Summary
Telomere length maintenance is a requisite feature of cellular immortalization and a hallmark of human cancer. While most human cancers express telomerase activity, approximately 10-15% employ a recombination-dependent telomere maintenance pathway known as Alternative Lengthening of Telomeres (ALT) that is characterized by multi-telomere clusters and associated promyelocytic leukemia protein bodies. Here, we show that a DNA double-strand break (DSB) response at ALT telomeres triggers long-range movement and clustering between chromosome termini, resulting in homology-directed telomere synthesis. Damaged telomeres initiate increased random surveillance of nuclear space before displaying rapid directional movement and association with recipient telomeres over micron-range distances. This phenomenon required Rad51 and the Hop2-Mnd1 heterodimer, which are essential for homologous chromosome synapsis during meiosis. These findings implicate a specialized homology searching mechanism in ALT dependent telomere maintenance and provide a molecular basis underlying the preference for recombination between non- sister telomeres during ALT.
Introduction
Homologous recombination (HR) is an evolutionarily conserved mechanism of DNA repair that is essential to genome integrity in meiotic and mitotic cells (Mazon et al., 2010; Moynahan and Jasin, 2010). This form of DNA double-strand break (DSB) repair necessitates an accurate search for homology into duplexed genomic regions by presynaptic RecA nucleoprotein filaments coating single stranded DNA. Successful capture of homology entails base pairing between invading single stranded DNA with the complementary strand of duplex DNA, forming a displacement loop (D-loop). Subsequent close association of the homologous strands (synapsis) and extension by DNA polymerases enables template directed DNA repair.
HR mediated DNA repair mechanisms are largely restricted to S and G2 phases in mitotic cells in eukaryotes when a sister chromosome is present and resection-promoting nucleases are more active (Aylon et al., 2004; Huertas et al., 2008; Ira et al., 2004). HR between sister chromatids rather than homologous chromosomes is thought to be the vastly preferred mechanism of HR in mitotic cells (Johnson and Jasin, 2000). Conversely, meiotic HR is not limited to sister chromatid recombination, but occurs extensively between sequences on homologous chromosomes (Neale and Keeney, 2006). Meiotic recombination involves a lineage restricted, programmatic form of HR that is initiated by Spo11 induced DSBs and culminates in synapsis of distant homologous loci. This process requires Rad51 and Dmc1 as well as the heterodimeric Hop2-Mnd1 proteins, which promote Rad51 and Dmc1 dependent D-loop formation in vitro and are epistatic to these RecA homologs during meiosis in yeast and in mammalian organisms (Bishop, 1994; Chi et al., 2007; Petukhova et al., 2003; Pezza et al., 2007). Thus, 3-dimensional genome organization during physiologic meiotic recombination is intimately linked to the repair mechanisms that execute homology searches between non-sister, homologous chromosomes.
Large stretches of homologous DNA sequences encase eukaryotic chromosome termini, composed of repetitive G-rich sequences contained within a specific nucleoprotein chromatin structure that protects against DNA damage responses. Deprotected chromosome ends exhibit both of the major DSB repair pathways, NHEJ and HR, depending on context. Loss of telomeric sequence due to replicative shortening, or deprotection in the absence of specific telomere binding proteins, results in a high incidence of NHEJ dependent telomere-telomere fusions (Counter et al., 1992; Dimitrova et al., 2008). Conversely, cells that use the alternative lengthening of telomeres (ALT) mechanism display elevated HR at telomeres (Dunham et al., 2000). A subset of ALT telomeres coalesces into characteristic ALT-associated PML Body (APB) structures that display multiple telomeres from different chromosomes in association with PML (Draskovic et al., 2009; Jegou et al., 2009; Molenaar et al., 2003; Yeager et al., 1999). These multi-telomere bodies are thought to be sites of homology-directed telomere synthesis. Mutations within certain chromatin associated genes are commonly found in ALT cancers (Heaphy et al., 2011; Schwartzentruber et al., 2012). However, the nature of the initiating stimulus for ALT is unclear, as are mechanisms that drive recombination within APBs.
One plausible mechanism is that DSB responses at a subset of ALT telomeres would represent a seminal event that initiates the search and capture of distant homologous DNA. Pairing and recombination between telomeres from different chromosomes during ALT would necessitate long range telomere movement. Damage dependent increases in local DNA mobility have been documented in both prokaryotes and eukaryotes, which may suggest that increased movement of broken chromosomes assists in repair of these loci (Aten et al., 2004; Chen et al., 2013; Dimitrova et al., 2008; Dion et al., 2012; Krawczyk et al., 2012; Mine-Hattab and Rothstein, 2012; Roukos et al., 2013). Interestingly, DNA damaging agents increase the prevalence of APBs in ALT cells, and a subset of ALT telomeres accumulates DNA repair proteins (Cesare et al., 2009; Fasching et al., 2007). Furthermore, while the majority of telomeres in ALT-positive osteosarcoma U2OS cells display relatively slow mobility confined to a radius of less than 0.5μm, up to 15% of telomeres show unusually high mobility (Jegou et al., 2009; Molenaar et al., 2003). Yet, how increased mobility would facilitate efficient associations between damaged DNA and homologous genomic regions remains enigmatic, as are molecular events underlying such migration of DNA across the nucleoplasm lacking canonical structures of cellular transport such as microtubules.
Here, we provide direct evidence that telomeric DSB responses drive inter-telomere associations in the context of ALT telomeric chromatin. Strikingly, increased ALT telomere mobility culminated in rapid and directional movement over micron distances toward a recipient telomere, providing a real-time cellular visualization of homology search and synapsis in a mammalian cell nucleus. This process required the HR machinery including Rad51, which could be directly visualized in between recombining telomeres, representing a putative recombination intermediate. Moreover, these studies reveal that ALT cells commandeer proteins critical for meiotic recombination searching mechanisms, providing insights into this specialized form of HR driven telomere maintenance.
Results
Telomere Double-Strand Breaks Increase the Hallmarks of ALT Recombination
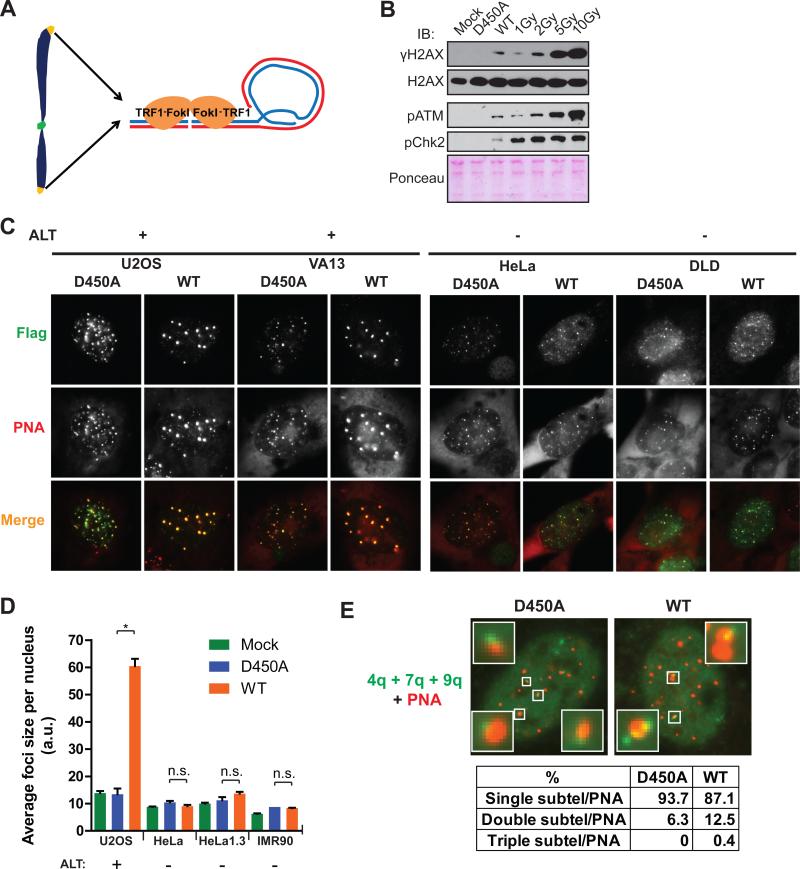
Telomeric chromatin is bound by a set of proteins that recognize double and single stranded repetitive telomere DNA, termed the Shelterin complex (Cesare and Karlseder, 2012; Palm and de Lange, 2008). Fusion of the telomere repeat binding factor, TRF1, to the FokI nuclease catalytic domain targets DSBs specifically at telomeres in both telomerase positive and ALT cells, leading to a robust induction of DSB responses that extend hundreds of kilobases into subtelomeric chromatin (Tang et al., 2013). Further characterization of TRF1-FokI expression revealed a DSB response equivalent to approximately 1-2 Gy ionizing radiation in U2OS cells as assessed by western blot using antibodies to γH2AX, and phosphorylated-ATM (Figures 1A and B). Notably, Chk2 phosphorylation was not increased to similar levels as phospho-ATM, consistent with prior reports that telomere damage signals are not efficiently transmitted to some ATM substrates (Cesare et al., 2013). Despite reduced transmission of ATM phosphorylation to Chk2, TRF1-FokI expression resulted in a nearly 2-fold increase in cells in the G2 phase of the cell cycle, consistent with the induction of a G2/M checkpoint (Figure S1A).
Figure 1. TRF1-FokI DSBs Promote Telomeric Clustering in ALT Cells.
(A) Schematic of telomere-specific DSB induction by the TRF1-FokI fusion protein.
(B) Comparison of DSB responses in U2OS cells between TRF1-FokI expression and escalating doses of IR. Immunoblot (IB) was performed at 30 minutes following irradiation. Mock indicates mock viral transduction; D450A and WT indicate nuclease inactive and wild type TRF1-FokI, respectively.
(C) Representative immuno-FISH images of TRF1-FokI (Flag) WT or D450A colocalized with telomeres (PNA) in ALT positive and negative cells.
(D) Average telomere foci size per nucleus after TRF1-FokI expression was calculated using ImageJ. Mean ± standard error of the mean (s.e.m.) for >50 cells in n = 3. * p<0.05, n.s. p>0.05.
(E) FISH was performed in cells expressing TRF1-FokI WT or D450A using a combination of chromosome-specific subtelomeric (subtel) probes and PNA. Percentages of colocalized subtel-PNA foci that contained one, two or three subtelomeric signals were quantified from >100 cells in n =2.
See also Figure S1.
Interestingly, TRF1-FokI expression resulted in up to 4-fold increases in average telomere foci size and reduced numbers of telomeres in each of 4 different ALT positive cell lines in comparison to cells expressing the nuclease inactive TRF1-FokI D450A mutant (Figures 1C, 1D and S1B). Telomere foci size increases did not occur in telomerase negative primary human IMR90 fibroblasts or 4 different telomerase positive cell lines. Telomere length difference between ALT and telomerase positive cells was not sufficient to explain foci size increases, as TRF1-FokI expression did not significantly increase telomere foci size in the telomerase positive HeLa 1.3 cells (Figure 1D), which have a mean telomere length comparable to ALT cell lines.
Telomeres within these larger foci in ALT cells contain chromosomally attached telomeres. This is supported by the observation that metaphase chromosome spreads from D450A and WT TRF1-FokI were not appreciably different with respect to the percentage of chromosome ends displaying telomeric signal, and by the presence of subtelomeric fluorescence in situ hybridization (FISH) signals or subtelomeric lac operator transgene repeats juxtaposing telomeres in interphase U2OS cells (Shanbhag et al., 2010) (Figures S1C-E). Furthermore, expression of TRF1-FokI increased the percentage of multiple subtelomeric FISH signals accumulating at a telomere cluster (Figure 1E). These data are in agreement with previous reports that APB bodies contain chromosomally attached telomeres (Draskovic et al., 2009). However, they do not exclude the possibility that extra-chromosomal telomeric repeats (ECTRs), which increase in response to DNA damage, are also present in these large telomere bodies (Cesare and Griffith, 2004; Fasching et al., 2007).
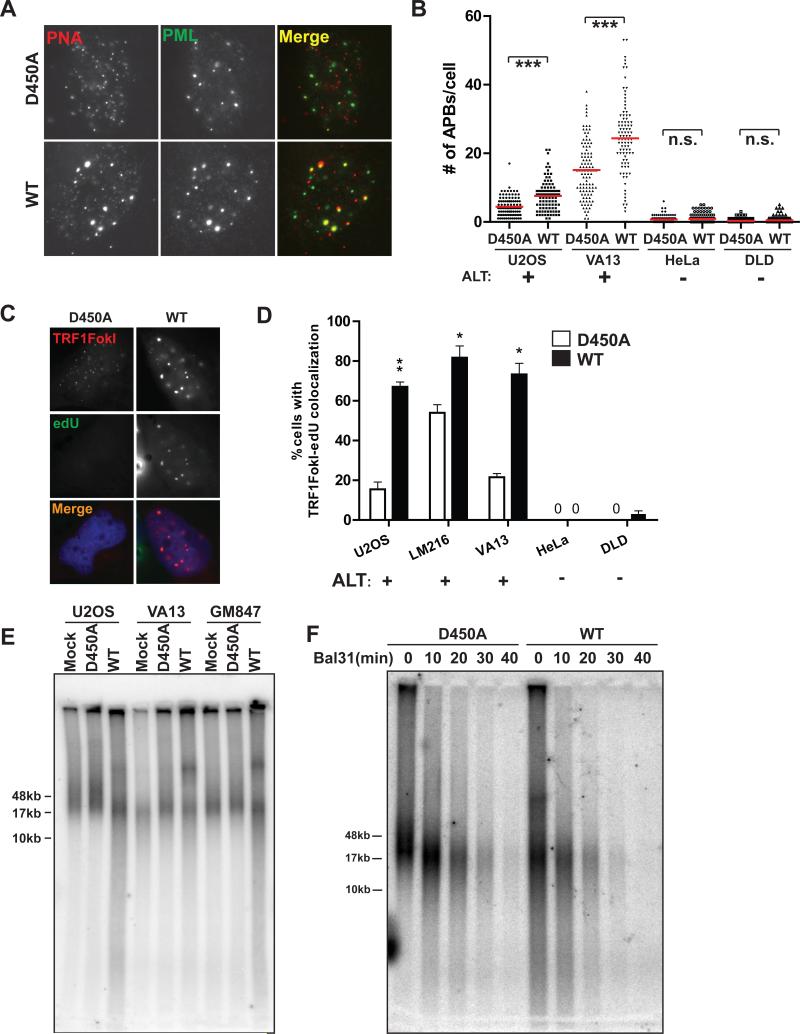
These findings suggest that DSB responses at ALT telomeric chromatin provide the initiating stimulus for telomere clustering. Consistent with this expectation, TRF1-FokI expression induced multiple hallmarks of ALT recombination, including significant increases in telomeres associated with promyelocytic leukemia bodies (APBs) and telomere associated DNA synthesis as evidenced by incorporation of thymidine analog 5-ethynyl-2’-deoxyuridine (edU) in non S-phase cells (Figures 2A-D and Figure S2A). Similar findings were not detectable in telomerase positive cells. Expression of TRF1-FokI also increased c-circle formation, a specific indicator of ALT activity (Figures S2B-E) (Henson et al., 2009). Moreover, TRF1-FokI expression increased telomere length heterogeneity by Terminal Restriction Fragment Analysis in 3 different ALT cell lines (Figure 2E). The increased heterogeneity could result from a combination of factors, including telomere cutting by TRF1-FokI, as well as ALT recombination associated length changes and ECTR generation. These telomeres were sensitive to digestion by Bal-31, an exonuclease that degrades duplex DNA from both 3’ and 5’ ends, indicating that the longer telomere fragments observable following TRF1-FokI WT expression were not a consequence of telomere-telomere end joining (Figure 2F).
Figure 2. TRF1-FokI DSBs Promote ALT Activity.
(A) Representative PNA/anti-PML immuno-FISH images of cells expressing TRF1-FokI WT or nuclease inactive D450A mutant.
(B) Number of PML-PNA colocalizations (APBs) per nucleus was quantified in cells expressing TRF1-FokI WT or D450A. Mean of >100 cells from three replicate experiments. *** p < 0.005, n.s. p > 0.05.
(C) Representative images of edU positive TRF1-FokI foci in VA13 cells.
(D) Fraction of nuclei with ≥ 3 TRF1-FokI telomere foci colocalizing with edU foci in non-S-phase cells was quantified after expression of TRF1-FokI as indicated. Mean ± s.e.m., >50 cells in n = 3. * p<0.05.
(E) Digested genomic DNA from cells expressing TRF1-FokI was resolved by pulsed-field gel electrophoresis (PFGE) and probed for total telomeric DNA.
(F) Digested genomic DNA from U2OS cells expressing TRF1-FokI WT or D450A were treated with Bal31 nuclease for the indicated durations, resolved by PFGE and probed for total telomeric DNA.
See also Figure S2.
Double-Strand Breaks Initiate Directional ALT Telomere Movement and Clustering
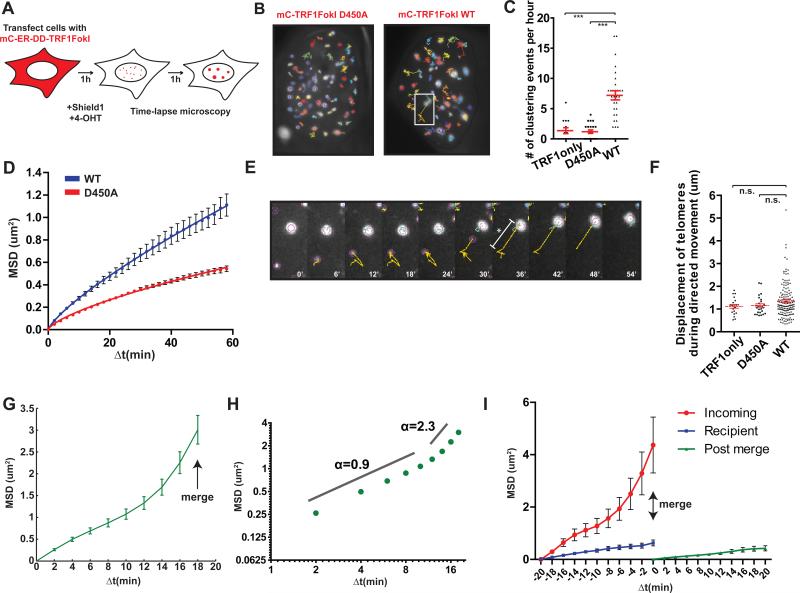
The presence of intense ALT-like telomere clusters suggests that DSB responses initiate a homology search process, followed by synapsis and recombination between distant telomeres. To directly test this hypothesis, we visualized telomere movement using an inducible mCherryTRF1-FokI fused to a modified estradiol receptor and destabilization domain, which allowed small molecule induction by administration of 4-hydroxytamoxifen and Shield1 ligand (Figure 3A). Following a 1-hour induction period, TRF1-FokI expressing cells were monitored over the following hour by capturing confocal z stacks of the entire nucleus every 2 minutes. Telomere foci were tracked in the z-projected plane, and a registration process (Thevenaz et al., 1998) assisted normalization for cellular movement.
Figure 3. ALT Telomere DSBs Rapidly Associate by Long Range, Directional Movement.
(A) Schematic of time-lapse imaging of telomeres in cells expressing TRF1-FokI. mC, mCherry; ER, modified estrogen receptor; DD, destabilization domain.
(B) Representative 1-hour traces of mCherryTRF1-FokI D450A or WT foci in U2OS nuclei. White box indicates region shown in (E).
(C) Quantification of telomere-telomere clustering where merged foci remain unseparated in 3 or more frames. In red, mean ± s.e.m. Each datapoint represents tracked nuclei from two independent experiments. *** p<0.0005. TRF1only represents mCherryTRF1 protein without C-terminal FokI.
(D) Mean squared displacement (MSD) analysis of telomere movement in U2OS cells expressing mCherryTRF1-FokI D450A or WT. Δt, time interval. Error bars, weighted s.e.m. and n = >700 tracks in two independent experiments. Fit was determined by a diffusion model, MSD = Γtα, where α is the time dependence coefficient. α WT = 0.8 and α D450A = 0.7.
(E) Expanded images of a tracing from (B) highlighting diffusive movement followed by directed movement toward another telomere. * and bar denotes displacement measured in (F). Yellow lines indicate the path traveled by the particle during the previous 10 frames.
(F) Quantification of telomere displacement during directed movement (mean ± s.e.m of tracks from 2 independent experiments; n.s. p>0.05). Directed phase was defined by consecutive motion toward the recipient telomere over 3 or more frames until the merge event.
(G) MSD analysis of clustering telomeres in U2OS cells expressing TRF1-FokI WT (see text). ΔT=18 represents the point of merge into a recipient telomere. n = 157 tracks from 2 independent experiments. Error bars, weighted s.e.m.
(H) Data from (G) is displayed on a log-log plot. Time dependence coefficient, α, for two phases of movement is indicated.
(I) MSD analysis of mobility before and after a merge event. Δt=0 represents the point of merge. Error bars, weighted s.e.m. and n = 46 tracks from two independent experiments.
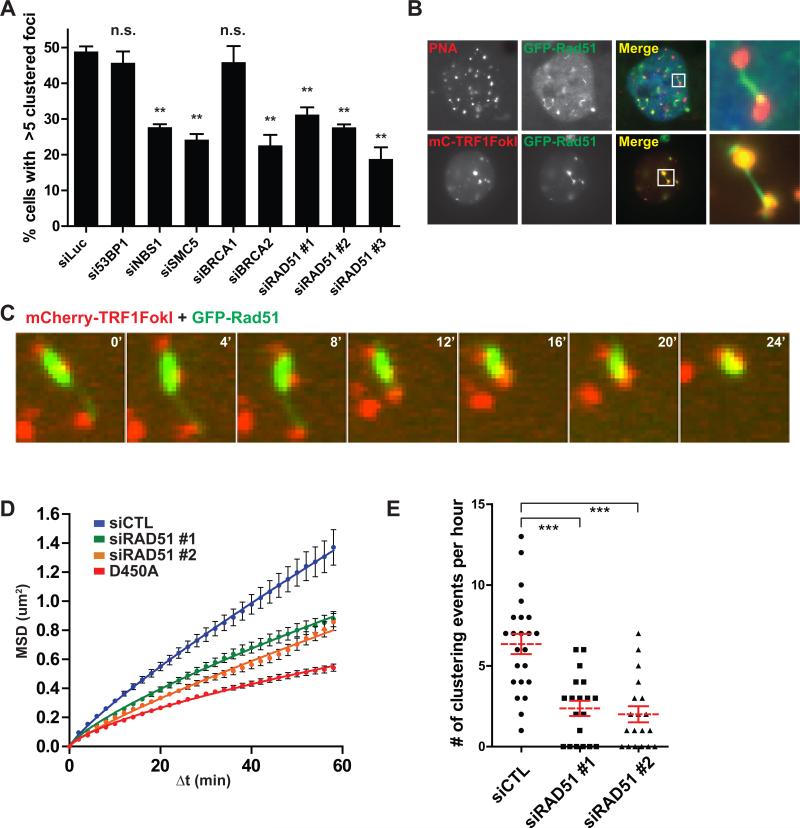
Strikingly, telomeres in TRF1-FokI WT expressing cells demonstrated increased mobility and an average of 7 telomere-telomere clustering events per hour between foci separated by up to 5μm (Figures 3B, C, E, F and Movie S1). Telomere clustering and movement were greatly diminished in the nuclease inactive D450A mutant or in cells expressing mCherry-TRF1 (Figures 3B-D and Movies S2 and S3). Importantly, less frequent instances of clustering were observed at mCherry-D450A and TRF1 containing telomeres, consistent with a previous report of an association of two telomeres in an unperturbed U2OS cell (Molenaar et al., 2003). TRF1-FokI induced DSBs at ALT telomeres greatly increase the frequency of telomere associations that normally occur in these cells.
To determine if DSBs at other regions of the genome in ALT cells would demonstrate similar movement and clustering as those observed at telomeres, we monitored targeted and random DSB positions at non-telomeric locations. Fusion of FokI to the Lac repressor (mCherryLacIFokI) enables efficient visualization of DSB responses at lac operator repeat sequences integrated into chromosome 1p36 (Shanbhag et al., 2010). mCherryLacIFokI DSBs did not display large increases in mobility at this locus in U2OS cells (Figures S3A and B). GFP-53BP1 movement and clustering was also minimal at most ionizing radiation induced foci during time lapse imaging in U2OS cells (Figure S3C). Conversely, telomeric DSBs moved coordinately with the sub-telomeric LacO locus in cells expressing both GFP-LacI and mCherryTRF1-FokI (Figure S3D). The lack of substantial DSB movement at non-telomeric regions of the genome is consistent with reports that ALT cells display elevated recombination at telomeres, but not elsewhere in the genome (Bechter et al., 2003; Dunham et al., 2000).
Importantly, the robust increase in ALT telomere movement allowed a quantitative analysis of this type of chromatin movement. Telomere tracks were subjected to a mean squared displacement (MSD) analysis, which plots the average squared displacements at each time interval, given by equation MSD = <(x(t+Δt)-x(t))2>, where x is the position of the focus and t is time. The MSD trajectories were then fitted to a single exponential time dependence diffusion model described by where is a generalized coefficient, and α is a time dependence coefficient which can be used to determine the type of motion. For α ~ 1, the particle is undergoing normal diffusion, and α < 1 represents sub-diffusion, also known as anomalous diffusion. Subdiffusive target searches in cells can result from molecular crowding of the nucleus and cytoplasm (Guigas and Weiss, 2008). Finally, α ≥ 2 represents an exponential dependence on time that indicates that the particle is moving in a directed manner, an example of which is active cellular transport.
A comparison of averaged MSD trajectories for all telomeres in TRF1-FokI WT or D450A expressing U2OS cells revealed that α = 0.8 for WT and α = 0.7 for D450A, both characteristic of subdiffusive motion (Figure 3D). The coefficient, which describes the magnitude of the behavior characterized by α, was greater for WT than for D450A, with values of 4.7×10−2μm2s−α and 3.3×10−2μm2s−α respectively. Calculation of time-dependent diffusion coefficient D(t) = MSD/t = Γtα−1 showed that the diffusion coefficient decreases linearly with time with a slope of α-1 when plotted on a log-log scale, consistent with subdiffusive motion (Saxton, 2007). For D450A, D(t) at 15 minutes was 1.4×10−2μm2min−1, consistent with values for normal U2OS telomeres (Molenaar et al., 2003), and decreased to 0.9×10−2μm2min−1 at the end of the observation period (Figure S3A). For WT, however, D(t) was consistently elevated at 2.6×10−2μm2min−1 and 1.9×10−2μm2min−1 at 15 minutes and 60 minutes respectively (Figure S3E). These results indicate that damaged telomeres move faster and roam a larger nuclear territory.
While all telomeres considered in sum demonstrated diffusive movement, it was readily apparent from imaging experiments that faster, “incoming” telomeres displayed a striking long-range directional movement prior to association with a comparatively slow-moving “recipient” telomere (Figure 3B, E and Movie S1). For a quantitative analysis of this observation, mobility data from telomeres that merged into a recipient telomere was isolated. The terminal behavior of such telomeres was characterized by MSD analysis of the last 10 timepoints of each track, with the ultimate timepoint representing the merge event. The shape of the resulting MSD trajectory suggested an initial, increased diffusive movement for Δt of up to 10 minutes, followed by a transition to directed movement at large Δt (Figure 3G) This change in behavior was clearly visualized on a log-log plot. The α coefficient for the initial portion of the clustering telomere trajectory was 0.9 suggestive of diffusive motion, but between Δt of 12-18 minutes, there was a clear transition of α to ~ 2.3, indicative of directed movement (Figure 3H).
The average displacement of telomeres during this directed phase was ~1.3μm with up to 4-5 μm observed for some tracks (Figure 3E, F). Following the clustering event, the merged telomere foci demonstrated reduced movement (Figure 3I), suggesting the searching process that underlies directional movement had concluded. Interestingly, the less mobile, “recipient” telomere was associated with PML in 85% of clustering events (Figure S3F). This supports a model in which PML promotes clustering and recombination of telomeres within APBs (Chung et al., 2011; Draskovic et al., 2009).
To further address whether the driving force behind ALT telomere movement is a DSB response, spontaneous telomere clustering events in mcherryTRF1 expressing VA13 cells were quantified with respect to colocalization of GFP-53BP1 as a marker of DSBs. Greater than 60% of clustering telomeres accumulated GFP-53BP1 prior to association, while 15% of all telomeres were associated with GFP-53BP1. This indicates that telomere movement and clustering is closely correlated with a local DNA damage response (Figure S3G, H).
Homologous Recombination Predominates at ALT Telomere DSBs
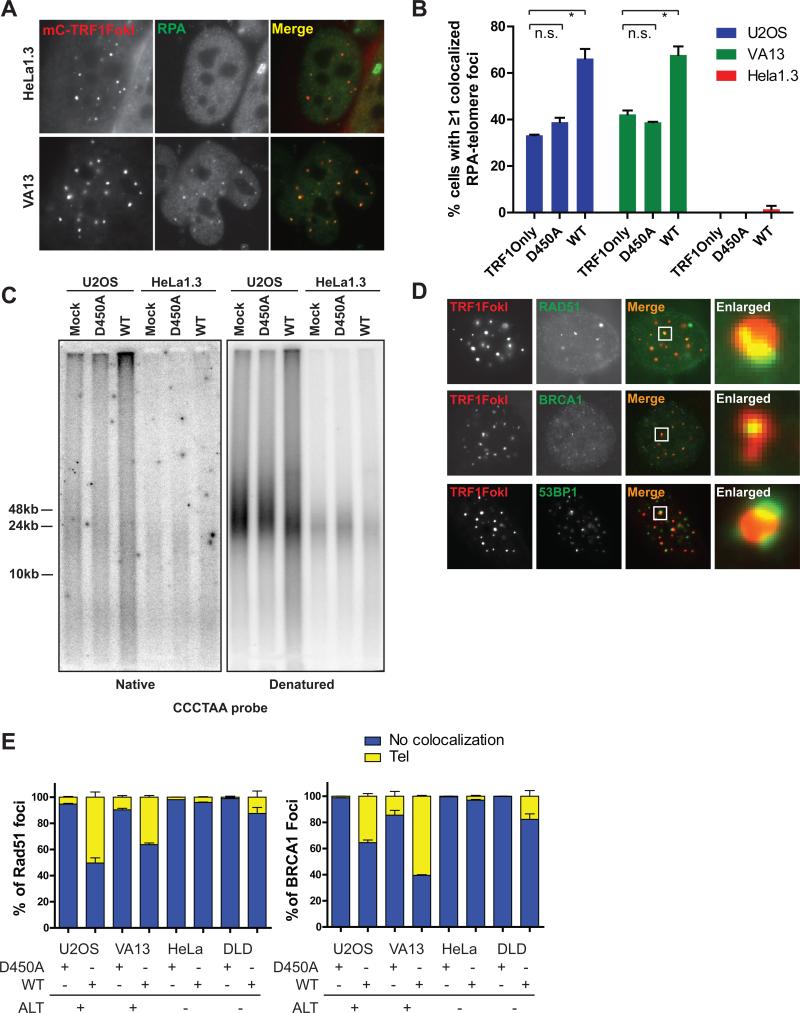
We postulated that the presence of random surveillance followed by directional DSB induced telomere movement is a consequence of a homology search and capture between distant telomeres. Resection of telomeric ends would be a critical determinant of this pathway choice. RPA localization was assessed at telomeres in cells expressing TRF1-FokI in ALT and telomerase positive cells. HeLa 1.3 did not significantly accumulate RPA at telomeres. Conversely, telomeres in both U2OS and VA13 cells were associated with RPA at baseline, which further increased in the presence of TRF1-FokI (Figures 4A, B). Furthermore, expression of TRF1-FokI in U2OS cells resulted in an increase in single-stranded telomeres as assessed by electrophoresis and hybridization of telomeric probes under native conditions (Figure 4C). The increased ssDNA was largely derived from telomeric overhangs, since the native single stranded telomeric signal was reduced following treatment with ExoI ssDNA exonuclease (Figure S4A).
Figure 4. Homologous Recombination Predominates at ALT Telomere DSBs.
(A) Representative immunofluorescence images of RPA2 and mCherry-TRF1-FokI WT in HeLa1.3 and VA13 cells.
(B) Quantification of RPA-telomere colocalization in ALT cell lines (U2OS and VA13) and a telomerase positive cell line (HeLa1.3).
(C) Telomeric DNA from U2OS and HeLa1.3 cells expressing TRF1-FokI D450A or WT was resolved by PFGE, and probed with p32 labeled oligos hybridizing to the G-rich single-stranded telomeres under native conditions. The gel was denatured and probed again for total telomeric signal.
(D) Representative immunofluorescence images of Rad51, BRCA1 and 53BP1 and mCherryTRF1-FokI WT and D450A positive telomeres in U2OS cells.
(E) Colocalization of Rad51 and BRCA1 to mCherryTRF1-FokI WT and D450A positive telomeres in ALT positive and negative cells. Tel, direct overlying colocalization to telomeres; Mean + s.e.m., n=2.
See also Figure S4.
Consistent with the observed increases in resection, homologous recombination proteins BRCA1 and Rad51 were present directly overlying 20-60% of telomeres in ALT cells and only 5-15% of telomeres in telomerase positive cells after TRF1-FokI DSB induction (Figures 4D and E). 53BP1 immunofluoresence juxtaposed BRCA1 in both ALT and telomerase positive cells, consistent with known differences of BRCA1 and 53BP1 chromatin localization adjacent to DSBs (Figures 4D and S4B) (Chapman et al., 2012; Tang et al., 2013).
Rad51 and the HR Machinery Control Directional ALT Telomere Movement and Clustering
These observations raise the possibility of homology directed telomere movement, analogous to the reported Rad51 dependency for DSB movement that occurs during homology searches in yeast (Dion et al., 2012; Kalocsay et al., 2009; Mine-Hattab and Rothstein, 2012; Oza et al., 2009). To test this hypothesis, we examined TRF1-FokI induced telomere clustering following siRNA-targeted depletion of factors involved in either HR or NHEJ (Figure 5A and S5A). Knockdown of NBS1 and SMC5 reduced telomere clustering in accord with their known involvement in ALT (Potts and Yu, 2007; Wu et al., 2003; Zhong et al., 2007). Similar reductions were observed in cells following knockdown of either BRCA2 or Rad51, but not 53BP1 (Figure 5A) (Jiang et al., 2007). Interestingly, ALT telomere clustering was independent of BRCA1, consistent with the HR competency of cells that exhibit extensive resection as a consequence of 53BP1 deficiency (Bouwman et al., 2010; Bunting et al., 2010).
Figure 5. Rad51 Promotes Diffusive and Directed ALT Telomere Movement.
(A) Quantification of TRF1-FokI induced telomere clustering at 72 hours following transfection of siRNA targeted to the indicated genes. Mean ± s.e.m., n=3. p values refer to tests between siLuciferase and indicated siRNA. ** p<0.005, n.s., p>0.05. See Experimental Procedures for analysis of clustering.
(B) Top panels, FISH image of a VA13 cell expressing GFP-Rad51, hybridized with telomeric PNA probe. Last panel shows an expanded area demarcated by the white box. Bottom panels, fluorescence image of a VA13 cell co-expressing mCherry-TRF1-FokI and GFP-Rad51.
(C) Representative montage of a live-cell telomere clustering event in a VA13 cell expressing mCherry-TRF1-FokI and GFP-Rad51.
(D) MSD analysis of telomere movement after Rad51 knockdown in U2OS cells. Nuclease inactive D450A is shown for reference. Fit determined by a diffusion model, MSD = Γtα. Error bars, weighted s.e.m. and n > 450 tracks from two independent experiments.
(E) Number of telomere clustering events that occur per nucleus following directed movement is quantified after Rad51 knockdown in U2OS cells. Mean ± s.e.m. from two independent experiments. *** p<0.0005.
Rad51 molecules nucleate onto RPA-coated ssDNA forming a dynamic nucleoprotein filament which mediates the presynaptic search for homology (Renkawitz et al., 2014). Remarkably, expression of GFP-tagged Rad51 in VA13 cells allowed visualization of GFP-Rad51 filaments that originate specifically at telomeres and extended to distant telomeres (Figure 5B). Live cell imaging revealed that clustering could proceed by rapid shortening of the GFP-Rad51 filament with synchronous directional movement of the incoming telomere (Figures 5C and Movie S4). Of 35 clustering events in cells in which a bridging filament formation was evident, 86% showed Rad51 localization. Rad51 filament could be directly visualized between recombining telomeres in approximately 46% of cases in which Rad51 was observable at telomeres (Figure S5B).
MSD analysis revealed that Rad51 knockdown restricted telomere mobility as well as telomere clustering events that occur as a result of directed movement (Figures 5D, E and Movies S5, S6). Interestingly, telomere clustering was decreased by expression of an ATPase defective dominant negative mutant of Rad51, K133R, which inhibits HR in mouse cells and has been reported to lock Rad51 filaments into an extended conformation that cannot transition to a compressed filament (Figure S5C) (Robertson et al., 2009; Stark et al., 2002).
Hop2-Mnd1 Regulate ALT Telomere Movement and Recombination
TRF1-FokI DSB-induced telomere recombination resembles certain aspects of recombination between homologous chromosomes during meiosis, which is also initiated by programmed DSBs and requires RecA homologs Rad51 and Dmc1. The Hop2-Mnd1 heterodimer is necessary for Dmc1 and Rad51 dependent inter-homolog recombination in vivo during gametogenesis in yeast and in mice (Leu et al., 1998; Petukhova et al., 2003), and strongly stimulates Rad51 or Dmc1 dependent D-loop formation in vitro (Bugreev et al., 2014; Chi et al., 2007; Petukhova et al., 2005; Pezza et al., 2007). Moreover, Hop2-Mnd1 or Dmc1 mutant yeast and mice display epistasis with respect to meiotic chromosome inter-homolog synapsis. Hop2-Mnd1 binds double stranded DNA and induces rapid condensation of large stretches of DNA in vitro, consistent with its requirement for homolog synapsis (Pezza et al., 2010).
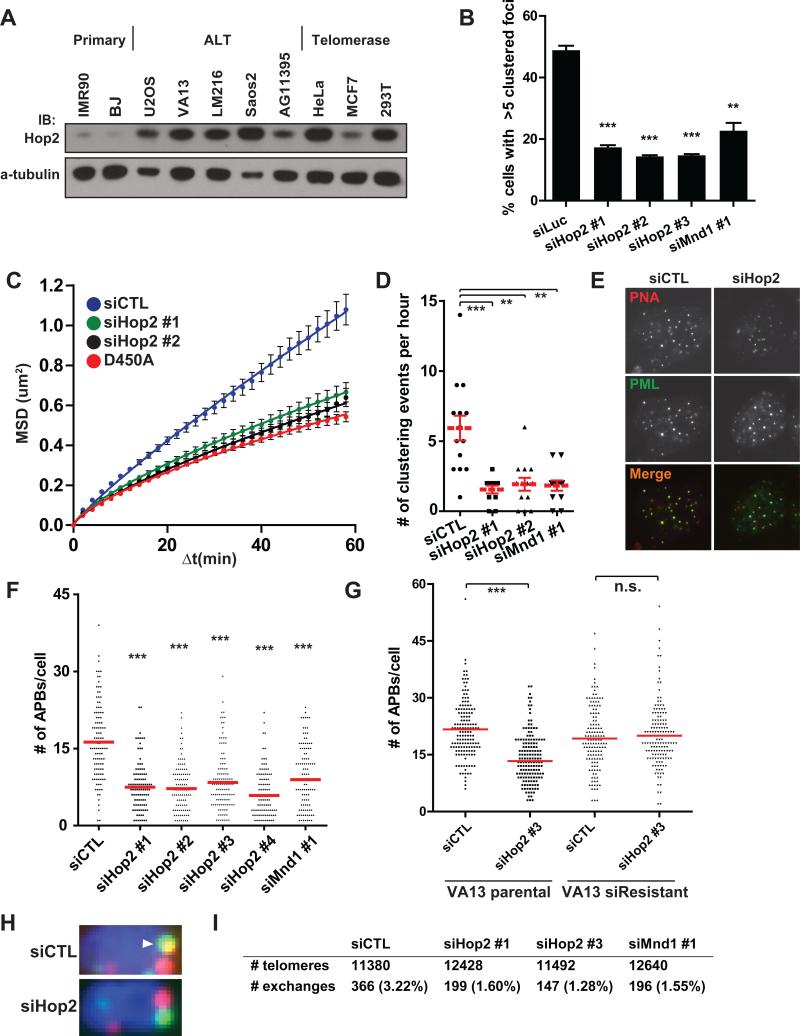
Hop2 protein was broadly expressed in all 16 different ALT cell lines and in telomerase positive cancer cell lines tested, with lower levels detected in primary human fibroblasts (Figures 6A and S6A-C). Endogenous Hop2 localized to approximately 10-20% of TRF1-FokI damaged telomeres in VA13 cells and at lower levels in the absence of TRF1-FokI (Figure S6E, F). GFP-Hop2 foci localized adjacent to telomeres in a subset of ALT cells and foci formation was completely ablated by an M110P point mutant within the Hop2 Leucine Zipper domain (Figures S6G, H). This domain is required for homolog pairing and recombination, with the Leucine Zipper also being necessary for Hop2 dependent D-loop formation in vitro (Pezza et al., 2006). Hop2 or Mnd1 knockdown strongly reduced telomere clustering, mobility and directional movement to levels observed in D450A control cells (Figures 6B-D, S6D and Movie S7). Hop2-Mnd1 depletion did not affect Rad51 localization to damaged telomeres (Figure S6I), in agreement with established roles for the heterodimer in meiotic inter-homolog pairing but not Rad51 or Dmc1 recruitment to Spo11 dependent DSBs (Petukhova et al., 2003).
Figure 6. Hop2-Mnd1 Regulate Telomere Clustering and Recombination in ALT.
(A) Western blotting was performed in the indicated cell lines using an antibody that recognizes the endogenous Hop2 protein.
(B) Telomere clustering in U2OS cells expressing TRF1-FokI was quantified as in (5A) after Hop2-Mnd1 knockdown. Mean ± s.e.m., n = 3. ** p<0.005, *** p<0.0005.
(C-D) Live-cell analysis of telomere movement in U2OS cells expressing mCherryTRF1-FokI WT was used to quantify telomere clustering after knockdown with the indicated siRNAs. D450A is shown for reference. Fit determined by MSD = Γtα. Error bars, weighted s.e.m. and n > 550 tracks from two independent experiments. ** p<0.005, *** p<0.0005.
(E) Representative images of telomere colocalization with PML foci in VA13 cells after control or Hop2 knockdown.
(F) Spontaneous APB formation was assessed in VA13 cells after serial knockdown of Hop2 or Mnd1 (Extended Experimental Procedures). Mean from >100 cells from 3 experiments.
(G) Spontaneous APB formation was assessed following Hop2 siRNA targeting the 3’ UTR region in parental VA13 cells and in VA13 cells stably expressing Hop2 cDNA. Mean from >100 cells from 3 experiments.
(H) Representative examples of telomere exchanges from CO-FISH are shown for cells with control or Hop2 knockdown (siRNA #1). The arrowhead reveals a T-SCE. Full images shown in Figure S7C.
(I) Quantification of total number of exchanges from CO-FISH assay after Control, Hop2 or Mnd1 knockdown. >50 metaphases in two independent experiments.
See also Figures S6, S7 and Movie S7.
To determine if these results would be recapitulated with respect to telomere clustering and recombination in ALT cell lines that did not express TRF1-FokI, we examined several different ALT lines for spontaneous APB formation following knockdown of Hop2 or Mnd1 with 5 different targeting siRNAs (Figures 6E, F and S7A, B). Knockdown of either Hop2 or Mnd1 significantly reduced APB formation in each of these lines. The reduction in APBs could be fully rescued by stable expression of full length Hop2, which is resistant to a siRNA targeted to the 3’UTR (Figure 6G). To assess the impact of Hop2-Mnd1 on ALT telomere recombination, telomere chromatid exchanges were assessed by chromosome orientation-FISH (CO-FISH). Knockdown of Hop2 or Mnd1 reduced telomere chromatid exchanges by 50% or greater in ALT cells (Figures 6H, I and S7C). Collectively, these data reveal that the forces driving directional telomere movement are intimately connected to the mechanism of ALT telomere recombination based lengthening (Figure 7).
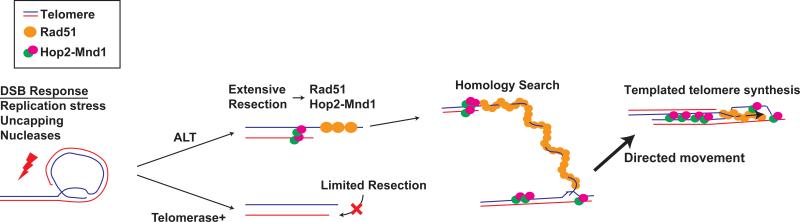
Figure 7. Model for a Specialized Homology Search Mechanism that drives ALT Telomere Recombination.
A specialized homology searching mechanism is required for synapsis between distant telomeres. Extensive end resection at ALT telomeres facilitates a Rad51 dependent homology search. Homology capture followed by synapsis and congression of homologously paired non-sister telomeres would be responsible for directional telomere movement. This ALT telomere recombination mechanism relies in part on Rad51 and Hop2-Mnd1 to promote synapsis between non-sister telomeres.
Discussion
The phenomenon of DSB movement has been described in prokaryotes, yeast and also in mammalian cells within distinct experimental contexts (Aten et al., 2004; Dimitrova et al., 2008; Dion et al., 2012; Kalocsay et al., 2009; Lesterlin et al., 2013; Mine-Hattab and Rothstein, 2012; Oza et al., 2009; Roukos et al., 2013). Telomeres appear to be a particularly predisposed genomic location to DNA damage induced mobility increases. Diffusive movement of damaged telomeres in telomerase positive cells has been reported in several independent studies (Chen et al., 2013; Dimitrova et al., 2008). Notably, the NHEJ promoting factor 53BP1 was required for movement of deprotected mouse telomeres. However, TRF1-FokI induced directional ALT telomere mobility required HR factors and was independent of 53BP1, indicative of distinct mechanisms underlying telomere mobility in each case. We postulate that extensive end resection and more prominent accumulation of HR factors at damaged ALT telomeric chromatin contributes to these differences (Figure 7).
In this study, expression of TRF1-FokI enabled quantitative characterization of an unanticipated type of chromatin movement. Damaged ALT telomeres initially roamed a larger nuclear territory at greater velocities than D450A controls, but notably, these movements culminated in rapid and directional movements of up to 5μm to synapse with a more stationary recipient telomere. These displacements were also much larger in magnitude and occurred over a longer time period than those of stochastic unidirectional “jumps” that could be seen in interphase chromatin (Levi et al., 2005). We note, however, that pre-selection of clustering tracks in our analysis introduces a bias of describing only highly mobile particles. We limit our analysis of directionality to clustering telomeres, and do not preclude the possibility that a proportion of non-clustering telomeres could move directionally.
To our knowledge, directional ALT telomere movement provides the first example of real time visualization of homology searches and synapsis in mammalian cells. Given our data, we favor a model in which Rad51 nucleoprotein filaments interrogate surrounding nuclear space, leading to homology capture of a non-sister telomere and subsequent directional movement during synapsis (Figure 7). Interestingly, dynamic formation of long stretches of prokaryotic RecA coated filaments mediated rapid associations between DSBs and homologous genomic regions that are separated by 1.3μm (Lesterlin et al., 2013), which are similar to the distances of directional phase movement we describe for ALT telomeres. The reported structure of ssDNA filaments in association with RecA reveals an extended conformation that is stretched to ~1.5 fold longer B-form DNA (Chen et al., 2008). Thus, it is predicted that 1.3μm of nuclear space connecting non-sister telomeres could theoretically require only ~2.5kb of Rad51 ssDNA filament for directional movement, which is well within the length possible for ALT telomeres. Furthermore, as vertebrate telomeres contain extensive regions of homology consisting of TTAGGG repeats, in effect every chromosome is a “homolog” with respect to telomere recombination. This feature of primary telomere sequence would be predicted to increase the probability of recombination between different chromosomes, enabling successful capturing of distant homology on timescales similar to those observed in much smaller genomes. It should also be noted that not all recombining telomeres displayed Rad51 foci, consistent with the presence of Rad51 independent mechanisms of ALT in type II survivors of telomerase deficiency in yeast (Chen et al., 2001).
Several obvious parallels exist between meiotic recombination and ALT. Both processes involve DSB responses to initiate recombination between homologous DNA sequences on non-sister chromatids. Hop2-Mnd1 uniquely contributes to chromosome pairing in meiotic recombination and ALT, but is not known to be important for sister chromatid recombination. Both constituents of this heterodimer are broadly expressed in ALT and telomerase positive cancers, yet appear to promote telomere recombination only in cells that use ALT. This may be a consequence of the known interaction of Hop2-Mnd1 with Rad51, which did not efficiently nucleate damaged telomeres in telomerase positive cells. It is also plausible that other factors related to the specific chromatin environment in ALT cells, such as the absence of ATRX and the association between ALT and defective histone chaperone activity (Heaphy et al., 2011; Lovejoy et al., 2012; O'Sullivan et al., 2014; Schwartzentruber et al., 2012), may promote inter-telomere recombination. Mechanistic studies into this process are warranted, as is the extent to which ALT recapitulates known mechanisms of meiotic recombination.
Experimental Procedures
Cell culture
Unless otherwise stated in the Extended Experimental Procedures, all cell lines were grown in DMEM (Invitrogen) with 10% calf serum and 1% penicillin/streptomycin. VA13 cell line refers to WI-38 VA-13 subline 2RA.
Transfections and lentiviral transductions
Transient plasmid transfections were carried out with LipoD293 (Signagen), and siRNA transfections with Lipofectamine RNAiMax (Invitrogen) according to manufacturer’s instructions. Concentrated TRF1-FokI lentivirus with polybrene (8ug/ml) diluted in media was added to cells at a minimum titer resulting in greater than 90% expression at 24 hours by immunofluorescence. Analyses were performed 16 hours after transfection of plasmids, and 48-72 hours after siRNA transfection. Analyses were performed 24 hours after transduction of cells with Flag-TRF1-FokI lentivirus.
Immunofluorescence, IF-PNA FISH and subtelomeric FISH
Cells grown on coverslips were fixed in 4% paraformaldehyde for 10 minutes at room temperature. Fixed coverslips were permeabilized in 0.5% Triton X100 for 5 minutes at 4°C. Primary antibody incubation was performed at 37°C in a humidified chamber for 20 minutes unless otherwise indicated. For immuno-FISH, coverslips were first stained with the primary antibody, fixed for 10 minutes at room temperature and dehydrated in ethanol series. After denaturation at 75°C for 5 minutes, coverslips were incubated with TelC-Cy3 PNA probe (Panagene) overnight at room temperature, then washed and mounted. For subtelomeric FISH, cells were fixed in freshly prepared 3:1 methanol:acetic acid for 5 minutes and dehydrated in ethanol series. Subtelomeric probes (Cytocell), were mixed with PNA probe in hybridization buffer (65% deionized formamide, 10% dextran sulfate, 2X SSC). Coverslips inverted onto the hyb + probe mix were denatured for 2 minutes at 75°C and incubated overnight at 37°C. Coverslips were washed once with Agilent FISH wash buffer 1 for 2 minutes at 65°C, washed once with wash buffer 2 for 1 minute and mounted.
Live cell imaging
Cells were transfected with mCherry-ER-DD-TRF1-FokI 16 hours prior to induction with 4-OHT and Shield1 ligand for 60 minutes. Confocal images were acquired under temperature controlled conditions calibrated to 37°C, using a 100× 1.4 NA objective on an inverted fluorescence microscope (DM6000, Leica Microsystems) equipped with an automated XYZ stage (Ludl Electronic Products), a charge-coupled device camera (QuantEM 512SC, Photometrics), an X-LIGHT Confocal Imager (Crisel Electrooptical Systems), and a SPECTRA X Light Engine (Lumencor), controlled by Metamorph Software (MDS Analytical Technologies). Images were collected as z stacks at 0.6 μm intervals that covered the entire nucleus, at 2-minute intervals for a total of 60 minutes. A detailed description of MSD analysis can be found in the Extended Experimental Procedures.
Analysis of telomere foci size and clustering
For measurements of telomeric foci size, ImageJ (NIH) was used to apply a constant threshold to images and subsequent binarization. Foci sizes were measured as square pixels for each telomeric focus within a nucleus and the average size was calculated for each nucleus analyzed. For analysis of clustering following siRNA transfection, a cluster was defined as a telomeric focus equal to or greater than fourfold the area as based on the radius, compared to the average size of undamaged telomeres.
Statistics
Unpaired t tests were used to generate two-tailed p values.
Supplementary Material
Highlights.
⍰ DSB responses initiate interchromosomal associations between ALT telomeres.
⍰ Homology search and synapsis can be visualized in real time in mammalian cells.
⍰ ALT telomeres associate via rapid directional movements of up to 5 ⍰m.
⍰ ALT telomere movement, synapsis and recombination require Rad51 and Hop2-Mnd1.
Acknowledgments
We thank Q. Jiang, G. Yuan, F.B. Johnson, R. Marmorstein, and D. Roth for critical discussion, and F.B. Johnson, R. Reddel, A. Sfeir and T. de Lange for sharing cell lines. We thank L. Kang and C. Yu for critical discussion of mobility data. This work was supported by NIH grants GM101149, CA138835, and CA17494, (to RAG) who is also supported by a DOD Breast Cancer Idea Award, a Harrington Discovery Institute Scholar-Innovator Award, a Pennsylvania Breast Cancer Coalition grant, and funds from the Abramson Family Cancer Research Institute and Basser Research Center for BRCA. NWC is supported by a HHMI International Student Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
N.W.C., M.A.L. and R.A.G. designed the study, analyzed the data and wrote the paper. N.W.C. and R.L.D. conducted the experiments.
The authors declare no conflict of interests. The authors declare no competing financial interest.
References
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter OE, Zou Y, Shay JW, Wright WE. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 2003;4:1138–1143. doi: 10.1038/sj.embor.7400027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK. RecA homologues Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, vander Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Huang F, Mazina OM, Pezza RJ, Voloshin ON, Daniel Camerini-Otero R, Mazin AV. HOP2-MND1 modulates RAD51 binding to nucleotides and DNA. Nat Commun. 2014;5:4198. doi: 10.1038/ncomms5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol Cell. 2013;51:141–155. doi: 10.1016/j.molcel.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Karlseder J. A three-state model of telomere control over human proliferative boundaries. Curr Opin Cell Biol. 2012;24:731–738. doi: 10.1016/j.ceb.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci. 2012;125:3529–3534. doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Leonhardt H, Rippe K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J Cell Sci. 2011;124:3603–3618. doi: 10.1242/jcs.084681. [DOI] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. Embo J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londono-Vallejo A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci. 2009;106:15726–15731. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Fasching CL, Neumann AA, Muntoni A, Yeager TR, Reddel RR. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 2007;67:7072–7077. doi: 10.1158/0008-5472.CAN-07-1556. [DOI] [PubMed] [Google Scholar]
- Guigas G, Weiss M. Sampling the cell with anomalous diffusion - the discovery of slowness. Biophys J. 2008;94:90–94. doi: 10.1529/biophysj.107.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegou T, Chung I, Heuvelman G, Wachsmuth M, Gorisch SM, Greulich-Bode KM, Boukamp P, Lichter P, Rippe K. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol Biol Cell. 2009;20:2070–2082. doi: 10.1091/mbc.E08-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Krawczyk PM, Borovski T, Stap J, Cijsouw T, ten Cate R, Medema JP, Kanaar R, Franken NA, Aten JA. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci. 2012;125:2127–2133. doi: 10.1242/jcs.089847. [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2013 doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLOS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G, Mimitou EP, Symington LS. SnapShot: Homologous recombination in DNA double-strand break repair. Cell. 2010;142:646, 646 e641. doi: 10.1016/j.cell.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14:510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- Molenaar C, Wiesmeijer K, Verwoerd NP, Khazen S, Eils R, Tanke HJ, Dirks RW. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J. 2003;22:6631–6641. doi: 10.1093/emboj/cdg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Ann Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Romanienko PJ, Camerini-Otero RD. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Camerini-Otero RD, Bianco PR. Hop2-Mnd1 condenses DNA to stimulate the synapsis phase of DNA strand exchange. Biophys J. 2010;99:3763–3772. doi: 10.1016/j.bpj.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza RJ, Petukhova GV, Ghirlando R, Camerini-Otero RD. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J Biol Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Lademann CA, Jentsch S. Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol. 2014;15:369–383. doi: 10.1038/nrm3805. [DOI] [PubMed] [Google Scholar]
- Robertson RB, Moses DN, Kwon Y, Chan P, Chi P, Klein H, Sung P, Greene EC. Structural transitions within human Rad51 nucleoprotein filaments. Proc Natl Acad Sci. 2009;106:12688–12693. doi: 10.1073/pnas.0811465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. A biological interpretation of transient anomalous subdiffusion. I. Qualitative model. Biophys J. 2007;92:1178–1191. doi: 10.1529/biophysj.106.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Hu P, Pierce AJ, Moynahan ME, Ellis N, Jasin M. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- Wu G, Jiang X, Lee WH, Chen PL. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589–2595. [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Zhong ZH, Jiang WQ, Cesare AJ, Neumann AA, Wadhwa R, Reddel RR. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.