Abstract
Serum and membrane proteins are two of the most attractive targets for proteomic analysis. Previous membrane protein studies tend to focus on tissue sample, while membrane protein studies in serum are still limited. In this study, an analysis of membrane proteins in normal human serum was carried out. Nano-liquid chromatography-electrospray ionization mass spectrometry (NanoLC-ESI-MS/MS) and bioinformatics tools were used to identify membrane proteins. Two hundred and seventeen membrane proteins were detected in the human serum, of which 129 membrane proteins have at least one transmembrane domain (TMD). Further characterizations of identified membrane proteins including their subcellular distributions, molecular weights, post translational modifications, transmembrane domains and average of hydrophobicity, were also implemented. Our results showed the potential of membrane proteins in serum for diagnosis and treatment of diseases.
Keywords: membrane proteins, proteomics, serum, NanoLC-ESI-MS/MS
Introduction
As the most easily obtained sample from patients, blood plasma is the primary specimen used to diagnose and monitor many diseases. Because changes in certain components of the plasma are indicative of abnormalities in the body system, many researches have been carried out in search for disease biomarkers in the plasma since the early history of clinical study. After proteomics emerged as a major discipline at the end of the 20th century, plasma study was launched in this new research direction, which could potentially analyze hundreds of proteins at the same time, instead of the one-protein study allowed by traditional genetic/biochemical approach.
The plasma contains thousands of proteins, including those originating from most, if not all kinds of cells and tissues.1 The plasma is considered one of the most promising proteomes for proteomic research, and many proteomic techniques and methods have been utilized and modified to analyze the human plasma.2 In an early study of human plasma, Pieper et al could identify 325 proteins using two-dimensional gel electrophoresis (2-DE) and mass spectrometry (MS).3 Advancements in the field of proteomics in the past decade have assisted plasma studies in both breadth and depth, exemplified by the forming of a human plasma Peptide Atlas by researchers participating in the Plasma Proteome Project of the Human Proteome Organization (HUPO).4 Comparative studies of the plasma have revealed many potential markers and some insights into the mechanisms of tissue specific diseases, such as cancers,5 Alzheimer’s disease,6 and myocardial infarction.7
Membrane proteomics is a highly focused branch of proteomics. Approximately 30% of proteins encoded by the mammalian genome are transmembrane proteins.8,9 Membrane proteins play an important part in many important cellular processes, including cross-membrane transportation, cell adhesion, signal transduction, immune response, etc.10 Due to their roles as transporters, receptors and structural proteins as well as their impacts on intracellular processes, they are the usual candidates for drug development: about 60% of approved drugs target membrane proteins.11 Many membrane proteins were also found to directly associate with various human diseases, such as Alzheimer’s disease, diabetes, Hodgkin’s disease and liver cirrhosis.12 Thus, researches on membrane proteins also hold promises for developments of diagnosis and disease treatment.
The recent studies on human and mammalian membrane proteins tend to focus on cell/tissue samples.13–15 Using carbonate extraction, trypsin digestion and NanoLC-MS/MS, a multilaboratory project has been found to profile membrane proteins from mouse liver.16,17 Another study based on similar approaches, more than 200 proteins were detected in pancreatic cancer cells.18 Furthermore, a large number (862) of membrane proteins were identified in the brain cortex.19 However, we have not found any independent research that focus only on membrane proteins in serum. As a subset of the plasma, serum contains thousands of proteins, including the proteins of membrane origin, even though their abundance can be low. In this study, some results of profiling and characterization of membrane proteins in human serum by using combination of NanoLC-ESI-MS/MS methods and bioinformatics tools are shown. The present study was approved by the Ethics Committee of the Institute of Biotechnology (IBT), Vietnam Academy of Science and Technology (VAST).
Materials and Methods
Materials
Dithiothreitol (DTT), iodoacetamide (IAA), ammonium bicarbonate, ammonium acetate, trypsin (proteomics sequencing grade), sodium bicarbonate and Triton X-100 were purchased from Sigma- Aldrich (St. Louis, MO, USA). Formic acid (FA) and triflouracetate (TFA) were obtained from Fluka (Fluka Chemie GmbH, Buchs, Switzerland). Acetonitrile (ACN, chromatogram grade) and other chemicals (analytical grade) were obtained from Barker (Pittsburgh, USA). The Bradford assay kit, Aurum serum protein mini kit, acrylamide, bis-acrylamide, urea, glycine, Tris, CHAPS, and SDS were purchased from Bio-Rad (Hercules, CA, USA). All equipment and standard reagents used directly should be clean as necessary.
Sample preparation
Sera from healthy middle-aged individuals (20–40 years old) were supplied by Bach Mai Hospital, Hanoi, Vietnam, and stored at −80 °C until analysis. After that, albumin and IgG were depleted from the samples using the Aurum serum protein mini kit (Bio-Rad Laboratories, Hercules, CA, USA). Briefly, serum protein column was washed with 1 ml of serum protein binding buffer. For each sample, 60 μl of serum was diluted with 180 μl serum protein binding buffer and then, 200 μl of diluted serum was loaded onto the column. Subsequently, the depleted serum – unbound fraction was collected by centrifugation at 10,000 × g for 20 sec. The removal of albumin and IgG was evaluated by 12.6% SDS-PAGE.
In-solution trypsin digestion
The depleted sera containing approximately 30 μg of total protein were dried by vacuum centrifugation and then dissolved in 50 mM NH4HCO3. The samples were reduced by 10 mM dithiothreitol (DTT) at 56°C for 30 min and alkylated by 5 mM iodoacetamide (IAA) at room temperature for 1 hr. Proteins in the samples were digested using trypsin (Sequencing grade, Sigma-Aldrich, St. Louis, MO, USA) at ratio of 1:50 (w/w) enzyme to protein at 37 °C for 12 hrs. The digestion was stopped by formic acid with final concentration of 0.1%.
Nano two-dimensional chromatography and mass spectrometry
Tryptic peptides were dissolved in 0.1% formic acid and loaded onto Strong Cation Exchange (SCX) Chromatography Column (LC Packing, Dionex, The Netherlands) for separation in the first dimension. The second dimension was performed using a C18 Reversed Phase (RP) Column (GraceVydac, Hesperia, CA, USA) with mobile phase consisting of 0.1% formic acid in water (A) and 0.1% formic acid in 85% acetonitrile (B). Peptides were eluted in a linear gradient from 0% to 100% mobile phase B at a flow rate of 0.2 μl/min for 90 min.
Tandem mass spectrometry analysis were performed using an ABI QSTAR®XL hybrid quadrupole/TOF MS/MS instrument (Applied Biosystems/MDS Sciex, Ontario, Canada) equipped with a nanoelectrospray source (Protana XYZ manipulator). Positive mode nanoelectrospray was generated from fused-silica PicoTip emitters with a 10 μm aperture (New Objective, Woburn, MA) at 2.5 kV. MS and MS/MS spectra were recorded and processed in IDA mode (Information Dependent Acquisition) controlled by Analyst QS software. The range of the MS full scan was from 200 to 1500 amu followed by MS/MS fragmentation of the three most intense precursor ions. The dynamic ion selection threshold for MS/MS experiments was set to 45 counts.
Identification of membrane proteins
The obtained MS and MS/MS spectra were searched against the NCBInr and the Swiss-Prot protein sequence database using Mascot™ V1.8 software (Matrix Science Ltd., London, UK). The parameters for searching were set as following: enzymatic digestion with trypsin with one potential missed cleavage; a peptide and fragment mass tolerance of ±0.5; carbamidomethyl (cysteine) as fixed modification; oxidation (methionine) as variable modification. Protein identifications were performed using a Mowse scoring algorithm with a confidence level of 95% and with at least two matched peptides. For further verification, proteins were validated using the open-source software MSQuant v 1.5 (http://msquant.sourceforge.net/).
Membrane proteins were sorted from total identified serum proteins based on UniProt protein database (http://www.uniprot.org). SOSUI prediction algorithm was used to predict transmembrane domains and average values of hydrophobicity of those membrane proteins.20
Results
Identification of membrane proteins in serum
The major aim of this study is to detect and profile membrane proteins from human serum. With the strategy and methods described above, 217 membrane proteins were detected from 2778 matched peptides (see Table 1, Supplementary Data). It is interesting to note that more than 90% of the identifications were based on 3 or more matched peptides. From the total of 217 identified membrane proteins, 129 proteins have at least one transmembrane domain (TMD) based on SOSUI prediction algorithm (see Table 1, Supplementary Data).
Evaluation of membrane protein molecular weight
The distribution of molecular weights of membrane proteins from serum was shown in Figure 1. Membrane proteins in serum have a wide range of molecular weights. Proteins with molecular weights from 100–200 kDa make up the largest group, accounting for 43% the total of 217 identified membrane proteins. The second most abundant is the group of proteins with molecular weights in the range of 200–300 kDa (26%). It is also notable that several membrane proteins (6%) have molecular weights above 500 kDa.
Figure 1.
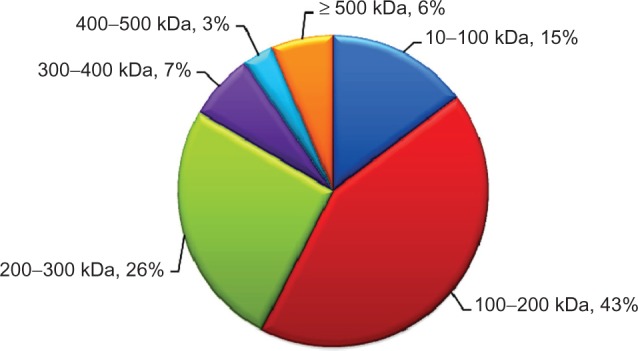
Distribution of molecular weights of membrane proteins found in human serum.
Note: Membrane protein identifications and molecular weights were obtained from MS and MS/MS data using Mascot software.
Transmembrane domains (TMDs) and average of hydrophobicity
TMD is one of the most distinguished features of membrane proteins. In our study, 129 (59.4%) proteins were predicted to have at least one TMD in the total of 217 membrane proteins according to SOSUI prediction algorithm. The majority of these 129 proteins have one TMD (25 proteins) and two TMDs (40 proteins), but a considerable number have more than 8 TMDs, which are often receptors, transporters or ion channels.
In this study, SOSUI prediction algorithm was also used to evaluate the hydrophobicity of membrane proteins from human serum. The average hydrophobicity values of all identified membrane proteins were calculated based on their amino acid sequences. It is notable that almost all proteins have average hydrophobicity values below zero. Only 17 proteins have positive average hydrophobicity values and most of them (16 proteins) are integral membrane proteins or transmembrane proteins. In addition, 7 UniProt annotated membrane proteins could not be analyzed by SOSUI prediction algorithm because their sizes have more than 5000 amino acids.
Subcellular distribution of membrane proteins
The subcellular distribution of 217 identified membrane proteins was determined using UniProt database according to their accession numbers. Analyzing the available information about the identified membrane proteins, we found that, these proteins have a certain distribution or are shuttled between organelles: 119 proteins are from plasma membrane and a range of proteins are from other cell components, such as nucleus (11 proteins), endoplasmic reticulum (16 proteins), Golgi apparatus (23 proteins), and mitochondrion (7 proteins).
Post-translational modifications of membrane proteins
In this study, 359 post-translational modifications (PTMs) of membrane proteins were found in 187 proteins based on UniProt database; the other 30 membrane proteins do not have post translational modifications. Among the total of membrane proteins having PTMs, proteins with one PTM (77 proteins – 41.2%) and two PTMs (66 proteins – 35.3%) are the majority, while proteins with 5 PTMs make up the smallest group (4 proteins – 2.1%).
Among 359 modifications found in the identified membrane proteins, phosphorylation was the most common modification, with 136 phosphoproteins. Follow-up was glycosylation, with 96 glycoproteins. The third and fourth common groups were proteins with dilsufide bonds (48 proteins) and acetylation sites (34 proteins). Lipoproteins, palmitoylated proteins, nitrated proteins, ubiquitinated proteins… contributed to a small fraction of proteins with modifications (Fig. 3).
Figure 3.
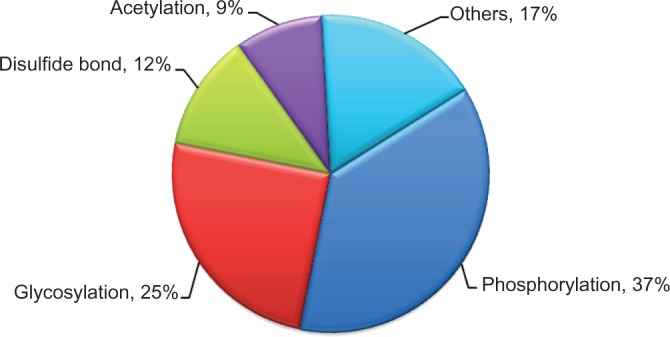
Components of post translational modifications (PTMs) of membrane proteins in human serum.
Notes: A total of 359 PTMs were found in 217 identified membrane proteins. The most common modifications are phosphorylation and glycosylation.
Discussions
In membrane proteomics, former studies are often inclined to use tissue/cell samples. Using membrane protein extraction by organic acid and NanoLC-MS/MS, Da Cruz et al found 182 membrane proteins from mouse liver mitochondrial inner membrane.21 Another study used membrane proteins extraction from pancreatic cancer cells, SDS-PAGE, in-gel digestion and NanoLC-ESI-MS/MS and detected more than 200 proteins.18 In addition, a large number (862) of membrane proteins were found in the brain cortex.19 The difference in the number of detected membrane proteins from various tissue types can be due to the specificity of each tissue, as well as fractionation and enrichment methods of each study. Combining data from four different sources, an early survey of the plasma by Anderson et al found 212 proteins with at least one TMD with multiple origins,1 which is similar to our results. Probably, there might be more membrane proteins in the serum that have not been detected yet because of their low abundance compared to other serum proteins.
Phosphorylation and glycosylation are the two most common modifications of protein in animal cells. Phosphorylation modifies the structure and function of many proteins. An upset balance between phosphorylation and dephosphorylation is the cause of many diseases.22 Glycosylation was estimated to be found in over 50% of human proteins,23 and also of interest because some diseases were found to be related to glycan structural alterations.24,25 Our results with the high proportion of phosphoproteins and glycoproteins showed the potential of membrane proteins in serum for diagnosis and treatment of disease.
Conclusion
By using NanoLC-ESI-MS/MS technologies and bioinformatics tools, a data set of 217 membrane proteins from normal human serum was identified. Some characterization such as subcellular distribution, molecular weigh, post translational modification, transmembrane domain (TMD) and average of hydrophobicity of the identified proteins were also given. 129 proteins (59%) have at least one transmembrane domain, and 187 proteins (86%) have post-translational modifications.
Supplementary Data
Table 1.
List of identified membrane proteins in human serum.
| No. | Accession number | Protein name | Score | Peptide match | Molecular weight | Subcellular location | PTM | Hydrophobicity value | TMDs |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Q8TC27 | A disintegrin and metalloprotease domain 32 | 43 | 5 | 90328 | Unclear | Disulfide bond, Glycoprotein, Phosphoprotein | −0.312452 | 2 |
| 2 | Q9H2U9 | A disintegrin and metalloproteinase 7 | 35 | 9 | 88036 | Unclear | Disulfide bond, Glycoprotein | −0.414589 | 2 |
| 3 | Q9BZC7 | ABC transporter ABCA2 | 61 | 16 | 272140 | Membrane | Glycoprotein, Phosphoprotein | −0.065216 | 14 |
| 4 | Q8IZY2 | ABC transporter ABCA7 | 46 | 6 | 236328 | Plasma membrane, Endosome membrane, Golgi apparatus membrane | Disulfide bond, Glycoprotein | 0.055080 | 12 |
| 5 | Q99758 | ABC3 | 41 | 23 | 192862 | Plasma membrane | Phosphoprotein | 0.089026 | 13 |
| 6 | P00519 | Abl protein | 46 | 3 | 123852 | Nucleus membrane | Lipoprotein, Myristate, Phosphoprotein | −0.597169 | 0 |
| 7 | O00763 | Acetyl-CoA carboxylase | 60 | 6 | 281569 | Endomembrane | Phosphoprotein | −0.227218 | 1 |
| 8 | O00763 | Acetyl-CoA carboxylase 2 | 54 | 6 | 278361 | Endomembrane | Phosphoprotein | −0.227218 | 1 |
| 9 | O14617 | Adapter-related protein complex 3 delta 1 subunit variant | 62 | 9 | 145094 | Golgi apparatus membrane | Phosphoprotein | −0.491761 | 1 |
| 10 | O95996 | Adenomatosis polyposis coli 2 | 49 | 24 | 245966 | Golgi apparatus membrane | Unknown | −0.529135 | 0 |
| 11 | Q13813 | Alpha II spectrin | 70 | 17 | 285689 | Extracellular space | Acetylation, Phosphoprotein | −0.790330 | 0 |
| 12 | P02763 | Alpha-1-acid Glycoprotein 1 precursor | 90 | 37 | 23579 | Unclear | Disulfide bond, Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.535821 | 1 |
| 13 | Q9UHC3 | Amiloride-sensitive cation channel 3 isoform c | 48 | 8 | 61376 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.265348 | 2 |
| 14 | Q07837 | Amino acid transport protein | 56 | 11 | 79240 | Plasma membrane | Disulfide bond, Glycoprotein | −0.441082 | 1 |
| 15 | Q07075 | Aminopeptidase A | 50 | 15 | 109689 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.390491 | 1 |
| 16 | Q5Y190 | Anchor protein | 53 | 7 | 458487 | Plasma membrane | Unknown | −0.188033 | 20 |
| 17 | Q96Q91 | Anion exchanger AE4 | 47 | 4 | 105149 | Plasma membrane | Glycoprotein, Phosphoprotein | 0.143947 | 11 |
| 18 | Q9P2R3 | ANKHZN | 54 | 7 | 129534 | Endosome membrane | Acetylation | −0.154490 | 0 |
| 19 | P02647 | Apolipoprotein A-I preproprotein | 245 | 45 | 30759 | Plasma membrane | Glycation, Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.717228 | 0 |
| 20 | Q12797 | Aspartyl(asparaginyl)beta-hydroxylase; HAAH | 42 | 3 | 86294 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.788128 | 1 |
| 21 | Q86UQ4 | ATP binding cassette, sub-family A (ABC1), member 13 | 50 | 12 | 580524 | Plasma membrane | Unknown | −0.010508 | 14 |
| 22 | Q96J66 | ATP-binding cassette protein C11 isoform A | 40 | 18 | 151566 | Plasma membrane | Glycoprotein | 0.191607 | 12 |
| 23 | Q96J66 | ATP-binding cassette transporter MRP8 | 47 | 5 | 155872 | Plasma membrane | Glycoprotein | 0.191607 | 12 |
| 24 | Q9HC28 | ATP-binding cassette, sub-family A, member 2 isoform b | 77 | 39 | 275244 | Lysosome membrane | Unknown | −0.064204 | 14 |
| 25 | Q92887 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 36 | 68 | 175237 | Plasma membrane | Glycoprotein, Phosphoprotein | 0.091068 | 13 |
| 26 | Q9UMD9 | Autoantigen | 55 | 8 | 154929 | Plasma membrane | Disulfide bond, Glycoprotein, Hydroxylation, Phosphoprotein | −0.573080 | 1 |
| 27 | Q13563 | Autosomal dominant polycystic kidney disease type II protein | 47 | 4 | 110462 | Endoplasmic reticulum membrane | Glycoprotein | −0.328512 | 8 |
| 28 | P50851 | Beige-like protein; CDC4L protein | 57 | 18 | 320076 | Plasma membrane | Acetylation, Phosphoprotein | −0.201817 | 0 |
| 29 | Q76KP1 | Beta-1,4-N-acetyl-galactosaminyl transferase 4 | 39 | 10 | 116954 | Golgi apparatus membrane | Glycoprotein | −0.597691 | 0 |
| 30 | Q01082 | Beta-spectrin | 43 | 10 | 275259 | Plasma membrane | Acetylation, Glycoprotein, Phosphoprotein | −0.766116 | 0 |
| 31 | Q01082 | Beta-spectrin 2 isoform 2 | 39 | 8 | 251948 | Plasma membrane | Acetylation, Glycoprotein, Phosphoprotein | −0.766116 | 0 |
| 32 | Q15413 | Brain ryanodine receptor | 73 | 24 | 556937 | Endoplasmic reticulum | Glycoprotein | −0.278727 | 5 |
| 33 | O14514 | Brain-specific angiogenesis inhibitor 1 | 51 | 14 | 176900 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.311806 | 8 |
| 34 | O60241 | Brain-specific angiogenesis inhibitor 2 | 70 | 11 | 174738 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.211924 | 9 |
| 35 | O60242 | Brain-specific angiogenesis inhibitor 3 | 59 | 7 | 176123 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.211863 | 9 |
| 36 | Q9HCU4 | Cadherin EGF LAG seven-pass G-type receptor 2 | 44 | 7 | 322214 | Plasma membrane | Disulfide bond, Glycoprotein, Hydroxylation | −0.270751 | 8 |
| 37 | Q9NYQ7 | Cadherin EGF LAG seven-pass G-type receptor 3 | 51 | 10 | 362937 | Plasma membrane | Disulfide bond, Glycoprotein, Hydroxylation | −0.271258 | 8 |
| 38 | Q9H251 | Cadherin-23 | 74 | 6 | 370095 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.142278 | 2 |
| 39 | Q59FJ3 | Calcium channel, voltage-dependent, N type, alpha 1B subunit | 49 | 8 | 264553 | Unclear | Unknown | −0.264964 | 12 |
| 40 | Q9HCF6 | Calcium-permeable store-operated channel TRPM3b | 72 | 11 | 180008 | Unclear | Unknown | −0.302195 | 6 |
| 41 | Q08499 | cAMP-specific phoshodiesterase HPDE4D3 variant | 42 | 6 | 76872 | Unclear | Phosphoprotein | −0.674043 | 0 |
| 42 | Q96P48 | Centaurin delta 2 isoform a variant | 41 | 9 | 166159 | Plasma membrane, Golgi apparatus, Membrane | Phosphoprotein | −0.422553 | 0 |
| 43 | Q15700 | Channel associated protein of synapse | 35 | 8 | 97896 | Plasma membrane | Lipoprotein, Palmitate, Phosphoprotein | −0.519310 | 0 |
| 44 | Q96QT4 | Channel-kinase 1 | 65 | 11 | 214583 | Plasma membrane | Phosphoprotein | −0.270456 | 6 |
| 45 | P51788 | Chloride channel protein 2 | 41 | 8 | 99402 | Plasma membrane | Unknown | 0.132517 | 11 |
| 46 | P00751 | Complement factor B preproprotein | 175 | 13 | 86847 | Plasma membrane | Acetylation | −0.501047 | 0 |
| 47 | P17927 | Complement receptor 1 | 38 | 8 | 230417 | Plasma membrane | Disulfide bond, Glycoprotein, Pyrrolidone carboxylic acid | −0.320010 | 3 |
| 48 | P10606 | COX5B | 64 | 178 | 13914 | Mitochondrion membrane | Acetylation | −0.297674 | 0 |
| 49 | Q7Z407 | CUB and Sushi multiple domains 3 isoform 1 | 36 | 13 | 414007 | Plasma membrane | Disulfide bond, Glycoprotein | −0.260239 | 1 |
| 50 | Q8IY37 | DEAH (Asp-Glu-Ala-His) box polypeptide 37 | 40 | 4 | 130547 | Unclear | Unknown | −0.415385 | 0 |
| 51 | Q6IC98 | Death-inducing-protein | 49 | 14 | 66766 | Mitochondrion membrane | Phosphoprotein | −0.431833 | 3 |
| 52 | Q14185 | Dedicator of cytokinesis 1 | 52 | 6 | 216208 | Unclear | Phosphoprotein | −0.430617 | 0 |
| 53 | Q5VWQ8 | Disabled homolog 2 interacting protein isoform 2 | 37 | 8 | 118547 | Plasma membrane | Phosphoprotein | −0.578302 | 0 |
| 54 | O75923 | Dysferlin isoform 8 | 43 | 4 | 239254 | Vesicle | Phosphoprotein | −0.400145 | 1 |
| 55 | P22413 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 36 | 6 | 107024 | Plasma membrane | Disulfide bond, Glycoprotein | −0.456000 | 1 |
| 56 | Q8WYP5 | ELYS transcription factor-like protein TMBS62 | 43 | 4 | 258191 | Nuclear membrane | Acetylation, Phosphoprotein | −0.448765 | 0 |
| 57 | Q541P7 | EPH receptor B4 precursor | 35 | 4 | 109741 | Plasma membrane | Unknown | −0.235562 | 2 |
| 58 | Q15375 | Ephrin receptor EphA7 | 55 | 10 | 113735 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.284268 | 2 |
| 59 | Q15375 | Ephrin receptor EphA7 variant | 61 | 10 | 113994 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.284268 | 2 |
| 60 | P54762 | Ephrin receptor EphB1 precursor | 45 | 20 | 111297 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.214431 | 2 |
| 61 | P29323 | Ephrin type-B receptor 2 | 36 | 11 | 119128 | Plasma membrane | Lipoprotein, Myristate,Palmitate, Phosphoprotein | −0.293460 | 2 |
| 62 | Q9NZJ5 | Eukaryotic translation initiation factor 2-alpha kinase 3 | 48 | 11 | 126095 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.450717 | 2 |
| 63 | Q16099 | Excitatory amino acid receptor 1; kainate receptor subunit EAA1 | 41 | 9 | 108529 | Plasma membrane | Glycoprotein | −0.046653 | 4 |
| 64 | Q86XX4 | Extracellular matrix protein FRAS1 | 45 | 10 | 453936 | Plasma membrane | Glycoprotein | −0.295608 | 2 |
| 65 | Q9NYQ8 | FAT tumor suppressor 2 precursor | 44 | 7 | 482097 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.249645 | 2 |
| 66 | Q6V0I7 | Fat-like cadherin FATJ protein | 43 | 10 | 354108 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.262940 | 2 |
| 67 | Q9NZM1 | Fer-1 like protein 3 | 66 | 11 | 234737 | Nuclear membrane, plasma membrane | Acetylation, Phosphoprotein | −0.456091 | 1 |
| 68 | Q59F30 | Fibroblast growth factor receptor 4 variant | 38 | 5 | 114772 | Unclear | Unknown | 0.017312 | 6 |
| 69 | P42345 | FK506 binding protein 12-rapamycin associated protein 1 | 40 | 10 | 290759 | Golgi membrane, endoplasmic reticulum membrane, mitochondrial outer membrane | Acetylation, Phosphoprotein | −0.192586 | 0 |
| 70 | Q9Y2I7 | FYVE finger-containing phosphoinositide kinase | 66 | 18 | 239581 | Endosome membrane | Acetylation, Phosphoprotein | −0.534366 | 0 |
| 71 | Q86SQ6 | G protein-coupled receptor 123 | 35 | 6 | 139868 | Plasma membrane | Glycoprotein | −0.317749 | 6 |
| 72 | Q5T848 | G protein-coupled receptor 158 | 53 | 8 | 136886 | Plasma membrane | Glycoprotein, Isopeptide bond, Phosphoprotein, Ubl conjugation | −0.480988 | 7 |
| 73 | O75899 | GABBR2 protein | 42 | 5 | 100861 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.149734 | 8 |
| 74 | Q14789 | Giantin | 44 | 13 | 377273 | Golgi apparatus membrane | Disulfide bond, Phosphoprotein | −0.936847 | 1 |
| 75 | P42261 | Glutamate receptor type 1 | 42 | 7 | 102270 | Plasma membrane, Endoplasmic reticulum membrane | Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.199227 | 5 |
| 76 | Q9Y3R0 | Glutamate receptor-interacting protein 1 | 46 | 5 | 123203 | Plasma membrane, Endoplasmic reticulum membrane | Phosphoprotein | −0.362500 | 0 |
| 77 | Q14789 | Golgi antigen gcp372 | 44 | 8 | 373440 | Golgi apparatus membrane | Disulfide bond, Phosphoprotein | −0.936847 | 1 |
| 78 | Q08378 | Golgi autoantigen, golgin subfamily a, 3 | 69 | 17 | 167765 | Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.834781 | 0 |
| 79 | Q08378 | Golgin-160 | 57 | 16 | 167810 | Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.834781 | 0 |
| 80 | Q6PRD1 | GPR158-like 1 receptor | 54 | 11 | 260635 | Plasma membrane | Glycoprotein | −0.583101 | 7 |
| 81 | Q8WXG9 | G-protein coupled receptor 98 | 69 | 21 | 694181 | Plasma membrane | Unknown | Too long | 0 |
| 82 | Q8IWJ2 | GRIP and coiled-coil domain-containing 2 isoform a | 53 | 15 | 196873 | Golgi apparatus membrane | Phosphoprotein | −0.918825 | 0 |
| 83 | O15068 | Guanine nucleotide exchange factor DBS | 37 | 4 | 129340 | Plasma membrane | Phosphoprotein | −0.552067 | 0 |
| 84 | P52272 | Heterogeneous nuclear ribonucleoprotein M isoform a | 44 | 13 | 77749 | Plasma membrane | Acetylation, Phosphoprotein, Ubl conjugation | −0.341643 | 0 |
| 85 | O43166 | High-risk human papilloma viruses E6 oncoproteins targeted protein E6TP1 alpha; putative GAP protei | 59 | 12 | 198570 | Plasma membrane | Phosphoprotein | −0.671840 | 0 |
| 86 | P35523 | Human ClC-1 muscle chloride channel | 37 | 7 | 109696 | Plasma membrane | Unknown | 0.034919 | 11 |
| 87 | Q14643 | Human type 1 inositol 1,4,5-trisphosphate receptor | 43 | 8 | 309942 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.315448 | 6 |
| 88 | Q9HCF6 | Hypothetical protein | 72 | 10 | 175211 | Unclear | Unknown | −0.300924 | 6 |
| 89 | Q9P1Z9 | Hypothetical protein | 43 | 14 | 199652 | Unclear | Phosphoprotein | −0.647996 | 2 |
| 90 | A6NI73 | Immunoglobulin-like transcript 11 protein | 39 | 4 | 27186 | Unclear | Disulfide bond, Glycoprotein | −0.267224 | 2 |
| 91 | Q14571 | Inositol 1,4,5-trisphosphate receptor type 2 | 50 | 6 | 311074 | Plasma membrane | Acetylation, Phosphoprotein | −0.266755 | 7 |
| 92 | Q6P9B9 | Integrator complex subunit 5 | 38 | 5 | 109239 | Nucleus membrane | Phosphoprotein | 0.109029 | 2 |
| 93 | Q15811 | Intersectin long form | 40 | 7 | 196293 | Plasma membrane | Acetylation, Phosphoprotein | −0.668043 | 0 |
| 94 | Q9H4E7 | IRF4-binding protein | 48 | 7 | 74422 | Plasma membrane | Acetylation, Phosphoprotein | −0.946435 | 0 |
| 95 | Q92508 | KIAA0233 | 50 | 17 | 234888 | Endoplasmic reticulum membrane | Glycoprotein | −0.002357 | 23 |
| 96 | Q13023 | KIAA0311 | 40 | 7 | 259702 | Nucleus membrane | Unknown | −0.675982 | 0 |
| 97 | Q96P48 | KIAA0782 protein | 45 | 4 | 145325 | Plasma membrane, Golgi apparatus membrane | Phosphoprotein | −0.422553 | 0 |
| 98 | Q9Y2H9 | KIAA0973 protein | 61 | 34 | 173192 | Plasma membrane | Phosphoprotein | −0.569363 | 0 |
| 99 | Q96RV3 | KIAA0995 protein | 42 | 7 | 109401 | Plasma membrane | Glycoprotein | −0.284538 | 13 |
| 100 | Q9BZ29 | KIAA1058 protein | 42 | 7 | 241774 | Plasma membrane | Phosphoprotein | −0.349204 | 0 |
| 101 | Q5T848 | KIAA1136 protein | 49 | 11 | 66642 | Plasma membrane | Glycoprotein, Isopeptide bond, Phosphoprotein, Ubl conjugation | −0.480988 | 7 |
| 102 | Q5T4S7 | KIAA1307 protein | 53 | 7 | 188524 | Unclear | Acetylation, Phosphoprotein | Too long | 0 |
| 103 | Q9BZ72 | KIAA1457 protein | 43 | 11 | 151395 | Plasma membrane | Phosphoprotein | −0.405561 | 0 |
| 104 | Q9P1Z9 | KIAA1529 protein | 46 | 6 | 196058 | Unclear | Phosphoprotein | −0.647996 | 2 |
| 105 | Q6KCM7 | KIAA1896 protein | 45 | 7 | 63937 | Mitochondrial membrane | Unknown | −0.099147 | 0 |
| 106 | Q16787 | Laminin alpha 3 subunit isoform 1 | 45 | 7 | 375652 | Unclear | Disulfide bond, Glycoprotein | −0.381368 | 0 |
| 107 | P11047 | Laminin B2 chain | 54 | 7 | 183195 | Unclear | Disulfide bond, Glycoprotein | −0.615042 | 0 |
| 108 | Q9Y6N6 | Laminin gamma 3 chain precursor | 40 | 6 | 177756 | Unclear | Disulfide bond, Glycoprotein | −0.465018 | 0 |
| 109 | Q13449 | LAMP | 41 | 13 | 37798 | Plasma membrane | Disulfide bond, GPI-anchor, Glycoprotein, Lipoprotein, Phosphoprotein | −0.278106 | 2 |
| 110 | P42704 | Leucine-rich PPR-motif containing protein | 52 | 7 | 146306 | Nuclear inner membrane | Acetylation | −0.206097 | 0 |
| 111 | Q86UK5 | Limbin | 41 | 5 | 148825 | Plasma membrane | Glycoprotein | −0.445185 | 2 |
| 112 | P50851 | Lipopolysaccharide-responsive and beige-like anchor protein | 63 | 10 | 321639 | Plasma membrane | Acetylation, Phosphoprotein | −0.201817 | 0 |
| 113 | Q9HCF6 | Long transient receptor potential channel 3 | 75 | 10 | 196157 | Plasma membrane | Unknown | −0.300924 | 6 |
| 114 | Q9NZR2 | Low density lipoprotein receptor related protein-deleted in tumor | 51 | 9 | 534844 | Plasma membrane | Disulfide bond, Glycoprotein | −0.489888 | 2 |
| 115 | Q5JRA6 | Melanoma inhibitory activity family, member 3 | 52 | 7 | 214255 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.887466 | 3 |
| 116 | Q86UL8 | Membrane associated guanylate kinase, WW and PDZ domain containing 2 | 39 | 5 | 159454 | Plasma membrane | Phosphoprotein | −0.668455 | 0 |
| 117 | Q13421 | Mesothelin | 45 | 4 | 68652 | Plasma membrane | Cleavage on pair of basic residues, GPI-anchor, Glycoprotein, Lipoprotein | −0.105714 | 2 |
| 118 | A1L467 | Met proto-oncogene isoform b precursor | 48 | 6 | 157779 | Plasma membrane | Unknown | −0.144388 | 2 |
| 119 | Q8IWA4 | Mitofusin 1 precursor | 38 | 10 | 84892 | Mitochondrial membrane | Unknown | −0.267476 | 2 |
| 120 | Q6UVY6 | Monooxygenase X | 36 | 9 | 63474 | Endoplasmic reticulum membrane | Disulfide bond, Glycoprotein | −0.260848 | 2 |
| 121 | Q8WXI7 | Mucin 16 | 38 | 7 | 747071 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | Too long | 1 |
| 122 | O75970 | Multiple PDZ domain protein | 60 | 7 | 222792 | Plasma membrane | Phosphoprotein | −0.233203 | 0 |
| 123 | Q9NZM1 | Myoferlin | 42 | 6 | 231092 | Vesicle membrane, nuclear membrane, plasma membrane | Acetylation, Phosphoprotein | −0.456091 | 1 |
| 124 | B2RTY4 | Myosin-IXa | 54 | 9 | 294918 | Unclear | Phosphoprotein | −0.623980 | 0 |
| 125 | Q8NFP9 | Neurobeachin | 53 | 6 | 330066 | Plasma membrane | Phosphoprotein | −0.188358 | 2 |
| 126 | P21359 | Neurofibromin isoform 1 | 43 | 10 | 322760 | Plasma membrane | Disulfide bond,GPI-anchor, Glycoprotein, Lipoprotein | −0.136950 | 4 |
| 127 | Q9H2E3 | Neuropilin-2b(5) | 53 | 6 | 102975 | Plasma membrane | Unknown | −0.440066 | 2 |
| 128 | Q13423 | Nicotinamide nucleotide transhydrogenase | 85 | 9 | 114564 | Mitochondrial membrane | Acetylation | 0.299079 | 13 |
| 129 | Q59GR1 | Niemann-Pick disease, type C1 variant | 36 | 5 | 145931 | Plasma membrane | Unknown | 0.216447 | 13 |
| 130 | Q13224 | N-methyl-D-aspartate receptor subunit NR3 | 53 | 16 | 168067 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.388342 | 6 |
| 131 | Q13813 | Nonerythroid alpha-spectrin | 59 | 7 | 284905 | Unclear | Acetylation, Phosphoprotein | −0.790330 | 0 |
| 132 | Q9UM47 | Notch homolog 3 | 43 | 8 | 256640 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.304826 | 3 |
| 133 | Q5STG5 | Notch homolog 4 (Drosophila) | 51 | 7 | 221594 | Unclear | Unknown | −0.287183 | 3 |
| 134 | P46531 | Notch1 preproprotein | 38 | 11 | 286350 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.420119 | 3 |
| 135 | Q92823 | NrCAM protein | 37 | 11 | 144138 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.379601 | 2 |
| 136 | Q8WXH0 | NUANCE | 104 | 29 | 801683 | Nuclear membrane | Acetylation, Phosphoprotein | Too long | 0 |
| 137 | Q5VU65 | Nucleoporin 210 kDa-like isoform 1 | 43 | 10 | 211668 | Unclear | Glycoprotein | 0.050265 | 3 |
| 138 | Q13017 | p190-B | 45 | 6 | 173000 | Unclear | Nitration, Phosphoprotein | −0.592345 | 0 |
| 139 | Q5T4S7 | p600 | 121 | 21 | 580607 | Unclear | Acetylation, Phosphoprotein | Too long | 0 |
| 140 | O60245 | PCDH7 (BH-Pcdh)c | 40 | 4 | 130999 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.390365 | 2 |
| 141 | Q9BXM0 | Periaxin | 59 | 8 | 155248 | Plasma membrane | Phosphoprotein | −0.225736 | 0 |
| 142 | O75747 | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide | 38 | 9 | 167814 | Unclear | Phosphoprotein | −0.322423 | 1 |
| 143 | O00443 | Phosphoinositide 3-kinase | 43 | 9 | 192271 | Plasma membrane | Acetylation, Phosphoprotein | −0.302136 | 0 |
| 144 | O00750 | PI-3 kinase | 46 | 5 | 184101 | Plasma membrane | Phosphoprotein | −0.363037 | 0 |
| 145 | Q6T4P5 | Plasticity-related protein 2 | 43 | 12 | 79523 | Membrane | Glycoprotein | −0.178273 | 6 |
| 146 | O75051 | Plexin A2 | 37 | 13 | 214220 | Cell membrane | Glycoprotein, Phosphoprotein | −0.181468 | 3 |
| 147 | Q9Y4D7 | Plexin-D1 | 48 | 9 | 215381 | Cell membrane, Membrane | Glycoprotein | −0.159013 | 2 |
| 148 | Q15142 | Polycystic kidney disease-associated protein | 57 | 5 | 396273 | Integral to membrane | Unknown | 0.022760 | 14 |
| 149 | Q8TDX9 | Polycystin-1 L1 | 35 | 4 | 319453 | Integral to membrane | Glycoprotein | −0.136856 | 7 |
| 150 | P13942 | Pro-a2(XI) | 51 | 10 | 160545 | Cell membrane | Disulfide bond, Glycoprotein, Sulfation | −0.805186 | 0 |
| 151 | P08123 | Pro-alpha 2(I) collagen | 51 | 9 | 129858 | Plasma membrane | Glycoprotein, Hydroxylation, Pyrrolidone carboxylic acid | −0.648098 | 0 |
| 152 | P02647 | Proapolipoprotein | 664 | 75 | 28944 | Plasma membrane | Glycation, Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.717228 | 0 |
| 153 | Q07954 | Prolow-density lipoprotein receptor-related protein 1 | 66 | 13 | 523119 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.510386 | 2 |
| 154 | Q05655 | Protein kinase C-delta 13 | 37 | 4 | 78624 | Plasma membrane | Phosphoprotein | −0.377958 | 0 |
| 155 | Q8TF72 | Protein Shroom 3 | 41 | 8 | 218125 | Plasma membrane | Phosphoprotein | −0.801652 | 0 |
| 156 | Q96QU1 | Protocadherin 15 | 59 | 13 | 217261 | Plasma membrane | Phosphoprotein | −0.245473 | 2 |
| 157 | Q96QU1 | Protocadherin 15 isoform CD1-4 precursor | 48 | 12 | 217303 | Plasma membrane | Phosphoprotein | −0.245473 | 2 |
| 158 | Q9UN70 | Protocadherin 43 | 45 | 5 | 98230 | Plasma membrane | Glycoprotein | −0.261670 | 2 |
| 159 | Q14517 | Protocadherin Fat 1 | 52 | 9 | 509384 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.294946 | 2 |
| 160 | Q9NYQ8 | Protocadherin Fat 2 | 59 | 7 | 482172 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.249645 | 2 |
| 161 | Q86V60 | PTPRM protein | 39 | 8 | 158434 | Unclear | Unknown | −0.385540 | 1 |
| 162 | Q8WZA2 | RAPGEF4 protein | 53 | 13 | 100339 | Unclear | Phosphoprotein | −0.333927 | 0 |
| 163 | P23467 | Receptor-type tyrosine-protein phosphatase beta | 50 | 26 | 225497 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.372159 | 2 |
| 164 | Q13332 | Receptor-type tyrosine-protein phosphatase S | 43 | 9 | 218155 | Plasma membrane | Disulfide bond, Glycoprotein | −0.401798 | 2 |
| 165 | Q86UR5 | Regulating synaptic membrane exocytosis 1 | 40 | 6 | 190154 | Plasma membrane | Phosphoprotein | −0.922459 | 0 |
| 166 | Q5T4S7 | Retinoblastoma-associated factor 600 | 124 | 20 | 580547 | Unclear | Acetylation, Phosphoprotein | Too long | 0 |
| 167 | Q5T5U3 | Rho GTPase-activating protein 21 | 41 | 8 | 218567 | Golgi apparatus membrane, vesicle membrane | Phosphoprotein | −0.827338 | 0 |
| 168 | Q13464 | Rho-associated, coiled-coil containing protein kinase 1 | 48 | 6 | 159102 | Golgi apparatus membrane | Unknown | −0.909454 | 0 |
| 169 | Q5T5U3 | Rho-GTPase activating protein 10 | 41 | 5 | 218563 | Golgi apparatus membrane, vesicle membrane | Phosphoprotein | −0.827338 | 0 |
| 170 | Q5JTH9 | Ribosomal RNA processing 12 homolog (S. cerevisiae) | 56 | 9 | 145067 | Nuclear membrane | Phosphoprotein | −0.225366 | 1 |
| 171 | Q86UR5 | RIM long form | 48 | 7 | 164927 | Plasma membrane | Phosphoprotein | −0.922459 | 0 |
| 172 | P21817 | Ryanodine receptor 1 | 61 | 11 | 55204 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.314300 | 7 |
| 173 | Q92736 | Ryanodine receptor 2 | 70 | 10 | 569626 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.324300 | 7 |
| 174 | Q15413 | Ryanodine receptor 3 | 71 | 24 | 557794 | Endoplasmic reticulum membrane | Glycoprotein | −0.278727 | 5 |
| 175 | O15027 | SEC16 homolog A | 36 | 5 | 115891 | Endoplasmic reticulum membrane, Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.606607 | 0 |
| 176 | A4QN19 | SEC16A protein | 40 | 2 | 116196 | Endoplasmic reticulum membrane, Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.606607 | 0 |
| 177 | Q9H2E6 | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 36 | 4 | 115608 | Plasma membrane | Disulfide bond, Glycoprotein | −0.448543 | 2 |
| 178 | O94921 | Serine/threonine protein kinase PFTAIRE-1 | 41 | 8 | 53609 | Plasma membrane | Phosphoprotein | −0.470362 | 0 |
| 179 | Q8NGB0 | Seven transmembrane helix receptor | 49 | 10 | 157779 | Unclear | Unknown | −0.275069 | 6 |
| 180 | Q9Y566 | SH3 and multiple ankyrin repeat domains protein 1 | 49 | 7 | 225738 | Plasma membrane | Phosphoprotein | −0.539239 | 0 |
| 181 | Q9UPX8 | SH3 and multiple ankyrin repeat domains protein 2 | 54 | 6 | 135115 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.608572 | 0 |
| 182 | Q7Z5N4 | Sidekick-like protein 1 | 39 | 4 | 242926 | Plasma membrane | Disulfide bond, Glycoprotein | −0.314144 | 1 |
| 183 | P21817 | Skeletal muscle ryanodine receptor | 78 | 9 | 570517 | Plasma membrane | Glycoprotein, Phosphoprotein, S-nitrosylation | Too long | 0 |
| 184 | O94813 | Slit homolog 2 | 47 | 13 | 175803 | Plasma membrane | Disulfide bond, Glycoprotein | −0.323414 | 0 |
| 185 | O94813 | SLIT2 | 40 | 7 | 175329 | Plasma membrane | Disulfide bond, Glycoprotein | −0.323414 | 0 |
| 186 | Q9C0H9 | SNAP-25-interacting protein | 42 | 6 | 127182 | Plasma membrane | Phosphoprotein | −0.644456 | 0 |
| 187 | Q9Y6M7 | Sodium bicarbonate cotransporter2 | 44 | 8 | 115215 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.130725 | 10 |
| 188 | Q53ZR1 | Solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | 64 | 13 | 132048 | Plasma membrane | Unknown | 0.069720 | 11 |
| 189 | Q9HBR0 | Solute carrier family 38, member 10 isoform b | 40 | 5 | 84069 | Unclear | Phosphoprotein | −0.363718 | 11 |
| 190 | Q9NRC6 | Spectrin beta chain, brain 4 | 72 | 14 | 419259 | Unclear | Unknown | −0.576588 | 0 |
| 191 | Q01082 | Spectrin, beta, non-erythrocytic 1 isoform 1 variant | 48 | 8 | 276797 | Plasma membrane | Acetylation, Glycoprotein, Phosphoprotein | −0.766116 | 0 |
| 192 | Q8WWQ8 | Stabilin 2 precursor | 57 | 9 | 288323 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein, Proteoglycan | −0.268563 | 2 |
| 193 | Q8WWQ8 | Stabilin-2 | 46 | 9 | 288329 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein, Proteoglycan | −0.268563 | 2 |
| 194 | Q8IVL0 | Steerin3 protein | 37 | 5 | 256885 | Nuclear membrane | Phosphoprotein | −0.638784 | 0 |
| 195 | Q4LDE5 | Sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 52 | 10 | 401915 | Unclear | Disulfide bond, Glycoprotein | −0.320528 | 1 |
| 196 | Q9Y4G6 | Talin-2 | 50 | 11 | 273723 | Plasma membrane | Acetylation, Phosphoprotein | −0.218412 | 0 |
| 197 | Q9H2K2 | Tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | 41 | 4 | 128492 | Golgi apparatus membrane | ADP-ribosylation | −0.317066 | 0 |
| 198 | O60343 | TBC1 domain family, member 4 | 36 | 5 | 148068 | Plasma membrane | Phosphoprotein | −0.553083 | 1 |
| 199 | Q15643 | Thyroid receptor-interacting protein 11 | 38 | 7 | 228184 | Golgi apparatus membrane | Phosphoprotein | −0.843506 | 0 |
| 200 | P12270 | Tpr | 54 | 10 | 265840 | Nuclear membrane | Acetylation, Phosphoprotein | −0.968726 | 0 |
| 201 | Q9BX84 | Transient receptor potential cation channel subfamily M member 6 variant c | 79 | 20 | 232975 | Unclear | Phosphoprotein | −0.369882 | 5 |
| 202 | Q8N6Q1 | Transmembrane and coiled-coil domains 5A | 47 | 5 | 34495 | Unclear | Unknown | −0.563888 | 1 |
| 203 | O95271 | TRF1-interacting ankyrin-related ADP-ribose polymerase | 47 | 12 | 143690 | Golgi apparatus membrane | ADP-ribosylation, Phosphoprotein | −0.327506 | 0 |
| 204 | Q15643 | Trip230 | 92 | 12 | 228116 | Unclear | Phosphoprotein | −0.843506 | 0 |
| 205 | Q9BX84 | Truncated transient receptor potential cation channel subfamily M member 6 variant a | 57 | 13 | 224977 | Plasma membrane | Phosphoprotein | −0.369882 | 5 |
| 206 | Q8NFA0 | Ubiquitin specific protease 32 | 47 | 8 | 183821 | Unclear | Lipoprotein, Phosphoprotein, Prenylation | −0.474689 | 0 |
| 207 | P55916 | Uncoupling protein 3 isoform UCP3S | 36 | 15 | 30219 | Mitochondrion membrane | Unknown | 0.074679 | 0 |
| 208 | P17927 | Unnamed protein product | 41 | 10 | 230401 | Plasma membrane | Disulfide bond, Glycoprotein, Pyrrolidone carboxylic acid | −0.320010 | 3 |
| 209 | P53420 | Unnamed protein product | 65 | 10 | 126774 | Vesicle membrane | Acetylation, Phosphoprotein | −0.659231 | 1 |
| 210 | P11717 | Unnamed protein product | 48 | 14 | 281089 | Plasma membrane | Acetylation, Disulfide bond, Glycoprotein, Phosphoprotein | −0.368528 | 3 |
| 211 | Q6ZUB1 | Unnamed protein product | 36 | 10 | 158870 | Unclear | Glycoprotein | −0.709205 | 1 |
| 212 | O75110 | Unnamed protein product | 39 | 10 | 108198 | Unclear | Phosphoprotein | 0.062178 | 9 |
| 213 | Q9NRW7 | Vacuolar protein sorting | 41 | 6 | 65388 | Endosome membrane, Golgi apparatus membrane | Unknown | −0.310526 | 0 |
| 214 | Q86Y38 | Xylosyltransferase I | 41 | 12 | 108357 | Endoplasmic reticulum membrane, Golgi apparatus membrane | Disulfide bond, Glycoprotein | −0.540564 | 0 |
| 215 | Q13433 | Zinc transporter ZIP6 | 41 | 5 | 84685 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.421987 | 6 |
| 216 | Q13439 | 256 kD golgin | 59 | 9 | 256666 | Golgi apparatus membrane | Phosphoprotein | −1.052981 | 0 |
| 217 | Q8N1I0 | Dedicator of cytokinesis protein 4 | 49 | 11 | 225206 | Endomembrane | Phosphoprotein | −0.371464 | 0 |
Figure 2.
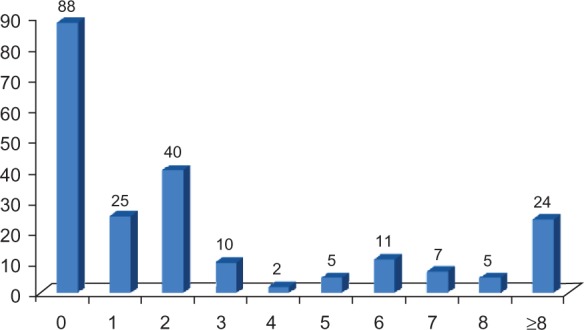
Prediction of transmembrane domains of identified membrane proteins.
Notes: Sequences of 217 membrane proteins indentified in human serum were processed by SOSUI prediction system for transmembrane domain (TMD) predictions. More than half (59.4%) of the identified membrane proteins have at least one TMD, and the rest (40.6%) with no TMD are membrane-associated proteins. The ordinate axis shows the protein number, whereas absciss axis shows the number of TMD.
Acknowledgments
The work was carried out at the National Key Laboratory of Gene Technology (NKLGT), Institute of Biotechnology (IBT), Vietnam Academy of Science and Technology (VAST).
Footnotes
Author Contributions
Conceived and designed the experiments: PVC. Analysed the data: NTD. Wrote the first draft of the manuscript: PVC, NTD. Contributed to the writing of the manuscript: PVC, NTD. Agree with manuscript results and conclusions: PVC, NTD. Jointly developed the structure and arguments for the paper: PVC, NTD. Made critical revisions and approved final version: PVC, NTD. All authors reviewed and approved of the final manuscript.
Funding
The work was funded by National Foundation for Science and Technology Development (NAFOSTED).
Competing Interests
The author declares no conflicts of interest
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Anderson NL, Polanski M, Peiper R, et al. The human plasma proteome: A nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3:311–26. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Pernemalm M, Lewensohn R, Lehtiö J. Affinity prefractionation for MS-based plasma proteomics. Proteomics. 2009;9(6):1420–7. doi: 10.1002/pmic.200800377. [DOI] [PubMed] [Google Scholar]
- 3.Pieper R, Gatlin CL, Makusky AJ, et al. The human serum proteome: Display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3(7):1345–64. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 4.Farrah T, Deutsch EW, Omenn GS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in Peptide Atlas. Mol Cell Proteomics. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–9. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 6.Hye A, Lynham S, Thambisetty M, et al. Proteome-based plasma biomarkers for Alzheimer’s disease. Brain. 2006;129(11):3042–50. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- 7.Marshall J, Kupchak P, Zhu W, et al. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J Proteome Res. 2003;2(4):361–72. doi: 10.1021/pr030003l. [DOI] [PubMed] [Google Scholar]
- 8.Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39(4):417–20. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 9.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean and eukaryotic organisms. Protein Sci. 1998;7(4):1029–38. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu CC, Yates JR. The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21(3):262–7. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 12.Fagerberg L, Jonasson K, von Heijne G, et al. Prediction of the human membrane proteome. Proteomics. 2010;10(6):1141–9. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 13.Josic D, Clifton JG. Mammalian plasma membrane proteomics. Proteomics. 2007;7(16):3010–29. doi: 10.1002/pmic.200700139. [DOI] [PubMed] [Google Scholar]
- 14.Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: Un amour impossible? Electrophoresis. 2000;21(6):1054–70. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Sprenger RR, Jensen ON. Proteomics and the dynamics plasma membrane: Quo Vadis? Proteomics. 2010;10(22):3997–011. doi: 10.1002/pmic.201000312. [DOI] [PubMed] [Google Scholar]
- 16.Peng L, Kapp EA, Fenyö D, et al. The Asia Oceania Human Proteome Organisation Membrane Proteomics Initiative. Preparation and characterisation of the carbonate-washed membrane standard. Proteomics. 2010;10(22):4142–8. doi: 10.1002/pmic.201000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng L, Kapp EA, McLauchlan D, et al. Characterization of the Asia Oceania Human Proteome Organisation Membrane Proteomics Initiative Standard using SDS-PAGE shotgun proteomics. Proteomics. 2011;11(22):4376–84. doi: 10.1002/pmic.201100169. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Zhang M, Go VL, et al. Membrane proteomic analysis of pancreatic cancer cells. J Biomed Sci. 2010;17(1):17–74. doi: 10.1186/1423-0127-17-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen PA, Olsen JV, Podtelejnikov AV, et al. Proteomic mapping of brain plasma membrane proteins. Mol Cell Proteomics. 2005;4(4):402–8. doi: 10.1074/mcp.T500002-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14(4):378–9. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 21.Da Cruz S Martinou JC. Purification and proteomic analysis of the mouse liver mitochondrial inner membrane. Methods Mol Biol. 2008;432:101–16. doi: 10.1007/978-1-59745-028-7_7. [DOI] [PubMed] [Google Scholar]
- 22.Cohen P. The role of protein phosphorylation in human health and disease. Eur J Biochem. 2001;268(19):5001–10. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 23.Wong CH. Protein glycosylation: new challenges and opportunities. J Org Chem. 2005;70(11):4219–25. doi: 10.1021/jo050278f. [DOI] [PubMed] [Google Scholar]
- 24.Durand G, Seta N. Protein glycosylation and diseases: blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin Chem. 2000;46(6):795–805. [PubMed] [Google Scholar]
- 25.Tian Y, Zhan H. Glycoproteomics and clinical applications. Proteomics Clin Appl. 2010;4(2):124–32. doi: 10.1002/prca.200900161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1.
List of identified membrane proteins in human serum.
| No. | Accession number | Protein name | Score | Peptide match | Molecular weight | Subcellular location | PTM | Hydrophobicity value | TMDs |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Q8TC27 | A disintegrin and metalloprotease domain 32 | 43 | 5 | 90328 | Unclear | Disulfide bond, Glycoprotein, Phosphoprotein | −0.312452 | 2 |
| 2 | Q9H2U9 | A disintegrin and metalloproteinase 7 | 35 | 9 | 88036 | Unclear | Disulfide bond, Glycoprotein | −0.414589 | 2 |
| 3 | Q9BZC7 | ABC transporter ABCA2 | 61 | 16 | 272140 | Membrane | Glycoprotein, Phosphoprotein | −0.065216 | 14 |
| 4 | Q8IZY2 | ABC transporter ABCA7 | 46 | 6 | 236328 | Plasma membrane, Endosome membrane, Golgi apparatus membrane | Disulfide bond, Glycoprotein | 0.055080 | 12 |
| 5 | Q99758 | ABC3 | 41 | 23 | 192862 | Plasma membrane | Phosphoprotein | 0.089026 | 13 |
| 6 | P00519 | Abl protein | 46 | 3 | 123852 | Nucleus membrane | Lipoprotein, Myristate, Phosphoprotein | −0.597169 | 0 |
| 7 | O00763 | Acetyl-CoA carboxylase | 60 | 6 | 281569 | Endomembrane | Phosphoprotein | −0.227218 | 1 |
| 8 | O00763 | Acetyl-CoA carboxylase 2 | 54 | 6 | 278361 | Endomembrane | Phosphoprotein | −0.227218 | 1 |
| 9 | O14617 | Adapter-related protein complex 3 delta 1 subunit variant | 62 | 9 | 145094 | Golgi apparatus membrane | Phosphoprotein | −0.491761 | 1 |
| 10 | O95996 | Adenomatosis polyposis coli 2 | 49 | 24 | 245966 | Golgi apparatus membrane | Unknown | −0.529135 | 0 |
| 11 | Q13813 | Alpha II spectrin | 70 | 17 | 285689 | Extracellular space | Acetylation, Phosphoprotein | −0.790330 | 0 |
| 12 | P02763 | Alpha-1-acid Glycoprotein 1 precursor | 90 | 37 | 23579 | Unclear | Disulfide bond, Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.535821 | 1 |
| 13 | Q9UHC3 | Amiloride-sensitive cation channel 3 isoform c | 48 | 8 | 61376 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.265348 | 2 |
| 14 | Q07837 | Amino acid transport protein | 56 | 11 | 79240 | Plasma membrane | Disulfide bond, Glycoprotein | −0.441082 | 1 |
| 15 | Q07075 | Aminopeptidase A | 50 | 15 | 109689 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.390491 | 1 |
| 16 | Q5Y190 | Anchor protein | 53 | 7 | 458487 | Plasma membrane | Unknown | −0.188033 | 20 |
| 17 | Q96Q91 | Anion exchanger AE4 | 47 | 4 | 105149 | Plasma membrane | Glycoprotein, Phosphoprotein | 0.143947 | 11 |
| 18 | Q9P2R3 | ANKHZN | 54 | 7 | 129534 | Endosome membrane | Acetylation | −0.154490 | 0 |
| 19 | P02647 | Apolipoprotein A-I preproprotein | 245 | 45 | 30759 | Plasma membrane | Glycation, Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.717228 | 0 |
| 20 | Q12797 | Aspartyl(asparaginyl)beta-hydroxylase; HAAH | 42 | 3 | 86294 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.788128 | 1 |
| 21 | Q86UQ4 | ATP binding cassette, sub-family A (ABC1), member 13 | 50 | 12 | 580524 | Plasma membrane | Unknown | −0.010508 | 14 |
| 22 | Q96J66 | ATP-binding cassette protein C11 isoform A | 40 | 18 | 151566 | Plasma membrane | Glycoprotein | 0.191607 | 12 |
| 23 | Q96J66 | ATP-binding cassette transporter MRP8 | 47 | 5 | 155872 | Plasma membrane | Glycoprotein | 0.191607 | 12 |
| 24 | Q9HC28 | ATP-binding cassette, sub-family A, member 2 isoform b | 77 | 39 | 275244 | Lysosome membrane | Unknown | −0.064204 | 14 |
| 25 | Q92887 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 36 | 68 | 175237 | Plasma membrane | Glycoprotein, Phosphoprotein | 0.091068 | 13 |
| 26 | Q9UMD9 | Autoantigen | 55 | 8 | 154929 | Plasma membrane | Disulfide bond, Glycoprotein, Hydroxylation, Phosphoprotein | −0.573080 | 1 |
| 27 | Q13563 | Autosomal dominant polycystic kidney disease type II protein | 47 | 4 | 110462 | Endoplasmic reticulum membrane | Glycoprotein | −0.328512 | 8 |
| 28 | P50851 | Beige-like protein; CDC4L protein | 57 | 18 | 320076 | Plasma membrane | Acetylation, Phosphoprotein | −0.201817 | 0 |
| 29 | Q76KP1 | Beta-1,4-N-acetyl-galactosaminyl transferase 4 | 39 | 10 | 116954 | Golgi apparatus membrane | Glycoprotein | −0.597691 | 0 |
| 30 | Q01082 | Beta-spectrin | 43 | 10 | 275259 | Plasma membrane | Acetylation, Glycoprotein, Phosphoprotein | −0.766116 | 0 |
| 31 | Q01082 | Beta-spectrin 2 isoform 2 | 39 | 8 | 251948 | Plasma membrane | Acetylation, Glycoprotein, Phosphoprotein | −0.766116 | 0 |
| 32 | Q15413 | Brain ryanodine receptor | 73 | 24 | 556937 | Endoplasmic reticulum | Glycoprotein | −0.278727 | 5 |
| 33 | O14514 | Brain-specific angiogenesis inhibitor 1 | 51 | 14 | 176900 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.311806 | 8 |
| 34 | O60241 | Brain-specific angiogenesis inhibitor 2 | 70 | 11 | 174738 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.211924 | 9 |
| 35 | O60242 | Brain-specific angiogenesis inhibitor 3 | 59 | 7 | 176123 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.211863 | 9 |
| 36 | Q9HCU4 | Cadherin EGF LAG seven-pass G-type receptor 2 | 44 | 7 | 322214 | Plasma membrane | Disulfide bond, Glycoprotein, Hydroxylation | −0.270751 | 8 |
| 37 | Q9NYQ7 | Cadherin EGF LAG seven-pass G-type receptor 3 | 51 | 10 | 362937 | Plasma membrane | Disulfide bond, Glycoprotein, Hydroxylation | −0.271258 | 8 |
| 38 | Q9H251 | Cadherin-23 | 74 | 6 | 370095 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.142278 | 2 |
| 39 | Q59FJ3 | Calcium channel, voltage-dependent, N type, alpha 1B subunit | 49 | 8 | 264553 | Unclear | Unknown | −0.264964 | 12 |
| 40 | Q9HCF6 | Calcium-permeable store-operated channel TRPM3b | 72 | 11 | 180008 | Unclear | Unknown | −0.302195 | 6 |
| 41 | Q08499 | cAMP-specific phoshodiesterase HPDE4D3 variant | 42 | 6 | 76872 | Unclear | Phosphoprotein | −0.674043 | 0 |
| 42 | Q96P48 | Centaurin delta 2 isoform a variant | 41 | 9 | 166159 | Plasma membrane, Golgi apparatus, Membrane | Phosphoprotein | −0.422553 | 0 |
| 43 | Q15700 | Channel associated protein of synapse | 35 | 8 | 97896 | Plasma membrane | Lipoprotein, Palmitate, Phosphoprotein | −0.519310 | 0 |
| 44 | Q96QT4 | Channel-kinase 1 | 65 | 11 | 214583 | Plasma membrane | Phosphoprotein | −0.270456 | 6 |
| 45 | P51788 | Chloride channel protein 2 | 41 | 8 | 99402 | Plasma membrane | Unknown | 0.132517 | 11 |
| 46 | P00751 | Complement factor B preproprotein | 175 | 13 | 86847 | Plasma membrane | Acetylation | −0.501047 | 0 |
| 47 | P17927 | Complement receptor 1 | 38 | 8 | 230417 | Plasma membrane | Disulfide bond, Glycoprotein, Pyrrolidone carboxylic acid | −0.320010 | 3 |
| 48 | P10606 | COX5B | 64 | 178 | 13914 | Mitochondrion membrane | Acetylation | −0.297674 | 0 |
| 49 | Q7Z407 | CUB and Sushi multiple domains 3 isoform 1 | 36 | 13 | 414007 | Plasma membrane | Disulfide bond, Glycoprotein | −0.260239 | 1 |
| 50 | Q8IY37 | DEAH (Asp-Glu-Ala-His) box polypeptide 37 | 40 | 4 | 130547 | Unclear | Unknown | −0.415385 | 0 |
| 51 | Q6IC98 | Death-inducing-protein | 49 | 14 | 66766 | Mitochondrion membrane | Phosphoprotein | −0.431833 | 3 |
| 52 | Q14185 | Dedicator of cytokinesis 1 | 52 | 6 | 216208 | Unclear | Phosphoprotein | −0.430617 | 0 |
| 53 | Q5VWQ8 | Disabled homolog 2 interacting protein isoform 2 | 37 | 8 | 118547 | Plasma membrane | Phosphoprotein | −0.578302 | 0 |
| 54 | O75923 | Dysferlin isoform 8 | 43 | 4 | 239254 | Vesicle | Phosphoprotein | −0.400145 | 1 |
| 55 | P22413 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 36 | 6 | 107024 | Plasma membrane | Disulfide bond, Glycoprotein | −0.456000 | 1 |
| 56 | Q8WYP5 | ELYS transcription factor-like protein TMBS62 | 43 | 4 | 258191 | Nuclear membrane | Acetylation, Phosphoprotein | −0.448765 | 0 |
| 57 | Q541P7 | EPH receptor B4 precursor | 35 | 4 | 109741 | Plasma membrane | Unknown | −0.235562 | 2 |
| 58 | Q15375 | Ephrin receptor EphA7 | 55 | 10 | 113735 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.284268 | 2 |
| 59 | Q15375 | Ephrin receptor EphA7 variant | 61 | 10 | 113994 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.284268 | 2 |
| 60 | P54762 | Ephrin receptor EphB1 precursor | 45 | 20 | 111297 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.214431 | 2 |
| 61 | P29323 | Ephrin type-B receptor 2 | 36 | 11 | 119128 | Plasma membrane | Lipoprotein, Myristate,Palmitate, Phosphoprotein | −0.293460 | 2 |
| 62 | Q9NZJ5 | Eukaryotic translation initiation factor 2-alpha kinase 3 | 48 | 11 | 126095 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.450717 | 2 |
| 63 | Q16099 | Excitatory amino acid receptor 1; kainate receptor subunit EAA1 | 41 | 9 | 108529 | Plasma membrane | Glycoprotein | −0.046653 | 4 |
| 64 | Q86XX4 | Extracellular matrix protein FRAS1 | 45 | 10 | 453936 | Plasma membrane | Glycoprotein | −0.295608 | 2 |
| 65 | Q9NYQ8 | FAT tumor suppressor 2 precursor | 44 | 7 | 482097 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.249645 | 2 |
| 66 | Q6V0I7 | Fat-like cadherin FATJ protein | 43 | 10 | 354108 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.262940 | 2 |
| 67 | Q9NZM1 | Fer-1 like protein 3 | 66 | 11 | 234737 | Nuclear membrane, plasma membrane | Acetylation, Phosphoprotein | −0.456091 | 1 |
| 68 | Q59F30 | Fibroblast growth factor receptor 4 variant | 38 | 5 | 114772 | Unclear | Unknown | 0.017312 | 6 |
| 69 | P42345 | FK506 binding protein 12-rapamycin associated protein 1 | 40 | 10 | 290759 | Golgi membrane, endoplasmic reticulum membrane, mitochondrial outer membrane | Acetylation, Phosphoprotein | −0.192586 | 0 |
| 70 | Q9Y2I7 | FYVE finger-containing phosphoinositide kinase | 66 | 18 | 239581 | Endosome membrane | Acetylation, Phosphoprotein | −0.534366 | 0 |
| 71 | Q86SQ6 | G protein-coupled receptor 123 | 35 | 6 | 139868 | Plasma membrane | Glycoprotein | −0.317749 | 6 |
| 72 | Q5T848 | G protein-coupled receptor 158 | 53 | 8 | 136886 | Plasma membrane | Glycoprotein, Isopeptide bond, Phosphoprotein, Ubl conjugation | −0.480988 | 7 |
| 73 | O75899 | GABBR2 protein | 42 | 5 | 100861 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.149734 | 8 |
| 74 | Q14789 | Giantin | 44 | 13 | 377273 | Golgi apparatus membrane | Disulfide bond, Phosphoprotein | −0.936847 | 1 |
| 75 | P42261 | Glutamate receptor type 1 | 42 | 7 | 102270 | Plasma membrane, Endoplasmic reticulum membrane | Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.199227 | 5 |
| 76 | Q9Y3R0 | Glutamate receptor-interacting protein 1 | 46 | 5 | 123203 | Plasma membrane, Endoplasmic reticulum membrane | Phosphoprotein | −0.362500 | 0 |
| 77 | Q14789 | Golgi antigen gcp372 | 44 | 8 | 373440 | Golgi apparatus membrane | Disulfide bond, Phosphoprotein | −0.936847 | 1 |
| 78 | Q08378 | Golgi autoantigen, golgin subfamily a, 3 | 69 | 17 | 167765 | Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.834781 | 0 |
| 79 | Q08378 | Golgin-160 | 57 | 16 | 167810 | Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.834781 | 0 |
| 80 | Q6PRD1 | GPR158-like 1 receptor | 54 | 11 | 260635 | Plasma membrane | Glycoprotein | −0.583101 | 7 |
| 81 | Q8WXG9 | G-protein coupled receptor 98 | 69 | 21 | 694181 | Plasma membrane | Unknown | Too long | 0 |
| 82 | Q8IWJ2 | GRIP and coiled-coil domain-containing 2 isoform a | 53 | 15 | 196873 | Golgi apparatus membrane | Phosphoprotein | −0.918825 | 0 |
| 83 | O15068 | Guanine nucleotide exchange factor DBS | 37 | 4 | 129340 | Plasma membrane | Phosphoprotein | −0.552067 | 0 |
| 84 | P52272 | Heterogeneous nuclear ribonucleoprotein M isoform a | 44 | 13 | 77749 | Plasma membrane | Acetylation, Phosphoprotein, Ubl conjugation | −0.341643 | 0 |
| 85 | O43166 | High-risk human papilloma viruses E6 oncoproteins targeted protein E6TP1 alpha; putative GAP protei | 59 | 12 | 198570 | Plasma membrane | Phosphoprotein | −0.671840 | 0 |
| 86 | P35523 | Human ClC-1 muscle chloride channel | 37 | 7 | 109696 | Plasma membrane | Unknown | 0.034919 | 11 |
| 87 | Q14643 | Human type 1 inositol 1,4,5-trisphosphate receptor | 43 | 8 | 309942 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.315448 | 6 |
| 88 | Q9HCF6 | Hypothetical protein | 72 | 10 | 175211 | Unclear | Unknown | −0.300924 | 6 |
| 89 | Q9P1Z9 | Hypothetical protein | 43 | 14 | 199652 | Unclear | Phosphoprotein | −0.647996 | 2 |
| 90 | A6NI73 | Immunoglobulin-like transcript 11 protein | 39 | 4 | 27186 | Unclear | Disulfide bond, Glycoprotein | −0.267224 | 2 |
| 91 | Q14571 | Inositol 1,4,5-trisphosphate receptor type 2 | 50 | 6 | 311074 | Plasma membrane | Acetylation, Phosphoprotein | −0.266755 | 7 |
| 92 | Q6P9B9 | Integrator complex subunit 5 | 38 | 5 | 109239 | Nucleus membrane | Phosphoprotein | 0.109029 | 2 |
| 93 | Q15811 | Intersectin long form | 40 | 7 | 196293 | Plasma membrane | Acetylation, Phosphoprotein | −0.668043 | 0 |
| 94 | Q9H4E7 | IRF4-binding protein | 48 | 7 | 74422 | Plasma membrane | Acetylation, Phosphoprotein | −0.946435 | 0 |
| 95 | Q92508 | KIAA0233 | 50 | 17 | 234888 | Endoplasmic reticulum membrane | Glycoprotein | −0.002357 | 23 |
| 96 | Q13023 | KIAA0311 | 40 | 7 | 259702 | Nucleus membrane | Unknown | −0.675982 | 0 |
| 97 | Q96P48 | KIAA0782 protein | 45 | 4 | 145325 | Plasma membrane, Golgi apparatus membrane | Phosphoprotein | −0.422553 | 0 |
| 98 | Q9Y2H9 | KIAA0973 protein | 61 | 34 | 173192 | Plasma membrane | Phosphoprotein | −0.569363 | 0 |
| 99 | Q96RV3 | KIAA0995 protein | 42 | 7 | 109401 | Plasma membrane | Glycoprotein | −0.284538 | 13 |
| 100 | Q9BZ29 | KIAA1058 protein | 42 | 7 | 241774 | Plasma membrane | Phosphoprotein | −0.349204 | 0 |
| 101 | Q5T848 | KIAA1136 protein | 49 | 11 | 66642 | Plasma membrane | Glycoprotein, Isopeptide bond, Phosphoprotein, Ubl conjugation | −0.480988 | 7 |
| 102 | Q5T4S7 | KIAA1307 protein | 53 | 7 | 188524 | Unclear | Acetylation, Phosphoprotein | Too long | 0 |
| 103 | Q9BZ72 | KIAA1457 protein | 43 | 11 | 151395 | Plasma membrane | Phosphoprotein | −0.405561 | 0 |
| 104 | Q9P1Z9 | KIAA1529 protein | 46 | 6 | 196058 | Unclear | Phosphoprotein | −0.647996 | 2 |
| 105 | Q6KCM7 | KIAA1896 protein | 45 | 7 | 63937 | Mitochondrial membrane | Unknown | −0.099147 | 0 |
| 106 | Q16787 | Laminin alpha 3 subunit isoform 1 | 45 | 7 | 375652 | Unclear | Disulfide bond, Glycoprotein | −0.381368 | 0 |
| 107 | P11047 | Laminin B2 chain | 54 | 7 | 183195 | Unclear | Disulfide bond, Glycoprotein | −0.615042 | 0 |
| 108 | Q9Y6N6 | Laminin gamma 3 chain precursor | 40 | 6 | 177756 | Unclear | Disulfide bond, Glycoprotein | −0.465018 | 0 |
| 109 | Q13449 | LAMP | 41 | 13 | 37798 | Plasma membrane | Disulfide bond, GPI-anchor, Glycoprotein, Lipoprotein, Phosphoprotein | −0.278106 | 2 |
| 110 | P42704 | Leucine-rich PPR-motif containing protein | 52 | 7 | 146306 | Nuclear inner membrane | Acetylation | −0.206097 | 0 |
| 111 | Q86UK5 | Limbin | 41 | 5 | 148825 | Plasma membrane | Glycoprotein | −0.445185 | 2 |
| 112 | P50851 | Lipopolysaccharide-responsive and beige-like anchor protein | 63 | 10 | 321639 | Plasma membrane | Acetylation, Phosphoprotein | −0.201817 | 0 |
| 113 | Q9HCF6 | Long transient receptor potential channel 3 | 75 | 10 | 196157 | Plasma membrane | Unknown | −0.300924 | 6 |
| 114 | Q9NZR2 | Low density lipoprotein receptor related protein-deleted in tumor | 51 | 9 | 534844 | Plasma membrane | Disulfide bond, Glycoprotein | −0.489888 | 2 |
| 115 | Q5JRA6 | Melanoma inhibitory activity family, member 3 | 52 | 7 | 214255 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.887466 | 3 |
| 116 | Q86UL8 | Membrane associated guanylate kinase, WW and PDZ domain containing 2 | 39 | 5 | 159454 | Plasma membrane | Phosphoprotein | −0.668455 | 0 |
| 117 | Q13421 | Mesothelin | 45 | 4 | 68652 | Plasma membrane | Cleavage on pair of basic residues, GPI-anchor, Glycoprotein, Lipoprotein | −0.105714 | 2 |
| 118 | A1L467 | Met proto-oncogene isoform b precursor | 48 | 6 | 157779 | Plasma membrane | Unknown | −0.144388 | 2 |
| 119 | Q8IWA4 | Mitofusin 1 precursor | 38 | 10 | 84892 | Mitochondrial membrane | Unknown | −0.267476 | 2 |
| 120 | Q6UVY6 | Monooxygenase X | 36 | 9 | 63474 | Endoplasmic reticulum membrane | Disulfide bond, Glycoprotein | −0.260848 | 2 |
| 121 | Q8WXI7 | Mucin 16 | 38 | 7 | 747071 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | Too long | 1 |
| 122 | O75970 | Multiple PDZ domain protein | 60 | 7 | 222792 | Plasma membrane | Phosphoprotein | −0.233203 | 0 |
| 123 | Q9NZM1 | Myoferlin | 42 | 6 | 231092 | Vesicle membrane, nuclear membrane, plasma membrane | Acetylation, Phosphoprotein | −0.456091 | 1 |
| 124 | B2RTY4 | Myosin-IXa | 54 | 9 | 294918 | Unclear | Phosphoprotein | −0.623980 | 0 |
| 125 | Q8NFP9 | Neurobeachin | 53 | 6 | 330066 | Plasma membrane | Phosphoprotein | −0.188358 | 2 |
| 126 | P21359 | Neurofibromin isoform 1 | 43 | 10 | 322760 | Plasma membrane | Disulfide bond,GPI-anchor, Glycoprotein, Lipoprotein | −0.136950 | 4 |
| 127 | Q9H2E3 | Neuropilin-2b(5) | 53 | 6 | 102975 | Plasma membrane | Unknown | −0.440066 | 2 |
| 128 | Q13423 | Nicotinamide nucleotide transhydrogenase | 85 | 9 | 114564 | Mitochondrial membrane | Acetylation | 0.299079 | 13 |
| 129 | Q59GR1 | Niemann-Pick disease, type C1 variant | 36 | 5 | 145931 | Plasma membrane | Unknown | 0.216447 | 13 |
| 130 | Q13224 | N-methyl-D-aspartate receptor subunit NR3 | 53 | 16 | 168067 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.388342 | 6 |
| 131 | Q13813 | Nonerythroid alpha-spectrin | 59 | 7 | 284905 | Unclear | Acetylation, Phosphoprotein | −0.790330 | 0 |
| 132 | Q9UM47 | Notch homolog 3 | 43 | 8 | 256640 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.304826 | 3 |
| 133 | Q5STG5 | Notch homolog 4 (Drosophila) | 51 | 7 | 221594 | Unclear | Unknown | −0.287183 | 3 |
| 134 | P46531 | Notch1 preproprotein | 38 | 11 | 286350 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.420119 | 3 |
| 135 | Q92823 | NrCAM protein | 37 | 11 | 144138 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.379601 | 2 |
| 136 | Q8WXH0 | NUANCE | 104 | 29 | 801683 | Nuclear membrane | Acetylation, Phosphoprotein | Too long | 0 |
| 137 | Q5VU65 | Nucleoporin 210 kDa-like isoform 1 | 43 | 10 | 211668 | Unclear | Glycoprotein | 0.050265 | 3 |
| 138 | Q13017 | p190-B | 45 | 6 | 173000 | Unclear | Nitration, Phosphoprotein | −0.592345 | 0 |
| 139 | Q5T4S7 | p600 | 121 | 21 | 580607 | Unclear | Acetylation, Phosphoprotein | Too long | 0 |
| 140 | O60245 | PCDH7 (BH-Pcdh)c | 40 | 4 | 130999 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.390365 | 2 |
| 141 | Q9BXM0 | Periaxin | 59 | 8 | 155248 | Plasma membrane | Phosphoprotein | −0.225736 | 0 |
| 142 | O75747 | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide | 38 | 9 | 167814 | Unclear | Phosphoprotein | −0.322423 | 1 |
| 143 | O00443 | Phosphoinositide 3-kinase | 43 | 9 | 192271 | Plasma membrane | Acetylation, Phosphoprotein | −0.302136 | 0 |
| 144 | O00750 | PI-3 kinase | 46 | 5 | 184101 | Plasma membrane | Phosphoprotein | −0.363037 | 0 |
| 145 | Q6T4P5 | Plasticity-related protein 2 | 43 | 12 | 79523 | Membrane | Glycoprotein | −0.178273 | 6 |
| 146 | O75051 | Plexin A2 | 37 | 13 | 214220 | Cell membrane | Glycoprotein, Phosphoprotein | −0.181468 | 3 |
| 147 | Q9Y4D7 | Plexin-D1 | 48 | 9 | 215381 | Cell membrane, Membrane | Glycoprotein | −0.159013 | 2 |
| 148 | Q15142 | Polycystic kidney disease-associated protein | 57 | 5 | 396273 | Integral to membrane | Unknown | 0.022760 | 14 |
| 149 | Q8TDX9 | Polycystin-1 L1 | 35 | 4 | 319453 | Integral to membrane | Glycoprotein | −0.136856 | 7 |
| 150 | P13942 | Pro-a2(XI) | 51 | 10 | 160545 | Cell membrane | Disulfide bond, Glycoprotein, Sulfation | −0.805186 | 0 |
| 151 | P08123 | Pro-alpha 2(I) collagen | 51 | 9 | 129858 | Plasma membrane | Glycoprotein, Hydroxylation, Pyrrolidone carboxylic acid | −0.648098 | 0 |
| 152 | P02647 | Proapolipoprotein | 664 | 75 | 28944 | Plasma membrane | Glycation, Glycoprotein, Lipoprotein, Palmitate, Phosphoprotein | −0.717228 | 0 |
| 153 | Q07954 | Prolow-density lipoprotein receptor-related protein 1 | 66 | 13 | 523119 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.510386 | 2 |
| 154 | Q05655 | Protein kinase C-delta 13 | 37 | 4 | 78624 | Plasma membrane | Phosphoprotein | −0.377958 | 0 |
| 155 | Q8TF72 | Protein Shroom 3 | 41 | 8 | 218125 | Plasma membrane | Phosphoprotein | −0.801652 | 0 |
| 156 | Q96QU1 | Protocadherin 15 | 59 | 13 | 217261 | Plasma membrane | Phosphoprotein | −0.245473 | 2 |
| 157 | Q96QU1 | Protocadherin 15 isoform CD1-4 precursor | 48 | 12 | 217303 | Plasma membrane | Phosphoprotein | −0.245473 | 2 |
| 158 | Q9UN70 | Protocadherin 43 | 45 | 5 | 98230 | Plasma membrane | Glycoprotein | −0.261670 | 2 |
| 159 | Q14517 | Protocadherin Fat 1 | 52 | 9 | 509384 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.294946 | 2 |
| 160 | Q9NYQ8 | Protocadherin Fat 2 | 59 | 7 | 482172 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein | −0.249645 | 2 |
| 161 | Q86V60 | PTPRM protein | 39 | 8 | 158434 | Unclear | Unknown | −0.385540 | 1 |
| 162 | Q8WZA2 | RAPGEF4 protein | 53 | 13 | 100339 | Unclear | Phosphoprotein | −0.333927 | 0 |
| 163 | P23467 | Receptor-type tyrosine-protein phosphatase beta | 50 | 26 | 225497 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.372159 | 2 |
| 164 | Q13332 | Receptor-type tyrosine-protein phosphatase S | 43 | 9 | 218155 | Plasma membrane | Disulfide bond, Glycoprotein | −0.401798 | 2 |
| 165 | Q86UR5 | Regulating synaptic membrane exocytosis 1 | 40 | 6 | 190154 | Plasma membrane | Phosphoprotein | −0.922459 | 0 |
| 166 | Q5T4S7 | Retinoblastoma-associated factor 600 | 124 | 20 | 580547 | Unclear | Acetylation, Phosphoprotein | Too long | 0 |
| 167 | Q5T5U3 | Rho GTPase-activating protein 21 | 41 | 8 | 218567 | Golgi apparatus membrane, vesicle membrane | Phosphoprotein | −0.827338 | 0 |
| 168 | Q13464 | Rho-associated, coiled-coil containing protein kinase 1 | 48 | 6 | 159102 | Golgi apparatus membrane | Unknown | −0.909454 | 0 |
| 169 | Q5T5U3 | Rho-GTPase activating protein 10 | 41 | 5 | 218563 | Golgi apparatus membrane, vesicle membrane | Phosphoprotein | −0.827338 | 0 |
| 170 | Q5JTH9 | Ribosomal RNA processing 12 homolog (S. cerevisiae) | 56 | 9 | 145067 | Nuclear membrane | Phosphoprotein | −0.225366 | 1 |
| 171 | Q86UR5 | RIM long form | 48 | 7 | 164927 | Plasma membrane | Phosphoprotein | −0.922459 | 0 |
| 172 | P21817 | Ryanodine receptor 1 | 61 | 11 | 55204 | Endoplasmic reticulum membrane | Glycoprotein, Phosphoprotein | −0.314300 | 7 |
| 173 | Q92736 | Ryanodine receptor 2 | 70 | 10 | 569626 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.324300 | 7 |
| 174 | Q15413 | Ryanodine receptor 3 | 71 | 24 | 557794 | Endoplasmic reticulum membrane | Glycoprotein | −0.278727 | 5 |
| 175 | O15027 | SEC16 homolog A | 36 | 5 | 115891 | Endoplasmic reticulum membrane, Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.606607 | 0 |
| 176 | A4QN19 | SEC16A protein | 40 | 2 | 116196 | Endoplasmic reticulum membrane, Golgi apparatus membrane | Acetylation, Phosphoprotein | −0.606607 | 0 |
| 177 | Q9H2E6 | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 36 | 4 | 115608 | Plasma membrane | Disulfide bond, Glycoprotein | −0.448543 | 2 |
| 178 | O94921 | Serine/threonine protein kinase PFTAIRE-1 | 41 | 8 | 53609 | Plasma membrane | Phosphoprotein | −0.470362 | 0 |
| 179 | Q8NGB0 | Seven transmembrane helix receptor | 49 | 10 | 157779 | Unclear | Unknown | −0.275069 | 6 |
| 180 | Q9Y566 | SH3 and multiple ankyrin repeat domains protein 1 | 49 | 7 | 225738 | Plasma membrane | Phosphoprotein | −0.539239 | 0 |
| 181 | Q9UPX8 | SH3 and multiple ankyrin repeat domains protein 2 | 54 | 6 | 135115 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.608572 | 0 |
| 182 | Q7Z5N4 | Sidekick-like protein 1 | 39 | 4 | 242926 | Plasma membrane | Disulfide bond, Glycoprotein | −0.314144 | 1 |
| 183 | P21817 | Skeletal muscle ryanodine receptor | 78 | 9 | 570517 | Plasma membrane | Glycoprotein, Phosphoprotein, S-nitrosylation | Too long | 0 |
| 184 | O94813 | Slit homolog 2 | 47 | 13 | 175803 | Plasma membrane | Disulfide bond, Glycoprotein | −0.323414 | 0 |
| 185 | O94813 | SLIT2 | 40 | 7 | 175329 | Plasma membrane | Disulfide bond, Glycoprotein | −0.323414 | 0 |
| 186 | Q9C0H9 | SNAP-25-interacting protein | 42 | 6 | 127182 | Plasma membrane | Phosphoprotein | −0.644456 | 0 |
| 187 | Q9Y6M7 | Sodium bicarbonate cotransporter2 | 44 | 8 | 115215 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.130725 | 10 |
| 188 | Q53ZR1 | Solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | 64 | 13 | 132048 | Plasma membrane | Unknown | 0.069720 | 11 |
| 189 | Q9HBR0 | Solute carrier family 38, member 10 isoform b | 40 | 5 | 84069 | Unclear | Phosphoprotein | −0.363718 | 11 |
| 190 | Q9NRC6 | Spectrin beta chain, brain 4 | 72 | 14 | 419259 | Unclear | Unknown | −0.576588 | 0 |
| 191 | Q01082 | Spectrin, beta, non-erythrocytic 1 isoform 1 variant | 48 | 8 | 276797 | Plasma membrane | Acetylation, Glycoprotein, Phosphoprotein | −0.766116 | 0 |
| 192 | Q8WWQ8 | Stabilin 2 precursor | 57 | 9 | 288323 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein, Proteoglycan | −0.268563 | 2 |
| 193 | Q8WWQ8 | Stabilin-2 | 46 | 9 | 288329 | Plasma membrane | Disulfide bond, Glycoprotein, Phosphoprotein, Proteoglycan | −0.268563 | 2 |
| 194 | Q8IVL0 | Steerin3 protein | 37 | 5 | 256885 | Nuclear membrane | Phosphoprotein | −0.638784 | 0 |
| 195 | Q4LDE5 | Sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 52 | 10 | 401915 | Unclear | Disulfide bond, Glycoprotein | −0.320528 | 1 |
| 196 | Q9Y4G6 | Talin-2 | 50 | 11 | 273723 | Plasma membrane | Acetylation, Phosphoprotein | −0.218412 | 0 |
| 197 | Q9H2K2 | Tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | 41 | 4 | 128492 | Golgi apparatus membrane | ADP-ribosylation | −0.317066 | 0 |
| 198 | O60343 | TBC1 domain family, member 4 | 36 | 5 | 148068 | Plasma membrane | Phosphoprotein | −0.553083 | 1 |
| 199 | Q15643 | Thyroid receptor-interacting protein 11 | 38 | 7 | 228184 | Golgi apparatus membrane | Phosphoprotein | −0.843506 | 0 |
| 200 | P12270 | Tpr | 54 | 10 | 265840 | Nuclear membrane | Acetylation, Phosphoprotein | −0.968726 | 0 |
| 201 | Q9BX84 | Transient receptor potential cation channel subfamily M member 6 variant c | 79 | 20 | 232975 | Unclear | Phosphoprotein | −0.369882 | 5 |
| 202 | Q8N6Q1 | Transmembrane and coiled-coil domains 5A | 47 | 5 | 34495 | Unclear | Unknown | −0.563888 | 1 |
| 203 | O95271 | TRF1-interacting ankyrin-related ADP-ribose polymerase | 47 | 12 | 143690 | Golgi apparatus membrane | ADP-ribosylation, Phosphoprotein | −0.327506 | 0 |
| 204 | Q15643 | Trip230 | 92 | 12 | 228116 | Unclear | Phosphoprotein | −0.843506 | 0 |
| 205 | Q9BX84 | Truncated transient receptor potential cation channel subfamily M member 6 variant a | 57 | 13 | 224977 | Plasma membrane | Phosphoprotein | −0.369882 | 5 |
| 206 | Q8NFA0 | Ubiquitin specific protease 32 | 47 | 8 | 183821 | Unclear | Lipoprotein, Phosphoprotein, Prenylation | −0.474689 | 0 |
| 207 | P55916 | Uncoupling protein 3 isoform UCP3S | 36 | 15 | 30219 | Mitochondrion membrane | Unknown | 0.074679 | 0 |
| 208 | P17927 | Unnamed protein product | 41 | 10 | 230401 | Plasma membrane | Disulfide bond, Glycoprotein, Pyrrolidone carboxylic acid | −0.320010 | 3 |
| 209 | P53420 | Unnamed protein product | 65 | 10 | 126774 | Vesicle membrane | Acetylation, Phosphoprotein | −0.659231 | 1 |
| 210 | P11717 | Unnamed protein product | 48 | 14 | 281089 | Plasma membrane | Acetylation, Disulfide bond, Glycoprotein, Phosphoprotein | −0.368528 | 3 |
| 211 | Q6ZUB1 | Unnamed protein product | 36 | 10 | 158870 | Unclear | Glycoprotein | −0.709205 | 1 |
| 212 | O75110 | Unnamed protein product | 39 | 10 | 108198 | Unclear | Phosphoprotein | 0.062178 | 9 |
| 213 | Q9NRW7 | Vacuolar protein sorting | 41 | 6 | 65388 | Endosome membrane, Golgi apparatus membrane | Unknown | −0.310526 | 0 |
| 214 | Q86Y38 | Xylosyltransferase I | 41 | 12 | 108357 | Endoplasmic reticulum membrane, Golgi apparatus membrane | Disulfide bond, Glycoprotein | −0.540564 | 0 |
| 215 | Q13433 | Zinc transporter ZIP6 | 41 | 5 | 84685 | Plasma membrane | Glycoprotein, Phosphoprotein | −0.421987 | 6 |
| 216 | Q13439 | 256 kD golgin | 59 | 9 | 256666 | Golgi apparatus membrane | Phosphoprotein | −1.052981 | 0 |
| 217 | Q8N1I0 | Dedicator of cytokinesis protein 4 | 49 | 11 | 225206 | Endomembrane | Phosphoprotein | −0.371464 | 0 |


