Abstract
Hypothalamic proopiomelanocortin (POMC) neurons, one of the major regulators of the HPA axis, immune functions, and energy homeostasis, are vulnerable to the adverse effects of fetal alcohol exposure (FAE). These effects are manifested in POMC neurons by a decrease in Pomc gene expression, a decrement in the levels of its derived peptide β-endorphin (β-EP) and a dysregulation of the stress response in the adult offspring. The HPA axis is a major neuroendocrine system with pivotal physiological functions and mode of regulation. This system has been shown to be perturbed by prenatal alcohol exposure. It has been demonstrated that the perturbation of the HPA axis by FAE is long-lasting and is linked to molecular, neurophysiological and behavioral changes in exposed individuals. Recently, we showed that the dysregulation of the POMC system function by FAE is induced by epigenetic mechanisms such as hypermethylation of POMC gene promoter and an alteration in histone marks in POMC neurons. This developmental programming of the POMC system by FAE altered the transcriptome in POMC neurons and induced a hyperresponse to stress in adulthood. These long-lasting epigenetic changes influenced subsequent generations via the male germline. We also demonstrated that the epigenetic programming of the POMC system by FAE was reversed in adulthood with the application of the inhibitors of DNA methylation or histone modifications. Thus, prenatal environmental influences such as alcohol exposure could epigenetically modulate POMC neuronal circuits and function to shape adult behavioral patterns. Identifying specific epigenetic factors in hypothalamic POMC neurons that are modulated by fetal alcohol and target Pomc gene could be potentially useful for the development of new therapeutic approaches to treat stress-related diseases in patients with Fetal Alcohol Spectrum Disorders.
INTRODUCTION
Prenatal alcohol exposure has long-lasting adverse effects on the functioning of the hypothalamic-pituitary-adrenal (HPA) axis (Helleman et al., 2010; Rachdaoui and Sarkar, 2013). Long-term alteration of the HPA axis function in response to fetal alcohol exposure (FAE) has been linked to a wide spectrum of molecular, neurophysiological and behavioral changes in exposed individuals. Specific endophenotypes such as behavioral deficits, hyperresponses to stress, altered metabolic functions and malfunctioning of the immune system are also observed in alcohol-exposed rodents in the adult stage (Table 1). Acute or chronic exposure to environmental factors such as drug of abuse or toxicants during critical periods of development has been shown to cause global or gene-specific alterations in histone modifications, chromatin remodeling and/or DNA methylation in different areas of the brain (Cummings et al., 2010). More importantly, there is now compelling evidence that prenatal exposure to these environmental factors including ethanol could incite epigenetic changes in the genome that could permanently modulate gene expression and function and adversely influence subsequent generations (Skinner, 2010; Govorko et al., 2012). In this review, we discuss the vulnerability of the POMC system, one of the important regulators of the HPA axis to FAE and describe how epigenetic changes such as histone modifications and DNA methylation modulate Pomc gene expression and function. We also summarize our recent findings from animal models and show that FAE programs the POMC system and the stress axis functions of subsequent generations via epigenetic mechanisms.
Table 1.
Consequences of the hypothalamic pituitary adrenal (HPA) axis alterations produced by fetal alcohol exposure on various physiological systems in offspring
| System | Effects |
|---|---|
| Central Nervous System |
|
| Neuroendocrine System |
|
| Metabolic System |
|
| Immune System |
|
| Cardiovascular System |
|
VULNERABILITY OF POMC SYSTEM TO FETAL ALCOHOL EXPOSURE
Alcohol is a toxic chemical and a drug of abuse that exhibits wide range of conspicuous effects on many systems including the central nervous system (CNS) (Zahr et al., 2011). The adverse effects of alcohol exposure on the brain are usually more prominent during fetal life and could be long-lasting (Haycock, 2009). β-endorphin-producing POMC neurons in the arcuate nucleus of the hypothalamus are particularly vulnerable to the neurotoxic action of alcohol during the developmental period (Rachdaoui and Sarkar, 2013). POMC system plays essential roles in many physiological processes including feeding circuitry (Parker and Bloom, 2012), stress axis regulation and immune system modulation (Sarkar and Zhang, 2013). It is not then suprising that the malfunctioning of this system by alcohol exposure could have wide range of effects and adverse consequences at the molecular, cellular, system and organismal levels. We focus here our discussion on the epigenetic effects of FAE on Pomc gene expression, β-endorphin peptide production and stress axis functioning.
POMC system
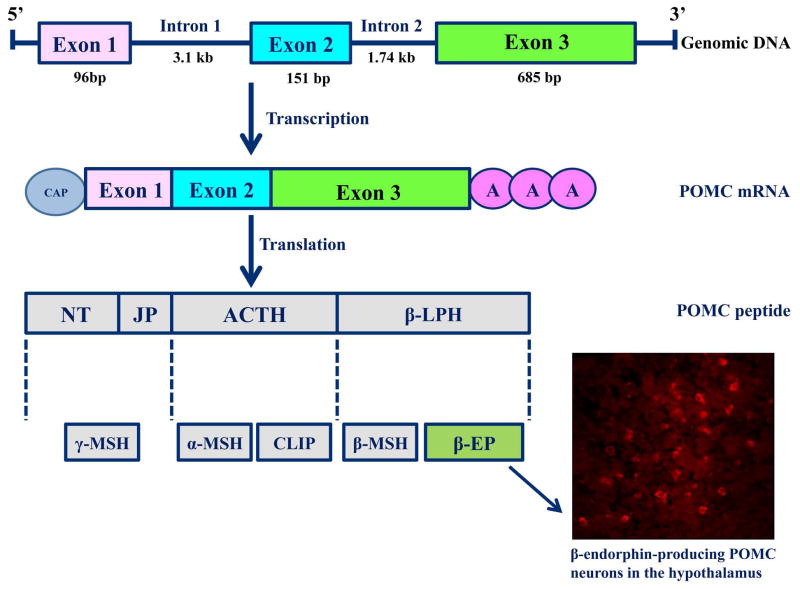
POMC is the common precursor for the melanocortin-related peptides (ACTH/α-melanocyte-stimulating hormones (MSH), β-MSH, and γ-MSH) and the opioid peptide β-endorphin (BEP). Pomc gene structure is highly conserved among mammalian species indicating that the peptides derived from this gene have physiological significance. In humans, POMC gene resides in chromosome 2p23, contains three exons and two introns (3708 and 2886 bp), and spans 7665 bp. It has three different promoters that regulate the differential transcription of this gene in different tissues. These promoters are embedded within a defined CpG island, and are methylated in normal non-expressing tissues, which is sufficient for silencing its expression. In tissues that are expressing Pomc gene, promoters are specifically unmethylated to allow the binding of essential transcription factors (Newell-Price, 2003). Pomc gene is expressed in the brain, the pituitary gland, and in various peripheral tissues. In the brain, this gene is primarily expressed by neurons in the arcuate nucleus of the hypothalamus, and is expressed in a lesser quantity in other areas of the brain such as the amygdala, the hippocampus, the cortex, and the nucleus tractus solitaries of the brainstem. In the pituitary, the Pomc mRNA level is highly expressed in the anterior and neurointermediate lobes. In the periphery, Pomc expression was found in detectable quantities in the semen and testes, ovaries and placenta, peripheral mononuclear cells, thymus and some tumors. The POMC polypeptide contains 241 amino acids, weighs 32 kDa, and can be cleaved into many biologically active neuropeptide hormones including ACTH, β-endorphin, lipotropins (LPHs) and MSHs by individual processing through a series of tissue-specific co- and post-translational modifications. POMC peptide is processed differentially in the pituitary: in the anterior lobe into ACTH and β-LPH, while in the intermediate lobe into α-MSH and β-endorphin. In the hypothalamus, POMC is processed into β-endorphin, γ-lipotropin and α-MSH (Fig. 1). Pomc gene plays an important role in the regulation of the HPA axis, adrenal development, depression, skin pigmentation and obesity. The transcription of POMC is stimulated by CRH and β-endorphin in turn inhibits CRH production as a negative feedback. In the anterior but not in the intermediate pituitary, glucocorticoids inhibit Pomc gene transcription (Wardlaw, 2010). Glucorticoid feedback on the CNS involves retrograde opioid signaling involving μ opioid receptors at the hypothalamic synapses (Wamsteeker Cusulin et al., 2013).
Fig. 1. POMC peptide processing in the hypothalamus.
Pomc gene (ENSRN000000012686) in rats is located on chromosone 6 (www.ensembl.org). It is transcribed into a mRNA (ENSRN000000016976), translated into a peptide that is proteolytically cleaved in a tissue specific manner into biologically-active peptides. This gene gives rise to two classes of peptides; the melanocortins and the endorphins. In the hypothalamus, POMC peptide processing yields primarily α-MSH, a negligeable amount of ACTH and β-EP (NT= N-terminal peptide, JP=Junctional peptide, ACTH= Adrenal corticotrophin thyroid hormone, β-LPH= Beta- lipotrophin hormone, α-MSH= Alpha melanocyte-stimulating hormone, CLIP= Corticotropin-like intermediate peptide, β-EP=Beta- endorphin). A representative figure of β-EP-producing POMC neurons in the arcuate nucleus of the hypothalamus is represented.
Developmental effect of alcohol exposure on hypothalamic POMC system
As discussed above, the HPA axis of fetal alcohol-exposed animals is often dysregulated. Studies using the transplantation of β-endorphin neurons into the hypothalamus provided evidence for a causative role for the loss of the opioid negative feedback on the HPA axis (Boyadjieva et al., 2009). Therefore, dysregulation in Pomc gene transcription and its peptide production (BEP) may be one of the causes for the induction of hyper-reactive HPA axis in fetal alcohol-exposed animals.
Extensive work has been done to determine the mechanism by which fetal alcohol exposure controls hypothalamic Pomc gene expression. Fetal alcohol exposed animals have decreased hypothalamic Pomc gene expression, decreased β-endorphin neuronal number and peptide production (Del Arbol et al, 2007; Boyadjiva et al, 2009). In vitro studies demonstrated that ethanol-induced apoptotic effect on BEP neurons involves a reduction in cAMP production and activation of transforming growth factor-β1 (TGF-β1)-linked signals, and enhancement of caspase 3 activity (Chen et al, 2006). In vivo studies also showed that FAE increased apoptotic death of β-endorphin neurons in association with increased TGF-β1 in the arcuate nucleus. The increased TGF-β1 in the arcuate tissue is correlated with a reduction of retinoblastoma protein (Rb) phosphorylation increased cyclin dependent kinase inhibitor p27/kip level and decrease of cyclin dependent kinase 4 and cyclin D3 levels (Kuhn and Sarkar, 2008).
In a postnatal alcohol-exposed rat model that mimics human exposure to ethanol in the third trimester of pregnancy, there was a significant reduction in Pomc mRNAs expression in the arcuate nucleus (Sarkar et al, 2007). These changes were observed shortly after ethanol ingestion by the pups. Collectively, these data may indicate that FAE causes apoptosis of β-endorphin producing POMC neurons through a c-AMP and TGF-β1 related pathway.
It is clear that ethanol exposure has toxic effects on hypothalamic fetal brain tissue and POMC neurons. It is quite intriguing that some of POMC neurons survive the alcohol’s insult during the developmental period but exhibit a dysregulation in the function of its gene and its derived peptide β-endorphin in the adult stage. Why those surviving POMC neurons exhibit such dysregulation in Pomc gene expression is an important question that is addressed in this review. Ethanol-induced physiological changes may have caused imprinting of the neuronal cells during critical periods of fetal development. Epigenetic modification is a highly possible imprinting mechanism for many genes. We will review the evidence for the epigenetic imprinting of FAE on Pomc gene.
EPIGENETIC INSIGHTS INTO DYSREGULATION OF POMC SYSTEM BY FETAL ALCOHOL EXPOSURE
Epigenetic changes are now considered potential mechanisms for the long-term effects of prenatal exposure to toxicants or drugs of abuse including alcohol. The epigenetic mechanisms include DNA methylation, histone modifications, chromatin remodeling and non-coding RNAs including microRNAs (miRNAs). Studies have demonstrated that ethanol exposure causes selective histone modifications in a tissue-specific manner and an alteration in neuronal gene expression and/or function (Moonat et al., 2010). Modulation in DNA methylation has also been linked to many neurological diseases or disorders including drug addiction (Nestler, 2014). Here we discuss the role of these epigenetic mechanisms in regulation of FAE-induced changes in Pomc gene expression.
Histone modifications
Histone proteins play important roles in modulation of gene expression by helping the organization of DNA into a chromatin state that is accessible or inaccessible to transcriptionally-active molecules. Histones are highly basic proteins that help in the packaging of DNA into nucleosomes, the building blocks of chromatin. These proteins are subject to epigenetic changes in response to internal or external factors including alcohol. These proteins are highly conserved and include: the linker histone H1, two copies of H3 and H4, two H2A/H2B dimers and histone variants such as H2AZ. The role of H1 histone in transcription has been recently demonstrated. It helps in the packaging of nucleosomes into a higher order chromatin structure and also acts as an “exit/entry” point to nucleosomes to impact gene transcription (Zlatanova et al., 2000). In the context of gene expression regulation, it has been shown that the histone variants H2AZ and the isoforms of H3 (H3.1/H3.3) play a role in gene activity by modulating nucleosome positioning along the DNA (Thakar et al., 2009).
Structurally, each histone consists of a globular domain and a charged amino NH2-terminus tail that protrudes out of each nucleosome. This histone tail undergoes malleable postranslational modifications and allows the chromatin to exist in a relaxation state (euchromatin) that is conducive for gene activation or in a compacted state (heterochromatin) that is conducive for gene repression. A wide variety of histone-modifying enzymes add a chemical group to the histone tail thus providing a “histone code” that will be read and translated by the epigenetic machinery to participate in the decision of the transcriptional outcome. These postranslational modifications include methylation, acetylation, phosphorylation, sumolylation or ubiquitination. Some of these modifications are mutually exclusive while other modifications coexist on the same nucleosome. Most of these modifications occur at specific amino acid residues on the tail such as lysine (K), arginine (R), serine (S) or threonine (T). Among these modifications, lysine methylation and acetylation of H3 and H4 are probably the best understood modifications in the context of transcriptional regulation. For example, histone acetylation such as acetylated H3K9 (AceH3K9) is mostly associated with transcriptional activation and is catalyzed by the activity of histone acetyltransferases (HATs). Histone methylation is one of the most complex histone marks and is catalyzed by the activity of histone methyltransferases (HMTs) that utilize S-adenosylmethionine (SAM) as a methyl (CH3) donor. This histone modification is considered an inert modification that modulates chromatin architecture and its accessibility to factors based on which residue of a specific histone is methylated and the number of methyl groups (mono (me1), di-(me2) or tri-(me3)) added on the amine of lysine residue (Strahl and Allis, 2000). We will focus our discussion on methylation of lysine 4 (K4) and lysine 9 (K9). These two histone methylation marks could have dual effects on gene transcription, such as gene activation or repression, depending on their location along the gene body, the type of effector proteins that will be recruited to the chromatin, their interactions with the DNA methylation machinery, and their interactions with other multiprotein complexes or ATPase chromatin-remodelers (Jenuwein, 2006).
We recently showed that FAE decreased the protein levels of the activation marks di-trimethylated H3K4 (H3K4me2,3), acetylated H3K9 (AceH3K9) and phosphorylated H3S10 (pH3S10) and increased the protein levels of the repressive mark dimethylated H3K9 (H3K9me2) in β-endorphin-producing POMC neurons. This modulation in protein levels correlated with changes in gene expression of several histone-modifying enzymes such as Set7/9, G9a/Setdb1 and CREB-binding protein (CBP) (Govorko et al., 2012; Bekdash, 2013) in these neurons. Several studies showed that changes in global or gene-specific histone marks are tissue- specific and most probably affected by the amount, duration and the developmental time of alcohol exposure. For example, Pal-Bhadra and colleagues (2007) showed that ethanol exposure increased H3K4 methylation and decreased H3K9 methylation in primary cultures of rat hepatocytes. In the developing rat cerebellum, perinatal ethanol exposure reduced CBP expression and the levels of acetylated H3 and H4 hence suggesting a potential role for the acetyltransferase CBP in ethanol-induced developmental defects in FASD subjects (Guo et al., 2011). Chronic alcohol exposure increased H3K4me3 levels in the cortex of chronic alcoholics (Ponomarev et al., 2012). These altered patterns in histone marks most often correlate negatively or positively with changes in gene expression and sometimes they do not cause any molecular change. Drug-induced neuroplasticity in gene expression and the modulation of downstream signaling mechanisms are not only mediated by changes in histone marks but most probably are influenced by the cumulative effects of many complex homotypic or heterotypic interactions between histone-methyltransferases (HMTs), histone deacetylases (HDACs), DNA methyltransferases (Dnmts), effector proteins, transcription factors (TFs), ATPase chromatin-remodeling complexes and RNA polymerase II and possibly other yet unidentified factors (Nestler, 2014).
DNA methylation
DNA methylation is another epigenetic mechanism that plays a critical role in modulating expression and/or function of many genes (Bird, 2001) including POMC gene (Govorko et al., 2012). DNA methylation has been historically considered as a mechanism that differentially modulates gene expression in both health and disease states of many systems including neural systems. It is a normal phenomenon that is precisely and temporally regulated in a tissue-specific manner and is vital to many physiological processes including DNA replication, gene repression, parental imprinting, cellular differentiation and normal brain development in mammals (Robertson and Wolffe, 2000). Abnormal methylation has been associated with many neurological disorders including Fetal Alcohol Spectrum Disorders (FASD) (Ungerer et al., 2013).
DNA methylation is an epigenetic mark that involves covalent modification of the cytosine residue in CpG dinucleotides in the promoter region to “lock in” the silent state of a gene (Deaton and Bird, 2011). Methylation is a covalent modification of the DNA that is catalyzed by the activity of DNA methyltransferases (Dnmts). DNA methyltransferases include Dnmt1, Dnmt2, Dnmt3a, Dnmt3b and Dnmt3L. In enkaryotes, these enzymes utilize SAM to methylate carbon C5 of cytosine (5-mC) that is located next to a guanine (G) in CpG dinucleotides in the promoter. It has been shown that there is a “cross-talk” between the DNA methylation machinery and the histone-modifiers (Bird, 2001). Methylated DNA most often recruits several methyl-binding proteins (MBDs), HDACs and other repressors and co-repressors to promote gene repression. 5-mC is now considered as the most stable heritable chromatin modification that is conducive for transcriptional inhibition. Another modification, 5-hydroxymethylcytosine (5-hmC) was recently identified in specific areas of the brain such as the cerebral cortex, brain stem and Purkinje neurons (Kriaucionis and Heintz, 2009). It is catalyzed by another group of enzymes known as the hydroxylase like ten-eleven translocation proteins (Tet1, Tet2 and Tet3). It has been suggested that the hydroxylation of 5-mC to 5-hmC modulates the binding of several effector proteins to the chromatin and facilitates DNA demethylation to promote gene transcription. There is now evidence that 5-hmC plays a role in both transcriptional activation and repression (Wu et al., 2011) but its role in DNA methylation is not yet fully understood. Although there is little evidence for a causative role of DNA methylation in gene silencing, CpG methylation in the promoter region of a gene most often correlates with silencing of its promoter activity (Deaton and Bird, 2011). A group of CpG dinucleotides in the promoter region or around the transcription start site (TSS) of a gene is often referred to as a “CpG islands” (CGIs) and are usually unmethylated. Abnormal methylation of the promoter CGIs in response to external or internal factors has been associated with gene silencing. For example, the methylation of CpGs in the binding site of certain transcription factors (TFs) such as the specificity protein 1 (Sp1), c-AMP responsive element binding protein (CREB) and CCCTC-binding factor (CTCF) has been demonstrated. The methylation of these binding sites could block the accessibility of these TFs to their regulatory sites along the DNA and modulate expression of the target gene (Blackledge and Klose, 2010). We have recently found that FAE increased the methylation of POMC gene promoter in the hypothalamus. One of the hypermethylated CpGs at position −62 upstream of POMC gene TSS coincided with the CCAAT box which is a binding site for TFs essential for transcriptional activation (Govorko et al., 2012). Another methylated CpG at position −216 contained an 11 bp sequence which is highly conserved in vertebrates and is important for Pomc gene expression in target tissues (Bumaschny et al., 2007). This could suggest that the hypermethylation of this site by FAE could be one of the causes of the decrease in Pomc gene expression and β-endorphin peptide production in hypothalamic POMC neurons.
The conventional view of linking DNA methylation in the promoter region to gene silencing has been recently challenged. The methylation of CpG dinucleotides in the gene body would also participate in determining the outcome of gene expression (Suzuki and Bird, 2008). CpG islands (CGIs) predominate in the promoter region of most genes and are also found in transcribed region of the gene (intragenic). On the other hand, intergenic regions (between genes) and intronic regions are usually CpG poor. Those CpGs that are located in intragenic or intergenic regions are referred to as “orphan CpG islands” due to their distant location relative to the promoter region and the uncertainty of their functions. However, their methylation status could provide additional information on target gene expression regulation. Transgenic studies demonstrated that methylated CpGs in intragenic regions impeded RNA Polymerase II elongation and decreased the occupancy of the activation marks di- and trimethylated H3K4 and acetylated H3K9. It was suggested that these orphan CGIs could: 1) function as promoter for the formation of short transcripts or small noncoding RNAs (ncRNAs), 2) alter the efficiency of transcriptional elongation by RNA polymerase II or 3) affect gene splicing (Kornblihtt, 2006). Orphan CGIs were identified in many studies and were found to generate non-coding RNAs with widespread effects on expression regulation of many genes (Deaton and Bird, 2011). In the context of POMC gene, the downstream 3′ CpG island in exon 3 generates short transcripts with unknown functions (Gardiner-Garden and Frommer, 1994).
Non-coding RNAs
Although DNA methylation and histone modifications are pivotal mechanisms in the modulation of gene expression, small noncoding RNAs (ncRNAs) including microRNAs (miRNAs) are now emerging as a group of intriguing regulatory elements with complex mode of gene expression regulation. These elements have been implicated in many diseases including alcohol-related diseases (Miranda et al., 2010) and emerged recently as potential therapeutic targets of many diseases of the CNS. MicroRNAs usually target the 3′-untranslated region (3′ UTR) of a target gene and regulate its expression at the posttranscriptional level. Depending on the complementarity between the seed region (2–7 nt) of miRNAs and the 3′ UTR of the target gene, these regulatory elements could cause mRNA cleavage, mRNA deadenylation or result in translational repression. Whether the 3′ UTR of Pomc gene could be targeted by specific miRNAs upon alcohol exposures and the functional consequences of such targeting are currently under investigation.
EPIGENETIC PROGRAMMING OF THE POMC SYSTEM BY FETAL ALCOHOL EXPOSURE IS REVERSED IN ADULTHOOD
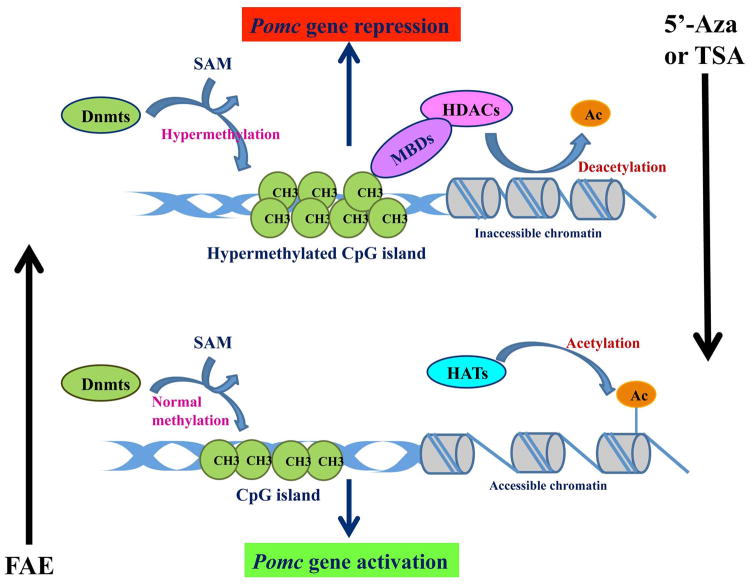
Inhibitors of histone deacetylases (HDACs) and DNA methyltransferases (Dnmts) are now considered potential therapeutic targets for the treatment of many diseases or disorders including neurological disorders. The injection of TSA, the inhibitor of HDACs, or 5′-Azacytidine (5′-Aza), the inhibitor of Dnmts, during the neonatal period normalized fetal alcohol-induced POMC gene hypermethylation in the adult stage. A conceptual diagram of the epigenetic effects of FAE on Pomc gene and its reversibility is presented in Fig. 2. This change correlated with normalization of Pomc gene expression, β-endorphin peptide production and ACTH/corticosterone response to lipopolysaccharide (LPS) immune challenge (Govorko et al., 2012). Pandey and colleagues (2008) demonstrated that TSA treatment reversed the molecular changes that were observed in alcohol-withdrawn rats in the amygdala and attenuated some of the behavioral manifestations that were associated with alcohol withdrawal such as anxiety. Collectively, these findings suggest that the components of the epigenetic machinery have potential medical applications for the treatment of some of the behavioral changes that are usually associated with alcohol exposure including the induction of an abnormal stress response. Behavioral tests should be performed in future studies to compare the stress response in control and fetal alcohol-exposed rats after inhibition of HDAC activity.
Fig. 2. Conceptual diagram of the epigenetic effects of fetal alcohol exposure on POMC gene in the hypothalamus.
Dnmt=DNA methyltransferase, MBD (methyl-binding domain proteins), HDAC=histone deacetylase, HAT= Histone acetyltransferase, Ac=acetylation, FAE=fetal alcohol exposure, TSA=trichostatin, 5′-Aza=5′-azacytidine, CH3=methyl group, SAM=S-adenosylmethionine.
MULTIGENERATIONAL TRANSFER OF EPIGENETIC MARKS VIA THE MALE GERMLINE IN POMC NEURONS
Transgenerational effect of fetal alcohol expsoure
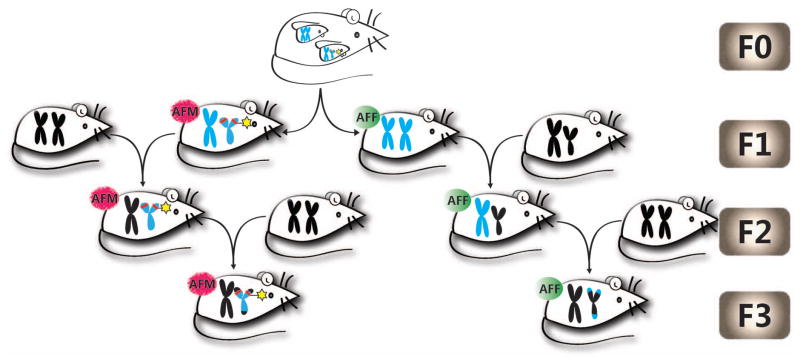
In addition to the direct effects of early life exposure to toxicants or drugs of abuse on adult onset disease, it has been recently shown that they also affect subsequent generations. Transgenerational inheritance of epigenetic changes in the genome is an interesting area of research that is still subject to debate and not fully understood. It is now considered an additional molecular mechanism, along with classic induction of genetic mutations, for the germline transmission of environmentally induced phenotypic changes (Skinner et al., 2010). To determine whether fetal alcohol effect on the stress axis is transmitted transgenerationally, we recently published results of a study where POMC gene methylation and expression as well as the endophenotypes related to Pomc gene expression defects following FAE were compared in F1–F3 male and female offsprings (Govorko et al., 2012). We produced two different germlines: 1) a male germline by breeding male fetal alcohol-exposed rats and their male offspring with normal females, and 2) a female germline by breeding female fetal alcohol- exposed rats and their female offspring with normal males (Fig. 3). We found that F1, F2 and F3 male progeny of male germline had a significant increase in DNA methylation levels in the proximal part of POMC gene promoter with the concomitant reduction in Pomc gene expression. DNA methylation or Pomc gene expression did not change in female F2 and F3 rats irrespective of their germline differences. Because the offspring (F1) and its germ cells (F2) were exposed to alcohol during development via the F0 female, the persistence of alcohol effects in the F3 male but not in the F3 female suggests that the transgenerational transmission of fetal alcohol effects on Pomc gene methylation and expression occurred via the male germline. The transgenerational effect of FAE was also noted to some endophenotypes of Pomc gene defect. We found an increase in the basal and immune stress-stimulated ACTH and corticosterone levels in both male and female offspring of F1 progeny and in males of male germline of F2 and F3 progeny. To further verify the male germline transmission of fetal alcohol effects on Pomc gene, we measured the DNA methylation status of its promoter in sperm of F1–F3 male rats that were derived from the male or female germline. We found that the methylation pattern of Pomc gene in sperm persisted in the F1 through F3 generations of male rats derived from the male germline. Overall, our findings provide the first direct evidence that fetal alcohol effects on Pomc gene hypermethylation and stress axis abnormalities persisted throughout adulthood and perpetuated into subsequent generations through the male germline.
Fig. 3. A schematic diagram describing how male and female germ lines are created to study transgeneational effects of fetal alcohol on stress axis.
Mmale germline (AFM) was created by breeding male fetal alcohol exposed rats and their male offspring with normal females, and a female germline (AFF) was created by breeding female fetal alcohol exposed rats and their female offspring with normal males. As fetal alcohol exposure causes heritable sex-linked changes in Pomc expression in male rats, the possibility is raised that a sex-linked decrease in Pomc expression caused by fetal alcohol exposure (indicated by the red line on the short p arms of the Y chromosome), creating the transgenerational pattern observed.
Hypothetical mechanism for the transgenerational effect of fetal alcohol exposure
Transgenerational epigenetic marks on genes are not very common as most of epigenetic signatures are typically lost during gametogenesis. In some cases, certain marks may be retained due to a bias in removal (Morrison and Reeve, 1998). Many epigenetic marks are removed at meiosis (Bond and Finnegan, 2007). Prior to fertilization, the male gamete carries the father’s germline epigenetic signature. When fertilization occurs, many of the male’s epigenetic marks are lost. Protamines which are necessary for stabilization and dense condensation of the male DNA in spermatozoa replace most of the histones in late spermatogenesis. During gamete fusion, protamines are exchanged with female histones, typically acetylated histones, and the male DNA is demethylated (Fulka et al, 2004). In the hypothalamus, it is known that the expression of β-endorphin is regulated in part by the non-pairing region of the Y chromosome (YNPAR) that contains the sex-determining region gene (Botbol et al, 2011). Additionally, transgenerational changes in the expression of Pomc in rats were detected in sperm (Govorko et al., 2012). As FAE causes heritable sex-linked changes in Pomc expression in male rats, the possibility is raised that a sex-linked decrease in Pomc expression is due in part to epigenetic changes in YNPAR caused by FAE, creating the transgenerational pattern observed. Current studies are underway to test this hypothesis. The novel transgenerational epigenetic effect of alcohol that we demonstrated in our study should be considered an important covariant with genetic factors in the determination of the longitudinal effects of FAE on behaviors and physiological processes.
CONCLUSION AND PROSPECTIVE
FAE has been shown to cause programming of the POMC system by inducing permanent changes in Pomc gene expression and function in the hypothalamus of adult exposed offspring. These changes were induced by epigenetic mechanisms such as hypermethylation of POMC gene promoter and an alteration of histone marks in β-endorphin-producing POMC neurons. At the physiological level, these alcohol-induced changes were manifested by reduced Pomc gene expression, β-endorphin peptide production and a hyperresponse of the HPA axis to a stress challenge in adult. More importantly, these fetal alcohol-induced long-lasting changes at the molecular, cellular and organismal levels inflicted subsequent generations via the male germline. These findings clearly indicate that the adverse effects of FAE on POMC system are inherited by the subsequent generations in a gender specific manner. The reversibility of the POMC system defect that we demonstrated via the modulation of the components of the epigenetic machinery may have therapeutic potentials. Future studies should aim at identifying specific epigenetic factors that are involved in modulation of Pomc gene expression and selectively target specifically these factors to attenuate some of the phenotypes that are usually associated with FASD individuals such as abnormal stress response.
Acknowledgments
This work is partly supported by a National Institute of Health grant R37AA08757 and R01AA016695
References
- Agapito MA, Zhang C, Murugan S, Sarkar DK. Fetal alcohol exposure disrupts metabolic signaling in hypothalamic proopiomelanocortin neurons via a circadian mechanism in male mice. Endocrinology. 2014 May 5;:en20132030. doi: 10.1210/en.2013-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjona A, Boyadjieva N, Kuhn P, Sarkar DK. Fetal ethanol exposure disrupts the daily rhythms of splenic granzyme B, IFN-gamma, and NK cell cytotoxicity in adulthood. Alcoholism Clin Exp Res. 2006;30(6):1039–1044. doi: 10.1111/j.1530-0277.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in β-endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin Exp Res. 2013;37(7):1133–42. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. Molecular biology. Methylation talk between histones and DNA. Science. 2001;7;294(5549):2113–5. doi: 10.1126/science.1066726. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Klose RJ. Histone lysine methylation: an epigenetic modification? Epigenomics. 2010;2(1):151–61. doi: 10.2217/epi.09.42. [DOI] [PubMed] [Google Scholar]
- Bond DM, Finnegan EJ. Passing the message on: inheritance of epigenetic traits. Trends Plant Sci. 2007;12:211–216. doi: 10.1016/j.tplants.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Botbol M, Roubertoux PL, Carlier M, Trabado S, Brailly-Tabard S, Perez-Diaz F, Bonnot O, Bronsard G, Tordjman S. Modulation of Brain b-Endorphin Concentration by the Specific Part of the Y Chromosome in Mice. PLoS ONE. 2011;6(3):e16704. doi: 10.1371/journal.pone.0016704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyadjieva NI, Ortigüela M, Arjona A, Cheng X, Sarkar DK. Beta-endorphin neuronal cell transplant reduces corticotropin releasing hormone hyperresponse to lipopolysaccharide and eliminates natural killer cell functional deficiencies in fetal alcohol exposed rats. Alcohol Clin Exp Res. 2009;33(5):931–7. doi: 10.1111/j.1530-0277.2009.00911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumaschny VF, de Souza FS, López Leal RA, Santangelo AM, Baetscher M, Levi DH, Low MJ, Rubinstein M. Transcriptional regulation of pituitary POMC is conserved at the vertebrate extremes despite great promoter sequence divergence. Mol Endocrinol. 2007;21(11):2738–49. doi: 10.1210/me.2006-0557. [DOI] [PubMed] [Google Scholar]
- Chen CP, Kuhn P, Chaturvedi K, Boyadjieva N, Sarkar DK. Ethanol induces apoptotic death of developing beta-endorphin neurons via suppression of cyclic adenosine monophosphate production and activation of transforming growth factor-beta1-linked apoptotic signaling. Mol Pharmacol. 2006;69(3):706–17. doi: 10.1124/mol.105.017004. [DOI] [PubMed] [Google Scholar]
- Cummings JA, Clemens LG, Nunez AA. Mother counts: how effects of environmental contaminants on maternal care could affect the offspring and future generations. Front Neuroendocrinol. 2010;31(4):440–51. doi: 10.1016/j.yfrne.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Arbol JL, Rico Irles J, Contreras I, Aguirre JC, Raya J, Ruiz Requena ME, Miranda MT. Plasma concentrations of beta-endorphins in the children of alcoholic patients. An Med Interna. 2007;24(6):273–7. [PubMed] [Google Scholar]
- Fulka H, Mrazek M, Tepla O, Fulka J. DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. Transcripts and CpG islands associated with the pro-opiomelanocortin gene and other neurally expressed genes. J Mol Endocrinol. 1994;12(3):365–82. doi: 10.1677/jme.0.0120365. [DOI] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry. 2012;72(5):378–88. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Crossey EL, Zhang L, Zucca S, George OL, Valenzuala CF, Zhao X. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PLOS ONE. 2011;6(5):e19351. doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper KM, Tunc-Ozcan E, Graf EN, Redei EE. Intergenerational effects of prenatal ethanol on glucose tolerance and insulin response. Epigenetics & Epigenomics. 2014;46:159–168. doi: 10.1152/physiolgenomics.00181.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley DW, Handmaker NS, Lowe J. Infant stress reactivity and prenatal alcohol exposure. Alcoholism Clin Exp Res. 2006;30:2055–2064. doi: 10.1111/j.1530-0277.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- Hellemans KG, Sliwowska JH, Verma P, Weinberg J. Prenatal alcohol exposure: fetal programming and later life vulnerability to stress, depression and anxiety disorders. Neurosci Biobehav Rev. 2010;34(6):791–807. doi: 10.1016/j.neubiorev.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biol Reprod. 2009;81(4):607–17. doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- Johnson S, Knight R, Marmer DJ, Steele RW. Immune deficiency in fetal alcohol syndrome. Pediatr Research. 1981;15:908–911. doi: 10.1203/00006450-198106000-00005. [DOI] [PubMed] [Google Scholar]
- Knee DS, Sato AK, Uyehara CF, Claybaugh JR. Prenatal exposure to ethanol causes partial diabetes insipidus in adult rats. Am J Physiol Regul Integr Comp Physiol. 2004;287(2):R277–283. doi: 10.1152/ajpregu.00223.2003. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13(1):5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P, Sarkar DK. Ethanol induces apoptotic death of beta-endorphin neurons in the rat hypothalamus by a TGF-beta 1-dependent mechanism. Alcohol Clin Exp Res. 2008;32(4):706–14. doi: 10.1111/j.1530-0277.2008.00627.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Choi I, Rivier C. Role of various neurotransmitters in mediating the long-term endocrine consequences of prenatal alcohol exposure. Ann N Y Acad Sci. 2008;1144:176–188. doi: 10.1196/annals.1418.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, Sampson W, Hereld D. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34(4):575–87. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Starkman BG, Sakharkar A, Pandey SC. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010;67(1):73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison IM, Reeve AE. A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Human Molecular Genetics. 1998;7 (10):1599–1609. doi: 10.1093/hmg/7.10.1599. [DOI] [PubMed] [Google Scholar]
- Murugan S, Zhang C, Mojtahedzadeh S, Sarkar DK. Alcohol exposure in utero increases susceptibility to prostate tumorigenesis in rat offspring. Alcohol Clin Exp Res. 2013;37:1901–1909. doi: 10.1111/acer.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76(Pt B):259–68. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Price J. Proopiomelanocortin gene expression and DNA methylation: implications for Cushing’s syndrome and beyond. J Endocrinol. 2003;177(3):365–72. doi: 10.1677/joe.0.1770365. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- and down-regulation of genes by ethanol in hepatocytes. Life Sci. 2007;81(12):979–87. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Rajesh U, Zhang H, Tang L, Prakash A. Brain Chromatin Remodeling: A novel Mechanism of Alcoholism. J Neuroscience. 2008;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JA, Bloom SR. Hypothalamic neuropeptides and the regulation of appetite. Neuropharmacology. 2012;63(1):18–30. doi: 10.1016/j.neuropharm.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Parkington HC, Coleman HA, Wintour EM, Tare M. Prenatal alcohol exposure: implications for cardiovascular function in the fetus and beyond. Clinical Experimental Pharmacology & Physiology. 2010;37:e91–e98. doi: 10.1111/j.1440-1681.2009.05342.x. [DOI] [PubMed] [Google Scholar]
- Polanco TA, Crismale-Gann C, Reuhl KR, Sarkar DK, Cohick WS. Fetal alcohol exposure increases mammary tumor susceptibility and alters tumor phenotype in rats. Alcohol Clin Exp Res. 2010;34:1879–1887. doi: 10.1111/j.1530-0277.2010.01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neuroscience. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachdaoui N, Sarkar DK. Effects of alcohol on the endocrine system. Endocrinol Metab Clin North Am. 2013;42(3):593–615. doi: 10.1016/j.ecl.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–9. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Sarkar DK, Zhang C. Beta-endorphin neuron regulates stress response and innate immunity to prevent breast cancer growth and progression. Vitam Horm. 2013;93:263–76. doi: 10.1016/B978-0-12-416673-8.00011-3. [DOI] [PubMed] [Google Scholar]
- Sarkar DK, Kuhn P, Marano J, Chen C, Boyadjieva N. Alcohol exposure during the developmental period induces beta-endorphin neuronal death and causes alteration in the opioid control of stress axis function. Endocrinology. 2007;148(6):2828–2834. doi: 10.1210/en.2006-1606. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;6;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, Fukui K, Tomschik M, Ausio J, Zlatanova J. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48(46):10852–7. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- Ungerer M, Knezovich J, Ramsay M. In utero alcohol exposure, epigenetic changes, and their consequences. Alcohol Res. 2013;35(1):37–46. [PMC free article] [PubMed] [Google Scholar]
- Wamsteeker Cusulin JI, Füzesi T, Inoue W, Bains JS. Glucocorticoid feedback uncovers retrograde opioid signaling at hypothalamic synapses. Nat Neurosci. 2013;16(5):596–604. doi: 10.1038/nn.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw SL. Hypothalamic proopiomelanocortin processing and the regulation of energy balance. Eur J Pharmacol. 2011;660(1):213–9. doi: 10.1016/j.ejphar.2010.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;1;25(7):679–84. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia LP, Shen L, Kou H, Zhang BJ, Zhang L, Wu Y, Li XJ, Xiong J, Yu Y, Wang H. Prenatal ethanol exposure enhances the susceptibility to metabolic syndrome in offspring rats by HPA axis-associated neuroendocrine metabolic programming. Toxicol Lett. 2014 Apr 7;226(1):98–105. doi: 10.1016/j.toxlet.2014.01.023. [DOI] [PubMed] [Google Scholar]
- Zahr NM, Kaufman KL, Harper CG. Clinical and pathological features of alcohol-related brain damage. Nat Rev Neurol. 2011;7:284–94. doi: 10.1038/nrneurol.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan N, Bach P, Nordstokke D, Yu W, Ellis L, Meadows GG, Weinberg J. Prenatal alcohol exposure alters the course and severity of adjuvant-induced arthritis in female rats. Brain Behav Immun. 2012;26(3):439–450. doi: 10.1016/j.bbi.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Caiafa P, Van Holde K. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J. 2011;14(12):1697–704. doi: 10.1096/fj.99-0869rev. [DOI] [PubMed] [Google Scholar]