Summary
Intermolecular RNA-RNA interactions are used by many noncoding RNAs (ncRNAs) to achieve their diverse functions. To aid in identifying these contacts, we developed a method based on RNA Antisense Purification to systematically map RNA-RNA interactions (RAP-RNA) and applied it to investigate two ncRNAs implicated in RNA processing: U1 snRNA, a component of the spliceosome, and Malat1, a lncRNA that localizes to nuclear speckles. U1 and Malat1 interact with nascent transcripts through distinct targeting mechanisms. Using differential crosslinking, we confirmed that U1 directly hybridizes to both 5’ splice sites and 5’-splice-site motifs throughout introns and found that Malat1 interacts with pre-mRNAs indirectly through protein intermediates. Interactions with nascent pre-mRNAs cause U1 and Malat1 to localize proximally to chromatin at active genes, demonstrating that ncRNAs can use RNA-RNA interactions to target specific pre-mRNAs and genomic sites. RAP-RNA is sensitive to lower abundance RNAs as well, making it generally applicable for investigating ncRNAs.
Introduction
Mammalian genomes encode thousands of noncoding RNAs (ncRNAs), many of which are implicated in diverse biological processes (Cabili et al., 2011; Carninci et al., 2005; Derrien et al., 2012; Guttman et al., 2009; 2010). While functional studies have begun to elucidate the cellular or organismal roles of many ncRNAs (Grote et al., 2013; Guttman et al., 2011; Sauvageau et al., 2013), the molecular mechanisms by which they accomplish these functions are harder to characterize. One general approach to gain insights into these mechanisms is to identify the cellular components with which ncRNAs interact, including proteins, DNA sites, and RNAs. Notably, many classical ncRNAs interact with other RNAs either directly through base-pairing (e.g., microRNA-mRNA or snRNA-pre-mRNA hybridization) or indirectly through protein intermediates (e.g., the multiple ncRNA components of the ribosome) (Figure S1A). In addition to these examples, numerous large ncRNAs (lncRNAs) associate with proteins that regulate RNA processing (Änkö et al., 2012; Tollervey et al., 2011; Wang et al., 2010), suggesting that they may target other RNAs as part of their regulatory function. These observations suggest that RNA-RNA interactions may represent a general strategy used by many ncRNAs and that comprehensively mapping these interactions may provide insight into ncRNA function and mechanism.
Recent RNA-centric biochemical purification techniques, such as RNA Antisense Purification (RAP) (Engreitz et al., 2013), have enabled the comprehensive mapping of RNA-DNA interactions in vivo (Chu et al., 2011; Engreitz et al., 2013; Simon et al., 2011). Despite progress in mapping RNA-DNA interactions, mapping RNA-RNA interactions remains challenging. Classical RNA-RNA interactions, such as U1 snRNA interactions with pre-mRNAs, were identified through observations of sequence complementarity (Lerner et al., 1980) followed by targeted genetics and in vitro affinity experiments (Mount et al., 1983; Zhuang and Weiner, 1986). Recently, computational and experimental methods have been developed to search more systematically for RNA-RNA interactions (Dieterich and Stadler, 2013; Helwak et al., 2013; Kretz et al., 2013; Kudla et al., 2011). However, these methods do not identify or distinguish between direct and indirect RNA-RNA interactions and have limited resolution for studying a specific RNA.
To address this challenge, we developed a general method based on RAP to identify the intermolecular RNA-RNA interactions of a target RNA (RAP-RNA). RAP-RNA identifies endogenous RNA-RNA complexes through in vivo crosslinking, RNA capture with antisense oligonucleotides, and high-throughput RNA sequencing. This approach provides a systematic view of other RNAs that interact with an RNA of interest, and furthermore can distinguish between direct and indirect RNA-RNA interactions through the use of crosslinking reagents with different specificity for proteins and nucleic acids.
To demonstrate the utility of this approach, we investigate the RNA-RNA and RNA-chromatin interactions of two ncRNAs implicated in RNA processing: U1 snRNA and Malat1 lncRNA. U1 is a core component of the spliceosome that makes direct base-pair contacts with 5’ splice sites (Mount et al., 1983; Zhuang and Weiner, 1986) and has also recently been reported to prevent premature cleavage and polyadenylation (PCPA) of nascent transcripts through interactions outside of 5’ splice sites (Kaida et al., 2010). Malat1 is a highly conserved lncRNA that localizes to nuclear speckles (Hutchinson et al., 2007) and interacts with multiple serine/arginine (SR) RNA splicing proteins (Änkö et al., 2012; Miyagawa et al., 2012; Sanford et al., 2009; Tripathi et al., 2010). While these data suggest that Malat1 may regulate RNA processing, the exact molecular function of Malat1 remains elusive (Gutschner et al., 2013).
Here, we show that U1 and Malat1 interact with many nascent pre-mRNAs through distinct mechanisms: U1 directly hybridizes to 5’ splice sites and at similar binding motifs throughout introns, whereas Malat1 interacts with pre-mRNAs indirectly via protein intermediates. These RNA-RNA interactions cause U1 and Malat1 to localize to chromatin at active genes. Both RNAs localize to DNA in a manner dependent on transcription, but U1 also contacts the 5’ end of active genes in the absence of active transcription. Our findings demonstrate that mapping RNA-RNA interactions provides insight into the regulatory roles of ncRNAs and reveal a strategy by which lncRNAs can localize to specific genomic sites.
Results
We set out to adapt RAP to identify direct and indirect RNA-RNA interactions. To this end, we developed three related protocols: RAP-RNA[AMT], RAP-RNA[FA], and RAP-RNA[FA-DSG] (Figure S1B). In the RAP-RNA[AMT] protocol, we fixed direct RNA-RNA hybrids in mouse embryonic stem (ES) cells with 4’-aminomethyltrioxalen (AMT), a psoralen-derivative crosslinker; AMT generates inter-strand crosslinks between uridine bases in RNA but does not react with proteins (Calvet and Pederson, 1979; Cimino et al., 1985). In the RAP-RNA[FA] protocol, we used a different crosslinking strategy to capture both direct and indirect RNA-RNA interactions: we fixed ES cells using formaldehyde (FA), which crosslinks protein-RNA and protein-protein interactions and thus can capture both indirect interactions as well as direct interactions that are caged or flanked by proteins. In the RAP-RNA[FA-DSG] protocol, we fixed with both FA and disuccinimidyl glutarate (DSG), a strong protein-protein crosslinker, to more efficiently capture RNAs linked indirectly through multiple protein intermediates (Figure S1B). In each protocol, we purified target RNAs with biotinylated antisense oligonucleotides and sequenced co-purifying RNAs (Experimental Procedures, Table S1).
RAP-RNA Captures U1 snRNA in Complex with 5’ Splice Sites
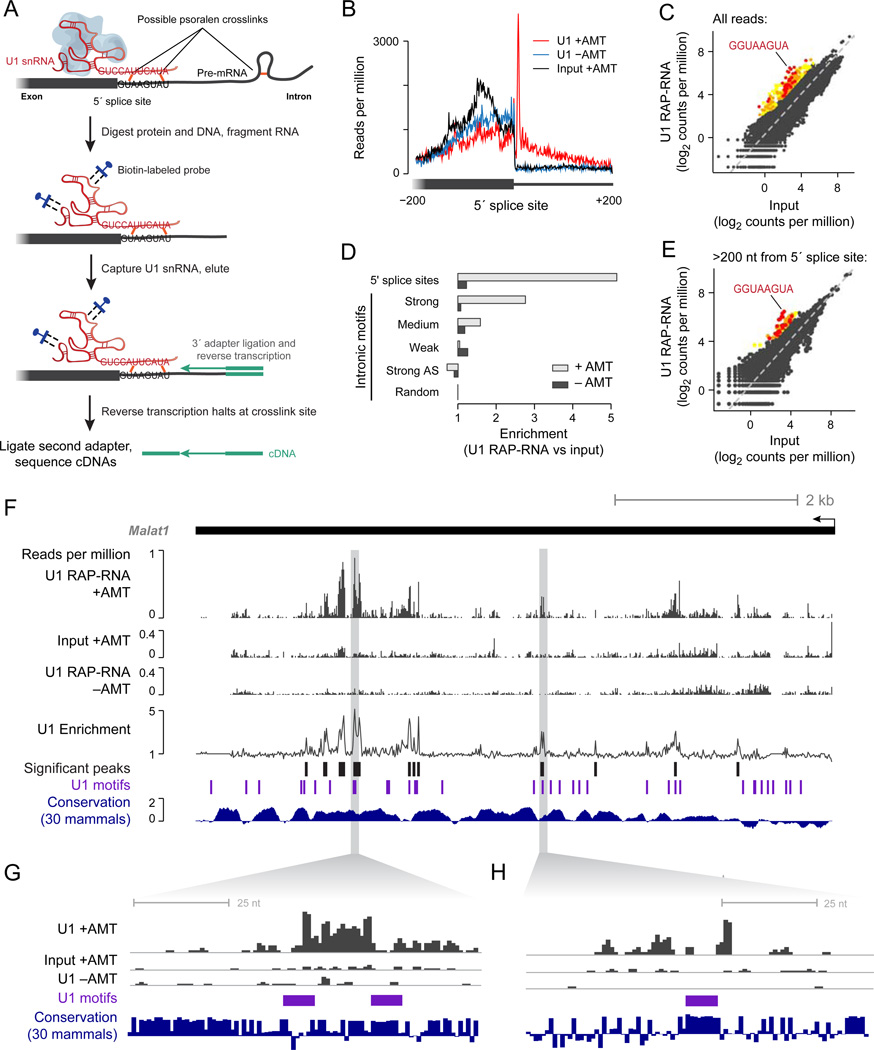
We first developed RAP-RNA[AMT] to identify direct RNA-RNA interactions at high resolution (Figure S1). To test this approach, we purified U1 snRNA, a core component of the spliceosome that makes base-pair contacts with pre-mRNAs at 5’ splice sites (Figure 1A). RAP-RNA[AMT] enriched for U1 by a factor of 54 versus input and strongly enriched for intronic sequences located 8 nucleotides (nt) downstream of 5’ splice sites (Figure 1B), precisely where an AMT crosslink would be expected to occur in a canonical U1-pre-mRNA interaction (Figure 1A). This enrichment extended 200 nt into the intron, likely caused by intramolecular crosslinks in the pre-mRNA that blocked reverse transcription before it reached the 5’ splice site (5’ss) (Figure 1A). To test whether RAP-RNA[AMT] could specifically detect these interactions de novo without prior knowledge of 5’ splice sites, we looked for enriched 8-mers in the pre-mRNA sequence upstream of each sequencing read: the most enriched 8-mer exactly matched the consensus 5’ss motif (17-fold enrichment versus input, Figure 1C). To confirm that RAP-RNA[AMT] did not capture indirect RNA-RNA interactions, we examined the enrichment of U2 snRNA, which indirectly interacts with U1 as part of the spliceosome: U2 was not enriched (0.93-fold versus input). Thus, RAP-RNA[AMT] can accurately and specifically identify RNA-RNA interactions mediated by direct hybridization.
Figure 1. U1 Binds Throughout Nascent Transcripts at 5’-Splice-Site Motifs.
(A) Schematic diagram of U1 RAP-RNA[AMT]. AMT forms covalent crosslinks (orange) between opposing uridine bases in the U1 snRNA (red) and target pre-mRNA (gray). Consensus sequences show canonical base-pairing interaction. To avoid capturing indirectly associated RNAs, we digested protein and DNA before purifying U1 with biotinylated antisense probes (blue). To map RNA-RNA interaction sites at high resolution, we fragmented RNA prior to capture and performed reverse transcription (RT) without reversal of crosslinks, leading to complementary DNA (cDNA, purple) that terminates at or near the site of a crosslink (see also Figure S1). Ligation of a second adapter to the 3’ end of the cDNA enabled sequencing and mapping the positions of RT termination.
(B) Read counts aggregated over all 5’ splice sites. Each read-pair contributes a count in the base corresponding to the 5’ end of the original RNA fragment. Cells were crosslinked with AMT (+AMT, red) or mock-crosslinked with DMSO (−AMT, blue). Crosslinked input RNA (black) is shown for comparison.
(C) Enrichment for every 8-mer RNA motif close to RAP-RNA sequencing reads. Colored dots represent 8-mers that are significantly enriched in U1 RAP-RNA versus input (P < 0.001 after Bonferroni correction, enrichment >= 4). Red, orange, and yellow dots correspond to 8-mers that have a Levenshtein edit distance from the 8-mer consensus motif of 1, 2, or >2, respectively.
(D) Enrichment for sequencing reads in U1 RAP-RNA[AMT] versus input at 5’ splice sites and at 5’ss motif matches >200 bases away from exons. Enrichments are normalized to the enrichment at random intronic sites >200 bases away from exons. 5’ss motif matches were classified as strong, medium, or weak as previously described (Almada et al., 2013). Strong AS = strong motifs on the antisense strand, which should not be bound by U1.
(E) Same as (C), but considering only reads that map to introns >200 bases away from exons.
(F) U1 RAP-RNA[AMT] coverage and enrichment across the Malat1 transcript (10-nt resolution), representing the ratio of U1 RAP +AMT and the maximum of U1 RAP −AMT and Input +AMT. Black bars represent significantly enriched windows (P < 0.001 after Bonferroni correction). Conservation represents the phyloP30wayPlacental track from the UCSC Genome Browser.
(G) and (H) Zoom-in on two significant U1 binding sites. Scales on y-axes are the same as in (F).
U1 snRNA Binds Throughout Nascent Transcripts at 5’-Splice-Site Motifs
In addition to its well-characterized role in splicing, U1 has also recently been implicated in preventing premature cleavage and polyadenylation (PCPA) of nascent transcripts. In particular, previous studies have reported that 5’ss mutations alter the location of transcription termination and that loss-of-function of U1 leads to premature termination of many pre-mRNAs (Kaida et al., 2010). These observations led to the hypothesis that U1 blocks PCPA by binding to pre-mRNAs at many locations in addition to 5’ splice sites. However, where U1 localizes on transcripts outside of 5’ splice sites and whether the same binding motif is responsible for this interaction remain unknown.
To address this question, we used RAP-RNA[AMT] to determine whether U1 interacts throughout nascent transcripts. Indeed, U1 RAP-RNA[AMT] enriched for 5’ss motifs in introns, with a level of enrichment that correlated with the strength of the motif match (Figure 1D). When we analyzed all U1 RAP-RNA[AMT] reads mapping within introns, the most enriched 8-mer near these reads exactly matched the 5’ss motif (14-fold enrichment, Figure 1E). To confirm that U1 interacts with these motifs independent of splicing, we examined U1 interactions with the single-exon Malat1 lncRNA, a 7-kilobase (kb) unspliced transcript that might require multiple exonic U1 interaction sites to block PCPA (Figure 1F–H, Experimental Procedures). Indeed, U1 RAP-RNA[AMT] enriched for numerous sites on Malat1, several of which occurred near conserved 5’ss motifs (e.g., Figure 1G). Because U1 co-localizes with Malat1 in nuclear speckles (Spector and Lamond, 2011), we note that these interactions might occur with both nascent Malat1 transcripts and mature transcripts that have been released from chromatin. Together, these results indicate that U1 binds directly to the 5’ss motif at many sites throughout introns and unspliced transcripts.
RAP-RNA Captures Direct and Indirect RNA-RNA Interactions
We next explored whether capturing both direct and indirect RNA-RNA interactions would yield broader information about RNA-RNA interactions. In particular, we aimed to use RAP-RNA to study lncRNAs whose molecular mechanisms are unknown and thus may interact with other RNAs indirectly. Accordingly, we developed RAP-RNA[FA-DSG] based on our previously published RAP-DNA protocol, using FA and DSG to crosslink molecular complexes that include both nucleic acids and proteins.
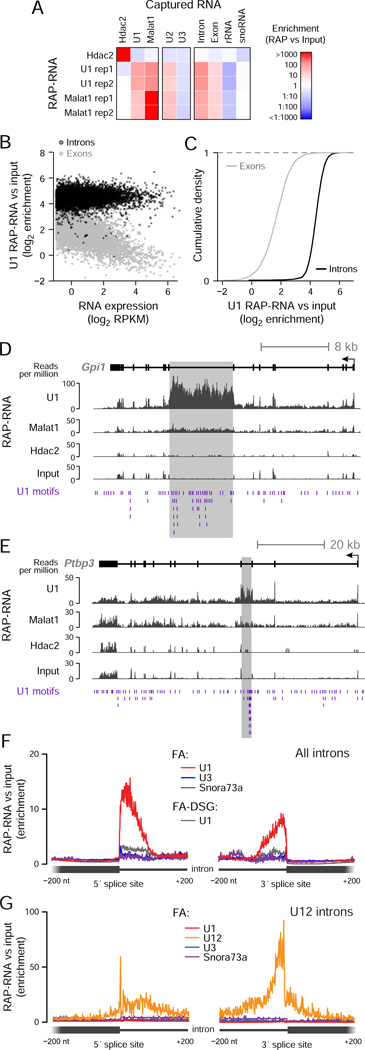
To test RAP-RNA[FA-DSG], we purified U1 complexes from ES cells crosslinked with FA and DSG. U1 RAP-RNA[FA-DSG] enriched for U2 snRNA (4-fold versus input) and Malat1 (20-fold versus input), but did not enrich for abundant RNAs, such as U3 snoRNA, that should not interact with U1 (Figure 2A). Consistent with the role of U1 in pre-mRNA splicing, U1 RAP-RNA[FA-DSG] enriched more strongly for introns than exons across nearly all genes in the genome (Figure 2B,C, Table S2). When we examined individual genes, U1 enrichment extended broadly across entire transcripts and showed only weak enrichment at 5’ splice sites (Figure 2D–F), likely because of the strong protein-protein crosslinking and large RNA fragment sizes (~1000 nt) in RAP-RNA[FA-DSG] (Figure S1B). Despite this, some of the most strongly enriched introns showed a very high density of U1 motifs (Figure 2D,E). Introns across the genome with the highest density of U1 motifs (95th percentile, >30.7 motifs per kb) had on average 14% stronger U1 RAP-RNA[FA-DSG] enrichment than other introns (P < 10−15, Mann-Whitney test). Together, these data show that RAP-RNA[FA-DSG] accurately identifies transcripts that interact both directly and indirectly with U1.
Figure 2. RAP-RNA Captures Both Direct and Indirect RNA-RNA Interactions.
(A) Enrichment for individual transcripts or classes of RNAs. RAP-RNA[FA-DSG] experiments include two replicate U1 and Malat1 purifications (rep1 and rep2) and one Hdac2 purification (negative control).
(B) Each point represents the average U1 RAP-RNA[FA-DSG] enrichment across all introns (black) or exons (gray) for one gene. The x-axis represents the expression of the mature transcript (exons) in input nuclear-enriched RNA in reads per kilobase per million (RPKM).
(C) Comparison of U1 RAP-RNA[FA-DSG] enrichment for the exons (gray) and introns (black) of all genes.
(D) RAP-RNA[FA-DSG] sequencing reads mapping to Gpi1 and (E) Ptbp3, two of the most highly enriched transcripts in U1 RAP-RNA[FA-DSG] (>100-fold versus input). Gray rectangles highlight enriched introns with many U1 motifs (purple).
(F) Enrichment of sequencing reads in RAP-RNA[FA] versus input aggregated across 5’ and 3’ splice sites for all introns (n > 180,000) or (G) U12 introns (n = 588). U1 RAP-RNA[FA] (red) more strongly enriches splice sites than U1 RAP-RNA[FA-DSG] (gray). Each read-pair contributes a count at the position corresponding to the 3’ end of the original RNA fragment for the 5’-splice-site panel and the 5’ end for the 3’-splice-site panel. See also Figure S2.
To map specific binding sites within interacting transcripts at higher resolution while capturing both direct and indirect interactions, we employed a third method, RAP-RNA[FA], which uses FA crosslinking without DSG and stronger RNA fragmentation prior to capture (~150 nt) (Figure S1B). Applying the RAP-RNA[FA] approach to U1, we observed strong enrichment at intronic sites immediately adjacent to 5’ splice sites (Figure 2F). In contrast to the AMT approach, U1 RAP-RNA[FA] also enriched for intronic sites adjacent to 3’ splice sites, which interact indirectly with U1 upon formation of the spliceosome complex. Because U1 binds nearly all nascent transcripts, we confirmed the specificity of the RAP-RNA[FA] approach by purifying U12 snRNA, a component of the minor spliceosome that processes a more limited class of ~500 introns with alternative splice-site recognition sequences (“U12 introns”). U12 RAP-RNA[FA] specifically enriched the 5’ and 3’ splice sites of U12 introns, while U1 RAP-RNA[FA] did not (Figure 2G). Thus RAP-RNA[FA] incorporates the strengths of both the RAP-RNA[AMT] and RAP-RNA[FA-DSG] methods: high resolution for interaction sites and the ability to identify both direct and indirect interactions.
To test the generality of RAP-RNA to study lower-abundance ncRNAs, we purified the snoRNAs U3 and Snora73a/U17a, which directly hybridize to specific sites on the pre-ribosomal 45S transcript to guide pre-rRNA processing. U3 and Snora73a are ~5- and 200-fold less abundant than U1, respectively (Figure S2A). RAP-RNA[FA] and RAP-RNA[AMT] correctly identified known interactions between these snoRNAs and 45S, including four sites for U3 and one site for Snora73a (Figure S2B,C). Interestingly, U3 RAP-RNA[FA] identified additional, previously unknown binding sites on the 45S pre-rRNA (Figure S2B), suggesting that U3 may form both direct and indirect contacts with specific sites on the ribosomal RNA precursor.
Together, these results highlight the utility of RAP-RNA for exploring different types of RNA-RNA interactions, with each of the three RAP-RNA protocols providing distinct but complementary information (Figure S1B). The AMT protocol provides information about direct RNA-RNA interactions at high resolution, but does not identify indirect interactions and is limited to direct interactions that contain nucleotides that can be crosslinked by psoralens. The FA protocol provides high resolution for both direct and indirect interactions using a broadly applicable crosslinker, but may not capture indirect interactions involving multiple protein intermediates. Finally, the FA-DSG protocol does not map binding sites at high resolution, but provides the best approach for identifying RNA transcripts that interact indirectly through multiple protein intermediates. More generally, the RAP-RNA method can be combined with additional crosslinking reagents or protocols to fix direct or indirect RNA-RNA interactions.
Malat1 Interacts with Pre-mRNAs Encoding RNA-Binding Proteins
Having developed three general methods for examining RNA-RNA interactions, we next used them to study Malat1, a highly conserved lncRNA that localizes to nuclear speckles (Hutchinson et al., 2007), interacts with multiple SR splicing proteins (Änkö et al., 2012; Miyagawa et al., 2012; Sanford et al., 2009; Tripathi et al., 2010) and can regulate gene expression and alternative splicing in some contexts (Eißmann et al., 2012; Nakagawa et al., 2012; Tripathi et al., 2010; 2013; Zhang et al., 2012). Although these data suggest that Malat1 may regulate RNA processing, it remains unclear whether this occurs indirectly through global modulation of SR protein activity (Tripathi et al., 2010) or whether Malat1 may itself interact with pre-mRNAs to more directly guide RNA processing. To test this, we purified Malat1 using RAP-RNA[AMT], RAP-RNA[FA], and RAP-RNA[FA-DSG], and achieved >1000-fold enrichment for Malat1 compared to input in all experiments (Figure 2A).
We first examined RAP-RNA[AMT] data for evidence of direct Malat1 interactions with other RNAs. Consistent with our finding that RAP-RNA[AMT] for U1 enriches for Malat1, RAP-RNA[AMT] for Malat1 significantly enriched for U1 snRNA in crosslinked (7-fold versus crosslinked input) but not mock-crosslinked RNA (0.06-fold versus mock-crosslinked input). We were unable to find any other enriched RNAs or sequence motifs using this method (Figure S3A). This suggests that either Malat1 does not interact directly with other RNAs (besides U1) through specific RNA elements or that these interactions do not contain the sequences required for psoralen crosslinking.
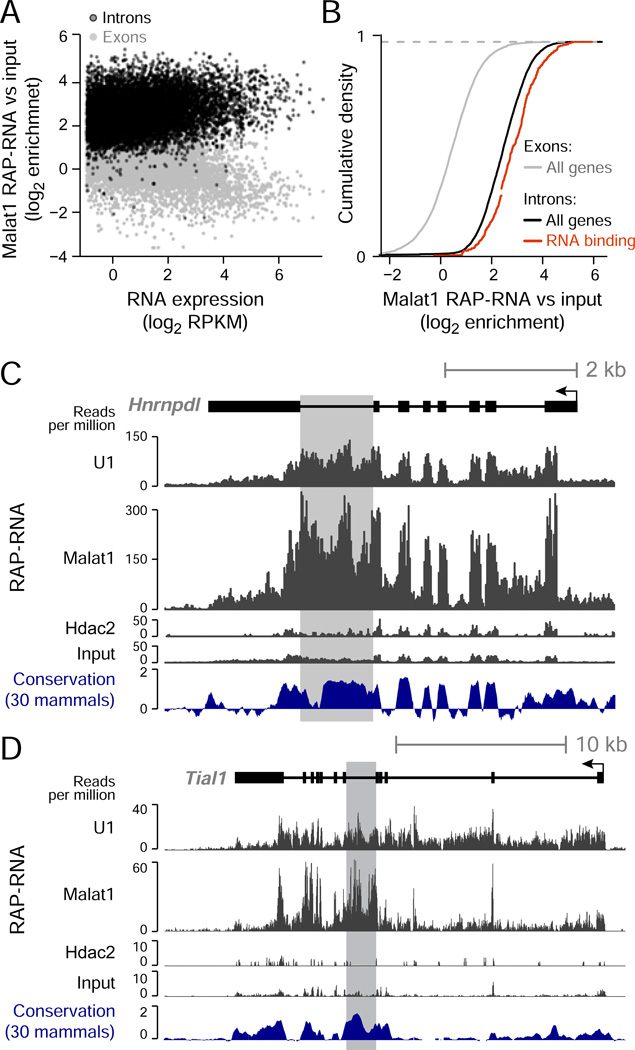
Accordingly, we focused on Malat1 interactions identified by the most inclusive protocol, RAP-RNA[FA-DSG]. Consistent with the localization of Malat1 in nuclear speckles (Spector and Lamond, 2011), we observed strong enrichments for both snRNAs and mRNAs (>5-fold), which are also present in nuclear speckles (Figure 2A). In contrast, Malat1 RAP-RNA[FA-DSG] strongly depleted other abundant RNAs, such as ribosomal RNAs (>4-fold), that are not localized in nuclear speckles. Because Malat1 associates with SR splicing proteins, we next asked whether Malat1 interacts with nascent pre-mRNAs. Indeed, introns showed stronger enrichment than coding exons (average 5.7-fold versus 1.6-fold, respectively) across the transcriptome (Figure 3A,B). We confirmed that this enrichment for introns versus exons did not result from differences in GC-content, which might affect crosslinking efficiency (Figure S3B). Notably, Malat1 RAP-RNA[FA] showed only weak enrichment for introns and snRNAs compared to Malat1 RAP-RNA[FA-DSG], suggesting that Malat1 may interact with these RNAs indirectly through multiple protein intermediates that are more efficiently crosslinked with DSG (Figure S1B).
Figure 3. Malat1 Interacts Indirectly with Nascent Transcripts Encoding RNA-Binding Proteins.
(A) Malat1 RAP-RNA[FA-DSG] enrichment at exons and introns, similar to Figure 2B.
(B) Comparison of Malat1 RAP-RNA[FA-DSG] enrichment for the exons (gray) and introns (black) of all genes or of significant gene sets. See also Figure S3.
(C) Malat1 RAP-RNA[FA-DSG] sequencing reads mapping to Hnrnpdl and (D) Tial1, two highly enriched genes encoding RNA-binding proteins. Gray rectangles highlight conserved introns containing RNA elements that control alternative splicing.
We next examined genes with the strongest enrichment (>14-fold versus input) in Malat1 RAP-RNA[FA-DSG] (Table S3). This gene set was enriched for transcripts encoding proteins involved in RNA binding and splicing (FDR < 0.001, Figure 3B, Figure S3C). Amongst these highly enriched genes, we observed several remarkable examples where the strongest enrichment occurred in introns near alternatively spliced exons (e.g., Figure 3C,D). These highly-enriched introns contained conserved noncoding elements that are known to control the generation of unproductive splice isoforms (i.e., isoforms containing premature stop codons that are subject to nonsense-mediated decay), a frequent mode of auto-regulation for RNA-binding proteins in which the protein regulates the alternative splicing of its own transcript (Lareau et al., 2007; Ni et al., 2007; Saltzman et al., 2008). Based on these examples, we explored whether Malat1 more generally enriches for alternatively spliced transcripts. Indeed, the introns of genes that have alternatively spliced isoforms were significantly more enriched for Malat1 binding than introns of genes that do not have alternative isoforms (22% higher enrichment, P < 10−16, Mann-Whitney test).
To determine whether these patterns were unique to Malat1, we directly compared the RNA interactions of Malat1 and U1. We ranked nascent transcripts based on their enrichment in Malat1 RAP-RNA[FA-DSG] versus U1 RAP-RNA[FA-DSG] and found that the most enriched transcripts were similarly enriched for genes encoding RNA-binding proteins (P < 0.001, Figure S3D,E). To further demonstrate that Malat1 had a unique pattern of RNA-RNA interactions that did not reflect nonspecific binding to other nuclear transcripts, we purified Xist, another abundant nuclear lncRNA. While Xist RAP-RNA[FA-DSG] strongly purified Xist, it did not show significant enrichment for nascent pre-mRNAs (1.03-fold, P = 0.17, Mann-Whitney test, Figure S3F). Together, these results demonstrate that Malat1 interactions are distinct from those of other nuclear RNAs. Thus, Malat1 specifically interacts with many pre-mRNAs and most strongly enriches for alternatively spliced transcripts, including those that encode RNA-binding proteins.
Malat1 Localizes to Chromatin at Active, Spliced Genes
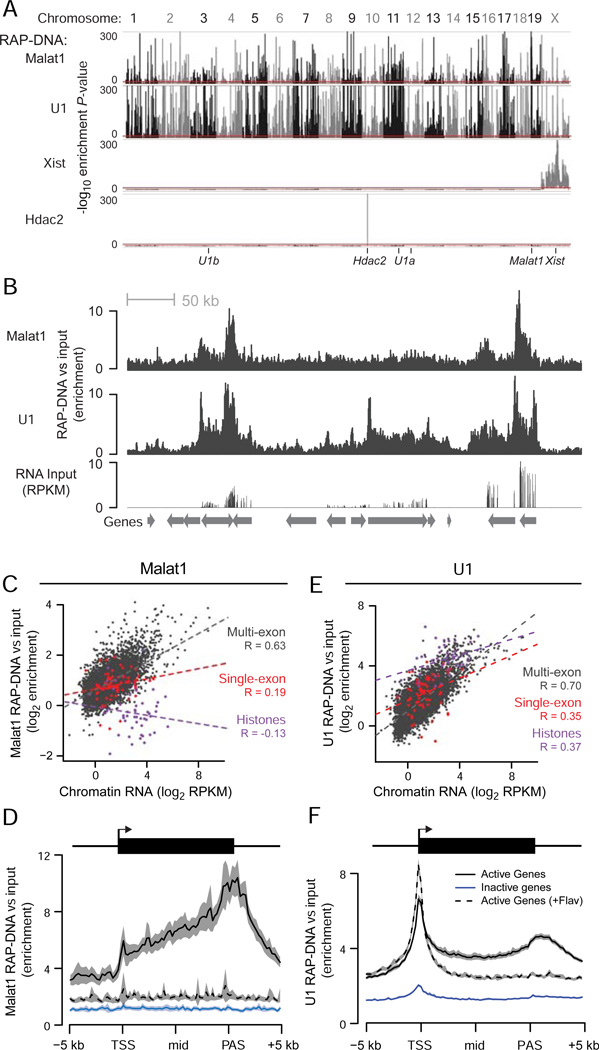
Because U1 and Malat1 both interact with nascent transcripts, we next explored the relationship between RNA-RNA interactions and RNA localization to chromatin. To map RNA-DNA interactions, we isolated and sequenced DNA that co-purified with Malat1 in cells crosslinked with FA and DSG (RAP-DNA). In a sliding window analysis at 10-kb resolution, Malat1 RAP-DNA significantly enriched for 4,177 sites across the genome (Figure 4A, Figure S4A). As negative controls, we performed the same analysis for Xist, which should only localize to the X-chromosome, and Hdac2 mRNA, which as a coding transcript is unlikely to interact with chromatin outside its own locus on chromosome 10. As expected, Xist RAP-DNA did not enrich for any regions outside of the X-chromosome and Hdac2 RAP-DNA did not enrich for any regions outside of the Hdac2 locus (Figure 4A), demonstrating the specificity of the RAP method and confirming that Malat1 localizes to thousands of sites across the genome.
Figure 4. Chromatin Localization of U1 and Malat1.
(A) Significance of enrichment across the genome in 10-kb windows for Malat1, U1, Xist, and Hdac2 RAP-DNA. Y-axis represents uncorrected binomial P-values, and the red lines mark the cutoff for genome-wide significance (P < 10−10). Ticks at the bottom denote the locations of the genes encoding each RNA. U1 is encoded on chromosomes 3, 11, and 12.
(B) RAP-DNA enrichment versus input across a region containing several active genes (chr18: 34,313,699–35,191,532). RNA sequencing of input total RNA shows relative gene expression.
(C) Scatterplot shows Malat1 RAP-DNA enrichment versus levels of chromatin-associated RNA for single-exon histone genes (purple), single-exon non-histone genes (red), and all other genes (dark gray). Dashed lines represent linear regressions. R = Pearson’s correlation.
(D) RAP-DNA enrichment averaged over active genes (black), inactive genes (blue), and active genes in cells treated with flavopiridol (black dashed line). Shaded regions represent 95% confidence intervals for the average enrichment. For Malat1, the averages represent the 5% of active genes with the highest Malat1 enrichment and an equal number of randomly selected inactive genes.
(E) Same as (C) for U1 RAP-DNA.
(F) Same as (D) for U1 RAP-DNA. For U1, average enrichments include all active and inactive genes. Notable peaks occur at the transcription start site (TSS) and at the polyadenylation site (PAS). See also Figure S4.
Upon examining the patterns of Malat1 localization to chromatin, we discovered a strong relationship between Malat1 DNA localization and active transcription. Malat1 was focally enriched over the bodies of genes that are actively transcribed (Figure 4B), and levels of Malat1 RAP-DNA enrichment correlated with levels of transcription as defined by the abundance of chromatin-associated RNA in cell fractionation experiments (Figure 4C, Pearson’s R = 0.63). Malat1 RAP-DNA enrichment similarly correlated with the amount of intron RNA captured in the Malat1 RAP-RNA[FA-DSG] experiment (Pearson’s R = 0.64). At these active genes, Malat1 was not uniformly enriched across gene bodies but rather showed enrichment that increased toward and peaked ~500 bases downstream of the annotated polyadenylation signal (PAS) (Figure 4D), similar to patterns seen for RNA polymerase II occupancy (Rahl et al., 2010).
Because Malat1 RAP-RNA highlighted Malat1 interactions with alternatively spliced transcripts, we wondered whether Malat1 localization to chromatin might depend not only on transcription but also on splicing. To study this, we compared Malat1 RAP-DNA enrichment at single-exon, multi-exon, and alternatively spliced genes. Because approximately half of the single-exon genes encode histones, which are processed differently from other RNAs, we considered histone and non-histone single-exon genes separately. As noted above, multi-exon genes displayed Malat1 RAP-DNA enrichment that correlated with the expression level of that gene. In contrast, single-exon histone genes were significantly depleted in Malat1 RAP-DNA compared to other genes, regardless of their expression level (Figure 4C, Figure S4B). Single-exon non-histone genes were more enriched than histone genes, but their enrichment levels did not strongly correlate with expression levels (Figure 4C). Similar to our observations for RAP-RNA[FA-DSG], Malat1 RAP-DNA preferentially enriched for genes annotated as having alternative isoforms (9% higher enrichment) and genes encoding proteins involved in RNA binding (20% higher enrichment) (P < 0.001, Mann-Whitney test), even after controlling for expression level (P < 10−5).
Malat1 Interactions with Chromatin Depend on Transcription at Its DNA Targets
We considered two models to explain these patterns of Malat1 RAP-DNA enrichment at active, spliced genes. First, Malat1 might localize to DNA through recognition of specific chromatin elements or DNA signals independent of the presence of the nascent transcript. In this model, the enrichment for nascent RNAs might simply reflect proximity to the DNA localization sites of Malat1. Alternatively, Malat1 might localize to chromatin as a secondary effect of interactions with the nascent pre-mRNA and associated RNA-binding proteins. We reasoned that halting transcription and repeating the RAP-DNA experiment would directly distinguish between these two possibilities. If the nascent pre-mRNA were a key intermediate, then Malat1 would lose its contacts with chromatin upon transcriptional inhibition. Conversely, if Malat1 localized to gene loci independent of the nascent transcript, then RAP-DNA enrichment at gene loci would be maintained.
To test this, we repeated the Malat1 RAP-DNA experiment after treating ES cells with an inhibitor of transcription elongation (flavopiridol) or a control (DMSO). After one hour of treatment, the abundance of introns in input nuclear-enriched RNA was reduced by on average 40% across all genes and up to 95% for some genes (Figure S4C,D), consistent with previous reports (Rahl et al., 2010). We note that the abundance of Malat1 itself did not measurably decrease during this one-hour interval. When we examined Malat1 localization across the genome, we found that Malat1 RAP-DNA enrichment at formerly active genes was significantly reduced (Figure 4D, Figure S4E–H). These results indicate that Malat1 localization to chromatin depends on transcription of its DNA targets.
These results demonstrate a strong relationship between the RNA and DNA interactions of Malat1 and suggest a model where Malat1 localization to chromatin occurs through interactions with nascent pre-mRNAs (Figure 5A). Through these interactions, Malat1 is brought in proximity to chromatin and thus purifies DNA at active gene loci. This model explains our observation of RAP-DNA enrichment accumulating over active gene loci, as the amount of Malat1 associated with a position along a gene locus would depend on the length of the pre-mRNA tethered to that location, which increases from the 5’ end to the 3’ end.
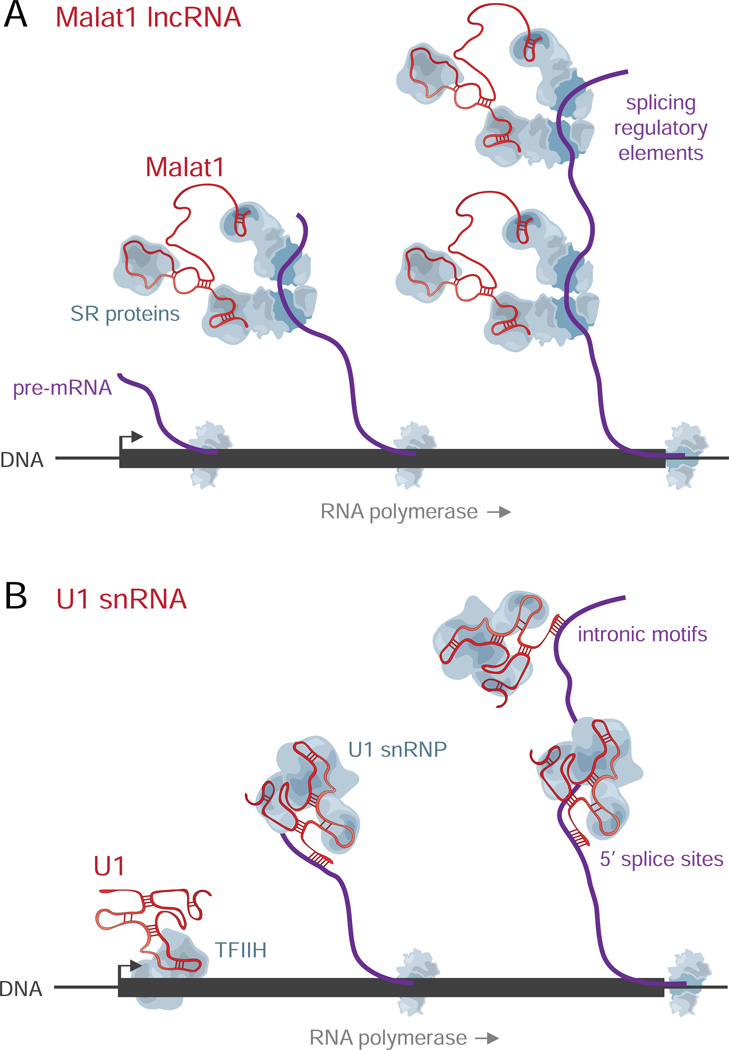
Figure 5. Malat1 and U1 Interact with Pre-mRNAs and Chromatin Through Different Mechanisms.
(A) Malat1 interacts indirectly with nascent RNAs at splicing regulatory elements, like the conserved noncoding elements in Hnrnpdl and Tial1, possibly through its known interactions with SR splicing proteins (Tripathi et al., 2010). These interactions enable Malat1 to localize to chromatin at active gene loci. Malat1 RAP may co-purify DNA through tethering of the nascent transcript to chromatin by RNA polymerase.
(B) U1 interacts with pre-mRNAs through direct hybridization, both at 5’ splice sites and throughout introns. U1 interacts with chromatin through two different mechanisms: (i) U1 localizes throughout active gene loci as a secondary result of its direct interactions with the nascent pre-mRNA, and (ii) U1 localizes at the 5’ ends of genes through a mechanism that does not depend on transcriptional elongation, perhaps via known interactions with the cyclin H subunit of TFIIH (Kwek et al., 2002). While these figures depict interactions with chromatin-associated pre-mRNAs, U1 and Malat1 may also interact with pre-mRNAs after their release from chromatin. snRNP: small nuclear ribonucleic particle.
U1 Localizes to Chromatin in Two Distinct Ways
We next explored whether the chromatin localization of U1 also depends on transcription of its DNA targets. We found that U1 was indeed enriched over the bodies of active genes and showed a level of enrichment that correlated with the abundance of chromatin-associated input RNA (Figure 4E, Pearson’s R = 0.70). At higher resolution across active genes, however, U1 RAP-DNA enrichment showed a striking bimodal pattern: U1 showed elevated enrichment both at the 5’ and 3’ ends of genes (Figure 4F, Figure S4E–G). Because these peaks appeared to extend over several kilobases and might be confounded by the inclusion of shorter genes, we performed the same analysis using only genes that are longer than 20 kb. In these genes, U1 RAP-DNA exhibited a sharp peak at the 5’ end (average of 0.8 kb) and a broader peak at the 3’ end (average of 4 kb) (Figure S4I).
Based on this bimodal pattern of enrichment, we hypothesized that U1 might interact with chromatin by two distinct mechanisms: one that would lead to enrichment at the 3’ ends of genes dependent on interactions with the nascent pre-mRNA, similar to Malat1; and one that would lead to its localization to DNA at the 5’ ends of genes near the transcription start site (TSS), possibly independent of interactions with the nascent transcript (Figure 5B). To gain insight into these mechanisms, we performed U1 RAP-DNA in cells treated with flavopiridol. Under these conditions, the U1 RAP-DNA peak at the 3’ ends of genes largely disappeared, similar to our results for Malat1 (Figure 4F, Figure S4E–G). At the 5’ ends of genes, however, U1 enrichment was maintained even in the absence of transcription elongation (Figure 4F), suggesting that U1 may interact with these sites through a mechanism that does not involve interactions with pre-mRNAs. Indeed, we found that U1 localization to the 5’ ends of genes did not depend on the presence of a 5’ss motif in the short nascent RNA produced by transcription initiation (Figure S4J,K). These results demonstrate that U1 localizes to chromatin in two ways (Figure 5B). First, U1 binds nascent RNAs and through these interactions co-purifies active gene loci. Second, U1 binds chromatin at the 5’ ends of genes through a mechanism that does not depend on transcription elongation or on 5’ss motifs near the TSS.
Discussion
Here we developed RAP-RNA, a method to comprehensively characterize in vivo RNA-RNA interactions, and applied it to investigate two ncRNAs implicated in RNA processing: U1 and Malat1. We show that RAP-RNA can map RNA-RNA interactions at high resolution and distinguish between hybridization- and protein-mediated binding, providing a powerful tool for exploring RNA function across a wide range of applications including identifying microRNAs that bind a specific mRNA, characterizing transcripts associated with ribosomes, or illuminating the regulatory functions of the many uncharacterized large and small ncRNAs implicated in gene regulation and human disease.
Using RAP, we find that U1 and Malat1 interact with many nascent RNAs and chromatin loci. U1 binding is pervasive throughout the transcriptome including introns and unspliced genes, and occurs through direct hybridization with the canonical 5’ss motif (Figure 5B). Together with the recent evidence that U1 acts to prevent PCPA (Kaida et al., 2010), our results suggest that U1 may play a ubiquitous role in protecting the integrity of the transcriptome through direct engagement of nascent transcripts. We also found that U1 can localize to DNA near TSSs independent of its interactions with nascent RNA. Notably, previous studies have reported that U1 directly binds TFIIH, a general transcription factor, and that this interaction enhances transcription initiation and reinitiation in reconstituted transcription assays in vitro (Kwek et al., 2002; O'Gorman et al., 2005). Furthermore, promoter-proximal 5’ splice sites can enhance transcription in vivo by recruiting basal transcription factors (Damgaard et al., 2008). In light of these previous observations, our results suggest that U1 may regulate transcription initiation throughout the genome.
Our data also provide new insight into the molecular function of Malat1. While some previous studies have shown that Malat1 depletion can affect alternative splicing (Tripathi et al., 2010) and reduce recruitment of SR proteins to a gene locus (Bernard et al., 2010), the molecular functions of Malat1 remain unclear (Gutschner et al., 2013). Our data demonstrate that Malat1 interacts with many nascent pre-mRNAs at active gene loci, raising the possibility that Malat1 may influence RNA processing through recruitment or modification of other proteins localized to these sites. Notably, Malat1 interacts indirectly with numerous alternatively spliced, auto-regulated transcripts encoding RNA-binding proteins, including some that are thought to physically interact with Malat1 (Änkö et al., 2012; Pandit et al., 2013). Based on these observations, we propose that Malat1 may interact with pre-mRNAs through its physical associations with SR splicing proteins (Figure 5A) (Tripathi et al., 2010). Indeed, previous work has shown that tethering SRSF1 to a transgenic chromatin locus is sufficient to recruit MALAT1 to that locus (Tripathi et al., 2012). Thus, interactions with sequence-specific SR splicing proteins may provide a mechanism for Malat1 recruitment to specific alternatively spliced pre-mRNAs and active gene loci on chromatin.
More generally, our observations suggest that nuclear-localized lncRNAs can recognize nascent pre-mRNAs to guide their regulatory functions. LncRNAs might achieve specificity for these nascent transcripts through direct hybridization or sequence-specific RNA-binding proteins, enabling localization to specific pre-mRNAs or chromatin sites. In some cases, these lncRNAs may directly target nascent RNAs to control RNA processing. In other cases, this recruitment may enable lncRNA-mediated regulation of chromatin or transcription at the gene locus. These two possibilities are not mutually exclusive: numerous lncRNAs interact with chromatin regulatory complexes and RNA processing proteins, suggesting that these lncRNAs may also associate with specific nascent RNAs to control pre-mRNA processing and/or regulate chromatin. In contrast to other mechanisms for lncRNA localization such as interactions with DNA-binding proteins (Soruco et al., 2013) or proximity-mediated search (Engreitz et al., 2013), lncRNA targeting through sequence-specific RNA-RNA interactions might enable integration of co-transcriptional (on-chromatin) and post-transcriptional (off-chromatin) regulatory signals. Future work will be required to determine the prevalence and functions of these lncRNA-RNA interactions. RAP-RNA provides a powerful tool to investigate these possibilities and to further dissect the relationship between RNA-RNA interactions, RNA localization to chromatin, and transcriptional regulation.
Experimental Procedures
Detailed RAP protocols are available at the authors’ web site: http://www.lncrna.caltech.edu/RAP. Sequencing data are available at the NCBI Gene Expression Omnibus (accession number GSE55914).
Cell Culture
For most experiments, we cultured V6.5 male mouse ES cells in 2i + LIF; for experiments involving Xist, we used pSM33 male mouse ES cells induced with doxycycline for three hours to activate Xist expression (Engreitz et al., 2013). For transcription inhibition, we treated cells with 1 µM flavopiridol or DMSO for one hour.
Probe Design and Generation
We captured target RNAs using pools of ssDNA biotinylated oligos. To generate tiled probesets to capture long RNAs (>250 nucleotides), we synthesized barcoded pools of oligos with 120-nt complementarity to the target RNA (Engreitz et al., 2013) (Table S4). We amplified specific probe subsets (Table S5) and generated ssDNA probes through in vitro transcription of dsDNA templates followed by reverse transcription with a 5’-biotinylated ssDNA primer. To capture short RNAs (<250 nucleotides), we designed and directly purchased three to four 50-nt 5’biotinylated ssDNA oligos (Table S4).
RAP with FA or FA-DSG Crosslinking
For the FA protocol, we crosslinked cells with 2% FA and solubilized chromatin with strong sonication, yielding RNA fragments of ~150 nt. For the FA-DSG protocol, we crosslinked cells with 2 mM DSG followed by 3% FA and solubilized chromatin with light sonication followed by treatment with DNase I to preserve RNA integrity. We performed RAP as previously described (Engreitz et al., 2013), with modifications (see Extended Experimental Procedures). We used 5 million cells and 50 pmol ssDNA probe per experiment, incubating in a highly denaturing buffer at 37 °C for 2 hours. We washed six times at 45 °C, then eluted the target RNA and co-purified complexes with RNase H. We digested protein with Proteinase K, isolated nucleic acids, and proceeded to RNA or DNA sequencing.
RAP with AMT Crosslinking
We resuspended cells in 0.5 mg/mL AMT solution (+AMT) or PBS only (−AMT) and irradiated the cells in a tissue culture dish for 7 minutes with long-wave ultraviolet light. We performed nuclear enrichment, isolated nucleic acids using TRIzol, fragmented the RNA to a median size of ~100 nt, and then digested residual DNA. We used 2 µg of the resulting purified RNA and 15 pmol ssDNA probe per purification. We used an alternate set of denaturing buffers for the AMT protocol hybridization and washes (see Extended Experimental Procedures).
Negative Controls for RAP-RNA
We performed numerous negative controls to eliminate potential sources of technical artifacts in the RAP-RNA protocols. For RAP-RNA[AMT], these include off-target hybridization between the probes and other RNAs, and interactions between other RNAs and the streptavidin beads. To control for these potential issues, we performed the experiment in parallel with a −AMT control, which was treated identically to the +AMT sample except that AMT was not added prior to UV irradiation. For RAP-RNA[FA-DSG], the potential sources of background additionally include off-target hybridization with DNA and nonspecific interactions between the probes and proteins that are crosslinked to RNA. To account for these potential issues, we performed two controls. First, we performed RAP-RNA experiments in non-crosslinked lysate. Second, we purified an mRNA as well as other abundant nuclear RNAs from crosslinked lysate. We searched for artificial peaks across the transcriptome in these control experiments and in some cases identified elevated signals at GC-rich simple or tandem repeats and the surrounding RNA sequences. These signals were clearly recognizable as probe-RNA hybridization artifacts by their characteristic read distributions, which result from RNase H cleavage during the elution step. We verified that these artificial signals did not affect our results and conclusions (see Extended Experimental Procedures).
Sequencing and Alignment
We sequenced each library to a depth of 10–50 million read-pairs, depending on the estimated library complexity. To analyze RNA sequencing data, we filtered reads aligning to highly abundant RNA transcripts, including rRNA, snRNA, and repeat elements defined by RepeatMasker. We aligned all remaining reads to the mouse transcriptome (RefSeq) and genome (mm9) using Tophat and discarded reads with MAPQ < 30. To analyze DNA sequencing data, we aligned reads to the mouse genome (mm9), removed duplicate read-pairs, and discarded reads with MAPQ < 30.
Enrichment Calculations
To calculate the enrichment for a target RNA, we divided the fraction of reads mapping to the target RNA in RAP-RNA by the same fraction in input. To calculate the enrichment for all other non-target RNAs or sets of RNAs, we performed the same calculation, except that we subtracted the number of reads mapping to the target RNA from the total read counts.
Peak Calling
We called peaks in RAP-RNA data for the AMT, FA, and non-crosslinked experiments using a sliding-window approach at 10 or 100-nucleotide resolution, comparing the number of reads in the purification sample with the number of reads in the input or control sample using a binomial test (see Extended Experimental Procedures). For RAP-RNA[FA-DSG], we focused on enrichments averaged across entire transcripts (exons or introns) rather than in smaller windows due the lower resolution of this approach for specific RNA interaction sites.
Analysis of 5’ Splice Sites
To relate RAP-RNA[AMT] reads to 5’ splice sites, we calculated the minimum distance from the 5’ base of the second read in each read-pair to any 5’ splice site (defined by RefSeq). To identify motifs enriched in RAP-RNA[AMT], we counted 8-mer sequences contained within 30 bases upstream of the second-of-pair read-ends and assessed significance with Fisher’s exact test (P < 0.001 after Bonferroni correction).
Supplementary Material
Highlights.
-
-
A general method to identify RNA-RNA interactions for many RNAs (>80 nucleotides)
-
-
Distinguishes direct and indirect RNA-RNA interactions using different crosslinkers
-
-
U1 snRNA interacts with pre-mRNAs directly while Malat1 lncRNA interacts indirectly
-
-
RNA-RNA interactions target U1 and Malat1 to chromatin at active gene loci
Acknowledgements
We thank Amy Pandya-Jones for chromatin fractionation protocols; Xuebing Wu for assistance with scoring 5’ss motif matches; Tarjei Mikkelsen for assistance with oligonucleotide synthesis; and Leslie Gaffney for assistance with figures. Supported by the Fannie and John Hertz Foundation (J.M.E.), National Defense Science and Engineering Graduate Fellowship (J.M.E.), NIH Director’s Early Independence Award DP5OD012190 (M.G.), the Broad Institute of MIT and Harvard (E.S.L., M.G.), and the California Institute of Technology (M.G.). J.M.E., E.S.L., and M.G. are inventors on a patent application from the Broad Institute that covers the selective purification of RNA-bound molecular complexes in cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Extended Experimental Procedures, four figures, and four tables and can be found with this article online.
Author Contributions
All authors designed experiments and analyses and approved the final manuscript. J.M.E. and K.S. performed experiments. J.M.E. performed computational analyses. J.M.E., P.M., A.Y.C., K.S., A.S., C.S., and M.G. developed experimental protocols. J.M.E., P.R., S.R.G., M.G., and E.S.L. developed analytical methods and tools. J.M.E. and M.G. conceived and designed the study. J.M.E., M.G., and E.S.L. wrote the manuscript.
References
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Änkö M-L, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet JP, Pederson T. Heterogeneous nuclear RNA double-stranded regions probed in living HeLa cells by crosslinking with the psoralen derivative aminomethyltrioxsalen. Proc. Natl. Acad. Sci. USA. 1979;76:755–759. doi: 10.1073/pnas.76.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell. 2011 doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5' splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C, Stadler PF. Computational biology of RNA interactions. Wiley Interdiscip. Rev. RNA. 2013;4:107–120. doi: 10.1002/wrna.1147. [DOI] [PubMed] [Google Scholar]
- Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun T, Zörnig M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature. 1980;283:220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell. 1983;33:509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman W, Thomas B, Kwek KY, Furger A, Akoulitchev A. Analysis of U1 small nuclear RNA interaction with cyclin H. J. Biol. Chem. 2005;280:36920–36925. doi: 10.1074/jbc.M505791200. [DOI] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M, Fu X-D. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell. 2013;50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soruco MML, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 2013;27:1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011;3:a000646–a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Song DY, Zong X, Shevtsov SP, Hearn S, Fu X-D, Dundr M, Prasanth KV. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol. Biol. Cell. 2012;23:3694–3706. doi: 10.1091/mbc.E12-03-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.