SUMMARY
Diet composition is a critical determinant of lifespan and nutrient imbalance is detrimental health. However, how nutrients interact with genetic factors to modulate lifespan remains elusive. We investigated how diet composition influences mitochondrial ATP synthase subunit d (ATPsyn-d) in modulating lifespan in Drosophila. ATPsyn-d knockdown extended lifespan in females fed low carbohydrate-to-protein (C:P) diets, but not the high C:P ratio diet. This extension was associated with increased resistance to oxidative stress, transcriptional changes in metabolism, proteostasis and immune genes, reduced protein damage and aggregation, and reduced phosphorylation of S6K and ERK in TOR and MAPK signaling, respectively. ATPsyn-d knockdown did not extend lifespan in females with reduced TOR signaling induced genetically by Tsc2 overexpression or pharmacologically by rapamycin. Our data reveal a link among diet, mitochondria, MAPK and TOR signaling in aging and stresses the importance of considering genetic background and diet composition in implementing interventions for promoting healthy aging.
Keywords: aging, diet composition, dietary restriction, electron transfer chain, oxidative stress, rapamycin, Target of rapamycin
INTRODUCTION
Dietary nutrients are among the most critical environmental factors that modulate healthspan and lifespan (Fontana et al., 2010). Nutrient imbalance is a major risk factor to human health and common among old people (Piper et al., 2011). Dietary restriction (DR), by reducing the amount of all or specific nutrients, is a potent non-genetic intervention that promotes longevity in many species (Fontana et al., 2010). In general, protein restriction is more effective in influencing lifespan than sugar or calorie restriction in Drosophila (Tatar, 2007). However, increasing evidence indicates that the composition of dietary nutrients, such as carbohydrate-to-protein (C:P) ratio, is more critical than individual nutrients in affecting health and lifespan (Lee et al., 2008; Piper et al., 2011; Skorupa et al., 2008). Optimal lifespan peaks at the C:P ratio 16:1 in Drosophila and 9:1 in Mexican fruit fly (Carey et al., 2008; Lee et al., 2008). A recent study in mice shows that lifespan is primarily regulated by the C:P ratio in the diet and tends to be longer with higher C:P ratios (Solon-Biet et al., 2014). Diet composition is also critical for DR to promote longevity in nonhuman primate rhesus monkeys (Colman et al., 2009; Mattison et al., 2012). Two major nutrient sensing pathways are known to modulate lifespan. One is target-of-rapamycin (TOR) signaling that mostly senses cellular amino acid content and the other is insulin/insulin-like signaling (IIS) that primarily responds to circulating glucose and energy levels (Fontana et al., 2010). Excessive carbohydrate and protein intake both contribute to development of insulin resistance and diabetes in animal models and humans (Ahima, 2009; Zoncu et al., 2011). Dietary macronutrients, such as sugar, protein and fat, may interact with each other to influence nutrient sensing pathways and consequently health outcome. It is, thus, critical to take into account diet composition in elucidating molecular mechanisms of aging and in developing effective interventions for promoting healthy aging.
Aging is associated with transcriptional and translational changes in many genes and proteins (Lee et al., 2000; Pletcher et al., 2002; Zahn and Kim, 2007; Zou et al., 2000). Some age-related changes are evolutionarily conserved and many function in nutrient metabolism, such as mitochondrial electron transfer chain (ETC) genes, many of which are down-regulated with age in worms, flies, rodents and humans (McCarroll et al., 2004; Zahn and Kim, 2007). Knocking-down ETC genes affects lifespan in yeast, worms and flies (Copeland et al., 2009; Kaeberlein et al., 2005; Lee et al., 2003; Zid et al., 2009). Mitochondrial genes also play a key role in numerous age-related diseases, such as Parkinson’s and Alzheimer’s disease (Fontana et al., 2010). However, how mitochondrial genes interact with nutrients to modulate lifespan and healthspan remains incompletely elucidated. Understanding gene-environment interactions will be a key to tackle aging and age-related diseases,
ATP synthase subunit d (ATPsyn-d) is a component of ATP synthase, ETC complex V (Wallace, 2005) and is known to modulate lifespan in C. elegans (Hansen et al., 2005). How ATPsyn-d modulates lifespan and whether it functions in modulating lifespan in other species remain to be determined. Given the importance of nutrients as environmental factors in modulating lifespan, here we have investigated whether and how ATPsyn-d interacts with dietary macronutrients to modulate lifespan in Drosophila. We have found that ATPsyn-d interacts with dietary macronutrients to influence accumulation of oxidative damage and protein aggregates, resistance to oxidative stress and expression of numerous genes involved in metabolism, proteolysis and innate immune response, and more importantly to modulate lifespan. Moreover, ATPsyn-d affects MAP kinase (MAPK) signaling, and genetically interacts with TOR signaling to influence lifespan of flies in a diet composition dependent manner. Our study reveals the critical interaction between mitochondrial genes and nutritional factors and the underlying mechanisms involving TOR signaling in modulating lifespan.
Results
ATPsyn-d modulates lifespan
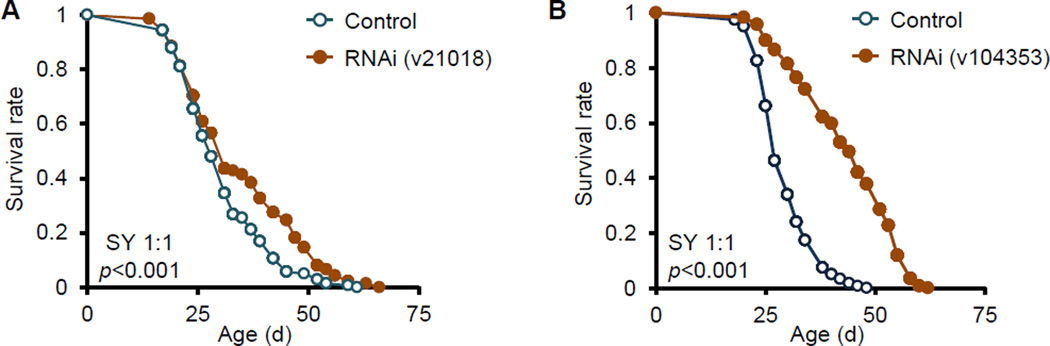
To determine the role of ATPsyn-d in modulating in lifespan, we first tested the effect of broad ATPsyn-d knockdown on lifespan in Drosophila by using a ubiquitously expressed Gal4 driver, daughterless-Gal4 (da-Gal4), and a UAS-ATPsyn-d-RNA interference (RNAi) line v21018. This knockdown resulted in lethality in flies before pupation, indicating that ATPsyn-d is essential for normal development. To bypass the lethality, we used the drug RU486 inducible gene switch Gal4 (GSG) system to knock down ATPsyn-d in adult flies (McGuire et al., 2004). This system also minimizes complications of genetic background, since all flies in principle have same genetic background and differ only in their exposure to RU486. Two different ubiquitously expressed GSG stocks, actin-GSG and daughterless-GSG (da-GSG), and two different UAS-ATPsyn-d-RNAi stocks, v21018 and v104353, were used to knockdown ATPsyn-d to further minimize complications of strain background. ATPsyn-d knockdown by using actin-GSG and UAS-ATPsyn-d-RNAi(v21018) extended both mean and maximal lifespan in females but not males fed SY1:1 with an equal amount of sugar (S) and yeast (Y) (SY1:1) (Figure 1A, Figure S1B and Table S1). The extent of ATPsyn-d knockdown at the transcript level induced by RU486 was similar in females and males on SY1:1 (Figure S1E and F), suggesting that the gender difference in lifespan may not be due to the difference in ATPsyn-d knockdown, but may be due to gender-specific physiology and response to nutrients (Tower, 2006). Lifespan extension for female flies fed SY1:1 was also observed using da-GSG and another UAS-ATPsyn-d-RNAi stock (v104353) to knock down ATPsyn-d (Figure 1B and Table S1). RU486 treatment alone did not increase the lifespan of male and female flies carrying both GSG driver and UAS-GFP-RNAi under SY1:1 (Figure S2 and Table S2). ATPsyn-d knockdown did not alter daily food intake in two RNAi strains fed SY1:1 (Figure S3A and B) and RU486 by itself did not significantly affect food intake in the UAS control flies (UAS-ATPsyn-d-RNAi(v104353)/+) (Figure S3B), suggesting that lifespan extension by ATPsyn-d knockdown is not due to any difference in food intake. ATPsyn-d knockdown did not substantially affect locomotor activity of flies, either, indicating that lifespan extension induced by ATPsyn-d knockdown is not necessarily associated with any improvement on motility, a healthspan-related parameter (Figure S3C).
Figure 1. The effect of ATPsyn-d knockdown on lifespan.
(A) Lifespan of actin-GSG>UAS-ATPsyn-d-RNAi(v21018) female flies on SY1:1. (B) Lifespan of da-GSG>UAS-ATPsyn-d-RNAi(v104353) females on SY1:1. “RNAi” indicates ATPsyn-d knockdown by 200 µM RU486. The UAS strain identification is shown in parentheses. “Control” represents genotype-matched flies without RU486 treatment. The p values were from logrank analysis between RNAi and control flies.
We next examined if ATPsyn-d overexpression with a UAS-ATPsyn-d line has any impact on lifespan. Feeding actin-GSG>UAS-ATPsyn-d overexpression flies with RU486 resulted in an increase of ATPsyn-d transcript level (Figure S1M and Q), but did not substantially change lifespan in both females and males fed SY1:1 relative to SY diet- and gender-matched no-RU486 controls (Figure S1N and R and Table S3). This suggests that ATPsyn-d overexpression does not have any significant impact on lifespan.
Lifespan extension by ATPsyn-d knockdown depends on diet composition
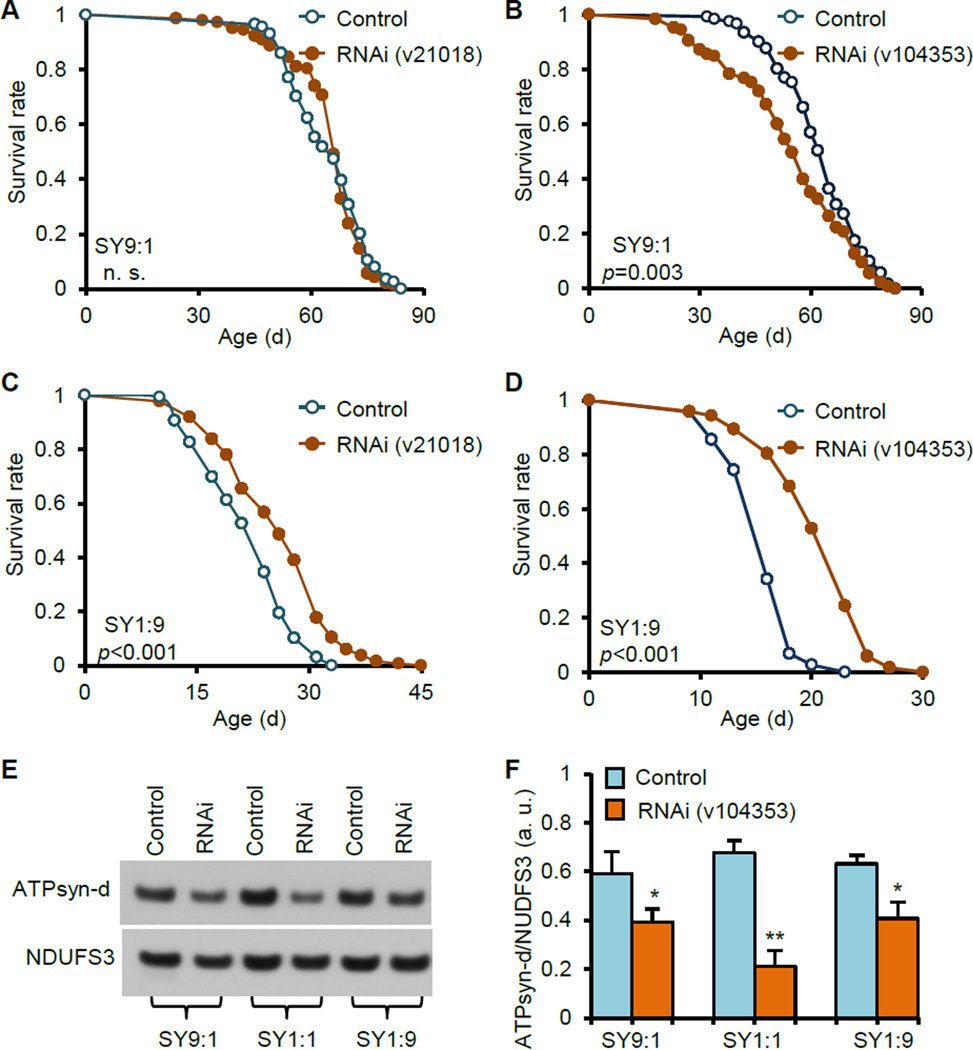
The cornmeal diet is another standard diet routinely used in fly culture and lifespan assay. Surprisingly, ATPsyn-d knockdown using actin-GSG>UAS-ATPsyn-d-RNAi(v21018) did not extend lifespan in females fed cornmeal diet (Figure S1A). Compared to SY1:1, cornmeal diet has higher sugar and lower protein contents (or C:P ratios) as well as different sugar types. Therefore, we postulated that macronutrient composition is critical in affecting the function of ATPsyn-d in modulating lifespan. To test this, we measured lifespan of flies fed SY1:9 or SY9:1, representing low sugar-high protein or high sugar-low protein diet, respectively. ATPsyn-d knockdown did not increase lifespan and appeared to slightly decrease lifespan of females fed SY9:1 using two different GSG drivers (actin-GSG and da-GSG) and two RNAi stocks (v21018 and v104353) (Figure 2A and B, and Table S1). This is consistent with findings from using cornmeal diet, which has similar C:P ratio as SY9:1. On the other hand, ATPsyn-d knockdown significantly increased lifespan of females fed SY1:9 using both RNAi stocks (Figure 2C and D, and Table S1). ATPsyn-d knockdown did not extend lifespan of males fed SY9:1 or SY1:9 (Figure S1C and D and Table S1). The extent of ATPsyn-d knockdown at the transcript level induced by RU486 was similar in females and males on SY1:9 or SY9:1 (Figure S1E and F), again suggesting that the gender difference in lifespan may not be due to the difference in ATPsyn-d knockdown. RU486 significantly reduced ATPsyn-d protein level by >25% in da-GSG>UAS-ATPsyn-d-RNAi(v104353) females fed SY9:1, SY1:1 or SY1:9 compared to diet-matched no-RU486 controls (Figure 2E and F). RU486 alone did not significantly increase lifespan of various control female and male flies, including actin-GSG>UAS-GFP-RNAi and da-GSG>UAS-GFP-RNAi fed SY1:9 or SY9:1 and actin-GSG/+ fed cornmeal, and in some cases might reduce lifespan of flies fed SY9:1 (Figure S2 and Table S2). These findings reveal a role of ATPsyn-d in modulating lifespan response to diet composition.
Figure 2. The impact of diet composition on the role of ATPsyn-d in modulating lifespan.
(A) Lifespan of actin-GSG>UAS-ATPsyn-d-RNAi(v21018) females on SY9:1. (B) Lifespan of da-GSG>UAS-ATPsyn-d-RNAi(v104353) females on SY9:1. (C) Lifespan of actin-GSG>UAS-ATPsyn-d-RNAi(v21018) females on SY1:9. (D) Lifespan of da-GSG>UAS-ATPsyn-d-RNAi(v104353) females on SY1:9. (E and F) ATPsyn-d protein levels in 14-d old da-GSG>UAS-ATPsyn-d-RNAi(v104353) females fed SY9:1, SY1:1 and SY1:9 with and without RU486 treatment. ATPsyn-d protein levels were normalized to NDUFS3 (n=3 biologically independent repeats). “RNAi” indicates ATPsyn-d knockdown by 200 µM RU486. The UAS strain identification is shown in parentheses. “Control” represents genotype-matched flies without RU486 treatment. The p values for lifespan were from logrank analysis between RNAi and control flies. For bar graphs, error bars represent standard errors and *p<0.05; **p<0.01 by Student’s t-test. n. s., not statistically significant. a.u., arbitrary unit.
ATPsyn-d knockdown did not affect food intake or locomotor activity of flies fed SY1:9, and RU486 alone did not affect food intake of flies, either (Figure S3A to C), indicating that lifespan extension induced by ATPsyn-d knockdown under S1:9 is not due to any change in food intake. ATPsyn-d overexpression did not dramatically increase lifespan of males and females fed SY9:1 or SY1:9 and in some cases for females on SY9:1 slightly decreased lifespan relative to gender- and diet-matched controls (Figure S1N to P and R to T and Table S3), suggesting that ATPsyn-d overexpression has no or little impact on lifespan.
Considering the relationship between reproduction and lifespan, we measured lifetime egg laying as reproductive output in da-GSG>UAS-ATPsyn-d-RNAi(v104353) flies with and without RU486 feeding. Lifetime egg laying was also measured in da-GSG driver-only and UAS-RNAi only flies to assess the effect of RU486 alone on reproduction. RU486 feeding significantly reduced lifetime egg laying in da-GSG>UAS-ATPsyn-d-RNAi flies by 54.8% on SY9:1, 83.4% on SY1:1 and 75.2% on SY1:9 relative to SY diet- and genotype- matched no-RU486 controls (Figure S3D). However, RU486 alone also significantly reduced lifetime egg laying in the driver control line da-GSG/+ by 53.6% on SY9:1, 72.1% on SY1:1 and 75.0% on SY1:9, and in the UAS control line UAS-ATPsyn-d-RNAi/+ by 25.3% on SY9:1, 75.4% on SY1:1 and 41.5% on SY1:9 relative to SY diet- and genotype- matched no-RU486 controls (Figure S3D). These indicate that RU486 by itself has a significant impact on reproduction. The decrease of reproduction in da-GSG>UAS-ATPsyn-d-RNAi flies is not necessarily severer than either driver-only or UAS-only control flies. These findings suggest that reproductive changes may not play a significant role in diet dependent lifespan extension induced by ATPsyn-d knockdown.
ATPsyn-d knockdown globally affects expression of genes in stress response and metabolic pathways
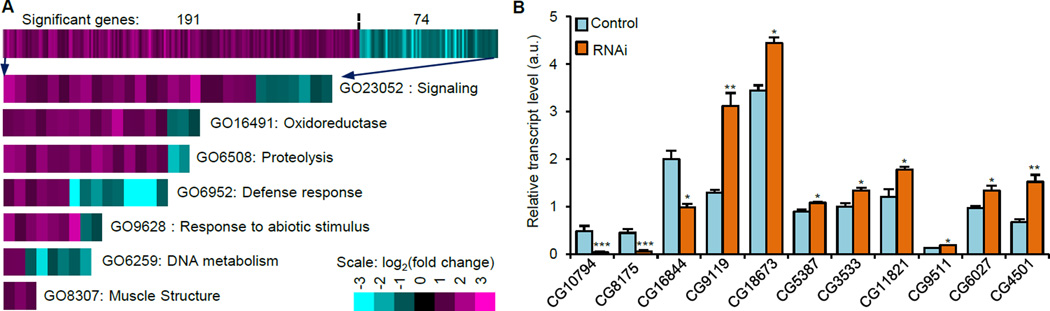
To determine molecular mechanisms by which ATPsyn-d knockdown promotes longevity, we examined global changes in gene expression induced by ATPsyn-d knockdown in females fed SY1:9. The transcript levels of 265 genes were significantly altered by ≥1.5 fold (p<0.01) (Figure 3A and Table S5), in which 191 were up-regulated and 74 were down-regulated by ATPsyn-d knockdown. Quantitative PCR (qPCR) confirmed transcript changes in 11 ATPsyn-d-regulated genes (Figure 3B). RU486 alone did not significantly alter the transcript levels of ATPsyn-d, Ucp5 and six ATPsyn-d-regulated genes in Canton S females relative to no-RU486 controls (Figure S4A). This suggests that RU486 alone does not have a substantial impact on gene expression although we cannot exclude the possibility that some putative ATPsyn-d-regulated genes are affected by RU486. Based on gene ontology (GO) analysis and information in the Flybase (www.flybase.org), ATPsyn-d knockdown affected expression of genes involved in diverse biological processes (Figure 3A and Table S6). One biological process affected by ATPsyn-d knockdown is defense response or innate immune response (GO6952), including antimicrobial peptide genes, drosomycin and diptericin. Second is energy metabolism regulated by oxidoreductases (GO16941), including genes encoding proteins with ATPase or ATP synthase activity. Third is proteolysis (GO6508), including genes encoding various proteases. Fourth is defense response to abiotic stimulus (GO9628), including detoxification-related cytochrome P450 genes, Cyp305a1 and Cyp4ac1. Moreover, ATPsyn-d knockdown altered expression of genes involved in DNA metabolism (GO6259), such as DNA damage response genes, WRN exonuclease and minichromosome maintenance 3. The global expression patterns suggest that ATPsyn-d knockdown affects innate immune response, energy production and utilization, proteostasis, detoxification and oxidative damage.
Figure 3. The effect of ATPsyn-d knockdown on global gene expression in female flies fed SY1:9.
(A) The first row of the heat map represents 265 genes whose transcription levels are significantly affected by ATPsyn-d knockdown in actin-GSG>UAS-ATPsyn-d-RNAi(v21018) females (n=6 biologically independent replicates). Among them, 191 are up-regulated and 74 are down-regulated. The other rows represent gene ontology categories that are affected by ATPsyn-d knockdown. Each column in the row represents one gene and its fold change is color-coded. (B) The transcript changes of 11 genes were confirmed by qPCR. “RNAi” indicates ATPsyn-d knockdown by 200 µM RU486 (n≥3). “Control” represents controls without RU486 treatment. Error bars represent standard errors. *p<0.05; **p<0.01; ***p<0.001 between RNAi and control by Student’s t-test.
ATPsyn-d knockdown increases resistance to oxidative stress and reduces oxidative damage
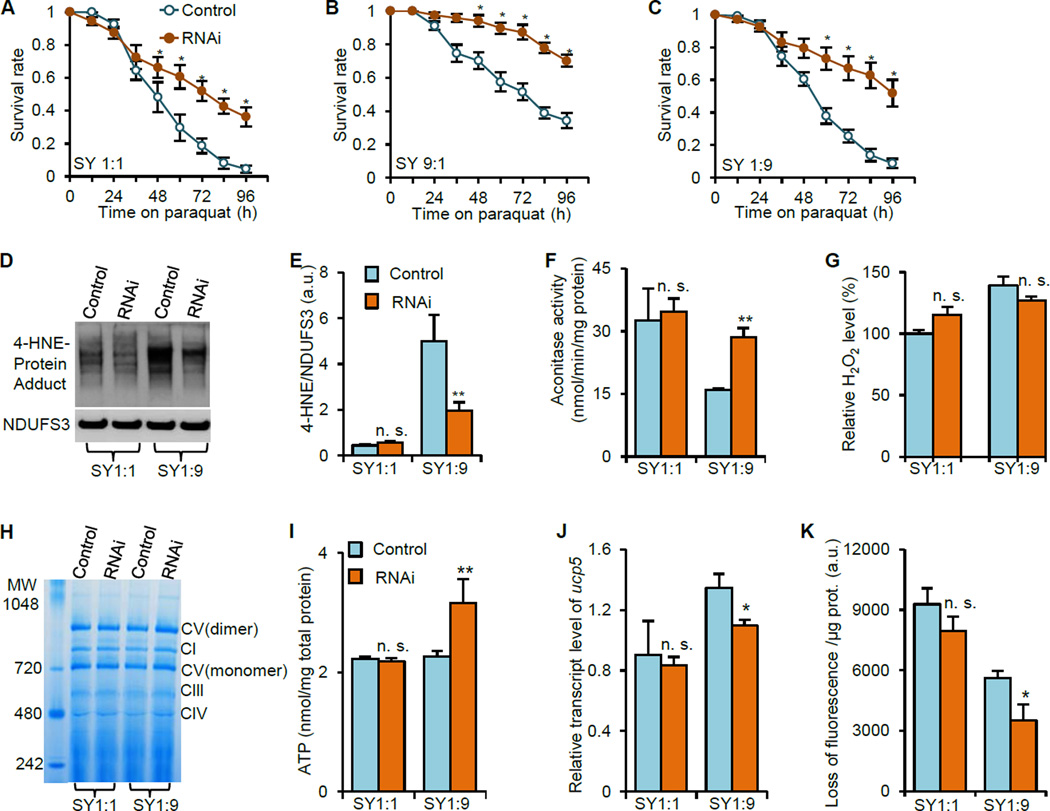
Lifespan extension is often associated with increased resistance to stresses (Fontana et al., 2010). We found that ATPsyn-d knockdown with RNAi strain v21018 increased resistance to paraquat-induced acute oxidative stress in females under any of the three SY diets (Figure 4A to C). ATPsyn-d knockdown using another RNAi strain v104353 also resulted in increased resistance to paraquat in females fed SY1:9 (Figure S5A). RU486 alone did not substantially increase resistance to paraquat in the driver only da-GSG/+ or wild type Canton S flies fed SY1:9 (Figure S5B and C). However, ATPsyn-d knockdown decreased the survival of females under starvation conditions (Figure S5D to F). ATPsyn-d knockdown did not affect sensitivity to oxidative stress or starvation in males (Figure S1G to L). These findings suggest that lifespan extension induced by ATPsyn-d knockdown is associated with increased resistance to oxidative stress but not starvation.
Figure 4. The effect of ATPsyn-d knockdown on stress resistance, oxidative damage and mitochondrial function.
(A to C) ATPsyn-d knockdown increased females’ resistance to paraquat-induced oxidative stress. The survival of 14-d old actin-GSG>UAS-ATPsyn-d-RNAi(v21018) females fed SY1:1, SY9:1 and SY1:9 were monitored on 20 mM paraquat. (D and E) The effect of ATPsyn-d knockdown on the level 4-HNE-protein adducts in mitochondria, which was normalized to NDUFS3. (F and G) The effect of ATPsyn-d knockdown on Aconitase activity and H2O2 production in mitochondria. (H to K) The effect of ATPsyn-d knockdown on mitochondrial complex formation, cellular ATP level, relative ucp5 transcript level and membrane potential, respectively. MW, molecular weight. 14-d old flies were collected for assays D to K. “RNAi” indicates that ATPsyn-d knockdown (actin-GSG>UAS-ATPsyn-d-RNAi(v21018)) is induced in by 200 µM RU486. “Control” represents genotype- and diet-matched controls without RU486 treatment. Error bars represent standard errors. n≥3 in each assay. *p<0.05; **p<0.01 by Student’s t-test.
To investigate mechanisms underlying oxidative stress resistance associated with lifespan extension, we examined the effect of ATPsyn-d knockdown on levels of 4-HNE-protein adducts and Aconitase activity, which are biomarkers for accumulated lipid and protein oxidation, respectively (Lind et al., 2006; Tsai et al., 1998). ATPsyn-d knockdown with RNAi line v21018 reduced 4-HNE-protein adducts level in mitochondria of females fed SY1:9, and to a lesser extent, of females fed SY1:1, and increased mitochondrial Aconitase activity in females fed SY1:9 but not significantly in flies fed SY1:1 (Figure 4D to F). The purity of mitochondrial and cytosolic fractions was verified by Western blot analysis with antibodies against cytosolic protein AKT and mitochondrial protein porin (Figure S5G). Consistently ATPsyn-d knockdown with another RNAi line v104353 also reduced 4-HNE-protein adducts level in mitochondria of females fed SY1:9 (Figure S6A and B). RU486 alone did not have any substantial effect on either 4-HNE or mitochondrial Aconitase activity level since these two parameters were not significantly different between UAS-ATPsyn-d-RNAi/+ control flies fed SY1:9 with and without RU486 (Figure S6C to E). We also measured other oxidative stress-related markers and found that ATPsyn-d knockdown did not affect H2O2 production in mitochondria or cytosolic Aconitase activity in females fed SY1:9 or SY1:1 (Figure 4G, S5H and S5I). These results suggest that ATPsyn-d knockdown selectively alleviates mitochondrial oxidative damage to maintain cellular homeostasis and ATPsyn-d responds differentially to acute and chronic oxidative stress and promotes longevity in a diet dependent manner.
ATPsyn-d knockdown affects mitochondrial function
ATPsyn-d is a key component of the ATP synthase complex responsible for ATP production in mitochondria (Wallace, 2005). We examined whether ATPsyn-d knockdown increases lifespan through modulating mitochondrial functions. ATPsyn-d knockdown with RNAi line v21018 did not significantly alter activities of Citrate synthase, a tricarboxylic acid (TCA) cycle component, mitochondrial density, a biomarker of mitochondrial biogenesis, Cytochrome c oxidoreductase, a complex III component, or Cytochrome C oxidase, a complex IV component (Figure S5J to N). ATPsyn-d knockdown did not significantly alter the formation of five oxidative phosphorylation complexes, either (Figure 4H). However, ATPsyn-d knockdown increased cellular ATP level in females fed SY1:9 but not SY1:1 (Figure 4I). To determine the mechanisms underlying the paradoxically increased ATP level by ATPsyn-d knockdown, we examined expression of mitochondrial uncoupling proteins (Ucps) and mitochondrial membrane potential, which involve in ATP production (Harper et al., 2004). ATPsyn-d knockdown with RNAi line v21018 reduced Ucp5 expression in females fed SY1:9 but not SY1:1 (Figure 4J). Consistently, ATPsyn-d knockdown with another RNAi line v104353 also reduced Ucp5 expression in females fed SY1:9 (Figure S4B). RU486 alone did not significantly affect Ucp5 expression or ATP level (Figure S4A and S6F). Moreover, ATPsyn-d knockdown reduced the loss of mitochondrial accumulation of JC-1, a fluorescent dye for membrane potential, in females fed SY1:9, indicating that ATPsyn-d knockdown increases membrane potential (Figure 4K). Reduced Ucp expression is known to be potentially associated with reduced proton leaking in mitochondria and increased ATP production (Wallace, 2005). We further measured whole body glucose and trehalose levels to determine the impact of ATPsyn-d knockdown on glucose homeostasis. The whole-body glucose and trehalose levels were not significantly different between ATPsyn-d knockdown and control females on SY1:9 (Figure S5O and P). Together our findings suggest that increased ATP production induced by ATPsyn-d knockdown is associated with reduced Ucp5 expression and increased membrane potential in a diet dependent manner without significantly changing mitochondrial biogenesis.
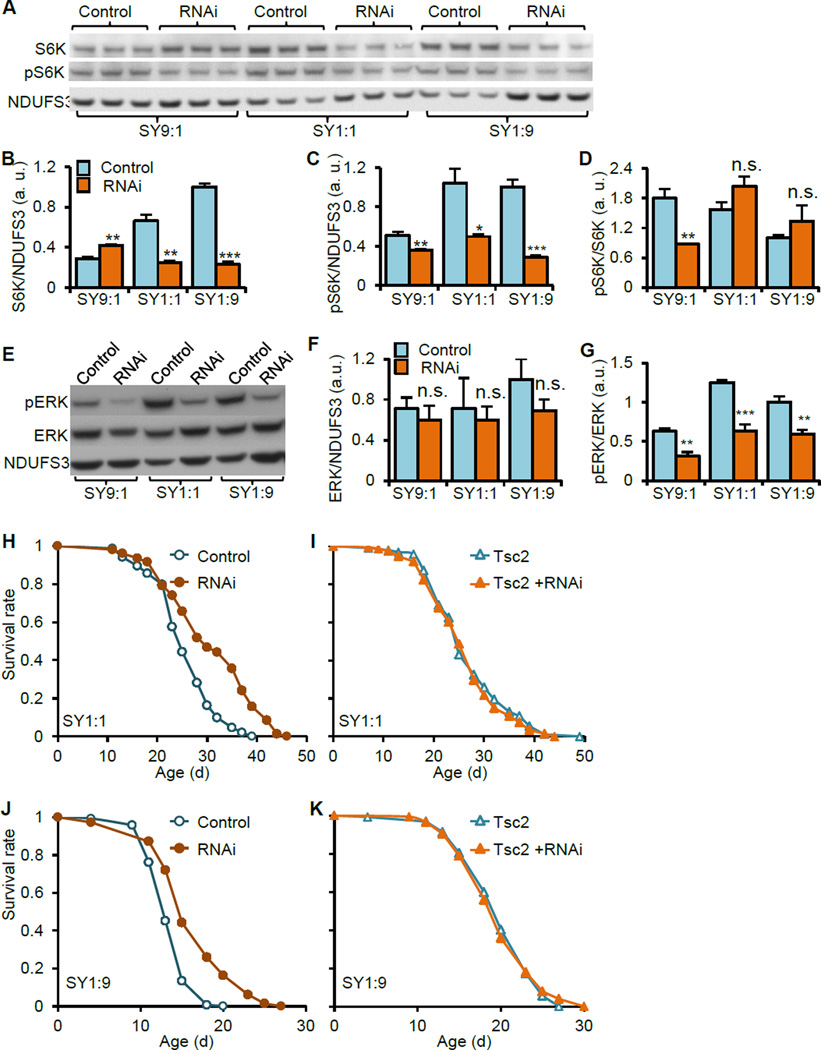
ATPsyn-d influences MAP kinase and TOR signaling pathways
Observation of the diet-dependent effect of ATPsyn-d knockdown on lifespan prompted us to examine the relationship between ATPsyn-d and TOR signaling, considering the critical role of TOR signaling in sensing nutrients especially amino acids (Zoncu et al., 2011). We first measured protein and phosphorylation levels of S6K, a TOR target involved in protein translation. ATPsyn-d knockdown with RNAi line v104353 reduced S6K protein level relative to total protein in females fed SY1:1 or SY1:9 but not SY9:1 compared to diet-matched controls (Figure 5A and B). S6K phosphorylation (pS6K) level relative to total protein was significantly reduced in ATPsyn-d knockdown females fed SY1:1 or SY1:9, too, and, to a lesser extent in flies fed SY9:1, compared to diet-matched controls (Figure 5A and C). The pS6K/S6K ratio was only reduced in ATPsyn-d knockdown females fed SY9:1 but not SY1:1 or SY1:9 compared to diet-matched controls (Figure 5D). Consistent with the literature that TOR signaling is typically reduced in animals fed low protein diets (Bjedov et al., 2010), both S6K and pS6K levels were reduced in no-RU486 control female flies fed the low protein diet SY9:1 relative to higher protein diet SY1:1 or SY1:9 (Figure 5B and C). These suggest that lifespan extension by ATPsyn-d knockdown is associated with reduced levels of both S6K and pS6K.
Figure 5. Genetic interaction between ATPsyn-d and TOR signaling.
(A to D) The effect of ATPsyn-d knockdown on S6K protein, its phosphorylation level (pS6K) and pS6K/S6K ratio in female flies fed SY9:1, SY1:1 and SY1:9. S6K and pS6K protein levels were normalized with NDUFS3. (E and G) The effect of ATPsyn-d knockdown on total ERK level and ERK phosphorylation as indicated by the pERK/ERK ratio in females fed SY9:1, SY1:1 and SY1:9. ERK protein levels were normalized with NDUFS3. (H and J) The lifespan of ATPsyn-d knockdown females and their controls under SY1:1 and SY1:9. (I and K) The lifespan of flies with Tsc2 overexpression and females with both Tsc2 overexpression and ATPsyn-d knockdown under SY1:1 or SY1:9. “RNAi” indicates ATPsyn-d knockdown (da-GSG>ATPsyn-d-RNAi(v104353)) by 200 µM RU486. “Control” represents genotype- and diet-matched controls without RU486 treatment. “Tsc2” represents Tsc2 overexpression. The values in Fig. 5B, C, D, F and G were normalized to the value of the control flies on SY1:9, which was set at 1. Error bars represent standard errors. a.u., arbitrary unit. *p<0.05; **p<0.01; ***p<0.001 by Student’s t-test.
To explore molecular mechanisms underlying pS6K changes, we examined phosphorylation levels of ERK1/2 (pERK). ERK1/2 is part of the MAP kinase (MAPK) pathway involved in stress response and is known to regulate TOR signaling through phosphorylation in mammalian models (Kim and Guan, 2011; Rubinfeld and Seger, 2005; Zoncu et al., 2011). ATPsyn-d knockdown did not significantly alter the total protein level of ERK, but reduced pERK/ERK ratio in females under any of the three SY diets relative to diet-matched controls (Figure 5E, F and G). RU486 alone did not change pERK/ERK ratio in driver only da-GSG/+ flies fed SY1:9 (Figure S6G and H), suggesting that RU486 alone has no or little impact on MAPK signaling. These findings suggest that ATPsyn-d knockdown reduces MAPK signaling, which may in turn reduce TOR signaling. We next examined phosphorylation level of AKT and found that pAKT/AKT ratio was not significantly different between ATPsyn-d knockdown and control females on SY1:1 or SY1:9(Figure S6I and J). These findings suggest that ATPsyn-d knockdown has no or little impact on insulin-like signaling and glucose homeostasis.
ATPsyn-d Genetically Interacts with TOR signaling to modulate lifespan
To determine the functional role of TOR signaling in lifespan extension induced by ATPsyn-d knockdown, we investigated whether overexpression of Tsc2, a genetic suppressor of TOR signaling (Zoncu et al., 2011), influences the effect of ATPsyn-d knockdown on lifespan. We confirmed that Tsc2 overexpression driven by da-Gal4 can promote longevity under SY1:1 (Figure S7A). Consistent with what described above, ATPsyn-d knockdown extended lifespan in females fed SY1:1 or SY1:9 (Figure 5H and J and Table S7). However, lifespan of females with both Tsc2 overexpression and ATPsyn-d knockdown was not significantly different from that of females with only Tsc2 overexpression under SY1:1 or SY1:9 (Figure 5I and K and Table S7). On the other hand, ATPsyn-d knockdown extended lifespan of females with or without overexpression of a dominant negative form of JNK under SY1:1 or SY1:9 (Figure S7B to E), suggesting that the failure of lifespan extension by ATPsyn-d knockdown in flies with Tsc2 overexpression is specific to Tsc2. Together, these findings suggest a genetic interaction between ATPsyn-d and Tsc2, supporting the existence of a genetic interaction between ATPsyn-d and TOR signaling.
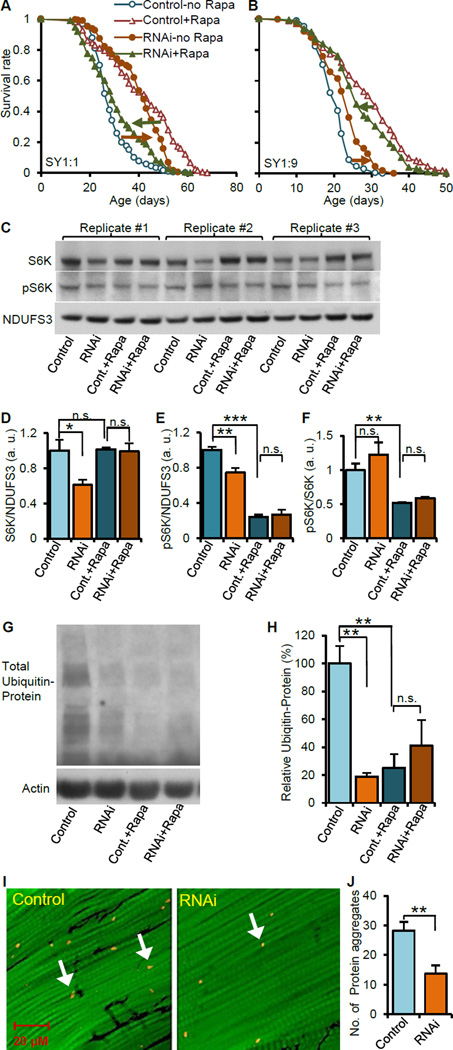
To further confirm the functional link between TOR signaling and ATPsyn-d, we measured lifespan of ATPsyn-d knockdown and their control female flies fed rapamycin. Rapamycin is known to suppress TOR signaling and promote longevity in yeast, worms, flies and mice (Bjedov et al., 2010; Harrison et al., 2009; Zoncu et al., 2011). We found that rapamycin feeding extended lifespan in flies without RU486 treatment, which have normal ATPsyn-d level, under SY1:1 or SY1:9, relative to SY diet-matched no-rapamycin groups (“Control-Rapa” vs. “Control-no Rapa” in Figure 6A and B, and Table S7). However, rapamycin feeding decreased instead of increasing lifespan of ATPsyn-d knockdown flies fed SY1:1 or SY1:9, relative to SY diet-matched no-rapamycin-fed knockdown flies (“RNAi-Rapa” vs. “RNAi-no Rapa in Figure 6A and B, and Table S7). Consistent with what was described above, without rapamycin feeding, ATPsyn-d knockdown extended lifespan of flies fed SY1:1 or SY1:9, relative to diet-matched controls (“RNAi-no Rapa” vs. “Control-no Rapa” in Figure 6A and B). However, ATPsyn-d knockdown did not further extend lifespan of flies fed rapamycin-supplemented SY1:1 or SY1:9 relative to rapamycin- and diet-matched control flies (“RNAi-Rapa” vs. “Control-Rapa” in Figure 6A and B). To assess the impact of food intake, we measured daily food intake for flies fed SY1:1 or SY1:9 with and without RU486 and/or rapamycin. Regardless of the presence of rapamycin, ATPsyn-d knockdown did not affect food intake of flies fed SY1:1 or SY1:9 (“+RU486” vs. “no RU486” groups in Figure S3E). Regardless of the presence of RU486, rapamycin feeding did not significantly affect food intake of flies fed SY1:9, while it reduced food intake of flies fed SY1:1 (“+rapamycin” vs. “no rapamycin” groups in Figure S3E). Therefore, food intake in general has no or little impact on lifespan patterns influenced by rapamycin feeding. Together these findings suggest that ATPsyn-d knockdown and rapamycin promote longevity through overlapping pathways, likely including TOR signaling (Figure 7).
Figure 6. The impact of ATPsyn-d knockdown on rapamycin feeding, TOR signaling and proteostasis.
(A and B) The effect of rapamycin (rapa) on the lifespan of ATPsyn-d knockdown females and their controls fed SY1:1 and SY1:9. (C to F) The effect of ATPsyn-d knockdown and rapamycin feeding on S6K and pS6K levels and pS6K/S6K ratios in females fed SY1:9. n≥3 biologically independent replicates for each Western blot assay. (G and H) The effect of ATPsyn-d knockdown and rapamycin feeding on total polyubiquitinated protein normalized with β-actin. (I and J) The impact of ATPsyn-d knockdown on the accumulation of polyubiquitinated protein aggregates in fly flight muscle. The scale bar is shown inside the image of I. “RNAi” indicates ATPsyn-d knockdown (da-GSG>ATPsyn-d-RNAi(v104353)) by 200 µM RU486. “Control” represents controls without RU486 treatment. “+Rapa” indicates rapamycin feeding. The values in the Fig. 6B to F were normalized to the value of the control flies, which was set at 1. Error bars represent standard errors. a.u., arbitrary unit. *p<0.05; **p<0.01; ***p<0.001 by Student’s t-test.
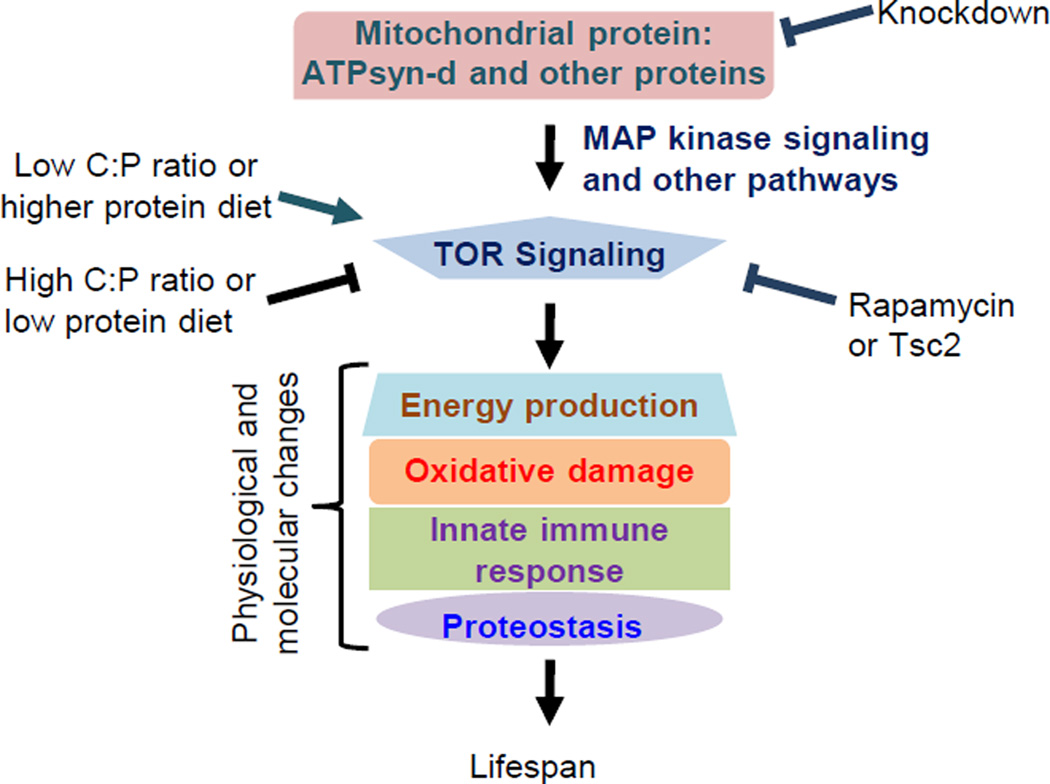
Figure 7. A speculated model on the role of ATPsyn-d in modulating lifespan in response to diet.
In this model, ATPsyn-d regulates TOR signaling probably through regulating ERK phosphorylation in MAPK signaling and modulating other pathways. Potential genetic interactions of ATPsyn-d and other mitochondrial proteins with TOR signaling induce molecular and physiological changes that modulate lifespan. These interactions can be disrupted by reduced TOR signaling, ATPsyn-d knockdown or a high C:P ratio diet and enhanced by a low C:P ratio diet.
To further investigate the relationship between ATPsyn-d knockdown and rapamycin, we measured S6K and pS6K protein levels. ATPsyn-d knockdown reduced of both S6K and pS6K levels but not pS6K/S6K ratio in female flies fed SY1:9 in consistent with the findings described above (“RNAi” vs. “Control” in Figure 6D to F). Rapamycin feeding reduced pS6K level and pS6K/S6K ratio but did not alter S6K level in control flies without ATPsyn-d knockdown (“Control-Rapa” vs. “Control” in Figure 6D to F). However, S6K or pS6K level or pS6K/S6K ratio in ATPsyn-d knockdown and control flies were not significantly different under rapamycin feeding (“RNAi+Rapa” vs. “Control+Rapa” in Figure 6D to F) These findings suggest that pS6K is a common downstream target of rapamycin and ATPsyn-d, further supporting that ATPsyn-d genetically interacts with TOR signaling to modulate lifespan.
ATPsyn-d knockdown improves protein homeostasis
TOR signaling is known to promote longevity through modulating protein homeostasis (Zoncu et al., 2011). In light of the link between ATPsyn-d and TOR signaling, we postulated that ATPsyn-d may play a critical role in maintaining proteostasis and consequently promote healthy aging. To test this hypothesis, we measured total polyubiquitinated protein level and polyubiquitinated protein aggregates, two biomarkers of proteostasis (Rana et al., 2013). Both ATPsyn-d knockdown and rapamycin feeding reduced the level of total polyubiquitinated protein in females fed SY1:9 (“Control” vs “RNAi” and “Control” vs “Cont.+Rapa” in Figure 6G, 6H, S7F and S7G). Reduced polyubiquitinated protein level by rapamycin feeding, was not further reduced by ATPsyn-d knockdown (“Cont.+Rapa” vs “RNAi+Rapa” in Figure 6G and H), further supporting a genetic interaction between ATPsyn-d and TOR signaling (Figure 7). Moreover, ATPsyn-d knockdown reduced the number of polyubiquitinated protein aggregates in flight muscle of females fed SY1:9 (Figure 6I and J). RU486 feeding by itself did not significantly change total polyubiquitinated protein level or polyubiquitinated protein aggregates (Figure S7H to K). Together these findings suggest that lifespan extension induced by ATPsyn-d knockdown is at least partially through modulating protein homeostasis.
Discussion
Dietary nutrients are important environmental factors that play a major role in modulating lifespan and healthspan (Fontana et al., 2010). However, how dietary nutrients affect the impact of genetic factors or interact with genetic factors to modulate lifespan and healthspan remains elusive. Considering the essential role of mitochondrial function in metabolism and aging, we have investigated how diet composition influences the function of ATPsyn-d, a component of mitochondrial ATP synthase, in aging and the underlying mechanisms. We have shown that ATPsyn-d knockdown extends lifespan in Drosophila under low sugar-high protein diets but not under a high sugar-low protein diet. Lifespan extension induced by ATPsyn-d knockdown is associated with increased resistance to oxidative stress and improved protein homeostasis. Furthermore, we have provided evidence suggesting ATPsyn-d modulates lifespan through genetically interacting with TOR signaling. Knocking-down of atp-5, the worm ATPsyn-d, extends lifespan in C. elegans, along with our data, suggesting a conserved role of ATPsyn-d in modulating lifespan (Hansen et al., 2005). Together, our findings reveal a novel connection among diet, mitochondrial ATP synthase, MAPK and TOR signaling in modulating lifespan, and shed light on the molecular mechanisms underlying the impact of diet composition on lifespan.
We propose the following model to explain how ATPsyn-d interacts with dietary macronutrients to modulate lifespan considering the genetic interaction between ATPsyn-d and TOR signaling, and the fact that suppression of TOR signaling by altering expression of its components, such as Tsc1/2, S6K and 4E-BP, activates autophagy, improves proteostasis and promotes longevity in high protein diets but not necessarily low protein diets (Kapahi et al., 2004; Zid et al., 2009). We postulate that TOR signaling is regulated by ATPsyn-d and perhaps other mitochondrial proteins (Figure 7). ATPsyn-d knockdown reduces MAPK signaling and probably affects other signaling pathways, which may consequently decrease TOR signaling to extend lifespan in Drosophila fed high protein diets, such as SY1:9 and SY1:1, but not low protein diets. It is possible that diet-dependent response is due to knockdown of ATPsyn-d protein to different extent by RNAi under different dietary conditions. This is not likely the case. The amount of ATPsyn-d knockdown is not much different between flies on SY1:9 and SY9:1, although lifespan is not increased by ATPsyn-d knockdown for flies under SY9:1. Therefore, variations in ATPsyn-d knockdown under current experimental conditions unlikely contribute significantly to diet-dependent lifespan extension. Consistent with this model, ATPsyn-d knockdown increases resistance to acute oxidative stress, reduces cellular oxidative damage and improves proteostasis in Drosophila. Reduced oxidative damage by ATPsyn-d knockdown may lead to decreased MAPK signaling, which in turn modulates TOR signaling and proteostasis.
Another likely scenario would be that ATPsyn-d and TOR signaling form a positive but vicious feedback loop through MAPK signaling to induce molecular, metabolic and physiological changes detrimental to lifespan. This vicious cycle can be disrupted by high C:P diet, knockdown of mitochondrial genes or suppression of TOR signaling pharmacologically by rapamycin or genetically by Tsc2 overexpression. Consistent with this possibility is that ATPsyn-d knockdown reduces phosphorylation of S6K, a component of TOR signaling, and increases expression of genes involved in maintaining proteostasis and possibly autophagy, which are regulated by TOR signaling. The level of pS6K reflects the strength of TOR signaling and reduction-of-function mutants of S6K are known to extend lifespan in several species (Zoncu et al., 2011). ATPsyn-d may genetically interact with TOR signaling to modulate lifespan by influencing protein levels of both S6K and pS6K, although it does not necessarily affect the pS6K/S6K ratio, which may not be a reliable indicator for the strength of TOR signaling under the three SY diets due to the change of S6K level. Furthermore, ATPsyn-d knockdown reduces oxidative damage and polyubiquitinated protein aggregates, which are biomarkers of aging. Rapamycin reduces lifespan extension induced by ATPsyn-d knockdown, which may be due to exacerbation of some detrimental effects of reduced TOR signaling (Zoncu et al., 2011). However, this observation further supports the connection between ATPsyn-d and TOR signaling. Although both rapamycin and ATPsyn-d knockdown reduce pS6K level, ATPsyn-d knockdown but not rapamycin decreases S6K level, suggesting ATPsyn-d knockdown and rapamycin affect TOR signaling in different manners. Further studies are warranted to clarify the epistatic relationship between ATPsyn-d and TOR signaling.
Increasing evidence has demonstrated the importance of diet composition or carbohydrate to protein ratio in modulating lifespan and health (Piper et al., 2011). Nutrient geometry studies conducted in Drosophila have shown that C:P ratio in the diet is far more important in determining lifespan than calorie content or single macronutrient (Lee et al., 2008; Skorupa et al., 2008). A recent tour de force nutrient geometry study in mice has confirmed and expanded the view on the critical role of C:P ratio in regulating lifespan and cardiometabolic health to mammals (Solon-Biet et al., 2014). An important implication from nutrient geometric studies is that diet composition would have a significant impact on the effectiveness of inventions for promoting healthy aging by genetic, pharmaceutical or nutraceutical approaches. This indeed is the case although evidence comes from only a handful of studies. Rapamycin feeding extends lifespan in yeast, worms, flies and mice (Bjedov et al., 2010; Zoncu et al., 2011). Although rapamycin feeding has been shown to extend lifespan of flies under a broad range of diets (Bjedov et al., 2010), some studies have shown that rapamycin feeding does not to extend lifespan in flies under high carbohydrate-low protein diets (Sun et al., 2012). Supplementation of a nutraceutical derived from cranberry extends lifespan in female flies under a high C:P ratio diet but not a low C:P ratio diet (Wang et al., 2013). Suppression of TOR signaling by overexpression of Tsc1/2 extends lifespan in flies under relatively higher protein diets but not under low protein diets although those studies focused on the variation of protein concentration instead of C:P ratio (Kapahi et al., 2004). Consistent with the link between ATPsyn-d and TOR signaling, ATPsyn-d knockdown extends lifespan in female flies under low sugar-high protein diets but not high sugar-low protein diet, likely due to the fact that TOR signaling is already low under the high sugar-low protein diet. We have further shown that rapamycin feeding extends lifespan in wild type female flies but not in ATPsyn-d knockdown flies. Our study on ATPsyn-d is one of the first studies that have investigated the impact of diet composition on lifespan modulation by genetic factors and aging interventions and the underlying mechanisms. These findings point out the importance of considering both genetic background and diet composition in implementing interventions for promoting healthy aging.
Aging is associated with profound decline in protein homeostasis, and many longevity-related pathways, such as TOR and Insulin-like signaling, modulate lifespan through improving proteostasis (Taylor and Dillin, 2011). Suppression of TOR signaling extends lifespan through decreasing protein translation and increasing autophagy, key processes for maintaining proteostasis (Zoncu et al., 2011). We have found that ATPsyn-d knockdown reduces the level of 4-HNE protein adducts, a biomarker for lipid protein oxidation (Tsai et al., 1998), and the level and aggregation of polyubiquitinated protein, a biomarker for proteostasis and aging (Rana et al., 2013). ATPsyn-d is a key component of mitochondrial ATP synthase complex. Along with the link between ATPsyn-d and TOR signaling, our data suggest that mitochondrial ATP synthase is critical for maintaining proteostasis and modulating lifespan. This notion is further supported by a recent study showing that α-ketoglutarate, an intermediate in the TCA cycle, suppresses mitochondrial ATP synthase probably by binding to ATP synthase subunit β (ATPsyn-β) and also inhibits TOR signaling to extend lifespan in C. elegans (Chin et al., 2014). However, it remains to be determined whether suppression of ATP synthase by α-ketoglutarate results in inhibition of TOR signaling in C. elegans or any other species. It is also likely that ATPsyn-d and ATPsyn-β influences ATP synthase and TOR signaling through different mechanisms, since α-ketoglutarate reduces cellular ATP level in C. elegans, while ATPsyn-d knockdown does not significantly change or even increases ATP level in Drosophila. This also suggests that lifespan extension is not necessarily associated with decreased ATP level, which is supported by a study in Drosophila showing that any change of ATP level is not correlated with any change of lifespan induced by knockdown of a number of mitochondrial genes (Copeland et al., 2009). Nevertheless, these studies suggest that ATP synthase is a key and conserved player linking dietary nutrients from TOR signaling to proteostasis and lifespan.
Similar to many longevity-related mutants (Salmon et al., 2010), lifespan extension induced by ATPsyn-d knockdown is associated with reduced oxidative damage and increased resistance to oxidative stress. ATPsyn-d knockdown increases lifespan and resistance to paraquat, an acute oxidative stress response, under SY1:9 or SY1:1. However, ATPsyn-d knockdown increases resistance to paraquat, but does not extend lifespan in female flies under SY9:1. In addition, ATPsyn-d knockdown decreases 4-HNE level, an indicator of accumulated oxidative damage, under SY1:9 but not SY1:1. These indicate that the effect of ATPsyn-d knockdown on oxidative damage and lifespan depends on diet composition, suggesting that oxidative stress resistance is at most partially responsible for lifespan extension. This should not be surprising since it is consistent with numerous studies in the literature showing that stress resistance does not always result in lifespan extension despite of the strong link between oxidative stress and aging (Salmon et al., 2010).
The role of mitochondrial genes in modulating lifespan is complex. Knockdown of some ETC genes increases lifespan while knockdown of others decreases or does not alter lifespan in C. elegans and Drosophila (Copeland et al., 2009; Lee et al., 2003; Zid et al., 2009). Our study reveals another layer of complexity regarding the role of ETC genes in lifespan modulation, namely the impact of diet composition. Our findings indicate that ATPsyn-d knockdown promotes longevity at least partially through TOR signaling. TOR Signaling senses cellular amino acid content and regulates numerous biological processes, including translation, autophagy and lifespan (Zoncu et al., 2011). 4E-BP, a translational repressor in TOR signaling, mediates lifespan extension induced by DR (Zid et al., 2009). Activated 4E-BP suppresses general translation, but selectively increases translation of some mitochondrial ETC genes (Zid et al., 2009), the latter of which results in increased mitochondrial biogenesis and potentially lifespan. Lifespan extension induced by DR is suppressed by knocking down ETC genes regulated by 4E-BP. The findings by Zid et al. suggest that increased protein expression of some ETC genes is associated with lifespan extension induced by DR. However, unlike those ETC genes, ATPsyn-d knockdown extends instead of decreases lifespan under high protein diets. Therefore, it is likely that ETC genes can be categorized into two groups, one selectively upregulated by activated 4E-BP and the other insensitive to activated 4E-BP, the latter of which may include ATPsyn-d. The two groups of ETC genes may interact with dietary macronutrients to modulate lifespan perhaps through different modes of action. Future studies are warranted to investigate the dichotomous role of translation of ETC genes in modulating lifespan.
Experimental Procedures
Fly media
Fly stocks were maintained on standard cornmeal medium at 25±1 °C, 60±5% humidity and a 12:12h light/dark cycle (Ashburner et al., 2005). SY1:9 contained 2% sugar and 18% autolyzed yeast (MP Biomedicals, cat. #103304), SY1:1 had 10% sugar and 10% yeast and SY9:1 had 18% sugar and 2% yeast.
Lifespan, food intake, lifetime reproduction and stress assays
Lifespan, food intake, lifetime reproduction and paraquat assays were conducted as previously described (Laslo et al., 2013; Mair et al., 2005; Sun et al., 2012). Cytosolic and mitochondrial Aconitase activities and mitochondrial H2O2 production were measured with commercially available kits.
Western blot analysis
Western blot analysis was performed as previously described (Sun et al., 2010).
Microarray assays and qPCR
Microarray assays were performed as previously described (Zhan et al., 2007) (n=4). Genes with p<0.01 and false discovery rate<0.08 and biological processes with p<0.05 were considered having significant changes. The microarray data were deposited in the NCBI GEO database (accession # GSE58778). qPCR was performed with Step-One plus system (Applied Biosystems) and relative transcript levels were normalized with rp49 (n≥3).
Mitochondrial assays
Blue Native Polyacrylamide Gel Electrophoresis was performed with mitochondrial protein as previously described (Copeland et al., 2009). ATP level was determined with whole-body lysates using ATP Bioluminescence Assay HS II kit (Roche Applied Science, Indianapolis, IN). Mitochondrial membrane potential was measured with isolated mitochondria using JC-1 fluorescent dye. Citrate synthase (CS) activity was measured in whole fly extracts and isolated mitochondrial fractions using Citrate Synthase Assay Kit (Sigma, St. Louis, MO). Cytochrome c oxidase (Complex IV) activities were performed with isolated mitochondria using cytochrome c oxidase assay kit (Sigma). All assays were repeated with ≥3 biological replicates.
Western blot and immunohistochemistry analyses of polyubiquitinated protein
Detergent-insoluble protein extracted from whole female flies was measured with anti-ubiquitin antibody by Western blot analysis as previously described (Rana et al., 2013). Protein aggregates were immunostained and quantified with anti-ubiquitin antibody followed by secondary Alexa-568 anti-mouse antibody and phalloidin staining by confocal analysis as previously described (Rana et al., 2013).
Statistical analysis
Statistical analyses were performed using PASW Statistics version 19 (IBM, Armonk, NY) and StatView Version 5 (SAS Institute, Cary, NC). Values were expressed as mean ± standard error. Survival data were analyzed by logrank test. All others were analyzed by Student’s t-test. A p<0.05 was considered statistically significant.
Supplementary Material
HIGHTLIGHTS.
ATPsyn-d knockdown extends lifespan and increases stress resistance.
Lifespan extension induced by ATPsyn-d knockdown depends on diet composition.
ATPsyn-d knockdown improves proteostasis.
ATPsyn-d interacts with TOR signaling to modulate lifespan.
Acknowledgements
We thank E. Spangler for critical reading of the manuscript, J. Tower, V. Monnier, D. Pan and T. P. Neufeld for reagents, Bloomington and Vienna stock centers for fly stocks. This study was supported by the Intramural Research Program of the NIA, NIH to S.Z. (Grant number: AG000365-09).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
X.S. and S.Z. conceived, designed and performed experiments and wrote the manuscript. All other authors performed experiments and edited the manuscript.
Conflict of Interests
All authors declare no conflict of interest.
References
- Ahima RS. Connecting obesity, aging and diabetes. Nat Med. 2009;15:996–997. doi: 10.1038/nm0909-996. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS, editors. Drosophila: A Laboratory Handbook. 2nd edn. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey JR, Harshman LG, Liedo P, Muller HG, Wang JL, Zhang Z. Longevity-fertility trade-offs in the tephritid fruit fly, Anastrepha ludens, across dietary-restriction gradients. Aging Cell. 2008;7:470–477. doi: 10.1111/j.1474-9726.2008.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin RM, Fu X, Pai MY, Vergnes L, Hwang H, Deng G, Diep S, Lomenick B, Meli VS, Monsalve GC, et al. The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014 doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New Genes Tied to Endocrine, Metabolic, and Dietary Regulation of Lifespan from a Caenorhabditis elegans Genomic RNAi Screen. PLoS Genet. 2005;1:e17. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper ME, Bevilacqua L, Hagopian K, Weindruch R, Ramsey JJ. Ageing, oxidative stress, and mitochondrial uncoupling. Acta Physiol Scand. 2004;182:321–331. doi: 10.1111/j.1365-201X.2004.01370.x. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guan KL. Amino Acid Signaling in TOR Activation. Annu Rev Biochem. 2011;80:1001–1032. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- Laslo M, Sun X, Hsiao CT, Wu WW, Shen RF, Zou S. A botanical containing freeze dried acai pulp promotes healthy aging and reduces oxidative damage in sod1 knockdown flies. Age (Dordrecht, Netherlands) 2013;35:1117–1132. doi: 10.1007/s11357-012-9437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Lee KP, Simpson SJ, Clissold FJ, Brooks R, Ballard JW, Taylor PW, Soran N, Raubenheimer D. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc Natl Acad Sci U S A. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lind MI, Missirlis F, Melefors O, Uhrigshardt H, Kirby K, Phillips JP, Soderhall K, Rouault TA. Of two cytosolic aconitases expressed in Drosophila, only one functions as an iron-regulatory protein. J Biol Chem. 2006;281:18707–18714. doi: 10.1074/jbc.M603354200. [DOI] [PubMed] [Google Scholar]
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:l6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Piper MD, Partridge L, Raubenheimer D, Simpson SJ. Dietary restriction and aging: a unifying perspective. Cell Metab. 2011;14:154–160. doi: 10.1016/j.cmet.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld H, Seger R. The ERK cascade: a prototype of MAPK signaling. Molecular biotechnology. 2005;31:151–174. doi: 10.1385/MB:31:2:151. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Richardson A, Perez VI. Update on the oxidative stress theory of aging: does oxidative stress play a role in aging or healthy aging? Free Radic Biol Med. 2010;48:642–655. doi: 10.1016/j.freeradbiomed.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 2008;7:478–490. doi: 10.1111/j.1474-9726.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi: 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Komatsu T, Lim J, Laslo M, Yolitz J, Wang C, Poirier L, Alberico T, Zou S. Nutrient-dependent requirement for SOD1 in lifespan extension by protein restriction in Drosophila melanogaster. Aging Cell. 2012;11:783–793. doi: 10.1111/j.1474-9726.2012.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Seeberger J, Alberico T, Wang C, Wheeler CT, Schauss AG, Zou S. Acai palm fruit (Euterpe oleracea Mart.) pulp improves survival of flies on a high fat diet. Exp Gerontol. 2010;45:243–251. doi: 10.1016/j.exger.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M. Diet restriction in Drosophila melanogaster. Design and analysis. Interdiscip Top Gerontol. 2007;35:115–136. doi: 10.1159/000096559. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J. Sex-specific regulation of aging and apoptosis. Mech Ageing Dev. 2006;127:705–718. doi: 10.1016/j.mad.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Tsai L, Szweda PA, Vinogradova O, Szweda LI. Structural characterization and immunochemical detection of a fluorophore derived from 4-hydroxy-2-nonenal and lysine. Proc Natl Acad Sci U S A. 1998;95:7975–7980. doi: 10.1073/pnas.95.14.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yolitz J, Alberico T, Laslo M, Sun Y, Wheeler CT, Sun X, Zou S. Cranberry Interacts With Dietary Macronutrients to Promote Healthy Aging in Drosophila. J Gerontol A Biol Sci Med Sci. 2013 doi: 10.1093/gerona/glt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Kim SK. Systems biology of aging in four species. Current opinion in biotechnology. 2007;18:355–359. doi: 10.1016/j.copbio.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan M, Yamaza H, Sun Y, Sinclair J, Li H, Zou S. Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome Res. 2007;17:1236–1243. doi: 10.1101/gr.6216607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.