Abstract
The homeodomain transcription factor CRX is a crucial regulator of mammalian photoreceptor gene expression. Mutations in the human CRX gene are associated with dominant inherited retinopathies Retinitis Pigmentosa (RP), Cone-Rod Dystrophy (CoRD) and Leber Congenital Amaurosis (LCA), of varying severity. In vitro and in vivo assessment of mutant CRX proteins have revealed pathogenic mechanisms for several mutations, but no comprehensive mutation-disease correlation has yet been reported. Here we describe four different classes of disease-causing CRX mutations, characterized by mutation type, pathogenetic mechanism, and the molecular activity of the mutant protein: I) hypomorphic missense mutations with reduced DNA binding, II) antimorphic missense mutations with variable DNA binding, III) antimorphic frameshift/nonsense mutations with intact DNA binding and IV) antimorphic frameshift mutations with reduced DNA binding. Mammalian models representing three of these classes have been characterized. Models carrying Class I mutations display a mild dominant retinal phenotype and recessive LCA, while models carrying Class III and IV mutations display characteristically distinct dominant LCA phenotypes. These animal models also reveal unexpected pathogenic mechanisms underlying CRX-associated retinopathies. The complexity of genotype-phenotype correlation for CRX-associated diseases highlights the value of developing comprehensive 'true-to-disease' animal models for understanding pathologic mechanisms and testing novel therapeutic approaches.
Keywords: neuronal degeneration, photoreceptors, transcription factors, human genetics, dominant-negative, antimorph, hypomorph, neural development, retina, gene expression, disease models
INTRODUCTION: CRX-ASSOCIATED RETINOPATHIES
The genetic basis for blinding retinopathies is incredibly diverse. Retinal diseases have been linked to mutations in >250 genes and mapped loci (https://sph.uth.edu/retnet/). This genetic heterogeneity has made it challenging to clearly define basic disease mechanisms for different disorders. Most retinopathies are currently untreatable and remain poorly understood, largely due to the lack of appropriate animal models. The inherited retinopathies Retinitis Pigmentosa (RP), Cone-Rod Dystrophy (CoRD) and Leber Congenital Amaurosis (LCA) all cause progressive loss of vision, each with distinct clinical features. RP is a ‘rod-centric’ disease with symptoms including night blindness and loss of peripheral vision, progressing into tunnel vision and loss of central visual field acuity (OMIM: 268000). CoRD is a ‘cone-centric’ disease with symptoms including loss of color vision and loss of central visual field acuity and progressing into night blindness and loss of peripheral vision (OMIM #120970). RP and CoRD vary in age of onset, rate of progression and severity. LCA is an early-onset disease with symptoms including vision loss, nystagmus, and severe retinal dysfunction (OMIM: #204000). RP, CoRD and LCA can exhibit either dominant or recessive inheritance. These retinopathies have each been linked to mutations in several different genes, but Cone-Rod Homeobox (CRX) (Accession: AAH53672.1) is the only gene associated with all three diseases (Berger et al. 2010). Mutations in CRX are primarily associated with dominant forms of RP, CoRD and LCA of variable severity and age of onset (Berger et al. 2010; Dharmaraj et al. 2000; Freund et al. 1997; Freund et al. 2001; Galvin et al. 2005; Hanein et al. 2004; den Hollander et al. 2008; Huang et al. 2012; Jacobson et al. 1998; Koenekoop et al. 2002; Lotery et al. 2000; Nakamura et al. 1998; Paunescu et al. 2007; Rivolta et al. 2001; Sohocki et al. 1998; Swain et al. 1997; Silva et al. 2000; Sohocki et al. 2001; Swaroop et al. 1999; Tzekov et al. 2000; Tzekov et al. 2001; Walia et al. 2010; Wang et al. 2007; Zhang et al. 2001). In this review, we summarize the effects of different classes of disease-causing CRX mutations on the function of CRX protein, based on in vitro experiments and currently available animal models (Figure 1). We demonstrate that animal models carrying mechanistically distinct Crx mutations exhibit different phenotypes that accurately represent the range of clinical features reported for human patients, and reveal unexpected pathogenic mechanisms. These animal models demonstrate the power of model organism research for studying complex human disease phenotypes and will facilitate the rational design and testing of therapeutic strategies.
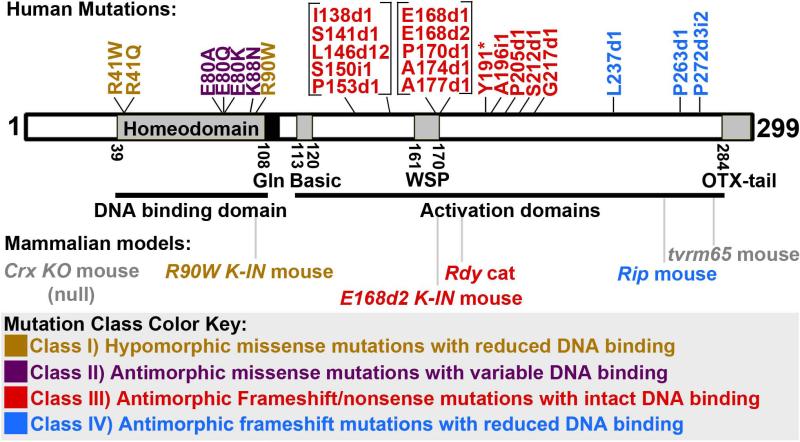
Figure 1. Schematic of CRX protein showing locations of co-segregating human mutations and mutations in associated animal models.
The CRX protein is represented by a rectangle, conserved domains are shaded. The locations of human mutations that co-segregate with disease are shown above the schematic. Based on mutation type and molecular function, mutations fall into four classes: I) hypomorphic missense mutations with reduced DNA binding (gold text), II) antimorphic missense mutations with variable DNA binding (purple text), III) antimorphic frameshift/nonsense mutations with intact DNA binding (red text) and IV) antimorphic frameshift mutations with reduced DNA binding (blue text). While both classes of frameshift mutations are predicted to have antimorphic activity, they act through different mechanisms. Currently, there are five mouse models and one cat model carrying mutations in Crx (bottom): 1) the Crx KO mouse (grey text) does not express CRX protein; 2) the R90W K-IN mouse (gold text) carries a Class I mutation; 3) the E168d2 K-IN mouse and Rdy cat (red text) both carry Class III mutations; 4) the Rip mouse (blue text) carries a Class IV mutation; 5) the tvrm65 mouse (grey text) carries a nonsense mutation that does not match any reported disease-causing human mutation classes.
MOLECULAR ROLE OF CRX WITHIN THE TRANSCRIPTIONAL REGULATORY NETWORKS OF THE RETINA AND PINEAL GLAND
CRX is an Otd/OTX-like ‘paired’ homeodomain (HD) transcription factor that is preferentially expressed in vertebrate rods and cones, the primary cells supporting vision in the retina, and pinealocytes, which regulate circadian rhythms in the brain (Chen et al. 1997; Furukawa et al. 1997). CRX plays an essential role in the establishment and maintenance of gene expression in mammalian rod and cone photoreceptors (Furukawa et al. 1999) and the pineal gland (Rovsing et al. 2011). The molecular function of CRX is highly conserved amongst mammals including humans, mice, and cats (Furukawa et al. 1997; Chen et al. 1997; Menotti-Raymond et al. 2010). Paralogues of CRX/OTX fulfill similar roles in the photoreceptors of the evolutionarily divergent vertebrate fish (Shen & Raymond 2004) and amphibians (Plouhinec et al. 2003) and the invertebrates Drosophila (Terrell et al. 2012) and Amphioxus (Vopalensky et al. 2012).
Mammalian CRX protein is 299 amino acids (aa) in length and contains several conserved functional domains including: the HD, glutamine rich (Gln), basic, WSP and OTX-tail motifs (Figure 1). The HD mediates DNA binding activity, while the carboxy-terminus (C-terminal) portion of CRX (from the basic to the OTX-tail domains) mediates transcriptional regulation (Chau et al. 2000; Chen et al. 1997; Fei & Hughes 2000). During mammalian retinal development, the establishment of the CRX transcriptional regulatory network requires the function of OTX2 (Accession: BAJ84003.1), another Otd/OTX family member. The HD's of OTX2 and CRX proteins are 86% homologous and maintain similar genomic DNA binding profiles in the neural retina (Corbo et al. 2010; Freund et al. 1997; Hennig et al. 2008; Samuel et al. 2014) but have distinct roles in the regulation of retinal gene expression (Chen et al. 1997; Furukawa et al. 1997; Hsiau et al. 2007; Housset et al. 2013; Nishida et al. 2003; Terrell et al. 2012). OTX2 is expressed earlier than CRX in the retinal progenitor cells that give rise to photoreceptors (Baas et al. 2000; Bovolenta et al. 1997; Nishida et al. 2003). In these cells, OTX2 is required for the activation of CRX expression followed by rod-specific (including RORβ, NRL and NR2E3 (Accessions: NP_001036819.1, NP_006168.1, and AAH41421.1, respectively) or cone-specific (including RORα, TRβ2, RXRγ and COUP TFs (Accessions: EDL26161.1, NP_033406.1, AAH58401.1, NP_034281.2, NP_033827.2, respectively)) transcription factors (Bovolenta et al. 1997; Nishida et al. 2003). As CRX expression increases in developing photoreceptors, OTX2 expression decreases in these cells but remains expressed in the inner nuclear layer and retinal pigmented epithelium (Housset et al. 2013; Nishida et al. 2003).
CRX functions synergistically with rod and cone transcription factors to coordinately control photoreceptor gene expression. CRX binds to photoreceptor target gene loci (Boatright et al. 1997; Bobola et al. 1999; Chau et al. 2000; Chen et al. 2002; Corbo et al. 2010; Fei et al. 1999; Furukawa et al. 1997; Livesey et al. 2000; Peng & Chen 2005) and can directly activate the promoters of target genes in vitro, including Rhodopsin (Accession: NP_001014890.1) and its own promoter (Chau et al. 2000; Chen et al. 1997; Kimura 2000; Langmann et al. 2008). CRX is thought to mainly act as a transcriptional activator in photoreceptors, based on expression profiling of the Crx knock-out (KO) mouse (Blackshaw et al. 2001; Furukawa et al. 1999; Hsiau et al. 2007), but can be associated with repression in certain genomic contexts (Corbo et al. 2010; Kwasnieski et al. 2012). CRX's function in transcriptional regulation requires specific interactions with co-factors including the histone acetyltransferases GCN5, CBP and p300 (Accessions: AAC50641.1, AAC17736.1, NP_001420.2, respectively) (Hennig et al. 2013; Peng & Chen 2007), promoting histone acetylation at target gene promoters (Hennig et al. 2008; Peng & Chen 2007) and mediating enhancer/promoter intrachromosomal looping interactions of target photoreceptor genes (Peng & Chen 2011). These events in chromatin remodeling and transcriptional activation are sequentially regulated during retinal development (Peng & Chen 2007). CRX also maintains specific interactions with transcriptional co-regulators including the rod-specific transcription factors NRL (Mitton et al. 2000; Nichols et al. 2010) and NR2E3 (Peng et al. 2005; Roduit et al. 2009) to coordinately control photoreceptor gene expression. NRL has been shown to activate rod gene expression (Mears et al. 2001; Rehemtulla et al. 1996) while NR2E3 has dual activator/repressor activity (Chen et al. 2005; Cheng et al. 2004; Cheng et al 2006; Peng et al. 2005; Webber et al. 2008). CRX, NRL and NR2E3 co-occupy target gene promoters and enhancers in vivo (Peng & Chen 2005), and synergistically promote rod gene expression (Mitton et al. 2000). Genome-wide profiling of CRX (Corbo et al. 2010) and NRL (Hao et al. 2012) reveal that a high percentage of genes bound by NRL are also bound by CRX. A mouse Knock-OUT (KO) of Nrl causes a fate switch of rods to a 'cone-like' state (Daniele et al. 2005; Yoshida et al. 2004), and the double KO of Crx and Nrl impairs the transcription of cone genes in this retina (Hsiau et al. 2007). Elsewhere in the brain, CRX broadly regulates transcription in the pineal gland (Rovsing et al. 2011) and loss of CRX attenuates circadian entrainment in the mouse (Furukawa et al. 1999).
CLASSIFICATION OF REPORTED HUMAN CRX MUTATIONS
CRX mutations associated with dominant RP, CoRD and LCA
Mutations in CRX have been reported in approximately 2% of LCA, 5% of CoRD and 1% of RP (Huang et al. 2012). To date, 50 different mutations have been reported, (Chen et al. 2013; Huang et al. 2012; Zou et al. 2013) and 25 of these mutations co-segregate with the disease phenotype (Figure 1, Table 1, Table 2). Reported mutations are composed of missense (39%), nonsense (4%), deletion (37%), insertion (16%) and indel (combination of insertion and deletion) (4%) mutations but the occurrence of each individual mutation is rare (Huang et al. 2012). Overall, mutations are spread evenly across the coding sequence but co-segregating missense mutations are restricted to the HD while frameshift (deletion, insertion and indel) and nonsense mutations are all found within the activation domains of CRX, C-terminal to the HD (Figure 1). Several missense mutations outside of the HD (Table 1) and one early insertion mutation (P9i1) (Table 2) have been reported but to this point have not been shown to co-segregate with the disease phenotype. This suggests that missense mutations and frameshift/nonsense mutations may affect discrete functional domains of the CRX protein. Missense mutations could affect the ability of CRX's HD to bind DNA or interact with co-factors (Chen et al. 2002; Mitton et al. 2000; Swaroop et al. 1999). In contrast, frameshift/nonsense mutations all have an intact HD but a disrupted C-terminal portion of the protein, which mediates CRX's transactivation activity. The next two sections summarize functional analyses of mutant CRX proteins, which suggest that these mutation types further subdivide into functional groups.
Table 1.
Summary of CRX missense mutations associated with retinopathy. Reported missense mutations from patients with retinopathy, showing effects of mutation on CRX's ability to bind DNA targets and transactivate gene expression. Gold text- hypomorphic missense mutations with reduced DNA binding and transactivation. Purple text- antimorphic missense mutations with variable DNA binding acitivity. Black text- missense mutations with uncharacterized molecular function. (n.d.- not determined)
| Protein | cDNA | Associated Phenotype | Cosegregation | DNA binding | Transactivation | Reference |
|---|---|---|---|---|---|---|
| p.R41W | c.121C>T | CoRD | yes | reduced | reduced | Swain et al. (1997); Chen et al. (2002) |
| p.R41Q | c.122G>A | RP/CoRD | yes | reduced | reduced | Swain et al. (1997); Sohocki et al. (1998); Rivolta et al. (2001); Chen et al. (2002) |
| p.E42K | c.124G>A | LCA | n.d. | n.d. | n.d. | Li et al. (2011) |
| p.A56T | c.166G>A | LCA | n.d. | normal | normal | Lotery et al. (2000); Chen et al. (2002) |
| p.D65H | c.193G>C | RP (homozygote) | n.d. | n.d. | n.d. | Jin et al. (2008) |
| p.V66I | c.196G>A | RP | n.d. | n.d. | n.d. | Vallespin et al. (2007) |
| p.E80K | c.238G>A | CoRD | yes | n.d. | n.d. | Sankila et al. (2000) |
| pE80Q | c.239A>G | CoRD | yes | n.d. | n.d. | Huang et al. (2012) |
| p.E80A | c.239A>C | CoRD | yes | normal | normal + antimorphic | Freund et al. (1997); Sohocki et al. (1998); Chen et al. (2002); Terrell et al. (2012) |
| p.K88N | c.264G>T | LCA | yes | reduced (predicted) | reduced + antimorphic | Nichols et al. (2010); Chen et al. (2002); Terrell et al. (2012) |
| p.R90W | c.268C>T | CoRD (heterozygote); LCA (homozygote) | yes | reduced | reduced | Swaroop et al. (1999); Chen et al. (2002) |
| p.A112V | c335C>T | LCA | n.d. | n.d. | n.d. | Zhang et al. (2002) |
| p.R115Q | c.344G>A | RP | n.d. | n.d. | n.d. | Sohocki et al. (2001) |
| p.G122D | c.365G>A | RP | n.d. | n.d. | n.d. | Jin et al. (2008) |
| p.Y142C | c.425A>G | LCA | n.d. | n.d. | n.d. | Vallespin et al. (2007) |
| p.S152Y | c.455C>A | RP | n.d. | n.d. | n.d. | Jin et al. (2008) |
| p.A158T | c.472G>A | LCA | no | n.d. | reduced | Zernant et al. (2005); Chen et al. (2002) |
| p.V242M | c.724G>A | CoRD | n.d. | n.d. | normal | Swain et al. (1997); Rivolta et al. (2001); Chen et al. (2002) |
| p.T273L | c.818C>T | LCA | no | n.d. | n.d. | Zernant et al. (2005) |
Table 2.
Summary of CRX frameshift and nonsense mutations associated with retinopathy. Reported frameshift and nonsense mutations from patients with retinopathy, showing the predicted stop codon position, size of the non-homologous Cterminus and total protein length. The nucleotide (nt) position in hCRX mRNA and frame for the predicted termination sites [bracketed text] are shown for each frameshift mutation. Text is color-coded based on Stop codon position, colors are matched with Figure 2. Nonsense mutations are shown in black text. (n.d.- not determined)
| Frameshift Mutation | Length (Amino Acids) | ||||||
| Protein | cDNA | Associated Phenotype | Co-segregation | Reference | Stop Codon (nt position [frame]) | Non- homologous C-terminus | Total Protein |
| p.P9i1 | c.24ins1 | LCA | no | Silva et al. (2000); Dharmaraj et al. (2000) | 207 [−2] | 61 | 69 |
| p.A117i1 | c.351dup1 | CoRD | n.d. | Huang et al. (2012) | 516 [−2] | 55 | 172 |
| p.I138d1 | c.413del1 | LCA | de novo | Nichols et al. (2010) | 557 [−1] | 48 | 185 |
| p.S141d1 | c.421del1 | LCA | yes | Zou et al. (2013) | 557 [−1] | 45 | 185 |
| p.P144d2i1 | c.429 430del2ins1 | LCA | n.d. | Huang et al. (2012) | 557 [−1] | 42 | 185 |
| p.L146d12 | c.436_447del12 | LCA | yes | Sohocki et al. (1998); Tzekov et al. (2001) | 898 [0] (in frame) | 0 | 295 |
| p.S150i1 | c.447ins1 | CoRD | Lines et al. (2002) | 516 [−2] | 23 | 172 | |
| p.P153d1 | c.458del1 | LCA/RP | yes | Wang et al. (2007); Ziviello et al. (2005) | 557 [−1] | 33 | 185 |
| p.T155ins4 | c.463 464ins4 | LCA | n.d. | Huang et al. (2012) | 516 [−2] | 19 | 173 |
| p.E168d1 | c.502del1 | CoRD | yes | Freund et al. (1997) | 557 [−1] | 18 | 185 |
| p.E168d2 | c.503_504del2 | LCA | de novo | Freund et al. (1998); Tran NM et al. (2014) | 516 [−2] | 4 | 171 |
| p.P170d1 | c.510del1 | LCA | yes | Perrault et al. (2003) | 557 [−1] | 15 | 185 |
| p.A174d1 | c.520del1 | LCA | de novo | Nakamura et al. (2002) | 557 [−1] | 12 | 185 |
| p.A177d1 | c.529del1 | LCA | yes | Koenkoop et al. (2002) | 557 [−1] | 9 | 185 |
| p.A181d1 | c.541del1 | LCA | n.d. | Zhang et al. (2001) | 557 [−1] | 5 | 185 |
| p.Y191i1 | c.570dup1 | LCA | n.d. | Huang et al. (2012) | 702 [−2] | 44 | 234 |
| p.Y191d1 | c.571del1 | LCA | de novo | Rivolta et al. (2001); Zou et al. (2013) | 578 [−1] | 2 | 192 |
| p.Y191* | c.572T>A Λ | LCA | yes | Chen et al. (2013) | novel | 0 | 191 |
| p.Y195* | c.585C>A Λ | LCA | n.d. | Huang et al. (2012) | novel | 0 | 195 |
| p.A196d4 | c.587_590del4 | CoRD | n.d. | Swain et al (1997); Sohocki et al. (2001) | 653 [−1] | 22 | 216 |
| p.A196i1 | c.590dup1 | CoRD | yes | Tzekov et al. (2000) | 702 [−2] | 38 | 234 |
| p.P205d1 | c.615del1 | CoRD | yes | Itabashi et al. (2004) | 653 [−1] | 12 | 217 |
| p.Y208* | c.624T>G Λ | LCA | n.d. | Huang et al. (2012) | novel | 0 | 208 |
| p.S212d1 | c.636del1 | CoRD | yes | Kitiratschky et al. (2008) | 653 [−1] | 5 | 217 |
| p.G217d1 | c.649del1 | LCA | de novo | Freund et al. (1998); Lotery et. Al (2000) | 653 [−1] | 1 | 217 |
| p.L237d1 | c.709del1 | LCA | de novo | Silva et al. (2000); Dharmaraj et al. (2000) | 653 [−1] | 133 | 369 |
| p.L237i1 | c.709dup1 | CoRD | n.d. | Huang et al. (2012) | 795 [−2] | 29 | 265 |
| p.S240i23 | c.720_742dup23 | LCA | n.d. | Huang et al. (2012) | 1109 [−1] | 137 | 377 |
| p.T251d1 | c.753del1 | LCA | n.d. | Huang et al. (2012) | 1109 [−1] | 118 | 369 |
| p.P263d1 | c.787del1 | LCA | yes | Rivolta et al. (2001) | 1109 [−1] | 106 | 369 |
| p.P272d3i2 | c.816_818del3ins2 | CoRD | yes | Paunescu et al. (2007) | 1109 [−1] | 97 | 369 |
| p.K295d8 | c.883 890del8 | CoRD | n.d. | Huang et al. (2012) | 951 [−2] | 20 | 314 |
Two functional classes of CRX missense mutations
In patients with retinopathies, 19 missense mutations have been reported to date, 7 of which co-segregate with the disease phenotype (Figure 1, Table 1). Co-segregating missense mutations are all located in the DNA binding domain of CRX. Phenotypic and functional analyses of representative mutations are summarized in Tran NM et al. (2014) and suggest that there are at least two classes of disease-causing missense mutations (Class I & II here): I) hypomorphic missense mutations that reduce DNA binding (Figure 1, gold text) and II) antimorphic missense mutations with either intact or uncharacterized DNA binding (Figure 1, purple text). Hypomorphic CRX mutations lose transactivation activity due to reduced DNA binding activity but do not interfere with the function of WT CRX. Antimorphic CRX mutations interfere with the transcriptional activity of WT CRX, either directly through protein/DNA interactions or indirectly through discrete cellular mechanisms. The mutations: p.R41W, p.R41Q, and p.R90W are predicted to be Class I type mutations (Figure 1; gold text). These three mutant proteins have reduced DNA binding and reduced ability to transactivate the promoter of the well-characterized CRX target gene Rhodopsin in vitro, and are thought to represent hypomorphic mutant proteins (Chen et al. 2002; Nichols et al. 2010; Swaroop et al. 1999; Tran et al. 2014). p.R90W protein also does not interfere with the transactivation activity of WT CRX (Tran et al. 2014). The mutations p.E80A, p.E80Q, p.E80K and p.K88N are predicted to be Class II type mutations (Figure 1, purple text). p.E80A, p.E80Q, and p.E80K all convert a negatively-charged glutamate to either a neutral or positively-charged aa and could have similar effects on CRX's protein function. Although p.E80A does not display reduced DNA binding or transactivation activity (Chen et al. 2002), it shows antimorphic activity on the expression of the photoreceptor-specific gene Rh5 in Drosophila in vivo rescue experiments (Terrell et al. 2012). p.K88N also shows reduced transactivation and has antimorphic activity on NRL expression in vitro (Nichols et al. 2010). However, in silico modeling of p.K88N protein predicts reduced DNA binding capability, suggesting it may act through a discrete antimorphic mechanism, although this has not yet been shown experimentally (Nichols et al. 2010). While the DNA binding properties and specific antimorphic mechanisms of the Class II mutations have not been fully characterized, these mutations clearly represent a distinct mutation class from the hypomorphic Class I based on molecular activity and disease phenotype. Class I mutations are all associated with late-onset CoRD (Huang et al. 2012; Paunescu et al. 2007; Rivolta et al. 2001; Sohocki et al. 1998; Swain et al. 1997), whereas Class II mutations are associated with early-onset adCoRD/LCA (den Hollander et al. 2008; Huang et al. 2012; Rivolta & Berson 2001; Sohocki et al. 1998).
Two functional classes of frameshift and nonsense CRX mutations
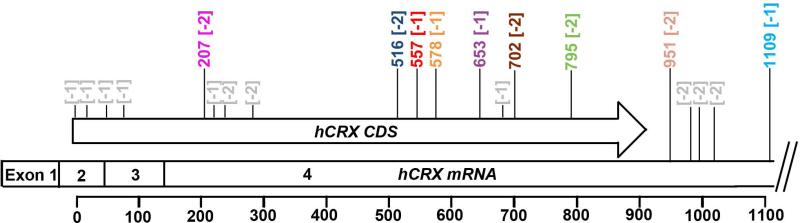
To date, 29 frameshift and 3 nonsense CRX mutations have been reported in patients with retinal diseases, 18 of which are known to co-segregate with the disease phenotype (Figure 1, Table 2). Phenotypic and functional analyses of representative mutations are summarized in Tran NM et al. (2014) and Roger et al. (2014) and suggest at least two functional classes (Class III and IV here): III) antimorphic frameshift/nonsense mutations with intact DNA binding (Figure 1, red text) and IV) antimorphic frameshift mutations that interfere with DNA binding (Figure 1, blue text). While all of the co-segregating mutations have an intact HD, the position and size of the frameshift mutation determines which out-of-frame stop codon will terminate translation. This new C-terminus could alter the function of the mutant protein and disrupt normal physiology. The antimorphic activity of mutant proteins that maintain DNA binding could stem from direct competition of mutant and wild-type (WT) CRX on target gene promoters, while mutant proteins that lose DNA binding likely act through a discrete mechanism. Table 2 lists all of the reported frameshift mutations, including the predicted stop codon position and length of the non-homologus C-terminal tail. Figure 2 shows the position of the out-of-frame stop codons in the human CRX mRNA with [-1] and [-2] being the frames 1 base pair (bp) and 2bp in the 5’ direction from the normal reading frame, respectively. The mutations in Table 2 are color-coded to match the corresponding predicted novel stop codons shown in Figure 2. The majority of the co-segregating frameshift/nonsense mutations result in the early truncation of CRX protein and carry a short non-homologous C-terminal tail that ranges in size from 0 to 55aa (Figure 1, red text; Table 2), which could have a minimal impact on DNA binding activity. The mutations p.E168d1, p.E168d2, p.A196d4, p.G217d1 and possibly several other frameshift/nonsense mutations are predicted to be Class III mutations (Figure 1, red text). The mutant proteins: p.E168d1, p.E168d2, p.A196d4 and p.G217d1 all lose the ability to transactivate the promoter of Rhodopsin (Chen et al. 2002). Additionally, p.E168d2 maintains DNA binding activity and interferes with WT CRX function (Tran NM et al. 2014). p.I138fs48 also shows antimorphic activity on target gene expression in Drosophila Rh5 rescue experiments (Terrell et al. 2012). These findings suggest that the Class III mutant CRX proteins maintain DNA binding, lose transactivation activity and possess antimorphic activity.
Figure 2. Schematic showing the position of out-of-frame stop codons in human Crx mRNA.
Human CRX coding sequence (hCRX CDS) and messenger RNA (hCRX mRNA) are shown. Scale below indicates the base pair position in hCRX CDS beginning at the start codon. All out-of-frame stop codons in hCRX mRNA coding region and its 3’UTR up to position 1109 are shown above the schematic. Nucleotide position and stop codon frame are shown in colored text (matched to Table 2) for all predicted termination sites for reported human mutations, non-utilized out-of-frame stop codons are shown in grey text. The positions of all out-of-frame stop codons are indicated in brackets. [-1] and [-2] indicate whether the stop codon is located 1 or 2 bases 5’ of the normal reading frame, respectively.
The co-segregating mutations: p.P237d2, p.P263d1 and p.P272d3i2 are predicted to be Class IV mutations (Figure 1, blue text, Table 2). These mutations each result in a long non-homologus C-terminal extension ranging from 97-133aa, increasing the total protein size by 70aa, which could greatly affect protein function and DNA binding. This extension occurs because following the [-1] stop codon at position 686bp, there is not another [-1] stop codon until 1109bp, which is 211bp after CRX's normal stop codon position (Figure 2). Therefore, any mutation after the 686bp position that shifts the reading frame 1bp in the 5’ direction would result in a long non-homologous C-terminal extension. This effect is demonstrated by the CrxRip mouse, which carries the spontaneous mutation p.G255d1 resulting in a mutant CRX protein with a 133aa nonhomologous C-terminal extension (Roger et al. 2014). While no direct human mutation equivalent to p.G255d1 has been reported, the mutations p.L237d1, p.S240i23, p.T251d1, p.P263d1 and p.P272d3i2 all have similar non-homologous C-terminal extensions and are associated with early onset CoRD/LCA (Table 2; blue text). CRXRIP protein does not bind CRX target DNA, although the early truncation protein CRX 1-254 maintains DNA binding (Roger et al. 2014). CRXRIP also loses transactivation activity but does not interfere with WT CRX function in vitro (Roger et al. 2014). The majority of both Class III and IV mutations are associated with early-onset (~0-20 years old) dominant CoRD/LCA (Dharmaraj et al. 2000; Freund et al. 1997; Galvin et al. 2005; Hanein et al. 2004; den Hollander et al. 2008; Huang et al. 2012; Jacobson et al. 1998; Lotery et al. 2000; Nakamura et al. 1998; Paunescu et al. 2007; Rivolta et al. 2001; Sohocki et al. 1998; Tzekov et al. 2000; Walia et al. 2010; Wang et al. 2007; Zhang et al. 2001). The severe dominant phenotypes associated with Class IV mutations suggest that they possess antimorphic activity despite the prediction that they reduce DNA binding activity. Collectively, these finding suggest that Class III and IV mutations are functionally distinct although both have antimorphic activity, and are associated with severe early-onset retinopathies.
MECHANISTICALLY DISTINCT ANIMAL MODELS FOR CRX-ASSOCIATED DISEASE
Non-mammalian animal models identify functional roles of CRX
The roles of CRX and the impact of mutant CRX proteins on retinal development have been studied in multiple animal models. While mammalian retinal development is guided by a trio of 'paired-like' homeodomain transcription factors: OTX1, OTX2 and CRX (Nishida et al. 2003; Furukawa et al. 1997; Chen et al. 1997; Plouhinec et al. 2003), Drosophila retinal development is guided by a single homeodomain transcription factor, Otd (Ranade et al. 2008; Vandendries et al. 1996). Photoreceptor-specific loss of Otd function in otduvi mutant flies results in a developmental defect and insensitivity to ultra violet light (Terrell et al. 2012; Vandendries et al. 1996). Human CRX and OTX2 rescue different aspects of the impaired photoreceptor development phenotype of otduvi flies, suggesting distinct, yet overlapping functions for CRX and OTX2 (Terrell et al. 2012). In addition, the CRX mutant proteins p.R90W (Class I), p.E80A (Class II) and p.I138fs48 (Class III) have partial and overlapping rescue function on Drosophila photoreceptor morphology and gene expression, while p.K88N (Class II) is unable to rescue the otduvi phenotype. Of these mutant proteins, only p.E80A and p.I138fs48 possess antimorphic activity in Drosophila, with p.I138fs48 having the strongest effect (Terrell et al. 2012). In other studies, a morpholino knockdown of Zebrafish Crx shows that CRX is critical for photoreceptor and bipolar cell development (Shen & Raymond 2004), and lipofection of Otx5b, a homologue of Crx, in Xenopus laevis biases retinal progenitors to the photoreceptor fate (Viczian et al. 2003). In summary, non-mammalian animal models demonstrate highly conserved roles for CRX and homeodomain transcription factor homologs in retinal development and photoreceptor gene expression.
Crx Knock-OUT mouse elucidates CRX's role in mammalian retinal development and serves as a non-functioning photoreceptor model for translational research
The role of Crx in mammalian retinal development was first characterized in vivo in a loss-of-function model, the Crx Knock-OUT (KO) mouse (Furukawa et al. 1999). In the homozygous Crx KO (“−/−“) mouse retina, photoreceptors fail to form outer segments, highly specialized organelles that contain visual pigment opsins and other proteins required for phototransduction. As a result, −/− photoreceptors do not function, form abnormal synapses, and undergo progressive degeneration (Furukawa et al. 1999; Morrow et al. 2005). Gene expression profile studies show that −/− mice have severely reduced expression of many photoreceptor-specific genes (Blackshaw et al. 2001; Hsiau et al. 2007; Livesey et al. 2000). Most of these genes are direct CRX targets as detected by ChIP-seq analyses of the genomic CRX binding profile in the mouse retina (Corbo et al. 2010).
Since −/− mice completely lack rod and cone function, they have served as an excellent animal model for developing translational approaches aimed at restoring photoreceptor function. Human embryonic stem cell-derived retinal cells or developing mouse rods transplanted into −/− retinas were able to integrate into the retina, differentiate into mature retinal cell types and restore some retinal function (Lamba et al. 2009, Homma et al. 2013). Recombinant adeno-associated virus (rAAV)-mediated gene therapy was also used to target gene delivery of WT CRX to immature photoreceptors of −/− mice. Cells expressing delivered CRX activate downstream transcription and restore some retinal function (Watanabe et al. 2013). These studies show that both cell-based and gene therapy approaches have potential for restoring vision in a retina with non-functioning photoreceptors.
The Crx −/− mouse demonstrates the central role of CRX in photoreceptor development, gene expression and the formation of vision. However, the heterozygous Crx KO mouse (“+/−“) only shows a slight delay in photoreceptor development and largely displays normal morphology, gene expression and retinal function in adulthood (Furukawa et al. 1999). Additionally, there are no co-segregating human mutations that result in the deletion of the CRX gene. Therefore, as a stand-alone model, the Crx KO mouse does not provide sufficient evidence that loss-of-function/null alleles cause dominant disease in humans, and also does not recapitulate severe dominant disease forms. In the next four sections, we will detail the findings from three mouse models and one cat model carrying CRX mutations equivalent to mutation types found in patients with CRX-associated diseases.
CrxR90W: A mouse model for recessive LCA caused by a Class I mutation
CrxR90W Knock-IN (K-IN) mice (“R90W”) carry the missense mutation p.R90W in the homeodomain of CRX (Figure 1, gold text bottom) (Tran NM et al. 2014). The p.R90W protein has reduced DNA binding and transactivation activity, categorizing R90W mouse as a Class I mutation (Figure 1; gold text). In humans, the p.R90W mutation is associated with a dominant mild late-onset CoRD and recessive LCA phenotype (Swaroop et al. 1999). Similarly, heterozygous R90W mice (R90W/+) show a very mild dominant cone phenotype, while homozygous mice (R90W/W) are completely blind (Tran et al. 2014). R90W/+ mice exhibit normal retinal morphology and gene expression in adulthood and only minor cone-driven functional deficits at 6 months (mo), as measured by electroretinogram (ERG). R90W/W mouse retinas display abnormal photoreceptor morphology lacking outer segments, highly disrupted gene expression and are completely blind. Heterozygous and homozygous R90W mice show phenotypes that only differ slightly from mice carrying a null Crx allele (Crx KO mice), suggesting that loss-of-function/null and hypomorphic CRX mutants produce mild dominant retinal phenotypes and recessive LCA.
CrxE168d2: A mouse model for dominant LCA caused by a Class III mutation
CrxE168d2 K-IN mice (“E168d2”) carry the frameshift deletion p.E168d2, which results in the early truncation of the CRX transactivation domains (Figure 1, red text bottom) (Tran NM et al. 2014). p.E168d2 protein retains the ability to bind target DNA, fails to activate target gene transcription and interferes with the transactivation activity of WT CRX in vitro (Tran NM et al. 2014), categorizing p.E168d2 as a Class III mutation (Figure 1, red text bottom). In humans, p.E168d2 is associated with dominant LCA (Freund et al. 2001; Jacobson et al. 1998). Heterozygous E168d2 mouse (E168d2/+) retinas are functionally impaired and homozygous E168d2 (E168d2/d2) mice are completely blind (Tran et al. 2014). E168d2/+ mice have 67% fewer cones than WT mice at 1mo. At this age, E168d2/+ mice have intact rods, suggesting cone degeneration occurs before rod degeneration, but rod outer segments are greatly shortened. The majority of E168d2/+ mouse rods are lost by 6mo. E168d2/+ retinas have reduced expression of several key phototransduction genes, both during development and adulthood. E168d2/d2 mouse photoreceptors do not form outer segments and have more severely impaired gene expression than Crx −/− mouse photoreceptors. The outer nuclear layer in E168d2/d2 retinas degenerates more rapidly than that of the −/− mouse, further highlighting the antimorphic function of the p.E168d2 protein. Unlike Crx KO, or R90W mice, E168d2 mice have an early-onset dominant LCA phenotype indicating that mutations that truncate the activation domain of CRX but maintain DNA binding act in an antimorphic fashion and are more damaging than loss-of-function or hypomorphic mutants. The Crx E168d2 mouse provides a model for CRX-associated early-onset LCA caused by an antimorphic frameshift mutation that maintains DNA binding activity.
The dominant E168d2 phenotype is also in contrast to the recessive phenotype of another Crx mutant mouse model, Crxtvrm65 (tvrm65), which carries a nonsense mutation at L277 (Won et al. 2011) (Figure 1, grey text). The truncation mutation in tvrm65 mice preserves the activation domains of CRX protein but removes the OTX tail domain, suggesting the presence or absence of the activation domains may be a critical determinant of the antimorphic properties of truncated proteins. Since there is no disease-causing mutation equivalent to the tvrm65 mutation, this model will not be discussed further in this review.
The CrxRdy cat provides a parallel large animal model with a Class III mutation to the E168d2 mouse
The CrxRdy cat (Rdy) carries a frameshift mutation in Crx, p.A182d1, generating a truncated CRX protein only 14aa C-terminal to the p.E168d2 mutation (Menotti-Raymond et al. 2010), suggesting it may also be a Class III mutation (Figure 1, red text). CrxRdy cats display severe dominant impairment of retinal function as measured by ERG, photoreceptor degeneration and abnormal Rhodopsin expression (Chong et al. 1999; Curtis et al. 1997; Leon et al. 1990; Leon et al. 1991; Menotti-Raymond et al. 2010). These findings are all consistent with the dominant LCA phenotype of the E168d2 mouse. While the molecular function of the Rdy protein has not yet been fully characterized, it appears to have similar antimorphic activity to the p.E168d2 mutant protein. The similarity of the Rdy cat and E168d2 mouse phenotypes suggest they are well-matched large and small animal models that likely share common disease mechanisms. The availability of both large and small animal models carrying the same class of Crx mutation could be advantageous for cross-validating conserved disease mechanisms and for pre-clinical testing of therapies in multiple animal systems.
CrxRip: A mouse model for dominant LCA caused by a Class IV mutation
CrxRip mice (Retina with Immature Photoreceptors, “Rip”) carry a frameshift deletion p.G255d1, which results in a 133aa non-homologous C-terminal extension of CRX protein (Figure 1, blue text) (Roger et al. 2014). Rip is a spontaneous mutation that was identified by linkage cross analysis and exome capture sequencing (Roger et al. 2014). The RIP protein does not bind target DNA, fails to activate target gene transcription and does not interfere with the transactivation activity of WT CRX in vitro (Roger et al. 2014). Unlike +/−, R90W/+ and E168d2/+ mice, however, heterozygous Rip mice (Rip/+) are completely blind, suggesting RIP protein has strong antimorphic activity in vivo. These findings categorize Rip as a Class IV mutation. Rip/+ mouse photoreceptors do not form outer segments and adult Rip/+ mouse retinas show even lower expression of several key phototransduction genes than the −/− mouse. The severe deficits in rod and cone gene expression suggests a photoreceptor differentiation defect in Rip/+ mice. Despite these strong expression changes, at 18mo the outer nuclear layer thickness in Rip/+ mouse retinas is roughly comparable to that of 6mo E168d2/+ mice. Similar to −/−, R90W/W and E168d2/d2 mice, homozygous Rip (Rip/Rip) mice do not form outer segments and are completely blind (Roger et al. 2014). These findings suggest that the RIP protein has a stronger antimorphic effect on photoreceptor gene expression and function but is less toxic to photoreceptor survival than the p.E168d2 protein. The Crx Rip mouse provides an animal model for CRX-associated dominant LCA caused by an antimorphic frameshift mutation with reduced DNA binding activity.
Discrete cellular mechanisms underlie the Crx E168d2 and Rip mouse phenotypes
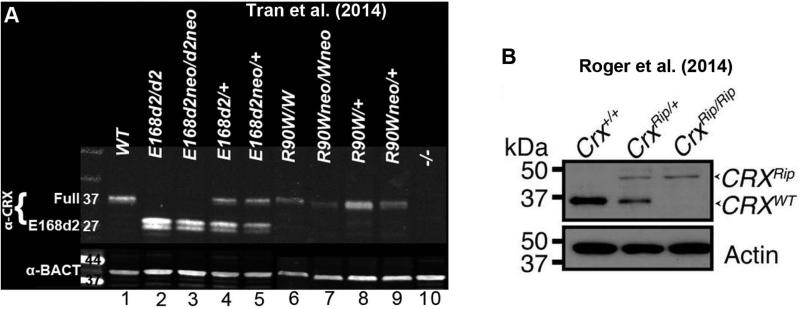
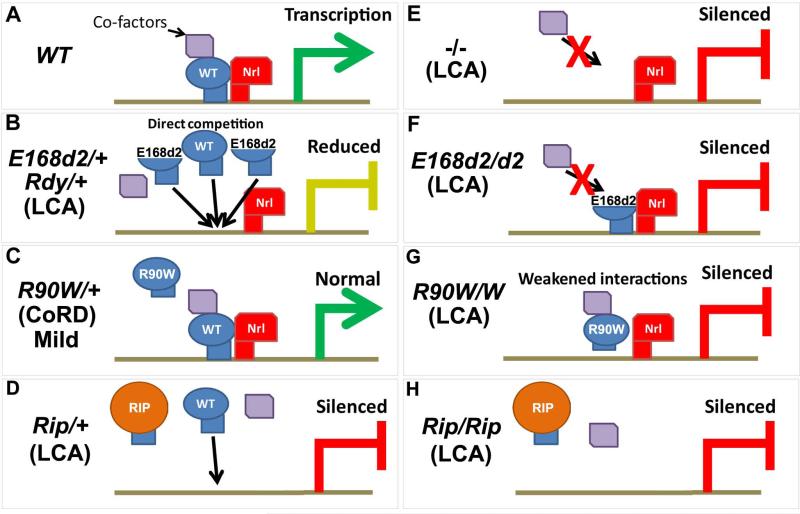
The Crx frameshift mutations p.E168d2 (Class III) and Rip (Class IV) produce mutant proteins with distinct molecular functions, and yield different retinal phenotypes in the mouse. These contrasting phenotypes are not simply gradations in disease severity but are caused by discrete cellular mechanisms. Assessment of CRX expression in heterozygous and homozygous R90W (Class I), E168d2 and Rip mouse retinas revealed allele-specific overexpression of the truncated p.E168d2 protein compared to WT CRX (Figure 3A) (Roger et al. 2014; Tran et al. 2014). This overexpression was not observed in either R90W or Rip mouse retinas (Figure 3A, B) (Roger et al. 2014; Tran et al. 2014). The p.E168d2 allele-specific overexpression was also seen at the mRNA level, suggesting an mRNA-based mechanism. Allele-specific overexpression of another predicted Class III CRX mutation, p.I138d1, was reported in in vitro expression assays and Drosophila in vivo rescue experiments (Table 3) (Nichols et al. 2010; Terrell et al. 2012;), suggesting the possibility of a conserved overexpression mechanism for this type of mutation. Unlike p.I138d1, the mutant proteins: p.R90W (Class I), p.K88N (Class II), and p.E80A (Class II) are not overexpressed in in vitro expression assays or Drosophila in vivo rescue experiments (Table 3) (Nichols et al. 2010; Terrell et al. 2012), suggesting allele-specific overexpression may be restricted to truncation mutations.
Figure 3. Expression of CRX protein in mutant mouse retinas.
A) Immunoblot analyses showing the expression of CRX protein in P10 E168d2, E168d2neo, R90W and R90Wneo mouse retinas using the rabbit polyclonal CRX 119b-1 antibody and mouse monoclonal anti-β-ACTIN antibody (α-BACT, Sigma-Aldrich) (Tran et al. 2014). Lanes are numbered below for reference. R90W mice express a normal length ~37kDA mutant CRX protein (lanes 6 & 8), E168d2 mice overexpress a truncated ~27kDA mutant CRX protein (lanes 2 & 4) and the intronic neo cassette results in reduced expression of both the R90W (lanes 7 & 9) and E168d2 (lanes 3 &5) proteins. B) Immunoblot analyses showing the expression of CRX protein in 1mo Rip mouse retinas using rabbit polyclonal CRX H-120 antibody (Santa Cruz) and mouse monoclonal anti-β-ACTIN antibody (Actin, Millipore) (Roger et al. 2014). Rip mice express an elongated ~44kDA mutant CRX protein.
Table 3.
Mutant CRX protein levels in alternate expression systems. The qualitative level of mutant CRX expression compared to WT CRX in either in vitro transient transfection of human retinal pigment epithelial-like cell (h-ARPE-19) or in vivo transgenic Drosophila otduvi rescue experiments.
| Protein | cDNA | System | Expression level | Reference |
|---|---|---|---|---|
| p.I138d1 | c.413del1 | in vitro | overexpressed | Nichols et al. (2010) |
| p.I138d1 | c.413del1 | Drosophila in vivo rescue | overexpressed | Terrell et al. (2012) |
| p.K88N | c.264G>T | in vitro | normal | Nichols et al. (2010) |
| p.K88N | c.264G>T | Drosophila in vivo rescue | normal | Terrell et al. (2012) |
| p.E80A | c.239A>C | Drosophila in vivo rescue | normal | Terrell et al. (2012) |
| p.R90W | c.268C>T | Drosophila in vivo rescue | reduced | Terrell et al. (2012) |
Since p.E168d2 is an antimorphic protein, its overexpression could be an important genetic modifier of disease severity. The E168d2neo mouse carries the same truncating CRX mutation but retains an intronic neomycin (neo) cassette, which was removed from the final E168d2 line. This neo cassette specifically interferes with the expression of the mutant allele but does not affect WT CRX expression (Tran et al. 2014), effectively shifting the balance of WT:mutant protein to an even ratio. E168d2neo/+ mice still have reduced retinal function compared to WT mice but have higher function than E168d2/+ mice, as measured by ERG. E168d2neo/+ mouse rods form normal-length outer segments and are intact through the first year of life, while cones are lost slowly over this time. E168d2neo/+ rod and cone gene expression remains reduced but is higher than in E168d2/+ mice. Overall, E168d2neo/+ mice display a much less severe phenotype than E168d2/+ and serve as a model for less severe CoRD. In summary, these findings highlight allele-specific overexpression of the truncated CRX protein p.E168d2 as a critical disease mechanism, which may be conserved for other truncating frameshift mutations.
Crx Rip mice do not overexpress mutant CRX protein (Figure 3B), yet they show an even stronger dominant visual phenotype than E168d2 mice. Additionally, the expression of key phototransduction genes gets progressively worse in Rip/+ mouse retinas from P2 to P21, while expression of the same genes improves from P10 to P21 in E168d2/+ mouse retinas (Roger et al. 2014, Tran et al. 2014). The Rip/+ mouse phenotype suggests that RIP protein has strong antimorphic activity. However, unlike p.E168d2 protein, RIP protein does not interfere with the transactivation activity of WT CRX in vitro (Roger et al. 2014), suggesting a distinct antimorphic mechanism. Roger et al. (2014) propose this mechanism involves OTX2, which plays a critical role in photoreceptor development. OTX2 is moderately overexpressed (~1.5-2.0 fold) in heterozygous and homozygous Rip, KO, R90W and E168d2 mouse retinas (Roger et al. 2014; Tran et al. 2014) and loss of OTX2 in Crx KO photoreceptors produces a stronger phenotype (Hsiau et al. 2007), suggesting OTX2 may play a compensatory role in these retinas. While OTX2 is expressed in Rip/Rip mice, it fails to bind target DNA including the promoter of Nrl (Roger et al. 2014). Unlike E168d2/+ mouse retinas, where NRL expression is maintained, Rip/+ mouse retinas initially express NRL at P6 but progressively lose expression by P21. Impaired CRX and OTX2 function and the loss of NRL expression results in incomplete photoreceptor differentiation and the severe impairment of rod and cone gene expression in Rip/+ mouse retinas. Further elucidating this mechanism, electroporation of either WT CRX or OTX2 conferred cell-autonomous rescue of Rhodopsin expression in Rip/+ mouse retinas, and transgenic expression of NRL under the Crx promoter was able to promote partial rescue of the Rip/+ phenotype (Roger et al. 2014). The differentiation defect of Rip/+ photoreceptors is not seen in E168d2/+ mice, which do express rod and cone specific genes, form outer segments and establish retinal function, albeit all at reduced levels compared to WT mice. In addition, the outer nuclear layer cells are preserved for a longer period of time in Rip/+ mice than in E168d2/+ mice, suggesting distinct developmental and degeneration phenotypes in each model. In summary, the photoreceptor transcription factor network is highly disrupted in Rip/+ mice leading to a defect in photoreceptor differentiation with distinct pathology from E168d2/+ mice. Collectively, these findings suggest that the antimorphic activities of Class III and IV CRX mutations are distinct and produce different phenotypes.
FUTURE DIRECTIONS
Caveats and remaining gaps in mammalian models of CRX-associated disease
The Crx KO, R90W, E168d2, Rip mice and Rdy cat serve as phenotypically distinct mammalian models for retinal diseases (Table 4) caused by CRX mutant proteins with different molecular functions (Figure 4). We propose that these mouse and cat lines serve as representative models for distinct classes of disease-causing CRX mutations, based on currently available genetic and molecular data. Further investigation of disease-causing mutations will likely reveal additional mutation classes with specific molecular defects. The rarity of each individual disease-causing mutation makes it impractical to model every one. Therefore, having paired models carrying similar mutations, like the E168d2 mouse and the Rdy cat, could be extremely valuable for cross-validating possible disease mechanisms and testing therapies designed to target that mutation class. Currently, there are no appropriate mammalian models for missense mutations with antimorphic activity (Class II). The mutant CRX proteins p.E80A and p.K88N have distinct antimorphic activities (Chen et al. 2002; Nichols et al. 2010; Terrell et al. 2012) but neither has been characterized in a mammalian system. Additionally, the available models do not address the phenotypic variation observed in patients with similar types of CRX mutations. For example, the mutation p.E168d1 is associated with a CoRD phenotype (Freund et al. 1997), while p.E168d2 is associated with a more severe LCA phenotype (Freund et al. 1998). It is unknown how genetic background or environmental factors contribute to these variable phenotypes. In summary, the currently available Crx mutant animal lines collectively serve as representative models for multiple forms of CRX-associated disease but other forms still remain unrepresented.
Table 4.
Summary of rod and cone phenotypes in mammalian mutant Crx models. Summary of the Crx E168d2, R90W, Rip and KO mice and Rdy cat phenotypes (Tran et al. 2014, Roger et al. 2014, Furukawa et al. 1999, Menotti-Raymond et al. 2010). Table shows the qualitative assessment of rod and cone outer segment morphology (OS), time-course of degeneration as determined by histology or immunohistochemistry, and function as determined by electroretinogram. The morphology and degeneration of cones was undetectable or unreliable in several models due to abnormalities in cone gene expression. The human disease represented by each genotype is also shown.
| Rod | Cone | Disease model | ||||||
|---|---|---|---|---|---|---|---|---|
| Genotype: | Species | Morphology | Degeneration | Function | Morphology | Degeneration | Function | |
| WT | normal | - | ++++ | normal | - | ++++ | ||
| E168d2/+ | mus musculus | shortened OS | 1-6mo | ++ | shortened OS; mislocalized nuclei | 1mo | + | LCA |
| Rdy/+ | felis catus | shortened OS | ~1-36mo | ++ | not determined | not determined | + | LCA |
| E168d2neo/+ | mus musculus | normal | - | +++ | shortened OS; mislocalized nuclei | 1yr+ | ++ | CoRD (moderate) |
| R90W/+ | mus musculus | normal | - | ++++ | normal | - | ++++ | CoRD (mild) |
| Rip/+ | mus musculus | no OS | 1-18mo+ | - | undetectable | undetectable | - | LCA |
| -/- | mus musculus | no OS | 1-3mo | - | undetectable | undetectable | - | LCA |
| E168d2/d2 | mus musculus | no OS | 1-3mo | - | undetectable | undetectable | - | LCA |
| R90W/W | mus musculus | no OS | 1-3mo | - | undetectable | undetectable | - | LCA |
| Rip/Rip | mus musculus | no OS | 1-9mo+ | - | undetectable | undetectable | - | LCA |
Figure 4. Proposed pathogenic mechanisms of mutant CRX proteins in mammalian models.
(Adapted from: Tran et al. 2014) A) In WT mice, CRX coordinately regulates transcription by interacting with target DNA and co-factors including the rod-specific transcription factor NRL. B) In E168d2/+ mice and possibly Rdy/+ cats, a Class III mutant CRX is overexpressed and directly competes with WT CRX, producing an LCA phenotype. C) In R90W/+ mice, a Class I mutant CRX is expressed and largely does not interfere with WT CRX producing only a mild late-onset CoRD phenotype. D) In Rip/+ mice, a Class IV mutant CRX is expressed and disrupts the photoreceptor transcription factor network including the loss of NRL expression, producing an LCA phenotype with distinct pathology from E168d2/+ and Rdy/+. E-H) Homozygous Crx KO, E168d2, R90W and Rip mice all produce a LCA phenotype and are completely blind from birth. CRX target gene expression is severely impaired in all homozygous mice with E168d2/d2 and Rip/Rip mice having the strongest changes.
Understanding the mechanisms of Crx E168d2 and Rip antimorphic activity
The Crx E168d2 and Rip mice show distinct dominant phenotypes caused by different underlying mechanisms. The p.E168d2 and RIP mutant proteins both have antimorphic activity but are functionally distinct (Figure 4) (Roger et al. 2014; Tran et al. 2014). In E168d2 mice, the dominant phenotype was linked to the allele-specific overexpression of the mutant CRX protein (Tran et al. 2014). Since allele-specific overexpression of p.E168d2 occurs at both the mRNA and protein level, it is thought to be caused by the intrinsic properties of p.E168d2 mRNA. While frameshift mutations have often been associated with mutant mRNA degradation through a process called nonsense mediated decay (NMD) (Schweingruber et al. 2013), there is little precedent for these mutations causing mRNA overexpression. The mechanisms mediating p.E168d2 overexpression are currently unknown and could involve increased p.E168d2 transcription, mRNA stability or a combination of both mechanisms. Although preliminary evidence suggests that this phenomenon is conserved for another Class III mutation, p.I138d1 (Nichols et al. 2010, Terrell et al. 2012), further investigation is required to determine if this phenomenon is conserved for all Class III mutations and if so what cellular mechanisms are involved. In Rip mice, the dominant phenotype is associated with the loss of OTX2 function and NRL expression (Roger et al. 2014). RIP protein has a long non-homologous C-terminal extension that affects protein structure and function. RIP does not bind DNA or interfere with WT CRX transactivation in vitro and thus it is not thought to directly interfere with target gene expression on target gene loci. It is possible that RIP protein could interact with OTX2, CRX or other co-factors in the cell, preventing them from being properly recruited to their DNA targets, or act indirectly through a different cellular mechanism. Further assessment of the molecular function of RIP protein in vivo is required to elucidate this mechanism. Additionally, photoreceptor cells in the outer nuclear layers of E168d2 and Rip mice are lost at different rates. Understanding the cellular mechanisms that lead to outer nuclear layer degeneration may be critical for designing therapies aimed at preserving photoreceptor survival.
Translating lessons from animal models to therapeutic approaches
Currently there are no treatment strategies for CRX-associated diseases. The availability of ‘true-to-disease’ animal models for distinct disease forms will facilitate the development and testing of novel therapeutic strategies. The Crx KO, R90W, E168d2 and Rip mice provide representative models for different classes of disease-causing mutations, each with unique molecular mechanisms and pathology (Figure 4, Table 4). Potential therapies could be tested in multiple models to determine their effectiveness for different types of CRX mutations. Additionally, the E168d2 mouse and CrxRdy cat provide matched small and large animal models to test the effectiveness of therapies for similar mutations in multiple animal systems. rAAV-mediated gene therapy approaches may have the greatest potential for rescuing retinal function for CRX-associated diseases since photoreceptors are efficiently transduced by rAAV. These approaches have successfully been used to improve retinal function in animal models of retinal dystrophies (Acland et al. 2001; Bennicelli et al. 2008; Boye et al. 2010; Jiang et al. 2011; Mancuso et al. 2009; O'reilly et al. 2007; Tam et al. 2008), as well as in human safety trials (Cideciyan et al. 2008; Colella et al. 2012; Macguire et al. 2008). Since defects in retinal function are detectable before photoreceptor degeneration in E168d2/+ and Rip/+ mice, it appears that there is a time window for effective gene therapy.
Antimorphic CRX mutant proteins have 'toxic' effects on the function of the WT protein. Theoretically, these ‘toxic’ effects can be reduced by shifting the ratio of the mutant to WT proteins. This effect is seen in E168d2/+ and E168d2neo/+ mice, where a reduction in the mutant protein level results in a dramatically less severe phenotype. The ratio of WT:mutant CRX protein could be shifted in multiple ways: 1) gene delivery of a WT allele to boost the overall level of WT CRX; 2) specific knock-down of the mutant allele without affecting WT levels; or 3) a combinatorial knock-down and WT gene delivery approach to simultaneously reduce mutant levels and boost WT levels. Specific knock-down of mutant alleles would be difficult to achieve as it would have to be designed for individual mutations, which are rare and the nucleotide differences from the WT mRNA can be as small as a single nucleotide. Combinatorial knock-down and gene delivery could be a more generalized approach. Crx short hairpin RNA (shRNA) knock-down of endogenous Crx expression (mutant+WT) could be combined with gene delivery of a shRNA-resistant recombinant Crx (containing synonymous mutations). However, the practical application and efficacy of this approach needs to be thoroughly tested.
CONCLUSION
The generation and characterization of mechanistically distinct animal models has greatly improved our understanding of CRX-associated disease. The phenotypic variation in human patients is, at least in part, driven by mutant proteins with different molecular properties. We have categorized four distinct classes of CRX mutations based on mutation type, the molecular activity of their mutant protein and associated phenotype. Currently, there are mammalian models carrying mutations from three of these four classes, all of which accurately reflect their associated human phenotypes. These models provide valuable insight into distinct CRX-associated disease pathologies and identify unexpected underlying disease mechanisms that could be targets for therapy. Additionally, these models demonstrate the importance of having comprehensive animal models to more fully represent the complex phenotypes of human diseases and will improve our ability to effectively test therapeutic strategies.
Key Findings.
Mechanism-based classification of human CRX mutations linked to blinding retinopathies.
Representative animal models for different mutation classes. • New insights into the molecular mechanisms underlying distinct phenotypes.
Future directions on basic mechanistic studies and translational research on therapy development.
ACKNOWLEDGEMENTS
This work was supported by grants from NIH EY012543-10S (to SC), EY002687 and EY013360 (to WU-DOVS), Research to Prevent Blindness (to SC and WU-DOVS) Foundation Fighting Blindness (to SC) and Hope for Vision (to SC).
We wish to thank Dr. Anne K. Hennig for critical reading of this manuscript. This work was supported by the U.S. National Institutes of Health [R01-EY012543-10S1 (to SC), T32-EY013360 (to WU), P30-EY02687 (to WU-Department of Ophthalmology and Visual Sciences)], by Research to Prevent Blindness [unrestricted funds (to WU-Department of Ophthalmology and Visual Sciences) and Lew Wasserman Merit Award (to SC)], and by Foundation Fighting Blindness (to SC) and Hope for Vision (to SC).
REFERENCES
- Acland GM, Aguirre GD, Ray J, Zhang Q, Aleman TS, Cideciyan AV, Pearce-Kelling SE, Anand V, Zeng Y, Maguire AM, Jacobson SG, Hauswirth WW, Bennett J. Gene therapy restores vision in a canine model of childhood blindness. Nat Genet. 2001;28:92–95. doi: 10.1038/ng0501-92. [DOI] [PubMed] [Google Scholar]
- Baas D, Bumsted KM, Martinez JA, Vaccarino FM, Wikler KC, Barnstable CJ. The subcellular localization of Otx2 is cell-type specific and developmentally regulated in the mouse retina. Brain Res Mol Brain Res. 2000;78:26–37. doi: 10.1016/s0169-328x(00)00060-7. [DOI] [PubMed] [Google Scholar]
- Bennicelli J, Wright JF, Komaromy A, Jacobs JB, Hauck B, Zelenaia O, Mingozzi F, Hui D, Chung D, Rex TS, Wei Z, Qu G, Zhou S, Zeiss C, Arruda VR, Acland GM, Dell'Osso LF, High KA, Maguire AM, Bennett J. Reversal of blindness in animal models of leber congenital amaurosis using optimized AAV2-mediated gene transfer. Mol Ther. 2008;16:458–465. doi: 10.1038/sj.mt.6300389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Progress in retinal and eye research. 2010;29(5):335–75. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Boatright JH, Borst DE, Peoples JW, Bruno J, Edwards CL, Si JS, Nickerson JM. A major cis activator of the IRBP gene contains CRX-binding and Ret-1 /PCE-I elements. Mol. Vis. 22. 1997;3:15. [PubMed] [Google Scholar]
- Bobola N, Briata P, Ilengo C, Rosatto N, Craft C, Corte G, Ravazzolo R. OTX2 homeodomain protein binds a DNA element necessary for interphotoreceptor retinoid binding protein gene expression. Mech. Dev. 1999;82:165–9. doi: 10.1016/s0925-4773(98)00162-2. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Mallamaci A, Briata P, Corte G, Boncinelli E. Implication of OTX2 in pigment epithelium determination and neural retina differentiation. J Neurosci. 1997;17:4243–4252. doi: 10.1523/JNEUROSCI.17-11-04243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye SE, Boye SL, Pang J, Ryals R, Everhart D, Umino Y, Neeley AW, Besharse J, Barlow R, Hauswirth WW. Functional and behavioral restoration of vision by gene therapy in the guanylate cyclase-1(GC1) knockout mouse. PLoS One. 2010;5:e11306. doi: 10.1371/journal.pone.0011306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau KY, Chen S, Zack DJ, Ono SJ. Functional domains of the cone-rod homeobox(CRX) transcription factor. J. Biol. Chem. 2000;275:37264–70. doi: 10.1074/jbc.M002763200. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins N. a, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–30. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang Q-L, Xu S, Liu I, Li LY, Wang Y, Zack DJ. Functional analysis of cone-rod homeobox(CRX) mutations associated with retinal dystrophy. Hum. Mol. Genet. 2002;11:873–84. doi: 10.1093/hmg/11.8.873. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang Q, Shen T, Xiao X, Li S, Guan L, Zhang J, Zhu Z, Yin Y, Wang P, Guo X, Wang J, Zhang Q. Comprehensive mutation analysis by whole-exome sequencing in 41 Chinese families with Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 2013;54:4351–7. doi: 10.1167/iovs.13-11606. [DOI] [PubMed] [Google Scholar]
- Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum. Mol. Genet. 2006;15:2588–2602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh ECT, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 2004;13:1563–75. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Aleman TS, Boye SL, Schwartz SB, Kaushal S, Roman AJ, Pang J-J, Sumaroka A, Windsor E. a M., Wilson JM, Flotte TR, Fishman G. a, Heon E, Stone EM, Byrne BJ, Jacobson SG, Hauswirth WW. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15112–7. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella P, Auricchio A. Gene therapy of inherited retinopathies: a long and successful road from viral vectors to patients. Hum. Gene Ther. 2012;23:796–807. doi: 10.1089/hum.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Lawrence KA, Karlstetter M, Myers CA, Abdelaziz M, Dirkes W, Weigelt K, Seifert M, Benes V, Fritsche LG, Weber BHF, Langmann T. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Research. 2010;20:1512–1525. doi: 10.1101/gr.109405.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN. Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest. Ophthalmol. Vis. Sci. 2005;46:2156–67. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmaraj SR, Silva ER, Pina a L., Li YY, Yang JM, Carter CR, Loyer MK, El-Hilali HK, Traboulsi EK, Sundin OK, Zhu DK, Koenekoop RK, Maumenee IH. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic Genet. 2000;21:135–50. [PubMed] [Google Scholar]
- Fei Y, Matragoon S, Smith SB, Overbeek P. a, Chen S, Zack DJ, Liou GI. Functional dissection of the promoter of the interphotoreceptor retinoid-binding protein gene: the cone-rod-homeobox element is essential for photoreceptor-specific expression in vivo. J. Biochem. 1999;125:1189–99. doi: 10.1093/oxfordjournals.jbchem.a022403. [DOI] [PubMed] [Google Scholar]
- Fei Y, Hughes TE. Nuclear trafficking of photoreceptor protein crx: the targeting sequence and pathologic implications. Invest. Ophthalmol. Vis. Sci. 2000;41:2849–56. [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene(CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Freund C, Wang QL, Chen S, Muskat B, Wiles C, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Galvin J. a, Fishman G. a, Stone EM, Koenekoop RK. Evaluation of genotype-phenotype associations in leber congenital amaurosis. Retina. 2005;25:919–29. doi: 10.1097/00006982-200510000-00016. [DOI] [PubMed] [Google Scholar]
- Hanein S, Perrault I, Gerber S, Tanguy G, Barbet F, Ducroq D, Calvas P, Dollfus H, Hamel C, Lopponen T, Munier F, Santos L, Shalev S, Zafeiriou D, Dufier J-L, Munnich A, Rozet J-M, Kaplan J. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum. Mutat. 2004;23:306–17. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- Hao H, Kim DS, Klocke B, Johnson KR, Cui K, Gotoh N, Zang C, Gregorski J, Gieser L, Peng W, Fann Y, Seifert M, Zhao K, Swaroop A. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012;8:e1002649. doi: 10.1371/journal.pgen.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig AK, Peng G-H, Chen S. Transcription coactivators p300 and CBP are necessary for photoreceptor-specific chromatin organization and gene expression. PLoS One. 2013;8:e69721. doi: 10.1371/journal.pone.0069721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Roepman R, Koenekoop RK, Cremers FPM. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Homma K, Okamoto S, Mandai M, Gotoh N, Rajasimha HK, Chang YS, Chen S, Li W, Cogliati T, Swaroop A, Takahashi M. Developing rods transplanted into the degenerating retina of Crx-knockout mice exhibit neural activity similar to native photoreceptors. Stem Cells. 2013;31(6):1149–59. doi: 10.1002/stem.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housset M, Samuel A, Ettaiche M, Bemelmans A, Béby F, Billon N, Lamonerie T. Loss of Otx2 in the adult retina disrupts retinal pigment epithelium function, causing photoreceptor degeneration. J. Neurosci. 2013;33:9890–904. doi: 10.1523/JNEUROSCI.1099-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One. 2007;2(7):e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Xiao X, Li S, Jia X, Wang P, Guo X, Zhang Q. CRX variants in cone-rod dystrophy and mutation overview. Biochem. Biophys. Res. Commun. 2012;426:498 –503. doi: 10.1016/j.bbrc.2012.08.110. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Huang Y, Hanna DB, Freund CL, Affatigato LM, Carr RE, Zack DJ, Stone EM, McInnes RR. Retinal degenerations with truncation mutations in the cone-rod homeobox(CRX) gene. Invest Ophthalmol Vis Sci. 1998;39:2417–2426. [PubMed] [Google Scholar]
- Jiang L, Zhang H, Dizhoor AM, Boye SE, Hauswirth WW, Frederick JM, Baehr W. Long-term RNA interference gene therapy in a dominant retinitis pigmentosa mouse model. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18476–81. doi: 10.1073/pnas.1112758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z-B, Mandai M, Yokota T, Higuchi K, Ohmori K, Ohtsuki F, Takakura S, Itabashi T, Wada Y, Akimoto M, Ooto S, Suzuki T, Hirami Y, Ikeda H, Kawagoe N, Oishi a, Ichiyama S, Takahashi M, Yoshimura N, Kosugi S. Identifying pathogenic genetic background of simplex or multiplex retinitis pigmentosa patients: a large scale mutation screening study. J. Med. Genet. 2008;45:465–72. doi: 10.1136/jmg.2007.056416. [DOI] [PubMed] [Google Scholar]
- Kimura A, Singh A, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. Both PCE-1/RX and OTX/CRX Interactions Are Necessary for Photoreceptor-specific Gene Expression. J. Biol. Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- Kitiratschky VBD, Nagy D, Zabel T, Zrenner E, Wissinger B, Kohl S, Jägle H. Cone and cone-rod dystrophy segregating in the same pedigree due to the same novel CRX gene mutation. Br. J. Ophthalmol. 2008;92:1086–91. doi: 10.1136/bjo.2007.133231. [DOI] [PubMed] [Google Scholar]
- Koenekoop RK, Loyer M, Dembinska O, Beneish R. Mutation report Visual improvement in Leber congenital amaurosis and the CRX genotype. 2002;23:49–60. doi: 10.1076/opge.23.1.49.2200. [DOI] [PubMed] [Google Scholar]
- Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis -regulatory element. Proc Natl Acad Sci U S A. 2012;109(47):19498–503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba D. a, Gust J, Reh TA. Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell. 2009;4:73–9. doi: 10.1016/j.stem.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmann T, Lai CCL, Weigelt K, Tam BM, Warneke-Wittstock R, Moritz OL, Weber BHF. CRX controls retinal expression of the X-linked juvenile retinoschisis(RS1) gene. Nucleic Acids Res. 2008;36:6523–34. doi: 10.1093/nar/gkn737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xiao X, Li S, Jia X, Wang P, Guo X, Jiao X, Zhang Q, Hejtmancik JF. Detection of variants in 15 genes in 87 unrelated Chinese patients with Leber congenital amaurosis. PLoS One. 2011;6:e19458. doi: 10.1371/journal.pone.0019458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines M. a, Hébert M, McTaggart KE, Flynn SJ, Tennant MT, MacDonald IM. Electrophysiologic and phenotypic features of an autosomal cone-rod dystrophy caused by a novel CRX mutation. Ophthalmology. 2002;109:1862–70. doi: 10.1016/s0161-6420(02)01187-9. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- Lotery a J., Namperumalsamy P, Jacobson SG, Weleber RG, Fishman G. a, Musarella M. a, Hoyt CS, Héon E, Levin a, Jan J, Lam B, Carr RE, Franklin a, Radha S, Andorf JL, Sheffield VC, Stone EM. Mutation analysis of 3 genes in patients with Leber congenital amaurosis. Arch. Ophthalmol. 2000;118:538–43. doi: 10.1001/archopht.118.4.538. [DOI] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr., Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell'Osso L, Hertle R, Ma JX, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso K, Hauswirth WW, Li Q, Connor TB, Kuchenbecker JA, Mauck MC, Neitz J, Neitz M. Gene therapy for red-green colour blindness in adult primates. Nature. 2009;461:784–787. doi: 10.1038/nature08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears a J., Kondo M, Swain PK, Takada Y, Bush R. a, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–52. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, Deckman KH, David VA, Myrkalo J, O'Brien SJ, Narfstrom K. Mutation discovered in a feline model of human congenital retinal blinding disease. Invest. Ophthalmol. Vis. Sci. 2010;51(6):2852–9. doi: 10.1167/iovs.09-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J. Biol. Chem. 2000;275:29794–9. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Raviola E, Cepko CL. Synaptogenesis and outer segment formation are perturbed in the neural retina of Crx mutant mice. BMC Neurosci. 2005;6:5. doi: 10.1186/1471-2202-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Ito S, Miyake Y. Novel de novo mutation in CRX gene in a Japanese patient with leber congenital amaurosis. Am. J. Ophthalmol. 2002;134(3):465–7. doi: 10.1016/s0002-9394(02)01542-8. [DOI] [PubMed] [Google Scholar]
- Nichols LL, Alur RP, Boobalan E, Sergeev YV, Caruso RC, Stone EM, Swaroop A, Johnson M. a, Brooks BP. Two novel CRX mutant proteins causing autosomal dominant Leber congenital amaurosis interact differently with NRL. Hum. Mutat. 2010;31:E1472–83. doi: 10.1002/humu.21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- O'Reilly M, Palfi A, Chadderton N, Millington-Ward S, Ader M, Cronin T, Tuohy T, Auricchio A, Hildinger M, Tivnan A, McNally N, Humphries MM, Kiang A-S, Humphries P, Kenna PF, Farrar GJ. RNA interference-mediated suppression and replacement of human rhodopsin in vivo. Am. J. Hum. Genet. 2007;81:127–35. doi: 10.1086/519025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunescu K, Preising MN, Janke B, Wissinger B, Lorenz B. Genotype phenotype correlation in a German family with a novel complex CRX mutation extending the open reading frame. Ophthalmology. 2007;114:1348–1357. e1. doi: 10.1016/j.ophtha.2006.10.034. [DOI] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis. Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum. Mol. Genet. 2007;16(20):2433–2452. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng GH, Chen S. Active opsin loci adopt intrachromosomal loops that depend on the photoreceptor transcription factor network. Proc. Natl. Acad. Sci. U. S. A. 2011;108(43):17821–6. doi: 10.1073/pnas.1109209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Hanein S, Gerber S, Barbet F, Dufier JL, Munnich A, Rozet JM, Kaplan J. Evidence of autosomal dominant Leber congenital amaurosis(LCA) underlain by a CRX heterozygous null allele. J. Med. Genet. 2003. 2003;40(7):e90. doi: 10.1136/jmg.40.7.e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouhinec JL, Sauka-Spengler T, Germot A, Le Mentec C, Cabana T, Harrison G, Pieau C, Sire JY, Véron G, Mazan S. The mammalian Crx genes are highly divergent representatives of the Otx5 gene family, a gnathostome orthology class of orthodenticle-related homeogenes involved in the differentiation of retinal photoreceptors and circadian entrainment. Mol. Biol. Evol. 2003;20:513–21. doi: 10.1093/molbev/msg085. [DOI] [PubMed] [Google Scholar]
- Ranade SS, Yang-Zhou D, Kong SW, McDonald EC, Cook TA, Pignoni F. Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol. 2008;315:521–534. doi: 10.1016/j.ydbio.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehemtulla a, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc. Natl. Acad. Sci. U. S. A. 1996;93:191–5. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, Berson EL, Dryja TP. Dominant Leber congenital amaurosis, cone-rod degeneration, and retinitis pigmentosa caused by mutant versions of the transcription factor CRX. Human mutation. 2001;18(6):488–98. doi: 10.1002/humu.1226. [DOI] [PubMed] [Google Scholar]
- Roduit R, Escher P, Schorderet DF. Mutations in the DNA-binding domain of NR2E3 affect in vivo dimerization and interaction with CRX. PLoS One. 2009;4:e7379. doi: 10.1371/journal.pone.0007379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger JE, Hiriyanna A, Gotoh N, Hao H, Cheng DF, Ratnapriya R, Kautzmann MI, Chang B, Swaroop A. OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. J. Clin. Invest. 2014;124(2):631–43. doi: 10.1172/JCI72722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovsing L, Clokie S, Bustos DM, Rohde K, Coon SL, Litman T, Rath MF, Møller M, Klein DC. Crx broadly modulates the pineal transcriptome. J. Neurochem. 2011;119:262–74. doi: 10.1111/j.1471-4159.2011.07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankila EM, Joensuu TH, Hamalainen RH, Raitanen N, Valle O, Ignatius J, Cormand B. A CRX mutation in a Finnish family with dominant cone-rod retinal dystrophy. Hum. Mutat. 2000;16:94. doi: 10.1002/1098-1004(200007)16:1<94::AID-HUMU25>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta. 2013;1829:612–23. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Shen Y, Raymond PA. Zebrafish cone-rod(crx) homeobox gene promotes retinogenesis. Developmental Biology. 2004;269(1):237–51. doi: 10.1016/j.ydbio.2004.01.037. [DOI] [PubMed] [Google Scholar]
- Silva E, Yang JM, Li Y, Dharmaraj S, Sundin OH, Maumenee IH. A CRX null mutation is associated with both Leber congenital amaurosis and a normal ocular phenotype. Invest. Ophthalmol. Vis. Sci. 2000;41:2076–9. [PubMed] [Google Scholar]
- Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, McInnes RR, Daiger SP. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am. J. Hum. Genet. 1998;63:1307–1315. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohocki MM, Daiger SP, Bowne SJ, Rodriquez J. a, Northrup H, Heckenlively JR, Birch DG, Mintz-Hittner H, Ruiz RS, Lewis R. a, Saperstein DA, Sullivan LS. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum. Mutat. 2001;17:42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–36. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Wang QL, Wu W, Cook J, Coats C, Xu S, Chen S, Zack DJ, Sieving PA. Leber congenital amaurosis caused by a homozygous mutation(R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum. Mol. Genet. 1999;8:299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- Tam LCS, Kiang A-S, Kennan A, Kenna PF, Chadderton N, Ader M, Palfi A, Aherne A, Ayuso C, Campbell M, Reynolds A, McKee A, Humphries MM, Farrar GJ, Humphries P. Therapeutic benefit derived from RNAi-mediated ablation of IMPDH1 transcripts in a murine model of autosomal dominant retinitis pigmentosa(RP10). Hum. Mol. Genet. 2008;17:2084–100. doi: 10.1093/hmg/ddn107. [DOI] [PubMed] [Google Scholar]
- Terrell D, Xie B, Workman M, Mahato S, Zelhof A, Gebelein B, Cook T. OTX2 and CRX rescue overlapping and photoreceptor-specific functions in the Drosophila eye. Dev. Dyn. 2012;241:215–28. doi: 10.1002/dvdy.22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Zhang A, Zhang X, Huecker JB, Hennig AK, Chen S. Mechanistically Distinct Mouse Models for CRX-Associated Retinopathy. PLoS Genet. 2014;10:e1004111. doi: 10.1371/journal.pgen.1004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzekov RT, Liu Y, Sohocki MM, Zack DJ, Daiger SP, Heckenlively JR, Birch DG. Autosomal dominant retinal degeneration and bone loss in patients with a 12-bp deletion in the CRX gene. Invest. Ophthalmol. Vis. Sci. 2001;42:1319–27. [PMC free article] [PubMed] [Google Scholar]
- Tzekov RT, Sohocki MM, Daiger SP, Birch DG. Visual phenotype in patients with Arg41Gln and ala196+1bp mutations in the CRX gene. Ophthalmic Genet. 2000;21:89–99. [PubMed] [Google Scholar]
- Vallespin E, Cantalapiedra D, Riveiro-Alvarez R, Wilke R, Aguirre-Lamban J, Avila-Fernandez A, Lopez-Martinez MA, Gimenez A, Trujillo-Tiebas MJ, Ramos C, Ayuso C. Mutation screening of 299 Spanish families with retinal dystrophies by Leber congenital amaurosis genotyping microarray. Invest. Ophthalmol. Vis. Sci. 2007;48:5653–61. doi: 10.1167/iovs.07-0007. [DOI] [PubMed] [Google Scholar]
- Vandendries ER, Johnson D, Reinke R. orthodenticle is required for photoreceptor cell development in the Drosophila eye. Developmental biology. 1996;173(1):243–55. doi: 10.1006/dbio.1996.0020. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Vopalensky P, Pergner J, Liegertova M, Benito-Gutierrez E, Arendt D, Kozmik Z. Molecular analysis of the amphioxus frontal eye unravels the evolutionary origin of the retina and pigment cells of the vertebrate eye. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15383–8. doi: 10.1073/pnas.1207580109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia S, Fishman G. a, Jacobson SG, Aleman TS, Koenekoop RK, Traboulsi EI, Weleber RG, Pennesi ME, Heon E, Drack A, Lam BL, Allikmets R, Stone EM. Visual acuity in patients with Leber's congenital amaurosis and early childhood-onset retinitis pigmentosa. Ophthalmology. 2010;117:1190–8. doi: 10.1016/j.ophtha.2009.09.056. [DOI] [PubMed] [Google Scholar]
- Wang P, Guo X, Zhang Q. Further evidence of autosomal-dominant Leber congenital amaurosis caused by heterozygous CRX mutation. Graefes Arch. Clin. Exp. Ophthalmol. 2007;245:1401–2. doi: 10.1007/s00417-007-0554-0. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Sanuki R, Ueno S, Koyasu T, Hasegawa T, Furukawa T. Tropisms of AAV for subretinal delivery to the neonatal mouse retina and its application for in vivo rescue of developmental photoreceptor disorders. PLoS One. 2013;8:e54146. doi: 10.1371/journal.pone.0054146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber AL, Hodor P, Thut CJ, Vogt TF, Zhang T, Holder DJ, Petrukhin K. Dual role of Nr2e3 in photoreceptor development and maintenance. Exp. Eye Res. 2008;87:35–48. doi: 10.1016/j.exer.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Won J, Shi LY, Hicks W, Wang J, Hurd R, Naggert JK, Chang B, Nishina PM. Mouse model resources for vision research. J. Ophthalmol. 2011;2011:391384. doi: 10.1155/2011/391384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum. Mol. Genet. 2004;13:1487–503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- Zernant J, Kulm M, Dharmaraj S, den Hollander AI, Perrault I, Preising MN, Lorenz B, Kaplan J, Cremers FP, Maumenee I, Koenekoop RK, Allikmets R. Genotyping Microarray(Disease Chip) for Leber Congenital Amaurosis: Detection of Modifier Alleles. Invest. Ophthalmol. Vis. Sci. 2005;46:3052– 3059. doi: 10.1167/iovs.05-0111. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li S, Guo X, Guo L, Xiao X, Jia X, Kuang Z. Screening for CRX gene mutations in Chinese patients with Leber congenital amaurosis and mutational phenotype. Ophthalmic Genet. 2001;22:89–96. doi: 10.1076/opge.22.2.89.2227. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li S, Xiao X. An A112V mutation in the CRX gene identified in 3 Chinese family with retinal degeneration other than LCA. Invest. Ophth. Vis. Sci. 43 Supplement. 2002;1:U173–U173. [Google Scholar]
- Ziviello C, Simonelli F, Testa F, Anastasi M, Marzoli SB, Falsini B, Ghiglione D, Macaluso C, Manitto MP, Garrè C, Ciccodicola a, Rinaldi E, Banfi S. Molecular genetics of autosomal dominant retinitis pigmentosa(ADRP): a comprehensive study of 43 Italian families. J. Med. Genet. 2005;42:e47. doi: 10.1136/jmg.2005.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Yao F, Liang X, Xu F, Li H, Sui R, Dong F. De novo Mutations in the Cone-rod Homeobox Gene Associated with Leber Congenital Amaurosis in Chinese Patients. Ophthalmic Genet. 2013:1–6. doi: 10.3109/13816810.2013.827219. [DOI] [PubMed] [Google Scholar]