Abstract
A missense mutation in the calsequestrin-1 gene (CASQ1) was found in a group of patients with a myopathy characterized by weakness, fatigue and the presence of large vacuoles containing characteristic inclusions resulting from the aggregation of sarcoplasmic reticulum (SR) proteins. The mutation affects a conserved aspartic acid in position 244 (p.Asp244Gly) located in one of the high-affinity Ca2+ binding sites of CASQ1 and alters the kinetics of Ca2+ release in muscle fibers. Expression of the mutated CASQ1 protein in COS-7 cells showed a markedly reduced ability in forming elongated polymers, while both in cultured myotubes and in in-vivo mouse fibers induced the formation of electron-dense SR vacuoles containing aggregates of the mutant CASQ1 protein that resemble those observed in muscle biopsies of patients. Altogether, these results support the view that a single missense mutation in the CASQ1 gene causes the formation of abnormal SR vacuoles containing aggregates of CASQ1 and other SR proteins, results in altered Ca2+ release in skeletal muscle fibers and, hence, is responsible for the clinical phenotype observed in these patients.
Keywords: aggregate myopathy, CASQ1, calsequestrin, sarcoplasmic reticulum, skeletal muscle
INTRODUCTION
Calsequestrin (CASQ), a high capacity, moderate-affinity Ca2+ binding protein, is the main Ca2+ buffer of the sarcoplasmic reticulum (SR) of cardiac and skeletal muscle (Damiani et al., 1990; MacLennan and Wong, 1971). CASQ exists in two isoforms coded by distinct genes: CASQ1 (MIM# 114250), predominant in fast skeletal muscle fibers, and CASQ2 (MIM# 114251), predominant in slow skeletal muscle fibers and in cardiac muscle (Schiaffino and Reggiani, 2011). Both in skeletal and cardiac muscles, CASQ is part of a multiprotein complex associated with the SR Ca2+ release channel, ryanodine receptor (Rossi et al., 2008), that participates in the mechanism of excitation-contraction coupling, which links T-tubule membrane depolarization to activation of Ca2+ release from the SR. In skeletal muscles, this mechanism occurs in specialized intracellular junctions named either triads or Ca2+ release units (Rossi et al., 2006). Mutations in the CASQ2 gene have been associated to an autosomal recessive form of catecholaminergic polymorphic ventricular tachycardia (CPVT) (Lahat et al., 2001) while, so far, no disease has been associated to CASQ1 mutations. The presence of vacuoles containing inclusions that stain positive for calsequestrin has been reported in muscle biopsies from patients with an uncharacterized form of myopathy (Tomelleri et al., 2006). More recently, a potential role of CASQ1 in human diseases has been proposed based on the evidence that CASQ1-null mice develop lethal episodes similar to Malignant Hyperthermia (MH) following heat-stress and halothane exposure (Protasi et al., 2011; Dainese et al., 2009). MH is an autosomal dominant pharmacogenetic disorder triggered by volatile anaesthetics, which in about 80% of the affected individuals is caused by mutations in the RYR1 gene (MIM# 180901) (Galli et al., 2006). Up to now, mutation screening in MH patients did not result in identification of causative mutations in CASQ1 (Kraeva et al., 2013;V.S. and L.G., unpublished data). Here we report the identification of the first mutation in the CASQ1 gene in patients with a myopathy characterized by protein aggregates containing CASQ1.
MATERIALS AND METHODS
Patients and ethics statement
Eight patients (six males and 2 females), age between 23 and 58, with mild myopathy participated in this study. Ethics committee approval and written informed consent was obtained for all patients. This study complies with the ethical standards laid down in the 1964 Declaration of Helsinki. All experiments that required animals followed the official guidelines laid down by the European Community Council (directive 86/609/EEC) incorporated into Italian Government Legislation. Experiments have been performed with the approval by the Local Ethical Committee and Ministero della Salute, Rome, Italy.
Genetic analysis
Genomic DNA was extracted from peripheral blood leucocytes by standard procedures (Galli et al., 2006). Primers were designed to amplify all exons of the CASQ1 gene (NC_000001.11) with primer-3 software (http://frodo.wi.mit.edu/cgibin/primer3/primer3_www.cgi). Amplified DNA fragments were directly sequenced using an ABI Prism 310 apparatus (Applied Biosystem) as previously described (Galli et al., 2006). DNA mutation numbering was based on cDNA reference sequence (NM_001231.4), taking nucleotide +1 as the A of the ATG translation initiation codon. The mutation nomenclature follows that described at the http://www.hgvs.org./mutnomen/. The CASQ1 variant (NM_001231.4:c.731A>G) has been submitted to the locus-specific database (www.lovd.nl/CASQ1). Mutation screening of the RyR1 gene was performed as previously described in Galli et al., 2006. Haplotype analysis was based on the following intragenic SNPs rs617698, rs61803844, rs617599, rs3747621, rs3747622, rs3747623 and rs822450.
Sequence alignment and protein structure
Sequence alignment was performed using the ClustalW software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). A 3D structure of CASQ1 p.Asp244Gly was predicted using the automated protein homology-modeling server SWISS-MODEL (http://swissmodel.expasy.org/). Chain A of human CASQ1 protein (PDB: 3UOM) was used as template. The comparison between wild type CASQ1 and the p.Asp244Gly CASQ1 variant was performed using the UCSF Chimera 1.9 software (University of California).
Histology and histochemistry of patients’ muscle biopsies
All patients underwent an open biopsy of vastus lateralis muscle. Muscle specimens were cryofixed in OCT compound (Tissue-Tek-Sakura) by immersion in liquid nitrogen-cooled isopentane and stored at 80°C. Serial 10 μm-thick transversal or longitudinal sections were stained with haematoxylin and eosin (H&E), Toluidine blue, modified Gomori trichrome and reduced nicotinamide adenine dinucleotide (NADH), as previously described (Tomelleri et al., 2006; Boncompagni et al., 2006).
Preparation of samples for electron microscopy
Muscle specimens were fixed in 3.5% glutaraldehyde in 0.1 M sodium-cacodylate buffer (pH 7.4, room temperature) for 2 hours followed by buffer rinse and post fixation for 1 hour in 2% osmium tetroxide. The specimens were rapidly dehydrated in graded ethanol and acetone, infiltrated with EPON-acetone (1:1) mixture for 2 hours, and embedded in EPON (Tomasi et al., 2012). Ultrathin sections (40–50 nm) were cut and, after staining with 4% uranyl acetate and lead citrate, examined with a Morgagni Series 268D electron microscope (FEI Company, Brno, Czech Republic), equipped with a Megaview III digital camera (Boncompagni et al., 2006 and Tomelleri et al, 2006). The quantitative analysis of Ca2+ release units was performed in a biopsy from a patient (family 3:II-3), whereas age-matched samples were used as controls (Boncompagni et al., 2006). Electron micrographs of non-overlapping regions were randomly collected at the following magnifications: 18,000X for qualitative analysis of Ca2+ release unit number and 28,000X for measurements of size of SR terminal cisternae. Frequency of triads and their morphology (discriminating between triads and dyads) and average size of SR terminal cisternae lumen were determined as previously described (Boncompagni et al., 2006).
Immunofluorescence
Serial 8 μm-thick transversal cryosections or single fibers from human skeletal muscle were fixed with 3% v/v paraformaldehyde (Sigma-Aldrich) in Phosphate Buffered Saline (PBS) and permeabilized in HEPES-Triton Buffer (20 mM HEPES pH 7.4, 300 mM Sucrose, 50 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100). Cryosections or single fibers were incubated with the following primary antibodies: anti-CASQ1 mouse monoclonal antibody diluted 1:300 (clone MA3-913, Thermo Scientific), anti-RyR mouse monoclonal antibody at a final concentration of 1 μg/ml (clone 34C, Thermo Scientific), anti-RyR1 rabbit polyclonal antibody (Rossi et al., 2014), anti-SERCA monoclonal antibody diluted 1:1 (clone Y/IF4 kindly provided by Prof. M. East). Cy3 or Cy2-conjugated anti-mouse or anti-rabbit secondary antibodies (Jackson Laboratories) were used for immunofluorescence detection. Images were acquired using a confocal laser scanning microscope (ZEISS LSM 510).
African green monkey kidney-derived COS-7 cells grown on glass coverslips and transfected with CASQ1MUT-GFP or CASQ1WT-GFP (see “Generation of CASQ1 expression vectors”) were fixed after 24 hours from transfection with 3% v/v paraformaldehyde in PBS. Myotubes expressing CASQ1MUT or CASQ1WT were fixed after 12 days of differentiation with 3% v/v paraformaldehyde in PBS and permeabilized in HEPES-Triton Buffer. Cells were incubated with the previously described primary antibodies.
Single fiber Ca2+ release and force measurement
Small fragments of biopsy specimens were immersed in glycerol-containing skinning solution (Salviati and Volpe 1988) for at least 24 hours. Single fibers were manually dissected and segments of approximately 1-mm length were cut and mounted with small aluminum clips (T clips) at both ends. The experimental setup was composed of a force transducer (AE 801 Sensonor, Horten, Norway), an electromagnetic puller to control fiber length and an aluminum plate, where seven small chambers (100 μl) were milled, as previously described (Doria et al., 2011). Fibers were mounted between the force transducer and the electromagnetic puller in the first chamber filled with relaxing solution. Next the SR was loaded by moving the fiber to a chamber containing solution at pCa 6.45 in the presence of 5 mM ATP. After the fibers were washed to remove excess EGTA, calcium release was induced by transferring the fibers to a release solution with low EGTA (0.1 mM) and low free Mg2+ containing caffeine in concentrations variable from 0.1 to 20 mM. The fibers were finally transferred to activating solution to measure their ability to develop force during a maximal activation (pCa 4.7) and quickly brought back to the relaxing solution to start a new cycle of loading and release. The release of calcium was detected by the transient tension development and quantified by measuring the tension-time area according to a method first developed by Endo and Iino, 1988 and widely used. The transient is determined by the combination of the calcium release from the SR with the calcium removal due to diffusion into the medium, buffering by EGTA, and reuptake in the SR (Makabe et al, 1996). Experimental and model analyses suggest that the area gives the best indication on the amount of calcium release, whereas other parameters such as the rate of tension rise is more related to the rates of calcium release and of tension generation by cross-bridges. The tension-time area was normalized to the tension developed during maximal activation (pCa 4.7) to account for the ability of the myofibrillar apparatus to develop force and thus expressed in seconds.
Generation of CASQ1 expression vectors
Human CASQ1 cDNA (NM_001231.4) was amplified from total RNA extracted from human skeletal muscle tissue using specific primers. The cDNA coding for the Asp244Gly CASQ1 mutant (CASQ1MUT) was generated by PCR using primers designed to introduce the adenosine to guanosine nucleotide change into the wild-type CASQ1 cDNA (Forward primer: 5’-gtg acc atc cca gGc aag ccc aat agc-3’; Reverse primer: 5’-gct att ggg ctt gCc tgg gat ggt cac-3’) and sequenced using an ABI Prism 310 apparatus (Applied Biosystems). The amplified sequences were cloned into the pEGFP-N3 vector (Clontech-Zymed Laboratories Inc.) using the EcoRI- BamHI sites to generate CASQ1WT-GFP and CASQ1MUT-GFP vectors. Both cDNAs were also inserted into the pCDNA3 vector (Invitrogen) using the HindIII-BamHI sites to generate the CASQ1WT and CASQ1MUT expression vectors.
Cell culture and DNA transfection
COS-7 cells were cultured in DMEM containing 2 mM L-glutamine (Sigma-Aldrich), 100 μg/ml streptomycin (Sigma-Aldrich), 100 U/ml penicillin (Sigma-Aldrich), 1 mM sodium pyruvate (Lonza), supplemented with 10% heat inactivated fetal bovine serum (Sigma-Aldrich). Cells were grown at 37°C at 5% CO2. COS-7 cells were plated on uncoated glass coverslips and transfected with X-tremeGENE HP DNA transfection Reagent (Roche Applied Science) according to manufacturer’s instruction.
Myoblasts were obtained from hind limb muscles of 2 day-old rats (Sprague-Dawley; Harlan Laboratories) as described in Rossi et al., 2014. Cell suspension was plated on 0.025% laminin (Sigma-Aldrich) coated glass coverslips. Cells were grown at 37°C at 5% CO2. After two days, cells were transfected with the Lipofectamine-Plus method (Life Technologies) following manufacturer’s instruction. Myoblasts were induced to differentiate with Minimum Essential Medium Alpha containing 2 mM L-glutamine (Sigma-Aldrich), 100 μg/ml streptomycin (Sigma-Aldrich), 100 U/ml penicillin (Sigma-Aldrich), 1 mM sodium pyruvate (Lonza), 1 mM dexamethasone (Sigma-Aldrich), 0.05 mM hydrocortisone, supplemented with 10% heat inactivated fetal bovine serum (Sigma-Aldrich) and 5% heat-inactivated horse serum (Sigma-Aldrich).
Flexor Digitorum Brevis (FDB) fibers electroporation with plasmids encoding CASQ1WT and CASQ1MUT proteins
Mice were anesthetized with 1.25% 2,2,2-Tribromoethanol (Avertin solution, Roche) and FDB muscles injected with 10 μg of DNA. Then, two electrodes were placed at each side of the muscle and electric pulses were delivered. Twenty 120 V electric pulses with fixed duration of 20 ms and interval of 1 s were delivered using an electric pulse generator (Electro Square Porator ECM830, BTX-Genetronics, Inc.).
RESULTS
Skeletal muscle fibers from patients present vacuoles containing aggregates of SR proteins
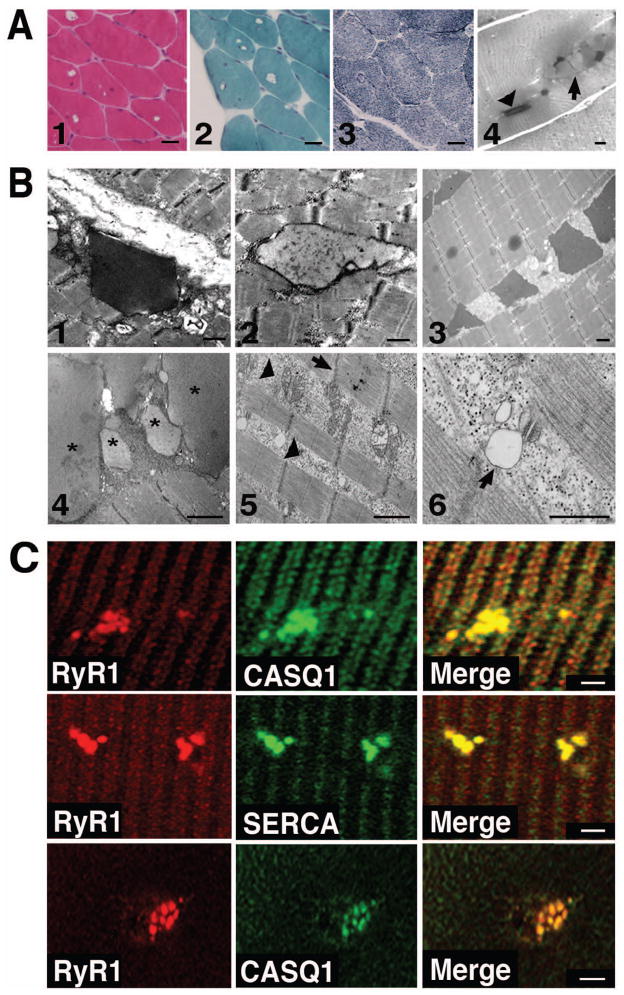
We analyzed 8 patients (7 belonging to 4 different families and 1 unrelated sporadic case) affected by a mild myopathy accompanied by one or more symptoms, including episodes of muscle cramping, reduced muscle strength, fatigue and elevated plasma creatine kinase (CK) levels. Light microscopy of muscle biopsies of all probands revealed, as a distinctive feature, the presence, mainly in type II fibers, of vacuoles that did not stain by routine histochemical analysis such as haematoxylin-eosin, Gomori-modified trichrome and reduced nicotinamide adenine dinucleotide (NADH-TR) (Fig. 1A). Occasional necrotic fibers were also observed (not shown). In longitudinal semi-thin sections stained with Toluidine Blue, two different types of inclusions with characteristic staining properties and shape were noted: type-1 inclusions, consisting of a dark-stained material with a rectangular shape (Fig. 1A, panel 4, arrowhead), and type-2 inclusions, characterized by pale staining and a rounded profile (Fig. 1A, panel 4, arrow). Electron microscopy analysis (EM) showed the presence of inclusions filled with electron-dense material that resembles CASQ accumulation in the SR (Fig. 1B) (Jones et al., 1998; Paolini et al., 2011). A significant percentage of fibers presented also areas rich in glycogen, various cytoplasmic debris and abnormal mitochondria. In some sarcomeres, Z disks appeared not parallel to the M lines (Fig. 1B). EM quantitative analysis of the density and morphology/orientation of Ca2+ release units in biopsies from one patient and two healthy individuals showed that the density of Ca2+ release units per unit area is slightly decreased in the patient’s fibers (24.1 ± 1.3 vs 30.3 ± 0.9 per 100 μm2, respectively). Also the general morphology of Ca2+ release units is changed: indeed a large percentage of triads (40.1 ± 2.6% in patient vs 5.8 ± 1.1% in controls) appears to miss one SR element. Similar alterations have been described in human biopsies from elderly individuals (Boncompagni et al., 2006). Finally, the average size of the SR terminal cisternae is slightly wider than in control (105.6 ± 1.9 nm vs. 86.4 ± 0.9 nm, respectively, p<0.01): this SR swelling could possibly represent an early step in the formation of inclusions. Occasional triads showing a bobbled junctional SR that appears clear and apparently empty were observed (Fig. 1B). Single fibers or cross sections from patients’ biopsies were stained with antibodies against CASQ1, RyR1 and SERCA. As shown in Fig. 1C, beside the cross striations typically observed with SR proteins, aggregates (consistent with inclusions found in EM), positive for all three antibodies were observed. Altogether, these results indicated that the inclusions present in muscle biopsies of the patients here reported are similar to those described in Tomelleri et al., 2006.
Figure 1. Morphological characterization of skeletal muscle fibers from patients.
A. Hystological analysis. Hematoxilin-eosin (1), Gomori-modified trichrome (2) and NADH-TR (3) stainings of muscle fibers of patient II:1 of family 2. Longitudinal sections from a biopsy of patient II:3 of family 3 stained with Toluidine Blue (4) showing rectangular dark inclusions defined as type 1 (arrowhead), and round pale inclusion defined as type 2 (arrow) (Tomelleri et al.,2006) . Scale bars: panels 1–3 = 40 μm, panel 4 = 5 μm.
B. Electron microscope analysis. Representative EM images of type 1 and type 2 inclusions from a biopsy of patient II:1 of family 2 (1 and 2) and of type 1 inclusions from a muscle biopsy of the sporadic patient (3). Representative EM images of patient II:3 of family 3 showing type 2 inclusions labelled with asterisks (4) and areas containing cytoplasmic debris and abnormal mitochondria (arrowheads in 5). The arrow (5) points to a misaligned Z disk. A vacuole clearly connected to the SR membrane of a Ca2+ release units is indicated with an arrow (6). Scale bars = 1 μm. C. Immunofluorescence analysis. Single muscle fibers (upper and middle panels) or cross sections (lower panels) of skeletal muscle biopsy of patient I:1 of family 2 stained with antibodies against CASQ1, SERCA andRyR1. Merged images show co-localization of CASQ1, RyR1 and SERCA in the aggregates. Scale bars = 2 μm.
A common variant in the CASQ1 gene is present in all patients
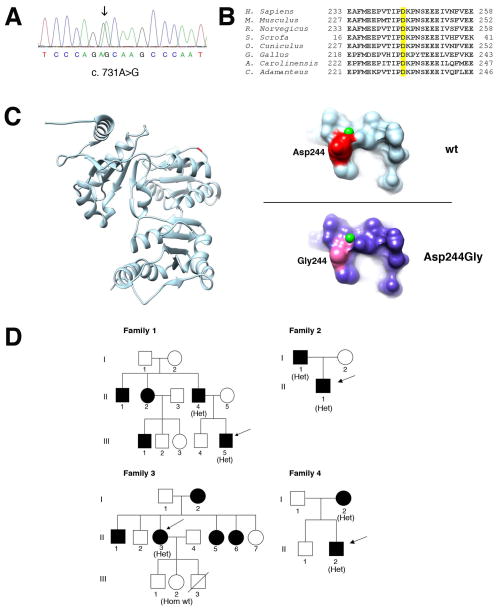
Sequence analysis of the CASQ1 gene (NC_000001.11) in five probands revealed an heterozygous variant c.731A>G (Fig. 2A) resulting in an amino acid change from aspartic acid to glycin (p.Asp244Gly). Each CASQ1 molecule can bind 70–80 Ca2+ ions via high- and low-affinity Ca2+ binding sites (Kumar et al., 2013; Sanchez et al., 2012). Of notice, the aspartic acid 244 is conserved among orthologs in species from snakes to humans (Fig. 2B) and contributes to form one of the high-affinity Ca2+ binding sites in CASQ1 (Sanchez et al., 2012). Indeed, substitution of the aspartic acid at codon 244 to glycin is predicted to result in a conformational change in this Ca2+ binding region (Fig. 2C). The variant segregated with the disease phenotype in the four families analyzed (Fig. 2D). The c.731A>G variant was found in the 33 years old proband of family 1, who had a history of mild myalgia, cramps and a 5-fold increase of serum CK levels. The same symptoms were also reported for several proband’s relatives, thus indicating recurrence of the disease. Unfortunately, DNA samples from other family members were not available for genetic analysis. The variant was also found in the 23 years old proband of family 2, who showed mild proximal weakness in the upper limbs and increased serum CK levels, and in his affected father, who reported onset of clinical symptoms in his sixties with a mild increase in serum CK levels. The same variant was present also in the 58 years old proband of family 3, who had a mild increase of serum CK levels (2- to 3-fold the upper normal limit). Two years before, she had suffered painful contractures in the posterior left thigh muscles that spontaneously disappeared within 12 months. One sister (II:5) had a diagnosis of vacuolar myopathy made elsewhere, but EM examination of muscle fibers was not performed. The same symptoms were also reported for other members of this family, but DNA samples were not available for genetic analysis, with the exception of the proband’s healthy daughter (III:2), who did not carry the c.731A>G variant. This family was already described in Tomelleri et al., 2006. The variant was detected in the 34 years old proband of family 4, who presented mild myalgia on thighs at rest and increased CK levels (about 6-fold the upper normal limit). The variant was also detected in his 57 years old mother that presented an asymptomatic increase of serum CK level of about 3-fold the upper normal value. The brother of this proband was apparently healthy and presented normal CK values. Finally, the same variant was detected in one patient of 44 years, who reported a ten months history of exercise-induced muscle pain, early fatigue in daily activities and increased CK levels, about 16-fold the upper normal value. Unfortunately, no clinical information was available about relatives of this patient. In spite of the evidence that Casq1 deletion in mice causes a MH-like phenotype, there was no family history of MH in these four families and screening of the RYR1 gene did not result in detection of MH causative mutations in the five probands. Furthermore, screening of additional 93 patients including individuals with Idiopathic Hyper CKemia and of 21 individuals with tubular-aggregate myopathy did not identify any variant in the CASQ1 gene. The c.731A>G variant was not found in more than 400 control DNAs and is not reported in the 1000 genomes database (http://www.1000genomes.org/), in NHLBI Exome Sequencing Project Exome Variant Server (evs.gs.washington.edu/EVS/) or in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Figure 2. Genetic and molecular analysis of the p.Asp244Gly mutation.
A. Electropherogram of the CASQ1 gene sequence of one patient. The patient is heterozygous for the c.731A>G variant (arrow).
B. Phylogenetic alignment of the CASQ1 orthologs. The aspartic acid in position 244 in the human CASQ1 is highlighted yellow and is conserved in species ranging from snakes (C. Adamanteus) to humans.
C. 3D structure of human CASQ1 protein. Ribbon representation of the model of chain A of the human CASQ1 3D structure (PDB:3UOM) with the Asp244 shown in red/dark (left). On the right, detailed views of predicted surface structure of the local molecular environments surrounding the Asp244 (upper panel) or the Gly244 residues (lower panel). The small (green) spheres represent Ca2+ ions. D. Pedigrees of families. Black filled symbols represent affected family members. The genotype of individuals is shown in parenthesis as follows: Het, heterozygous for the c.731A>G variant; Hom wt, homozygous for the wild-type allele. Arrows indicate probands.
Haplotype investigation was performed analyzing 7 intragenic SNPs (Supp. Table S1). Concerning families 2, and 4, homozygosity for all SNPs in one of the two analyzed individuals allowed us to reconstruct haplotypes, evidencing the presence of the same GGATGTA haplotype carrying the mutation. Conversely, in families 1 and 3, it was not possible to determine the correct phase for all SNPs. However, considering the high level of linkage disequilibrium present in this genomic region (HapMap data, Release 27, phase II+III, Feb 09; http://hapmap.ncbi.nlm.nih.gov/), the observed genotypes at the polymorphic loci may be compatible with the same haplotype associated with the mutation in families 2 and 4 (Supp. Table S1), thus suggesting the existence of a common ancestor. We also analyzed the same SNPs in the sporadic patient; in this case, however, two SNPs (rs3747623 and rs822450) resulted to be incompatible with the haplotype associated with the mutation in families 2 and 4, thus suggesting the existence of a different haplotype. Noteworthy, the four families are from two neighboring cities in the north east of Italy, while the sporadic patient is originally from an island in the south of Italy.
Single muscle fibers from two patients with the p.Asp244Gly mutation show altered Ca2+ release kinetics from the SR
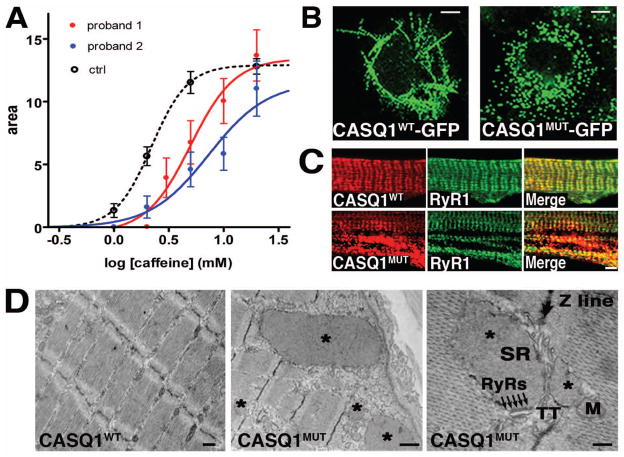
To correlate SR alterations to the myopathic phenotype, the ability of taking up and releasing Ca2+ from the SR was studied in 15 single muscle fibers isolated from a biopsy of patient I:1 of family 2 (Fig. 2D) and in 21 fibers from a biopsy of the sporadic case, and compared with fibers dissected from 4 healthy subjects of comparable age. A protocol of cyclic SR loading in the presence of ATP at low pCa2+ and releasing Ca2+ with increasing concentrations of caffeine in low Mg2+, low EGTA medium was applied (Salviati and Volpe, 1988). Tension-time area was adopted as a reliable indicator of the amount of Ca2+ released (Lamb et al., 2001; Endo and Iino, 1988; Salviati and Volpe, 1988). As can be seen in Fig. 3A, average dose-response curves show that the amplitude of the maximal release (20 mM caffeine) was not significantly different in controls’ and patients’ fibers, while the responsiveness to caffeine was significantly reduced in a balanced pool of fast and slow fibers of both patients in comparison to a pool of healthy control fibers (Lamb et al., 2001; Salviati and Volpe, 1988). While the similar maximal release in the probands’ fibers and in the control fibers plays against reduced Ca2+ content of the SR, the difference in EC50 points to an altered regulatory action of CASQ1 on RyR1 opening.
Figure 3. Ca2+ release kinetics in muscle fibers of two patients and expression of recombinant CASQ1WT and CASQ1MUT proteins.
A. Ca2+ release kinetics in muscle fibers of two patients. Dose-response curves of permeabilized muscle fibers from control healthy subjects (black dashed line, average of 48 fibers from 4 subjects), patient I:1 of family 2 (continuous red line, average of 15 fibers) and from the sporadic case (continuous blue line, average of 21 fibers) exposed to increasing concentrations of caffeine (from 0.1 to 20 mM). Curves are interpolated by a sigmoidal Hill equation: . The parameter logEC50 is significantly greater (p<0.01) in the patient I:1 of family 2 (0.697+/− 0.062) and in the sporadic case (0.863+/− 0.101) than in controls (0.343 +/− 0.006). B. CASQ1WT-GFP and CASQ1MUT-GFP were expressed in COS-7 cells. Living cells were imaged by confocal laser scan microscopy. Scale bars = 7 μm. C. Rat myotubes expressing recombinant CASQ1WT and CASQ1MUT were immunostained with antibodies against CASQ1 and RyR1. Scale bar = 2 μm. D. EM analysis of CASQ1WT and CASQ1MUT in mouse FDB fibers. Fibers expressing CASQ1MUT show the presence of vacuoles filled with electron-dense material (asterisks). These vacuoles were not present in fibers expressing CASQ1WT. Higher magnification shows enlarged vesicles derived from the SR terminal cisternae: arrows point to RyR-feet between an enlarged SR vesicle and a T-tubule. M, mitochondrion; TT, transverse tubule; SR, sarcoplasmic reticulum. Scale bars in left, middle and right panels = 0.5, 1 and 0.1 μm, respectively.
Expression of recombinant mutated CASQ1 protein causes the formation of large intracellular aggregates
A cDNA coding for CASQ1 containing the Asp244Gly mutation was cloned in frame with the Green Fluorescent Protein (GFP) sequence (CASQ1MUT-GFP). As control, a GFP-tagged wild-type CASQ1 protein was generated (CASQ1WT-GFP). Vectors expressing CASQ1MUT-GFP and CASQ1WT-GFP were transfected in COS-7 cells and fusion proteins visualized by confocal laser scan microscopy. As shown in Fig. 3B, CASQ1WT-GFP formed linear aggregates, which were never seen in cells expressing CASQ1MUT-GFP protein (Fig. 3B). Generally, the CASQ molecule exists as either a monomer or a wide range of high molecular mass polymeric structures, depending on the ionic environment. Formation of polymers is mediated by front-to-front and back-to-back contacts between the CASQ monomers and is promoted by Ca2+ (Kim et al., 2007; Park et al., 2003). The regular linear structures observed in cells expressing CASQ1WT-GFP thus appear to reflect the assembly of large CASQ1 polymers in physiological Ca2+ concentration. On the contrary, lack of these linear structures in cells expressing CASQ1MUT-GFP proteins suggests that the mutation affects CASQ1 polymerization. We next expressed the recombinant CASQ1WT and CASQ1MUT proteins, without the GFP tag, in cultured primary rat myoblasts that were induced to differentiate for 12 days. Staining with a monoclonal antibody against CASQ1 indicated that both proteins co-localized with RyR1 at the junctional regions of the SR. However, CASQ1MUT expression resulted also in the appearance of large intracellular aggregates strongly positive for CASQ1 (Fig. 3C), similar to those found in muscle biopsies of patients. Finally, EM analysis of mouse Flexor Digitorum Brevis (FDB) fibers electroporated with vectors expressing CASQ1MUT (Fig. 3D), showed the presence of inclusions containing electron-dense material similar to those observed in patients carrying the p.Asp244Gly mutation. The size of these inclusions was quite variable and a higher magnification revealed that i) these are enlarged vesicles of SR origin as they are surrounded by a membrane and bear RyRs in the sites of contact with T-tubule (Fig. 3D), and ii) they are filled with an electron-dense matrix, which is identical to accumulation of over-expressed CASQ (Tomasi et al., 2012). Similar aggregates were not found in fibers transfected with the CASQ1WT protein (Fig. 3D).
DISCUSSION
Protein aggregate myopathies (PAM) are an emerging group of diseases characterized by structural abnormalities resulting from the aggregation of intrinsic proteins within muscle fibers. These aggregates are likely started by the aggregation of a mutant protein (e.g. desmin) that may also recruit additional normal proteins (Goebel and Blaschek, 2011). The principles governing protein aggregation and induction of the structural alterations found within muscle fibers are still largely unknown. However, the recent identification of mutations in causative genes is shedding light on the molecular mechanisms that result in aggregate formation in these myopathies (Schroeder, 2013; Goebel and Blaschek, 2011; North, 2008).
The p.Asp244Gly mutation in CASQ1, found in patients with a vacuolar myopathy characterized by distinctive SR protein aggregates in muscle fibers, represents the first mutation in CASQ1 linked to an inherited human muscle disease. This mutation affects the aspartic acid at position 244 in CASQ1 that, in addition to form a high-affinity Ca2+ binding site, is also close to a region of the protein proposed to be relevant for the interaction between adjacent monomers (Kim et al., 2007). In close analogy with our results, the p.Arg33Gln mutation in CASQ2, identified in a patient with CPVT, appears also to affect a high-affinity Ca2+ binding site and to interfere with assembly of longitudinal polymers, resulting in increased aggregate formation (Kim et al., 2007). Indeed, several studies have been performed on both CASQ2 carrying mutations found in patients with CPVT and on CASQ1 containing selected mutations addressing specific structural domains: these mutated forms of CASQ showed an anomalous polymerization behavior, increased propensity to form aggregates and displayed altered subcellular localization with reduced Ca2+ binding ability (Subra et al., 2012; Cho et al., 2007; Kim et al., 2007; Park et al., 2003; Wang et al., 1998). It would then appear that mutations that alter Ca2+ binding ability and/or polymerization may reduce the ability of CASQ proteins to remain in solution and to efficiently store Ca2+ (Kumar et al., 2013; Kim et al, 2007). Considering that CASQ1 polymers have a role in sustaining the rate of Ca2+ release in muscle contraction (Rossi and Dirksen, 2006), the reduced ability to polymerize and the increased tendency to form large aggregates observed in the CASQ1 mutant protein might explain the altered Ca2+ release kinetics observed in muscle fibers from two patients with the p.Asp244Gly mutation. These functional modifications, together with the overall structural alterations observed in muscle fibers from patients’ biopsies, may then result in the myopathic phenotype observed. These results indicate that, similarly to what observed with other aggregate myopathies, the mutated CASQ1 might form the seed for aggregation of the other SR proteins detected in the inclusions observed in patients with this myopathy. Thus, in agreement with previous terminology (Schröder, 2013; Goebel and Blaschek, 2011), we propose that this myopathy should be referred to as Calsequestrin Aggregate Myopathy.
Supplementary Material
Acknowledgments
Contract grant sponsors: Telethon (GGP13213), U.S. National Institute of Health (AR059646 and AR053349); Ministero dell’ Istruzione, dell’ Università e della Ricerca, Fondo Innovazione in Ricerca.
This work was supported by Telethon grant GGP13213 to FP, VS, and CR; by U.S. National Institute of Health Grants AR059646 and AR053349 (subcontracts to FP) and a by MIUR-FIR grant to EP. "The EuroBioBank and Telethon Network of Genetic Biobanks (GTB07001D) are gratefully acknowledged for providing biological samples". We thank Dr. Rosanna Asselta (University of Milan) for helpful advice and assistance in haplotype analysis.
References
- Boncompagni S, d’Amelio L, Fulle S, Fanò G, Protasi F. Progressive disorganization of the excitation-contraction coupling apparatus in ageing human skeletal muscle as revealed by electron microscopy: a possible role in the decline of muscle performance. J Gerontol A Biol Sci Med Sci. 2006;61:995–1008. doi: 10.1093/gerona/61.10.995. [DOI] [PubMed] [Google Scholar]
- Cho JH, Ko KM, Singaruvelu G, Lee W, Kang GB, Rho SH, Park BJ, Yu JR, Kagawa H, Eom SH, Kim do H, Ann J. Functional importance of polymerization and localization of calsequestrin in C. elegans. J Cell Sci. 2007;120:1551–1558. doi: 10.1242/jcs.001016. [DOI] [PubMed] [Google Scholar]
- Dainese M, Quarta M, Lyfenko AD, Paolini C, Canato M, Reggiani C, Dirksen RT, Protasi F. Anesthetic- and heat-induced sudden death in calsequestrin-1-knockout mice. FASEB J. 2009;23:1710–1720. doi: 10.1096/fj.08-121335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani E, Volpe P, Margreth A. Coexpression of two isoforms of calsequestrin in rabbit slow-twitch muscle. J Muscle Res Cell Motil. 1990;11:522–530. doi: 10.1007/BF01745219. [DOI] [PubMed] [Google Scholar]
- Doria C, Toniolo L, Verratti V, Cancellara P, Pietrangelo T, Marconi V, Paoli A, Pogliaghi S, Fano G, Reggiani C, Capelli C. Improved VO2 uptake kinetics and shift in muscle fiber type in high altitude trekkers. J Appl Physiol. 2011;111:1597–605. doi: 10.1152/japplphysiol.01439.2010. [DOI] [PubMed] [Google Scholar]
- Endo M, Iino M. Measurement of Ca2+ release in skinned fibers from skeletal muscle. Methods Enzymol. 1988;157:12–26. doi: 10.1016/0076-6879(88)57064-7. [DOI] [PubMed] [Google Scholar]
- Galli L, Orrico A, Lorenzini S, Censini S, Falciani M, Covacci A, Tegazzin V, Sorrentino V. Frequency and localization of mutations in the 106 exons of the RYR1 gene in 50 individuals with malignant hyperthermia. Hum Mutat. 2006;27:830–839. doi: 10.1002/humu.9442. [DOI] [PubMed] [Google Scholar]
- Goebel HH, Blaschek A. Protein aggregation in congenital myopathies. Semin Pediatr Neurol. 2011;18:272–276. doi: 10.1016/j.spen.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Jones LR, Suzuki YJ, Wang W, Kobayashi YM, Ramesh V, Franzini-Armstrong C, Cleemann L, Morad M. Regulation of Ca2+ signaling in transgenic mouse cardiac myocytes overexpressing calsequestrin. J Clin Invest. 1998;101:1385–1393. doi: 10.1172/JCI1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Youn B, Kemper L, Campbell C, Milting H, Varsanyi M, Kang C. Characterization of human cardiac calsequestrin and its deleterious mutants. J Mol Biol. 2007;373:1047–1057. doi: 10.1016/j.jmb.2007.08.055. [DOI] [PubMed] [Google Scholar]
- Kraeva N, Zvaritch E, Frodis W, Sizova O, Kraev A, MacLennan DH, Riazi S. CASQ1 gene is an unlikely candidate for malignant hyperthermia susceptibility in the North American population. Anesthesiology. 2013;118:344–349. doi: 10.1097/01.anes.0000530185.78660.da. [DOI] [PubMed] [Google Scholar]
- Kumar A, Chakravarty H, Bal NC, Balaraju T, Jena N, Misra G, Bal C, Pieroni E, Periasamy M, Sharon A. Identification of calcium binding sites on calsequestrin 1 and their implications for polymerization. Mol Biosyst. 2013;9:1949–1957. doi: 10.1039/c3mb25588c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahat H, Pras E, Olender T, Avidan N, Ben-Asher E, Man O, Levy-Nissenbaum E, Khoury A, Lorber A, Goldman B, Lancet D, Eldar M. A missense mutation in a highly conserved region of CASQ2 is associated with autosomal recessive catecholamine-induced polymorphic ventricular tachycardia in Bedouin families from Israel. Am J Hum Genet. 2001;69:1378–1384. doi: 10.1086/324565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb GD, Cellini MA, Stephenson DG. Different Ca2+ releasing action of caffeine and depolarisation in skeletal muscle fibres of the rat. J Physiol. 2001;531:715–728. doi: 10.1111/j.1469-7793.2001.0715h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan DH, Wong PT. Isolation of a calcium-sequestering protein from sarcoplasmic reticulum. Proc Natl Acad Sci U S A. 1971;68:1231–1235. doi: 10.1073/pnas.68.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makabe M, Werner O, Fink RH. The contribution of the sarcoplasmic reticulum Ca2+-transport ATPase to caffeine-induced Ca2+ transients of murine skinned skeletal muscle fibres. Pflugers Arch. 1996;432:717–726. doi: 10.1007/s004240050190. [DOI] [PubMed] [Google Scholar]
- North K. What’s new in congenital myopathies? Neuromuscul Disord. 2008;18:433–442. doi: 10.1016/j.nmd.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Paolini C, Quarta M, D'Onofrio L, Reggiani C, Protasi F. Differential effect of calsequestrin ablation on structure and function of fast and slow skeletal muscle fibers. J Biomed Biotech. 2011 doi: 10.1155/2011/634075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Wu S, Dunker AK, Kang C. Polymerization of calsequestrin. J Biol Chem. 2003;278:16176–16182. doi: 10.1074/jbc.M300120200. [DOI] [PubMed] [Google Scholar]
- Protasi F, Paolini C, Canato M, Reggiani C, Quarta M. The lesson of Calsequestrin-1 ablation in vivo: much more that a buffer, after all. J Mus Res Cell Mot. 2011;32:257–270. doi: 10.1007/s10974-011-9277-2. [DOI] [PubMed] [Google Scholar]
- Rossi AE, Dirksen RT. Sarcoplasmic reticulum: the dynamic calcium governor of muscle. Muscle Nerve. 2006;33:715–31. doi: 10.1002/mus.20512. [DOI] [PubMed] [Google Scholar]
- Rossi D, Barone V, Giacomello E, Cusimano V, Sorrentino V. The sarcoplasmic reticulum: an organized patchwork of specialized domains. Traffic. 2008;9:1044–1049. doi: 10.1111/j.1600-0854.2008.00717.x. [DOI] [PubMed] [Google Scholar]
- Rossi D, Bencini C, Maritati M, Benini F, Lorenzini S, Pierantozzi E, Scarcella AM, Paolini C, Protasi F, Sorrentino V. Distinct regions of triadin are required for targeting and retention at the junctional domain of the sarcoplasmic reticulum. Biochem J. 2014;458:407–417. doi: 10.1042/BJ20130719. [DOI] [PubMed] [Google Scholar]
- Salviati G, Volpe P. Ca2+ release from sarcoplasmic reticulum of skinned fast- and slow-twitch muscle fibers. Am J Physiol. 1988;254:C459–C465. doi: 10.1152/ajpcell.1988.254.3.C459. [DOI] [PubMed] [Google Scholar]
- Sanchez EJ, Lewis KM, Danna BR, Kang C. High-capacity Ca2+ binding of human skeletal calsequestrin. J Biol Chem. 2012;287:11592–11601. doi: 10.1074/jbc.M111.335075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Schröder R. Protein aggregate myopathies: the many faces of an expanding disease group. Acta Neuropathol. 2013;125:1–2. doi: 10.1007/s00401-012-1071-8. [DOI] [PubMed] [Google Scholar]
- Subra AK, Nissen MS, Lewis KM, Muralidharan AK, Sanchez EJ, Milting H, Kang C. Molecular mechanisms of pharmaceutical drug binding into calsequestrin. Int J Mol Sci. 2012;13:14326–14343. doi: 10.3390/ijms131114326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi M, Canato M, Paolini C, Dainese M, Reggiani C, Volpe P, Protasi F, Nori A. Calsequestrin (CASQ1) rescues function and structure of calcium release units in skeletal muscles of CASQ1-null mice. Am J Physiol Cell Physiol. 2012;302:C575–586. doi: 10.1152/ajpcell.00119.2011. [DOI] [PubMed] [Google Scholar]
- Tomelleri G, Palmucci L, Tonin P, Mongini T, Marini M, L'Erario R, Rizzuto N, Vattemi G. SERCA1 and calsequestrin storage myopathy: a new surplus protein myopathy. Brain. 2006;129:2085–2092. doi: 10.1093/brain/awl128. [DOI] [PubMed] [Google Scholar]
- Wang S, Trumble WR, Liao H, Wesson CR, Dunker AK, Kang CH. Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat Struct Biol. 1998;5:476–483. doi: 10.1038/nsb0698-476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.