Abstract
Background
The yst gene that encodes the production of Yst enterotoxins is one of the most important and reliable virulence markers. Its ability to produce Yst has been demonstrated in pathogenic strains isolated from clinical cases of yersiniosis with diarrhea. However, not all yst positive strains produce enterotoxins. According to some authors, Yst production can be restored in a silent strain by ymoA mutation. In this study, the HRM method was applied to identify ymoA single nucleotide polymorphism with the aim of evaluating their influence on the enterotoxic properties of Y. enterocolitica strains.
Results
Two genotypes (A and G) of the examined nucleotide sequence and some variations were detected in the HRM analysis. A phylogenetic analysis of 10 genotype A nucleotide sequences revealed 100% similarity with the Yersinia enterocolitica subsp. enterocolitica 8081 genome NCBI Acc. No. AM286415. An analysis of 10 genotype G nucleotide sequences and 3 variations sequences revealed two point mutations in the examined region: transition A3387326G and insertion A in position 3387368. However, no mutations were observed in the coding region of any of the examined ymoA gene fragments. Genotype G was identified in nearly all Y. enterocolitica strains isolated from pigs. Only 4 nucleotide sequences were similar to AM286415 and did not feature point mutations. In case of human Y. enterocolitica strains 31 were classified as belonging to genotype A, the remaining 59 belonged to genotype G and were characterized by the presence of point mutations.
Conclusions
No correlations were observed between enterotoxic properties and the presence of mutations in the ymoA gene region of Y. enterocolitica strains isolated from both humans and pigs.
Keywords: Yersinia enterocolitica, ymoA gene, Yst enterotoxins, HRM, SNP
Background
Yersinia (Y.) enterocolitica is an etiological agent of yersiniosis, a zoonotic disease that poses a growing threat for public health and produces a variety of clinical symptoms. However, not all Y. enterocolitica strains are pathogenic for humans and animals. Until recently, biotype and serotype diversity and the presence of pYV (plasmid Yersinia Virulence) were the key criteria for pathogenicity evaluation. Strains belonging to biotypes 1B and 2–5 Y. enterocolitica with pYV and chromosomal virulence markers were considered as pathogenic for humans and animals, but biotype 1A without classical virulence markers was regarded as non-pathogenic [1,2]. The most recent reports concerning clinical cases of yersiniosis in patients infected with Y. enterocolitica biotype 1A [3–5] indicate that the previous pathogenicity criteria should be revised.
The yst gene that encodes the production of Yst enterotoxins (Yersinia stable toxins) is one of the most important and reliable virulence markers. Its ability to produce Yst has been demonstrated in pathogenic strains isolated from clinical cases of yersiniosis with diarrhea. The mechanism of Yst action is based on guanylate cyclase activation, which results in increased cGMP levels in enterocytes and extracellular accumulation of liquid in the intestines [6]. However, not all yst positive strains produce enterotoxins. According to some authors, Yst production can be restored in a silent strain by ymoA mutation [7,8]. The ymoA gene encodes the YmoA (Yersinia modulator) protein, a member of the growing family of conservative Hha (hemolysin expression modulating) proteins with low molecular mass that are similar to H-NS (histone-like nucleoid structuring) group proteins [9]. H-NS proteins play an important role in intestinal microflora, both as structural proteins and gene expression modulators, including virulence markers [10]. The expression of genes directly responsible for the pathogenicity of Y. enterocolitica has been researched extensively. It is generally believed that YmoA is the key modulator of gene expression in response to environmental factors, including temperature [11,12]. The YmoA protein has also been shown to inhibit the expression of the inv gene encoding invasion, an important virulence factor responsible for the transport of bacterial cells across M cells [13] and participating in temperature-dependent production of Yersinia outer proteins (Yops) and Yersinia adhesin (YadA) – plasmid virulence markers [7]. The role of YmoA and related proteins (H-NS) in the regulation of gene expression in various microorganisms, including E. coli, has been established by numerous studies, whereas there are only few reports about Y. enterocolitica [7,9,11,13–15].
Single nucleotide polymorphism (SNP), referred to as the new generation genetic marker, is a DNA sequence polymorphism caused by single nucleotide (A, G, C, T) variation. The genotype distribution of the SNP site related to disease susceptibility can be analyzed to determine the correlation between a given genotype and susceptibility to disease, which provides a theoretical basis for disease prevention, personalized diagnosis and treatment [16]. Several SNP genotyping techniques have been developed, including polymerase chain reaction (PCR) followed by restriction fragment length polymorphism (RFLP), direct sequencing or PCR-based TaqMan chemistry. Some of those techniques are expensive, laborious and time-consuming because they support the analysis of only one SNP per reaction. High-resolution melting (HRM) analysis is a new and rapid method for the detection of mutations, in which PCR and mutation scanning are carried out simultaneously in a single procedure [17]. The detection of genetic variations in PCR amplicons requires HRM fluorescent dye and direct melting after PCR. The amplicon melting profile is determined by amplicon length, GC content, sequence characteristics and heterozygosity. This approach is a simple, closed-tube detection method that facilitates the identification of heteroduplexes and homoduplexes based on their characteristic melting curve profiles [18].
In this study, the HRM method was applied to identify ymoA SNP with the aim of evaluating their influence on the enterotoxic properties of Y. enterocolitica strains.
Methods
An ethical approval was not required as this study was performed retrospectively. Samples of diseased humans were routinely submitted to the diagnostic laboratories (National Institute of Public Health – the National Institute of Hygiene in Poland and Department of Microbiology, Ludwik Rydygier Collegium Medicum in Bydgoszcz). The collection of Y. enterocolitica strains isolated from pigs was obtained from previous studies, performed in adherence to the Local Ethics Committee of the University of Warmia and Mazury in Olsztyn.
Experimental material
The material for the study consisted of 90 Y. enterocolitica strains isolated from clinical cases of human yersiniosis accompanied by diarrhea (mostly from the collection of the National Institute of Public Health – the National Institute of Hygiene in Poland) and 90 Y. enterocolitica strains isolated from clinically healthy fattening pigs. Both human and pig strains of Y. enterocolitica had been previously biotyped, serotyped and molecularly examined (multiplex PCR – ystA, ystB, ystC, ymoA) [19]. Determination of enterotoxic properties Y. enterocolitica isolated from pigs was done using suckling mouse bioassay, described previously [19]. Enterotoxin production has been evaluated semi quantitatively by the measurement of the ratio of intestinal mass to the rest of the body mass in the group of three examined sucklings. According to Gianella [20] ratio ≤ 0.074 was regarded as a negative result, 0.075 – 0,082 as a doubtful result and ratio ≥ 0.083 was regarded as a positive result. In this study 14 positive, 33 doubtful and 43 negative Y. enterocolitica strains were used. All the Y. enterocolitica strains isolated from clinical cases of yersiniosis produced enterotoxin, what resulted in diarrhea.
HRM analysis
The HRM analysis was performed in the Rotor-Gene 6000™ real-time analyzer (Corbett Life Science, Sydney, Australia) using the PCR HRM curve analysis assay. PCR HRM was conducted with the use of the Eva Green saturating dye (Type-it HRM PCR Kit, Qiagen, Hilden, Germany). Sequences of primers used in the reaction: ymoA-1 (Forward) 5′GACTTTTCTCAGGGGAATAC3′ and ymoA-2 (Reverse) 5′GCTCAACGTTGTGTGTCT3′ were previously published by Grant et al. [8]. PCR was performed in 25 μl reaction volumes containing 12.5 μl of 2× HRM PCR Master Mix, 10.15 μl of RNase-free water, 1.75 μl of the primer mix (final concentration 0.7 μM each) and 0.6 μl of DNA (50 ng/reaction). PCR amplification was performed with initial denaturation at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s and 55°C for 30 s. HRM ramps were generated by acquiring fluorescence data at the temperature ramp of 65°C to 90°C at 0.1°C intervals. HRM curves were normalized, and the genotype was assigned based on the shape of the HRM curve with the use of the Rotor-Gene software and by visual examination.
Data analysis
The observed melting curves were analyzed using RotorGene 6000 Series Software 1.7. Ten samples of each of the two SNP genotypes and 3 variations were randomly chosen for sequencing to verify genotyping results (Genomed Sp. z o.o., Warsaw, Poland). Sequence data from the examined Y. enterocolitica strains were compared with the nucleotide sequence of the previously identified ymoA gene, lodged in National Centre for Biotechnology Information (NCBI, position 3387372–3387575 in the Y. enterocolitica Acc. No. AM286415) using BLASTN version 2.2.18. [21]. Multiple sequence alignment was carried out in ClustalW [22] incorporated in the freeware Computational Evolutionary Biology package MEGA version 5.2.1. [23]. Nucleotide sequences were demonstrated using BioEdit v.7.2.0. software.
Results
Two genotypes (A and G) of the examined nucleotide sequence and some variations were detected in the HRM analysis (e.g. Figure 1). Direct sequencing revealed that examined nucleotide sequences had the length of 319 bp (base pairs) according to the NCBI. They were linked in position 3387283–3387600 in the Yersinia enterocolitica subsp. enterocolitica 8081 genome NCBI Acc. No. AM286415, where the gene responsible for the production of a conserved hypothetical protein (NCBI, Protein ID CAL13149) is encoded in position 3386958–3387326, and the ymoA gene responsible for the production of histone-like protein YmoA (NCBI, Protein ID CAL13150) is encoded in position 3387372–3387575. A phylogenetic analysis of 10 genotype A nucleotide sequences revealed 100% similarity with the Y. enterocolitica AM286415 sequence. An analysis of 10 genotype G nucleotide sequences and 3 variations sequences revealed two point mutations in the examined region: transition A3387326G and insertion A in position 3387368 (Figure 2). Transition A3387326G was related to the stop codon (TAA»TAG) of the gene encoding conserved hypothetical protein CAL13149. Insertion A was noted in the non-coding region of the investigated area, and it did not change the reading frame for the start codon of the ymoA gene in position 3387372. No mutations were observed in the coding region of any of the examined ymoA gene fragments.
Figure 1.
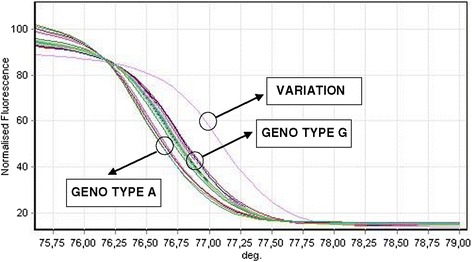
HRM normalized curve of Y. enterocolitica ymoA gene. Figure shows 6 genotypes A, 17 genotypes G and one variation of the examined nucleotide sequence ymoA gene Y. enterocolitica strains isolated from human cases of yersiniosis detected using HRM.
Figure 2.
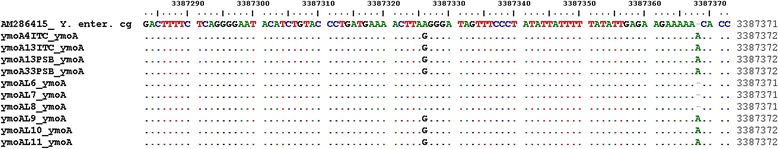
Single nucleotide polymorphism of Y. enterocolitica ymoA gene . A phylogenetic analysis of 3 genotype A nucleotide sequences ymoA gene Y. enterocolitica strains isolated from human cases of yersiniosis (L6, L7, L8) revealed 100% similarity with the Y. enterocolitica AM286415 sequence. An analysis of 7 genotype G nucleotide sequences ymoA gene (3 Y. enterocolitica strains isolated from human cases of yersiniosis – L9, L10, L11 and 4 isolated from pigs – 4ITC, 13ITC, 13PSB, 33PSB) revealed two point mutations in the examined region: transition A3387326G and insertion A in position 3387368.
Two SNPs (transition A3387326G and insertion A3387368) were identified in the examined sequence in nearly all Y. enterocolitica strains isolated from pigs. Only 4 (3.6%) nucleotide sequences were similar to AM286415 and did not feature point mutations. Genotype A strains belonged to bioserotype 4/O:3 – one of them was isolated from the toxin-producing Y. enterocolitica strain, two were isolated from doubtful strains, and one – from a strain that did not produce toxins in a study of suckling mice. No correlations were observed between enterotoxic properties and the presence of mutations in the examined sequence of Y. enterocolitica strains isolated from pigs. More than 27.0% of Y. enterocolitica strains from clinical cases of human yersiniosis were similar to nucleotide sequences of ymoA gene Y. enterocolitica AM286415. A total of 31 human Y. enterocolitica strains were classified as belonging to genotype A, including 14 strains belonging to bioserotype 1B/O:8 and 17 strains belonging to bioserotype 4/O:3. The remaining 59 human Y. enterocolitica strains of bioserotype 4/O:3 belonged to genotype G and were characterized by the presence of point mutations.
Discussion
In the present study, potential mutations in the ymoA gene associated with the pathogenesis of yersiniosis were identified by HRM analysis. Genotype data from Y. enterocolitica strains with confirmed enterotoxic properties were compared to data from Y. enterocolitica strains that do not produce enterotoxins. Amplicon genotyping by HRM analysis supported rapid identification of the frequencies of ymoA SNPs suspected of influencing the yst gene function.
The ymoA mutation and its possible effects on the yst gene were first described by Cornelis et al. [7] who identified two Tn5-Tc1 chromosomal insertion mutants of W22703 transcribing classical virulence markers at low temperature. Those mutants also resumed their production of Yst, with its typical temperature dependence. Both mutations were insertions in the same gene called ymoA for ‘Yersinia modulator’. The cloned ymoA gene fully complemented the two mutations. The cited authors suggested that the ymoA mutation unblocks the silencing of the yst gene and stimulates enterotoxin production and that the ymoA gene without mutation is responsible for negative regulation of yst gene expression.
In 1994, Mikulskis et al. [24] suggested the presence of a mechanism switching the expression of yst to a silent state. According to the above authors, gene silencing was triggered by changes in the status of bacterial host factors rather than modifications in the yst gene. They also noted that negative regulator YmoA participated in yst silencing and temperature regulation of yst. YmoA was regarded as necessary for growth-phase regulation of yst, although it was not the only factor involved in the regulation process.
In 1998, Grant et al. [8] also demonstrated that yst gene carriage is not always correlated with the presence of the toxin in culture supernatants, and similar results were noted in our previous studies [19]. Both cited studies suggested that the absence of enterotoxic properties could result from reduced gene expression in in vitro cultures, possibly by ymoA.
The cited results differs from observations made in this study. Instead of the two insertions reported by Cornelis et al. [7], this study revealed transition A3387326G in the stop codon of the gene fragment encoding protein CAL13149 and insertion A3387368 in the non-coding region of the ymoA gene. No mutations were observed in the coding region of any of the examined ymoA gene fragments.
Two SNPs detected in this study were observed in Y. enterocolitica strains, regardless of their enterotoxic properties, isolated from both humans and clinically healthy pigs. Genotype G characterized by those SNPs accounted for 96.4% of strains isolated from pigs and 72.1% of strains isolated from humans. Genotype A without point mutations was observed far less frequently, but was detected in 100% bioserotype 1B/O:8 strains isolated from humans, which are believed to be the most pathogenic causative agents of acute yersiniosis [25].
Conclusions
The results of this study indicate that two point mutations in the analyzed nucleotide sequences do not affect the toxic properties of the examined strains. It should also be noted that the absence of mutations in the coding region of the ymoA gene does not confirm their influence on ystA gene silencing, which was postulated by other authors. The possible role of YmoA in the negative regulation of yst genes should be analyzed through simultaneous measurements of yst and ymoA gene expression levels.
Acknowledgements
This study was supported by the National Science Centre (NCN, grant No. N N308 609338).
Abbreviations
- Y. enterocolitica
Yersinia enterocolitica
- pYV
Plasmid Yersinia Virulence
- Yst
Yersinia stable toxins
- cGMP
Cyclic guanosine monophosphate
- YmoA
Yersinia modulator
- Hha
Hemolysin expression modulating protein
- H-NS
Histone-like nucleoid structuring protein
- Inv
Yersinia enterocolitica invasin
- Yops
Yersinia outer proteins
- YadA
Yersinia adhesin
- E. coli
Escherichia coli
- SNP
Single nucleotide polymorphism
- PCR
Polymerase chain reaction
- RFLP
Restriction fragment length polymorphism
- HRM
High-resolution melting
- NCBI
National Centre for Biotechnology Information
- bp
Base pair
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ABK designed the study, performed SNP detection using the HRM method, interpreted the results and wrote the manuscript. KL participated in SNP detection with the HRM method and in drafting the manuscript. AST developed genotype models based on direct sequencing and participated in drafting the manuscript. APS contributed to the experimental design and participated in drafting the manuscript. EG selected Y. enterocolitica 1B/O:8 strains from clinical cases of yersiniosis and participated in drafting the manuscript. WS contributed to the elaboration of the experimental design, interpretation of the results and manuscript drafting. All authors have read and approved the final manuscript.
Contributor Information
Agata Bancerz-Kisiel, Email: a.bancerz-kisiel@uwm.edu.pl.
Karolina Lipczyńska, Email: karolina.lipczynska@uwm.edu.pl.
Anna Szczerba-Turek, Email: a.szczerba@uwm.edu.pl.
Eugenia Gospodarek, Email: gospodareke@cm.umk.pl.
Aleksandra Platt-Samoraj, Email: platt@uwm.edu.pl.
Wojciech Szweda, Email: szweda@uwm.edu.pl.
References
- 1.Singh I, Virdi JS. Production of Yersinia stable toxin (YST) and distribution of yst genes in biotype 1A strains of Yersinia enterocolitica. J Med Microbiol. 2004;53:1065–1068. doi: 10.1099/jmm.0.45527-0. [DOI] [PubMed] [Google Scholar]
- 2.Tennant SM, Skinner NA, Joe A, Robins-Browne RM. Homologues of insecticidal toxin complex genes in Yersinia enterocolitica biotype 1A and their contribution to virulence. Infect Immun. 2005;73:6860–6867. doi: 10.1128/IAI.73.10.6860-6867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNally A, Dalton T, La Ragione RM, Stapleton K, Manning G, Newell DG. Yersinia enterocolitica isolates of differing biotypes from humans and animals are adherent, invasive and persist in macrophages, but differ in cytokine secretion profiles in vitro. J Med Microbiol. 2006;5:1725–1734. doi: 10.1099/jmm.0.46726-0. [DOI] [PubMed] [Google Scholar]
- 4.Huovinen E, Sihvonen LM, Virtanen MJ, Haukka K, Siitonen A, Kuusi M. Symptoms and sources of Yersinia enterocolitica-infection: a case–control study. BMC Infect Dis. 2010;10:122. doi: 10.1186/1471-2334-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batzilla J, Heesemann J, Rakin A. The pathogenic potential of Yersinia enterocolitica 1A. Int J Med Microbiol. 2011;301:556–561. doi: 10.1016/j.ijmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Revell PA, Miller VL. Yersinia virulence: more than a plasmid. FEMS Microbiol Lett. 2001;205:159–164. doi: 10.1111/j.1574-6968.2001.tb10941.x. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis GR, Sluiters C, Delor I, Geib D, Kaniga K, Lambert de Rouvroit C, Sory MP, Vanooteghem JC, Michiels T. YmoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 8.Grant T, Bennett-Wood V, Robins-Browne RM. Identification of virulence-associated characteristics in clinical isolates of Yersinia enterocolitica lacking classical virulence markers. Infect Immun. 1998;66:1113–1120. doi: 10.1128/iai.66.3.1113-1120.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison DW, Miller VL. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J Bacteriol. 2006;188:5101–5112. doi: 10.1128/JB.00862-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 11.Banos RC, Pons JI, Madrid C, Juarez A. A global modulatory role for the Yersinia enterocolitica H-NS protein. Microbiology. 2008;154:1281–1289. doi: 10.1099/mic.0.2007/015610-0. [DOI] [PubMed] [Google Scholar]
- 12.Madrid C, Balsalobre C, García J, Juárez A. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol. 2007;63:7–14. doi: 10.1111/j.1365-2958.2006.05497.x. [DOI] [PubMed] [Google Scholar]
- 13.Ellison DW, Young B, Nelson K, Miller VL. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J Bacteriol. 2003;185:7153–7159. doi: 10.1128/JB.185.24.7153-7159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brzostek K, Brzóstkowska M, Bukowska I, Karwicka E, Raczkowska A. OmpR negatively regulates expression of invasion in Yersinia enterocolitica. Microbiology. 2007;153:2416–2425. doi: 10.1099/mic.0.2006/003202-0. [DOI] [PubMed] [Google Scholar]
- 15.Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr Opin Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi LP, He Y, Liu ZD. Correlation between single nucleotide polymorphism of rs3811047 in IL-1 F7 gene and rheumatoid arthritis susceptibility among Han population in central plains of China. Asian Pac J Trop Dis. 2013;1:73–75. doi: 10.1016/S1995-7645(12)60204-1. [DOI] [PubMed] [Google Scholar]
- 17.Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 18.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 19.Bancerz-Kisiel A, Szczerba-Turek A, Platt-Samoraj A, Szweda W. Distribution of the ymoA and ystA genes and enterotoxins Yst production by Yersinia enterocolitica strains isolated from humans and pigs. Pol J Vet Sci. 2012;15:609–614. doi: 10.2478/v10181-012-0096-1. [DOI] [PubMed] [Google Scholar]
- 20.Giannella RA. Suckling mouse model for detection of heat-stable Escherichia coli enterotoxin: characteristics of the model. Infect Immun. 1976;14:95–99. doi: 10.1128/iai.14.1.95-99.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikulskis AV, Delor I, Ha Thi V, Cornelis GR. Regulation of the Yersinia enterocolitica enterotoxin Yst gene. Influence of growth phase, temperature, osmolarity, pH and bacterial host factors. Mol Microbiol. 1994;14:905–915. doi: 10.1111/j.1365-2958.1994.tb01326.x. [DOI] [PubMed] [Google Scholar]
- 25.Rastawicki W, Szych J, Gierczyński R, Rokosz N. A dramatic increase of Yersinia enterocolitica serogroup O:8 infections in Poland. Eur J Clin Microbiol Infect Dis. 2009;28:535–537. doi: 10.1007/s10096-008-0647-7. [DOI] [PubMed] [Google Scholar]


