT; 20 mM NaHCO3; 2 mM Na2HPO4; Sigma plant protease inhibitor cocktail (diluted 1:100); 1 mM PMSF; 2 mM benzamidine; 5 mM ε-aminocaproic acid). Rubisco carboxylase activity was measured immediately in 500 μL of assay buffer containing 100 mM EPPS-NaOH pH8.0, 20 mM MgCl2, 0.8 mM RuBP and 10 mM, 20 mM or 50 mM NaH14CO3 (18.5 kBq. mol−1) at room temperature (22°C). The assay was initiated by the addition of 20 μL of the leaf homogenate, and was quenched after 2, 4, 6 or 10 minutes, by the addition of 100 μL of 10M formic acid. The samples were oven dried and the acid stable 14C determined by liquid scintillation counting, following residue rehydration (400 μL H2O) and the addition of 3.6 mL liquid scintillation cocktail (Ultima Gold, PerkinElmer, UK).
For Rubisco inhibition using the tight binding Rubisco inhibitor, 2-carboxy-D-arabinitol-1,5-bisphosphate (CABP), leaf homogenates were incubated on ice for 15 minutes in the presence of 50 μM CABP25. Residual carboxylase activity (if any) was then measured as described above.
Extended Data
Extended Data Figure 1. Rubisco and CcmM35 content of SeLSM35 tobacco leaves.
The stated concentrations of purified Se Rubisco (a) and CcmM35 (b) proteins were used as standards. a, Immunoblot using an antibody against cyanobacterial LSU (top) and the standard curve used to estimate the amount of cyanobacterial Rubisco in uk1-uk3 samples extracted from SeLSM35 tobacco leaves (bottom). b, Immunoblot using an antibody against CcmM (top) and the standard curve used to estimate the amount of CcmM35 in uk4-uk6 samples extracted from SeLSM35 tobacco leaves (bottom). The band intensities in the two standard curves were obtained with ImageJ software (NIH, USA) and the standard curves with Microsoft Excel. c, The absolute and relative amounts (mean ± standard deviation) of CcmM35 and cyanobacterial Rubisco in SeLSM35 tobacco line from two separate measurements. Each Rubisco holoenzyme is assumed to be composed of 8 LSU and an unknown quantity of SSU.
Extended Data Figure 2. Electron micrographs of ultrathin sections of leaf mesophyll cells from the chloroplast transformant SeLSM35 showing big compartments (black arrows) containing cyanobacterial Rubisco and CcmM35 in the chloroplast stroma.
Leaf tissues were prepared by high pressure freeze fixation (HPF) in combination with immunogold labeling using an antibody against CcmM. A secondary antibody conjugated with 10 nm gold particles was used for the labelling. Scale bars = 500 nm.
Extended Data Figure 3. Rubisco-specific 14CO2 fixation by crude leaf homogenates from tobacco lines expressing cyanobacterial Rubisco (SeLSX and SeLSM35) and wild type tobacco (wt).
a, Carboxylase activity assayed with [+] and without [−] RuBP. b, Carboxylase activity assayed with [+] and without [−] the inhibitor CABP. The rates of carboxylase activity (mols fixed. mol act sites−1. s−1) are the means ± standard deviation of the 2,4 & 10 min data obtained in assays at 125 μM steady-state [14CO2] (i.e. 10 mM NaH14CO3, at pH 8.0).
Table 1.
Rubisco, total soluble protein and chlorophyll content of the wild-type and transformed homoplastomic tobacco leaves of similar size, development and canopy position. The wild-type plants were grown in air and the transformants in air supplemented with 0.9% (v/v) or 9000 ppm CO2. Fresh 4 cm2 leaf samples were homogenised in (1 ml) ice-cold extraction buffer. The crude homogenate was used for determination of chlorophyll and Rubisco content. The total soluble protein was determined by the Bradford method following extract clarification (13,200xg, 5 min, 4°C). Values are means ± standard deviation from 3 different leaves per sample.
| Sample | Rubisco (mg/dm2) |
Total soluble protein (mg/dm2) |
Chlorophyll a & b (mg/dm2) |
|---|---|---|---|
| Wild-type | 9.06 ± 0.90 | 37.4 ± 0.60 | 3.17 ± 0.21 |
| SeLSX | 1.07 ± 0.14 | 18.5 ± 0.20 | 2.13 ± 0.08 |
| SeLSM35 | 1.64 ± 0.07 | 14.6 ± 0.90 | 1.75 ± 0.10 |
Table 2.
Oligonucleotides used in the construction of chloroplast transformation vectors, DNA blot analyses of the tobacco chloroplast rbcL locus and RT-PCR analyses of the tobacco chloroplast rbcL gene and transgenes introduced in the transplastomic lines. The restriction sites are underlined.
| Primers | Nucleotide sequences |
|---|---|
| F1OLrbcLfor | CATGAGTTGTAGGGAGGGATTTATGCCTAAAACCCAAAGTGCTG |
| 4RErbcLrev | ATAACGCGTCTGCAGGGCAGGCGGCCGCCGCGCGCGTTAAAGCTTATCCATTGTCTCAAA |
| F1for | GGCCCCCACTATGTGGACCTTGAACTACC |
| F1for2 | AGCTCGGGCCCCAAATAATGATT |
| F1rev | AAATCCCTCCCTACAACTCATG |
| F2for | ATGCCTGCAGATGCAGGTCGAGCATATGAAACAGTAGACATTAGCAGATAAATTAG |
| F2rev | TCCAACGCGTTGGAAATAATCAACATTACTGCAACTAGAATTG |
| SMOfor | CTATTGCTCCTTTCTTTTTCTGCAG |
| SMOrev | ATGCCTGCAGGATAACTTCGTATAGCATACATTATACG |
| TrbcLfor | AGATCGCGCGCGAAACAGTAGACATTAGCAGATAAATTAG |
| TrbcLrev | AGATGGGCCCTTCAAATCTTGTATATCTAGGTAAGTATATAC |
| IEESDrev | GTATATCTCCTTCTTGAGATCTGTTGACTTTGTATACCATTCCGTTGTAAATAAATGATC |
| IEESD18rev | CCCATATGTATATCTCCTTCTCCCATATGTATATCTCCTTCTTGAGATCTGTTGAC |
| SD18rev2 | CATGGGTATATCTCCTTCTCCCATATGTATATCTCCTTCTCCCATATGTA |
| TpetDAtfor | AGATGGGCCCACGCGTCGCGCGCGTTCAATTTATTCAATTGTAAAATAAACGACG |
| TpetDAtrev | CCATTCCGTTGTAAATAAATGATCTTAACCCATTTTAATTAATTAATTAAATTAATTAG |
| TpsbAAtfor | AGATGGGCCCACGCGTCGCGCGCGTTCGTTAGTGTTAGTCTAGATCTAG |
| TpsbAAtrev | CCATTCCGTTGTAAATAAATGATCTTAAATATGATACTCTATAAAAATTTGCTC |
| Trpsl6Atfor | AGATGGGCCCACGCGTCGCGCGCGAGTCTTACTAAAACGAAATGAAATTAATG |
| Trps16Atrev | CCATTCCGTTGTAAATAAATGATCTTACAAAATAAATATGATGGAAGTGAAAGAG |
| rbcSfor | GTCAACAGATCTCAAGAAGGAGATATACCCATGAGTATGAAAACCTTGCCAAAAG |
| rbcSrev | AGATGCGGCCGCACGCGTTTAATATCTTCCAGGTCGATGCAC |
| rbcXfor: | GTCAACAGATCTCAAGAAGGAGATATACCCATGGCGTCAACGCAGAGG |
| rbcXrev: | AGATGCGGCCGCACGCGTTCAATCCGCATGGGAGGCATTAG |
| M35for: | GTCAACAGATCTCAAGAAGGAGATATACCCATGAGCGCTTATAACGGCCAAGG |
| M35rev: | AGATGCGGCCGCACGCGTTTACGGCTTTTGAATCAACAGTTCAGC |
| aadAfor: | ATGGCTCGTGAAGCGGTTATCG |
| aadArev: | TTATTTGCCAACTACCTTAGTGATCTCG |
| SBprbfor: | ACCATGCAATTGAACCGATTCAATTG |
| SBprbrev: | TGTATACTCTTTCATATATATAGCGCAAC |
| Nt-rbcLfor: | ATGTCACCACAAACAGAGACTAAAG |
| Nt-rbcLrev: | TTACTTATCCAAAACGTCCACTGCTG |
Supplementary Material
Acknowledgements
We thank Cheryl Kerfeld (Michigan State University) for helpful discussion and providing us with the Se7942 genomic DNA and purified, His-tagged CcmM protein, WenFang Li (Cornell University) for technical assistance in generating, selecting and analyzing the tobacco chloroplast transformants and Muhammad Waqar Hameed (Cornell University) for the codon-optimized cyanobacterial Rubisco genes. This material is based upon work supported by the National Science Foundation under grant number EF-1105584 to M.R.H., Biotechnology and Biological Sciences Research Council under grant number BB/I024488/1 to M.A.J.P. and the National Institute of General Medical Sciences of the National Institutes of Health under award number F32GM103019 to M.T.L. P.J.A. and M.A.J.P. also acknowledge support from the 20:20 Wheat® Institute Strategic Program (BBSRC BB/J/00426X/1).
Footnotes
Online Content
Methods, along with any additional Extended Data display items and SourceData, are available in the online version of the paper; references unique to these sections appear only in the online paper.
References
- 1.Andersson I, Backlund A. Structure and function of Rubisco. Plant Physiol. Biochem. 2008;46:275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Whitney SM, Houtz RL, Alonso H. Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011;155:27–35. doi: 10.1104/pp.110.164814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parry MAJ, et al. Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 2013;64:717–730. doi: 10.1093/jxb/ers336. [DOI] [PubMed] [Google Scholar]
- 4.Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 2013;64:787–798. doi: 10.1093/jxb/ers294. [DOI] [PubMed] [Google Scholar]
- 5.McGrath JM, Long SP. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol. 2014;164:2247–2261. doi: 10.1104/pp.113.232611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price GD, et al. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 2013;64:753–768. doi: 10.1093/jxb/ers257. [DOI] [PubMed] [Google Scholar]
- 7.Whitney SM, Baldet P, Hudson GS, Andrews TJ. Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J. 2001;26:535–547. doi: 10.1046/j.1365-313x.2001.01056.x. [DOI] [PubMed] [Google Scholar]
- 8.Kanevski I, Maliga P, Rhoades DF, Gutteridge S. Plastome engineering of ribulose-1,5-bisphosphate carboxylase/oxygenase in tobacco to form a sunflower large subunit and tobacco small subunit hybrid. Plant Physiol. 1999;119:133–141. doi: 10.1104/pp.119.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saschenbrecker S, et al. Structure and function of RbcX, an assembly chaperone for hexadecameric rubisco. Cell. 2007;129:1189–1200. doi: 10.1016/j.cell.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 10.Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- 11.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell. 2013;155:1131–1140. doi: 10.1016/j.cell.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 12.Chen AH, Robinson-Mosher A, Savage DF, Silver PA, Polka JK. The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PLoS One. 2013;8:e76127. doi: 10.1371/journal.pone.0076127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parry MAJ, Andralojc PJ, Mitchell RAC, Madgwick PJ, Keys AJ. Manipulation of Rubisco: the amount, activity, function and regulation. J. Exp. Bot. 2003;54:1321–1333. doi: 10.1093/jxb/erg141. [DOI] [PubMed] [Google Scholar]
- 14.Zhu XG, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
- 15.Dhingra A, Portis AR, Daniell H. Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6315–6320. doi: 10.1073/pnas.0400981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Caemmerer S, Quick WP, Furbank RT. The development of C4 rice: current progress and future challenges. Science. 2012;336:1671–1672. doi: 10.1126/science.1220177. [DOI] [PubMed] [Google Scholar]
- 17.Whitney SM, Andrews TJ. Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14738–14743. doi: 10.1073/pnas.261417298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maliga P, Tungsuchat-Huang T. Plastid transformation in Nicotiana tabacum and Nicotiana sylvestris by biolistic DNA delivery to leaves. Methods Mol. Biol. 2014;1132:147–163. doi: 10.1007/978-1-62703-995-6_8. [DOI] [PubMed] [Google Scholar]
- 19.Zhou F, Karcher D, Bock R. Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 2007;52:961–972. doi: 10.1111/j.1365-313X.2007.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drechsel O, Bock R. Selection of Shine-Dalgarno sequences in plastids. Nucleic Acids Res. 2011;39:1427–1438. doi: 10.1093/nar/gkq978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long BM, Rae BD, Badger MR, Price GD. Over-expression of the beta-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth. Res. 2011;109:33–45. doi: 10.1007/s11120-011-9659-8. [DOI] [PubMed] [Google Scholar]
- 22.Long BM, Tucker L, Badger MR, Price GD. Functional cyanobacterial b-Carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 2010;153:285–293. doi: 10.1104/pp.110.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokota A, Canvin DT. Ribulose bisphosphate carboxylase/oxygenase content determined with [14C]carboxypentitol bisphosphate in plants and algae. Plant Physiol. 1985;77:735–739. doi: 10.1104/pp.77.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller-Cajar O, Whitney SM. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 2008;414:205–214. doi: 10.1042/BJ20080668. [DOI] [PubMed] [Google Scholar]
- 25.Parry MAJ, Keys AJ, Madgwick PJ, Carmo-Silva AE, Andralojc PJ. Rubisco regulation: a role for inhibitors. J. Exp. Bot. 2008;59:1569–1580. doi: 10.1093/jxb/ern084. [DOI] [PubMed] [Google Scholar]
- 26.Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM. RbcX can function as a Rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell Physiol. 2006;47:1630–1640. doi: 10.1093/pcp/pcl028. [DOI] [PubMed] [Google Scholar]
- 27.Lin MT, et al. β-carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 2014;79:1–12. doi: 10.1111/tpj.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray BN, Yang H, Ahner BA, Hanson MR. An efficient downstream box fusion allows high-level accumulation of active bacterial beta-glucosidase in tobacco chloroplasts. Plant Mol. Biol. 2011;76:345–355. doi: 10.1007/s11103-011-9743-7. [DOI] [PubMed] [Google Scholar]
- 29.Bainbridge G, et al. Engineering rubisco to change its catalytic properties. J. Exp. Bot. 1995;46:1269–1276. [Google Scholar]
- 30.Wintermans JF, de Mots A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta. 1965;109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.