Abstract
The long-term risk of malignancy associated with stem cell therapies is a significant concern in the clinical application of this exciting technology. We report a cancer-selective strategy to enhance the safety of stem cell therapies. Briefly, using a cell engineering approach we show that aggressive cancers derived from human or murine iPS and ES cells are strikingly sensitive to temporary MYC blockade. On the other hand, differentiated tissues derived from human or mouse iPS cells can readily tolerate temporary MYC inactivation. In cancer cells, endogenous MYC is required to maintain the metabolic and epigenetic functions of the embryonic and cancer-specific pyruvate kinase M2 isoform (PKM2) (Vander Heiden et al., 2009; Yang et al., 2012). In summary, our results implicate PKM2 in cancer’s increased MYC dependence and indicate dominant MYC inhibition as a cancer-selective failsafe for stem cell therapies.
INTRODUCTION
Tissues derived from pluripotent stem cells (PSCs) cells have great potential in regenerative medicine and can, in principle, replace any differentiated tissue (Hanna, 2007; Takahashi and Yamanaka, 2006). Recent successes include the generation of retinal cells (Osakada et al., 2009), functional liver tissue (Takebe et al., 2013), and dopaminergic neurons (Kriks et al., 2011). These strategies are approaching clinical testing however the risk of iatrogenic malignancy remains a significant concern (Goldring et al., 2011; Lee et al., 2013). For example, cancers develop with increased frequency in iPS-chimeric animals (Carey et al., 2010; Okita et al., 2007; Stadtfeld et al., 2010b), neuronal tumors occur in primates injected with PSC-derived neurogenic cells (Doi et al., 2012). Most dramatically, an ataxia telangiectasia patient developed multifocal aggressive brain cancer following administration of neurogenic stem cells (Amariglio et al., 2009). These citations illustrate a need for effective and cancer-selective fail-safe mechanisms.
The causes of malignancy are not entirely clear. Reactivation of reprogramming factors, especially the MYC oncogene, has been implicated (Okita et al., 2007). However cancers also occurred, albeit with lower frequency, when MYC was omitted form reprogramming protocols (Miura et al., 2009; Nakagawa, 2008; Werbowetski-Ogilvie et al., 2009). Notably, malignant and pluripotent cells show increased genomic instability, frequent, non-random chromosomal aberrations, and recurrent inactivation of canonical tumor suppressors genes (Hussein et al., 2011; Marion et al., 2009; Mayshar et al., 2010). These findings suggest that initial barriers to transformation may be fortuitously inactivated in PSC and derived tissues.
Improved reprogramming procedures have greatly reduced, but not eliminated, the risk of cancer (Lee et al., 2013). These include non-integrating and excisable vectors, the exclusion of MYC, and reprogramming by RNA, protein, or small molecules (Carey et al., 2010; Kaji et al., 2009; Stadtfeld et al., 2010a; Wernig et al., 2008). Additional strategies seek to purge residual PSCs, genomic surveys for somatic mutations, and conventional suicide genes (Choo et al., 2008; Tan et al., 2009). In this study we explore a strategy based on recent insight into cancer’s “oncogene dependence” (Jain, 2002; Soucek et al., 2008; Weinstein, 2002). We show that introduction of a dominant negative MYC construct and temporary MYC inactivation can destroy aggressive iPS and ES derived cancers while sparing healthy PSC-derived tissues.
RESULTS
To explore the MYC dependence of PSC-derived tissues we introduced a dominant negative MYC allele into karyotypically normal human and murine pluripotent stem cells (Figure 1A). Briefly, OmomycER is an inducible dominant negative MYC allele that is uniquely able to form inactive dimers with all three endogenous MYC proteins and does not bind other helix-loop-helix factors(Savino et al., 2011; Soucek, 1998). We reprogrammed human and murine fibroblasts using a single excisable polycistronic construct or four separate vectors, respectively (Papapetrou et al., 2011). We confirmed reprogramming by immunofluorescence for NANOG and showed loss of the exogenous construct by FACS and PCR (Figure S1A–C). We isolated karyotypically normal clones and introduced Omomyc along with a citrine reporter into both human iPS and murine iPS and ES cells (Figure S1D–E).
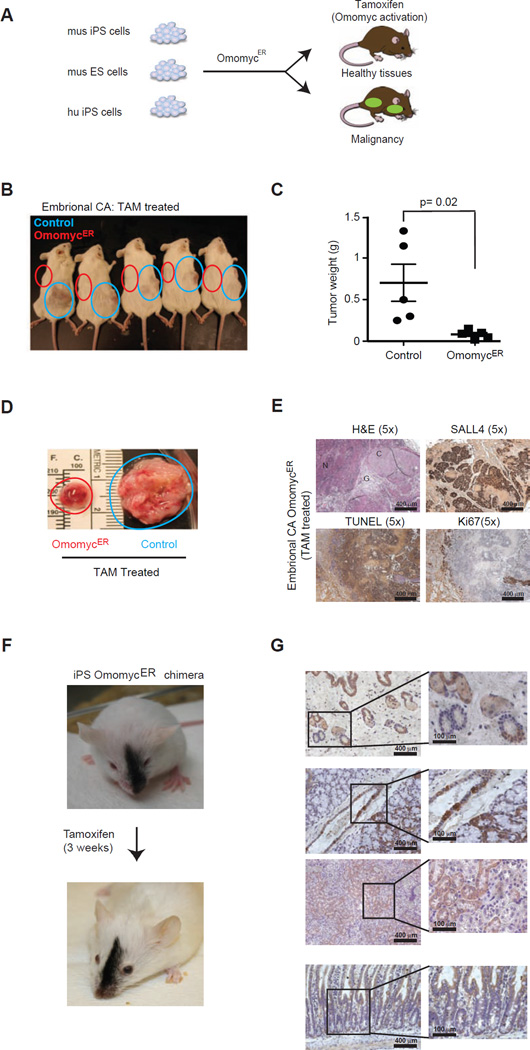
Figure 1. Aggressive embryonal carcinomas are sensitive to OmomycER treatment.
A, Schematic of mouse iPS (mu iPS), ES (mu ES) and human iPS (hu iPS) engineer with and without inducible dominant negative MYC allele OmomycER. B, Animals bearing embryonal carcinomas derived either from iPS cells expressing OmomycER (red circle) or control vector (blue circle) and treated with tamoxifen (TAM). C and D, Comparison of tumor weights following TAM treatment. The blue and red circles highlight the position (b) and the size (c) of the xenografted tumors. E, Histopathology of primitive embryonal carcinomas (Embryonal CA) stained as indicated; F, Representative chimeric animal before and after a 3 week course of tamoxifen (TAM) treatment to activate the dominant negative MYC construct in iPS cell derived tissues; (the same schedule was used in tumor treatment studies); G, Immunohistochemical stain for citrine identifies tissues derived from OmomycER expressing iPS cells in different organ sites (panels from top): hair follicles, upper gastrointestinal tract and adjacent thyroid gland, kidney, lower gastrointestinal tract and mucosal villi.
Murine iPS cells deficient for the p53 tumor suppressor give rise to aggressive embryonal carcinomas. Briefly, the p53 tumor suppressor restricts reprogramming and p53 deficient murine fibroblast formed iPS colonies faster than wild type cells (Figure S1F)(Hong, 2009; Marion et al., 2009). Upon transplantation the p53−/− iPS cells rapidly formed aggressive cancers (p53+/+: n = 5; p53−/−: n = 5; latency to 1cm3 tumor p < 0.001) (Figure S1G). Pathologically, these cancers resembled primitive embryonal carcinomas (EC), composed of immature OCT4 and CD30 negative tissues with some SALL4 expression (28% ± 11% [mean/SD]), a high proliferation index by Ki67 (34.6% ± 6%), and little apoptosis by TUNEL (14.2% ± 10%) (Figure S1H). Notably, the human MYC transgene was not reactivated in these cancers, and instead we observed elevated expression of the endogenous Myc mRNA (Figure S1I).
Temporary MYC blockade produced dramatic regression in aggressive iPS-derived embryonal carcinomas. We initiated tamoxifen (TAM) treatment when tumors reached 1cm3 (TAM: 10 mg/ml, alternate days for 2 weeks). This treatment caused the OmomycER expressing cancers (left flank) to collapse whereas control tumors (right flank) continued to grow (nOmo = 5, nControl = 5, p < 0.005) (Figure 1B–D). After TAM treatment we retrieved a residual cystic mass containing cartilaginous material, large areas of TUNEL positive apoptosis, and some SALL4 positive and Ki67 negative cells indicating yolk sac differentiation and absence of proliferation (OmomycER versus control: SALL4: 92.3% ± 19% versus 28.7% ± 14%, p < 0.05; TUNEL: 41.2% ± 13% versus 14.2% ± 10%; p < 0.05; Ki67: 34.6% ± 6% versus 24.2% ± 10%) (Figure 1E, Figure S1J).
We confirmed these observations using murine ES cells expressing a p53 short-hairpin RNA to ensure these results did not reflect specific properties of the iPS cells. Briefly, the ES-derived cancers were pathologically indistinguishable form the iPS-derived cancers (Figure S1K–M). They responded in exactly the same manner to OmomycER activation (Figure S1N–P). Hence, aggressive and p53-deficient cancers derived from pluripotent stem cells strictly depend on the continuous activity of endogenous MYC.
Next, we tested the effect of MYC inactivation on well-differentiated, benign teratomas. Briefly, p53 wild type iPS and ES cells give rise to typical teratomas composed from several different germ cell layers including cartilage, neuronal and glial tissues that lacked the pluripotency markers CD30, OCT4 and contained a few SALL4 positive cells indicating early yolk sac differentiation (11.8% ± 7% [mean/SD]) (Figures S1Q and S1U/V). As seen with the aggressive embryonal carcinomas the iPS-derived teratomas showed no expression of the exogenous MYC and instead the endogenous Myc mRNA was modestly increased (Figure S1R/S). OmomycER induction using the same protocol as before produced no measurable effect on iPS or ES cell derived teratomas (nOmo = 6, ncontrol =6; p = 0.7) (Figures S1T/U; Figure S1W–Z), and showed no difference in proliferation or apoptosis by Ki67 and TUNEL stains, respectively (Control versus Omomyc: Ki67: 8.7% ± 5% vs. 9.1% ± 2.5%; p > 0.05; TUNEL 5.6% ± 3.6% vs. 3.6% ± 2%; p > 0.05). Hence, differentiated teratomas no longer depend on MYC and are insensitive to MYC blockade.
Potentially, MYC blockade can be used to purge in vitro cultures form residual iPS cells and to prevent tumor growth. To test these possibilities we first activated omomyc in iPS cultures and observed rapid induction of cell death consistent with the notion that MYC is required to maintain these pluripotent cells (Figure S1A1/B1) (Cartwright, 2005). We also tested whether Omomyc might be able to delay or prevent tumor formation in vivo. Briefly, we activated OmomycER shortly following transplantation and this could significantly reduce teratoma formation in vivo (iPS OmomycER untreated n=4, iPS OmomycER tamoxifen (10 mg/ml) n=4, p=0.02) (Figure S1C1/D1). Hence, OmomycER can effectively purge residual tumorigenic iPS cells.
Next, we wanted to explore the effect of temporary MYC inactivation on healthy iPS-derived tissues in vivo. We generated chimeric animals from iPS cells transduced with OmomycER to test the effect of temporary MYC inactivation in a physiological context. Briefly, we injected 2 clones of karyotypically normal, iPS cells of C57B1/6J background into C57Bl/6J (B6(Cg)-Tyrc-2J/J blastocysts and confirmed chimerism by the presence of black coat derived from donor cells. Tamoxifen treatment (TAM, 10 mg/ml on alternate days for 3 weeks) had no untoward effects on animal health, weight or behavior (not shown) or skin and hair (Figure 1F). OmomycER expressing tissues were readily detectable by the co-expressed citrine reporter, and immunohistochemistry confirmed significant contributions of iPSOMO cells before and after tamoxifen treatment to various organs. For example, hair follicles, dermal and epidermal structures, the upper and lower gastrointestinal tract, the kidney, red and white pulp of spleen, and thyroid gland showed persistent and extensive contribution of iPSOMO derived and citrine positive cells and histologically intact organs after a three week course of systemic tamoxifen treatment (Figure 1G). Hence, temporary MYC blockade is tolerated and does not cause permanent disruption of iPS-mosaic organs and tissues.
We now wanted to test this new strategy in human iPS-derived tissues to model a clinically relevant setting. Briefly, we engineered human iPS cells to express OmomycER and differentiated these cells into neurogenic precursors (Rosette stage) and midbrain dopaminergic (DA) neurons according to published protocols (Chambers et al., 2009; Kriks et al., 2011). Intracerebral injection of 2 × 105 neurogenic precursors into the striatum of NOD/SCID animals resulted in the development of brain tumors within 4 weeks (Tumor incidence: 9/9; vector n = 4 and OmomycER n = 5) (Figure 2A). Histologically, these tumors resembled primitive neuroectodermal tumors (PNETs) with islands of highly proliferative and Ki67 positive cells, invasive spread across the brain, and expression of primitive markers (SOX2, NESTIN) in the absence of differentiation markers (OLIG2, NeuN, synaptophysin or GFAP) (Figure S2A/B). Hence, we can model primitive iPS-derived brain tumors in vivo and recapitulate the pathology previously seen in a patient treated with neurogenic precursors (Amariglio et al., 2009; Doi et al., 2012).
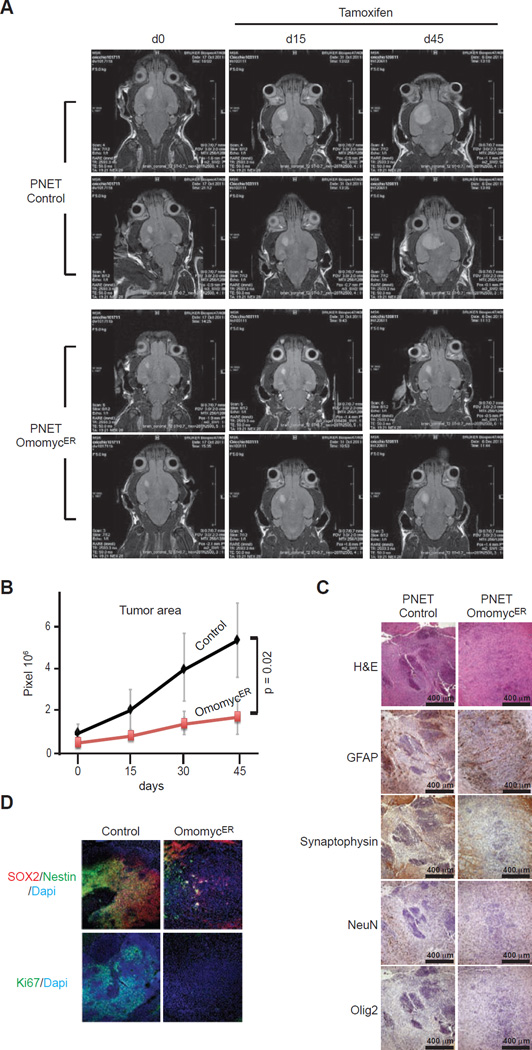
Figure 2. Primitive neuroectodermal tumors (PNETs) derived from human iPS cells respond to OmomycER activation in vivo.
A, Magnetic resonance imaging (MRI) of vector control (Control) and OmomycER expressing PNETs before tamoxifen treatment and at the indicates times after treatment; B, Tumor size measured as pixel counts on MRI images of control and OmomycER PNETs; C, Histology and indicated immunohistochemical stains on control and OmomycER PNETs after tamoxifen; D, Immunfluorescence stains for Ki67 and SOX2/Nestin on OmomycER and control PNETs after treatment.
OmomycER arrests the growth of brain cancers caused by human iPS-derived neurogenic precursors. We initiated systemic tamoxifen treatment to induce the OmomycER construct once brain tumors were diagnosed by magnet resonance imaging (MRI) (here ‘d0’ represents the start of TAM treatment at 10 mg/ml, 2×/week for 5 weeks). This treatment blocked the expansion of OmomycER expressing PNETs while PNETs expressing a control vector continued to grow (n = 4; p = 0.02) (Figure 2A/B). Histology and immunohistochemistry revealed complete disappearance of the Ki67 positive proliferative compartment of these cancers; the treatment did not alter tumor cell differentiation indicated by unchanged surface marker expression (Figure 2C/D). Hence, MYC blockade eliminates the growth fraction in iPS-derived PNETs.
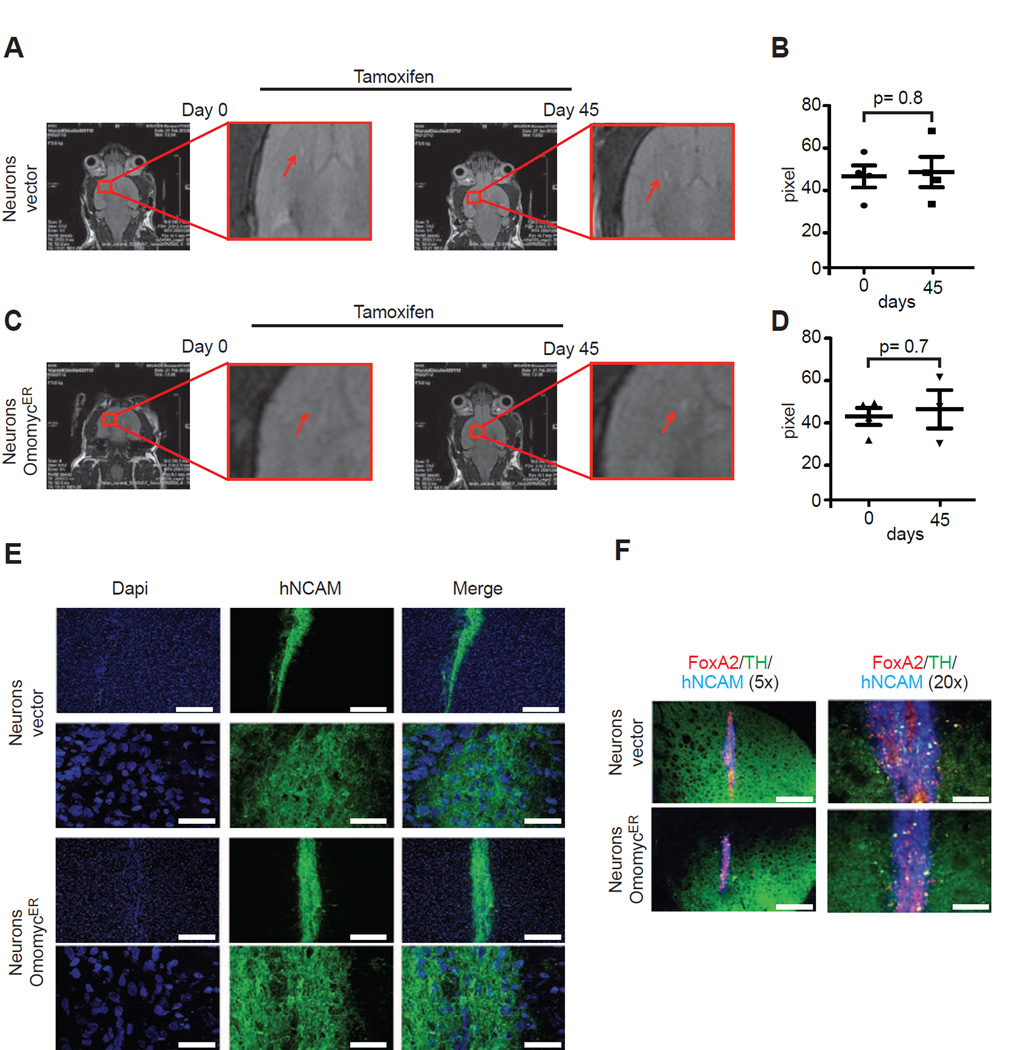
Next, we wondered how MYC blockade would affect differentiated midbrain dopaminergic (DA) neurons in vivo. The engraftment of 1.5 × 105 iPS-derived neurons injected into the striatum of NOD/SCID animals was readily discernible by MRI and subsequent tamoxifen treatment (using the same schedule as above) to activate OmomycER did not affect the radiological appearance control or OmomycER expressing neurons (Figure 3A–D). Microscopy and immunohistochemical stains for neuronal markers (TH, human NCAM), and a transcription factor characteristic of DA midbrain neurons (FoxA2) confirmed equal engraftment of morphologically intact OmomycER and controls dopaminergic neurons (Figure 3E/F). Together, these data reveal a differential requirement for MYC activity in iPS-derived brain cancers (PNETs) that renders these tumors sensitive to an inhibitory MYC allele.
Figure 3. Human iPS-derived dopaminergic (DA) neurons engraft in vivo and tolerate OmomycER activation.
A and C, Magnetic resonance imaging (MRI) showing engraftment of control (b) and OmomycER (d) neurons following injected into the midbrain (striatum) under tamoxifen treatment (same schedule as in tumor studies); B and D, Quantification of engraftment for control (c) and OmomycER (e) expressing neurons by MRI and pixel count to determine area of engraftment; E, Immunofluorescence stains of murine brain sections stained for human-NCAM (green) and Dapi (blue); F, Immunohistochemical stains for human DA neuron markers including FOXA2, TH, and hNCAM on control and OmomycER expressing neurons.
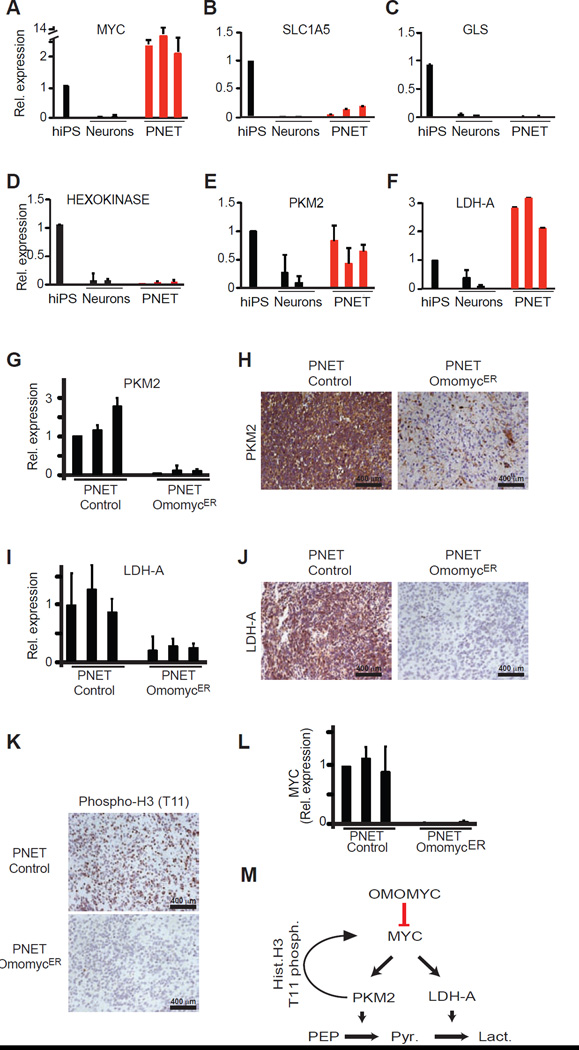
What may cause this differential MYC requirement between normal and malignant tissues? We examined how MYC contributes to the hallmark differences between normal neurons and malignant PNETS (Figure 4A). For example, MYC has been implicated in cancer-specific changes in glutamine metabolism and mitochondrial biology (Gao et al., 2009; Zhang et al., 2012). However, however we saw only modest or no difference in expression of the glutamine transporter (SLC1A5), or the glutaminase enzyme (GLS) between differentiated neurons and PNETs (Figure 4A/B/C, Figure S3A). Similarly, the ratio of mitochondrial to nuclear DNA, the expression of the mitogenesis factor TFAM (Li et al., 2005), or the cancer-associated uncoupling protein UCP2 (Zhang et al., 2011) revealed no cancer-specific changes (Figure S3B–D).
Figure 4. OmomycER sensitive changes in PKM2 and LDH-A expression distinguish PNETs from differentiated neurons.
A, Quantitative RT-PCR measuring relative expression of MYC in human iPS cells (hiPS), dopaminergic neurons (Neurons), and brain tumors (PNETs); B–F, Quantitative RT-PCR measuring relative expression of the indicated genes (glutamine transporter SLC1A5) (B), glutaminase (GLS) (C), hexokinase (D), pyruvate kinase M2 isoform (PKM2) (E), lactate dehydrogenase A (LDH-A) (F); G and H, Quantitative RT-PCR (G) and immunohistochemistry (H) measuring LDH-A mRNA and protein expression in control and OmomycER PNETs following in vivo tamoxifen treatment; I and J, Quantitative RT-PCR (I) and immunohistochemistry for LDH-A in control and OmomycER PNETs following treatment; K, Immunohistochemical stain for histone H3 (T-11 phosphorylation) in control and OmomycER PNETs after tamoxifen treatment in vivo; L, Quantitative RT-PCR measuring endogenous MYC expression in control and OmomycER PNETs after tamoxifen treatment in vivo. M, Diagram describing that OmomycER disrupts MYC and PKM2 dependent metabolic and transcriptional changes in cancer.
MYC has been implicated in the Warburg effect and aerobic glucose metabolism (Dang et al., 2009). While we found no change in hexokinase expression (Figure 4D), both the pyruvate kinase isoform M2 (PKM2) and lactate dehydrogenase A (LDH-A) were strikingly increased in PNETS compared to neurons (Figure 4E/F). Importantly, their expression was strictly dependent on MYC and OmomycER activation resulted in near complete loss of PKM2 and LDH-A mRNA and protein expression in PNETs (Figure 4G–J, Fig. S3E). In addition to its role in aerobic glycolysis PKM2 has a surprising nuclear function and phosphorylates histone H3 to control MYC and CCND1 expression (Yang et al., 2012). Indeed, we confirm this novel PKM2 function in OmomycER expressing PNETs in vivo, where TAM treatment causes rapid loss of histone H3 (T11) phosphorylation and reduction in the expression of the c-MYC mRNA (Figure 4K/L). Hence, the differential response of normal and malignant cells to MYC inactivation reflects, at least in part, the disruption of a metabolic and epigenetic feed-forward program (Figure 4M).
DISCUSSION
We report a new strategy to enhance the safe use of stem cells in regenerative medicine. The propensity of PSC-derived tissues for malignant transformation is a significant clinical concern as highlighted by recent reports of cancer development following the administration of neurogenic cells (Amariglio et al., 2009; Doi et al., 2012). Continuing improvements in reprogramming procedures, the selection of genetically suitable clones, and different purging strategies will clearly help attenuate the risk (Lee et al., 2013). We report an additional strategy that is rooted in the concept of cancer’s “oncogene dependence” (Jain, 2002; Soucek et al., 2008; Weinstein, 2002). Specifically, the dominant negative OmomycER allele provides a unique tool to block transcriptional activation by all three MYC proteins (Savino et al., 2011; Soucek, 1998, 2002; Soucek et al., 2008). Engineering PSCs that harbor an inducible MYC inhibitory allele provides a failsafe mechanism that can be used to i) purge residual iPS cells (before and after engraftment), and ii) to treat aggressive cancers with a high degree of selectivity for the malignant and rapidly proliferating cells and little or no effect on differentiated tissues.
Our study also explore the causes underlying the differential MYC requirement in cancer (von Eyss and Eilers, 2011). Prior work has emphasized MYC’s role maintaining angiogenesis (Sodir et al., 2011), and its ability to restrain cellular senescence or differentiation (Lin et al., 2009; Varlakhanova et al., 2011; Wu et al., 2007). Our results indicate that the differential MYC requirement reflects, at least in part, the need to maintain the expression of the cancer-specific isoforms of pyruvate kinase (PKM2) and lactate dehydrogenase (LDH-A) (Christofk et al., 2008; Clower et al., 2010; David et al., 2010; Shim et al., 1997; Vander Heiden et al., 2009). Moreover, PKM2 is required to maintain MYC expression and PKM2 loss disrupts a cancer-specific epigenetic feed-forward mechanism (Yang et al., 2012).
Experimental Procedures (see Suppl. Experimental procedure for details)
Generation and characterization of iPS cells
Murine iPS cells were derived from MEFs infected with four lentiviral vectors (pLM-OCT4, pLM-Sox2, pLM-Myc and pLM-Klf4) expressing OCT4, SOX2, KLF4 and MYC, plated on feeder layers of mitomycin–C treated embryonic fibroblast (GlobalCells) after 96h. Single ES-like colonies were picked and expanded after 10–25d and cultivated on mitomycin-C treated MEFs in KnockOut™ DMEM with high glucose (GIBCO), supplemented with 15% Fetal Bovine Serum (Hyclone), 0.1mM b-mercaptoethanol, 4mM L-glutamine, 1× Non-essential amino acids and 1000U/ml of LIF. Media was changed every day. The human iPS cell line thal5.10 was generated with a single, excisable polycistronic vector expressing OCT4, KLF4, MYC and SOX2 (Papapetrou et al., 2011). The media were changed 24 h later and replaced every day thereafter with hES cell media supplemented with 6 ng/ml FGF2 (R&D Systems) and 0.5 mM VPA (Sigma). Fifteen to 25 days after transduction, colonies with hES cell morphology were mechanically dissociated and transferred into plates pre-seeded with mitomycin C–treated mouse embryonic fibroblasts (MEFs) (GlobalStem). Cells were thereafter passaged with dispase and expanded to establish iPS cell lines.
iPS clones were characterized by immunofluorescence using anti-Nanog-Alexa Fluor 488 conjugated (eBioscience) and anti-SSEA-1 Alexa Fluor 647 conjugated (eBioscience). Gene expression of endogenous and transgenic Oct4, Myc, Sox2 and Klf4 was by TaqMan qRT-PCR assay.
Tumour analysis
1*106 iPS cells expressing omomycER or controls were injected s.c. into NOD/MrkBomTac-Prkdcscid (Taconic) mice. When the tumors were well palpable (~1 cm3), the mice were treated with 100 µl of tamoxifen (TAM, i.p, 10mg/ml in peanut oil). Tumors were collected one week after the last treatment, weighed and fixed in 4% formaldehyde for subsequent stains including haematoxylin/eosin and immunohistochemistry. SALL4 and OCT4 were performed on automated platforms (Ventana, Tucson AZ) per manufacturer’s instructions.
Generation of chimeric mice
Single cell suspensions of exponentially growing iPS cells were used for blastocysts injection by standard procedure. Blastocysts were obtained at day 3.5 of gestation from superovulated albino C57Bl/6J (B6(Cg)-Tyrc-2J/J, Jax # 000058) female mice. For each blastocyst, an average of either 8 or 12 iPS cells were injected. After injection, blastocysts were returned to optimization medium and placed at 37 °C until transferred to recipient females. 10–15 injected blastocysts were transferred to each uterine horn of 2.5-d-postcoitum-pseudopregnant females.
Supplementary Material
Acknowledgments
We thank Gerard Evan and Laura Soucek for the OmomycER construct, Zhimin (James) Lu, Martin Eilers, Hank Kung, and Craig Thompson for helpful discussions. D.P. Winkleman and C.C. Le for the help with MRI imaging, the MSK research animal facility and the MSK Mouse Genetics Core, the MSKCC Pathology Core Facility. This work is supported by grants from the NCI (R01-CA142798-01), and a P30 supplemental award (HGW), the Leukemia Research Foundation (HGW), the Louis V. Gerstner Foundation (HGW), the WLBH Foundation (HGW), the Society of MSKCC (HGW and JTH), the Starr Cancer Consortium grant I4-A410 (HGW), The NYSTEM grant #C028131 (HGW), NCI Carrier Developmental Award 1K99CA175179-01A1 (EO) Leukemia and Lymphoma Society fellowship (AOM), the Doris Duke Charitable Foundation (JTH), the Sidney Kimmel Foundation (JTH), the AACR/Landon Foundation (JTH), the MSKCC Brain Tumor Center (JTH) MSKCC Core Grant NIH (P30-CA 008748) (MHW), the NINDS (R01NS066390; LS)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary information accompanies the paper.
Author contributions E.O. designed, conducted, analyzed data; E.P., S.K., F.L, Y.G and A. O-M conducted experiment and analyzed experiments; J.T.F., V.R., and JTH tumor pathology, W.H.M. performed blastocyst injections, L.S. and M.S. design and data analysis, H.G.W. and E.O. designed the study and wrote the paper.
The authors declare no competing financial interests.
References
- Amariglio N, Hirshberg A, Scheithauer BW, Cohen Y, Loewenthal R, Trakhtenbrot L, Paz N, Koren-Michowitz M, Waldman D, Leider-Trejo L, et al. Donor-derived brain tumor following neural stem cell transplantation in an ataxia telangiectasia patient. PLoS medicine. 2009;6:e1000029. doi: 10.1371/journal.pmed.1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nature methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature biotechnology. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AB, Tan HL, Ang SN, Fong WJ, Chin A, Lo J, Zheng L, Hentze H, Philp RJ, Oh SK, et al. Selection against undifferentiated human embryonic stem cells by a cytotoxic antibody recognizing podocalyxin-like protein-1. Stem Cells. 2008;26:1454–1463. doi: 10.1634/stemcells.2007-0576. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi D, Morizane A, Kikuchi T, Onoe H, Hayashi T, Kawasaki T, Motono M, Sasai Y, Saiki H, Gomi M, et al. Prolonged Maturation Culture Favors a Reduction in the Tumorigenicity and the Dopaminergic Function of Human ESC-Derived Neural Cells in a Primate Model of Parkinson's Disease. Stem Cells. 2012;30:935–945. doi: 10.1002/stem.1060. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring CE, Duffy PA, Benvenisty N, Andrews PW, Ben-David U, Eakins R, French N, Hanley NA, Kelly L, Kitteringham NR, et al. Assessing the safety of stem cell therapeutics. Cell stem cell. 2011;8:618–628. doi: 10.1016/j.stem.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Hanna J. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hong H. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narva E, Ng S, Sourour M, Hamalainen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Jain M. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Molecular and cellular biology. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. The EMBO journal. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti J-C, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and Classification of Chromosomal Aberrations in Human Induced Pluripotent Stem Cells. Cell stem cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, et al. Variation in the safety of induced pluripotent stem cell lines. Nature biotechnology. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- Nakagawa M. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Osakada F, Ikeda H, Sasai Y, Takahashi M. Stepwise differentiation of pluripotent stem cells into retinal cells. Nature protocols. 2009;4:811–824. doi: 10.1038/nprot.2009.51. [DOI] [PubMed] [Google Scholar]
- Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A, et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nature biotechnology. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, Soucek L, Nasi S. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PloS one. 2011;6:e22284. doi: 10.1371/journal.pone.0022284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Gene Dev. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17:2463–2472. doi: 10.1038/sj.onc.1202199. [DOI] [PubMed] [Google Scholar]
- Soucek L. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res. 2002;62:3507–3510. [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010a;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nature methods. 2010b;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013 doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Tan HL, Fong WJ, Lee EH, Yap M, Choo A. mAb 84, a cytotoxic antibody that kills undifferentiated human embryonic stem cells via oncosis. Stem Cells. 2009;27:1792–1801. doi: 10.1002/stem.109. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova N, Cotterman R, Bradnam K, Korf I, Knoepfler PS. Myc and Miz-1 have coordinate genomic functions including targeting Hox genes in human embryonic stem cells. Epigenetics & chromatin. 2011;4:20. doi: 10.1186/1756-8935-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eyss B, Eilers M. Addicted to Myc-but why? Gene Dev. 2011;25:895–897. doi: 10.1101/gad.2053311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes: the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Werbowetski-Ogilvie TE, Bosse M, Stewart M, Schnerch A, Ramos-Mejia V, Rouleau A, Wynder T, Smith MJ, Dingwall S, Carter T, et al. Characterization of human embryonic stem cells with features of neoplastic progression. Nature biotechnology. 2009;27:91–97. doi: 10.1038/nbt.1516. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell stem cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. The EMBO journal. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell stem cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.