Abstract
While hydrophobic small molecules often can freely permeate a lipid bilayer, ions and other polar molecules cannot and require transporters to mediate their transport. Recently, a number of important structures have been reported which have advanced our understanding of how membrane protein transporters function to transport small molecules. Structures of TbpA/B and HmuUV provided new insight into iron uptake by pathogenic bacteria while the structures of NarK, ASBT, and VcINDY revealed molecular details about the transport of nitrate, bile acids and dicarboxylates, respectively. The structure of the folate ECF transporter indicated that the S component likely undergoes a large conformational shift to mediate folate transport, while the cellulose synthase/transporter contains an elongated translocation pore for passage through the inner membrane.
Small molecule transport systems
The transport of small molecules across membranes is essential for the import of nutrients and other energy sources into the cell and for the export of waste and other potentially harmful byproducts out of the cell [1–3]. While hydrophobic molecules are permeable to membranes, ions and other small polar molecules require transport via specialized membrane transport proteins. The two major classes of membrane transport proteins are carrier proteins (transporters) and channel proteins [4] (Figure 1A). With our focus here on transporters, we will briefly highlight some recent structural biology reports that have substantially contributed to our understanding of the various mechanisms that mediate the transport of small molecules across membranes. We also discuss the structure of the cellulose synthase/transporter since its substrates can vary significantly in size. The studies discussed here represent only a fraction of the exciting structures that have been reported recently, all of which collectively advance our overall understanding of these small molecule transport systems.
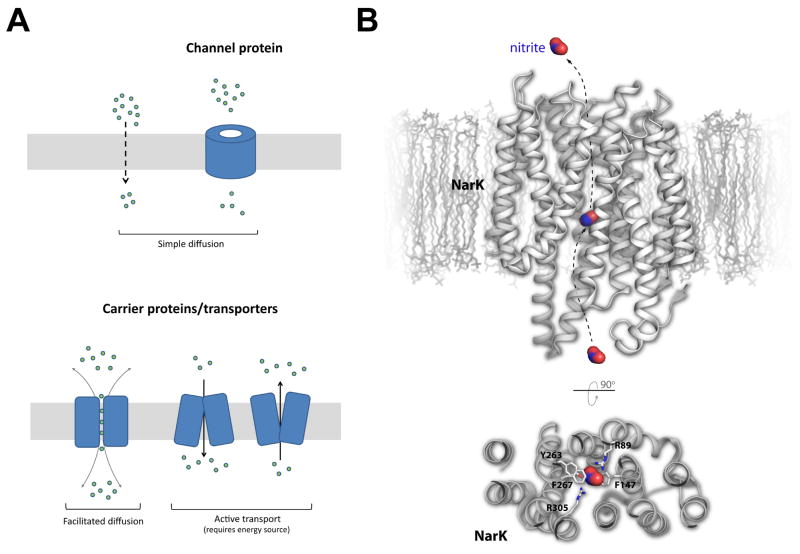
Figure 1. Small molecule transport systems and the structure of NarK.
A. The two major classes of membrane transport proteins are carrier proteins (transporters) and channel proteins. The focus of this review is on the protein transporters which require energy for function. B. The structure of NarK, a nitrate/nitrite exchanger, is shown in gray within a membrane bilayer (top). A cutaway view from the periplasmic side is also shown (bottom) illustrating the interactions of NarK residues with the bound nitrite molecule, which is shown in red/blue surface. The proposed exchange pathway is indicated by dashed arrows (nitrate import is in the opposite direction as shown for nitrite export).
Nitrate/nitrite transport by NarK in E. coli
Nitrate (NO3−) is a key source of mineral nitrogen uptake for bacteria. However, nitrogen metabolism leads to an abundance of cellular nitrite (NO2−), which can eventually lead to the accumulation of nitric oxide, a cytotoxic free radial that can lead to DNA damage and degradation of iron-sulfur centers [5,6]. The exact mechanism for how bacteria manage nitrate import versus nitrite export has remained elusive until recently with the report of the crystal structure of the nitrate/nitrite transporter NarK from E. coli [7]s. NarK belongs to the major facilitator superfamily (MFS) of secondary transporters, containing a conserved twelve transmembrane helices structure consisting of two structurally conserved N- and C-terminal subdomains of six transmembrane helices each related by a pseudo twofold symmetry. The NarK structure was solved in apo form and in complex with sodium nitrite, with both structures found in the inward-facing conformation. The putative substrate transport pathway is formed along the interface between the two subdomains (Figure 1B). Based on the electrostatics of the substrate translocation pathway and the lack of classical proton transporting residues (Glu, Asp, and/or His), it was concluded that NarK is in fact a nitrate/nitrite exchanger which relies on a cyclic rocker switch mechanism, allowing nitrate binding (and nitrite release) in the outward facing conformation and nitrate release (and nitrite binding) in the inward facing conformation [7].
Iron transport in Neisseria meningitidis
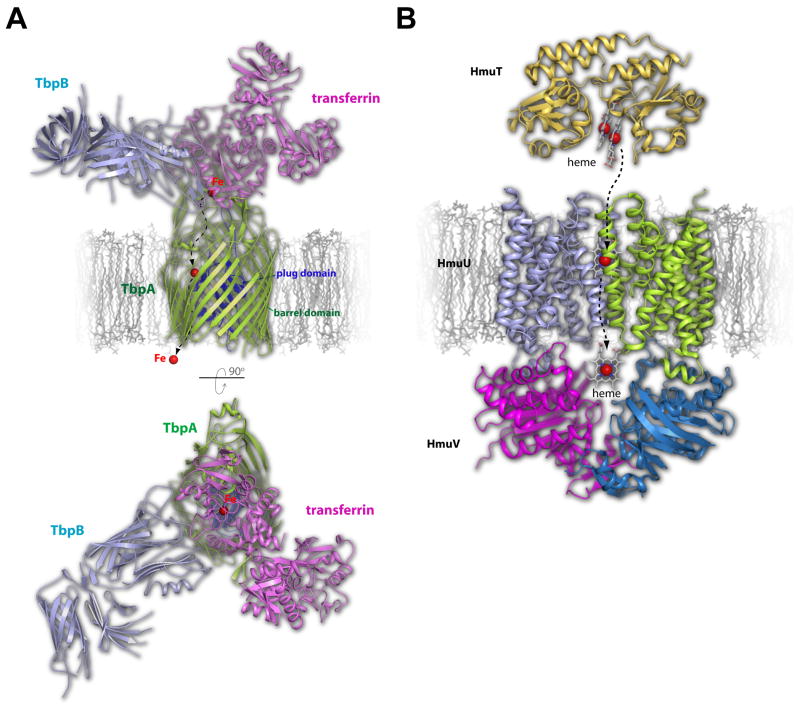
Iron is essential for survival and therefore pathogenic bacteria have developed complex machineries to scavenge iron from the host environment or even steal iron directly from host proteins [8]. For example, Neisseria meningitidis has specialized surface receptors that have evolved to acquire iron from specific host sources, such as heme, lactoferrin, and transferrin (Tf) [9–12]. Recently, the crystal structures of the Neisserial Tf-binding proteins A (TbpA, a TonB-dependent transporter) and B (TbpB, a lipoprotein co-receptor) were each reported in complex with human Tf, providing molecular insight into how Neisseria are able to steal iron from Tf and import it across the outer membrane for survival and virulence [10,11]. TbpA, which binds both apo and diferric Tf, consists of a large N-terminal plug domain tucked inside a 22-stranded β-barrel transmembrane domain, while TbpB, which only bind to diferric Tf, is anchored to the surface of the cell via an N-terminal lipid anchor and consists of two structurally similar domains, each containing an eight stranded β-barrel with an adjacent four stranded β-rich handle domain. Tf binding to TbpA and TbpB was mediated exclusively along the C-lobe of Tf at non-overlapping binding sites (Figure 2A). Identification of a conserved lysine residue found on the helix finger in loop 3 of TbpA provided a clue suggesting that TbpA itself may be hijacking the pH sensor of Tf, which is typically utilized to mediate iron release at low pH within the human host, to catalytically trigger iron release for import through the barrel domain of TbpA.
Figure 2. Structural insights into iron transport by TbpA/B and HmuUV in pathogenic bacteria.
A. Iron (red sphere) is extracted from transferrin (magenta) by a concerted effort from TbpA (green), a TonB-dependent transporter, and its lipoprotein co-receptor TbpB (cyan), both found in the outer membrane. Based on the crystal structures of transferrin with TbpA and TbpB separately, a model is shown here of the TbpA-TbpB-transferrin triple complex. The proposed pathway for iron import is indicated by dashed arrows. A view in the membrane is shown on top while a view from the surface is shown on the bottom. B. Heme can also be utilized as an iron source which is mediated by the inner membrane protein HmuUV, which is composed of two membrane integrated HmuU subunits (cyan and green) and two periplasmic nucleotide exchange subunits called HmuV (magenta and blue). The periplasmic heme carrier protein, HmuT (gold), delivers heme to HmuUV for import which follows the proposed pathway indicated by the dashed arrows. Heme is shown in stick and iron as a red sphere.
The structure of HmuUV, a heme transporter from Y. pestis
As discussed earlier, iron is essential for survival and therefore pathogenic bacteria have evolved systems to acquire iron from the host environment. Another such iron source that is targeted is heme, which is the most abundant source of iron in the human body [13]. While numerous studies have described how heme may be captured and imported across the outer membrane by TonB-dependent transporters [14], less is known about how heme makes its way across the inner membrane into the cytoplasm. Recently, the crystal structure of the heme transporter HmuUV from Yersinia pestis was reported and found to belong to the type II ABC transporters/importers, consisting of two copies of each component HmuU (transmembrane domain) and HmuV (nucleotide-binding domain) (HmuU2V2) and related by a twofold symmetry axis [15] (Figure 2B). Each HmuU subunit contained ten transmembrane helices with HmuV interacting non-covalently via a conserved cytoplasmic coupling helix. Unlike type I ABC transporters, an outward facing conformation was observed in the absence of nucleotide [16,17]. A cavity, which is open to the periplasm but closed to the cytoplasm, was found at the interface between the two HmuU subunits, revealing the site where HmuT, the periplasmic heme-escort protein, interacts with HmuUV for heme exchange and subsequent translocation [18]. Mutagenesis of the cavity forming transmembrane helix 5 of HmuU and functional analysis using a novel in vitro heme transport assay, allowed further characterization of heme transport and the heme translocation pathway, with the conclusion that while type I and type II ABC transporters may be conserved structurally, their mechanisms for translocating substrate may vary significantly [15].
Structural and mechanistic insight into bile acid transporters
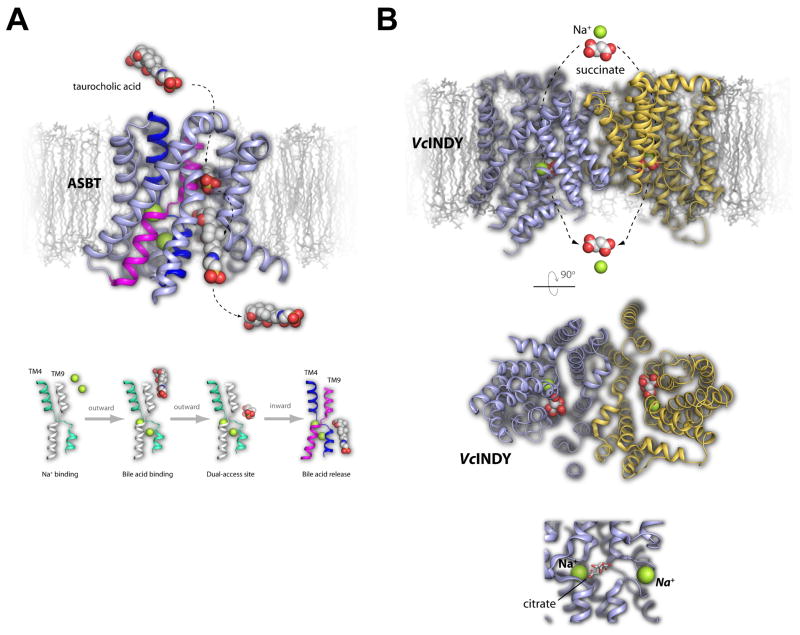
Bile acids serve essential functions in digestion and in the absorption of lipids and fat-soluble vitamins in the small intestine [19]. Via enterohepatic recirculation, the bile acids eventually cycle back to the liver for resecretion, a process known to involve two Na+-dependent transporters, one of which is called the apical sodium-dependent bile acid transporter (ASBT) [20,21]. Inhibition of ASBT has been shown to increase bile acid synthesis which leads to an increase in cholesterol consumption, making it an ideal drug target against elevated cholesterol levels [22]. Recently, the crystal structures of ASBT homologues from Neisseria meningitidis (ASBTNm) and Yersinia frederiksenii (ASBTYf) were reported providing valuable structural insight into the transport of bile acids [23,24]. The ASBT structure contains ten transmembrane helices which form two subdomains termed the panel domain (TM1, 2, 6 and 7) and the core domain (TM3–5, and 8–10) and both ASBTNm, which contained two sodium ions and taurocholic acid (TCA, bile acid substrate), and ASBTYf, in apo form, were found in both an inward-open conformation (Figure 3A). However, an outward-open conformation was also solved of ASBTYf by mutating a highly conserved glutamate (Glu254) found at one of the sodium binding sites, allowing a comparison of the two conformational states and identification of a an important helical crossover region (along TM4 and 9) within the core domain [23,24]. Together, structural and functional studies led to the conclusion that bile acid transport may be mediated by a putative dual-accessible binding site along the crossover region, which may undergo localized conformational changes in response to sodium binding/release [23,24].
Figure 3. The transport of bile acids by ASBT and dicarboxylates by an INDY homologue dicarboxylate transporter.
A. Bile acids such as taurocholic acid (spheres) are transported by the ASBT transporter (top, light blue/magenta/dark blue). The structure led to the suggested mechanism where taurocholic acid binds ASBT in the outward facing conformation, undergoes a horizontal positioning along a putative dual-access binding site, and is then imported and released following a conformational change to the inward facing conformation. The pathway for import is indicated by dashed arrows. This has been proposed to be dependent on Na+ binding and requires slight changes in both TM4 and TM9. B. VcINDY (top, blue/gold) is a dicarboxylate transporter that was solved in the presence of citrate (red/gray spheres) and Na+ (green spheres). The import pathway is indicated by dashed arrows, with a periplasmic view of the structure shown in the middle panel. The bottom panel is a zoomed view of the citrate binding pocket along with the observed Na+ site (left, green sphere) and the putative Na+ binding site (right, green sphere), both of which are important for transport.
The crystal structure of a bacterial INDY homologue from V. cholerae
Tricarboxylates such as citrate and dicarboxylates such as succinate and fumarate serve many essential roles within the cell including the synthesis of fatty acids and cholesterol and the regulation and generation of energy [25,26]. Import of these small molecules relies on direct import into the cytoplasm by specialized Na+-dependent transporters, all of which belong to the divalent anion/Na+ symporter (DASS) family and includes the homologous fly protein INDY, as well as, several bacterial homologs [27,28]. While transport is known to be Na+ driven with 2–4 Na+ ions co-transported per each substrate molecule, the exact mechanism for specificity and transport has remained largely unknown. However, the crystal structure of an INDY homologue from Vibrio cholera (VcINDY) was recently reported in complex with Na+ and citrate, providing insight into the transport mechanism of this family of transporters [29]. While VcINDY was found to function as a dicarboxylate transporter, early experiments showed it could be significantly thermostabilized by the presence of citrate, which contributed to crystallization and structure determination. VcINDY is a dimer representing an inward-facing conformation with each subunit consisting of eleven transmembrane helices and the interface between the subunits being formed by TM3, TM4a and TM9b (subunit A) and TM4b, TM8 and TM9a (subunit B) (Figure 3B). Each monomer consists of structurally conserved N- and C-terminal subdomains which are related by an inverted twofold symmetry parallel to the membrane plane. One Na+ ion and one citrate molecule was bound to each monomer within a cleft along the cytoplasmic face. Identification of a putative second Na+ binding site and subsequent mutagenesis and functional studies provided the necessary insight to postulate a mechanism for dicarboxylate transport by VcINDY whereby Na+ binding is coupled to substrate binding (outward-facing conformation) which triggers a conformational switch driving substrate transport across the membrane (inward-facing conformation) and followed by Na+ and substrate release into the cytoplasm [29].
The structure of the folate ECF transporter from L. brevis
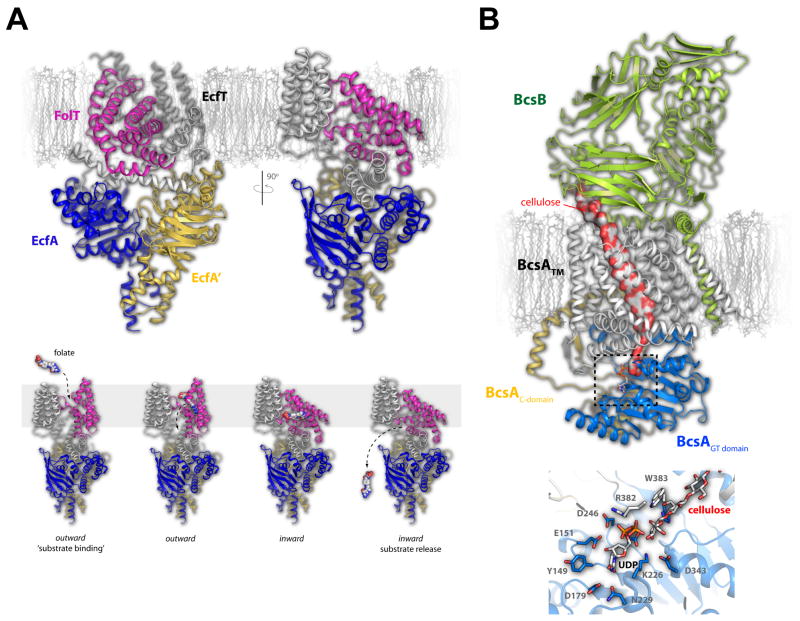
Energy-coupling factor (ECF) transporters are a new class of ABC transporters that consist of four components including the membrane spanning components (1) energy coupling module (EcfT) and (2) substrate-specific binding S protein (EcfS), and (3 and 4) two nucleotide-binding proteins (EcfA and EcfA’) [30–32]. ECF transporters lack solute-binding proteins and have different subunit organization than classical ABC transporters [33]. To better understand this new family of transporters, the crystal structure of a fully assembled folate ECF-transporter complex from Lactobacillus brevis was recently reported [34]. The structure lacked nucleotides or substrates and was found in an inward-facing conformation, providing a first look at the overall organization and interactions of the components within the complex. The folate-specific binding protein FolT consists of a six helix bundle that sits nearly horizontal within the membrane and interacts almost exclusively with EcfT, which contains five transmembrane helices along with three cytoplasmic helices which are directly coupled to the nucleotide-binding proteins EcfA and EcfA’[34–36] (Figure 4A). Based on the structure, a working model was proposed where in an outward-facing conformation, FolT adopts a conformation perpendicular to the membrane where it can bind folate. ATP hydrolysis then triggers a conformational switch within the EcfA and EcfA’ subunits that is relayed to FolT and EcfT via the coupling helices, reorienting the FolT domain nearly horizontal to the membrane and resulting in the inward-facing conformation for folate release [34].
Figure 4. Folate transport by the ECF transporter and the structure of the cellulose synthase/transporter complex.
A. The fully assembled folate ECF transporter is shown in the inward facing conformation with the FolT (magenta), EcfT (gray), EcfA (blue), and EcfA’ (gold) subunits show in cartoon (top). The substrate-specific subunit, FolT, was found nearly parallel with the membrane masking the substrate binding site, leading to the proposed mechanism where this subunit must undergo a dramatic conformational shift to allow folate to bind and be transported across the membrane (bottom). B. The cellulose transporter is shown here with BcsA in gray (transmembrane domain), blue (glycosyltransferase domain), and gold (C-terminal domain) and the BcsB subunit in green. Cellulose is shown as a continuous surface (red/gray) while UDP is shown in stick. The dashed box indicates the region depicted at the bottom which is the site of glycosyltransferase activity and cellulose formation. Cellulose, UDP, and the interacting residues are shown in stick.
The crystal structure of the BcsA/B cellulose transporter complex from R. sphaeroides
Bacterial biofilms and plant cell walls are composed of cellulose, the most abundant organic polymer, which consists of a linear chain of β-1,4-linked D- glucose subunits that can range from a few hundred to many thousands of units long [37,38]. It is known that bacteria secrete cellulose using machinery found in both the inner membrane (BcsA and BcsB) and the outer membrane (BcsC), however, the mechanism for synthesis and secretion has remained unknown [39–41]. While BcsC is predicted to be an 18-stranded βbarrel membrane protein, BcsA is an α-helical membrane protein that forms a complex with BcsB, which is anchored to the periplasmic face of the inner membrane via a single C-terminal transmembrane helix. Recently, the crystal structure of the BcsA/BcsB complex from Rhodabacter sphaeroides was reported in an intermediate state during synthesis and translocation containing bound cellulose and UDP [42].
Containing a single copy of each component, the complex measured ~60 Å in width by ~150 Å in height with the interface burying ~4500 Å2 of surface area. BcsA consists of eight transmembrane helices with a large cytoplasmic domain forming the conserved glycosyltransferase (GT) domain, while BcsB has an overall dome-shape consisting of primarily β-strands with its transmembrane helix interacting directly with the membrane embedded helices of BcsA. Within the interior of the complex was found a cellulose fragment containing approximately 18 glucose molecules extending from the center of the GT domain, where a UDP molecule was also found, to the BcsB subunit of the complex (Figure 4B). The structural analysis of the BcsA/BcsB complex provided the molecular insight to propose a model for cellulose synthesis and translocation where only one glucose subunit is added at a time with rotational steric constraints (following glucose transfer) and/or binding of a new donor UDP-glucose molecule facilitating translocation across the membrane [42].
Conclusions
Structure determination of small molecule membrane protein transporters has benefited significantly in the past decade from improved methodologies in sample preparation (new contructs, thermostability mutagenesis, improved expression systems/protocols), crystallization (bicelles, lipic cubic phase, fusion tags), data collection (microfocus beams, FEL sources, rastering and vector collection methods, SONICC, improved detectors), and structure determination (SAD phasing, software automation, improved algorithms). It is often the case that crystals of these transporters are being solved at resolutions between 3 – 4 angstroms on a routine basis. Even with these tools available, structure determination of these membrane protein transporters is still far from being trivial and almost always requires vast amounts of energy, resources, support, and perseverance. Further, while the structures being reported are significantly advancing our overall knowledge of the various mechanisms that these membrane protein transporters use for their function, most of the targets are from bacterial sources with a clear lack of crystal structures representing targets from higher eukaryotes. While bacterial homologs can provide clues towards function and mechanism, membrane protein transporters from eukaryotic systems are often more complex and merit their own structure determination in order to fully understand their mechanisms. As technology and methodologies continue to improve in NMR, crystallography [43] and high resolution electron microscopy [44,45 ], the anticipation is that structure determination of more difficult targets from eukaryotes, such as large multi-component complexes, will provide a more detailed understanding of small molecule transport across membranes.
Highlights.
NarK, a nitrate/nitrite exchanger, uses a rocker switch mechanism for transport
Translocation mechanisms for type I and type II ABC transporters can vary
Na+ binding along the crossover domain of ASBT mediates bile acid transport
The VcINDY transporter requires prior Na+ binding to assist substrate binding
A large shift of the S component of ECF transporters mediates substrate transport
Acknowledgments
NN and SKB are supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
(•) of special interest (••) of outstanding interest
- 1••.Bulut H, Ma Q, Moniot S, Saenger W, Schneider E, Vahedi-Faridi A. Crystal structures of receptors involved in small molecule transport across membranes. Eur J Cell Biol. 2012;91:318–325. doi: 10.1016/j.ejcb.2011.12.004. This review provides an overview of the protein crystallography relating to transporters and highlights and discusses a number of ABC transporters. [DOI] [PubMed] [Google Scholar]
- 2.Gale PA. From anion receptors to transporters. Acc Chem Res. 2011;44:216–226. doi: 10.1021/ar100134p. [DOI] [PubMed] [Google Scholar]
- 3.Koepsell H. The SLC22 family with transporters of organic cations, anions and zwitterions. Mol Aspects Med. 2013;34:413–435. doi: 10.1016/j.mam.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Alberts B. Molecular biology of the cell. 4. New York: Garland Science; 2002. [Google Scholar]
- 5.Martinez-Espinosa RM, Cole JA, Richardson DJ, Watmough NJ. Enzymology and ecology of the nitrogen cycle. Biochem Soc Trans. 2011;39:175–178. doi: 10.1042/BST0390175. [DOI] [PubMed] [Google Scholar]
- 6.Einsle O, Kroneck PM. Structural basis of denitrification. Biol Chem. 2004;385:875–883. doi: 10.1515/BC.2004.115. [DOI] [PubMed] [Google Scholar]
- 7.Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497:647–651. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins HL. The role of iron in infections with intracellular bacteria. Immunol Lett. 2003;85:193–195. doi: 10.1016/s0165-2478(02)00229-8. [DOI] [PubMed] [Google Scholar]
- 9.Noinaj N, Buchanan SK, Cornelissen CN. The transferrin-iron import system from pathogenic Neisseria species. Mol Microbiol. 2012;86:246–257. doi: 10.1111/mmi.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calmettes C, Alcantara J, Yu RH, Schryvers AB, Moraes TF. The structural basis of transferrin sequestration by transferrin-binding protein B. Nat Struct Mol Biol. 2012;19:358–360. doi: 10.1038/nsmb.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noinaj N, Easley NC, Oke M, Mizuno N, Gumbart J, Boura E, Steere AN, Zak O, Aisen P, Tajkhorshid E, et al. Structural basis for iron piracy by pathogenic Neisseria. Nature. 2012;483:53–58. doi: 10.1038/nature10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noinaj N, Cornelissen CN, Buchanan SK. Structural insight into the lactoferrin receptors from pathogenic Neisseria. J Struct Biol. 2013;184:83–92. doi: 10.1016/j.jsb.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Wilks A, Burkhard KA. Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep. 2007;24:511–522. doi: 10.1039/b604193k. The authors present a thorough review describing the battle between bacteria and their host for heme as an iron source for survival and virulence. [DOI] [PubMed] [Google Scholar]
- 14.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo JS, Zeltina A, Goetz BA, Locher KP. X-ray structure of the Yersinia pestis heme transporter HmuUV. Nat Struct Mol Biol. 2012;19:1310–1315. doi: 10.1038/nsmb.2417. [DOI] [PubMed] [Google Scholar]
- 16.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 17.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 18.Mattle D, Zeltina A, Woo JS, Goetz BA, Locher KP. Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J Mol Biol. 2010;404:220–231. doi: 10.1016/j.jmb.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Dawson PA. Role of the intestinal bile acid transporters in bile acid and drug disposition. Handb Exp Pharmacol. 2011:169–203. doi: 10.1007/978-3-642-14541-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Claro da Silva T, Polli JE, Swaan PW. The solute carrier family 10 (SLC10): beyond bile acid transport. Mol Aspects Med. 2013;34:252–269. doi: 10.1016/j.mam.2012.07.004. This review provides a comprehensive overview of the family of carrier proteins that are involved in bil acid transport and the diseases and therapies associated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alrefai WA, Gill RK. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm Res. 2007;24:1803–1823. doi: 10.1007/s11095-007-9289-1. [DOI] [PubMed] [Google Scholar]
- 22.West KL, Ramjiganesh T, Roy S, Keller BT, Fernandez ML. 1-[4-[4[(4R,5R)-3,3-Dibutyl-7-(dimethylamino)-2,3,4,5-tetrahydro-4-hydroxy-1,1-di oxido-1-benzothiepin-5-yl]phenoxy]butyl]-4-aza-1-azoniabicyclo[2.2. 2]octane methanesulfonate (SC-435), an ileal apical sodium-codependent bile acid transporter inhibitor alters hepatic cholesterol metabolism and lowers plasma low-density lipoprotein-cholesterol concentrations in guinea pigs. J Pharmacol Exp Ther. 2002;303:293–299. doi: 10.1124/jpet.102.038711. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X, Levin EJ, Pan Y, McCoy JG, Sharma R, Kloss B, Bruni R, Quick M, Zhou M. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature. 2014;505:569–573. doi: 10.1038/nature12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011;478:408–411. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer AF, Lowenstein JM. The supply of precursors for the synthesis of fatty acids. J Biol Chem. 1962;237:3640–3648. [PubMed] [Google Scholar]
- 26.Vance DE, Vance JE. Biochemistry of lipids, lipoproteins and membranes. 5. Amsterdam; Boston: Elsevier; 2008. [Google Scholar]
- 27.Pajor AM. Molecular properties of the SLC13 family of dicarboxylate and sulfate transporters. Pflugers Arch. 2006;451:597–605. doi: 10.1007/s00424-005-1487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prakash S, Cooper G, Singhi S, Saier MH., Jr The ion transporter superfamily. Biochim Biophys Acta. 2003;1618:79–92. doi: 10.1016/j.bbamem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Mancusso R, Gregorio GG, Liu Q, Wang DN. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature. 2012;491:622–626. doi: 10.1038/nature11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, et al. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol. 2009;191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.ter Beek J, Duurkens RH, Erkens GB, Slotboom DJ. Quaternary structure and functional unit of energy coupling factor (ECF)-type transporters. J Biol Chem. 2011;286:5471–5475. doi: 10.1074/jbc.M110.199224. This work contributes significantly to what is currently known about the overall organization of ECF transporters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Erkens GB, Berntsson RP, Fulyani F, Majsnerowska M, Vujicic-Zagar A, Ter Beek J, Poolman B, Slotboom DJ. The structural basis of modularity in ECF-type ABC transporters. Nat Struct Mol Biol. 2011;18:755–760. doi: 10.1038/nsmb.2073. Here, the authors report the crystal structure of the S component ThiT which allows them to propose a mechanism for how this ECF transport systems functions in thiamine uptake. [DOI] [PubMed] [Google Scholar]
- 33.Eitinger T, Rodionov DA, Grote M, Schneider E. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol Rev. 2011;35:3–67. doi: 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 34.Xu K, Zhang M, Zhao Q, Yu F, Guo H, Wang C, He F, Ding J, Zhang P. Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature. 2013;497:268–271. doi: 10.1038/nature12046. [DOI] [PubMed] [Google Scholar]
- 35.Zhang P, Wang J, Shi Y. Structure and mechanism of the S component of a bacterial ECF transporter. Nature. 2010;468:717–720. doi: 10.1038/nature09488. [DOI] [PubMed] [Google Scholar]
- 36•.Song J, Ji C, Zhang JZ. Unveiling the gating mechanism of ECF Transporter RibU. Sci Rep. 2013;3:3566. doi: 10.1038/srep03566. The authors here use computation methods to study the conformational changes of the S component RibU within an ECF transporter system to determine what changes are necessary for function in riboflavin transport. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 38.Nishiyama Y, Langan P, Chanzy H. Crystal structure and hydrogen-bonding system in cellulose Ibeta from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc. 2002;124:9074–9082. doi: 10.1021/ja0257319. [DOI] [PubMed] [Google Scholar]
- 39.Romling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 40.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu SQ, Gao YG, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, et al. Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci U S A. 2010;107:17957–17961. doi: 10.1073/pnas.1000601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. Published in the same issue with the high resolution cryo-EM structure, the authors characterize the conformational dynamics of the TRPV1 channel and correlate these back to its function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. This work reports the structure of the TRPV1 channel to 3.4 angstroms resolution using single-particle electron cryomicroscopy, a significant accomplishment for a sample this small. [DOI] [PMC free article] [PubMed] [Google Scholar]