Abstract
Activation of Toll-like receptor (TLR) signaling which stimulates inflammatory and proliferative pathways is the key element in the pathogenesis of Mallory-Denk bodies (MDBs) in mice fed DDC. However, little is known as to how TLR signaling is regulated in MDB formation during chronic liver disease development. The first systematic study of TLR signaling pathway transcript regulation in human archived formalin-fixed, paraffin-embedded (FFPE) liver biopsies with MDB formation is presented here. When compared to the activation of Toll-like signaling in alcoholic hepatitis (AH) and Non alcoholic steatohepatitis (NASH) patients, striking similarities and obvious differences were observed. Similar TLRs (TLR3 and TLR4, etc.), TLR downstream adaptors (MyD88 and TRIF, etc.) and transcript factors (NFκB and IRF7, etc.) were all up regulated in the patients’ livers. MyD88, TLR3 and TLR4 were significantly induced in the livers of AH and NASH compared to normal subjects, while TRIF and IRF7 mRNA were only slightly up regulated in AH patients. This is a different pathway from the induction of the TLR4-MyD88-independent pathway in the AH and NASH patients with MDBs present. Importantly, chemokine receptor 4 and 7 (CXCR4/7) mRNAs were found to be induced in the patients livers in FAT10 positive hepatocytes. The CXCR7 pathway was significantly up regulated in patients with AH and the CXCR4 was markedly up regulated in patients with NASH, indicating that CXCR4/7 is crucial in liver MDB formation. This data constitutes the first demonstration of the up regulation of the MyD88-dependent TLR4/NFκB pathway in AH and NASH where MDBs formed, via the NFκB-CXCR4/7 pathway, and provides further insight into the mechanism of MDB formation in human liver diseases.
Keywords: Toll-like receptor (TLR) signaling, Mallory-Denk bodies (MDBs), chemokine receptor 4 and 7 (CXCR4/7), MyD88-dependent pathway, liver disease development
Introduction
Mallory-Denk Bodies (MDBs), which are an intracellular deposition of misfolded protein in ballooned hepatocytes, consist of abnormally phosphorylated, ubiquitylated, and cross-linked keratins or non-keratin K18 and 8, and p62 components (Zatloukal et al., 2007; Caldwell et al., 2010; Basaranoglu et al., 2011; Haybaeck et al., 2012). Ballooning of hepatocytes is induced by oxidative stress, and both ballooning of hepatocytes and MDBs are two characteristics of ongoing inflammation (Basaranoglu et al., 2011). MDBs are prevalent in various hepatic diseases including the hepatitis B and C virus (HBV and HCV), alcoholic hepatitis (AH), non-alcoholic steatohepatitis (NASH), drug injuries and hepatocellular carcinoma (HCC) (Zatloukal et al., 2007; Basaranoglu et al., 2011). Balloon hepatocytes forming MDBs are progenitor preneoplasia cells (French et al., 2011). In the DDC fed mouse model where liver cells proliferate, MDBs form and later, after DDC withdrawal, HCCs develop (Oliva et al., 2008). Three new mechanisms of MDB formation (epigenetic mechanisms, shift from the 26S proteasome to the immunoproteasome and activation of Toll-like signaling) have recently been identified (French et al., 2010). However, the detailed molecular events involved in liver MDB formation, especially in human liver disease development, remains undetermined.
Toll-like receptors (TLRs) are a family of pattern-recognition receptors (PRRs) that play a critical role in chronic liver disease by recognizing pathogen-associated molecular patterns (PAMPs) (Roh & Seki, 2013). By recognizing corresponding ligands (bacteria, viruses, fungi, parasites, etc.), the TLRs signaling pathways are orchestrated via myeloid differentiation factor 88 (MyD88)-dependent or the TRIF-dependent pathway which initiates innate immune responses and priming antigen-specific acquired immunity (Takeuchi & Akira, 2010). Except for TLR3, all TLRs activate the MyD88-dependent pathway, recruit the IRAK family of protein kinases, and lead to the activation of TRAF6 (Akira & Takeda, 2004). TRAF6 induces the activation of the IKK complex and leads to activation of NFκB, or activates AP-1 by TAK1 (Roh & Seki, 2013). In contrast, TLR3 and TLR4 can utilize the MyD88-independent, TRIF-dependent pathway. Subsequently, TRIF associates with TRAF 3 and TRAF 6 to activate the TBK1 and IKK complex, which results in the activation of interferon inducible genes and NFκB respectively (Takeuchi & Akira, 2010). These transcription factors induce the expression of various inflammatory cytokines (TNF-α and IL-6, etc.), interferons, and chemokines. TLRs play key roles in the pathogenesis of a variety of liver diseases (Seki & Brenner, 2008), including the formation of HCC when mice are fed ethanol and LSP (French et al., 2012; Machida et al., 2012; Chen et al., 2013). Activation of the resident liver cells, such as Kupffer cells, is one of the key elements in the pathogenesis of AH and NASH, producing inflammatory cytokines and chemokines to initiate the inflammatory cascade (Petrasek et al., 2013a). Kupffer cells express all TLRs with the exception of TLR5 at mRNA and protein levels while hepatic stellate cells also express all TLRs at transcriptional levels in quiescent and activated states (Wang et al., 2009). Hepatocytes express all TLRs at the transcriptional level, while the expression levels of TLR2, TLR3, TLR4 and TLR5 are very low in vivo (Isogawa et al., 2005) except in the Machida model (Machida et al., 2009; Machida et al., 2012). However, gene expression of Toll-signaling in ballooned hepatocytes (MDB-forming cells) is still unknown.
The Chemokine CXCL12 (SDF-1α) is a broadly expressed CX chemokine, and the predominant CXCL12 receptor is the CX chemokine receptor 4 (CXCR4), a protein frequently over-expressed on the surface of human tumor cells of epithelial origin (Kaifi et al., 2005; Schimanski et al., 2005). CXCL12 can also bind another chemokine receptor CXCR7, which is present on the surface of many different malignant cell types (Burns et al., 2006), and tumor-associated blood vessels (Miao et al., 2007). Recently FAT10, which is over-expressed in various cancers including liver tumors (Lee et al., 2003; Qing et al., 2011), was found to activate NFκB which in turn up regulates CXCR4/7 in NeHepLxHT and HCT116 cells (Gao et al., 2014). This raises an interest for us to explore the relationship of NFκB with CXCR4/7 induced by Toll-like signaling in liver patients with MDBs presents.
In this study, gene expression of TLRs signaling was investigated for the first time in the livers of AH and NASH patients’ biopsies. Striking similarities and obvious differences in the activation of Toll-like signaling in AH and NASH patients were studied. Interestingly, CXCR4 and CXCR7 were both significantly up regulated in the patients’ livers in FAT10 over-expressing hepatocytes.
Materials and Methods
Biopsies
Human archived formalin-fixed paraffin-embedded (FFPE) liver biopsies from patients who had alcoholic hepatitis (AH; n=3) and non-alcoholic steatohepatitis (NASH; n=3) were obtained from Harbor UCLA hospital archives. In all the cases liver forming MDBs were present except in the normal control livers (Control; n=3). The biopsy sections were cut 4 μm thick.
RNA isolation and cDNA synthesis
RNA isolation of FFPE sections of human liver biopsies was performed as we previously described (Liu et al., 2014). The quality and yield of the resulting total RNAs were assessed with an absorbance reading at 260 nm (A260) using a spectrophotometer (Thermo). Synthesis of first-strand cDNAs was performed with the above mentioned total RNA (250ng), and random hexamer primers using SuperSript III First-Strand Synthesis SuperMix following the instruction (Invitrogen).
Quantitative Real-time PCR analysis
Real-time PCR was performed using the Fast SYBR Green Master Mix on a StepOnePlus™ Real-time PCR System (Applied Biosystems) with a primer concentration of 200 nM. Primer sequences and the related gene Accession Number are listed in Table I. Reaction conditions consisted of 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec, 60 °C for 30 sec. Single PCR product was confirmed with the heat dissociation protocol at the end of the PCR cycles. Human α-tubulin was used as control to normalize the starting quantity of RNA. The target mRNA abundance in each sample was normalized to its endogenous control level and the relative mRNA expression levels were analyzed using the ΔΔCT method. Reaction of each sample was performed in triplicate.
Table I.
Sequences of the forward and reverse real-time PCR primers of human TLR signaling
| Gene name (Species) | Accession Number | Sequences of primer |
|---|---|---|
| TLR1 (Human) | NM_003263 | Forward Primer:: 5′-ATT CCG CAG TAC TCC ATT CC-3′ Reverse Primer: 5′-TTT GCT TGC TCT GTC AGC TT -3′ |
| TLR2 (Human) | NM_003264 | Forward Primer: 5′-ATG CCT ACT GGG TGG AGA AC -3′ Reverse Primer: 5′-TGC ACC ACT CAC TCT TCA CA -3′ |
| TLR3 (Human) | NM_003265 | Forward Primer: 5′-GAA AGG CTA GCA GTC ATC CA -3′ Reverse Primer: 5′-CAT CGG GTA CCT GAG TCA AC -3′ |
| TLR4 (Human) | NM_138554 | Forward Primer: 5′-CAG CTC TTG GTG GAA GTT GA -3′ Reverse Primer: 5′-GCA AGA AGC ATC AGG TGA AA -3′ |
| TLR5 (Human) | NM_003268 | Forward Primer: 5′-GGA ACC AGC TCC TAG CTC CT -3′ Reverse Primer: 5′-AAG AGG GAA ACC CCA GAG AA -3′ |
| TLR7 (Human) | NM_016562 | Forward Primer: 5′-TTG CAA AAC ACA ACT GCC TA -3′ Reverse Primer: 5′-AAA CCC CAT CTT TCC AAC TC -3′ |
| TLR8 (Human) | NM_138636 | Forward Primer: 5′-GGG CAT TGC ATT TAA GAG GT -3′ Reverse Primer: 5′-TCC GGA TAT GAC GTT GAA AA -3′ |
| TLR9 (Human) | NM_017442 | Forward Primer: 5′-CTG CGA GAG CTC AAC CTT AG -3′ Reverse Primer: 5′-CTC CAG CAG GAA GTC CAT AA -3′ |
| MyD88 (Human) | NM_002468 | Forward Primer: 5′-GCA CAT GGG CAC ATA CAG AC -3′ Reverse Primer: 5′-GAC ATG GTT AGG CTC CCT CA -3′ |
| TRIF (Human) | NM_182919 | Forward Primer: 5′-GGA ATC ATC ATC GGA ACA GA-3′ Reverse Primer: 5′-TCG AAG TTG GAG GTG AGA AG-3′ |
| IRAK1 (Human) | NM_001569 | Forward Primer: 5′-CCA AAC ATT GTG GAC TTT GC -3′ Reverse Primer: 5′-GGC TGT ACC CAG AAG GAT GT -3′ |
| IRAK4 (Human) | NM_001114182 | Forward Primer: 5′-CTG AGG CAG GAG ACT AGC TG -3′ Reverse Primer: 5′-AAT GTG GGC AAA ACC TGT AA -3′ |
| TAK1 (Human) | NM_003188 | Forward Primer: 5′-AGA GGA GCC TTT GGA GTT GT-3′ Reverse Primer: 5′-CCA TCA CAA GAC ACA CTG GA-3′ |
| TRAF3 (Human) | NM_145725 | Forward Primer: 5′-CAG AGG TTG TGC AGA GCA GT -3′ Reverse Primer: 5′-CCG GTA TTT ACA CGC CTT CT -3′ |
| TRAF6 (Human) | NM_145803 | Forward Primer: 5′-GGA AGA TTG GCA ACT TTG GA -3′ Reverse Primer: 5′-CGT GGT TTT GCC TTA CAG GT -3′ |
| IRF3 (Human) | NM_001571 | Forward Primer: 5′-TCG AGG TGA CAG CCT TCT AC -3′ Reverse Primer: 5′-GCC TCA CGT AGC TCA TCA CT -3′ |
| IRF7 (Human) | NM_001572 | Forward Primer: 5′-TAC CAT CTA CCT GGG CTT CG -3′ Reverse Primer: 5′-GCT CCA TAA GGA AGC ACT CG -3′ |
| CXCR4 (Human) | NM_001008540 | Forward Primer: 5′-GGT GGT CTA TGT TGG CGT CT -3′ Reverse Primer: 5′-TGG AGT GTG ACA GCT TGG AG -3′ |
| CXCR7 (Human) | NM_020311 | Forward Primer: 5′-TCT GGG ACG GGT TTA CTT GT -3′ Reverse Primer: 5′-CTG CCT GTT GCA AAA CTG TC -3′ |
| P65 (Human) | NM_021975 | Forward Primer: 5′-CTC TGC TTC CAG GTG ACA GT -3′ Reverse Primer: 5′-TCC TCT TTC TGC ACC TTG TC -3′ |
Immunohistochemical staining
Formalin fixed, paraffin embedded tissue slides were double stained for CXCR4 (Abcam Inc., Cambridge MA) and Ubiquitin (Millipore, Temecula CA). A second set of slides was double stained for CXCR7 (Millipore, Temecula CA) and Ubiquitin (Millipore, Temecula, CA. CXCR4 and CXCR7 were detected using the second antibody donkey anti rabbit Alexa Fluor 488 (Jackson Immuno Research Laboratories Inc. West Grove, PA). Ubiquitin was detected using the second antibody donkey anti mouse Alexa Fluor 594 (Jackson Labs. West Grove, PA). All slides were stained with the nuclear stain DAPI (Molecular Probes, Eugene, OR). The slides were examined with a Nikon 400 fluorescent microscope.
Statistical analysis
Statistical significance was determined using the t-test and One Way ANOVA test with SigmaStat software. P values less than 0.05 were considered statistically significant. Regression plots were constructed using the Sigmaplot software. All data were presented as the mean ± S.E.M and were representative of at least two-independent experiments done in triplicate.
Results
TLR3 and TLR4 are significantly up regulated both in the livers of AH and NASH patients
An increased mRNA expression of TLR2 and TLR4 were observed in mice re-fed DDC (Bardag-Gorce et al., 2010), implying the involvement of TLR signaling in liver MDB formation. To further examine how gene expression of Toll-like signaling was changed in human chronic liver diseases, where MDB formation progresses to tumor, TLRs were measured using real-time PCR analysis. Human liver biopsies, including normal liver tissue, AH and NASH biopsies were utilized in this assay. As expected, all TLRs mRNA tested were up regulated both in the livers of AH and NASH biopsies (Figure 1). Among them, approximately 3- and 7-fold (respectively, p<0.05) up regulation levels of TLR3 and TLR4 mRNAs were observed in the livers of AH biopsies when compare to other TLR mRNAs (Fig. 1A). In contrast to AH, TLR3 and TLR4 mRNAs were induced to 3- and 2.6-fold levels (respectively, p<0.05) in the livers of NASH patients (Fig. 1B). These results suggest that both TLR3 and TLR4 play an important role in the pathogenesis in AH and NASH patients. Other TLR mRNAs, including TLR2, TLR5 and TLR8, were also up regulated to various degrees in the livers of AH and NASH with MDBs present (Figure 1).
Figure 1.
Gene expression of TLRs signaling in the livers of AH and NASH biopsies. mRNA levels of different TLRs in the livers of AH (A) and NASH (B) biopsies are shown. Data represents mean values ±S.E.M. Statistical significance was determined using the t-test with SigmaStat software. * p<0.05, **p<0.01, and ***p<0.001 by t-test.
The MyD88-dependent pathways were markedly activated in the livers of both AH and NASH biopsies
Gene expression of TLRs signaling downstream molecules were further examined by real-time PCR analysis. Similar to some previous observations in mice (Bardag-Gorce et al., 2010), the mRNA levels of MyD88, TRIF, IRAK1, IRAK4, TAK1, TRAF3, TRAF6 and IRF7 were all up regulated to various degrees in the AH patients liver biopsies (Fig. 2A). The MyD88 mRNA in the AH biopsies was induced by up to a 15-fold expression, while IRF3 mRNA showed almost no changes. This might suggest that the MyD88-dependent pathway is the main pathway in AH patients with MDBs present. In contrast to AH, mRNAs of MyD88, IRF3 and IRF7 were all significantly up regulated in the livers of NASH biopsies (Fig. 2B), suggesting both MyD88-dependent and MyD88-independent pathways are up regulated in the NASH patients. These results clearly indicated that MyD88-dependent pathway is markedly up regulated both in the livers of AH and NASH patients with MDB formation.
Figure 2.
Induction of TLRs downstream components in the livers of AH (A) and NASH (B) biopsies. Quantification of mRNA was carried out by SYBR real-time PCR assays. * p<0.05 by t-test with SigmaStat software.
Liver MDB formation which may progress to HCC is involved in the NFκB-CXCR4/7 pathway in FAT10 over-expression hepatocytes
Recent research showed that FAT10 is markedly up regulated in the livers of AH, cirrhosis and NASH biopsies with MDBs present (Liu et al., 2014). FAT10 activates nuclear factor κB (NFκB), resulting in up regulation of the chemokine receptors CXCR4 and CXCR7 in NeHepLxHT cells (Gao et al., 2014). Therefore, the mRNA levels of NFκB p65, CXCR4 and CXCR7 were measured in the livers of AH and NASH biopsies by real-time PCR. As expected, the up regulation of mRNA expression of p65, CXCR4 and CXCR7 were observed in the patients’ livers (Figure 3). Interestingly, the CXCR7 pathway was significantly up regulated in patients with AH and the CXCR4 and p65 expression were significantly up regulated in patients with NASH (Figure 3), indicating that CXCR4/7 plays a role in liver MDB formation. Immunohistochemical staining showed that MDB forming balloon hepatocytes in AH liver biopsies were expressing CXCR7 in the liver cytoplasm and co-localizing with ubiquitin in MDBs (Figure 4). These results indicate that the NFκB-CXCR4/7 pathway is involved in liver MDB formation.
Figure 3.
Gene transcript expression of human p65, CXCR4 and CXCR7 in patient livers from human archived biopsies with AH, NASH and normal liver tissue. Data represent mean values ±S.E.M. * p<0.05 by t-test with SigmaStat software.
Fig. 4.
IHC double stain of Alcoholic Hepatitis biopsy compared to Control Liver showing positive staining of CXCR7 (A-FITC) including MDB positive Balloon hepatocytes (Texas Red and Tricolor) compared to Control x 606
Discussion
Chronic liver disease such as alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH) represents a significant cause of morbidity and mortality worldwide (Burroughs & McNamara, 2003; Minino et al., 2007). Recognition of Toll-like receptors (TLR) signaling, which produce inflammatory cytokines and chemokines to initiate the inflammatory cascade, is the key component involved in activation of innate immunity in AH and NASH (Petrasek et al., 2013a). The TLR signaling pathways are up regulated in various chronic liver diseases. Different cell types in the liver express different TLRs (Seki et al., 2011). TLR2 and TLR4 mRNA are also up regulated in the livers of DDC re-fed mice (Bardag-Gorce et al., 2010) and in primary hepatocyte cultures where MDBs form (Yuan et al., 2000; Nan et al., 2005; Nan et al., 2006). TLRs signaling are up regulated in rats fed ethanol intragastrically, especially when the blood alcoholic levels are high (Khachatoorian et al., 2013). However, information about the pathologic significance of TLRs signaling in AH and NASH patients with MDBs present is still unknown.
To provide clinical evidence of gene expression of the TLRs signaling in the livers of AH and NASH patients with MDB-forming hepatocytes, biopsies from these patients were examined by real-time PCR analysis. An interesting observation is that the mRNA levels of TLR3 and TLR4 were much higher than other TLRs both in the livers of AH and NASH biopsies. Why TLR3 and TLR4 are significantly up regulated in the AH and NASH patients and what are the biological functions or mechanism of up regulation of these two TLRs in liver MDB formation? A possible reason is that TLR3 and TLR4 recognize different ligands where TLR3 recognizes viral double-strand RNA and poly (I: C) while TLR4 recognizes the endotoxin from Gram-negative bacteria, which can both induce an inflammatory response and antiviral immune response (Zhao et al., 2008; Ganz & Szabo, 2013). Ample epidemiological evidence suggests that there is a strong connection between hepatitis C virus (HCV) and AH, where the prevalence of HCV is significantly higher among alcoholics than in the normal population (Heintges & Wands, 1997). Second, alcohol may enhance the replication of HCV and thus increase the expression of viral RNA and proteins, resulting in more severe HCV-induced liver injury (Brechot et al., 1996). So the increase of viral RNA and protein levels might induce the up regulation expression of TLR3 mRNA to initiate the antiviral immune response. Additionally, we noticed that TLR7 and TLR8 whose ligands are viral single-stranded RNA, were also slightly up regulated in AH biopsies.
The activation of both MyD88-dependent and MyD88-independent pathways were reported in mouse models of AH and NASH (Hritz et al., 2008; Csak et al., 2011a). The downstream molecules of TLRs in AH and NASH were further investigated by real-time PCR analysis. A significantly up regulated expression of MyD88 mRNA (15 fold, p<0.05) was observed in the livers of AH patients, indicating that the MyD88-dependent pathway was activated. To understand liver tumorigenesis, it is very important to analyze which cell types are involved in the process. Kupffer cells may be the major cells expressing TLRs in the liver (Roh & Seki, 2013). A recent study showed that TLR4 signaling in AH is mediated via the TRIF/IRF3-dependent, MyD88-independent pathway using the Lieber-DeCarli model of AH (Petrasek et al., 2013b). However, we did not observe the obvious changes of IRF3 and IRF7 mRNA, nor TRIF mRNA in the livers of AH biopsies. Our observation suggests that the MyD88-dependent pathway is the main pathway in AH patients with MDBs present, different from the observation in Kupffer cells. The possible cause may be the differences of cell types involved, since all cases studied by immunohistochemistry showed an increased expression of TLR4 in the cytoplasm of the liver cells (Lee et al., 2014), including the balloon cells forming MDBs, and including incorporation of TLR4 within the MDB. We also noticed that the mRNA level of TLR4 was higher than TLR3 in the livers of AH patients, probably suggesting that inflammatory response is the main pathway involved compared to antivirus immune response in the hepatocytes.
It was observed that MyD88, IRF3 and IRF7 mRNAs were all significantly up regulated in the livers of NASH biopsies, which was different from AH. Also it was noted that the mRNA level of TLR3 is slightly higher than TLR4 in the livers of NASH patients, indicating both TLR4 MyD88-dependent and TLR3 MyD88-independent pathways were activated in the pathogenesis of NASH development in MDB formation. The differential induction of MyD88-dependent and MyD88-independent pathway in AH and NASH patients may relate to the differences in dynamics between these two disease (Petrasek et al., 2013a). This is because excessive consumption of alcohol may induce liver inflammation in a shorter period of time than the consumption of a steatogenic diet, which results in the development of NASH (Csak et al., 2011a; Csak et al., 2011b). Secondly, the development of NASH involves insulin resistance and an endocrine crosstalk between adipose tissue and the liver, which inhibits the TLR4/MyD88-dependent pathway in macrophages (Mandal et al., 2010). However, it is still uncertain as to how adiponectin, an anti-inflammatory cytokine secreted by adipose tissue, contributes to the MyD88-dependent pathways of NASH in MDB-forming hepatocytes. Strangely, we did not observe the significant up regulation of TRIF mRNA in the livers of AH and NASH patients, although there was a marked increase of IRF3 and IRF7 mRNAs in the livers of NASH biopsies.
After activation by ligands, TLRs triggers a cascade of downstream adaptor signaling and ultimately activate the transcription factors NFκB and chemokines. The chemokine CXCL12 appears to be of particular importance in tumor biology, and interaction of CXCL12 with its receptor CXCR4 and CXCR7 is found to be highly expressed in tumor and tumor-endothelial cells (Miao et al., 2007; Burger & Peled, 2009; Hattermann et al., 2010). The CXCL12/CXCR4 and CXCL12/CXCR7 axis are important factors in tumor initiation, promotion, progression, metastasis and cancer cell survival (Vandercappellen et al., 2008; Marechal et al., 2009). However, the biological significance of CXCR4 and CXCR7 in liver MDB-forming cells is still unknown. Therefore, the transcript regulation of CXCR4 and CXCR7, with NFκB p65 were investigated by real-time PCR analysis in the livers of AH and NASH biopsies. In our series, CXCR4 mRNA was significantly up regulated in patients with NASH, while CXCR7 mRNA was significantly up regulated in the patients with AH, suggesting a different role played by these two receptors in NASH and AH. The reasons for the differences between CXCR4 and CXCR7 mRNA levels in AH and NASH patients probably result from CXCR4 and CXCR7 having different CXCL12 binding domains and distinct roles during disease development (Sierro et al., 2007), although both of them can bind to CXCL12. Another reason may be because unlike CXCR4, CXCL12 activation of CXCR7 does not induce calcium mobilization and cell migration but rather results in a proliferative effect and increased adhesion properties (Burns et al., 2006; Wang et al., 2008). Although we observed the differential expression of CXCR4 and CXCR7 in the livers of AH and NASH patients, whether they are involved in MDB-forming hepatocytes needs to be determined.
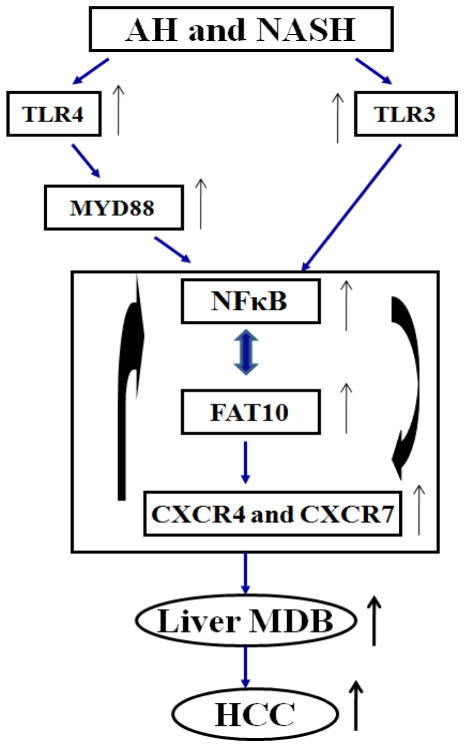
Recently, we observed the marked up regulation expression of FAT10 in the livers of AH and NASH patients (Liu et al., 2014). FAT10 expression is induced by interferon (IFN)-γ and tumor necrosis factor α (TNFα) in tumor cells (Lukasiak et al., 2008; Ren et al., 2011) and activates NFκB, which in turn un regulates CXCR4/7 in NeHepLxHT and HCT116 cells (Gao et al., 2014). Interestingly, the NFκB binding site was found at the FAT10 promoter region (Zhang et al., 2006), while TNFα-dependent induction of FAT10 expression requires NFκB activation (Ren et al., 2011). The interferon sequence response element (ISRE) located on the FAT10 promoter region activates NFκB in response to IFNγ (Oliva et al., 2010). These findings indicate a potential feedback system in chronic inflammatory-associated microenvironments such as in AH and NASH. The cytokine TNF-α or chemokines induced by the activation of TLRs signaling induce FAT10, which activates NFκB. This in turn promotes the FAT10 transcription by binding to FAT10 promoter sites (Oliva et al., 2010). NFκB has recently been found to bind a transcript factor for both CXCR4 and CXCR7 receptors (Tarnowski et al., 2010), and CXCR4 and CXCR7 can also activate NFκB (Huang et al., 2009), further suggesting CXCR4 and CXCR7 may sustain NFκB activity by a feedback system. We confirmed that the CXCR4 and CXCR7 mRNAs were up regulated in MDB-forming (FAT10-over-expressing) hepatocytes within this NFκB network. Based on these findings, transcription up regulation of TLR signaling and CXCR4/7 which results in liver MDB formation is shown in Figure 5. The gene expression changes induced by TLR signaling, inducing the up regulation of FAT10 and CXCR4/7, may be a novel mechanism in liver MDB formation. The further elucidation of the relationship of FAT10 and CXCR4/7 in the inflammatory pathway including TLR pathway will provide a better understanding of chronic liver disease pathogenesis and MDB formation.
Figure 5.
Proposed model of gene expression up regulation of TLR signaling and CXCR4/7 in human liver MDB formation. Alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH) transcript up regulation of TLR signaling activation and CXCR4/7 are presented.
In summary, our data demonstrates for the first time the gene expression changes of TLR signaling in the livers of AH and NASH patients where MDBs formed. Striking similarities and obvious differences were observed in the TLR signaling activation, where the MyD88-dependent pathway was significantly up regulated in patients with AH, while both MyD88-dependent and MyD88-independent pathways were up regulated in patients with NASH. The prominent transcription up regulation expression of CXCR4 and CXCR7 were observed in FAT10 over expressing hepatocytes, indicating that CXCR4 and CXCR7 may be attractive targets for AH and NASH therapy. The data provide evidence to further understand human liver disease and MDB formation.
Acknowledgments
This work was supported by grants from NIH (AAU01021898-02) and P50-11999 Morphology Core. Some results were submitted in a Poster Abstract (2027981) in AASLD LIVER MEETING Nov 2014, BOSTON
Abbreviations
- AH
alcoholic hepatitis
- CXCR4/7
Human chemokine (C-X-C motif) receptor 4/7
- DDC
diethyl 1, 4-dehydro-2, 4, 6-trimethyl-3, 5-pyridine-dicarboxylate
- FFPE
formalin-fixed, paraffin-embedded
- HCC
hepatocellular carcinoma
- MDB
Mallory-Denk body
- MyD88
Myeloid differentiation primary response gene 88
- NASH
non-alcoholic steatohepatitis
- TLR
Toll-like receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Lin A, Li J, French BA, French SW. SAMe prevents the up regulation of toll-like receptor signaling in Mallory-Denk body forming hepatocytes. Exp Mol Pathol. 2010;88:376–379. doi: 10.1016/j.yexmp.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaranoglu M, Turhan N, Sonsuz A, Basaranoglu G. Mallory-Denk Bodies in chronic hepatitis. World J Gastroenterol. 2011;17:2172–2177. doi: 10.3748/wjg.v17.i17.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechot C, Nalpas B, Feitelson MA. Interactions between alcohol and hepatitis viruses in the liver. Clin Lab Med. 1996;16:273–287. [PubMed] [Google Scholar]
- Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23:43–52. doi: 10.1038/leu.2008.299. [DOI] [PubMed] [Google Scholar]
- Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ, Wei K, McMaster BE, Wright K, Howard MC, Schall TJ. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs A, McNamara D. Liver disease in Europe. Aliment Pharmacol Ther. 2003;18(Suppl 3):54–59. doi: 10.1046/j.0953-0673.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- Caldwell S, Ikura Y, Dias D, Isomoto K, Yabu A, Moskaluk C, Pramoonjago P, Simmons W, Scruggs H, Rosenbaum N, Wilkinson T, Toms P, Argo CK, Al-Osaimi AM, Redick JA. Hepatocellular ballooning in NASH. J Hepatol. 2010;53:719–723. doi: 10.1016/j.jhep.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Tsukamoto H, Liu JC, Kashiwabara C, Feldman D, Sher L, Dooley S, French SW, Mishra L, Petrovic L, Jeong JH, Machida K. Reciprocal regulation by TLR4 and TGF-beta in tumor-initiating stem-like cells. J Clin Invest. 2013;123:2832–2849. doi: 10.1172/JCI65859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Csak T, Dolganiuc A, Kodys K, Nath B, Petrasek J, Bala S, Lippai D, Szabo G. Mitochondrial antiviral signaling protein defect links impaired antiviral response and liver injury in steatohepatitis in mice. Hepatology. 2011a;53:1917–1931. doi: 10.1002/hep.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology. 2011b;54:133–144. doi: 10.1002/hep.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW, Bardag-Gorce F, French BA, Li J, Oliva J. The role of innate immunity in the pathogenesis of preneoplasia in drug-induced chronic hepatitis based on a mouse model. Exp Mol Pathol. 2011;91:653–659. doi: 10.1016/j.yexmp.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW, Bardag-Gorce F, Li J, French BA, Oliva J. Mallory-Denk body pathogenesis revisited. World J Hepatol. 2010;2:295–301. doi: 10.4254/wjh.v2.i8.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW, Lee J, Zhong J, Morgan TR, Buslon V, Lungo W, French BA. Alcoholic liver disease - Hepatocellular carcinoma transformation. J Gastrointest Oncol. 2012;3:174–181. doi: 10.3978/j.issn.2078-6891.2012.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz M, Szabo G. Immune and inflammatory pathways in NASH. Hepatol Int. 2013;7:771–781. doi: 10.1007/s12072-013-9468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Theng SS, Zhuo J, Teo WB, Ren J, Lee CG. FAT10, an ubiquitin-like protein, confers malignant properties in non-tumorigenic and tumorigenic cells. Carcinogenesis. 2014;35:923–934. doi: 10.1093/carcin/bgt407. [DOI] [PubMed] [Google Scholar]
- Hattermann K, Held-Feindt J, Lucius R, Muerkoster SS, Penfold ME, Schall TJ, Mentlein R. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010;70:3299–3308. doi: 10.1158/0008-5472.CAN-09-3642. [DOI] [PubMed] [Google Scholar]
- Haybaeck J, Stumptner C, Thueringer A, Kolbe T, Magin TM, Hesse M, Fickert P, Tsybrovskyy O, Muller H, Trauner M, Zatloukal K, Denk H. Genetic background effects of keratin 8 and 18 in a DDC-induced hepatotoxicity and Mallory-Denk body formation mouse model. Lab Invest. 2012;92:857–867. doi: 10.1038/labinvest.2012.49. [DOI] [PubMed] [Google Scholar]
- Heintges T, Wands JR. Hepatitis C virus: epidemiology and transmission. Hepatology. 1997;26:521–526. doi: 10.1002/hep.510260338. [DOI] [PubMed] [Google Scholar]
- Hritz I, Mandrekar P, Velayudham A, Catalano D, Dolganiuc A, Kodys K, Kurt-Jones E, Szabo G. The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology. 2008;48:1224–1231. doi: 10.1002/hep.22470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Lee CY, Chen MY, Yang WH, Chen YH, Chang CH, Hsu HC, Fong YC, Tang CH. Stromal cell-derived factor-1/CXCR4 enhanced motility of human osteosarcoma cells involves MEK1/2, ERK and NF-kappaB-dependent pathways. J Cell Physiol. 2009;221:204–212. doi: 10.1002/jcp.21846. [DOI] [PubMed] [Google Scholar]
- Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol. 2005;79:7269–7272. doi: 10.1128/JVI.79.11.7269-7272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifi JT, Yekebas EF, Schurr P, Obonyo D, Wachowiak R, Busch P, Heinecke A, Pantel K, Izbicki JR. Tumor-cell homing to lymph nodes and bone marrow and CXCR4 expression in esophageal cancer. J Natl Cancer Inst. 2005;97:1840–1847. doi: 10.1093/jnci/dji431. [DOI] [PubMed] [Google Scholar]
- Khachatoorian R, Dawson D, Maloney EM, Wang J, French BA, French SW. SAMe treatment prevents the ethanol-induced epigenetic alterations of genes in the Toll-like receptor pathway. Exp Mol Pathol. 2013;94:243–246. doi: 10.1016/j.yexmp.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL, Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, Choti M, Lee LA. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22:2592–2603. doi: 10.1038/sj.onc.1206337. [DOI] [PubMed] [Google Scholar]
- Lee J, French B, Morgan T, French SW. The liver is populated by a broad spectrum of markers for macrophages. In alcoholic hepatitis the macrophages are M1 and M2. Exp Mol Pathol. 2014;96:118–125. doi: 10.1016/j.yexmp.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li J, Tillman B, French BA, French SW. Ufmylation and FATylation pathways are downregulated in human alcoholic and nonalcoholic steatohepatitis, and mice fed DDC, where Mallory-Denk bodies (MDBs) form. Exp Mol Pathol. 2014;97:81–88. doi: 10.1016/j.yexmp.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasiak S, Schiller C, Oehlschlaeger P, Schmidtke G, Krause P, Legler DF, Autschbach F, Schirmacher P, Breuhahn K, Groettrup M. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27:6068–6074. doi: 10.1038/onc.2008.201. [DOI] [PubMed] [Google Scholar]
- Machida K, Chen CL, Liu JC, Kashiwabara C, Feldman D, French SW, Sher L, Hyeongnam JJ, Tsukamoto H. Cancer stem cells generated by alcohol, diabetes, and hepatitis C virus. J Gastroenterol Hepatol. 2012;27(Suppl 2):19–22. doi: 10.1111/j.1440-1746.2011.07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, Liu HM, Asahina K, Govindarajan S, Ray R, Ou JH, Seki E, Deshaies R, Miyake K, Lai MM. Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci U S A. 2009;106:1548–1553. doi: 10.1073/pnas.0807390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P, Roychowdhury S, Park PH, Pratt BT, Roger T, Nagy LE. Adiponectin and heme oxygenase-1 suppress TLR4/MyD88-independent signaling in rat Kupffer cells and in mice after chronic ethanol exposure. J Immunol. 2010;185:4928–4937. doi: 10.4049/jimmunol.1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal R, Demetter P, Nagy N, Berton A, Decaestecker C, Polus M, Closset J, Deviere J, Salmon I, Van Laethem JL. High expression of CXCR4 may predict poor survival in resected pancreatic adenocarcinoma. Br J Cancer. 2009;100:1444–1451. doi: 10.1038/sj.bjc.6605020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Luker KE, Summers BC, Berahovich R, Bhojani MS, Rehemtulla A, Kleer CG, Essner JJ, Nasevicius A, Luker GD, Howard MC, Schall TJ. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minino AM, Heron MP, Murphy SL, Kochanek KD. Deaths: final data for 2004. Natl Vital Stat Rep. 2007;55:1–119. [PubMed] [Google Scholar]
- Nan L, Dedes J, French BA, Bardag-Gorce F, Li J, Wu Y, French SW. Mallory body (cytokeratin aggresomes) formation is prevented in vitro by p38 inhibitor. Exp Mol Pathol. 2006;80:228–240. doi: 10.1016/j.yexmp.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Nan L, Wu Y, Bardag-Gorce F, Li J, French BA, Wilson LT, French SW. The p105/50 NF-kappaB pathway is essential for Mallory body formation. Exp Mol Pathol. 2005;78:198–206. doi: 10.1016/j.yexmp.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, McPhaul L, Amidi F, Dedes J, Habibi A, Nguyen S, French SW. Fat10 is an epigenetic marker for liver preneoplasia in a drug-primed mouse model of tumorigenesis. Exp Mol Pathol. 2008;84:102–112. doi: 10.1016/j.yexmp.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Lin A, French BA, French SW. The role of cytokines in UbD promoter regulation and Mallory-Denk body-like aggresomes. Exp Mol Pathol. 2010;89:1–8. doi: 10.1016/j.yexmp.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Csak T, Ganz M, Szabo G. Differences in innate immune signaling between alcoholic and non-alcoholic steatohepatitis. J Gastroenterol Hepatol. 2013a;28(Suppl 1):93–98. doi: 10.1111/jgh.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt-Jones EA, Fitzgerald KA, Szabo G. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci U S A. 2013b;110:16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing X, French BA, Oliva J, French SW. Increased expression of FAT10 in colon benign, premalignant and malignant epithelial neoplasms. Exp Mol Pathol. 2011;90:51–54. doi: 10.1016/j.yexmp.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wang Y, Gao Y, Mehta SB, Lee CG. FAT10 mediates the effect of TNF-alpha in inducing chromosomal instability. J Cell Sci. 2011;124:3665–3675. doi: 10.1242/jcs.087403. [DOI] [PubMed] [Google Scholar]
- Roh YS, Seki E. Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. J Gastroenterol Hepatol. 2013;28(Suppl 1):38–42. doi: 10.1111/jgh.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski CC, Schwald S, Simiantonaki N, Jayasinghe C, Gonner U, Wilsberg V, Junginger T, Berger MR, Galle PR, Moehler M. Effect of chemokine receptors CXCR4 and CCR7 on the metastatic behavior of human colorectal cancer. Clin Cancer Res. 2005;11:1743–1750. doi: 10.1158/1078-0432.CCR-04-1195. [DOI] [PubMed] [Google Scholar]
- Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48:322–335. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- Seki E, Park E, Fujimoto J. Toll-like receptor signaling in liver regeneration, fibrosis and carcinogenesis. Hepatol Res. 2011;41:597–610. doi: 10.1111/j.1872-034X.2011.00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro F, Biben C, Martinez-Munoz L, Mellado M, Ransohoff RM, Li M, Woehl B, Leung H, Groom J, Batten M, Harvey RP, Martinez AC, Mackay CR, Mackay F. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tarnowski M, Grymula K, Reca R, Jankowski K, Maksym R, Tarnowska J, Przybylski G, Barr FG, Kucia M, Ratajczak MZ. Regulation of expression of stromal-derived factor-1 receptors: CXCR4 and CXCR7 in human rhabdomyosarcomas. Mol Cancer Res. 2010;8:1–14. doi: 10.1158/1541-7786.MCR-09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–244. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- Wang B, Trippler M, Pei R, Lu M, Broering R, Gerken G, Schlaak JF. Toll-like receptor activated human and murine hepatic stellate cells are potent regulators of hepatitis C virus replication. J Hepatol. 2009;51:1037–1045. doi: 10.1016/j.jhep.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Wang J, Shiozawa Y, Wang Y, Jung Y, Pienta KJ, Mehra R, Loberg R, Taichman RS. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- Yuan QX, Nagao Y, French BA, Wan YJ, French SW. Dexamethasone enhances mallory body formation in drug-primed mouse liver. Exp Mol Pathol. 2000;69:202–210. doi: 10.1006/exmp.2000.2320. [DOI] [PubMed] [Google Scholar]
- Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Zhang DW, Jeang KT, Lee CG. p53 negatively regulates the expression of FAT10, a gene upregulated in various cancers. Oncogene. 2006;25:2318–2327. doi: 10.1038/sj.onc.1209220. [DOI] [PubMed] [Google Scholar]
- Zhao XJ, Dong Q, Bindas J, Piganelli JD, Magill A, Reiser J, Kolls JK. TRIF and IRF-3 binding to the TNF promoter results in macrophage TNF dysregulation and steatosis induced by chronic ethanol. J Immunol. 2008;181:3049–3056. doi: 10.4049/jimmunol.181.5.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]