Summary
Lipoate scavenging from the human host is essential for malaria parasite survival. Scavenged lipoate is covalently attached to three parasite proteins: the H-protein and the E2 subunits of branched chain amino acid dehydrogenase (BCDH) and α-ketoglutarate dehydrogenase (KDH). We show mitochondrial localization for the E2 subunits of BCDH and KDH, similar to previously localized H-protein, demonstrating that all three lipoylated proteins reside in the parasite mitochondrion. The lipoate ligase 1, LipL1, has been shown to reside in the mitochondrion and it catalyzes the lipoylation of the H-protein; however, we show that LipL1 alone cannot lipoylate BCDH or KDH. A second mitochondrial protein with homology to lipoate ligases, LipL2, does not show ligase activity and is not capable of lipoylating any of the mitochondrial substrates. Instead, BCDH and KDH are lipoylated through a novel mechanism requiring both LipL1 and LipL2. This mechanism is sensitive to redox conditions where BCDH and KDH are exclusively lipoylated under strong reducing conditions in contrast to the H-protein which is preferentially lipoylated under less reducing conditions. Thus, malaria parasites contain two different routes of mitochondrial lipoylation, an arrangement that has not been described for any other organism.
Introduction
Plasmodium falciparum (Pf) is the deadliest of five apicomplexan parasite species capable of causing malaria in humans (Okiro et al., 2009a, Okiro et al., 2009b, Mudhune et al., 2011, Okiro et al., 2011). Plasmodium spp. first invade the liver and undergo a single round of replication. This phase is asymptomatic, in contrast to the erythrocytic stage of the parasite life cycle, which is characterized by a cyclical fever corresponding to multiple rounds of parasite replication within red blood cells. Increasing resistance to current drugs that target erythrocytic stage parasites makes it necessary to characterize pathways that are essential for parasite survival (Fidock et al., 2008, Hay et al., 2009, Dondorp et al., 2011, Phyo et al., 2012). One such pathway believed to be essential for malaria parasites is lipoate metabolism (Allary et al., 2007, Spalding et al., 2010b, Storm et al., 2012). The parasite contains two organelles that harbor non-redundant pathways for lipoate metabolism: the apicoplast -a plastid organelle- and the mitochondrion (Gunther et al., 2005, Allary et al., 2007, Spalding et al., 2010b). The apicoplast contains a lipoate biosynthesis pathway that is critical for liver stage parasite development, but is dispensable in the erythrocytic stage (Gunther et al., 2007, Pei et al., 2010, Falkard et al., 2013). The mitochondrion relies on lipoate scavenging from the host, which has been proposed to be essential for parasite survival during the erythrocytic stage (Allary et al., 2007, Gunther et al., 2009, Storm et al., 2012).
Lipoate is an essential cofactor for aerobic metabolism in oxidative decarboxylation reactions of 2-oxoacid complexes. Each complex is comprised of three subunits named E1, E2 and E3, except for glycine cleavage system which does not form a stable complex (Reed et al., 1958). Lipoate is attached to a conserved lysine residue of a small lipoyl domain (LD) through an amide bond, serving as a swinging arm to shuffle reaction intermediates between active site s (Reche et al., 1999, Perham, 2000). LDs are typically found at the N-terminus of the E2-subunit of the 2-oxoacid dehydrogenase complexes. The H-protein of the glycine cleavage system (GCV) functions as a LD. Lipoate shuffles the decarboxylated acid from the E1 subunit through a thioester linkage to the core of the E2-subunit where the acyl thioester is converted to the corresponding Coenzyme-A (CoA) thioester by thioester exchange. The dihydrolipoamide is oxidized to lipoamide by the dihydrolipoamide dehydrogenase (E3 subunit) to reset the catalytic cycle. The glycine cleavage system works in a similar fashion, but instead of three subunits it is composed of four loosely associated proteins: the P-protein (corresponding to the E1 subunit), the H-protein (lipoyl domain), the T-protein (core of E2-subunit) and the L-protein, which is the E3 subunit (Spalding et al., 2010a).
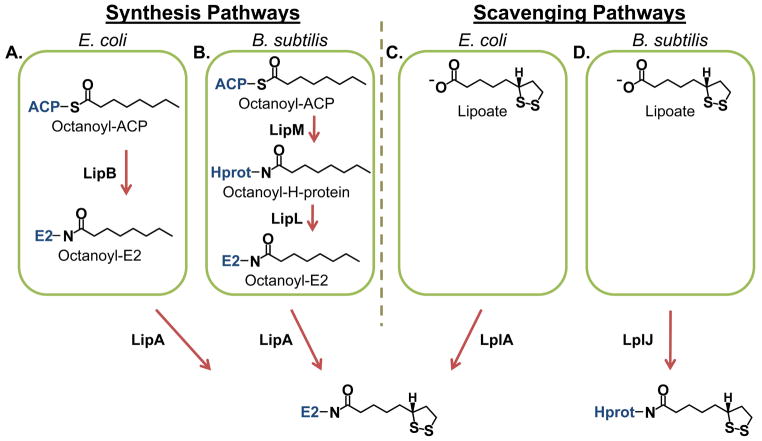
Lipoate metabolism has been best characterized in Escherichia coli (E. coli), which has both biosynthetic and scavenging pathways (Fig. 1A and 1C). In E. coli, unlike in Plasmodium spp., these two pathways have been shown to be redundant (Morris et al., 1995). Lipoate biosynthesis starts with an octanoyltransferase (EcLipB), which transfers the octanoyl group from octanoyl-Acyl Carrier Protein (octanoyl-ACP) to a conserved lysine of the lipoate requiring proteins (Jordan et al., 2003). Then a lipoate synthase (EcLipA) inserts two sulfur atoms in positions C6 and C8 of the octanoyl group to produce the lipoyl moiety (Fig. 1A) (Miller et al., 2000). The scavenging pathway uses a lipoate ligase (EcLplA) to attach lipoate to LDs in an ATP-dependent reaction (Fig. 1C) (Morris et al., 1995, Fujiwara et al., 2010).
Fig. 1.
Lipoate synthesis and scavenging pathways.
(A) Lipoate synthesis as observed in E. coli. Octanoyl groups from octanoyl-ACP (acyl carrier protein) are transferred to E2 proteins (and the H-protein) by an octanoyltransferase (LipB) followed by the insertion of sulfur atoms by a lipoate synthase (LipA).
(B) Lipoate synthesis as observed in B. subtilis. Octanoyl groups are transferred specifically to the H-protein by octanoyltransferase (LipM) and are subsequently distributed to the E2 proteins by an amidotransferase (LipL). As in E. coli, sulfur insertion is catalyzed by a lipoate synthase (LipA).
(C) Lipoate scavenging as observed in E. coli. Lipoate scavenged from the external environment is attached to E2 proteins (and the H-protein) by an ATP-dependent lipoate ligase (LplA).
(D) Lipoate scavenging as observed in B. subtilis. The lipoate ligase (LplJ) has specificity for the H-protein.
Lipoate synthesis in the apicoplast of Pf follows the same pathway as the biosynthetic pathway described for E. coli (Foth et al., 2005, Spalding et al., 2010b). However, less is known about the mechanism of lipoate metabolism in the parasite mitochondrion, which relies on lipoate scavenged from the host (Allary et al., 2007). Once lipoate is imported into the mitochondrion, there are two candidate enzymes, lipoate ligase 1 (LipL1) and lipoate ligase 2 (LipL2), which could catalyze the attachment of free lipoate to the lipoate requiring proteins. LipL1 and LipL2, which have both been localized to the mitochondrion (Wrenger et al., 2004, Gunther et al., 2007), share about 21% sequence identity and are unlikely to be paralogs (Allary et al., 2007). Three lipoate requiring proteins have been localized or predicted to be in the mitochondrion: branched chain amino acid dehydrogenase (BCDH), α-ketoglutarate dehydrogenase (KDH), and the H-protein of the GCV (Gunther et al., 2005). The H-protein has been previously localized to the mitochondrion (Spalding et al., 2010a) and LipL1 can lipoylate the H-protein in an ATP-dependent reaction (Allary et al., 2007). There is no known activity for LipL2 in the Pf mitochondrion, and no mechanism to lipoylate E2-BCDH or E2-KDH has yet been described.
It has been proposed that lipoate scavenged from red blood cells is essential to the parasite (Storm et al., 2012). The halogenated compound 8-bromooctanoate (8-BrO) inhibits parasite growth and reduces lipoylation of lipoate-requiring proteins in the mitochondrion (Allary et al., 2007). The reliance on scavenged lipoate makes this an attractive process to target with therapeutics. Yet, several questions remain unanswered: what is the compartmentalization of BCDH and KDH substrates; why does the parasite possess two non-paralogous lipoate ligases; what is their activity and substrate specificity against the three possible lipoate requiring substrates in the mitochondrion; and last, how does 8-BrO inhibit parasite growth?
Here we present mitochondrial localization data for the E2 subunits of BCDH and KDH in Pf. We show that LipL1 is the only active lipoate ligase in the parasite mitochondrion and is capable of forming the lipoylation reaction intermediate lipoyl-AMP. LipL1 modifies the H-protein through an ATP-dependent reaction; however LipL1 does not recognize the other substrates, BCDH and KDH. LipL2 does not exhibit ligase activity against any of the mitochondrial substrates, nor is it capable of synthesizing lipoyl-AMP. Interestingly, both LipL1 and LipL2 are required in an ATP-dependent reaction to lipoylate BCDH and KDH. These two mitochondrial lipoylation pathways show dependence on the lipoate redox state. The H-protein can be lipoylated under all conditions tested, but increased lipoylation is observed under less reducing conditions. In contrast to the H-protein, BCDH and KDH are only lipoylated under strong reducing conditions. Last, a lipoate analog that mimics the reduced form, 6,8-dichlorooctanoate (6,8-diClO), causes blood stage parasite growth inhibition similar to 8-BrO. 6,8-diClO can be scavenged by the parasite and attached to BCDH and KDH in the mitochondrion. Modification with 6,8-diClO or 8-BrO leads to irreversible inactivation of these mitochondrial enzymes and subsequent parasite death.
Results
The E2 subunits of BCDH and KDH localize to the parasite mitochondrion
To better understand lipoylation in the mitochondrion of Plasmodium falciparum (Pf), we first investigated the localization of the two lipoylated substrates predicted to be mitochondrial: the E2 subunits of BCDH and KDH (Gunther et al., 2005). The E1β subunit of BCDH and the shared E3 subunit have been previously localized to the mitochondrion in Pf, showing that components of 2-oxoacid dehydrogenase reside in this compartment (Gunther et al., 2005). The E2-BCDH of Plasmodium berghei has been localized to the mitochondrion (Falkard et al., 2013). To confirm the localization of the Pf E2 proteins, we generated constructs in which the first 30 amino acids containing the predicted mitochondrial targeting sequence of E2-BCDH and E2-KDH were fused to green fluorescent protein (GFP). The transgene was inserted into the Pf cg6 locus by mycobacteriophage integrase mediated recombination (Nkrumah et al., 2006, Spalding et al., 2010a) and GFP fluorescence from these parasites was analyzed by live microscopy. For both E2-GFP transgenic parasite lines, we observed GFP fluorescence in an organelle with branched-like morphology that co-localized with the mitochondrial marker MitoTracker (Fig. 2A). We confirmed expression of these constructs by western blot (Fig. 2B). These results demonstrate that the lipoylated E2 subunits of BCDH and KDH, in addition to the H-protein, reside in the mitochondrion with the previously localized lipoate attachment enzymes, LipL1 and LipL2 (Wrenger et al., 2004, Gunther et al., 2007).
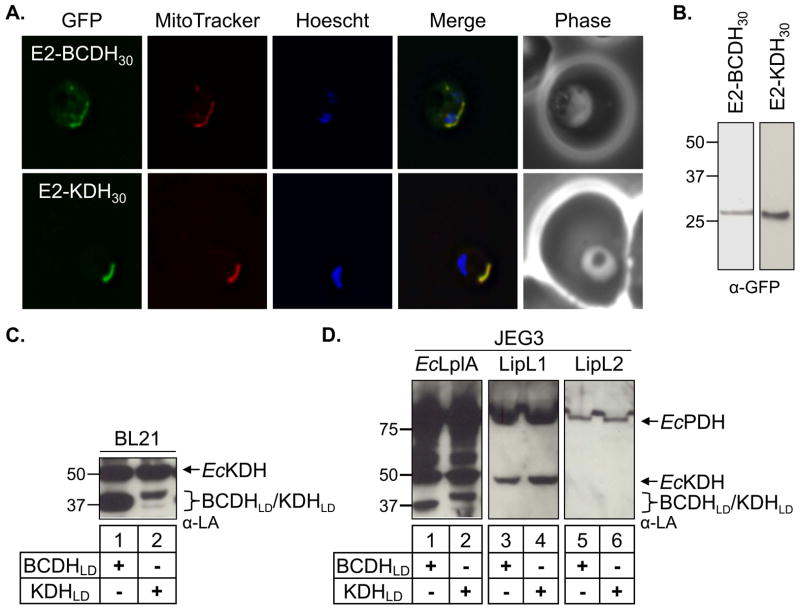
Fig. 2.
BCDH and KDH localization and cell-based lipoylation assays.
(A) Epifluorescent images of live P. falciparum erythrocytic-stage parasites expressing GFP fused to either the BCDH leader peptide (E2-BCDH30) or the KDH leader peptide (E2-KDH30). The parasites were stained with MitoTracker to identify mitochondria and Hoescht to identify nuclei. Image z-stacks were deconvolved and then presented as a single combined image. In both cases, GFP fluorescence co-localizes with MitoTracker to the mitochondrion.
(B) Expression of the E2-BCDH30-GFP and E2-KDH30-GFP constructs. An α-GFP western blot of parasite lysates identifies both proteins with the predicted molecular mass.
(C) Lipoylation of BCDHLD and KDHLD in BL21 E. coli. The lipoylation domains (LD) of both the E2-BCDH and the E2-KDH are lipoylated in anti-lipoate (α–LA) western blots of bacterial lysate, verifying that these constructs act as lipoylation substrates.
(D) Cell-based assay of lipoate ligase activity. Lipoylation deficient JEG3 cells were used to test the activity of three candidate lipoate ligases (EcLplA, LipL1 and LipL2) with two lipoylation substrates (BCDHLD and KDHLD). E. coli lipoate ligase (EcLplA) is sufficient to lipoylate BCDHLD and KDHLD, whereas neither LipL1 nor LipL2 lipoylate either parasite substrate.
Neither LipL1 nor LipL2 lipoylate BCDHLD or KDHLD in a cell-based lipoylation assay
The lipoate attachment enzymes LipL1 and LipL2 are predicted to be lipoate ligases based on sequence homology to the E. coli lipoate ligase (EcLplA) (Allary et al., 2007). LipL1 can complement a lipoylation deficient E. coli cell line (TM136) in minimal media by adding exogenous lipoate, demonstrating its activity as a ligase (Allary et al., 2007). LipL2 has been shown to have similar activity in TM136 cells but cell growth required four days of incubation, making this datum less conclusive (Allary et al., 2007). An additional factor complicating these experiments involves the low level of endogenous lipoylation that remains in the TM136 line. Apparently, transposon disruption of the lplA gene does not completely ablate lipoate ligase activity in TM136 (Fig. S1). Therefore, we used a lambda red recombination system (Datsenko et al., 2000, Baba et al., 2006) to generate an E. coli lplA and lipB deletion line (lplA-/lipB-) that is completely deficient in lipoylation activity (Fig. S1). We used this new strain (JEG3) to conduct cell-based lipoylation assays. The lipoylation domains of BCDH and KDH (BCDHLD and KDHLD, respectively) were subcloned into a pGEX vector expressing GST (glutathione S-transferase) fusion proteins. To validate that BCDHLD and KDHLD are competent lipoylation substrates, we expressed each domain in wild type BL21 cells and found that they were lipoylated by E. coli lipoylation enzymes when visualized by western blot using an antibodies specific for lipoylated proteins (α-LA) (Fig. 2C). We next subcloned LipL1, LipL2 and EcLplA into a pMAL vector expressing MBP (maltose binding protein) fusion proteins. We co-expressed each LD in the JEG3 cell line with either Pf ligase or EcLplA. We found that the EcLplA was sufficient to lipoylate BCDHLD and KDHLD (Fig. 2D, lanes 1–2), but neither substrate was lipoylated in our in vitro cell-based assay by either putative Pf ligase when visualized by western blot (Fig. 2D, lanes 3–6). The lack of activity against BCDHLD and KDHLD was surprising since LipL1 is competent to lipoylate EcKDH and EcPDH (Fig. 2D, lanes 3–4) and LipL2 apparently lipoylates EcPDH, albeit poorly (Fig. 2D, lanes 5–6).
LipL1 has substrate specific lipoate ligase activity
To verify the unexpected behavior of LipL1 and LipL2, we decided to study the activity of purified recombinant enzymes. The BCDHLD and KDHLD lipoylation domains and truncated H-protein, lacking the N-terminal transit peptide, were expressed as GST fusion proteins in the lipoylation deficient JEG3 cell line and purified in the apo form. We tested recombinant LipL1 and LipL2 alone against each apo substrate in the presence or absence of ATP, and visualized lipoylation by western blot. As shown in Figure 3A, LipL1 is an active ligase only against the H-protein in an ATP-dependent reaction (top panel, lanes 1–2) in agreement with a previous report in which full length H-protein was used (Allary et al., 2007). We also found that ATP is the preferred nucleotide used by LipL1 with minor activity observed with GTP (Fig. S2A). Consistent with results observed in the cell-based assay, LipL1 did not catalyze the lipoylation of BCDHLD or KDHLD (Fig. 3A, top panel, lanes 3–6). Similarly, we found that LipL2 did not lipoylate any of the Pf substrates in our in vitro ligation assay (Fig. 3A, lower panel). These results, along with those from the in vitro cell-based assay, show that BCDHLD and KDHLD are lipoylated by E. coli lipoylation enzymes, but not by either candidate Pf lipoate ligase. LipL1 appears to be the only lipoate ligase with the H-protein as its only parasite substrate.
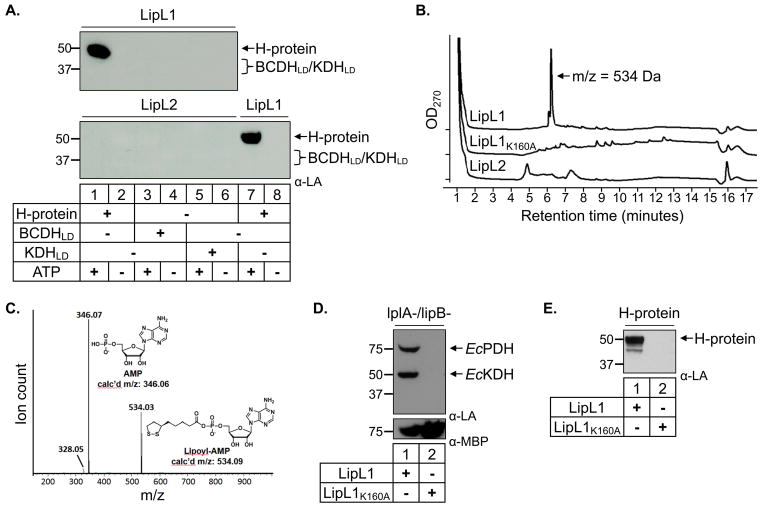
Fig. 3.
In vitro ligation assays.
(A) Lipoate ligase activity. Western blot analysis shows that LipL1 is an ATP-dependent lipoate ligase only when H-protein is used as substrate, while LipL2 does not display lipoate ligase activity with any substrate.
(B) HPLC traces for LipL1, LipL1K160A and LipL2 adenylation reactions. Each enzyme was reacted with lipoate and ATP for 10 minutes prior to HPLC analysis with absorbance monitored at 270 nm. The LipL1 reaction produced a major product eluting at 6.2 minutes with a mass of 534 Da as determined by ESI-MS.
(C) Identification of Lipoyl-AMP produced by LipL1. The major product generated by LipL1 was subjected to collision induced dissociation MS/MS, demonstrating that this product contains AMP.
(D) JEG3 complementation by LipL1 and LipL1K160A. LipL1 is able to lipoylate endogenous E. coli substrates in lipoylation deficient strain JEG3, whereas LipL1K160A is unable to catalyze this activity. Expression of both proteins was confirmed by stripping and re-probing the blot with α-MBP (lower panel).
(E) Recombinant LipL1K160A cannot lipoylate the H-protein in an in vitro lipoylation assay.
LipL1 is the only active ligase
Lipoate ligases typically catalyze two sequential reactions. First, lipoate is activated in an adenylation reaction forming lipoyl-AMP; then the lipoyl moiety is transferred to protein substrates (Fujiwara et al., 2010). Based on the results shown in Figure 3A, LipL1 appears to catalyze both reactions when the H-protein is used as a substrate. We examined LipL1 incubated with lipoate and ATP for formation of the lipoyl-AMP intermediate. Quenched reactions were separated by HPLC and the major product peak was analyzed by mass spectrometry and found to have a mass of 534 Da, consistent with that of lipoyl-AMP (Fig. 3B). To further confirm the identity of this 534 Da product, we subjected it to collision induced dissociation (CID) MS/MS which produced a peak at 346 m/z corresponding to an AMP fragment (Fig. 3C). Similar experiments with LipL2 showed that this enzyme is not capable of synthesizing lipoyl-AMP (Fig. 3B); minor peaks in the LipL2 reaction can also be found in controls (Fig. S3). These results demonstrate that LipL2 cannot form the active lipoyl-AMP intermediate and that only LipL1 is a lipoate ligase.
We next decided to interfere with the ligase activity of LipL1 through site directed mutagenesis. Mutation of an active site lysine residue in E. coli lipoate ligase (EcLplA) abolishes both the adenylation and transfer reactions (Fujiwara et al., 1994, Fujiwara et al., 2010). Using multiple sequence alignments, we were able to identify the corresponding lysine in LipL1 (K160), but not in LipL2. We proceeded to mutate Lys160 in LipL1 (LipL1K160A) and express it in our lipoylation deficient JEG3 cell line to assess its activity. We found that LipL1K160A is not able to lipoylate E. coli proteins even though we can detect the expression of soluble mutant protein by anti-MBP western blot (Fig. 3D). We then expressed and purified LipL1K160A, and tested its activity against the H-protein in our in vitro assay. As expected, this mutant is unable to lipoate the H-protein (Fig. 3E). We also tested the ability of LipL1K160A in forming the reaction intermediate lipoyl-AMP as done for LipL1 and LipL2. As expected, we found that LipL1K160A is unable to form lipoyl-AMP (Fig. 3B). Taken together, we found that LipL1 lipoate ligase activity depends on an active site lysine for the adenylation reaction, and possibly for the transfer reaction, similar to what has been described for EcLplA.
Neither LipL1 nor LipL2 acts as an amidotransferase
Lipoate metabolism has also been described in other systems such as Bacillus subtilis (Christensen et al., 2011, Martin et al., 2011). In B. subtilis, an enzyme similar to E. coli LipB (BsLipM) specifically octanoylates the H-protein, but does not modify other protein substrates (Fig. 1B). An amidotransferase is then required to transfer octanoyl (or possibly lipoyl) groups from the H-protein to the remaining four lipoate-dependent proteins in B. subtilis (Christensen et al., 2011, Martin et al., 2011). We tested whether LipL1 or LipL2 could act as an amidotransferase in an assay in which the only lipoate source is the holo H-protein. We generated holo H-protein by expressing the Pf H-protein and LipL1 in the E. coli lipoylation deficient JEG3 cell line. The H-protein was then purified in the holo form by GST affinity chromatography. We tested the transferase activity of either LipL1 or LipL2 and found that neither enzyme can act as an amidotransferase (Fig. 4A, lanes 1–6). Even when LipL1 and LipL2 are combined in the same reaction, they are unable to transfer lipoyl groups from the H-protein to either BCDHLD or KDHLD (Fig. 4A, lanes 7–9). Thus, neither enzyme appears to be a lipoyl amidotransferase. We did not test LipL1 or LipL2 for the octanoyl amidotransferase activity observed in B. subtilis because there is no known source of octanoyl groups (fatty acid biosynthesis or degradation pathways) in the parasite mitochondrion.
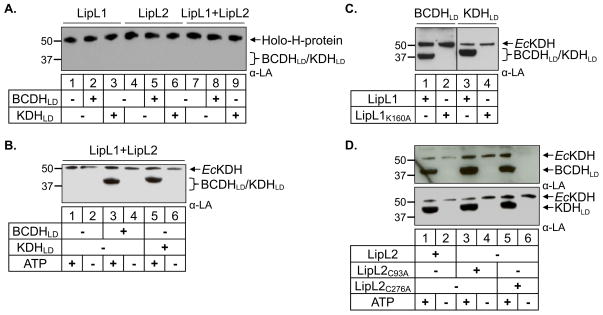
Fig. 4.
LipL1 and LipL2 are required for BCDHLD and KDHLD lipoylation in an ATP-dependent reaction.
(A) Lipoyl amidotransferase activity. Western blot analysis shows that LipL1 and LipL2 are not able to transfer lipoyl groups from Holo-H-protein to either BCDHLD or KDHLD.
(B) Combined LipL1 + LipL2 lipoate ligase activity. Both LipL1 and LipL2 are needed for lipoylation of BCDHLD and KDHLD in an ATP-dependent lipoate ligase reaction. Note a minor population of E. coli E2-KDH found in purified LipL2.
(C) Catalytically active LipL1 is required for the lipoylation of both BCDHLD and KDHLD.
(D) Mutation of both conserved cysteine residues in LipL2 (C93A and C276A) did not prevent lipoylation of BCDHLD and KDHLD, showing that lipoylation does not proceed through a thioester intermediate.
LipL1 and LipL2 are both required to lipoylate BCDHLD and KDHLD in a ligation reaction
We next tested whether both LipL1 and LipL2 are required to ligate free lipoate to BCDHLD or KDHLD in our in vitro ligation assay. In these reactions, equimolar amounts of LipL1 and LipL2 were combined with substrate in the presence or absence of ATP. As shown in Fig. 4B, both enzymes are required to lipoylate BCDHLD and KDHLD in an ATP-dependent ligation reaction (Fig. S2B). Thus, LipL1 and LipL2 function together as a lipoate ligase for both substrates. Taken together, our results suggest two different routes by which the mitochondrial substrates of Pf are lipoylated: one in which LipL1 alone is sufficient to lipoylate the H-protein, and a second route in which both LipL1 and LipL2 are necessary to lipoylate BCDHLD and KDHLD.
LipL1 activity is essential for lipoylation of BCDHLD and KDHLD
We next addressed the role of LipL1 in the lipoylation of BCDHLD and KDHLD catalyzed by LipL1 and LipL2. We found that substituting LipL1 with the LipL1K160A mutant abolished lipoylation of both substrates (Fig. 4C), showing that LipL1 catalytic activity is essential for the lipoylation of these substrates. We know that LipL1 is needed to catalyze the formation of the lipoyl-AMP intermediate. LipL2 could then transfer the lipoyl group to protein substrates, or it could have a non-catalytic role in recruiting these substrates for lipoylation by LipL1. In an attempt to understand LipL2 activity we decided to mutate residues that could be essential for its activity. As previously mentioned, we found no conserved lysine residue in LipL2 corresponding to LipL1 K160. A second possibility is that LipL2 could transfer lipoyl groups using a cysteine residue, as observed in Bacillus subtilis (Christensen et al., 2011, Martin et al., 2011). We mutated two conserved cysteine residues based on alignment against other Plasmodium LipL2 enzymes. We expressed and purified these two mutants (LipL2C93A and LipL2C256A) and tested their activity in our in vitro assay. We found that both mutants were still active in our assays, showing that LipL2 activity does not proceed through a thioester intermediate (Fig. 4D). Although LipL2 is required for the lipoylation of BCDHLD and KDHLD, its role in the reaction remains unclear.
The two lipoylation routes differ in their redox dependence
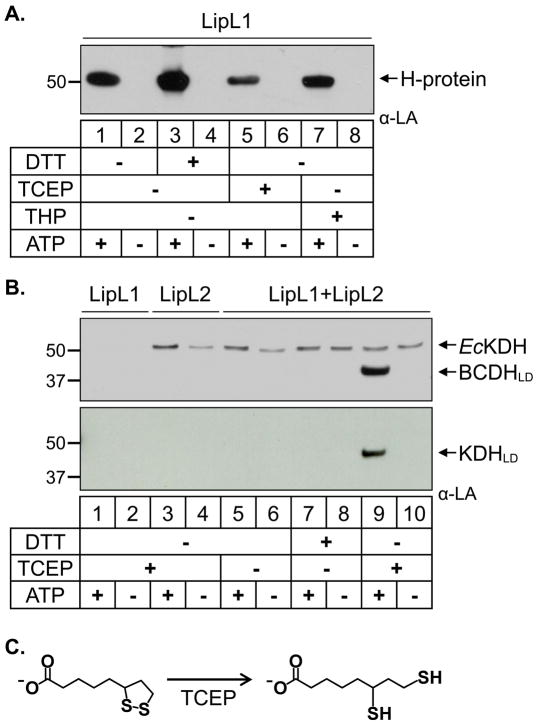
The lipoylation assays described in the previous sections were performed using tris(2-carboxyethyl)phosphine (TCEP) as reducing agent. As observed, all substrates can be modified in the presence of TCEP, but we found that the two lipoylation routes differed in their redox dependence. In order to better understand this effect we tested three reducing agents, dithiothreitol (DTT), TCEP, and tris(hydroxypropyl)phosphine (THP) in our in vitro ligation assay. We first tested the lipoylation of the H-protein by LipL1 in the presence and absence of the reducing agents. As seen in Fig. 5A, the H-protein can be lipoylated by LipL1 under oxidizing or reducing conditions, albeit at different levels. There was enhanced activity of LipL1 under reducing conditions with DTT as compared to no reducing agent, perhaps because reducing agents in general help to protect lipoate from oxidative damage under aerobic conditions. However, the strong reducing agents TCEP and THP resulted in lower LipL1 activity compared to the weaker reducing agent DTT. The lower activity of LipL1 in the phosphine reducing agents may have to do with the ability of agents like TCEP to reduce the dithiolane ring of lipoate (Burns et al., 1991).
Fig. 5.
Redox dependence of the two lipoylation routes.
(A) Redox dependence of H-protein lipoylation. Western blot analysis shows that lipoylation of the H-protein by LipL1 is enhanced in the presence of DTT.
(B) Dependence of the LipL1 + LipL2 reaction on strong reducing conditions. Western blots show that lipoylation of BCDHLD and KDHLD occur only in the presence of the strong reducing agent TCEP. Note a minor population of E. coli E2-KDH found in purified LipL2.
(C) Redox states of lipoate. The oxidized ring form of lipoate is reduced to the dihydrolipoate form by treatment with TCEP or THP.
We next tested the activity of both LipL1 and LipL2 in lipoylating BCDHLD and KDHLD under different reducing environments. In contrast to the H-protein, BCDHLD and KDHLD are not lipoylated in the absence of reducing agents or under reducing conditions with DTT (Fig. 5B, lanes 5–8). The LipL1/LipL2 reaction only occurs in the presence of the strong reducing agents TCEP (Fig. 5B, lane 9) or THP (Fig. S4, top panel). However, it is not clear from these results whether these strong reducing agents are acting on the lipoate redox state (Fig. 5C) or affecting the enzymes (LipL1 and/or LipL2) and/or protein substrates (BCDHLD and KDHLD).
6,8-dichlorooctanoate (6,8-diClO) is a non-redox sensitive lipoate analog
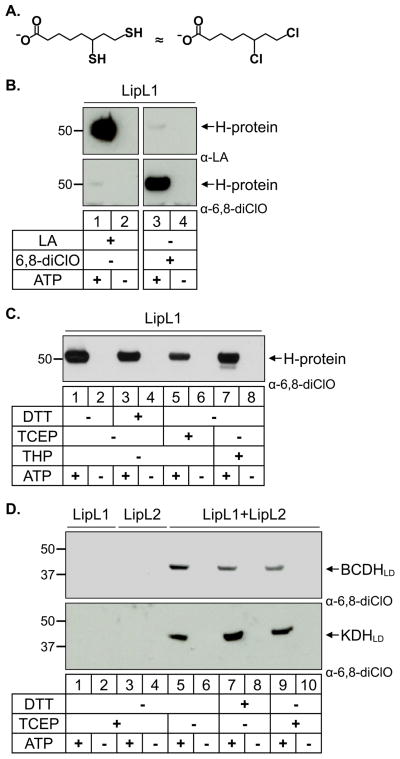
To further probe the requirements of the Pf lipoylation pathways, we synthesized a lipoate analog, 6,8-dichlorooctanoate (6,8-diClO), which mimics the reduced form of lipoate but is not redox active (Fig. 6A). We generated antiserum that is specific for proteins modified with 6,8-diClO and showed that LipL1 can use 6,8-diClO as a substrate to modify the H-protein (Fig. 6B, lower panel). As shown in Figure 6B, α-LA antibodies and the α-6,8-diClO antiserum are specific for their respective substrates, even when the western blots are significantly overexposed. Using these reagents we tested the different lipoylation routes and their redox dependence in our in vitro assay.
Fig. 6.
6,8-dichlorooctanoate is attached to all substrates independent of redox environment.
(A) 6,8-Dichlorooctanoate (6,8-diClO) is a structural analog of reduced lipoate (dihydrolipoate).
(B) Modification of the H-protein by lipoate and 6,8-diClO. LipL1 catalyzes the attachment of lipoate as shown in the α-LA western blot (top panels) and 6,8-diClO as shown in the α-6,8-diClO western blot (bottom panels). The α-LA antibodies and the α-6,8-diClO antiserum show very little cross-reactivity.
(C) Effect of reducing conditions on the enzymatic activity of LipL1 against the H-protein. The H-protein is modified by 6,8-diClO independent of the redox environment.
(D) Effect of reducing conditions on the enzymatic activity of LipL1 + LipL2. BCDHLD and KDHLD are modified by 6,8-diClO independent of the redox environment.
When 6,8-diClO was used as a substrate to modify the H-protein, DTT was no longer the most favorable reducing agent and LipL1 activity appeared to be independent of redox conditions (Fig. 6C). The lower activity with TCEP (also observed in Fig. 5A) probably results from the instability of TCEP in phosphate buffer rather than a redox effect (Hansen et al., 2009, Svagera et al., 2012). The other strong reducing agent THP only decreased LipL1 activity relative to DTT when lipoate, but not 6,8-diClO was used as a substrate, indicating a redox-specific effect on lipoate itself (compare lanes 3 & 7 from Figs. 5A and 6C). A similar phenomenon was observed for the lipoylation of BCDHLD and KDHLD. LipL1 and LipL2 modified these substrates with 6,8-diClO independent of the redox environment (Fig. 6D and Fig. S4, bottom panel). These results demonstrate that the requirement for the strong phosphine reducing agents in the LipL1/LipL2 lipoylation reactions is actually a requirement for reduced lipoate in these reactions and that the proteins themselves do not need to be reduced in order to be functional (Fig. 5C). The redox state of lipoate appears to be important for the lipoylation of the different mitochondrial substrates. BCDHLD and KDHLD are only modified when lipoate is in the reduced state, while the H-protein shows increased lipoylation under less reducing conditions.
Lipoate analogs are scavenged and attached to mitochondrial substrates
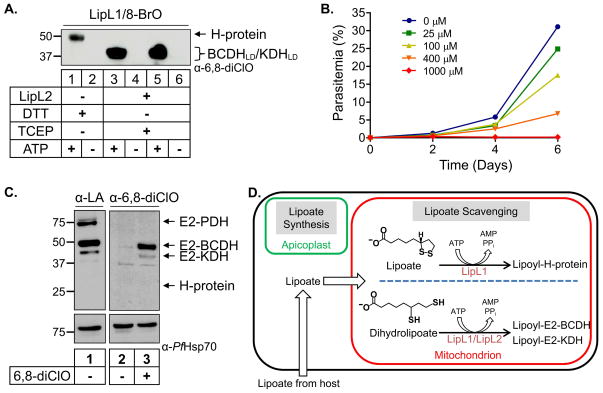
We have previously shown that the lipoate analog 8-BrO inhibits the growth of blood stage Pf and that treatment with 8-BrO reduces the lipoylation of the protein substrates found in the mitochondrion of Pf (Allary et al., 2007). From these results, it was not clear whether 8-BrO blocked lipoate import, activation and transfer of lipoate; or if attachment of 8-BrO to the mitochondrial substrates inhibited their activity. We know from the above experiments that 6,8-diClO can be attached to all mitochondrial substrates in our in vitro assay (see Fig. 6C and D). To determine if 8-BrO can be attached to the parasite proteins, we used the α-6,8-diClO antiserum, which also recognizes proteins modified with 8-BrO. As shown in Fig. 7A, 8-BrO can be attached to all three mitochondrial substrates in an ATP-dependent reaction in our in vitro assay, suggesting that both lipoate analogs share a similar mode of action in malaria parasites.
Fig. 7.
Modification of mitochondrial proteins with lipoate analogs.
(A) Attachment of 8-bromooctanoate (8-BrO) to lipoylation substrates in vitro. An α-6–8-diClO western blot shows that LipL1 can attach 8-BrO to the H-protein and that LipL1+LipL2 can attach 8-BrO to BCDHLD and KDHLD.
(B) Growth inhibition of P. falciparum treated with 6,8-diClO. The lipoate analog 6,8-diClO inhibits blood stage parasite growth in a dose-dependent manner.
(C) Uptake and attachment of LA and 6,8-diClO in parasite culture. Western blot analysis of parasite lysate shows that treatment of P. falciparum parasites with either 6,8-diClO or LA results in the modification of E2-BCDH and E2-KDH. Lipoylated E2-PDH is derived from a synthesis pathway in the apicoplast and not from lipoate scavenging.
(D) Mitochondrial lipoylation pathways in Plasmodium falciparum. The mitochondrion relies on lipoate scavenging from the host while the apicoplast contains an isolated lipoate synthesis pathway. Our data suggest that there are two routes for lipoylation of protein substrates in the mitochondrion. The H-protein is solely lipoylated by LipL1 in an ATP-dependent reaction; this activity is enhanced under less reducing conditions. Both LipL1 and LipL2 are needed for lipoylation of the E2 domains of BCDH (E2-BCDH) and KDH (E2-KDH) in an ATP-dependent reaction which only occurs under conditions capable of reducing lipoate to dihydrolipoate.
We also treated asexual blood stage parasites with 6,8-diClO at different concentrations as done previously for 8-BrO (Allary et al., 2007). We found that 6,8-diClO significantly inhibits parasite growth at a concentration of 100 μM (Fig. 7B), similar to growth inhibition results observed for 8-BrO. To determine whether this lipoate analog is attached to mitochondrial proteins in the parasites, we treated cultured parasites with 0 or 10 μM 6,8-diClO and probed the lysates with α-6,8-diClO antiserum. Both E2-BCDH and E2-KDH were modified with 6,8-diClO, demonstrating that this compound can be imported into parasites and used by the mitochondrial lipoylation enzymes (Fig. 7C). In parallel experiments, we were able to detect lipoylated E2-BCDH, E2-KDH, and E2-PDH in parasites treated with lipoate (Fig. 7C). Lipoylation of E2-PDH is independent of lipoate scavenging, consistent with the finding that lipoate is synthesized in the apicoplast (Gunther et al., 2005). We were unable to detect modification of the H-protein by either substrate, possibly due to low expression levels of this protein in the parasite.
Discussion
The mitochondrion of Plasmodium falciparum (Pf) relies on lipoate scavenging from the host. Treatment of parasites with the lipoate analog 8-bromooctanoate (8-BrO) inhibited parasite growth and specifically blocked the lipoylation of mitochondrial proteins (Allary et al., 2007). These results indicated that lipoate scavenging is essential for the survival of malaria parasites and that the lipoylation machinery in the parasite mitochondrion would likewise be essential. Plasmodium spp. possess two putative lipoate ligases in the genome (LipL1 and LipL2) that are believed to catalyze the attachment of free lipoate to the lipoate requiring proteins in an ATP-dependent reaction. In Pf, LipL1 was localized to the mitochondrion (Wrenger et al., 2004) while LipL2 was reportedly found in the mitochondrion as well as the apicoplast organelle (Gunther et al., 2007). Although the role of LipL2 in the apicoplast is not well defined, both LipL1 and LipL2 are involved in the lipoylation of three proteins in the mitochondrion of malaria parasites. We have previously shown that one of these proteins, the H-protein, is mitochondrial (Spalding et al., 2010a), and in this work we confirmed the mitochondrial localization of the E2 subunits of BCDH and KDH (Fig. 2A). Thus, the mitochondrion of erythrocytic stage Pf contains two putative lipoate ligases responsible for the lipoylation of three protein substrates.
It was previously shown that LipL1 can catalyze the attachment of lipoate to full length H-protein in an ATP-dependent reaction (Allary et al., 2007) and it was assumed that LipL1 could lipoylate all of the mitochondrial substrates. Attempts to disrupt Pf LipL1 and the P. berghei ortholog failed (Gunther et al., 2009) reinforcing the idea that lipoate scavenging is essential for blood stage malaria parasites and that LipL1 plays a key role in this process. In the present study, we observed lipoylation of the H-protein by LipL1, and found that it preferentially uses ATP as a nucleotide substrate (Fig. S2A). Furthermore, we were able to demonstrate that LipL1 catalyzes formation of the reaction intermediate, lipoyl-AMP, and that LipL1 adenylation activity depends on a conserved lysine residue (Fig. 3). However, LipL1 is unable to lipoylate Pf BCDH and Pf KDH (Figs. 2D and 3A), while EcLplA is able to lipoylate both parasite proteins (Fig. 2D). It appears that LipL1 can distinguish between the H-protein and the other protein substrates found in malaria parasites. This may be because H-protein sequences are divergent from other E2 lipoyl domains (Christensen et al., 2011) and seem to represent a distinct clade of lipoate-dependent proteins. Indeed, phylometabolomic analysis places the H-protein (and the glycine synthesis pathway) at the base of the tree of life, suggesting that the other lipoyl domains arose subsequently (Braakman et al., 2013).
The specificity of LipL1 for the H-protein is reminiscent of the lipoylation pathways described for B. subtilis (Fig. 1B). In B. subtilis, an amidotransferase (LipL) with homology to lipoate ligases shuffles the lipoyl or octanoyl moiety from the H-protein to the other protein substrates (Christensen et al., 2011, Martin et al., 2011). We tested LipL1 or LipL2 alone, or in complex in an amidotransferase assay using lipoyl-H-protein as substrate (parasites lack a pathway to generate octanoyl-H-protein), but did not detect any transfer of lipoyl groups to either BCDH or KDH (Fig. 4A). We also investigated whether conserved cysteine residues are essential for LipL2 activity, as has been shown for the B. subtilis LipL. Mutation of the only two cysteine residues conserved in Plasmodium LipL2 sequences did not affect LipL2 activity (Fig. 4D), further highlighting the differences between LipL2 and BsLipL. We also considered a recent report showing that a lipoate ligase homolog in Saccharomyces cerevisiae (Lip3) transfers octanoyl groups from octanoyl-CoA to protein substrates followed by sulfur insertion by a lipoate synthase (Lip5) (Schonauer et al., 2009). We did not test LipL1 or LipL2 for this type of octanoyltransferase activity since the mitochondrion of malaria parasites does not contain a lipoate synthase or a biochemical pathway likely to produce octanoyl-CoA.
The lipoylation of BCDH and KDH occurs through a novel mechanism requiring both LipL1 and LipL2 (Fig. 4B). We demonstrated that LipL1 is responsible for catalyzing activation of lipoate, forming lipoyl-AMP, and that LipL2 is not capable of catalyzing this reaction (Fig. 3B and 3C). We generated a catalytically inactive mutant of LipL1 unable to form lipoyl-AMP by substituting an active site lysine (K160) with alanine, as has been done previously with EcLplA (Fujiwara et al., 1994, Fujiwara et al., 2010). The catalytic activity of LipL1 is essential for the lipoylation of the H-protein (Fig. 3E) as well as for the combined LipL1/LipL2 lipoylation of BCDH and KDH (Fig. 4C). The role of LipL2 is less clear. It is possible that LipL2 catalyzes the transfer of lipoyl groups from lipoyl-AMP to BCDH and KDH. It is also possible that LipL2 does not have a catalytic role, but instead acts as an effector helping to recruit protein substrates that would otherwise not be recognized by LipL1. In either case, the LipL1/LipL2 reaction defines a second lipoylation pathway in the parasite mitochondrion (Fig. 7D). Attempts to disrupt the gene encoding Pf LipL2 have so far been unsuccessful (Storm et al., 2012), suggesting that this second lipoylation pathway is essential for the survival of blood stage malaria parasites.
The two mitochondrial lipoylation pathways are sensitive to redox conditions. Lipoylation of H-protein by LipL1 in the first pathway is significantly enhanced in the presence of DTT, but is less active in the presence of strong reducing agents such as TCEP and THP (Fig. 5A). The decreased lipoylation of H-protein in the presence of TCEP as compared to THP is likely due to the reactivity of TCEP in phosphate buffer (Hansen et al., 2009, Svagera et al., 2012). In contrast to the first pathway, the lipoylation of BCDH and KDH by the second pathway only occurs in the presence of TCEP or THP (Fig. 5B and S4). The redox potential for both DTT and lipoate has been measured to be −0.33 V and −0.32 V, respectively (Haramaki et al., 1997, Hanson et al., 2004). Currently, there is no measurement of redox potential for TCEP or THP, but experimental data show that TCEP readily reduces both DTT and lipoate (Burns et al., 1991). In the absence of reducing agent, lipoate is found in the oxidized, closed-ring form, but upon reduction with TCEP or THP it presumably adopts the open-ring dihydrolipoate form (Fig. 5D). We synthesized a structural analog (6,8-diClO) of dihydrolipoate and used this compound to probe the redox requirements of the second lipoylation pathway. Using 6,8-diClO as a substrate, all three proteins were modified regardless of the redox conditions (Fig. 6C and 6D). This demonstrates that the second pathway requires dihydrolipoate, and that the LipL1 and LipL2 enzymes are not themselves sensitive to redox conditions. These results have interesting connotations for how lipoylation may work and why two different routes are needed in the mitochondrion of a malaria parasite.
Our results suggest that dihydrolipoate is needed in the mitochondrion for the lipoylation of BCDH and KDH. Therefore, the redox potential of the parasite mitochondrion needs to be highly reducing or the parasites must employ a mechanism to reduce lipoate to dihydrolipoate. While there is no measurement of the redox potential for the Plasmodium mitochondrion, the redox potential of the mitochondrion of HeLa cells has been reported to be −0.36 V (Hanson et al., 2004). This value is more reducing than that of lipoate, and it might be sufficient to reduce lipoate to dihydrolipoate and/or maintain lipoate in the reduced state. Pf has two redox systems: the gluthathione and thioredoxin systems (Jortzik et al., 2012). Six proteins from these systems have been localized to the mitochondrion: thioredoxin reductase (TrxR - dually localized to cytosol and mitochondrion), thioredoxin-like protein 2 (Tlp2), peroxiredoxin (Prx1m), glutaredoxin-like protein (Glp3), and peroxidase-like thioredoxin peroxidase (TPxG1 – localized to the cytosol, mitochondria and apicoplast) (Chaudhari et al., 2012, Jortzik et al., 2012). These proteins are likely involved in maintaining the redox potential of the mitochondrion and/or defending against oxidative stress. It would be interesting to determine whether any of these proteins have the additional role of reducing lipoate. It has been shown that the thioredoxin reductase can reduce lipoate in an in vitro system (Snider et al., 2014). In addition to these redox systems, the dihydrolipoamide dehydrogenase (LipDH, PF3D7_1232200) likely recognizes free lipoate and reduces it. This has been observed for mammalian systems, in which lipoate is converted to dihydrolipoate in a NADH-dependent reaction in isolated mitochondrion (Constantinescu et al., 1995, Arner et al., 1996, Haramaki et al., 1997).
In the current study we show that lipoate metabolism in the mitochondrion of Pf follows two lipoylation routes (Fig. 7D). In the first route, lipoylation of H-protein is catalyzed solely by the lipoate ligase LipL1. In the second route, both LipL1 and LipL2 are required to lipoylate the BCDH and KDH. LipL1 catalyzes the adenylation reaction, while LipL2 could act as a lipoyl transferase or an effector protein. The two lipoylation routes are dependent on the redox state of lipoate, a phenomenon that has not been previously observed in any other system. Furthermore, we show that all three mitochondrial substrates can be modified by lipoate analogs (8-BrO and 6,8-diClO) suggesting that the lethality of these compounds results from their attachment to essential parasite proteins by lipoylation enzymes. Our results suggest that one or more of the mitochondrial protein substrates and by extension, LipL1 and LipL2, are essential for parasite survival making mitochondrial lipoylation in Plasmodium an attractive target for therapeutic intervention.
Experimental Procedures
Materials
The reducing agents used in this study were obtained from Fluka (TCEP; tris(2-carboxyethyl)phosphine), Calbiochem (THP; tris(hydroxypropyl)phosphine), and Sigma-Aldrich (DTT; dithiothreitol and DTBA; (S)-2-Aminobutane-1,4-dithiol). Antibodies were purchased from Calbiochem (rabbit α-LA polyclonal antibodies), New England Biolabs (murine α-MBP monoclonal antibody HRP conjugated) and GE Healthcare (donkey α-Rabbit IgG HRP conjugate). The following chemicals were purchased from Sigma-Aldrich: 8-BrO, NADH, NAD+ and R-LA. The compound 6,8-diClO and the α-6,8-diClO antiserum were synthesized as described below.
Expression plasmids
The malaria genome resource PlasmoDB (Bahl et al., 2003) was used to identify the genes encoding the E2 subunits of BCDH (PF3D7_0303700) and KDH (PF3D7_1320800). Nucleotides encoding the first 30 amino acids of both proteins were amplified from cDNA extracted from erythrocytic stage Plasmodium falciparum 3D7 parasites using primers BCDH30.AvrII.F and BCDH30.BsiWI.R or KDH30.AvrII.F and KDH30.BsiWI.R (Table S1). The BCDH30 and KDH30 amplicons were digested with AvrII and BsiWI and ligated into parasite transfection plasmid pRL2 (Gisselberg et al., 2013) in frame with GFP, generating plasmids pRL2-BCDH30GFP and pRL2-KDH30GFP. Plasmid pRL2 is a variant of pLN (Nkrumah et al., 2006) in which the calmodulin promoter has been replaced with the weaker ribosomal protein L2 promoter (Balabaskaran Nina et al., 2011).
Ampicillin-resistant plasmid pGEXT (Delli-Bovi et al., 2010) was used to express lipoylation domains with an amino-terminal glutathione S-transferase (GST) tag. The nucleotides encoding the lipoylation domain of BCDH (residues V34-L139) and KDH (residues I35-N152) were amplified using primers BCDH. BamHI.F100 and BCDH. EcoRI.R417 or KDH.BamHI.F103 and KDH.EcoRI.R456 (Table S1). The BCDHLD and KDHLD amplicons were digested with EcoRI and BamHI and ligated into pGEXT, generating plasmids pGEXT-BCDHLD and pGEXT-KDHLD.
A kanamycin-resistant expression plasmid, pGEXTK, was generated by subcloning the kanamycin cassette from plasmid pRK586 (Kapust et al., 2000) into the BspHI sites of pGEXT using primers Kan.BspHI.F and Kan.BspHI.R (Table S1). The nucleotides encoding BCDHLD and KDHLD were subcloned from pGEXT to pGEXTK using BamHI and SalI, generating plasmids pGEXTK-BCDHLD and pGEXTK-KDHLD. Nucleotides encoding residues E35-K200 (lacking the predicted N-terminal mitochondrial transit peptide) of the H-protein (PF3D7_1132900) were amplified from an expression plasmid similar to pMA007 (Allary et al., 2007) with primers Hprot.BamHI.F and Hprot.SalI.R (Table S1). The resulting amplicon was digested with BamHI and SalI, and ligated into pGEXTK, generating plasmid pGEXTK-Hprot.
Expression plasmid pMALcHT (Muench et al., 2003, Allary et al., 2007) was used to express lipoylation enzymes as maltose binding protein (MBP) fusion proteins with a linker region composed of a tobacco etch virus (TEV) protease cut site followed by a six histidine affinity tag. Plasmid pMA006 (Allary et al., 2007) expressing LipL1 (PF3D7_1314600) was renamed pMALcHT-LipL1 for this study. The LipL1K160A mutant created by site-directed mutagenesis of pMALcHT-LipL1 using primers LipL1K160A.F and LipL1K160A.R (Table S1) generating plasmid pMALcHT-LipL1K160A. A codon harmonized gene (Fig. S5) encoding LipL2 (PF3D7_0923600) was synthesized by GeneArt and ligated to pMALcHT using the EcoRI and HindIII sites, generating pMALcHT-LipL2. Both LipL2 mutants, C93A and C276A, were created by site-directed mutagenesis of pMALcHT-LipL2 generating pMALcHT-LipL2C93A and pMALcHT-LipL2C276A (See Table S1 for primer sequences). The gene encoding E. coli liopate ligase, EcLplA, was amplified from BL21 gDNA with primers LplA.BamHI.F and LplA.SalI.R (Table S1), digested with BamHI and SalI, and ligated to pMALcHT, generating pMALcHT-LplA.
Construction of lplA-/lipB- E. coli (JEG3 strain)
In order to construct the JEG3 cell line a lipB- E. coli strain was obtained from the Keio deletion collection (Keio JW5089). The kanamycin resistance cassette was removed by transforming JW5089 cells with the ampicillin resistant pCP20 Flp recombinase plasmid (Cherepanov et al., 1995). Colonies were selected on kanamycin and ampicillin and incubated at 30 degrees. One colony was transferred to liquid LB medium without antibiotic and incubated overnight at 42 degrees in order to induce the loss of the pCP20 plasmid. Colonies were selected on LB agar plates followed by replica plating on LB plates containing LB alone, ampicillin, or kanamycin to screen for colonies which had successfully lost both the kanamycin resistance cassette and the pCP20 plasmid. This kanamycin sensitive lipB- strain was called JEG1. Primers LplA.H.F and LplA.H.R (Table S1) were designed based on the homologous regions flanking the lplA gene originally described by Baba and coworkers (Baba et al., 2006). These primers were used to amplify the kanamycin cassette flanked by FRT sites which had previously been inserted into the lplA locus in lplA - Keio strain JW4349. JEG1 cells were transformed with plasmid pKD46 which expresses λ Red recombinase (GenBank Accession number AY048746) (Datsenko et al., 2000). Chemically competent JEG1/pKD46 cells were prepared and transformed with 400 ng of the PCR product generated above and grown in SOC medium with 1 mM L-arabinose for 2 hours at 37 degrees, after which they were selected with kanamycin on LB agar plates. Three colonies were screened for the lplA deletion by PCR using primers specific for the kanamycin cassette (K1.int.F and K2.int.R, Table S1) in combination with primers upstream and downstream of lplA (LplA.U and LplA.D, Table S1). This lipB-: KanS, lplA-: KanR line was called JEG2. A kanamycin sensitive line was generated as described above using the pCP20 plasmid and called JEG3. Primers LipB.U and LipB.D (Table S1) flanking the lipB locus were used in conjunction with primers LplA.U and LplA.D to verify the deletion of both genes in strain JEG3.
Parasite culture for protein localization
P. falciparum asexual blood stage cultures were maintained at 2% hematocrit in complete medium consisting of RPMI 1640 medium with L-glutamine (Gibco) supplemented with 10% human serum (Interstate Blood Bank, Memphis, TN), 25 mM HEPES, 0.24% NaHCO3, and 12.5 μg/ml hypoxanthine (Trager et al., 1997). Site-specific integration mediated by the mycobacteriophage Bxb1 integrase was used to generate cell lines with the pRL2-BCDH30GFP and pRL2-KDH30GFP plasmids described above that were integrated into the attB locus of Dd2attB parasites (Nkrumah et al., 2006). Transfection of parasites was conducted as described previously (Spalding et al., 2010a). Cell lines were analyzed for integration at the attB site by PCR analysis of genomic DNA using primers that flank the 5′ and 3′ integration sites. Parasites were imaged on a Nikon Eclipse E800 epifluorescence microscope. For live fluorescence, cells were incubated with 100 nM MitoTracker CMX-Ros (Invitrogen) and 1 μg/ml Hoechst 33258 to visualize parasite mitochondria and nuclei.
Protein expression and purification
Plasmids pMALcHT-LipL1 and pMALcHT-LipL1K160A were transformed into BL21-Star (DE3) cells (Invitrogen) and co-transformed with the pRIL plasmid isolated from BL21-CodonPlus-RIL cells (Agilent) and plasmid pRK586 encoding the Tobacco Etch Virus (TEV) protease (Kapust et al., 2000), as previously described (Allary et al., 2007). These cells produce LipL1 or LipL1K160A fused to an amino-terminal six histidine-tag. Transformed cells were grown to mid-log phase, and the expression of recombinant proteins was induced by the addition of 0.4 mM IPTG. Cells were harvested after growth for 10 h at 20°C. LipL1 and LipL1K160A were purified by metal chelate chromatography followed by cation exchange chromatography; LipL1 was further purified by gel filtration chromatography. Plasmids pMALcHT-LipL2, pMALcHT-LipL2C93A or pMALcHT-LipL2C276A were transformed into BL21-Star (DE3) and co-transformed with the pRIL plasmid. These cells produce LipL2, LipL2C93A or LipL2C276A fused to an amino-terminal MBP tag. LipL2, LipL2C93A and LipL2C276A were affinity purified using amylose resin (NEB #E8021).
The plasmids pGEXT-BCDHLD, pGEXT-KDHLD or pGEXTK-Hprot were transformed into JEG3 cells containing the pRIL plasmid. These cells produced BCDHLD, KDHLD or the H-protein in the apo-form (non-lipoylated) with an amino-terminal GST fusion protein. The proteins were purified by GSTrap FF chromatography (GE Healthcare). All cultures were grown and harvested as described for LipL1, with the exception that JEG3 cultures were supplemented with 5 mM sodium succinate, 5 mM sodium acetate and 1% glucose. All purifications were conducted with 20 mM HEPES, 100 mM NaCl at pH 7.5 as the base buffer. For GST purifications, 1 mM DTT was used during the steps of lysis and purification. The concentration of proteins after purification was determined by Bradford assay.
Cell-based lipoylation assay
The JEG3 lipoylation deficient E. coli strain was transformed with the pRIL plasmid and grown in LB medium supplemented with 1% glucose as a carbon source and 35 μg ml−1 chloramphenicol. Sodium succinate (5 mM) and sodium acetate (5 mM) were added to bypass the requirement for KDH and PDH activity respectively. JEG3/pRIL was transformed with a plasmid expressing a candidate lipoate ligase (either pMALcHT-LipL1, pMALcHT-LipL2, pMALcHT-EcLplA or pMALcHT-LipL1K160A) and/or a plasmid expressing a lipoylation substrate (pGEXTK-BCDHLD or pGEXTK-KDHLD) and selected using 100 μg ml−1 ampicillin and/or 50 μg ml−1 kanamycin. Transformants were grown in LB medium and supplemented as described above with the addition of 200 μM R-lipoic acid (Sigma). Plasmids pGEXTK-BCDHLD and pGEXTK-KDHLD were also transformed into BL21-Star (DE3) cells containing the pRIL plasmid and maintained in LB medium containing 35 μg ml−1 chloramphenicol and 50 μg ml−1 kanamycin. For the cell-based assay 20 mL cultures were grown to mid-log phase at 37 degrees and induced with 0.4 mM IPTG for 10 hours at 20 degrees. Cells were harvested by centrifugation and resuspended with 0.5 mL of buffer containing 20 mM HEPES, 100 mM NaCl at pH 7.5 and lysed by sonication. The cell lysates were clarified by centrifugation at 16,000 g and the supernatants were collected and resolved by SDS-PAGE (Invitrogen). Lipoylated proteins were visualized by western blot as described in lipoate ligase assay section. Blots were stripped by using a buffer containing 100 mM BME, 2% SDS in 62.5 mM Tris-HCl pH 7.5 at 55 degrees. After stripping, blots were blocked with 5% milk/PBS and probed with 1:5,000 murine α-MBP monoclonal antibody (HRP conjugated) in 1% milk/PBS for 1 hour at room temperature. The membranes were visualized with enhanced chemiluminescence (ECL) western substrate (Pierce) and exposed to film.
HPLC and Mass Spectroscopy analysis of LipL1 and LipL2
HPLC analyses were performed on a Beckman Gold Nouveau System with a Grace Alltima 3 μ C18 Analytical Rocket column (53 mm×7 mm). Electrospray ionization mass spectrometry (ESI-MS) was performed on a Thermos linear triple quadrupole (LTQ) XL mass spectrometer. Samples were analyzed in negative ion mode.
Reaction mixtures containing 2 mM ATP, 2 mM MgCl2, 1 mM R-LA, 5 mM TCEP in 100 mM phosphate buffer 150 mM NaCl pH 7.5 were initiated by addition of LipL1, LipL1K160A or LipL2 (3.2 mg/mL) and incubated at room temperature for 10 minutes. Reactions were quenched with ice cold methanol (1:1 v/v with reaction mixture). Precipitated protein was removed by centrifugation at 15,000 g for 15 minutes at 4°C. The supernatant was then subjected to HPLC analysis with UV detection using the following conditions: flow rate of 3 mL/min; solvent A: Et3NHOAc (50 mM, pH 6); solvent B: acetonitrile; method: 5% B for 3 minutes followed by a gradient of 5% to 100% B over 7 minutes. Peaks of interest were collected manually and analyzed by ESI-MS in negative ionization mode. MS/MS analysis was performed by selecting 535 ± 2 m/z for collision induced dissociation (CID) at 13% normalized collision energy.
Synthesis of 6,8-dichlorooctanoic acid (6,8-diClO)
The preparation of 6,8-diClO was carried out using the ester hydrolysis conditions previously reported for similar analogs (Woster et al., 1989). All reagents were obtained from commercial sources. A solution of ethyl 6,8-dichlorooctanoate (500 mg, 2 mmol) in EtOH (1 mL) was added to a suspension of LiOH (72 mg, 3.0 mmol) in 50:50 EtOH/water (1 mL) at room temperature. The mixture was stirred at room temperature and monitored by thin layer chromatography (CH2Cl2) until the ester hydrolysis reaction appeared to be complete (2 h). Water was added (20 mL), and the resulting mixture was extracted with EtOAc (3 × 10 mL). The aqueous layer containing the product was acidified to pH 2 using 1N HCl, and extracted with EtOAc (3 × 20 mL). 6,8-Dichlorooctanoic acid was obtained as an oil (386 mg, 88%, Rf = 0.76 in CH2Cl2). NMR characterization was performed on a 400 MHz Varian instrument. Chemical shifts are reported in parts per million from tetramethylsilane. HRMS characterization was carried out at the University of Illinois Mass Spectrometry Lab using ES ionization. 1H NMR (CDCl3) δ 4.10 (m, 1H); 3.70 (m, 2H); 2.37 (t, 2H); 2.11 (m, 2H); 1.76 (q, 2H); 1.65 (m, 3H), 1.50 (m, 1H). HRMS: calcd. for C8H14O2Cl2: m/z 235.0277 [M + Na]+; found: 235.0269.
Lipoate ligase assay
Purified LipL1, LipL1K160A, LipL2, LipL2C93A or LipL2C276A (1 μM) was incubated in reaction buffer (100 mM Na/K Phosphate buffer, 150 mM NaCl at pH 7.5) containing 2 mM ATP, 2 mM MgCl2, 5 mM TCEP, 200 μM R-lipoic acid and 10 μM apo-protein substrate: H-protein, BCDHLD, or KDHLD. After incubation at 37 degrees for 1 hour, the reactions were terminated with the addition of gel loading buffer and analyzed by SDS-PAGE followed by transfer to nitrocellulose membrane. The membranes were blocked with 5% milk in PBS for 30 minutes, and probed with 1:5000 rabbit polyclonal α-LA (Calbiochem) for 2 hours in 1% milk/PBS at room temperature. The membrane was washed with 1xPBS three times and then probed with 1:5000 donkey α-Rabbit IgG horseradish peroxidase (HRP) secondary antibody (GE Healthcare) in 1% milk/PBS overnight at 4 degrees. The membranes were visualized with enhanced chemiluminescence (ECL) western substrate (Pierce) and exposed to film.
All lipoylation assays were conducted similarly using the same protein concentrations and base buffer. Where indicated, ATP was omitted from the reaction and 5 mM of DTT, THP or water was used instead of TCEP. R-lipoic acid was substituted with 200 μM 6,8-dichlorooctanoate or 200 μM 8-bromooctanoate where indicated, and in these reactions 1:5000 rabbit polyclonal α-6,8-diClO was used as the primary probe for western blots.
Amidotransferase assay
Lipoylated H-protein (Holo H-protein) was generated by co-transforming pGEXTK-Hprot and pMALcHT-LipL1 in JEG3 cells containing the pRIL plasmid. These cells express the H-protein with an amino-terminal GST tag and LipL1 with an amino-terminal MBP. One liter cultures were grown to mid-log phase and induced with 0.4 mM IPTG for 10 hours at 20 degrees. Cells were harvested as described above and Holo H-protein was purified by affinity chromatography using a GSTrap FF column (GE Healthcare), followed by anion exchange using a MonoQ column (Pharmacia). The protein concentration was determined by Bradford assay. A final concentration of 10 μM of Holo H-protein was used in all assays, in the presence of 1 μM of LipL1 and/or LipL2 and 10 μM of BCDHLD or KDHLD, 2 mM MgCl2, 5 mM TCEP in 100 mM Na/K Phosphate buffer, 150 mM NaCl at pH 7.5. The reactions were incubated and analyzed as described in the lipoate ligase assay.
Production of rabbit α-6,8-diClO antiserum
Compound 6,8-diClO was conjugated to keyhole limpet hemocyanin using the Imject EDC mcKLH Spin Kit (Pierce). Antibodies to the conjugate were generated in rabbits using the standard protocol of the custom antibody service Cocalico Biologicals (Reamstown, PA). The resulting antiserum was used in western blots where indicated with the label α-6,8-diClO to probe for proteins modified with either 8-bromooctanoate or 6,8-dichlorooctanoate.
Parasite culture for 6,8-diClO growth inhibition assay and labeling
All Plasmodium falciparum parasite lines for these experiments were grown as previously described (Allary et al., 2007) with the following exceptions: complete media was supplemented with 10% Albumax (Gibco) and parasites were maintained under a 3% O2 and 3% CO2 gas mixture. To assess parasite growth in the presence of 6,8-diClO, Dd2attB parasites were synchronized with 5% sorbitol (Lambros et al., 1979) and seeded into 6-well plates at 0.05% parasitemia. Parasites were treated with 0, 25, 100, 400, and 1000 μM 6,8-diClO. All dilutions of 6,8-diClO were prepared in DMSO so that the final amount of DMSO in the parasite culture was 0.05%. For all experiments culture media and drug were added fresh each day. Blood smears were made every other day to monitor parasitemia.
For western blot analysis, asynchronous 3D7 parasites were seeded at 1% parasitemia and treated with 10 μM 6,8-diClO, 2 μM R-lipoic acid in 10% EtOH or vehicle for three days. Parasites were isolated from red blood cells using 0.5% saponin in PBS as previously described (Allary et al., 2007). Parasite pellets were resuspended in RIPA Buffer (Boston BioProducts) supplemented with protease inhibitors (Complete Mini EDTA-free, Roche) and lysed by vigorous vortexing. Insoluble debris was pelleted at 10,000 g and total protein of the supernatant was measured by Bradford assay. Loading dye was added to the supernatant and samples were boiled for 5 min at 95 degrees and separated on a 4–12% gradient gel (Invitrogen) by SDS-PAGE. Western blots were conducted as described above using 1:5000 rabbit α-6,8-diClO or 1:1000 rabbit α-LA (Calbiochem) as the primary probes. Membranes were stripped and reprobed with 1:10,000 mouse α-PfHsp70 (Kumar et al., 1998) as a loading control.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R56 AI065853 to STP and R01 GM084998 to CLFM and AI099704 DB), and the Johns Hopkins Malaria Research Institute and the Bloomberg Family Foundation. This work was also made possible by UL1 TR001079 from the National Institutes of Health (NIH) National Center for Research Resources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Swetha Velivela and Marina Allary for help with the initial cloning of the lipoylation substrates. We also thank John Cronan for E. coli strain TM136 and the National BioResource Project (NIG, Japan) for strains JW5089 and JW4349. Plasmids pKD46 and pCP20 were provided by the Yale University CGSC (Coli Genetic Stock Center) which is supported by NSF program DBI-0742708. We also thank Roger McMacken for the gift of rabbit α-EcHsp70 antiserum and Nirbhay Kumar for mouse α-PfHsp70 antiserum.
References
- Allary M, Lu JZ, Zhu L, Prigge ST. Scavenging of the cofactor lipoate is essential for the survival of the malaria parasite Plasmodium falciparum. Mol Microbiol. 2007;63:1331–1344. doi: 10.1111/j.1365-2958.2007.05592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner ES, Nordberg J, Holmgren A. Efficient reduction of lipoamide and lipoic acid by mammalian thioredoxin reductase. Biochem Biophys Res Commun. 1996;225:268–274. doi: 10.1006/bbrc.1996.1165. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100050. 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl A, Brunk B, Crabtree J, Fraunholz MJ, Gajria B, Grant GR, et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabaskaran Nina P, Morrisey JM, Ganesan SM, Ke H, Pershing AM, Mather MW, Vaidya AB. ATP synthase complex of Plasmodium falciparum: dimeric assembly in mitochondrial membranes and resistance to genetic disruption. J Biol Chem. 2011;286:41312–41322. doi: 10.1074/jbc.M111.290973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman R, Smith E. The compositional and evolutionary logic of metabolism. Phys Biol. 2013;10:011001. doi: 10.1088/1478-3975/10/1/011001. [DOI] [PubMed] [Google Scholar]
- Burns JA, Butler JC, Moran J, Whitesides GM. Selective Reduction of Disulfides by Tris(2-Carboxyethyl)Phosphine. Journal of Organic Chemistry. 1991;56:2648–2650. [Google Scholar]
- Chaudhari R, Narayan A, Patankar S. A novel trafficking pathway in Plasmodium falciparum for the organellar localization of glutathione peroxidase-like thioredoxin peroxidase. FEBS J. 2012;279:3872–3888. doi: 10.1111/j.1742-4658.2012.08746.x. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Christensen QH, Martin N, Mansilla MC, de Mendoza D, Cronan JE. A novel amidotransferase required for lipoic acid cofactor assembly in Bacillus subtilis. Mol Microbiol. 2011;80:350–363. doi: 10.1111/j.1365-2958.2011.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, et al. Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in Plasmodium falciparum. J Biol Chem. 2013 doi: 10.1074/jbc.M113.503557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu A, Pick U, Handelman GJ, Haramaki N, Han D, Podda M, et al. Reduction and transport of lipoic acid by human erythrocytes. Biochem Pharmacol. 1995;50:253–261. doi: 10.1016/0006-2952(95)00084-d. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delli-Bovi TA, Spalding MD, Prigge ST. Overexpression of biotin synthase and biotin ligase is required for efficient generation of sulfur-35 labeled biotin in E. coli. BMC Biotechnol. 2010;10:73. doi: 10.1186/1472-6750-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Fairhurst RM, Slutsker L, Macarthur JR, Breman JG, Guerin PJ, et al. The threat of artemisinin-resistant malaria. N Engl J Med. 2011;365:1073–1075. doi: 10.1056/NEJMp1108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkard B, Kumar TR, Hecht LS, Matthews KA, Henrich PP, Gulati S, et al. A key role for lipoic acid synthesis during Plasmodium liver stage development. Cell Microbiol. 2013 doi: 10.1111/cmi.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Eastman RT, Ward SA, Meshnick SR. Recent highlights in antimalarial drug resistance and chemotherapy research. Trends Parasitol. 2008;24:537–544. doi: 10.1016/j.pt.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Mol Microbiol. 2005;55:39–53. doi: 10.1111/j.1365-2958.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Maita N, Hosaka H, Okamura-Ikeda K, Nakagawa A, Taniguchi H. Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A. J Biol Chem. 2010;285:9971–9980. doi: 10.1074/jbc.M109.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Okamura-Ikeda K, Motokawa Y. Purification and characterization of lipoyl-AMP:N epsilon-lysine lipoyltransferase from bovine liver mitochondria. J Biol Chem. 1994;269:16605–16609. [PubMed] [Google Scholar]
- Gisselberg JE, Dellibovi-Ragheb TA, Matthews KA, Bosch G, Prigge ST. The Suf Iron-Sulfur Cluster Synthesis Pathway Is Required for Apicoplast Maintenance in Malaria Parasites. PLoS Pathog. 2013;9:e1003655. doi: 10.1371/journal.ppat.1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther S, Matuschewski K, Muller S. Knockout studies reveal an important role of Plasmodium lipoic acid protein ligase A1 for asexual blood stage parasite survival. PLoS One. 2009;4:e5510. doi: 10.1371/journal.pone.0005510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther S, McMillan PJ, Wallace LJ, Muller S. Plasmodium falciparum possesses organelle-specific alpha-keto acid dehydrogenase complexes and lipoylation pathways. Biochem Soc Trans. 2005;33:977–980. doi: 10.1042/BST20050977. [DOI] [PubMed] [Google Scholar]
- Gunther S, Wallace L, Patzewitz EM, McMillan PJ, Storm J, Wrenger C, et al. Apicoplast lipoic acid protein ligase B is not essential for Plasmodium falciparum. PLoS Pathog. 2007;3:e189. doi: 10.1371/journal.ppat.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- Haramaki N, Han D, Handelman GJ, Tritschler HJ, Packer L. Cytosolic and mitochondrial systems for NADH- and NADPH-dependent reduction of alpha-lipoic acid. Free Radic Biol Med. 1997;22:535–542. doi: 10.1016/s0891-5849(96)00400-5. [DOI] [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Gething PW, Patil AP, Tatem AJ, Noor AM, et al. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 2009;6:e1000048. doi: 10.1371/journal.pmed.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SW, Cronan JE. The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein : protein transferase. Journal of Bacteriology. 2003;185:1582–1589. doi: 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jortzik E, Becker K. Thioredoxin and glutathione systems in Plasmodium falciparum. Int J Med Microbiol. 2012;302:187–194. doi: 10.1016/j.ijmm.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr Purif. 2000;19:312–318. doi: 10.1006/prep.2000.1251. [DOI] [PubMed] [Google Scholar]
- Kumar N, Zheng H. Evidence for epitope-specific thymus-independent response against a repeat sequence in a protein antigen. Immunology. 1998;94:28–34. doi: 10.1046/j.1365-2567.1998.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- Martin N, Christensen QH, Mansilla MC, Cronan JE, de Mendoza D. A novel two-gene requirement for the octanoyltransfer reaction of Bacillus subtilis lipoic acid biosynthesis. Mol Microbiol. 2011;80:335–349. doi: 10.1111/j.1365-2958.2011.07597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan PJ, Stimmler LM, Foth BJ, McFadden GI, Muller S. The human malaria parasite Plasmodium falciparum possesses two distinct dihydrolipoamide dehydrogenases. Mol Microbiol. 2005;55:27–38. doi: 10.1111/j.1365-2958.2004.04398.x. [DOI] [PubMed] [Google Scholar]
- Miao J, Lawrence M, Jeffers V, Zhao F, Parker D, Ge Y, et al. Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development. Mol Microbiol. 2013;89:660–675. doi: 10.1111/mmi.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, et al. Escherichia coli LipA is a lipoyl synthase: In vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry. 2000;39:15166–15178. doi: 10.1021/bi002060n. [DOI] [PubMed] [Google Scholar]
- Morris TW, Reed KE, Cronan JE. Lipoic Acid Metabolism in Escherichia-Coli - the Lpla and Lipb Genes Define Redundant Pathways for Ligation of Lipoyl Groups to Apoprotein. Journal of Bacteriology. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhune SA, Okiro EA, Noor AM, Zurovac D, Juma E, Ochola SA, Snow RW. The clinical burden of malaria in Nairobi: a historical review and contemporary audit. Malar J. 2011;10:138. doi: 10.1186/1475-2875-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench SP, Rafferty JB, McLeod R, Rice DW, Prigge ST. Expression, purification and crystallization of the Plasmodium falciparum enoyl reductase. Acta Crystallogr D Biol Crystallogr. 2003;59:1246–1248. doi: 10.1107/s0907444903008813. [DOI] [PubMed] [Google Scholar]
- Nickel C, Rahlfs S, Deponte M, Koncarevic S, Becker K. Thioredoxin networks in the malarial parasite Plasmodium falciparum. Antioxid Redox Signal. 2006;8:1227–1239. doi: 10.1089/ars.2006.8.1227. [DOI] [PubMed] [Google Scholar]
- Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR, Jr, Fidock DA. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiro EA, Al-Taiar A, Reyburn H, Idro R, Berkley JA, Snow RW. Age patterns of severe paediatric malaria and their relationship to Plasmodium falciparum transmission intensity. Malaria Journal. 2009a:8. doi: 10.1186/1475-2875-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiro EA, Bitira D, Mbabazi G, Mpimbaza A, Alegana VA, Talisuna AO, Snow RW. Increasing malaria hospital admissions in Uganda between 1999 and 2009. Bmc Medicine. 2011:9. doi: 10.1186/1741-7015-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiro EA, Mutheu J, Gething PW, Juma E, Snow RW. The Changing Patterns of Malaria Admissions since 1999 at 18 Hospitals across Kenya. American Journal of Tropical Medicine and Hygiene. 2009b;81:54–54. [Google Scholar]
- Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, Kappe SH. Plasmodium pyruvate dehydrogenase activity is only essential for the parasite’s progression from liver infection to blood infection. Mol Microbiol. 2010;75:957–971. doi: 10.1111/j.1365-2958.2009.07034.x. [DOI] [PubMed] [Google Scholar]
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche P, Perham RN. Structure and selectivity in post-translational modification: attaching the biotinyl-lysine and lipoyl-lysine swinging arms in multifunctional enzymes. Embo Journal. 1999;18:2673–2682. doi: 10.1093/emboj/18.10.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Koike M, Levitch ME, Leach FR. Studies on the nature and reactions of protein-bound lipoic acid. J Biol Chem. 1958;232:143–158. [PubMed] [Google Scholar]
- Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- Salcedo E, Sims PFG, Hyde JE. A glycine-cleavage complex as part of the folate one-carbon metabolism of Plasmodium falciparum. Trends in Parasitology. 2005;21:406–411. doi: 10.1016/j.pt.2005.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonauer MS, Kastaniotis AJ, Kursu VA, Hiltunen JK, Dieckmann CL. Lipoic acid synthesis and attachment in yeast mitochondria. J Biol Chem. 2009;284:23234–23242. doi: 10.1074/jbc.M109.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends in Parasitology. 2008;24:468–478. doi: 10.1016/j.pt.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Snider GW, Dustin CM, Ruggles EL, Hondal RJ. A mechanistic investigation of the C-terminal redox motif of thioredoxin reductase from Plasmodium falciparum. Biochemistry. 2014;53:601–609. doi: 10.1021/bi400931k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MD, Allary M, Gallagher JR, Prigge ST. Validation of a modified method for Bxb1 mycobacteriophage integrase-mediated recombination in Plasmodium falciparum by localization of the H-protein of the glycine cleavage complex to the mitochondrion. Mol Biochem Parasitol. 2010a;172:156–160. doi: 10.1016/j.molbiopara.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MD, Prigge ST. Lipoic acid metabolism in microbial pathogens. Microbiol Mol Biol Rev. 2010b;74:200–228. doi: 10.1128/MMBR.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm J, Muller S. Lipoic acid metabolism of Plasmodium--a suitable drug target. Curr Pharm Des. 2012;18:3480–3489. doi: 10.2174/138161212801327266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svagera Z, Hanzlikova D, Simek P, Husek P. Study of disulfide reduction and alkyl chloroformate derivatization of plasma sulfur amino acids using gas chromatography-mass spectrometry. Anal Bioanal Chem. 2012;402:2953–2963. doi: 10.1007/s00216-012-5727-y. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Continuous culture of Plasmodium falciparum: its impact on malaria research. Int J Parasitol. 1997;27:989–1006. doi: 10.1016/s0020-7519(97)00080-5. [DOI] [PubMed] [Google Scholar]
- Woster PM, Black AY, Duff KJ, Coward JK, Pegg AE. Synthesis and biological evaluation of S-adenosyl-1,12-diamino-3-thio-9-azadodecane, a multisubstrate adduct inhibitor of spermine synthase. J Med Chem. 1989;32:1300–1307. doi: 10.1021/jm00126a026. [DOI] [PubMed] [Google Scholar]
- Wrenger C, Muller S. The human malaria parasite Plasmodium falciparum has distinct organelle-specific lipoylation pathways. Mol Microbiol. 2004;53:103–113. doi: 10.1111/j.1365-2958.2004.04112.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.