Abstract
RNA-based mechanisms of regulation represent a ubiquitous class of regulators that are associated with diverse processes including nutrient sensing, stress response, modulation of horizontal gene transfer, and virulence factor expression. While better studied in Gram-negative bacteria, the literature is replete with examples of the importance of RNA-mediated regulatory mechanisms to the virulence and fitness of Gram-positives. Regulatory RNAs are classified as cis-acting, e.g. riboswitches, which modulate the transcription, translation, or stability of co-transcribed RNA, or trans-acting, e.g. small regulatory RNAs, which target separate mRNAs or proteins. The group A Streptococcus (GAS, Streptococcus pyogenes) is a Gram-positive bacterial pathogen from which several regulatory RNA mechanisms have been characterized. The study of RNA-mediated regulation in GAS has uncovered novel concepts with respect to how small regulatory RNAs may positively regulate target mRNA stability, and to how CRISPR RNAs are processed from longer precursors. This review provides an overview of RNA-mediated regulation in Gram-positive bacteria, and is highlighted with specific examples from GAS research. The key roles that these systems play in regulating bacterial virulence are discussed and future perspectives outlined.
Keywords: control, sRNA, CRISPR, S. pyogenes
Introduction
The idea that bacterial RNA molecules could serve a regulatory role was proposed more than 50 years ago (Jacob & Monod, 1961), although the multiple mechanisms by which this can occur have only recently begun to be appreciated. For example, there have been bacterial RNA molecules described that (i) regulate target mRNAs at the levels of transcription, mRNA stability, and translation (Storz et al., 2004, Cavanagh & Wassarman, 2014), (ii) regulate the activity of target proteins through binding and sequestration (Babitzke & Romeo, 2007), and (iii) regulate the ability of horizontally transferred nucleic acids (e.g. bacteriophage and plasmids) to be maintained within the cell (Brouns et al., 2008). A common method by which regulatory RNAs are classified is based upon whether the regulatory activity is targeted to the same RNA molecule (cis-acting RNAs) or to different RNAs, DNA, or proteins (trans-acting RNAs). The best described class of cis-acting RNAs are riboswitches, RNA elements typically located in the 5′-untranslated regions (5′-UTRs) of select mRNAs that respond to effector molecules (e.g. cyclic di-GMP) (Sudarsan et al., 2008) or a physical parameter (e.g. temperature) (Johansson et al., 2002), and modulate transcription, translation, or cleavage of the associated mRNA (Henkin, 2008, Serganov & Nudler, 2013). The best described class of trans-acting RNAs are the small regulatory RNAs (sRNAs, also known as non-coding RNAs [ncRNAs]) that function by binding to target mRNAs and/or proteins to modify their expression or activity (Storz et al., 2011). In general, research into the RNA-mediated regulation of Gram-positive bacterial species lags behind that of Gram-negative species. The importance of RNA-mediated regulatory mechanisms in the virulence of Gram-positive pathogens (Brantl & Bruckner, 2014, Johansson et al., 2002, Mann et al., 2012), as well as apparent differences between Gram-positive and -negative bacteria, such as many Gram-positive genomes lacking a gene encoding the RNA chaperone protein Hfq (Nielsen et al., 2010, Sun et al., 2002), warrant the study of RNA-mediated regulation in these organisms.
The group A Streptococcus (GAS; Streptococcus pyogenes) is a Gram-positive pathogen that causes a range of human infections, from mild, self-limiting infections such as pharyngitis (a.k.a. strep throat), to severe invasive infections such as necrotizing fasciitis (a.k.a. the flesh-eating disease) (Cunningham, 2000). GAS has a large repertoire of virulence factors that promote colonization, immune evasion, and the disruption of host tissue barriers (Reglinski & Sriskandan, 2014, Thomas & Lee, 2012). The coordinated expression of these virulence factors is believed to be critical to the ability of GAS to cause distinct human infections. RNA-mediated regulatory mechanisms are an important component of the ability of GAS to regulate gene expression (Liu et al., 2012, Ramirez-Pena et al., 2010, Fuchs et al., 2006). Throughout this review, we will use examples from GAS research to highlight the mechanisms and activity of different categories of regulatory RNAs. Please note that due to space constraints we will not discuss house-keeping small RNA molecules that provide critical cell functions such as the 4.5S and 6S RNAs (Steuten et al., 2014, Trevino et al., 2010).
Trans-acting RNAs
The trans-acting RNAs include (a) sRNAs, which modify the expression or function of target molecules, (b) CRISPR RNAs, which contribute to a form of immunity that reduces acquisition of horizontally transferred elements, and (c) plasmid-encoded RNAs that contribute to plasmid maintenance and stability.
Highlights of sRNA-mediated regulation in Gram-positive bacteria
Similar to their Gram-negative counterparts, most sRNAs from Gram-positive species range from 50 to 250 nucleotides in length and, with notable exceptions (Balaban & Novick, 1995, Gimpel et al., 2010), do not encode for any proteins; rather, the RNA molecules themselves have intrinsic regulatory activity. For the most part, given the absence of the regulator-protein sequestering CsrB-like sRNAs from Gram-positive genomes (Babitzke & Romeo, 2007), sRNAs from Gram-positive organisms fulfill their regulatory activity by base-pairing to one or more target mRNAs, leading to an alteration in the stability and/or translation of the hybridized mRNAs.
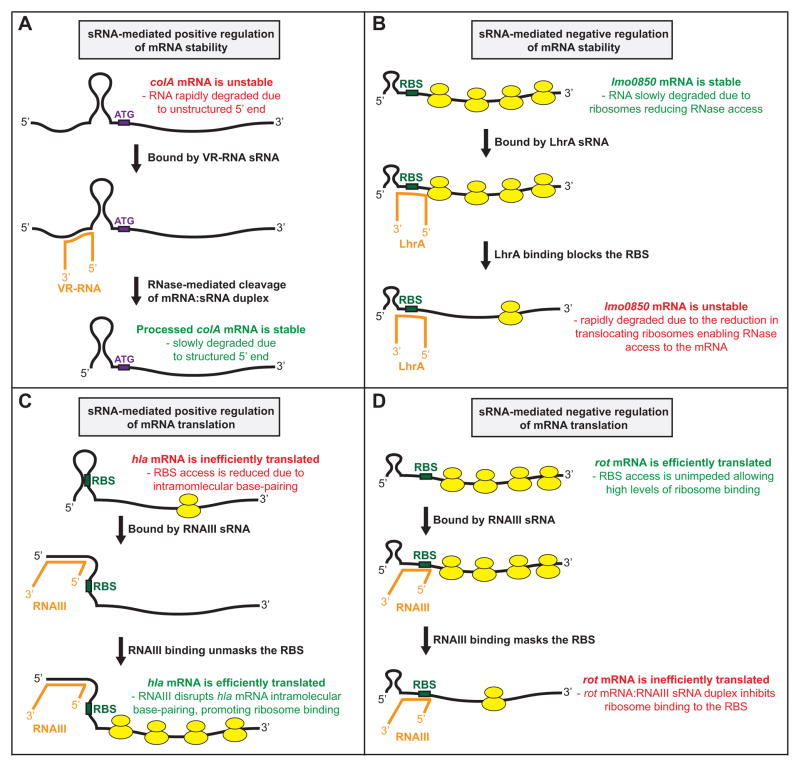
The stability of an sRNA-targeted mRNA can be increased or decreased depending upon multiple factors, in particular the location of the sRNA:mRNA interaction relative to the 5′ end, the Shine-Dalgarno ribosome binding site (RBS), and/or RNase cleavage sites, of the mRNA molecule. For example, the Clostridium perfringens sRNA VR-RNA increases the stability of colA mRNA, encoding the toxin collagenase, after binding to the mRNA 5′-UTR between the 5′ end and RBS (Obana et al., 2010). VR-RNA:colA mRNA duplex formation leads to a single cleavage event within the colA mRNA 5′-UTR, which generates a new 5′ end (Figure 1A). Crucially, the nucleotides of the processed colA mRNA generate a stem-loop structure at the 5′ end, unlike the situation with the full-length transcript (Obana et al., 2010). Secondary structure at the 5′ end of an mRNA molecule promotes stability through the inhibition of RNA pyrophosphorylase binding as this enzyme converts 5′ tri-phosphate ends into mono-phosphate ends; the preferred substrates of several ribonucleases (Condon & Bechhofer, 2011, Lehnik-Habrink et al., 2012). An example of an mRNA with decreased stability following sRNA binding is the lmo0850 mRNA following LhrA sRNA binding in Listeria monocytogenes (Nielsen et al., 2010). While the exact mechanism by which lmo0850 mRNA stability is reduced has not been investigated, it is believed to be a consequence of reduced ribosome activity on the mRNA (Figure 1B). Translocating ribosomes enhance the stability of mRNAs by inhibiting ribonuclease access (Deana & Belasco, 2005).
Figure 1. Mechanisms by which sRNAs from Gram-positive pathogens regulate the stability or translation of target mRNAs.
(A) The Clostridium perfringens sRNA VR-RNA positively regulates the stability of colA mRNA (Obana et al., 2010). The colA mRNA (black), colA mRNA start codon (ATG; purple), and VR-RNA sRNA (orange) are shown. (B) The Listeria monocytogenes sRNA LhrA negatively regulates the stability of lmo0850 mRNA (Nielsen et al., 2010). The lmo0850 mRNA (black), lmo0850 mRNA RBS (green), LhrA sRNA (orange), and ribosomes (yellow) are shown. (C) The Staphylococcus aureus sRNA RNAIII positively regulates the translation of hla mRNA (Morfeldt et al., 1995). The hla mRNA (black), hla mRNA RBS (green), RNAIII sRNA (orange), and ribosomes (yellow) are shown. (D) The Staphylococcus aureus sRNA RNAIII negatively regulates the translation of rot mRNA (Boisset et al., 2007). The rot mRNA (black), rot mRNA RBS (green), RNAIII sRNA (orange), and ribosomes (yellow) are shown.
The translation of an sRNA-targeted mRNA can also be increased or decreased depending upon the location and nature of the sRNA:mRNA interaction. For example, the best described sRNA from a Gram-positive pathogen, the 514 nt RNAIII from Staphylococcus aureus (Novick et al., 1993), positively regulates translation of hla mRNA encoding α-hemolysin and negatively regulates translation of rot mRNA encoding the repressor-of-toxins protein Rot. RNAIII positively regulates hla mRNA translation by disrupting intramolecular base-pairing in the hla mRNA that ordinarily blocks ribosome access to the RBS (Morfeldt et al., 1995) (Figure 1C). Conversely, RNAIII negatively regulates rot mRNA translation by binding to the RBS and occluding ribosome access (Geisinger et al., 2006) (Figure 1D).
Candidate sRNAs are encoded throughout the GAS genome
Multiple distinct approaches have been used to identify sRNAs expressed by bacterial pathogens. In GAS, these include the use of tiling microarrays (Perez et al., 2009, Patenge et al., 2012), RNAseq analysis (Deltcheva et al., 2011), and bioinformatics (Livny et al., 2006, Tesorero et al., 2013, Raasch et al., 2010). In combination, over 100 candidate sRNAs have been predicted to be encoded within the GAS genome. Importantly, multiple of the candidate sRNAs are differentially expressed in serotype, growth phase, and/or growth media-specific fashion. Thus, it has been proposed that GAS differentially expresses sRNAs during infection and that this influences GAS virulence. While there are many candidate sRNAs within the GAS genome the only one thus far characterized is FasX.
The GAS sRNA FasX differentially regulates virulence factor expression
The fibronectin/fibrinogen-binding/hemolytic activity/streptokinase-regulator (fas) locus was discovered in GAS, based upon modest homology to the S. aureus Agr and Streptococcus pneumoniae Com systems (Kreikemeyer et al., 2001). The fas locus consists of the fasBCAX genes which encode for two putative membrane-spanning histidine kinases (FasB and FasC), one putative DNA-binding response regulator (FasA), and a 205 nt sRNA (FasX). While the FasBCA proteins have not been studied in detail, it is hypothesized that they function similar to classical two-component systems. FasX abundance increases during the exponential phase of growth and is significantly decreased in stationary phase, similar to the abundance of RNAIII from the agr quorum sensing system of S. aureus (Boisset et al., 2007). However, while auto-inducing peptides have been detected in GAS culture supernatants none function with the FasBCA system (Chang et al., 2011, Belotserkovsky et al., 2009), reducing the possibility that the Fas locus functions as part of a quorum sensing mechanism (Kreikemeyer et al., 2001).
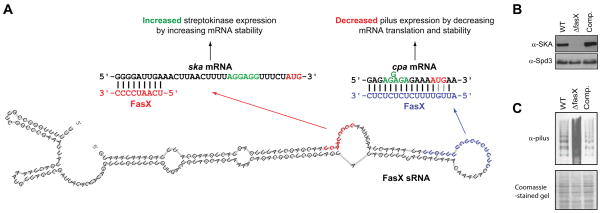
Two virulence factor-encoding mRNAs have been confirmed as targets of FasX and the molecular mechanisms behind the regulation delineated. The first confirmed FasX-regulated GAS virulence factor was the thrombolytic agent streptokinase (Ramirez-Pena et al., 2010, Kreikemeyer et al., 2001). Streptokinase is a secreted virulence factor that promotes the conversion of the human protein plasminogen into the protease plasmin (McArthur et al., 2012). Plasmin has multiple activities, including degrading blood clots (directly by degrading the fibrin fibers within a blood clot) and tissue barriers (directly by degrading extracellular matrix components and indirectly by activating collagenases and metalloproteases) (Syrovets et al., 2012). FasX base-pairs to the first nine nucleotides of the streptokinase-encoding (ska) mRNA, an interaction that increases the stability of the mRNA leading to a ten-fold increase in ska mRNA abundance and streptokinase protein expression (Figures 2A and 2B) (Ramirez-Pena et al., 2010). The increase in ska mRNA stability following FasX binding is a consequence of the creation of secondary structure at the 5′ end of ska mRNA, a heretofore unrecognized mechanism of positive regulation by an sRNA (Podkaminski & Vogel, 2010). Thus, although the mechanism differs between the FasX-mediated regulation of ska mRNA stability in GAS and the VR-RNA-mediated regulation of colA mRNA stability in C. perfringens, both ultimately promote stability through formation of 5′ end secondary structure (Obana et al., 2010, Ramirez-Pena et al., 2010). Differences between the two mechanisms include that FasX must remain bound to its mRNA target to enhance stability while VR-RNA does not. The reversible nature of the FasX:ska mRNA interaction implies that the positive regulation afforded by FasX could be removed by decreasing FasX transcription, or increasing FasX turnover.
Figure 2. The FasX sRNA positively and negatively regulates GAS virulence factor expression.
(A) Schematic showing the putative FasX secondary structure with nucleotides involved in the positive regulation of ska mRNA (red) and the negative regulation of cpa mRNA (blue) highlighted. Only the 5′ ends of the ska and cpa mRNAs are shown, with the Shine-Dalgarno ribosome binding sites (green) and AUG start codons (red) highlighted. This panel is a modified version of a previously published figure (Liu et al., 2012). (B + C) Western blot analyses of secreted (B) or cell wall (C) protein fractions from a parental GAS strain (WT), an isogenic fasX mutant (ΔfasX), and a complemented mutant derivative (Comp). The secreted protein Westerns used an anti-SKA antibody as the test antibody and an anti-Spd3 antibody as a loading control. The cell wall protein Westerns used an anti-pilus antibody as the test antibody, with the gel coomassie-stained prior to performing the Western as a loading control. Note the characteristic laddering pattern of pili.
It has previously been proposed that the hybridization of an sRNA to an mRNA between nucleotides −35 to +15, relative to the A of the ATG start codon, leads to inhibition of mRNA translation (Frohlich & Vogel, 2009, Sharma et al., 2007, Storz et al., 2004). However, this does not appear to be the case for the FasX:ska mRNA interaction, which occurs at ska mRNA nucleotides −32 to −24. This is supported by the fact that the increase in streptokinase expression mirrors the increase in ska mRNA abundance (Ramirez-Pena et al., 2010). Therefore, the consequences to translation with respect to sRNA:mRNA interactions must be determined on a case-by-case basis.
The second confirmed FasX-regulated GAS virulence factor is the pilus (Liu et al., 2012). GAS pili bind to collagen and promote the ability of this pathogen to adhere to host cells, as well as to form biofilms (Abbot et al., 2007, Lizano et al., 2007, Manetti et al., 2007). FasX base pairs to 16 of the first 17 nucleotides of cpa mRNA, the first gene in the pilus biosynthesis operon that encodes the collagen-binding minor pilus protein located at the pilus tip (Quigley et al., 2009). In contrast to the FasX:ska mRNA interaction, the FasX:cpa mRNA interaction negatively regulates expression (Figures 2A and 2C). The main mechanism by which FasX negatively regulates pilus expression is through the inhibition of cpa mRNA translation, reducing access of ribosomes to the cpa mRNA RBS and consequently, translation of the pilus biosynthesis genes (Figure 2A) (Liu et al., 2012). By reducing expression of adhesins and enhancing expression of streptokinase, which aids GAS spread, FasX is proposed to be a key regulator in the transition of GAS from the colonization to the dissemination stages of infection.
The FasX sequences complementary to ska and cpa mRNAs are UCAAUCCCC and CUCUCUCUCUUUUGUU, respectively. The concentrations of U and C nucleotides in these regions are reminiscent of the conserved UCCC sequence motif recently identified in 11 previously uncharacterized S. aureus sRNAs (Geissmann et al., 2009). Similarly, three UCCC sequences present in the S. aureus RNAIII sRNA are known to interact with target mRNA sequences (Boisset et al., 2007). It has been proposed that the UCCC motif highlights a novel subset of S. aureus sRNAs that function by inhibiting translation of target mRNAs through sRNA:mRNA interactions (Geissmann et al., 2009), and we propose that similarly functioning sRNAs are also present in other low GC% Gram-positive pathogens such as GAS.
Highlights of CRISPR/Cas systems in Gram-positive bacteria
CRISPR/Cas (clustered, regularly interspaced short palindromic repeat/CRISPR-associated proteins) together constitute an adaptive immune system which reduces the ability of bacteriophage and plasmids to be maintained in a recipient cell following transfer (Brouns et al., 2008). Thus, this activity, first described in Streptococcus thermophilus (Barrangou et al., 2007), represents a major impediment to horizontal gene transfer. CRISPRs consist of repeat sequences 24–28 bp in size that are separated by unique ‘spacer’ regions which vary in size (26 to 72 bp), with the majority showing homology to foreign DNA, such as bacteriophages and plasmids (Bolotin et al., 2005, Jansen et al., 2002, Mojica et al., 2005). CRISPR/Cas systems enable the bacterial cell to develop an ‘immune response’ against horizontally transferred DNA that is complementary to one or more spacer sequences.
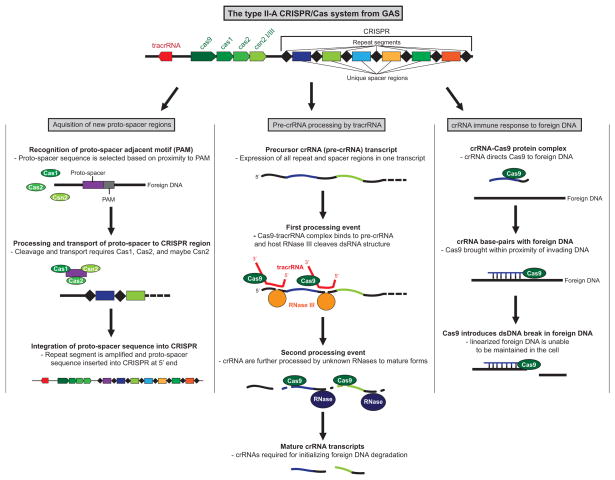
CRISPR elements are transcribed as a single precursor transcript (pre-CRISPR RNA [pre-crRNA]). Subsequently, the pre-crRNA undergoes a maturation process whereby each repeat-spacer region is cleaved into a separate crRNA. How the pre-crRNA is processed into mature crRNAs provides the basis of a classification system (Makarova et al., 2011). In Type I CRISPR/Cas systems, the pre-crRNA is cleaved by a Cas endonuclease that is part of a larger complex termed Cascade (CRISPR-associated complex for antiviral defense) (Sinkunas et al., 2013, Brouns et al., 2008). In Type II CRISPR/Cas systems, the generation of crRNAs requires a small RNA molecule termed tracrRNA (trans-activating CRISPR RNA) which base-pairs to the repeat units in pre-crRNA and promotes their cleavage via the double-stranded RNA-cleaving enzyme RNase III (Deltcheva et al., 2011) (Figure 3). In Type III CRISPR/Cas systems, the pre-crRNA is cleaved by a single Cas endonuclease prior to being transferred to a Cas protein complex for additional processing (Carte et al., 2008).
Figure 3. Schematic of how a type II-A CRISPR/Cas system generates new spacer sequences, processes pre-crRNA into mature crRNA transcripts, and targets foreign DNA for cleavage.
The three functions associated with CRISPR/Cas systems are shown. The conserved repeat segments of the CRISPR region are represented by black diamonds, while unique spacers are represented by colored rectangles.
A key factor in the functionality of CRISPR/Cas systems is their ability to integrate new information in the form of additional spacer sequences, thus enabling the constant monitoring of horizontally transferred elements. Typically, only a single new spacer is inserted following plasmid or bacteriophage transfer, with the integration also resulting in the duplication of a repeat sequence to create a novel spacer-repeat unit (Figure 3). The region of bacteriophage or plasmid DNA that forms the spacer sequences is not randomly selected, rather the selection of spacer precursors (proto-spacers) is determined by adjacent sequences termed proto-spacer-adjacent motifs (PAMs) (Shah et al., 2013, Heler et al., 2014). While the exact mechanism by which new spacers are acquired is not known, the Cas1 and Cas2 proteins appear to play key roles (Nunez et al., 2014).
The “immune” activity of generated crRNAs occurs through the ability of crRNA/Cas ribonucleoprotein complexes to target and cleave bacteriophage or plasmid DNA at sites complementary to the crRNA sequences (Figure 3). While all Type I and II CRISPR/Cas systems target DNA, Type III systems can be subdivided into Type III-A or III-B, where Type III-A systems also target DNA, while Type III-B systems target RNA (Staals et al., 2013). The site-specific targeting achieved by CRISPR/Cas systems has led to their exploitation as a tool for genome engineering, with a Type II system from GAS being the most extensively used, in part due to this being the first Type II system characterized (Cong et al., 2013, Wang et al., 2013, Hwang et al., 2013, Bassett et al., 2013, Deltcheva et al., 2011).
Do CRISPR-Cas systems influence GAS virulence?
GAS strains are poly-lysogenic such that between ~5 to 10% of any one genome is attributable to integrated bacteriophage, most of which encode one or more virulence factors (Banks et al., 2002). The assortment of bacteriophage present within GAS strains is highly variable and, given the importance of phage-encoded virulence factors to GAS pathogenicity (e.g. the superantigen SpeA and the immune modulating DNase SdaD2) (Sumby et al., 2005, Kasper et al., 2014), mechanisms that influence bacteriophage acquisition such as CRISPR systems are believed to have an indirect effect on GAS virulence (Nozawa et al., 2011). Given that antibiotic resistance genes are commonly associated with plasmids (e.g. erm(T)) (DiPersio et al., 2011, Woodbury et al., 2008), CRISPR systems may also impact the ability of GAS to gain antibiotic resistances. Finally, at least one CRISPR spacer identified in GAS shares homology not with mobile genetic elements, but with a chromosomally-encoded gene (Nozawa et al., 2011). This, plus similar observations in other pathogens (Westra et al., 2014), has led to the hypothesis that CRISPR/Cas systems may be able to influence the expression of targeted chromosomal genes, and hence would represent a novel regulatory mechanism.
RNA-based regulation of plasmid maintenance and stability
The regulatory RNA RNAI is encoded on the E. coli plasmid pColEI and is involved in modulating plasmid replication (Dooley et al., 1985). Multiple plasmids from Gram-positive bacteria also have a regulatory RNA component controlling plasmid replication. For example, the GAS plasmid pSM19035 has at least three genes that modulate replication, the protein-encoding genes copS and repS, and the regulatory RNA-encoding RNAIII (Brantl et al., 1993, Lioy et al., 2010) (not to be confused with the RNAIII sRNA from S. aureus) (Figure S1A). RepS mediates plasmid synthesis and is regulated by both the transcription factor CopS and RNAIII. RNAIII and repS are divergently transcribed from one another and are arranged such that the 5′ ends of the two RNA molecules are complementary. As a consequence of the sequence complementarity between RNAIII and repS mRNA, there is base-pairing between the two 5′ ends which sequesters the repS mRNA RBS and inhibits translation. Deletion of either CopS or RNAIII results in a 10–20 fold increase in plasmid copy number. This tight regulation of RepS controls the rate of plasmid replication, maintaining a low plasmid count per cell (Lioy et al., 2010).
In some cases, the maintenance and propagation of low-copy plasmids are also controlled through the action of an RNA molecule. Toxin-antitoxin (TA) systems contain a stable, protein-encoded toxin which is repressed by either an unstable RNA antitoxin (Type I TA systems) or by an unstable protein antitoxin (Type II TA systems) (Van Melderen, 2010). As the cells replicate and segregate, the stable toxin molecule remains present in the daughter cells. If the daughter cell loses the plasmid, it loses the ability to produce the antitoxin and is killed by the toxin. The Enterococcus faecalis plasmid pAD1 has a well characterized Type I TA system termed the par cassette (Figure S1B). The par region harbors two convergently transcribed RNA molecules, RNA I (which encodes the 33 amino acid toxin) and RNA II (the RNA antitoxin) (Weaver, 2012). Due to complementarity between RNA I/II molecules they are able to hybridize to one another, resulting in the inhibition of RNA I translation (Greenfield et al., 2000). While the GAS plasmid pSM19035 harbors a Type II TA system no Type I systems have thus far been described in this pathogen (Brzozowska et al., 2012, Lioy et al., 2010). A chromosomally-encoded homolog of the Type I TA system par cassette has been identified in Streptococcus pneumoniae (Fozo et al., 2010). Chromosomally-encoded TA systems appear, in part, to function in the general stress response (Durand et al., 2012).
Cis-acting RNAs
Cis-acting RNAs are sequences that are co-transcribed with their respective mRNA regulatory targets, the best described class of which are riboswitches.
Riboswitches trigger structural rearrangement of mRNA
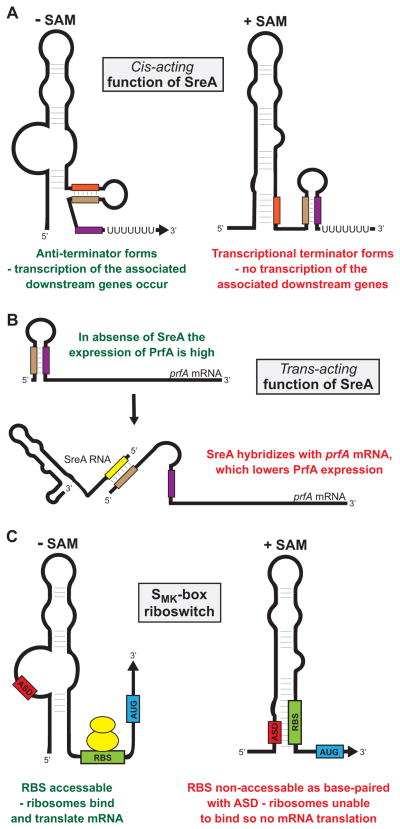
Riboswitches are RNA motifs located almost exclusively in the 5′-UTR of select mRNAs that, either through binding effector molecules or sensing a physical parameter, modulate the transcription, translation, or stability of the associated mRNA (Serganov & Nudler, 2013, Breaker, 2012). Regulation occurs as a consequence of conformational changes in the secondary RNA structure of the riboswitch following activation. This leads to the formation of transcriptional terminators or anti-terminators, the sequestrating or unmasking of the RBS altering mRNA translation, or the exposing or masking of RNase cleavage sites altering mRNA stability (Caron et al., 2012, Hollands et al., 2012) (Figure 4A). Recently, several S-box riboswitches, which bind the metabolite S-adenosylmethionine (SAM), have been identified that not only regulate in cis but also regulate in trans. For example, two S-box riboswitches (SreA/B) from Listeria monocytogenes regulate in trans by binding the 5′-UTR of the mRNA encoding the major virulence factor regulator PrfA (Loh et al., 2009) (Figure 4B). Thus, while regulatory RNAs are commonly classified into different groups they should be viewed as fluid classifications, not rigid, as new information is gathered and new functions discovered.
Figure 4. Examples of regulatory mechanisms by SAM-binding riboswitches from Gram-positive bacteria.
(A) The Listeria monocytogenes S-box riboswitch SreA functions in cis to terminate transcription of the downstream genes lmo2417–2419 in the presence, but not in the absence, of SAM (Loh et al., 2009). In the presence of SAM the rearrangement of secondary structure results in the formation of the terminator hairpin through nucleotides highlighted as brown and purple rectangles. In the absence of SAM the anti-terminator forms through nucleotides highlighted as red and brown rectangles. (B) The Listeria monocytogenes S-box riboswitch SreA can also function in trans to reduce expression of the virulence factor regulatory protein PrfA (Loh et al., 2009). SreA RNA can bind near the 5′ end of prfA mRNA. Through unknown mechanisms the interaction between SreA RNA and prfA mRNA lowers PrfA expression levels. (C) The SMK-box riboswitch from lactic acid bacteria negatively regulates the translation of its associated gene in the presence, but not in the absence, of SAM (Fuchs et al., 2006). The anti-RBS sequence (ASD), the RBS (green) and start codon (AUG; blue) of the associated gene, and ribosomes (yellow) are shown. In the absence of SAM the ASD nucleotides do not base-pair the RBS, enabling ribosome binding and translation of the gene. In the presence of SAM the rearrangement of secondary structure results in base-pairing between the ASD and RBS sequences, occluding the RBS and preventing mRNA translation.
The investigation of riboswitches in GAS has thus far been limited to a SAM-binding riboswitch termed the SMK-box riboswitch (Fuchs et al., 2007), which shares little sequence homology with the classic S-box riboswitch. The SMK-box riboswitch is found in the 5′-UTR of metK genes, encoding SAM synthetase, in lactic acid bacteria, including Streptococcal and Enterococcal species (Grundy & Henkin, 1998). The SMK-box regulates metK mRNA translation by sequestering the RBS and inhibiting ribosome binding in the presence of SAM (Fuchs et al., 2006, Fuchs et al., 2007) (Figure 4C). Following SAM binding, the SMK-box also regulates metK transcription by stabilizing a 5′ terminator and disrupting RNA polymerase activity.
Putative regulatory roles for the 5′-UTRs of the C5a peptidase and CovR-encoding mRNAs
While no GAS riboswitches have been confirmed to regulate virulence factors or their regulators, there are three 5′-UTRs that have been proposed to have regulatory activity, with riboswitch-like activity being one of several possibilities. The three 5′-UTRs are located in the scpA mRNA encoding the C5a peptidase immune evasion protein (Pritchard & Cleary, 1996), the rivR mRNA encoding the virulence factor regulatory protein RivR (Trevino et al., 2013), and the covR mRNA encoding the response regulator component of the major virulence factor regulatory two component system CovR/S (Sumby et al., 2006). A characteristic shared by the scpA and rivR mRNAs is that their transcription can terminate prematurely within the 5′-UTR sequence, leading to 170 nt and 140 nt RNA truncation products, respectively (Pritchard & Cleary, 1996, Trevino et al., 2013). In both instances, the transcriptional termination occurs at the site of an inverted repeat, which is hypothesized to form a hairpin and terminate transcription in a Rho-independent manner (Figures S2A and S2B). Removal of the inverted repeat region upstream of the scpA and rivR genes enhances transcript abundance four and three-fold, respectively (Pritchard & Cleary, 1996, Trevino et al., 2013). Whether the rate of transcriptional termination within the 5′-UTRs is regulated is hypothesized but remains untested.
Bioinformatic analysis of the 196 nt covR 5′-UTR is consistent with this region possessing a high degree of secondary structure that includes the RBS (Figure S2C). If correct, then the sequestering of the RBS in a secondary structure would be expected to reduce ribosome binding and therefore, inhibit covR mRNA translation. Additional support for a regulatory role for the covR 5′-UTR comes from related pathogens. For example, the covR 5′-UTR from Streptococcus mutans shares only 45% nucleotide identity with that of GAS (Chong et al., 2008), but is predicted to form a similar secondary structure (data not shown). Given the critical contribution of the CovR/S regulatory system to GAS pathogenicity (Sumby et al., 2006, Federle et al., 1999, Hollands et al., 2010, Trevino et al., 2009), the investigation of how this system is regulated is currently under investigation.
Conclusions and Future Prospects
In general, tools for the genetic manipulation of Gram-positive bacteria trail those of Gram-negatives, and is likely a contributing factor into why mechanisms of RNA-mediated regulation have been better studied in Gram-negative bacteria. Another contributing factor is that most Gram-positive bacteria lack a homologue of the RNA chaperone protein Hfq, a protein which binds sRNAs and has been exploited by Gram-negative researchers to identify candidate sRNAs via pull-down assays (Pfeiffer et al., 2007, Sittka et al., 2009, Sonnleitner et al., 2008, Zhang et al., 2003). While RNA-mediated regulation may be better studied in Gram-negatives, this does not mean that this large class of regulators are any less important in Gram-positives. Indeed, multiple studies, from the RNAIII sRNA in S. aureus (Novick et al., 1993), to the SreA/B riboswitches in L. monocytogenes (Loh et al., 2009), to our own work with the FasX sRNA in GAS (Ramirez-Pena et al., 2010), all highlight the critical importance of RNA-mediated regulation to the virulence of Gram-positive pathogens. Furthermore, Gram-positive regulatory RNA research has uncovered novel aspects of this class of regulators. For example, GAS regulatory RNA research has led to the discovery of a novel positive regulatory mechanism by an sRNA (FasX regulating streptokinase by formation of 5′ end secondary structure) (Ramirez-Pena et al., 2010), and the discovery of a novel mechanism to generate crRNAs from pre-crRNA precursors (via tracRNA and RNase III; thus describing the prototypical Type II CRISPR/Cas system) (Deltcheva et al., 2011). These and other findings highlight the importance of continuing the investigation of RNA-mediated regulatory mechanisms in Gram-positive pathogens. Giving credence to the term “be careful what you wish for”, the current bottleneck to characterizing novel mechanisms of RNA-mediated regulation comes not from identifying candidate regulatory RNAs, but rather from the testing of the many candidates identified from recent next-generation sequencing, tiling microarray, and bioinformatic approaches. Improvements in the accuracy of bioinformatic-based analyses and the speed of lab-based confirmation of candidate regulatory RNA activities are crucial to future efforts in delineating the overall contribution of this fundamental class of regulatory system in Gram-positives.
Supplementary Material
Figure S1. Plasmid-based RNA regulators of replication and stability. (A) Regulation of pSM19035 replication is achieved by the concerted action of the RepS and CopS proteins and the RNA RNAIII (Lioy et al., 2010). (B) Regulation of pAD1 stability is controlled by a type I toxin-antitoxin system (Weaver, 2012).
Figure S2. The putative secondary structures of the 5′-UTRs from the GAS mRNAs covR, rivR, and scpA suggest regulatory roles for these UTRs. The 5′-UTR nucleotide sequences from the (A) rivR, (B) scpA, and (C) covR mRNAs, from the transcriptional start site to the last base before the ATG start codon, were used with the bioinformatic program RNAfold to determine their putative secondary structure. Nucleotides highlighted in green represent the putative ribosome binding sites of each mRNA. For the rivR and scpA 5′-UTRs the relative location of the early termination site is shown (Pritchard & Cleary, 1996, Trevino et al., 2013).
Acknowledgments
This review was made possible in part by grant AI087747 from The National Institute of Allergy and Infectious Diseases (NIAID, NIH; to P.S.).
References
- Abbot EL, Smith WD, Siou GP, Chiriboga C, Smith RJ, Wilson JA, Hirst BH, Kehoe MA. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cellular microbiology. 2007;9:1822–1833. doi: 10.1111/j.1462-5822.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Current opinion in microbiology. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Balaban N, Novick RP. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′-end deletion. FEMS microbiology letters. 1995;133:155–161. doi: 10.1111/j.1574-6968.1995.tb07877.x. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends in microbiology. 2002;10:515–521. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, NY) 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovsky I, Baruch M, Peer A, Dov E, Ravins M, Mishalian I, Persky M, Smith Y, Hanski E. Functional analysis of the quorum-sensing streptococcal invasion locus (sil) PLoS pathogens. 2009;5:e1000651. doi: 10.1371/journal.ppat.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes & development. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading, England) 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brantl S, Birch-Hirschfeld E, Behnke D. RepR protein expression on plasmid pIP501 is controlled by an antisense RNA-mediated transcription attenuation mechanism. Journal of bacteriology. 1993;175:4052–4061. doi: 10.1128/jb.175.13.4052-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S, Bruckner R. Small regulatory RNAs from low-GC Gram-positive bacteria. RNA biology. 2014:11. doi: 10.4161/rna.28036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches and the RNA world. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science (New York, NY) 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowska I, Brzozowska K, Zielenkiewicz U. Functioning of the TA cassette of streptococcal plasmid pSM19035 in various Gram-positive bacteria. Plasmid. 2012;68:51–60. doi: 10.1016/j.plasmid.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Masse E, Lafontaine DA. Dual-acting riboswitch control of translation initiation and mRNA decay. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3444–3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes & development. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, Wassarman KM. 6S RNA, A Global Regulator of Transcription in Escherichia coli, Bacillus subtilis, and Beyond. Annual review of microbiology. 2014 doi: 10.1146/annurev-micro-092611-150135. [DOI] [PubMed] [Google Scholar]
- Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS pathogens. 2011;7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong P, Drake L, Biswas I. Modulation of covR expression in Streptococcus mutans UA159. Journal of bacteriology. 2008;190:4478–4488. doi: 10.1128/JB.01961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Bechhofer DH. Regulated RNA stability in the Gram positives. Current opinion in microbiology. 2011;14:148–154. doi: 10.1016/j.mib.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes & development. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPersio LP, DiPersio JR, Beach JA, Loudon AM, Fuchs AM. Identification and characterization of plasmid-borne erm(T) macrolide resistance in group B and group A Streptococcus. Diagnostic microbiology and infectious disease. 2011;71:217–223. doi: 10.1016/j.diagmicrobio.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Dooley TP, Tamm J, Polisky B. Isolation and characterization of mutants affecting functional domains of ColE1 RNAI. Journal of molecular biology. 1985;186:87–96. doi: 10.1016/0022-2836(85)90259-1. [DOI] [PubMed] [Google Scholar]
- Durand S, Jahn N, Condon C, Brantl S. Type I toxin-antitoxin systems in Bacillus subtilis. RNA biology. 2012;9:1491–1497. doi: 10.4161/rna.22358. [DOI] [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. Journal of bacteriology. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic acids research. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich KS, Vogel J. Activation of gene expression by small RNA. Current opinion in microbiology. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nature structural & molecular biology. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4876–4880. doi: 10.1073/pnas.0609956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Molecular microbiology. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, Romby P. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic acids research. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpel M, Heidrich N, Mader U, Krugel H, Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Molecular microbiology. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Molecular microbiology. 2000;37:652–660. doi: 10.1046/j.1365-2958.2000.02035.x. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Molecular microbiology. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- Heler R, Marraffini LA, Bikard D. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Molecular microbiology. 2014 doi: 10.1111/mmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes & development. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands A, Pence MA, Timmer AM, Osvath SR, Turnbull L, Whitchurch CB, Walker MJ, Nizet V. Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A streptococcus M1T1 clone. The Journal of infectious diseases. 2010;202:11–19. doi: 10.1086/653124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. Journal of molecular biology. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular microbiology. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Kasper KJ, Zeppa JJ, Wakabayashi AT, Xu SX, Mazzuca DM, Welch I, Baroja ML, Kotb M, Cairns E, Cleary PP, Haeryfar SM, McCormick JK. Bacterial Superantigens Promote Acute Nasopharyngeal Infection by Streptococcus pyogenes in a Human MHC Class II-Dependent Manner. PLoS pathogens. 2014;10:e1004155. doi: 10.1371/journal.ppat.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Molecular microbiology. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Lewis RJ, Mader U, Stulke J. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Molecular microbiology. 2012;84:1005–1017. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- Lioy VS, Pratto F, de la Hoz AB, Ayora S, Alonso JC. Plasmid pSM19035, a model to study stable maintenance in Firmicutes. Plasmid. 2010;64:1–17. doi: 10.1016/j.plasmid.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Liu Z, Trevino J, Ramirez-Pena E, Sumby P. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Molecular microbiology. 2012;86:140–154. doi: 10.1111/j.1365-2958.2012.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic acids research. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizano S, Luo F, Bessen DE. Role of streptococcal T antigens in superficial skin infection. Journal of bacteriology. 2007;189:1426–1434. doi: 10.1128/JB.01179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nature reviews. Microbiology. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti AG, Zingaretti C, Falugi F, Capo S, Bombaci M, Bagnoli F, Gambellini G, Bensi G, Mora M, Edwards AM, Musser JM, Graviss EA, Telford JL, Grandi G, Margarit I. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Molecular microbiology. 2007;64:968–983. doi: 10.1111/j.1365-2958.2007.05704.x. [DOI] [PubMed] [Google Scholar]
- Mann B, van Opijnen T, Wang J, Obert C, Wang YD, Carter R, McGoldrick DJ, Ridout G, Camilli A, Tuomanen EI, Rosch JW. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS pathogens. 2012;8:e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JD, Cook SM, Venturini C, Walker MJ. The role of streptokinase as a virulence determinant of Streptococcus pyogenes--potential for therapeutic targeting. Current drug targets. 2012;13:297–307. doi: 10.2174/138945012799424589. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. The EMBO journal. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic acids research. 2010;38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. The EMBO journal. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PloS one. 2011;6:e19543. doi: 10.1371/journal.pone.0019543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nature structural & molecular biology. 2014 doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obana N, Shirahama Y, Abe K, Nakamura K. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Molecular microbiology. 2010;77:1416–1428. doi: 10.1111/j.1365-2958.2010.07258.x. [DOI] [PubMed] [Google Scholar]
- Patenge N, Billion A, Raasch P, Normann J, Wisniewska-Kucper A, Retey J, Boisguerin V, Hartsch T, Hain T, Kreikemeyer B. Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC genomics. 2012;13:550. doi: 10.1186/1471-2164-13-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez N, Trevino J, Liu Z, Ho SC, Babitzke P, Sumby P. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PloS one. 2009;4:e7668. doi: 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Molecular microbiology. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- Podkaminski D, Vogel J. Small RNAs promote mRNA stability to activate the synthesis of virulence factors. Molecular microbiology. 2010;78:1327–1331. doi: 10.1111/j.1365-2958.2010.07428.x. [DOI] [PubMed] [Google Scholar]
- Pritchard KH, Cleary PP. Differential expression of genes in the vir regulon of Streptococcus pyogenes is controlled by transcription termination. Molecular & general genetics: MGG. 1996;250:207–213. doi: 10.1007/BF02174180. [DOI] [PubMed] [Google Scholar]
- Quigley BR, Zahner D, Hatkoff M, Thanassi DG, Scott JR. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Molecular microbiology. 2009;72:1379–1394. doi: 10.1111/j.1365-2958.2009.06727.x. [DOI] [PubMed] [Google Scholar]
- Raasch P, Schmitz U, Patenge N, Vera J, Kreikemeyer B, Wolkenhauer O. Non-coding RNA detection methods combined to improve usability, reproducibility and precision. BMC bioinformatics. 2010;11:491. doi: 10.1186/1471-2105-11-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Molecular microbiology. 2010;78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reglinski M, Sriskandan S. The contribution of group A streptococcal virulence determinants to the pathogenesis of sepsis. Virulence. 2014;5:127–136. doi: 10.4161/viru.26400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA biology. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes & development. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. The EMBO journal. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA biology. 2009;6:266–275. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Sorger-Domenigg T, Madej MJ, Findeiss S, Hackermuller J, Huttenhofer A, Stadler PF, Blasi U, Moll I. Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology (Reading, England) 2008;154:3175–3187. doi: 10.1099/mic.0.2008/019703-0. [DOI] [PubMed] [Google Scholar]
- Staals RH, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, Masuda A, Dohmae N, Schaap PJ, Doudna JA, Heck AJ, Yonekura K, van der Oost J, Shinkai A. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Molecular cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuten B, Hoch PG, Damm K, Schneider S, Kohler K, Wagner R, Hartmann RK. Regulation of transcription by 6S RNAs: Insights from the Escherichia coli and Bacillus subtilis model systems. RNA biology. 2014:11. doi: 10.4161/rna.28827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Current opinion in microbiology. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Molecular cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science (New York, NY) 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, Deleo FR, Musser JM. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS pathogens. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic acids research. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrovets T, Lunov O, Simmet T. Plasmin as a proinflammatory cell activator. Journal of leukocyte biology. 2012;92:509–519. doi: 10.1189/jlb.0212056. [DOI] [PubMed] [Google Scholar]
- Tesorero RA, Yu N, Wright JO, Svencionis JP, Cheng Q, Kim JH, Cho KH. Novel regulatory small RNAs in Streptococcus pyogenes. PloS one. 2013;8:e64021. doi: 10.1371/journal.pone.0064021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Lee SW. Knowing is half the battle: targeting virulence factors of group A Streptococcus for vaccine and therapeutics. Current drug targets. 2012;13:308–322. doi: 10.2174/138945012799424679. [DOI] [PubMed] [Google Scholar]
- Trevino J, Liu Z, Cao TN, Ramirez-Pena E, Sumby P. RivR is a negative regulator of virulence factor expression in group A Streptococcus. Infect Immun. 2013;81:364–372. doi: 10.1128/IAI.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino J, Perez N, Ramirez-Pena E, Liu Z, Shelburne SA, 3rd, Musser JM, Sumby P. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun. 2009;77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino J, Perez N, Sumby P. The 4.5S RNA component of the signal recognition particle is required for group A Streptococcus virulence. Microbiology (Reading, England) 2010;156:1342–1350. doi: 10.1099/mic.0.036558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L. Toxin-antitoxin systems: why so many, what for? Current opinion in microbiology. 2010;13:781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver KE. The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA biology. 2012;9:1498–1503. doi: 10.4161/rna.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nature reviews. Microbiology. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- Woodbury RL, Klammer KA, Xiong Y, Bailiff T, Glennen A, Bartkus JM, Lynfield R, Van Beneden C, Beall BW. Plasmid-Borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrobial agents and chemotherapy. 2008;52:1140–1143. doi: 10.1128/AAC.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Molecular microbiology. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Plasmid-based RNA regulators of replication and stability. (A) Regulation of pSM19035 replication is achieved by the concerted action of the RepS and CopS proteins and the RNA RNAIII (Lioy et al., 2010). (B) Regulation of pAD1 stability is controlled by a type I toxin-antitoxin system (Weaver, 2012).
Figure S2. The putative secondary structures of the 5′-UTRs from the GAS mRNAs covR, rivR, and scpA suggest regulatory roles for these UTRs. The 5′-UTR nucleotide sequences from the (A) rivR, (B) scpA, and (C) covR mRNAs, from the transcriptional start site to the last base before the ATG start codon, were used with the bioinformatic program RNAfold to determine their putative secondary structure. Nucleotides highlighted in green represent the putative ribosome binding sites of each mRNA. For the rivR and scpA 5′-UTRs the relative location of the early termination site is shown (Pritchard & Cleary, 1996, Trevino et al., 2013).