Abstract
The cohesin complex is at the heart of many chromosomal activities, including sister chromatid cohesion and transcriptional regulation1-3. Cohesin loading onto chromosomes depends on the Scc2/Scc4 cohesin loader complex4-6, but the chromatin features that form cohesin loading sites remain poorly understood. Here, we show that the RSC chromatin remodeling complex recruits budding yeast Scc2/Scc4 to broad nucleosome-free regions, that the cohesin loader itself helps to maintain. Consequently, inactivation of the cohesin loader or RSC complex have similar effects on nucleosome positioning, gene expression and sister chromatid cohesion. These results reveal an intimate link between local chromatin structure and higher order chromosome architecture. Our findings pertain to the similarities between two severe human disorders, Cornelia de Lange syndrome, caused by mutations in the human cohesin loader, and Coffin-Siris syndrome, resulting from mutations in human RSC complex components7-9. Both could arise from gene misregulation due to related changes in the nucleosome landscape.
The Scc2/Scc4 complex loads cohesin onto DNA in vitro in a sequence-independent manner10. In vivo, the cohesin loader associates with actively transcribed genomic regions11-15, but the cause or possible consequences of this distribution remain poorly understood. To identify determinants of Scc2/Scc4 localization, we generated high resolution genome-wide binding maps of the budding yeast cohesin loader using chromatin immunoprecipitation (ChIP) against its two subunits, fused to Pk-epitope tags at their endogenous gene loci (Fig. 1a). This allowed us to assign 423 peaks that were enriched in intergenic regions (Supplementary Fig. 1a). We have previously observed Scc2/Scc4 binding at ribosomal protein (RP) genes and tRNA genes12,14. These features were also seen in our new map, where the improved resolution showed that Scc2/Scc4 preferentially binds within RP gene promoters (Fig. 1a and Supplementary Fig. 1b). When we compiled mRNA levels of the genes nearest to each Scc2/Scc4 binding site, we found a bimodal distribution of expression levels with overall transcript levels greater than the genome average (Supplementary Fig. 1c). We validated five of the detected Scc2/Scc4 peaks using ChIP followed by quantitative real time PCR and performed a control ChIP experiment using a strain lacking an epitope tag to confirm the specificity of our detection (Supplementary Fig. 1d,e).
Figure 1. Scc2/Scc4 associates with promoters characterized by broad nucleosome-free regions.
a, ChIP-sequencing analysis of Scc2 and Scc4, three representative regions on chromosome 2 are shown. b, Correlation of Scc2/Scc4 binding sites with transcription factor binding. The total number of bound promoters16 and those with an Scc2/Scc4 peak within 1 kb of the TSS are listed, together with the false discovery rate (FDR, see Methods for details). c, Sequence motifs found at Scc2/Scc4 binding sites. The number of occurrences and the average distance from the Scc2/Scc4 peak summit ± its variance are shown. d, The cohesin loader binds broad nucleosome-free regions. Shown is the averaged nucleosome profile at Scc2/Scc4-bound genes, centered at their TSS (black). The nucleosome profiles of four gene clusters21 in our experimental strain is shown, together with the relative enrichment of Scc2/Scc4-bound genes in these clusters.
To identify chromosomal features that could explain Scc2/Scc4 binding, we compared its pattern to published genome-wide distributions of chromatin landmarks16-18. This revealed colocalization with the transcription factor Fhl1 and the high mobility group protein Hmo1, known regulators of RP gene promoters (Fig. 1b and Supplementary Table 1). When we excluded Scc2/Scc4 peaks at RP genes from the analysis, these associations lost significance, suggesting that Fhl1 and Hmo1 do not globally explain the Scc2/Scc4 binding pattern. Correlations with additional transcription factors extended to only a small number of Scc2/Scc4 peaks (Fig. 1b). When comparing Scc2/Scc4 binding with histone occupancy and modifications19, we found a negative correlation with histone occupancy and the histone variant Htz1, as well as significant colocalization with H3K9 and H4 acetylation marks (Supplementary Table 1). These features are suggestive of Scc2/Scc4 binding to promoters of active genes, however, they do not explain which of the many active promoters are chosen to become Scc2/Scc4 binding sites.
We next took an unbiased approach to look for DNA sequence elements at Scc2/Scc4 binding sites20. This identified the RNA polymerase III B-Box promoter element, restricted to binding sites at tRNA genes (Fig. 1c and Supplementary Fig. 2). Notably, an oligo(A) tract was present at over half of cohesin loader binding sites. Oligo(A) tracks are thought to disfavor nucleosome binding due to their relative stiffness. Prompted by this, we compared the Scc2/Scc4 binding pattern with the genome-wide nucleosome distribution.
Nucleosome signatures at budding yeast transcriptional start sites (TSSs) have been grouped into four clusters that correlate with gene function and transcript abundance21. Scc2/Scc4 binding sites were strikingly enriched at TSSs characterized by broad and shallow nucleosome-free regions (“cluster 2”; Fig. 1d and Supplementary Table 2). The correlation with broad shallow nucleosome free regions held up for all subsets of Scc2/Scc4 binding sites at tRNA, RP and all other genes. We visualized nucleosome distribution at Scc2/Scc4 binding sites by averaging the nucleosome profiles around TSSs of all Scc2/Scc4 bound genes. This confirmed that Scc2/Scc4 peaks are associated with broad, shallow nucleosome-free regions (Fig. 1d).
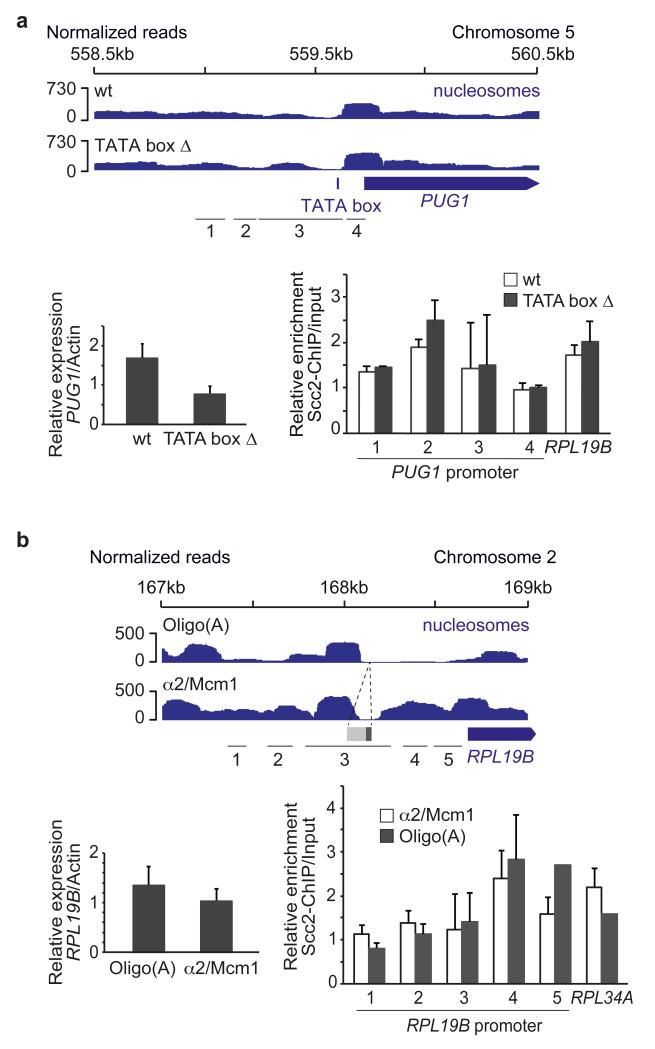
Determinants of the nucleosome landscape at TSSs are as yet poorly understood22-24. Because transcription has been previously linked to cohesin loading11,13,15, we first tested whether active transcription promoted Scc2/Scc4 recruitment. We deleted the TATA box core promoter element of the non-essential PUG1 gene25. PUG1 expression was downregulated following the TATA box deletion, while the nucleosome pattern in its promoter remained unchanged, as did Scc2/Scc4 binding (Fig. 2a). This rules out active transcription or core promoter binding proteins as a major driving force for Scc2/Scc4 recruitment.
Figure 2. Scc2/Scc4 recruitment is independent of an active, open promoter.
a, Scc2/Scc4 binding persists following transcriptional downregulation by TATA box deletion. Nucleosome profiles, PUG1 mRNA levels and cohesin loader binding at the PUG1 promoter, and the RPL19B promoter as a control, are compared. b, Cohesin loader recruitment is independent of a nucleosome free region. Nucleosome profiles, RPL19B mRNA levels and Scc2/Scc4 binding are shown at the RPL19B promoter, and the RPL34A promoter as a control, following replacement of the oligo(A) sequence within the RPL19B promoter by an α2/Mcm1 binding site. The means and standard error of at least 3 independent experiments are shown.
We next turned to the RP gene transcription factor Fhl1. As reported26, its deletion resulted in marked downregulation of RP gene transcription, but this did not change Scc2/Scc4 binding, at least at the RPL34A and RPL19B promoters (Supplementary Fig. 3). This confirms that Scc2/Scc4 recruitment is independent of strong transcription and suggests that it is also independent of a main gene-specific transcription factor, at least at RP genes.
To directly evaluate the importance of a nucleosome-free region for Scc2/Scc4 recruitment, we replaced the oligo(A) stretch in the RPL19B promoter with an α2/Mcm1 repressor binding site, which is thought to act as nucleosome positioning signal27. In response, the nucleosome-free region in the RPL19B promoter was filled, accompanied by a small decrease in RPL19B transcription, yet cohesin loader binding at this locus remained unchanged (Fig. 2b). We conclude that Scc2/Scc4 binding sites are characterized by their nucleosome signature, but being nucleosome free is not what determines Scc2/Scc4 recruitment.
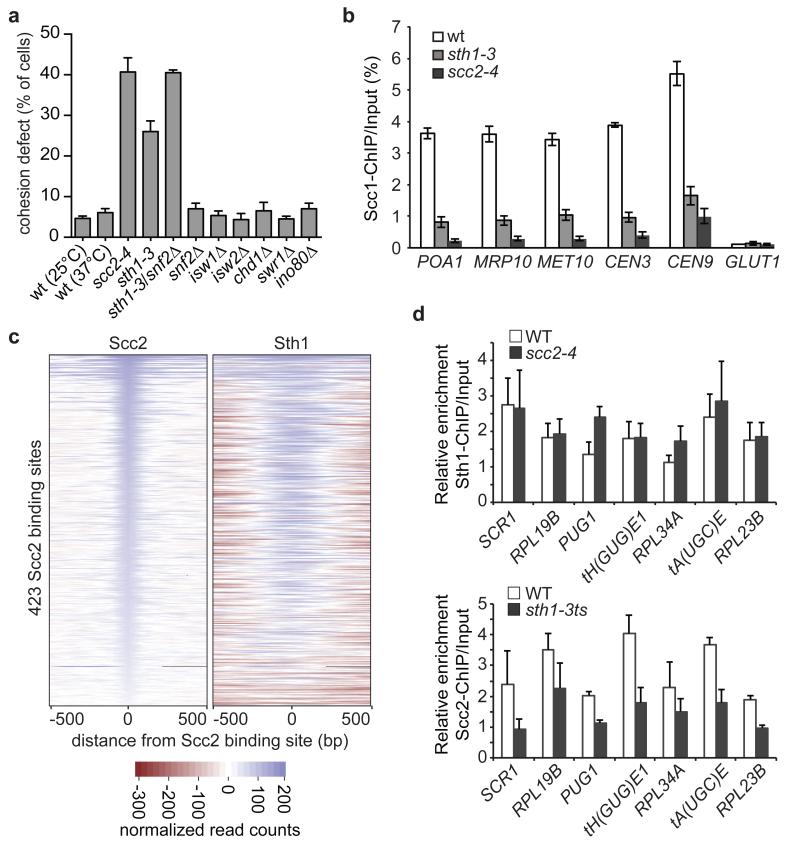
We addressed whether a common upstream regulator is responsible for both the nucleosome landscape at cohesin loading sites and for recruiting the Scc2/Scc4 complex. Chromatin remodeling complexes are prime candidates for this role, and SWI/SNF, ISWI and CHD family chromatin remodelers have variably been linked to cohesin function28-31. To gain insight into which chromatin remodelers contribute to sister chromatid cohesion we inactivated each of the eight known budding yeast remodeler ATPases32. RSC complex inactivation using the temperature sensitive sth1-3 allele33 led to a marked loss of sister chromatid cohesion, while deletion of snf2, isw1, isw2, chd1, swr1 or ino80 did not cause detectable defects (Fig. 3a). Sth1 and Snf2 are two SWI/SNF-family chromatin remodeling ATPase in yeast. While snf2 deletion by itself did not compromise sister chromatid cohesion, its deletion in the sth1-3 background increased the cohesion defect to levels comparable to those seen after Scc2/Scc4 inactivation by the scc2-4 allele5. This suggests that the two budding yeast SWI/SNF chromatin remodelers together are of equal importance for sister chromatid cohesion as the Scc2/Scc4 cohesin loader.
Figure 3. Scc2/Scc4 is recruited by the RSC chromatin remodeling complex.
a, Sister chromatid cohesion at the GFP-marked URA3 locus was examined in mitotically arrested cells of the indicated genotypes. Cells containing the scc2-4 or sth1-3 alleles were observed at 37°C, all others at 25°C. The means and standard error of 3 independent experiments are shown. b, Cohesin levels detected by ChIP against its Scc1 subunit followed by qPCR at 5 cohesin binding sites37 and a negative control site in the indicated strains arrested in mitosis at 37°C. The means and standard error of 3 independent experiments are shown. c, Colocalization of Scc2 and Sth1. Heatmaps of the Scc2 and Sth1 ChIP-Seq counts, centered on the 423 Scc2 binding sites. d, Interdependence of Scc2 and Sth1 binding at 7 of their binding sites analyzed by ChIP-qPCR analysis. The means and standard error of 3 independent experiments are shown.
In previous studies, no effect or only a partial effect on cohesin loading at chromosome arms, but not centromeres, was observed after RSC inactivation29,30. To clarify the role of RSC in cohesin loading, we quantitatively analyzed cohesin levels on chromosomes following sth1-3 inactivation. (Fig. 3b). This revealed marked cohesin loss at both centromeres and along chromosomes arms, close to what is observed after cohesin loader inactivation. We were unable to combine sth1-3 with snf2 deletion in this assay because of the poor growth of the double mutant strain. These results suggest that chromatin remodeling plays an integral part in loading cohesin onto chromosomes.
To investigate a possible relationship between RSC and Scc2/Scc4, we performed ChIP against Sth1. This revealed significant genome-wide colocalization of Sth1 with Scc2 (Fig. 3c and Supplementary Fig. 4), as well as a shared preference with the cohesin loader for “cluster 2” nucleosome-free regions (Supplementary Table 2). Quantitative analysis of Sth1 chromatin immunoprecipitates confirmed RSC binding to tRNA genes22 as well as SCR1, PUG1 and ribosomal protein gene promoters (Fig. 3d>). To analyze the hierarchy of binding, we repeated Sth1 ChIP after cohesin loader inactivation. This did not alter RSC occupancy at its binding sites. In contrast, Scc2 levels at the same sites were markedly reduced after RSC inactivation (Fig. 3d). These findings suggest that the RSC complex acts upstream to recruit Scc2/Scc4 to chromosomes.
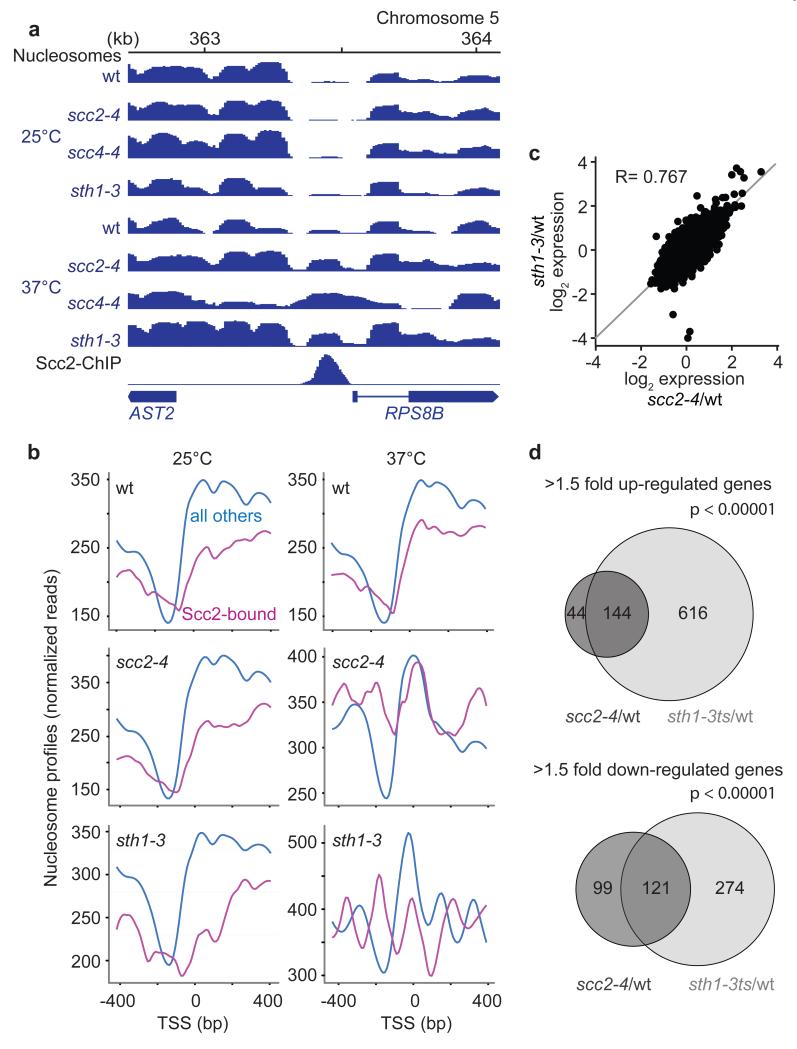
Having identified RSC as determinant for Scc2/Scc4 binding, we asked whether the relationship between the two protein complexes extended beyond a mere recruitment role. We analyzed the consequences of RSC or cohesin loader inactivation on nucleosome positioning at the TSSs of Scc2/Scc4-bound genes. After shift of sth1-3, scc2-4 or scc4-4 strains to a non-permissive temperature, these nucleosome-free regions were filled by a nucleosome, while they remained unchanged in wild type cells at this temperature. This is shown at the RPS8B and RPL19B genes, as examples (Fig. 4a and Supplementary Fig. 5), and in the average nucleosome profile at all Scc2/Scc4-bound promoters (Fig. 4b). The nucleosome free region at other promoters remained unaffected by RSC or cohesin loader inactivation. This reveals that the Scc2/Scc4 complex cooperates with RSC to maintain nucleosome depletion at its binding sites.
Figure 4. Shared roles of Scc2/Scc4 and RSC in chromatin remodeling and transcriptional regulation.
a, Nucleosome profiles and the Scc2-ChIP profile at the RPS8B promoter in the indicated strains at 25°C and 37°C. b, Average plot of the nucleosome profiles at promoters of Scc2/Scc4-bound genes, compared to all other promoters, under the conditions in a. c, Comparison of genome wide expression changes in scc2-4 and sth1-3 strains, each compared to wild type, at the restrictive temperature. The Pearson correlation coefficient (R) is given. d, Overview of gene expression changes following Scc2 or Sth1 inactivation, compared to wild type. A hypergeometric test confirmed significant coregulation.
Cornelia de Lange syndrome, caused most often by mutations of the human cohesin loader subunit NIPBLScc2, is thought of as a ‘cohesinopathy’, based on the tacit assumption that cohesin’s role in mediating long range chromatin interactions underlies transcriptional changes that characterize the disease3,8,34. Our finding that the cohesin loader controls nucleosome positioning opens an alternative explanation for its role in transcriptional regulation. To investigate this, we compared transcriptional changes in budding yeast cells following inactivation of the cohesin loader or the RSC complex. The gene expression changes in scc2-4 and sth1-3 cells at a restrictive temperature, compared to identically treated wild type cells, were strikingly similar (Fig. 4c). 188 genes were up-regulated upon Scc2/Scc4 inactivation by greater than 1.5-fold. Over 76% of these were also up-regulated following Sth1 inactivation (Fig. 4d). Similarly, 55% of the 220 genes down-regulated in scc2-4 cells were also down in sth1-3 cells. As expected, genes whose transcription was affected by cohesin loader and RSC complex inactivation often contained an Scc2/Scc4 binding site in their promoters (Supplementary Fig. 6). Some cases of Cornelia de Lange syndrome have been difficult to discern from Coffin-Siris syndrome7, a related developmental disorder caused by mutations in human orthologs of RSC complex components9,35. Our findings offer a molecular explanation for the similarities between these two disorders and open up the possibility that gene expression changes in both disorders are due to related nucleosome positioning defects. Both diseases might therefore be more aptly known as ‘nucleosome disorders’.
How the tight spatial control over nucleosome removal by the RSC complex is achieved is incompletely understood22-24. Our identification of Scc2/Scc4 as a cofactor provides an unexpected advance, the mechanistic implications of which will be important to explore. The cohesin loader’s activities towards catalyzing topological cohesin loading onto DNA are confined to its Scc2 subunit10, making the essential Scc4 subunit a candidate for functional interaction with a chromatin remodeler. The Scc4 subunit is also implicated in cohesin loader recruitment to prereplicative complexes in Xenopus oocyte extract36, suggesting that this subunit of the complex engages with a chromatinized DNA template. In this way, the cohesin loader both creates an accessible DNA template and performs the cohesin loading reaction. Our findings highlight the importance of the nucleosome landscape in determining the higher order chromosome architecture rendered by the cohesin complex and open a new window for understanding the molecular basis of severe human developmental disorders.
Online Methods
Yeast strains and culture
All strains used in this study were derivatives of W303, a list of strains can be found in Supplementary Table 3. Gene deletions and epitope tagging of endogenous genes were performed by gene targeting using polymerase chain reaction (PCR) products38,39. Deletion of the TATA box was achieved by replacing the TATAAAAG sequence in the PUG1 promoter (coordinates 559328 to 559335 on chromosome 5 as annotated in the Saccharomyces Genome Database) with a URA3 auxotrophic marker flanked by 142 bp direct repeats40. The URA3 insertion was then lost after counter selection on 5-fluoroorotic acid (5-FOA), leaving one repeat behind. As a control, a strain containing the 142 bp repeat insertion in front of the TATA box was created. A similar strategy was followed to replace the oligo(A) tract within the RPL19B promoter with an α2/MCM1 binding site. The (A)16 sequence between positions 167978 and 167993 on chromosome 2 was replaced with the URA3 marker, flanked by the repeats appended to the α2/MCM1 binding site41 from the STE6 promoter, TGTAATTACCTAATAGGGAAATTTACA. The URA3 insertion was again lost after counter selection on 5-FOA, leaving one repeat and the α2/MCM1 recognition sequence behind. The scc2-4 and scc4-4 temperature sensitive alleles were previously described5,42, the sth1-3 allele was recreated based on its published mutant sequence33. Cells were grown in YPD medium at the indicated temperatures43, if not indicated otherwise.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described11. The antibodies used for ChIP were α-Pk (clone SV5-Pk1, Serotec), α-myc (clone 9E10) and α-HA (clone 12CA5). The chromatin immunoprecipitates were sequenced on a Genome Analyzer IIx (Illumina), sequencing libraries were prepared according to the manufacturer’s protocol. The datasets were aligned using Eland (version 1.4) to the Saccharomyces cerevisiae genome (version sacCer2) with the default settings. To identify protein enriched regions, the genomic distance between every pair of forward and reverse strand mapped reads was calculated and the average fragment size was taken to be the median of these distances. Half of this value was used with MACS version 1.3.7.1 in NOMODEL mode to identify regions enriched in the aligned data over a whole genome input DNA sample that was processed and sequenced in parallel44. Default settings were used apart from ‘model fold = 32’ and an effective genome size of 1.2 × 108.
To generate heat maps of Scc2 and Sth1 binding, we extracted the sequence counts within 500 bases of each peak summit. The counts were then adjusted so that in each sample, the average number of counts across all of that sample’s peaks and all loci be zero. We then plot heatmaps for each sample-peakset combination, where the horizontal coordinate represents the distance from the peak in bases, and the rows are in descending order of the count exactly over the peak summit. Loci that had no counts for a particular sample are colored grey.
Quantitative analyses of the chromatin immunoprecipitates were performed using an 7900HT Fast Real-Time PCR System (Applied Biosystems). All primer sequences are contained in Supplementary Table 4. Occupancies at individual genomic regions were calculated by dividing the amount of DNA recovered in the immunoprecipitate with the DNA level in the input sample. To make repeat experiments comparable with each other, amplification levels were then normalized by dividing each relative enrichment by the average of the relative enrichments at three negative control sites. At least three independent experiments were performed for each condition.
Scc2/Scc4 peak correlation with other genomic features
To compare the Scc2/Scc4 peak pattern with that of known transcription factors, we used the published list of ‘single-input modules’ as a source for 89 transcription factors and their respective targets16. We calculated the proportion of transcription factor targets whose TSS was within 1 kb of an Scc2/Scc4 peak summit. To simulate a null, we chose random TSSs from SGD, with the constraint that the TSS should be chosen from the same chromosome as the original target and that each transcription factor should have the correct number of random targets. For each transcription factor, we determined the expected proportion as the mean across 10,000 simulations of the null proportions, and the p-value was empirically determined by the frequency with which the actual proportion was exceeded by a null proportion. To account for multiple testing, we have provided false discovery rates calculated across p values corresponding to all 89 TFs.
To assess colocalization with other features, a threshold-based method was used. For two sets of loci A (Scc2/Scc4 peaks) and B (another feature), we calculated, for each locus in A, the distance to the nearest locus in B to give a set of distances. We then derived four statistics based on these distances: the mean, the median, the proportion that were under 2kb, and the 30th centile. To assess the relative size and significance of the observed statistics, we randomized the set B repeatedly (10,000 times). Randomization consisted of breaking each chromosome into 20 random pieces, and reconstructing them in random order, so that local clusters of loci within B would be preserved. This manipulation was applied to both in each pair of datasets symmetrically, so that we generated two sets of distances, one containing the distances of each Scc2/Scc4 peak to its nearest feature, and one containing the distance of each feature to its nearest Scc2/Scc4 peak. We report those distances that gave the more conservative result. P represents the proportion of simulations that generated greater mean distances than observed. A value of P close to 1 therefore indicates that the distances in the observed distribution are smaller than expected by chance and that therefore features A and B co-localize. Two unrelated features are expected to generate a P score of about 0.5, while a score close to 0 indicates that the features exclude each other and are found at greater distance from each other than expected by chance. Analyses using the different measures, mean or median distances, proportion under 2 kb and 30th centile, in all cases revealed qualitatively similar correlations. We report results obtained using mean distances.
Sequence motif calling
Sequence data was obtained from the Saccharomyces Genome Database S288C reference genome. A window of +/− 500 bp around the center of each peak was taken, and the 1001 bp fragments of sequence were fed into the AlignACE motif discovery algorithm using its default parameters20. Weights were assigned to the sequences based on which quantile the peak-height belonged to, and then further subdivided so that peaks within 2 kb of a tRNA were down-weighted, to give 8 equally spaced weights. The generated motifs were then ordered by the number of occurrences, the average distance of the motif to the center of the relevant peak and the variance of the position of the motif, to assess consistency of whether a motif occurred up- or downstream of the peak.
Nucleosome position analysis
The preparation of mono-nucleosomal DNA followed a published protocol45. Sequencing of MNase resistant DNA was performed on Illumina GAIIx, HiSeq 2000 or HiSeq 2500 platforms to generate at least 11 million paired-end reads. Read alignment was performed using the Burrows-Wheeler Aligner, bwa (version 0.5.9-r16)46. If required, prior to alignment, the sacCer2 release of the yeast genome was modified to reflect genome alterations that were introduced for the experiment. Discordantly mapped read-pairs were removed, leaving only those that were paired, had a maximum of 2 mismatches in any given read and an insert size between 140 -170 bp. Subsequently, genome-wide wig files were generated, treating each pair of reads as one single fragment.
To generate the average plot of the nucleosome profiles at promoters of Scc2/Scc4-bound genes, we mapped nucleosome reads for each sample around each TSS from 500 bp upstream to 500 bp downstream of the start site. We associated to each Scc2 peak summit its nearest gene. We then discarded all tRNA genes and calculated the mean read depth per locus across all remaining Scc2-associated genes, and correspondingly for all remaining unassociated genes, and plotted these for loci within 400 bp of the TSS.
Gene expression analysis
Cells from 25 ml of exponentially growing cultures were harvested by centrifugation. Total RNA was extracted using RNeasy reagents (Quiagen) following the manufacturer’s instructions. Double stranded cDNA (dscDNA) was synthesized from 10 μg of RNA using SuperScript™ II Reverse Transcriptase (Invitrogen). Random primers were used. dscDNA was coupled to biotin (Enzo Life Sciences) using Terminal Transferase (Roche) and hybridized to a GeneChip Yeast Genome 2.0 Array (Affymetrix). Two independent experiments were averaged and combined in the analysis. The raw microarray data were processed using Bioconductor47. First the data were normalized using the RMA procedure and then log fold changes and their moderated p-values were calculated using the ‘limma’ package48,49. Only probe sets that map to a verified ORF in the Saccharomyces Genome Database were included in the analysis. For the correlation of Scc2/Scc4 peaks with expression levels along budding yeast chromosomes shown in Supplementary Figure 1c, we made use of previously published expression values50.
Sister chromatid cohesion assay
To analyze sister chromatid cohesion, cells were first synchronized in G1 using α-factor. Cells were released from the α-factor block by filtration to pass through the cell cycle until arrest in metaphase, which was achieved by addition of 5 μg/ml nocodazole to the culture from a 2 mg/ml stock solution in DMSO. To inactivate the temperature sensitive scc2-4 and sth1-3 alleles, the cultures were shifted to 37°C 30 minutes before the release from the α-factor block in G1. The sister chromatid cohesion status was analyzed in the metaphase arrested cells by visualizing the GFP-marked URA3 locus using the tetOs/tetR-GFP system51. Cells from 2 ml of culture were harvested by centrifugation and resuspended in 1 ml ice cold 100% ethanol. After fixation for 1 hour on ice, cells were stored at -20°C. Cells were mounted on slides that were covered with a thin layer of 1% UltraPure agarose (Life technologies). Cells were imaged using an Axioplan 2 Imaging microscope (Zeiss) equipped with a 100x/1.45 NA objective. 200 cells were scored per sample. Cells containing two separated GFP dots were scored as defective in sister chromatid cohesion. Cells lacking Ino80 were diploid at the outset of the experiment, consistent with published observations52. All cells displayed 2 GFP dots in G1 so that more than 2 GFP dots in metaphase was taken to reflect a sister chromatid cohesion defect.
Supplementary Material
Acknowledgements
We thank P. Chambers at the Cancer Research UK Genome Variation Laboratory at St. James’s University Hospital, Leeds and N. Matthews from the Advanced Sequencing Facility at the Cancer Research UK London Research Institute for high throughput sequencing. We thank C. Esnault, A. Lengronne and our laboratory members for discussions and comments on the manuscript. This work was supported by a Beca Postdoctoral del Ministerio de Ciencia y Tecnologia (Spain) and a Marie Curie Intra-European Fellowship (L. L.-S.) and the European Research Council (L. L.-S. and F. U.).
Footnotes
Database accession numbers: The processed Scc2/Scc4 and Sth1 ChIP sequencing data, nucleosome position data, as well as gene expression microarray data have been deposited in the gene expression omnibus (GEO) database, accession number (GSE56994).
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Ann. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 2.Ocampo-Hafalla MT, Uhlmann F. Cohesin loading and sliding. J. Cell Sci. 2011;124:685–691. doi: 10.1242/jcs.073866. [DOI] [PubMed] [Google Scholar]
- 3.Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 6.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 7.Fryns J-P. On the nosology of the Cornelia de Lange and Coffin-Siris syndromes. Clin. Genet. 1986;29:263–264. doi: 10.1111/j.1399-0004.1986.tb00824.x. [DOI] [PubMed] [Google Scholar]
- 8.Krantz ID, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsurusaki Y, et al. Coffin-Siris syndrome is a SWI/SNF complex disorder. Clin. Genet. 2014;85:548–554. doi: 10.1111/cge.12225. [DOI] [PubMed] [Google Scholar]
- 10.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505:367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misulovin Z, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt CK, Brookes N, Uhlmann F. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 2009;10:R52. doi: 10.1186/gb-2009-10-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasahara K, et al. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 20.Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- 21.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1242. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 22.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badis G, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 26.Schawalder SB, et al. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- 27.Morohashi N, et al. Effect of sequence-directed nucleosome disruption on cell-type-specific repression by α2/Mcm1 in the yeast genome. Eukaryot. Cell. 2006;5:1925–1933. doi: 10.1128/EC.00105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakimi M-A, et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–997. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 29.Baetz KK, Krogan NJ, Emili A, Greenblatt J, Hieter P. The ctf13- 30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 2004;24:1232–1244. doi: 10.1128/MCB.24.3.1232-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Hsu J.-m., Laurent BC. The RSC nucleosome-remodelling complex is required for cohesin’s association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 31.Fasulo B, et al. The Drosophila Mi-2 chromatin-remodeling factor regulates higher-order chromatin structure and cohesin dynamics in vivo. PLoS Genet. 2012;8:e1002878. doi: 10.1371/journal.pgen.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 33.Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santen GW, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat. Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7- Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ocampo-Hafalla MT, Katou Y, Shirahige K, Uhlmann F. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma. 2007;116:531–544. doi: 10.1007/s00412-007-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional Methods References
- 38.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 39.Knop M, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 40.Reid RJD, Sunjevaric I, Kedacche M, Rothstein R. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast. 2002;19:319–328. doi: 10.1002/yea.817. [DOI] [PubMed] [Google Scholar]
- 41.Zhong H, McCord R, Vershon AK. Identification of target sites of the a2-Mcm1 repressor complex in the yeast genome. Genome Biol. 1999;9:1040–1047. doi: 10.1101/gr.9.11.1040. [DOI] [PubMed] [Google Scholar]
- 42.Tóth A, et al. Yeast Cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose MD, Winston F, Hieter P. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- 44.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lantermann A, Strålfors A, Fagerström-Billai F, Korber P, Ekwall K. Genome-wide mapping of nucleosome positions in Schizosaccharomyces pombe. Methods. 2009;48:218–225. doi: 10.1016/j.ymeth.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucl. Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 50.David L, et al. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 52.Chambers AL, et al. The INO80 chromatin remodeling complex prevents polyploidy and maintains normal chromatin structure at centromeres. Genes Dev. 2012;26:2590–2603. doi: 10.1101/gad.199976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.