Abstract
AMPK exerts pro-longevity effects in diverse species; however, the tissue-specific mechanisms involved are poorly understood. Here, we show that up-regulation of AMPK in the adult Drosophila nervous system induces autophagy both in the brain and also in the intestinal epithelium. Induction of autophagy is linked to improved intestinal homeostasis during aging and extended lifespan. Neuronal up-regulation of the autophagy-specific protein kinase Atg1 is both necessary and sufficient to induce these inter-tissue effects during aging and to prolong lifespan. Furthermore, up-regulation of AMPK in the adult intestine induces autophagy both cell autonomously and non-autonomously in the brain, slows systemic aging and prolongs lifespan. We show that the organism-wide response to tissue-specific AMPK/Atg1 activation is linked to reduced insulin-like peptide levels in the brain and a systemic increase in 4E-BP expression. Together, these results reveal that localized activation of AMPK and/or Atg1 in key tissues can slow aging in a cell-non-autonomous manner.
Keywords: Autophagosome, DILP2, intestinal integrity, longevity, protein homeostasis, S6K, TOR
Reduced nutrient intake (dietary restriction or DR) or genetic/pharmacological interventions that mimic energy deprivation can extend lifespan in diverse species (Fontana et al., 2010). Macroautophagy (hereafter referred to as autophagy) is a process in which cytoplasmic substrates are degraded under conditions of nutrient limitation, allowing cellular macromolecules to be catabolized and recycled. In recent years, the induction of autophagy has emerged as a unifying downstream feature of several evolutionarily conserved anti-aging interventions (Gelino and Hansen, 2012; Rubinsztein et al., 2011), including both DR and reduced target of rapamycin (TOR) signaling (Alvers et al., 2009; Bjedov et al., 2010; Hansen et al., 2008; Jia and Levine, 2007; Matecic et al., 2010; Toth et al., 2008). However, fundamental questions remain regarding the relationships between cellular energy homeostasis, tissue-specific autophagy induction and lifespan determination.
AMP-activated protein kinase (AMPK) is the principal energy sensor in eukaryotic cells and functions to maintain cellular energy homeostasis (Hardie et al., 2012). The functional AMPK is a heterotrimer consisting of a catalytic alpha (α), a regulatory gamma (γ), and a scaffolding beta subunit (β) and is activated by low cellular energy status. Upon activation, AMPK promotes ATP production by increasing the activity or expression of proteins involved in catabolism while switching off biosynthetic pathways (Hardie et al., 2012). More specifically, AMPK has been shown to directly phosphorylate the mammalian TOR (mTOR) binding partner raptor thereby inhibiting mTOR pathway activity (Gwinn et al., 2008). It was originally assumed that AMPK activation induces autophagy indirectly via reduced TOR signaling (Hardie, 2011). However, more recently it has been shown that AMPK also mediates autophagy by direct phosphorylation of the protein kinase that initiates autophagy, ULK1 (one of the two mammalian orthologs of Atg1) (Egan et al., 2011; Kim et al., 2011). In addition to its role in metabolic regulation, AMPK plays a key role in aging and lifespan determination (Burkewitz et al., 2014). In C elegans, increased gene dosage of the AMPK catalytic subunit, aak-2, increases lifespan (Apfeld et al., 2004; Mair et al., 2011), several DR protocols require aak-2 to prolong lifespan (Greer and Brunet, 2009; Greer et al., 2007a) and the AMPK complex is the key mediator of the synergistically prolonged longevity produced by reduced TOR and insulin-like signaling (Chen et al., 2013). Recently, it was reported that muscle- or fat body-specific up-regulation of AMPK can extend lifespan in the fruit fly Drosophila (Stenesen et al., 2013). However, the driver lines used to up-regulate AMPK in this study are both expressed in the intestine as well as the fat body/muscle (Poirier et al., 2008; Rera et al., 2013) and the impact of neuron-specific AMPK activation on lifespan has not been reported in any species. Therefore, the tissue-specific requirements for AMPK-mediated lifespan extension remain unclear.
In this study, we have investigated the impact of tissue-restricted expression of an AMPKα transgene on AMPK activity, autophagy, tissue homeostasis and lifespan in Drosophila. We show that neuronal AMPK activation inhibits TOR and induces autophagy in the brain and, at the same time, prolongs lifespan. To determine whether neuronal AMPK-mediated lifespan extension proceeds via a cell autonomous or non-autonomous mechanism, we examined markers of aging in distal tissues upon neuronal activation of AMPK. Importantly, we show that neuronal activation of AMPK leads to improved tissue homeostasis in the aging intestine, which is linked to the cell non-autonomous induction of autophagy in the intestinal epithelium. Furthermore, we show that up-regulation of Atg1, a direct initiator of autophagy (Scott et al., 2007), in adult neurons is both necessary and sufficient to improve intestinal homeostasis during aging and prolong lifespan. Interestingly, intestine-specific AMPK up-regulation can also modulate aging; both in a cell-autonomous and non-autonomous manner. Indeed, intestine-specific AMPK up-regulation activates autophagy in the intestine and also produces a cell-non-autonomous induction of autophagy in the brain, reduces proteotoxicity in aged muscle tissue and prolongs lifespan. We show that the inter-tissue effects conferred by AMPK/Atg1 are linked to altered insulin-like peptide signaling. Our findings suggest that tissue-specific induction of AMPK and/or autophagy could slow aging in a cell-non-autonomous manner in other species, including humans.
Results
Neuronal up-regulation of AMPK induces autophagy in the brain and increases lifespan
To better understand the relationship between neuronal energy homeostasis and organismal aging, we sought to examine the impact of neuron-specific up-regulation of the catalytic (α) AMPK subunit (hereafter referred to as AMPK) on Drosophila lifespan. To do so, we used the RU486-inducible, pan-neuronal Elav-Gene-Switch (GS) driver line (Poirier et al., 2008) to activate a wild-type UAS-AMPK transgene created by (Lee et al., 2007), specifically in adult neurons. To confirm that transgenic expression of AMPK can increase AMPK activity in nervous tissue, we measured phosphorylation of the catalytic subunit of AMPK at Thr184 in head samples. Western blot analysis using a phosphospecific antibody revealed a significant increase in phospho-Thr184-AMPK levels in head tissue lysates from ElavGS>UAS-AMPK flies upon RU486 treatment compared to uninduced controls (Fig. 1A). Control flies fed RU486 showed no difference in phosphorylation of AMPK (Fig. S1A). Next, we examined whether neuronal AMPK activation is sufficient to extend lifespan. Adult-onset, neuronal up-regulation of AMPK resulted in increases in median lifespan in female flies and had variable effects on male lifespan (Figure 1B, Table S1). Using an independently generated UAS-AMPK transgene that is tagged with the red fluorescent protein mCherry (Mirouse et al., 2007), we also observed increased female lifespan upon RU486 feeding (Figure 1C, Table S1). No lifespan increase was observed in control flies exposed to RU486 (Fig. S1B, Table S1). As both the tagged and untagged AMPK transgenes can prolong lifespan when induced in adult neurons, we focused on the mCherry (mCh)-tagged AMPK as it facilitates the detection of transgene (as opposed to the endogenous gene) expression in different tissues. As the AMPK-mediated longevity effects were stronger in females, we focused on female flies throughout the rest of this study.
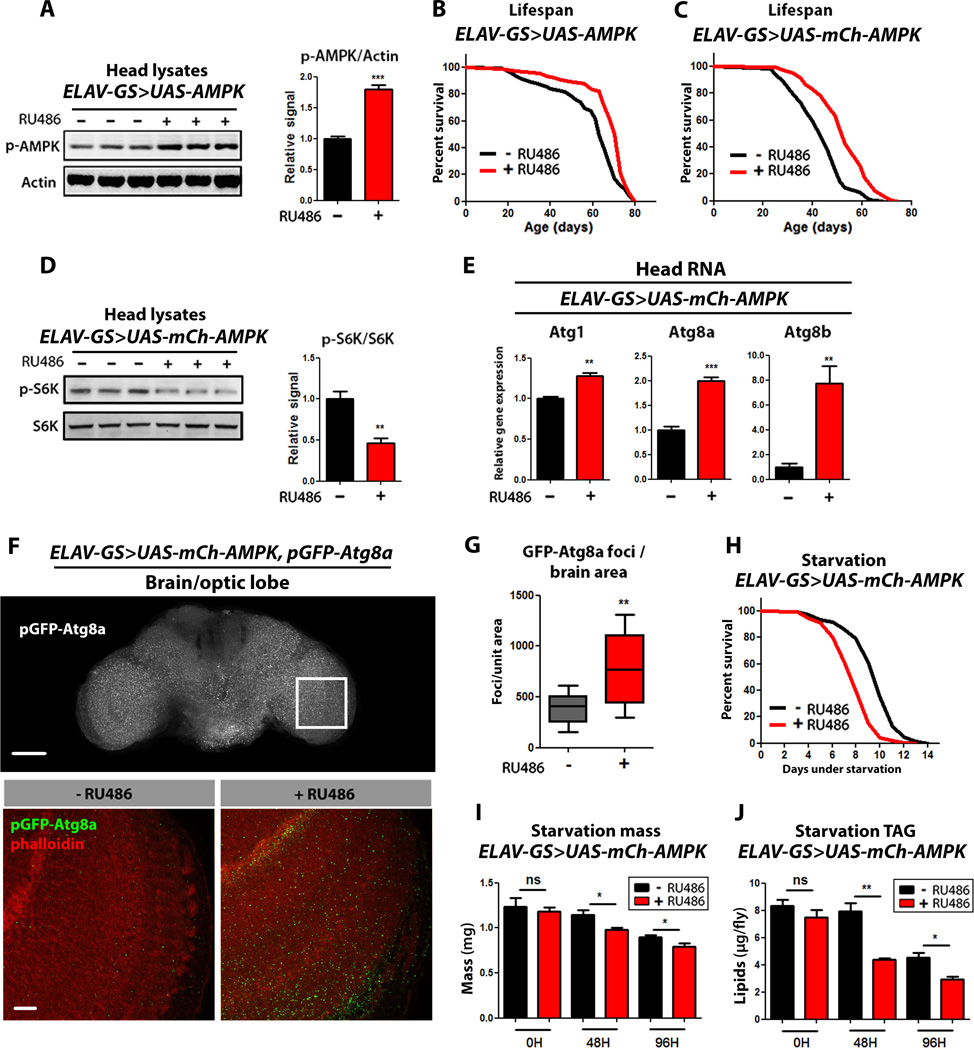
Figure 1. Neuronal AMPK activation extends lifespan.
(A) Western blot analysis of AMPK phosphorylated on T184 (p-AMPK) and loading control (actin) from head lysates of 10 day old ELAV-GS>UAS-AMPK female flies with or without RU486-mediated transgene induction. Densitometry quantification (right) (p<0.0001; t-test; n=3 replicates; 10 heads/replicate).
(B) Survival curves of ELAV-GS>UAS-AMPK females with or without RU486-mediated transgene induction (p<0.0001; log-rank test; n> 106 flies).
(C) Survival curves of ELAV-GS>UAS-mCh-AMPK females with or without RU486-mediated transgene induction (p<0.0001 log-rank test; n> 159 flies).
(D) Western blot analysis of S6K phosphorylated at T398 and total S6K from head lysates of 10 day old ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. Densitometry quantification (right) (p<0.0098; t-test; n=3 replicates; 10 heads/replicate).
(E) Expression of autophagy genes in head tissue of 10 day old ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (t-test; n=3 of RNA extracted from 10 heads/replicate).
(F) GFP-Atg8a localization in adult brain with 10X objective (above) and representative images (below) from optic lobes of 10 day old ELAV-GS>UAS-mCh-AMPK, pGFP-Atg8a female flies with or without RU486-mediated transgene induction (red channel-phalloidin, green channel-GFP-Atg8a, upper scale bar represents 50µm, lower scale bar represents 10µm).
(G) Quantification of brain GFP-Atg8a foci (p<0.004; t-test; n>10 confocal stacks from optic lobes/condition; one brain/replicate stack).
(H) Survival curves without food of ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction (p<0.0001; log-rank; n>122 flies/condition).
(I) Body mass during starvation of ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction (p<0.05, at 48hours and 96 hours of starvation; t-test; n>6 samples/condition; 10 flies weighed/sample).
(J) Whole body lipid stores during starvation of ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.01 at 48 hours, and p<0.05 at 96 hours of starvation; t-test; n>3 samples/condition/timepoint; lipids extracted from 5 flies per sample).
Data are represented as mean ± SEM. RU486 was provided in the media after eclosion at a concentration of 50 µg/ml (A, B) and 25µg/ml (C–J)
To explore the effects of increased AMPK activity in the adult nervous system on downstream pathways, we first measured phosphorylation of the S6 ribosomal subunit Kinase (S6K), a well characterized downstream target of TOR kinase, by western blotting using a phospho-Thr398-dependent S6K antibody. We observed reduced levels of phospho-T398-S6K in head lysates of ElavGS>UAS-mCh-AMPK flies upon RU486 treatment compared to uninduced controls (Fig. 1D) suggesting that TOR signaling is down regulated in adult neurons upon AMPK activation. Control flies exposed to RU486 showed no difference in S6K phosphorylation (Fig. S1C). TOR and AMPK act in concert to control autophagy induction (Alers et al., 2012). As the induction of autophagy can be accompanied by an increase in mRNA levels of certain autophagy-related genes (ATGs) (Fullgrabe et al., 2014), we examined the transcript levels of ATGs in response to AMPK activation. Indeed, Atg1, Atg8a, and Atg8b mRNA levels were significantly increased in head tissue of ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 1E). To further investigate the impact of neuronal AMPK activation on autophagy, we utilized a transgenic autophagosome marker, GFP-tagged Atg8a under the control of its endogenous promoter (pGFP-Atg8a), generated by (Denton et al., 2009). A characteristic of autophagy is the formation of the autophagosome, and this can be monitored by the association of GFP-Atg8a with autophagosomal membranes observed as GFP puncta (Denton et al., 2009; Klionsky et al., 2012). We observed a significant increase in GFP puncta in brain tissue of ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 1F, quantification Fig. 1G). Control flies exposed to RU486 showed no difference in autophagy markers in head tissue (Fig. S1D–F). Together, these data indicate that activation of AMPK in the adult nervous system can induce autophagy in the target tissue and prolong lifespan.
Long-lived neuronal AMPK flies show normal feeding behavior and fecundity, but sensitivity to starvation
To better understand neuronal AMPK-mediated lifespan extension, we examined a number of behavioral and physiological parameters in long-lived flies. As a reduction in food intake can extend organismal lifespan, we first set out to determine whether neuronal AMPK activation affects feeding behavior. Using two independent methods, a capillary feeding (CAFE) assay (Ja et al., 2007) and a dye-tracking assay (Wong et al., 2009), we failed to observe alterations in feeding behavior in ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. S1G, H). Furthermore, long-lived flies with increased neuronal AMPK activity showed normal fecundity (Fig. S1I) and spontaneous physical activity (Fig. S1K). As AMPK is activated by energy deprivation (Hardie et al., 2012), we hypothesized that increased AMPK activity in the nervous system could affect organismal survival under starvation conditions. Indeed, neuronal activation of AMPK conferred a decrease in survival when flies were maintained on an agar-only diet to induce starvation (Fig. 1H). To better understand this observation, we examined mass and triglyceride (TAG) levels in neuronal AMPK flies and controls in response to starvation. Interestingly, we observed rapid weight loss (Fig. 1I) and depletion of TAG stores (Fig. 1J), in response to starvation, in ElavGS>UAS-mCh-AMPK flies upon RU486 treatment. In contrast, neuronal AMPK activation confers a moderate increase in resistance to both hyperoxia (Fig. S1O) and heat stress (Fig. S1P). Feeding RU486 to control flies did not alter feeding behavior, stress resistance, mass, or TAG levels in response to starvation and produced a moderate decrease in fecundity (Fig. S1G, H, J, L–N, Q, R). Taken together, these data show that neuronal AMPK-mediated lifespan extension is not linked to reduced food consumption, physical activity or reproductive output. However, increased AMPK activity in the adult nervous system, which extends lifespan when food is present, leads to rapid loss of TAG stores and early onset mortality in the absence of food. Alterations in lipid metabolism have previously been implicated in Drosophila aging (Katewa et al., 2012).
Neuronal AMPK up-regulation induces autophagy cell non-autonomously and slows intestinal aging
Neuron-specific activation of AMPK could prolong organismal survival by slowing aging exclusively within the nervous system. Alternatively, neuronal AMPK activation could impact systemic aging via cell non-autonomous effects in distal, non-neuronal cells. In recent years, maintenance of intestinal homeostasis has been shown to play a key role in lifespan determination in Drosophila (Biteau et al., 2010; Rera et al., 2013; Rera et al., 2011; Rera et al., 2012). Indeed, loss of intestinal integrity in aged flies is linked to multiple markers of organismal aging and, critically, is a harbinger of death (Rera et al., 2012). To determine whether neuronal AMPK-mediated lifespan extension proceeds via a cell autonomous or non-autonomous mechanism, we examined intestinal integrity during aging in response to up-regulation of AMPK in adult neurons. Loss of intestinal integrity can be assayed in living flies by monitoring the presence of non-absorbed dyes (e.g., FD&C blue No. 1) outside of the digestive tract post-feeding (Rera et al., 2011; Rera et al., 2012). Remarkably, we observed a delay in the onset of intestinal barrier dysfunction in ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 2A), indicating a delay in intestinal aging at the tissue level. Feeding RU486 to control flies did not affect intestinal integrity during aging (Fig. S2A). To extend these findings, we used the constitutive ELAV-GAL4 to both knock-down and up-regulate AMPK in the nervous system and examined the impact on intestinal aging. Using this approach, we observed that neuron-specific RNAi of AMPK accelerated intestinal aging, while neuron-specific up-regulation of AMPK delayed the onset of intestinal aging (Fig. S2B).
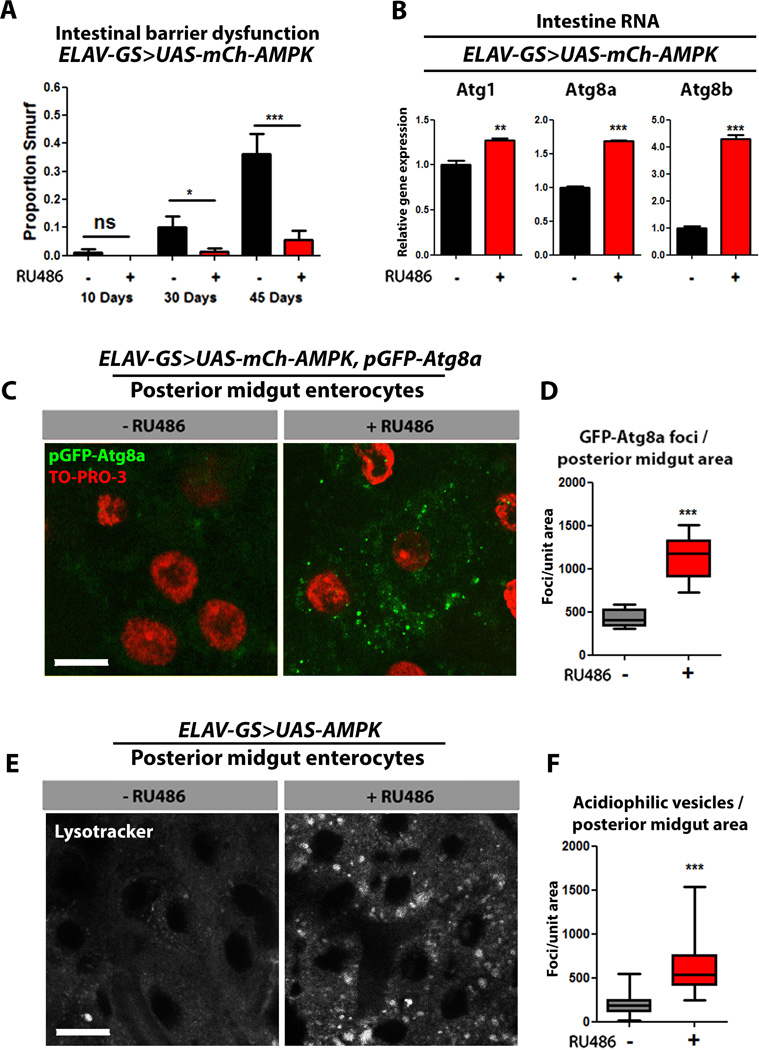
Figure 2. Neuronal AMPK activation maintains intestinal homeostasis during aging.
(A) Intestinal integrity during aging in ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction (p<0.05; binomial test, at 30 days of age, and p<0.01 at 45 days; n> 60 flies/condition).
(B) Intestinal expression of autophagy genes in 10 day old ELAV-GS>UAS-mCh-AMPK female flies (p<0.01 for Atg1, p<0.001 for Atg8a and Atg8b; t-test; n=3 of RNA extracted from 15 intestines/replicate).
(C) GFP-Atg8a staining. Representative images of enterocytes from the posterior midgut of 10 day old ELAV-GS>UAS-mCh-AMPK, pGFP-Atg8a female flies with or without RU486-mediated transgene induction (red channel-TO-PRO-3 DNA stain, green channel-GFP-Atg8a, scale bar represents 10µm).
(D) Quantification of posterior midgut GFP-Atg8a foci (p<0.0001; t-test; n>10 confocal stacks from posterior midgut/condition; one fly per replicate stack).
(E) Lysotracker Red staining. Representative images of posterior midgut enterocytes from 10 day old ELAV-GS>UAS-AMPK female flies with or without RU486-mediated transgene induction (scale bars represent 10µm).
(F) Quantification of acidophilic vesicles (p<0.0001; t-test; n>19 confocal stacks from posterior midgut/condition; one fly per replicate stack).
Data are represented as mean ± SEM. RU486 was provided in the media after eclosion at a concentration of 50µg/ml (E, F) and 25µg/ml in all remaining figures.
Intrigued by the manner in which neuronal AMPK can modulate intestinal aging, we hypothesized that AMPK may be acting in a cell-non-autonomous fashion. Previous studies have reported that ElavGS displays RU486-dependent gene expression exclusively in the nervous system (Poirier et al., 2008; Shen et al., 2009). To validate the tissue-specificity of ElavGS, we examined expression of the exogenous AMPK transgene in heads, thoraces, and intestines, in ElavGS>UAS-mCh-AMPK flies with and without RU486 induction. Only RNA isolated from head tissue showed a significant RU486-dependant increase in exogenous AMPK (Fig. S2C). Furthermore, Western blot analysis of lysates from different tissues of ElavGS>UAS-eGFP also showed a visible RU486-dependent increase in GFP levels in head tissue (Fig. S2D). To better understand how neuron-specific expression of an AMPK transgene could impact intestinal aging, we assayed markers of autophagy in the intestine upon neuronal AMPK activation. Interestingly, we observed increased mRNA levels of Atg1, Atg8a, and Atg8b, in intestinal tissue from ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 2B). Consistently, using the transgenic autophagosome marker, pGFP-Atg8a, we observed a significant increase in GFP puncta in posterior mid-gut enterocytes in ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 2C, quantification Fig. 2D). Up-regulation of AMPK in neurons also significantly increased the amount of lysosomal foci found in the mid-gut enterocytes as marked by the acidophilic dye lysotracker (Fig. 2E, quantification Fig. 2F). Feeding RU486 to control flies did not affect ATG transcript levels, autophagosome formation or lysosomal foci in intestinal tissue (Fig. S2E–I). Taken together, our findings support a model whereby neuronal AMPK activation modulates intestinal aging via a cell-non-autonomous mechanism.
Neuronal AMPK up-regulation maintains protein homeostasis during muscle aging
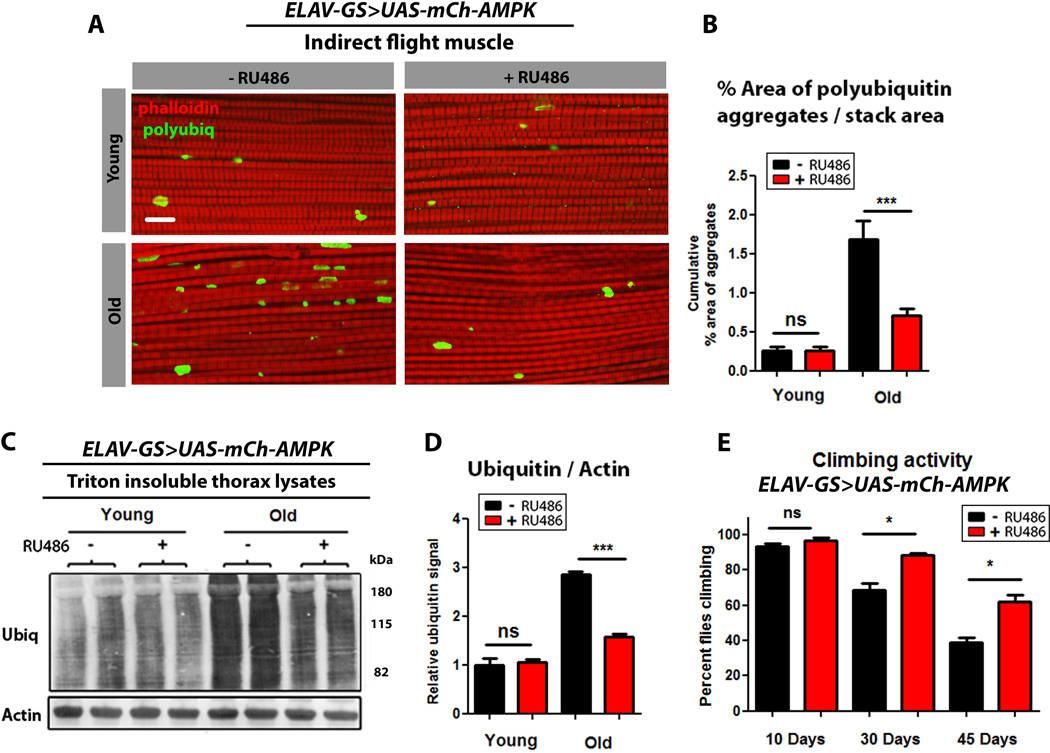
An age-related loss of muscle structure or function has been reported in diverse species including worms, flies and humans (Herndon et al., 2002; Nair, 2005; Zheng et al., 2005). In Drosophila, muscle aging is associated with a loss of protein homeostasis, leading to the accumulation of insoluble protein aggregates (proteotoxicity) (Demontis and Perrimon, 2010; Rana et al., 2013). To better understand the cell-non-autonomous nature of AMPK-mediated lifespan extension, we set out to determine whether neuronal AMPK activation can influence protein homeostasis during muscle aging. Firstly, we characterized the age-related deposition of protein aggregates in thoracic muscles by immunofluorescence. Importantly, neuronal AMPK activation lead to reduced levels of protein aggregates in aged muscles (Fig. 3A and quantification Fig. 3B). In a complementary approach, we examined the levels of insoluble ubiquitinated proteins, by Western blotting, in thoraces of neuronal AMPK flies and controls during aging. Consistent with the immunofluorescence data, we observed that neuronal up-regulation of AMPK reduced levels of insoluble ubiquitinated proteins in aged muscle tissue (Fig. 3C and quantification Fig. 3D). Moreover, neuronal up-regulation of AMPK improved climbing ability during aging (Fig. 3E). Feeding RU486 to controls did not alter markers of muscle aging (Fig. S3A–E). Interestingly, the mRNA levels of Atg1, Atg8a, and Atg8b, were significantly increased in thoracic tissue from ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. S3F). Control flies fed RU486 showed no changes in ATG transcript levels in thoracic tissue (Fig. S3G). Taken together, our findings indicate that neuron-specific AMPK activation leads to an increase in autophagy gene expression in muscle tissue, which is associated with reduced proteotoxicity and improved muscle function in aged flies.
Figure 3. Neuronal AMPK activation maintains protein homeostasis during muscle aging.
(A) Confocal images of indirect flight muscle, in ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction, showing protein polyubiquitinated aggregates at young (10 days), and aged (30 days) timepoints (red channel-phalloidin/F-actin, green channel- anti-polyubiquitin, scale bar represents 10µm).
(B) Quantification of polyubiquitin aggregates in muscle (p<0.001; t-test; n>10; one fly/replicate stack).
(C) Western blot detection of total ubiquitin-conjugated proteins from thorax detergent-insoluble extracts of young (10 days) and aged (30 days) ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction.
(D) Densitometry of ubiquitin blots from thoraces of flies (p<0.001; t-test; n=4 samples/condition; 10 thoraces/sample).
(E) Climbing activity of ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.05; t-test; n>6 vials/condition; 30 flies/vial).
Data are represented as mean ± SEM. RU486 was provided in the media after eclosion at a concentration of 25µg/ml.
Neuronal Atg1 up-regulation induces autophagy and slows aging in a cell-non-autonomous manner
To seek evidence for a causal role for autophagy in mediating the anti-aging effects of AMPK activation, we set out to directly manipulate Atg1; a Ser/Thr protein kinase involved in the initiation of autophagosome formation (Nakatogawa et al., 2009). Firstly, we asked whether autophagy was required for the extended lifespan that results from neuronal AMPK activation. To investigate this, we used the ElavGS driver line to inhibit Atg1 by RNAi in the adult nervous system of control flies and flies with increased neuronal expression of AMPK (Fig. S4A, B) and compared survivorship to flies with increased neuronal AMPK expression alone. We found that induced RNAi of Atg1 in adult neurons suppressed the lifespan extension associated with neuronal up-regulation of AMPK (Fig. 4A, B) but did not shorten lifespan in control flies (Fig. 4C). This result indicates that neuronal AMPK-mediated longevity is dependent on Atg1 gene activity. In addition, the ability of neuronal AMPK to slow markers of systemic aging was also dependent on neuronal Atg1 gene activity (Fig. S4C–F) as was sensitivity to starvation (Fig. S4G–I).
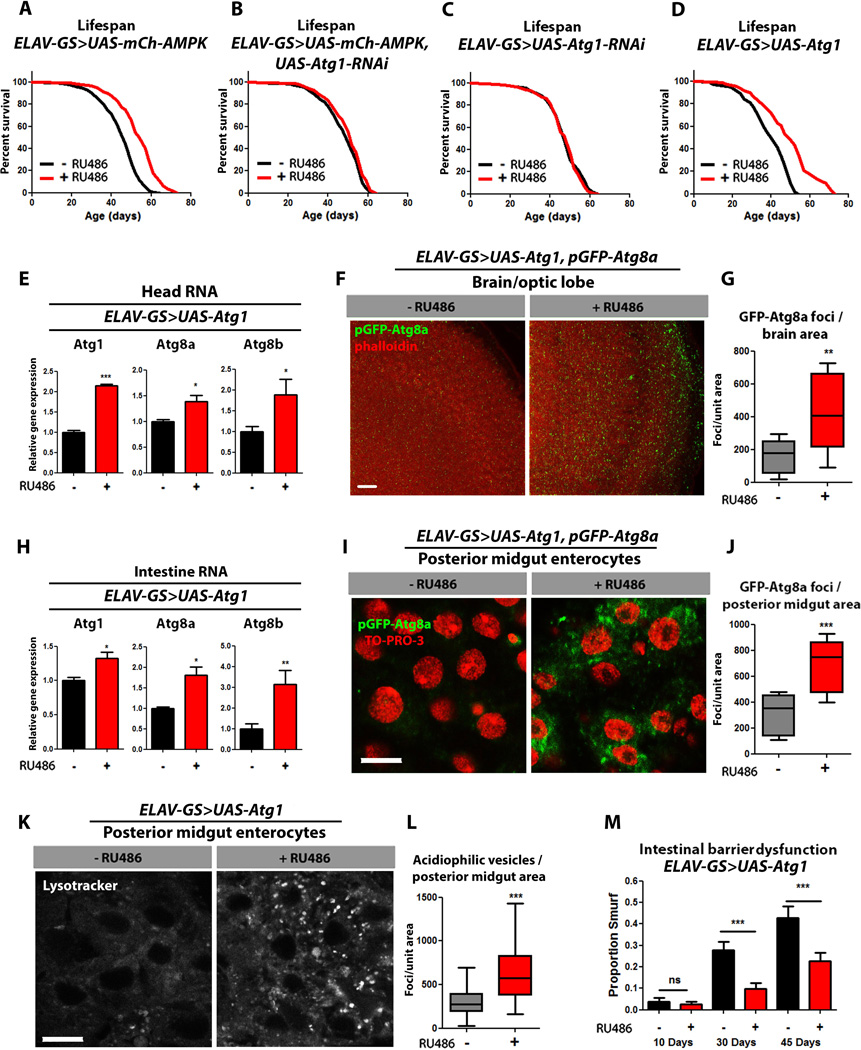
Figure 4. Neuronal Atg1 up-regulation maintains intestinal homeostasis during aging and extends lifespan.
(A) Survival curves of ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction (p<0.0001; log-rank test; n> 138 flies).
(B) Survival curves of ELAV-GS>UAS-mCh-AMPK, UAS-Atg1-RNAi female flies with or without RU486-mediated transgene induction (p>0.0284; log-rank test; n>210 flies).
(C) Survival curves of ELAV-GS>UAS-Atg1-RNAi female flies with or without RU486-mediated transgene induction (p>0.05; log-rank test; n>111 flies).
(D) Survival curves of ELAV-GS>UAS-Atg1 female flies with or without RU486-mediated transgene induction (p>0.0001; log-rank test; n> 151 flies).
(E) Expression levels of autophagy genes in heads of 10 day old ELAV-GS>UAS-Atg1 female flies with or without RU486-mediated transgene induction (t-test; n>3 of RNA extracted from 10 heads/replicate).
(F) Brain GFP-Atg8a Staining. Representative images from optic lobes of 10 day old ELAV-GS>UAS-Atg1, pGFP-Atg8a females with or without RU486-mediated transgene expression (red channel-phalloidin, green channel-GFP-Atg8a, scale bar represents 10µm).
(G) Quantification of brain GFP-Atg8a foci (p<0.0082; t-test; n>10 confocal stacks from optic lobes/condition; one brain/replicate stack).
(H) Expression levels of autophagy genes in the intestines of 10 day old ELAV-GS>UAS-Atg1 female flies with or without RU486-mediated transgene induction (t-test; n>3 of RNA extracted from 15 intestines/replicate).
(I) GFP-Atg8a staining. Representative images of enterocytes from the posterior midgut of 10 day old ELAV-GS>UAS-Atg1, pGFP-Atg8a female flies with or without RU486-mediated transgene induction in neurons (red channel-TO-PRO-3 DNA stain, green channel-GFP-Atg8a, scale bar represents 10µm).
(J) Quantification of posterior midgut GFP-Atg8a foci (p<0.0001; t-test; n>10 confocal stacks from posterior midgut/condition; one fly per replicate stack).
(K) Lysotracker Red staining. Representative images of posterior midgut enterocytes from 10 day old ELAV-GS>UAS-Atg1 female flies with or without RU486-mediated transgene induction stained with the acidophilic dye (scale bar represents 10µm).
(L) Quantification of acidophilic vesicles (p<0.0001; t-test; n>25 confocal stacks from posterior midgut/condition; one fly per replicate stack).
(M) Intestinal integrity during aging in ELAV-GS>UAS-Atg1 females with or without RU486-mediated transgene induction (p<0.001; binomial test, at 30 and 45 days; n>91 flies/condition).
Data are represented as mean ± SEM. RU486 was provided in the media after eclosion at a concentration of 50µg/ml for all figures.
In previous work, Atg1 overexpression has been shown to directly induce autophagy in Drosophila (Liu and Lu, 2010; Scott et al., 2007). However, the question of whether increased Atg1 expression can slow aging or extend animal lifespan has not been addressed. Given our findings with neuronal AMPK activation, we sought to examine the impact of neuron-specific up-regulation of Atg1 on Drosophila aging. To do so, we used the ElavGS driver to activate a UAS-Atg1 transgene created by (Scott et al., 2007). Adult-onset, neuronal up-regulation of Atg1 resulted in increases in median and maximum lifespan in female flies (Figure 4D, Table S2) and the mRNA levels of Atg1, Atg8a, and Atg8b, were significantly increased in head tissue (Fig. 4E). Moreover, using the transgenic autophagosome marker, pGFP-Atg8a, we observed a significant increase in GFP puncta in the brain tissue of ElavGS>UAS-Atg1 flies upon RU486 treatment (Fig. 4F, quantification Fig. 4G). Together, these data indicate that up-regulation of Atg1 in the adult nervous system can induce autophagy in the target tissue and prolong lifespan.
Next, we set out to determine whether Atg1 can induce autophagy in a cell-non-autonomous manner. Remarkably, up-regulation of Atg1 in adult neurons significantly increased mRNA levels of Atg1, Atg8a, and Atg8b in intestinal tissue (Fig. 4H) and using the transgenic autophagosome marker, pGFP-Atg8a, we observed a significant increase in GFP puncta in posterior mid-gut enterocytes of ElavGS>UAS-Atg1 flies upon RU486 treatment (Fig. 4I, quantification Fig. 4J). Up-regulation of Atg1 in neurons also significantly increased the amount of lysosomal foci found in the mid-gut enterocytes as marked by the acidophilic dye lysotracker (Fig. 4K, quantification Fig. 4L). Importantly, the cell-non-autonomous induction of autophagy, mediated by neuronal Atg1, was associated with improved intestinal homeostasis during aging (Fig. 4M) and a delay in the onset of muscle aging (Fig. S4J–L). To determine whether altered feeding behavior may play a role in mediating these effects, we assayed feeding in response to neuron-specific Atg1 up-regulation. As was the case with neuronal AMPK-mediated longevity, long-lived flies overexpressing Atg1 in the adult nervous system did not show altered feeding behavior (Fig.S4M). However, up-regulation of Atg1 in adult neurons did confer sensitivity to starvation conditions, including early-onset mortality (Fig. S4N), rapid loss of body weight (Fig. S4O) and TAG stores (Fig. S4P) and a moderate increase in resistance to both hyperoxia (Fig. S4Q) and heat stress (Fig. S4R). Together, these data indicate that direct activation of the autophagy pathway in adult neurons can impact systemic physiology in a similar fashion to neuronal AMPK activation.
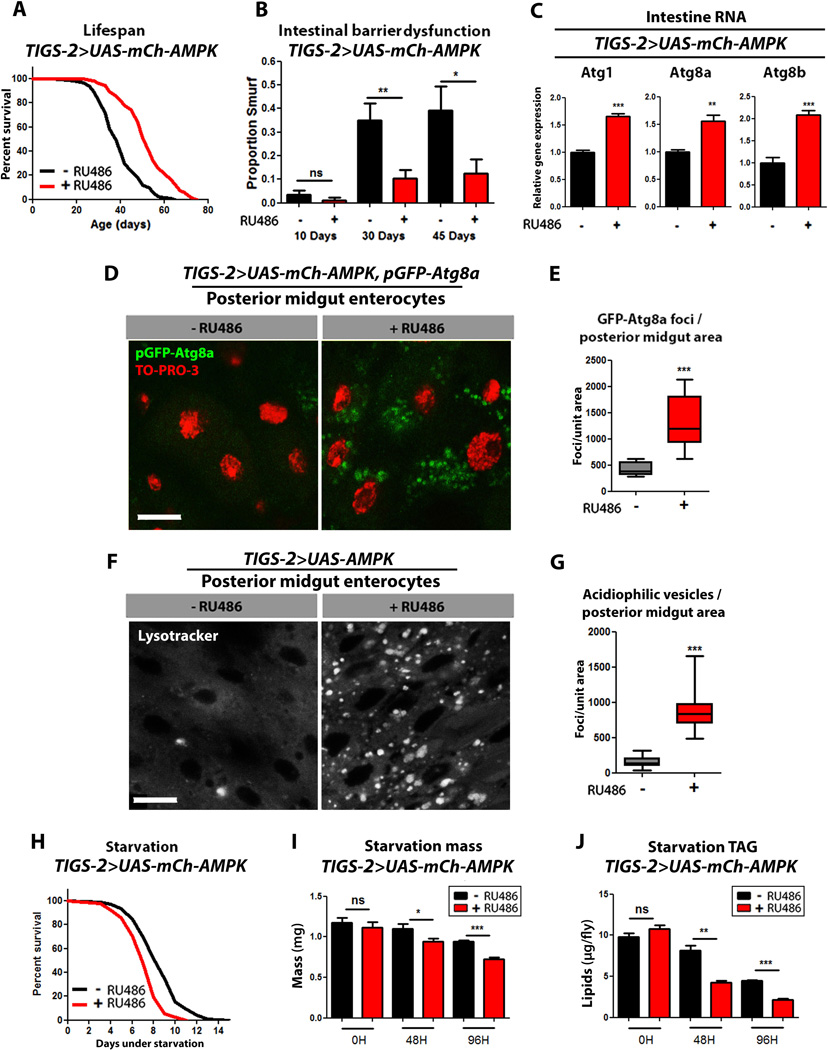
Intestine-specific AMPK up-regulation slows intestinal aging and extends lifespan
Previous work has shown that Drosophila AMPK mutants show impaired intestinal peristalsis and decreased nutrient absorption (Bland et al., 2010). To better understand the relationships between AMPK, intestinal homeostasis and organismal aging, we set out to determine whether up-regulation of AMPK in the intestine was sufficient to activate autophagy in the target tissue and/or prolong lifespan. To do so, we utilized the intestine-specific Gene-Switch (GS) driver line TIGS-2 (Poirier et al., 2008) to activate UAS-mCh-AMPK. To validate the tissue-specificity of TIGS-2, we examined the expression of the exogenous AMPK transgene in heads, thoraces, and intestines, in TIGS-2>UAS-mCh-AMPK flies with and without RU486 induction. Only RNA isolated from intestinal tissue showed a significant RU486-dependant increase in exogenous AMPK transcript (Fig. S5A). Furthermore, Western blot analysis of lysates from different tissues of TIGS-2>UAS-eGFP also showed a visible RU486-dependent increase in GFP levels in intestinal tissue (Fig S5B). Adult-onset, intestine-specific up-regulation of AMPK resulted in increased median and maximum lifespan in female flies (Fig. 5A, Table S3) and a smaller lifespan increase in male flies (Table S3). Feeding RU486 to control flies did not produce major changes in lifespan (Fig. S5C). To determine whether AMPK up-regulation was sufficient to improve tissue homeostasis in the aging intestine, we examined intestinal integrity as a function of age. Importantly, we observed a delay in the onset of intestinal barrier dysfunction in TIGS-2>UAS-mCh-AMPK flies upon RU486 treatment compared to uninduced controls (Fig. 5B). Feeding RU486 to control flies did not affect intestinal integrity during aging (Fig. S5D).
Figure 5. Intestinal AMPK activation maintains intestinal homeostasis during aging and extends lifespan.
(A) Survival curves of TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction (p<0.0001; log-rank test; n>116 flies).
(B) Intestinal integrity during aging in TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction (p<0.01, at 30 days, p<0.05 at 45 days; binomial test; n>127 flies/condition).
(C) Expression levels of autophagy genes in from intestines of TIGS-2>UAS-mCh-AMPK female flies at 10 days of adulthood with or without RU486-mediated transgene induction. (t-test; n>3 of RNA extracted from 15 intestines/replicate).
(D) GFP-Atg8a staining. Representative images of enterocytes from the posterior midgut of 10 day old TIGS-2>UAS-mCh-AMPK, pGFP-Atg8a females with or without RU486-mediated transgene expression (red channel-TO-PRO-3 DNA stain, green channel-GFP-Atg8a, scale bar represents 10µm).
(E) Quantification of posterior midgut GFP-Atg8a foci (p<0.0001; t-test; n>10 confocal stacks from posterior midgut/condition; one fly per replicate stack).
(F) Lysotracker Red staining. Representative images of posterior midgut enterocytes from 10 day old TIGS-2>UAS-AMPK females with or without RU486-mediated transgene induction (scale bar represents 10µm).
(G) Quantification of acidophilic vesicles (p<0.0001; t-test; n>10 confocal stacks from posterior midgut/condition; one fly per replicate stack).
(H) Survival curves without food of TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction (p<0.001; log-rank; n>257 flies).
(I) Body mass during starvation of TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction (p<0.05, at 48hours and p<0.01 at 96 hours of starvation; t-test; n>6 samples/condition; 10 flies weighed/sample).
(J) Whole body lipid stores during starvation of TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction (p<0.01 at 48 hours, and p<0.001 at 96 hours of starvation; t-test; n>3 samples/condition/timepoint; lipids extracted from 5 flies/sample).
Data are represented as mean ± SEM. RU486 was provided in the media after eclosion at a concentration of 25µg/ml (E, F) and 100µg/ml for all other figures.
Next, we examined markers of autophagy in response to up-regulation of AMPK in the intestine. The mRNA levels of Atg1, Atg8a, and Atg8b, were significantly increased in intestinal tissue from TIGS-2>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 5C). Moreover, using pGFP-Atg8a, we observed a significant increase in GFP puncta in posterior mid-gut enterocytes in TIGS-2>UAS-mCh-AMPK flies upon RU486 treatment compared to uninduced controls (Fig. 5D, quantification 5E). Up-regulation of AMPK in the intestine also significantly increased the amount of lysosomal foci found in the mid-gut enterocytes as marked by the acidophilic dye lysotracker (Fig. 5F, quantification 5G). Feeding RU486 to control flies did not affect ATG transcript levels in intestinal tissue, autophagosome formation or lysosomal foci (Fig. S5E–I).
As was the case with neuronal AMPK-mediated longevity, long-lived flies overexpressing AMPK in the intestine did not show reduced feeding behavior (Fig. S5J, and K) or fecundity (Fig. S5L). However, intestine-specific AMPK overexpression did confer sensitivity to starvation conditions, including early-onset mortality (Fig. 5H), rapid loss of body weight (Fig. 5I) and TAG stores (Fig. 5J) and, in contrast, increased tolerance to both hyperoxia (Fig. S5Q) and heat stress (Fig. S5R). Feeding RU486 to control flies did not affect fecundity or sensitivity to stress conditions (Fig. S5M–P, S, T). Taken together, our findings indicate that intestine-specific up-regulation of AMPK induces autophagy and maintains intestinal homeostasis during aging, which is associated with increased lifespan at the organismal level.
Intestine-specific AMPK up-regulation induces autophagy cell non-autonomously and slows muscle aging
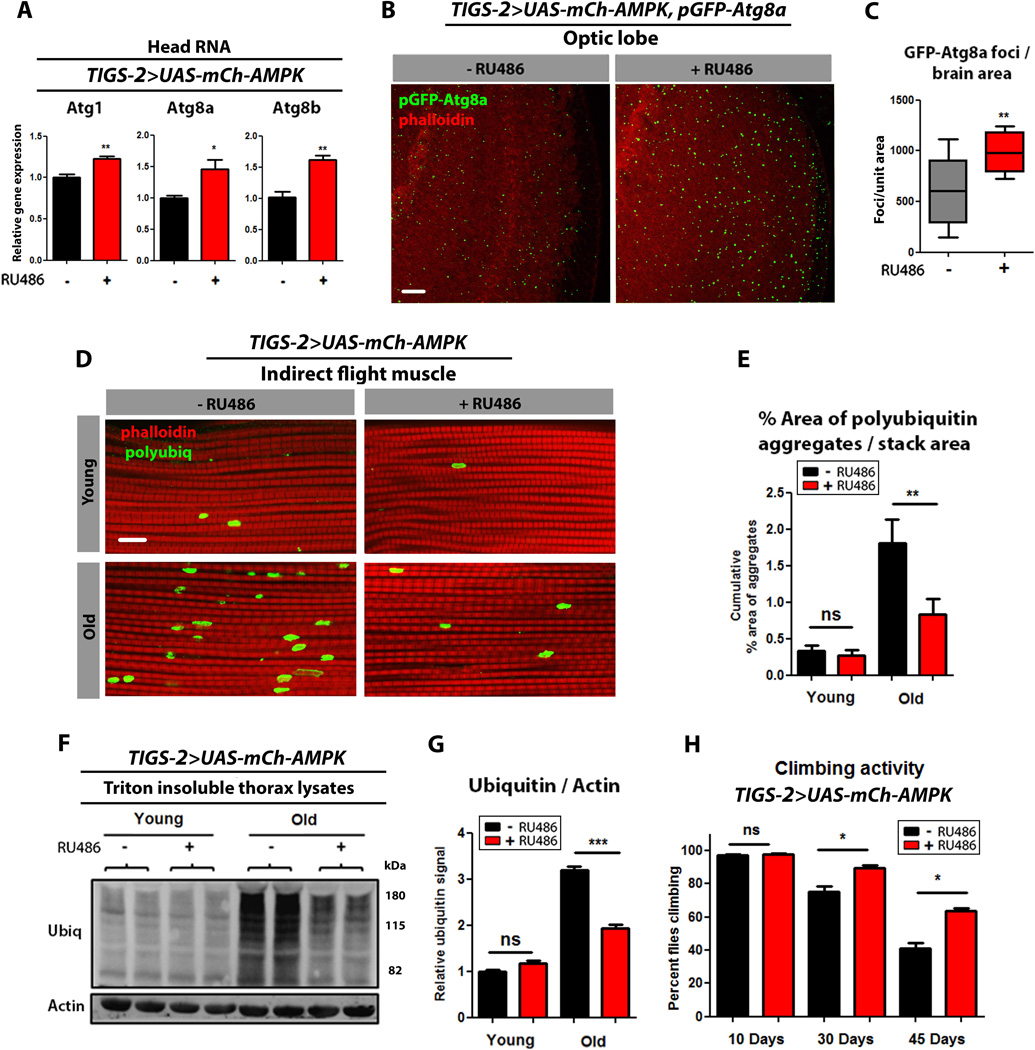
As transgenic up-regulation of AMPK in the adult nervous system can increase markers of autophagy in the intestine, we wanted to assess the converse by up-regulating AMPK in the intestine and monitoring autophagy markers in the brain. Interestingly, Atg1, Atg8a, and Atg8B mRNA levels were moderately increased in head tissue upon intestine-specific up-regulation of AMPK (Fig. 6A). Moreover, using the transgenic autophagosome marker, pGFP-Atg8a, we observed a significant increase in GFP puncta in brain tissue of TIGS-2>UAS-mCh-AMPK flies upon RU486 treatment compared to uninduced controls (Fig. 6B, quantification Fig. 6C). Feeding RU486 to control flies did not affect autophagy markers in head tissue (Fig S6A–C). These findings indicate that targeted up-regulation of AMPK in adult intestinal cells can induce autophagy in brain tissue.
Figure 6. Intestinal AMPK activation induces autophagy in the brain and slows muscle aging.
(A) Expression levels of autophagy genes in heads of 10 day old TIGS-2>UAS-mCh-AMPK flies at with or without RU486-mediated transgene induction. (t-test; n>3 of RNA extracted from 10 heads/replicate).
(B) Brain GFP-Atg8a Staining. Representative images from optic lobes of 10 day old TIGS-2>UAS-mCH-AMPK, pGFP-Atg8a females with or without RU486-mediated transgene expression (red channel-phalloidin, green channel-GFP-Atg8a, scale bar represents 10µm).
(C) Quantification of brain GFP-Atg8a foci (p<0.007; t-test; n>10 confocal stacks from optic lobes/condition; one brain/replicate stack).
(D) Confocal imaging of flight muscle of TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction showing protein polyubiquitinated aggregates at young (10 days), and old (30 days) timepoints (red channel-phalloidin/F-actin, green channel- anti-polyubiquitin, scale bar represents 10µm).
(E) Quantification of polyubiquitin aggregates in muscle (p<0.01; t-test; n>10; one fly/replicate stack).
(F) Western blot detection of total ubiquitin-conjugated proteins from thorax detergent-insoluble extracts of young (10 days) and aged (30 days) TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction.
(G) Densitometry of ubiquitin blots (p<0.001; t-test; n=4 samples/condition; 10 thoraces/sample).
(H) Climbing activity of TIGS-2>UAS-mCh-AMPK females with or without RU486-mediated transgene induction. (p<0.05; t-test; n=6 vials/condition; 30 flies/vial).
Data are represented as mean ± SEM. RU486 was provided in the media after eclosion at a concentration of 100µg/ml.
To further explore the cell-non-autonomous consequences of up-regulating AMPK in the intestine, we set out to examine markers of muscle aging. Interestingly, up-regulation of AMPK in the intestine reduced levels of protein aggregates during muscle aging (Fig. 6D, quantification Fig. 6E). Consistent with the immunofluorescence data, we observed reduced levels of insoluble ubiquitinated proteins in aged muscle of flies with intestinal AMPK activation (Fig.6F, and quantification Fig. 6G) and improved climbing ability during aging (Fig. 6H). Feeding RU486 to control flies did not alter markers of muscle aging (Fig. S6D–H). Additionally, the mRNA levels of Atg1, Atg8a, and Atg8b, were significantly increased in thoracic tissue from TIGS-2>UAS-mCh-AMPK flies upon RU486 treatment (Fig. S6I). Feeding RU486 to control flies did not affect protein homeostasis during muscle aging, or ATG transcript levels in thoracic tissue (Fig. S6J). Taken together, our findings indicate that intestine-specific AMPK up-regulation leads to an increase in autophagy gene expression in brain and muscle tissue, which is associated with delayed systemic aging.
Inter-tissue effects of AMPK/Atg1 are linked to altered insulin-like signaling
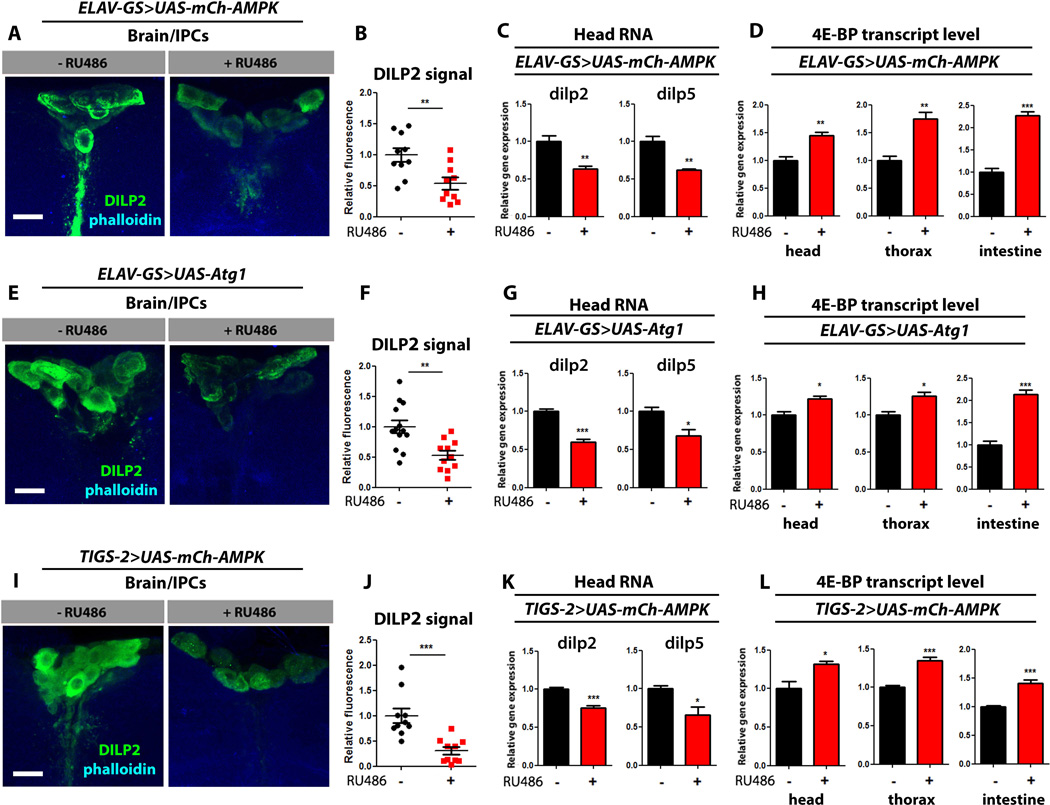
The insulin/IGF-1 signaling (IIS) pathway modulates lifespan through interacting autonomous and non-autonomous actions (Kenyon, 2010; Taylor et al., 2014). To explore a potential role for IIS in mediating the systemic effects associated with tissue-specific AMPK/Atg1 activation, we first examined whether neuronal AMPK activation affects Drosophila insulin-like peptide (DILP) levels in the brain. Indeed, we observed a significant decrease in DILP2 levels in the insulin producing cells (IPCs) of ElavGS>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 7A, quantification Fig. 7B) and a decrease in both dilp2 and dilp5 mRNA levels in head tissue (Fig. 7C). The translational regulator 4E-BP is a direct transcriptional target of Drosophila FOXO (dFOXO) that is induced when IIS is repressed (Puig et al., 2003) and has previously been implicated in mediating the anti-aging effects of DR in Drosophila (Zid et al., 2009). Importantly, 4E-BP transcript levels were increased in the head and non-autonomously in both the thorax and intestine upon neuronal AMPK activation, consistent with a systemic reduction in IIS (Fig. 7D). In a similar fashion, neuronal up-regulation of Atg1 reduced DILP levels in the brain (Fig. 7E–G) and was associated with a systemic increase in 4E-BP expression (Fig. 7H). Finally, we asked whether intestinal AMPK activation could also impact DILP levels in the brain and/or systemic 4E-BP expression. We observed a significant decrease in DILP2 levels in the IPCs of TIGS-2>UAS-mCh-AMPK flies upon RU486 treatment (Fig. 7I, quantification Fig. 7J) and a decrease in both dilp2 and dilp5 mRNA levels in head tissue (Fig. 7K). Furthermore, 4E-BP transcript levels were increased in the head, thorax and intestine upon intestinal AMPK up-regulation (Fig. 7L). Feeding RU486 to control flies did not affect DILP levels or 4E-BP expression (Fig S7A–H). Taken together, these findings suggest that the whole-body effects associated with localized AMPK/Atg1 up-regulation may be mediated by altered DILP signaling.
Figure 7. Inter-tissue effects of AMPK/Atg1 are linked to altered insulin-like signaling.
(A) Representative images of DILP2 antibody stained insulin producing cells (IPCs) from 10 day old ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction (green channel - Dilp2 antibody, blue channel phalloidin, scale bars represent 10µm).
(B) Quantification of DILP2 signal from IPCs of 10 day old ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.01; t-test; n>10 brains/condition).
(C) Expression level of dilp genes from dissected heads of 10 day old ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.01; t-test; n>3 of RNA extracted from 10 heads /replicate).
(D) Expression level of 4E-BP from dissected body parts of 10 day old ELAV-GS>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.01; t-test; n=3 of RNA extracted from 10 body parts/replicate).
(E) Representative images of DILP2 antibody stained IPCs from 10 day old ELAV-GS>UAS-Atg1 female flies (green channel - Dilp2 antibody, blue channel phalloidin, scale bars represent 10µm).
(F) Quantification of DILP2 signal from IPCs of 10 day old ELAV-GS>UAS-Atg1 female flies with or without RU486-mediated transgene induction. (p<0.01; t-test; n>10 brains/condition).
(G) Expression level of dilp genes from dissected heads of 10 day old ELAV-GS>UAS-Atg1 female flies with or without RU486-mediated transgene induction. (p<0.05; t-test; n>3 of RNA extracted from 10 heads /replicate).
(H) Expression level of 4E-BP from dissected body parts of 10 day old ELAV-GS>UAS-Atg1 female flies with or without RU486-mediated transgene induction. (p<0.05; t-test; n=3 of RNA extracted from 10 body parts/replicate).
(I) Representative images of DILP2 antibody stained IPCs from 10 day old TIGS-2>UAS-mCh-AMPK female flies (green channel - Dilp2 antibody, blue channel -phalloidin, scale bars represent 10µm) with or without RU486-mediated transgene induction.
(J) Quantification of DILP2 signal from IPCs of 10 day old TIGS-2>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.001; t-test; n>10 brains/condition).
(K) Expression level of dilp genes from dissected heads of 10 day old TIGS-2>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.05; t-test; n=3 of RNA extracted from 10 heads /replicate).
(L) Expression level of 4E-BP from dissected body parts of 10 day old TIGS-2>UAS-mCh-AMPK female flies with or without RU486-mediated transgene induction. (p<0.05; t-test; n=3 of RNA extracted from 10 body parts/replicate).
Data are represented as mean ± SEM. RU486 was provided in the media after eclosion at the following concentrations (A-D 25µg/ml) (E-H 50µg/ml)(I-L 100µg/ml).
Discussion
The cellular recycling process of autophagy has been proposed to exert anti-aging effects in diverse species (Gelino and Hansen, 2012; Rubinsztein et al., 2011). Although AMPK, a key regulator of autophagy (Alers et al., 2012), has also been linked to aging and lifespan determination (Burkewitz et al., 2014), little was known about the relationships between AMPK and autophagy in the coordination of tissue and organismal aging. In this study, we show that adult-onset up-regulation of AMPK in either the nervous system or intestine stimulates autophagy in the target tissue and is sufficient to prolong Drosophila lifespan. Interestingly, neuronal AMPK activation also induces autophagy cell non-autonomously in the intestinal epithelium and delays the onset of intestinal aging. Consistent with a causal role for autophagy in mediating these inter-tissue and organism-level effects, we show that adult-onset, neuron-specific up-regulation of Atg1 is both necessary and sufficient to improve intestinal homeostasis during aging and prolong lifespan. Importantly, we find that intestine-specific AMPK activation also induces autophagy cell non-autonomously in the brain and maintains protein homeostasis during muscle aging. Finally, we show that the cell-non-autonomous effects of localized AMPK/Atg1 activation are linked to a systemic increase in 4E-BP expression and reduced DILP levels in the brain.
Our work, coupled with that of others, strengthens the emerging concept that stress responses are coordinated across tissues (Taylor et al., 2014). Notably, muscle-specific dFOXO/4E-BP/activin signaling can induce autophagy autonomously and, thereby, retard muscle aging; these effects in muscle are linked to altered DILP levels in the brain, dampened systemic IIS and extended organismal lifespan (Bai et al., 2013; Demontis and Perrimon, 2010). Our findings are consistent with a model where tissue-specific AMPK/Atg1 up-regulation antagonizes DILP signaling leading to a systemic activation of dFOXO that, in turn, slows organismal aging. As AMPK has been shown to directly activate FOXO activity in C. elegans and mammalian cells (Greer et al., 2007a; Greer et al., 2007b), it is tempting to speculate that an AMPK-FOXO-autophagy circuit may play a role in our findings. Interestingly, however, it has recently been reported that up-regulation of dFOXO in the intestine/fat body of flies can slow markers of neuromuscular aging in the absence of dFOXO in the responding tissue (Alic et al., 2014). In a similar fashion, it will be interesting to determine whether dFOXO is required for the changes in gene expression, physiology and lifespan observed upon tissue-specific AMPK/Atg1 induction. It is important to note, however, that although we show that AMPK/Atg1 can antagonize DILP signaling and induce autophagy cell-non-autonomously, it is not yet known whether these two phenomena are causally linked. Indeed, our work raises several questions concerning the regulation of autophagy by AMPK both autonomously and non-autonomously. As we observe increased mRNA levels of several autophagy genes upon AMPK activation, an interesting area of future investigation will be to explore the transcriptional control of autophagy in AMPK overexpressing flies. It will be of particular interest to identify the mechanisms by which intestinal AMPK up-regulation leads to changes in DILP levels and autophagy in the brain. In any case, a plausible interpretation of our findings is that the AMPK/Atg1-mediated induction of autophagy slows aging (both autonomously and non-autonomously) by increasing the turnover of damaged macromolecules and/or organelles. In this model, a pressing challenge will be to identify the relevant autophagic cargo in the context of aging.
While tissue-specific up-regulation of either AMPK or Atg1 can slow Drosophila aging in the constant presence of food, these same interventions sensitize flies to starvation conditions. This information could prove useful when designing therapeutic interventions based around AMPK and/or autophagy induction. Indeed, AMPK (Martin-Montalvo et al., 2013) and autophagy (Bjedov et al., 2010; Rubinsztein et al., 2011) have been proposed to mediate the anti-aging effects of metformin and rapamycin respectively, both of which can prolong lifespan in mice (Harrison et al., 2009; Martin-Montalvo et al., 2013). It will be interesting to determine whether activation of AMPK and/or autophagy in a single tissue, such as the nervous system or intestine, can induce an organism-wide response to slow systemic aging in mammals.
Materials and Methods
Fly Strains
UAS-AMPK flies were obtained from Jongkyeong Chung. pGFP-Atg8a flies were provided by Eric Baehrecke. UAS-Atg1-RNAi line was received from the Vienna Drosophila RNAi Center (VDRC). All other stocks were obtained from the Bloomington Stock Center. A complete and detailed list of all fly strains and methods used can be found in the supplemental information.
Immunostaining procedure for DILP2, and pGFP-Atg8a
Briefly, female flies were anesthetized on ice and intestines/brains were dissected in cold PBS. Samples were then fixed in 4% formaldehyde in PBS at room temperature for 30 minutes and rinsed three times in PBS + 0.2% Triton X-100 (PBS-Tx) for 10 minutes. Blocking was performed in 5% BSA in PBS-Tx. Primary antibody, rabbit anti-DILP2 (a generous gift from Dr. Seung Kim) or anti-GFP (Cell Signaling, D5.1, XP), was added 1:250 and incubated overnight at 4° C. After washing, secondary antibody was added and incubated for 4 hours at room temperature. Intestines/brains were then mounted and imaged. Detailed quantification methods can be found in the supplemental information.
Supplementary Material
Highlights.
-
-
Neuronal AMPK induces autophagy in brain/gut and slows systemic aging
-
-
Neuronal Atg1 induces autophagy in brain/gut and slows systemic aging
-
-
Intestinal AMPK induces autophagy in gut/brain and slows systemic aging
-
-
Inter-tissue effects of AMPK/Atg1 linked to altered insulin-like signaling
Acknowledgments
We thank T. Neufeld, E. Baehrecke, L. Seroude, S. Pletcher, J. Chung, the Vienna Drosophila RNAi Center and the Drosophila Stock Center (Bloomington) for fly stocks, S. Kim for antibodies and the L. Jones, M. Frye and D. Simmons labs for use of their equipment. DWW is supported by the National Institute on Aging (R01 AG037514, R01 AG040288). MU was supported by a Ruth L. Kirschstein National Research Service Award (GM07185), and Eureka and Hyde fellowships from the UCLA department of Integrative Biology and Physiology. JG was funded by the National Institutes of Health/National Institute of General Medical Sciences (grant number NIH MARC T34 GM008563).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Molecular and cellular biology. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N, Tullet JM, Niccoli T, Broughton S, Hoddinott MP, Slack C, Gems D, Partridge L. Cell-nonautonomous effects of dFOXO/DAF-16 in aging. Cell reports. 2014;6:608–616. doi: 10.1016/j.celrep.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr, Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes & development. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Kang P, Hernandez AM, Tatar M. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS genetics. 2013;9:e1003941. doi: 10.1371/journal.pgen.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS genetics. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell metabolism. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland ML, Lee RJ, Magallanes JM, Foskett JK, Birnbaum MJ. AMPK supports growth in Drosophila by regulating muscle activity and nutrient uptake in the gut. Developmental biology. 2010;344:293–303. doi: 10.1016/j.ydbio.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K, Zhang Y, Mair WB. AMPK at the Nexus of Energetics and Aging. Cell metabolism. 2014 doi: 10.1016/j.cmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Li PW, Goldstein BA, Cai W, Thomas EL, Chen F, Hubbard AE, Melov S, Kapahi P. Germline signaling mediates the synergistically prolonged longevity produced by double mutations in daf-2 and rsks-1 in C. elegans. Cell reports. 2013;5:1600–1610. doi: 10.1016/j.celrep.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Current biology : CB. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nature reviews. Molecular cell biology. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- Gelino S, Hansen M. Autophagy - An Emerging Anti-Aging Mechanism. Journal of clinical & experimental pathology. 2012;(Suppl 4) doi: 10.4172/2161-0681.s4-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Current biology : CB. 2007a;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. The Journal of biological chemistry. 2007b;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS genetics. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMPK and autophagy get connected. The EMBO journal. 2011;30:634–635. doi: 10.1038/emboj.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature reviews. Molecular cell biology. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, Kapahi P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell metabolism. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee SH, Shong M, Kim JM, Kim J, et al. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- Liu S, Lu B. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS genetics. 2010;6:e1001237. doi: 10.1371/journal.pgen.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, et al. Metformin improves healthspan and lifespan in mice. Nature communications. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. A microarray-based genetic screen for yeast chronological aging factors. PLoS genetics. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouse V, Swick LL, Kazgan N, St Johnston D, Brenman JE. LKB1 and AMPK maintain epithelial cell polarity under energetic stress. The Journal of cell biology. 2007;177:387–392. doi: 10.1083/jcb.200702053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nair KS. Aging muscle. The American journal of clinical nutrition. 2005;81:953–963. doi: 10.1093/ajcn/81.5.953. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature reviews. Molecular cell biology. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Poirier L, Shane A, Zheng J, Seroude L. Characterization of the Drosophila gene-switch system in aging studies: a cautionary tale. Aging cell. 2008;7:758–770. doi: 10.1111/j.1474-9726.2008.00421.x. [DOI] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes & development. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Azizi MJ, Walker DW. Organ-specific mediation of lifespan extension: more than a gut feeling? Ageing research reviews. 2013;12:436–444. doi: 10.1016/j.arr.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell metabolism. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Current biology : CB. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Curtis C, Tavare S, Tower J. A screen of apoptosis and senescence regulatory genes for life span effects when over-expressed in Drosophila. Aging. 2009;1:191–211. doi: 10.18632/aging.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenesen D, Suh JM, Seo J, Yu K, Lee KS, Kim JS, Min KJ, Graff JM. Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell metabolism. 2013;17:101–112. doi: 10.1016/j.cmet.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Berendzen KM, Dillin A. Systemic stress signalling: understanding the cell non-autonomous control of proteostasis. Nature reviews. Molecular cell biology. 2014;15:211–217. doi: 10.1038/nrm3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Wong R, Piper MD, Wertheim B, Partridge L. Quantification of food intake in Drosophila. PloS one. 2009;4:e6063. doi: 10.1371/journal.pone.0006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Edelman SW, Tharmarajah G, Walker DW, Pletcher SD, Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.