Abstract
Inflammasomes are innate immune sensors that respond to pathogen and damage-associated signals with caspase-1 activation, IL-1β and IL-18 secretion, and macrophage pyroptosis. The discovery that dominant gain-of-function mutations in NLRP3 cause the Cryopyrin Associated Periodic Syndromes (CAPS) and trigger spontaneous inflammasome activation and IL-1β oversecretion, led to successful treatment with IL-1 blocking agents1. Herein, we report a de novo missense mutation, c.1009A>T, p.Thr337Ser, in the nucleotide-binding domain of inflammasome component NLRC4 (IPAF/CARD12) that causes early-onset recurrent fever flares and Macrophage Activation Syndrome (MAS). Functional analyses demonstrated spontaneous inflammasome formation and production of the inflammasome-dependent cytokines IL-1β and IL-18, the latter exceeding levels in CAPS. The NLRC4 mutation caused constitutive caspase-1 cleavage in transduced cells and increased production of IL-18 by both patient and NLRC4 mutant macrophages. Thus, we describe a novel monoallelic inflammasome defect that expands the monogenic autoinflammatory disease spectrum to include MAS and suggests novel targets for therapy.
Inflammasomes are platforms that integrate danger recognition with the production of the potent proinflammatory cytokines IL-1β and IL18. The discovery that dominant mutations in the NOD-Like Receptor (NLR) NLRP3 cause CAPS linked constitutive activation of the NLRP3 inflammasome to human disease1–3. At least two other NLR proteins are known to directly assemble IL-1β/IL-18 activating inflammasomes, NLRC4 and NLRP1, but have not been linked to monogenic diseases. Upon activation, these NLR sensors oligomerize and enable the proteolytic activation of caspase-1 through homotypic CARD-CARD interactions4–6. Active caspase-1 then cleaves proIL-1β and proIL-18 to their active forms and induces pyroptotic cell death through caspase-dependent but IL-1β/IL-18-independent mechanisms1,7.
Dominant mutations in two NLR genes, NLRP3 and NOD2, have previously been associated with the autoinflammatory diseases CAPS and Blau Syndrome, respectively. CAPS presents with fever (often cold-induced), neutrophilic urticaria, and inflammatory hearing loss (familial cold induced autoinflammatory syndrome (FCAS): MIM 120100; and Muckle Wells Syndrome (MWS): MIM 191100). The most severe form of CAPS, known as Neonatal-Onset Multisystem Inflammatory Disease (NOMID: MIM 607115), is often caused by de novo mutations in NLRP3, and presents with continuous flares of fever and urticaria, early-onset hearing loss, aseptic meningitis, optic nerve atrophy, and bony overgrowth8. The dramatic response of CAPS symptoms to IL-1β blocking therapies validated the pivotal role of IL-1β in driving the phenotype9,10, and guided the successful use of IL-1 inhibition in other autoinflammatory syndromes8. Blau Syndrome (a.k.a. Pediatric Granulomatous Arthritis, MIM 186580) is an early-onset autoinflammatory disorder characterized by granulomatous inflammation of the skin, synovium, and uveal tract11, and is most commonly associated with inappropriate NF-kB signaling rather than inflammasome activation12.
In our protocol investigating and treating patients with early-onset autoinflammatory diseases (NCT00059748), we evaluated a seven-year-old Caucasian female with recurrent episodes of fever, malaise, splenomegaly, vomiting, loose stools with mild duodenitis, and occasional rash beginning at six months of age (Fig. 1a–d). Fever episodes were initially attributed to recurrent viral infections, but prolonged flares requiring corticosteroid and colchicine treatment and negative evaluations for infection, immunodeficiency, and malignancy suggested an autoinflammatory syndrome (Supplementary Note and data not shown). Over time, chronic inflammation and corticosteroid dependence contributed to poor growth (Supplementary Fig. 1a). On laboratory evaluation, the combination of elevated inflammatory markers, chronic anemia, transaminitis, hypertriglyceridemia, hyperferritinemia, leucopenia and thrombocytopenia with severe flares were not consistent with CAPS but suggestive of Macrophage Activation Syndrome (MAS) (Fig. 1e and Supplementary Fig. 1b).
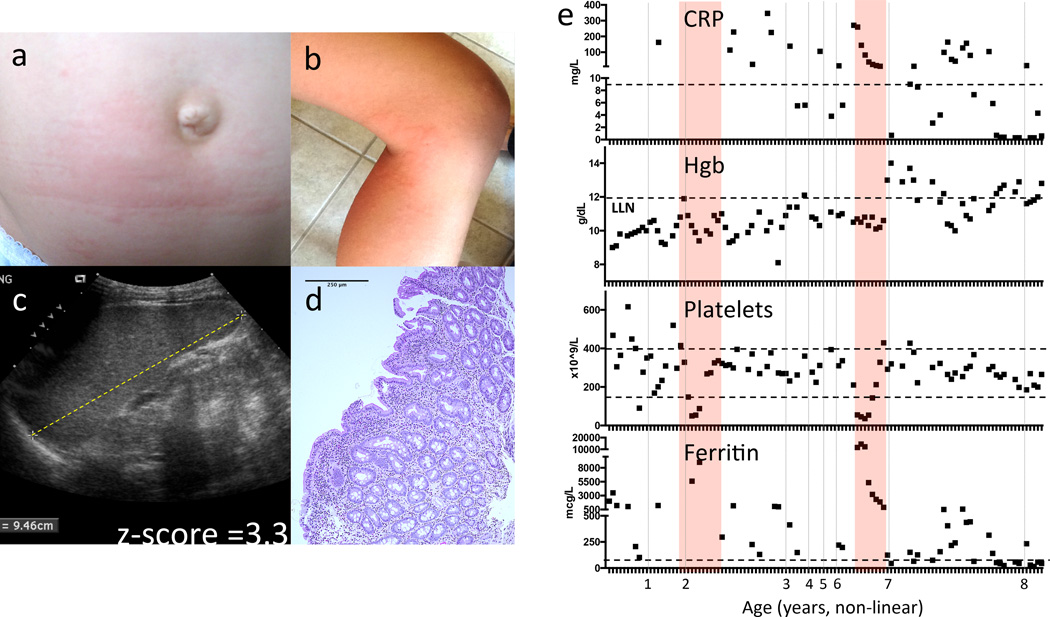
Figure 1. Severe flares in a patient with an NLRC4 mutation are consistent with Macrophage Activation Syndrome.
(a) and (b) Evanescent, urticaria-like rashes with dermographism during flares. (c) Sonographic measurement of splenomegaly with longest dimension equivalent to an age-matched z-score of 3.3239. (d) H&E micrograph of duodenal mucosal biopsy obtained at 18 months of age for vomiting and poor weight gain (20× magnification) showing non-specific mixed inflammatory infiltrate in the lamina propria and mild villous blunting. (e) Laboratory markers used to monitor Macrophage Activation Syndrome (MAS). The C-reactive Protein (CRP) is elevated, Hemoglobin (Hgb) is depressed, Platelet count is depressed, and serum ferritin rises during flares of MAS. All of the patient’s available laboratory data since birth are plotted (see also Supplementary Table 4). Pink bars indicate severe disease flares. Dashed lines indicate the normal ranges. The x-axis indicates the patient’s age in years (non-linear).
MAS and the related entity Hemophagocytic Lymphohistiocytosis (HLH) are life-threatening systemic immune dysregulatory conditions associated with uncontrolled macrophage activation and hemophagocytosis13,14. MAS/HLH flares present with fevers, pancytopenia, elevated triglycerides, impaired NK cell killing, and very high serum ferritin, and if left untreated can progress to coagulopathy, organ failure, and death. MAS is a known complication of some rheumatic diseases (including Systemic Lupus Erythematosus, Adult-onset Still’s Disease (AOSD), and systemic Juvenile Idiopathic Arthritis (JIA)), but is rarely observed in CAPS or other periodic fever syndromes8,9,15. MAS/HLH can also be triggered by infections (e.g. Epstein-Barr Virus) and malignancies. Insights into the pathogenesis of familial HLH came from genetic discoveries that loss-of-function mutations in PRF (encoding perforin) or other genes necessary for perforin/granzyme-mediated killing result in unchecked activation and survival of macrophages and dendritic cells13,16,17. This association has shaped the concept that impaired cytotoxic cell killing drives the pathogenesis of HLH and possibly MAS.
NK cell function was normal in our patient, and genetic testing for HLH and for periodic fever syndromes was negative (see Supplementary Note). To pursue a molecular diagnosis, she underwent whole exome sequencing (WES), which revealed a heterozygous de novo mutation resulting in a p.Thr337Ser substitution in a highly conserved region of the NLRC4 nucleotide-binding domain (NBD) (Fig. 2a,b, Supplementary Fig. 2a). The variant was confirmed by Sanger sequencing and predicted to be pathogenic on the basis of conservation, pathogenicity prediction packages18–21, and absence from the Exome Sequencing Project (ESP) database of more than 6500 control alleles22 and an in-house collection of >150 exomes (Fig. 2b and Supplementary Fig. 2a,b). Conformational analysis based on the crystal structure of the highly homologous murine NLRC4 (recently solved by Hu et al.6) suggested that this mutation may destabilize Helical Domain 1 interactions with NBD residues 170 and/or 173, or directly affect ADP binding, any of which may be essential for maintaining NLRC4 in an auto-inhibited conformation (Fig. 2c)6. A nearby heterozygous NLRC4 mutation, p.Val341Ala, is reported by Romberg et al. to present similarly as recurrent MAS23.
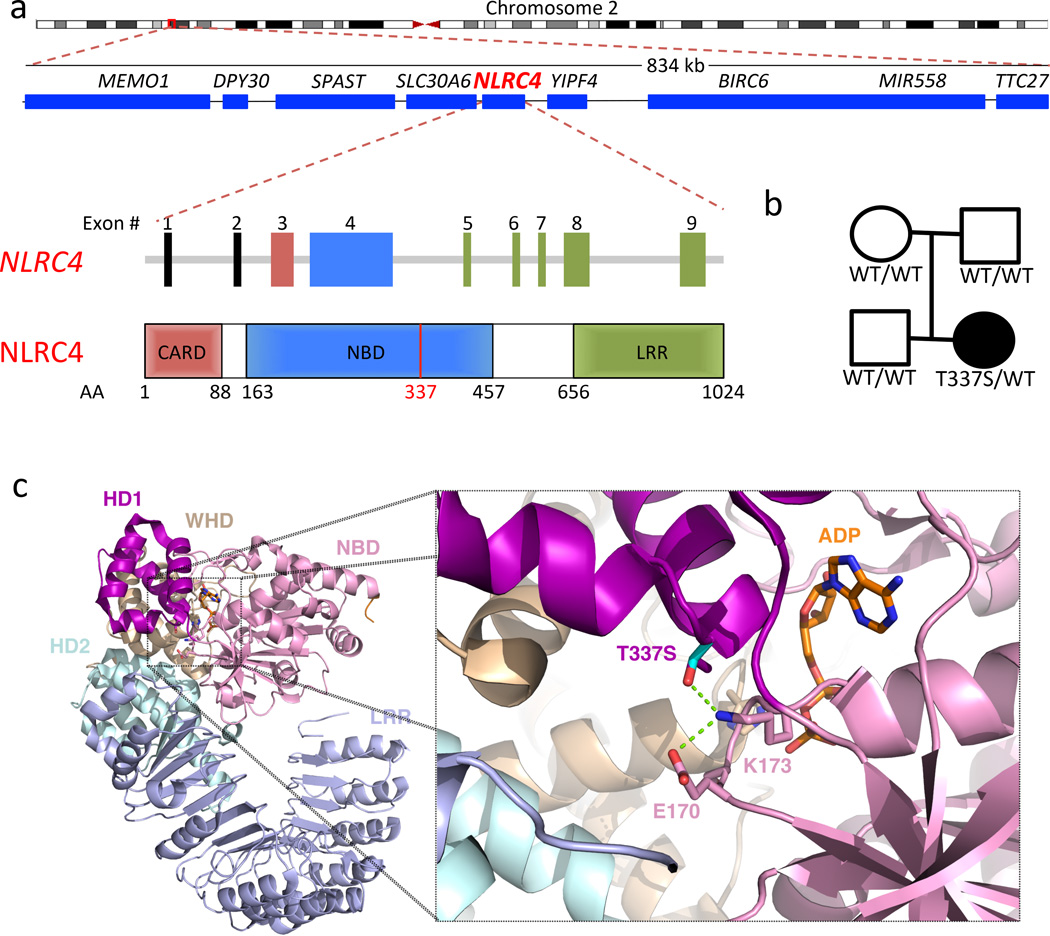
Figure 2. The NLRC4 mutation occurs de novo in a highly conserved area of the nucleotide binding domain.
(a) Chromosomal location, adjacent genes, and exon structure of the NLRC4 gene and domain structure of the NLRC4 protein. AA = Amino Acid number. (b) Pedigree showing a de novo heterozygous mutation. The affected patient is denoted by a filled circle. (c) Location of amino acid substitution from threonine to serine at position 337 and R-group interactions with adjacent residues in the NBD which are predicted to be important for stabilizing ADP-binding (based on the crystal structure of the murine protein6). NBD = Nucleotide Binding Domain, WHD = Winged-Helix Domain, HD = Helical Domain, LRR = Leucine Rich Repeat Domain. Green dashed line indicates predicted side chain interactions.
We hypothesized that this mutation may result in increased NLRC4 inflammasome activity and NLRC4-mediated MAS (subsequently referred to as NLRC4-MAS). Corroborating this hypothesis, we found a cluster of serum cytokines elevated in NLRC4-MAS but not controls or NOMID patients9,24, including the inflammasome-activated cytokine IL-18 (8,316 to 17,355 pg/mL in NLRC4-MAS, 102 to 1,281 pg/mL in NOMID, and 56 to 105 pg/mL in controls, Fig. 3a). Comparably high levels of serum IL-18 have also been associated with MAS in systemic JIA25, AOSD26, infection27, and XIAP deficiency28. Other serum markers that clustered distinctly in NLRC4-MAS included macrophage stimulating cytokines (M-CSF, IL-12p4029), apoptotic factors (TRAIL, LTα), chemokines (CCL7, CXCL12, IL-16), and hematopoietic growth factors (SCF, IL-3) (Fig.3a and Supplementary Fig. 3). Transcriptome analysis comparing the NLRC4-MAS patient with NOMID patients and healthy controls similarly showed upregulation of genes associated with apoptosis and dysregulation of genes associated with macrophage activation (Supplementary Table 1 and Supplementary Fig. 4), particularly in an NLRC4-MAS disease flare sample. These findings are consistent with macrophage stimulation/activation and upregulation of apoptosis and hematopoiesis30. We also found the calgranulins S100A8, S100A9, and S100A12 were among the most highly upregulated genes during an NLRC4-MAS flare, but were minimally upregulated in active NOMID patients (Supplementary Table 2, Fig. 3b). Increased calgranulin expression was not associated with granulocytosis (Supplementary Fig. 5). When calgranulins are released from activated monocytes and neutrophils, they act as damage-associated signals to drive pro-inflammatory responses31. They are particularly elevated in sepsis32, systemic JIA/AOSD, and Familial Mediterranean Fever, but not CAPS33, suggesting induction of the calgranulin damage response may help differentiate NLRC4-MAS from NLRP3-driven inflammation.
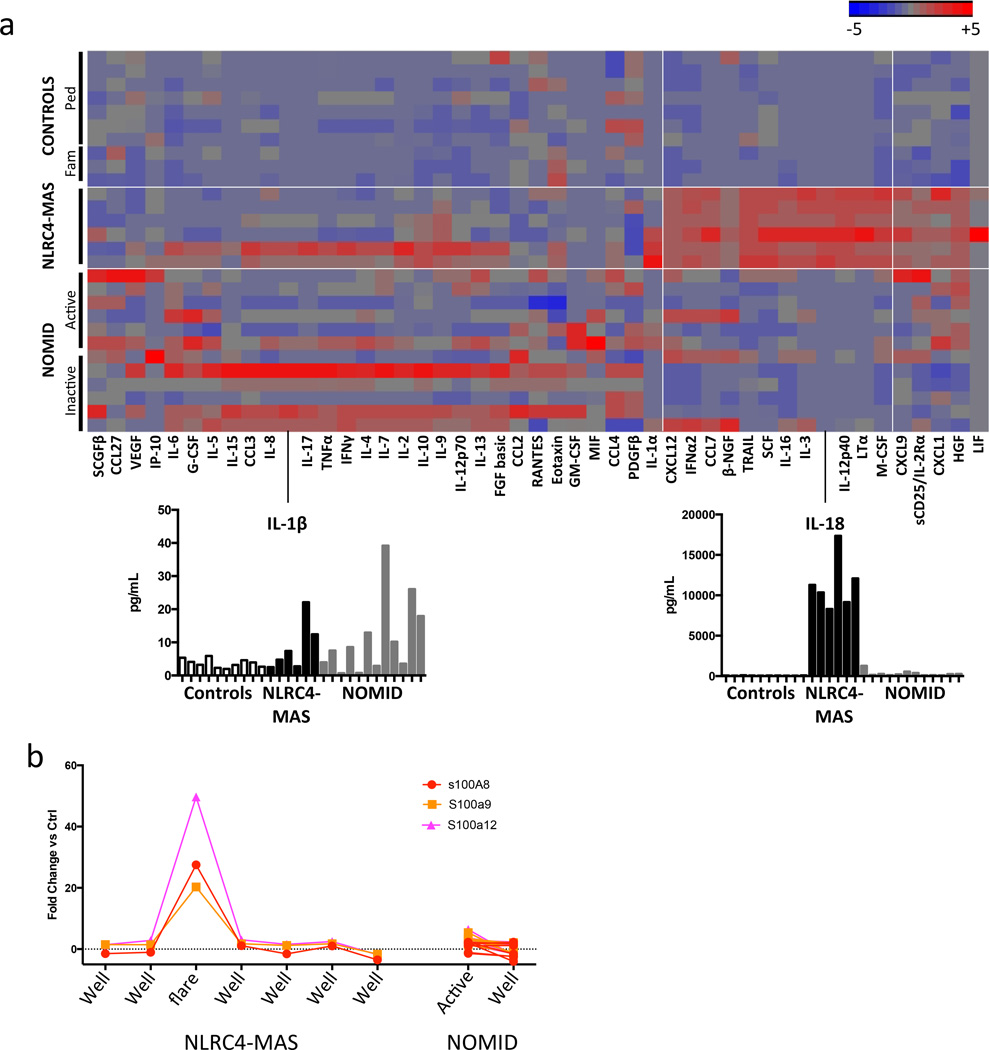
Figure 3. Peripheral blood signatures differentiate NLRC4-MAS from healthy controls and NOMID patients.
(a) We performed multiplex cytokine analysis on serum from the NLRC4-MAS patient, healthy pediatric controls, the patient’s family, and NOMID patients. NLRC4-MAS patient samples were obtained at seven distinct timepoints over the course of 10 months. Paired NOMID samples were obtained from six patients during both clinically active and inactive disease. No serum sample during a clinical flare from the NLRC4-MAS patient was available for cytokine analysis. Values represent the mean of two technical replicates per sample. Hierarchical clustering was performed on cytokine values normalized to the mean value across all samples. The magnitude of serum IL-1β and IL-18 concentrations are shown below. Horizontal white lines separate disease groups and vertical white lines indicate cytokines that form the NLRC4-MAS signature. (b) Whole blood transcriptional analysis of S100A genes from the NLRC4-MAS patient and matched samples from seven NOMID patients during active or inactive disease. All patient values are expressed as Fold Change (FC) above or below the average expression in 5 healthy controls.
Because NLRP3 mutations lead to increased inflammasome activation and IL-1β/IL-18 secretion from monocytes and macrophages, we assessed these responses in our patient’s cells. Whereas ATP is a canonical trigger of the NLRP3 inflammasome, murine studies suggest that NLRC4 inflammasome activation is triggered by cytosolic flagellin. Unlike Toll-like Receptor 5, which recognizes flagellin at the cell surface, the NLRC4 inflammasome recognizes a distinct region of flagellin that has been injected through a bacterial secretion system (as seen in S. typhimurium or L. pneumophila infection)34. We assessed spontaneous and stimulated responses in patient monocytes and monocyte-derived macrophages after priming with lipopolysaccharide (LPS) and stimulating with triggers of NLRP3 (ATP) and NLRC4 (intracellular flagellin, IC-FLA) inflammasome activation.
As expected, monocytes from the NLRC4-MAS patient and from a NOMID patient (NLRP3, G569R) produced significantly more IL-1β and IL-18 with all stimuli than healthy controls, but no difference in IL-1β secretion was seen between NOMID and NLRC4-MAS monocytes (Fig. 4a). However, NLRC4-MAS macrophages secreted more IL-1β than NOMID macrophages. Interestingly, IL-18 was constitutively secreted from NLRC4-MAS monocytes and macrophages in contrast to NOMID and healthy controls (Fig. 4a). Upon stimulation, IL-18 was not significantly further upregulated in monocytes but it was significantly upregulated in stimulated macrophages. IL-18 levels were 4–5 times higher in stimulated macrophages than monocytes, pointing to activated macrophages as an important source of IL-18. (Fig. 4a and Supplementary Fig. 6). IC-FLA-induced cytokine production was sustained with prolonged stimulation, suggesting that the mutation affects both the kinetic and amplitude of the cytokine response (Supplementary Fig. 7). Production of the inflammasome-independent cytokines IL-6, TNFα, and IL-10 was elevated in NLRC4-MAS monocytes compared to healthy and NOMID controls, particularly prior to IL-1 blocking therapy (Supplementary Figs. 6 and 8). However, these cytokines were not overproduced by matched monocyte-derived macrophages from the NLRC4-MAS patient (Fig. 4a, Supplementary Figs. 6, 7, and 8). Together, these results suggest increased responsiveness of NLRC4-MAS monocytes, but specific over-production of IL-1β and IL-18 by NLRC4-MAS macrophages, and differential roles for the NLRP3 and NLRC4 inflammasomes in inducing IL-18 activation and secretion.
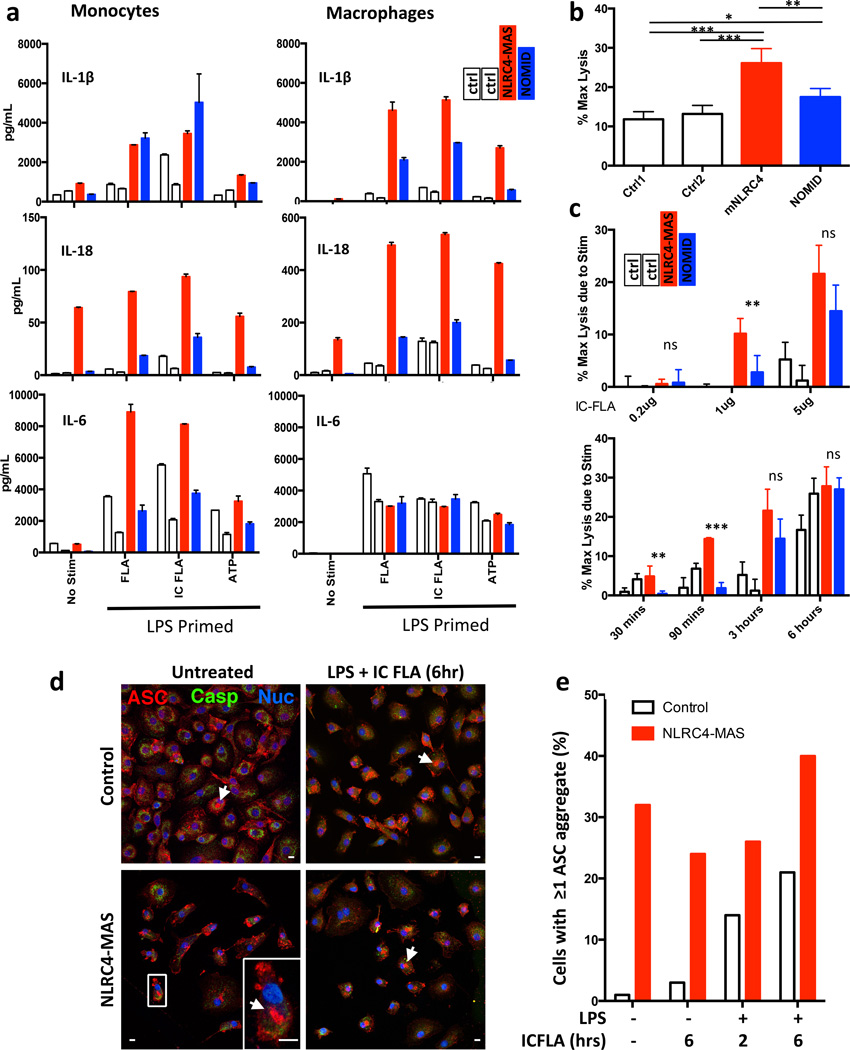
Figure 4. NLRC4-MAS monocytes and macrophages have increased inflammasome-related cytokine secretion, cell death, and ASC aggregate formation.
(a) Monocytes and monocyte-derived macrophages from healthy controls (white bars) NLRC4-MAS (red) and NOMID patients (blue) were primed with LPS and stimulated with purified flagellin (FLA), liposomal flagellin (IC-FLA), or ATP. Secreted cytokines were measured by Luminex. Columns represent mean and SD of technical duplicates. Monocytes and macrophages represent matched samples. In LPS-primed cells, the addition of ATP did not alter cytokine production (data not shown). (b and c) LDH release as a marker of cell death, presented as the percentage of maximum lysis induced by 0.8% Triton X-100, was measured from macrophages after resting for six hours in serum-free medium alone (b) or after stimulation (c) with varying doses of IC-FLA for three hours (top) or with 5µg/mL IC-FLA for indicated time periods (bottom). Cell death assays were performed in biological quadruplicates. Mean and SD are representative of two independent experiments. Statistical comparisons were done by one-way ANOVA with Tukey’s Post-test. *p<0.05, **p<0.01, ***p<0.0001. Comparisons between NLRC4-MAS and NOMID are depicted. (d) and (e) Macrophages were stimulated as noted and imaged by confocal immunofluorescence microscopy. Red staining represents ASC, Green indicates caspase-1, and blue indicates nuclei. Representative images (d) with a detail of a representative ASC aggregate (arrowheads and inset) were taken and ASC aggregate formation (e) was quantitated. A 10µm scale bar is shown. All samples from NLRC4-MAS and NOMID patients (a through e) were obtained after IL-1 receptor antagonist treatment for at least 3 months.
Inflammasome activation also results in an inflammatory cell death termed pyroptosis35. To evaluate spontaneous and stimulated cell death, human monocyte-derived macrophages were cultured with or without IC-FLA stimulation and monitored for lactate dehydrogenase (LDH) release. Spontaneous cell death was significantly higher in patient macrophages than in healthy controls or NOMID macrophages (Fig. 4b). Similarly, cell death specifically attributable to IC-FLA stimulation was substantially higher in the patient’s macrophages than controls (Fig. 4c).
Inflammasome activation triggers the oligomerization of Apoptosis-associated Speck-like Protein with CARD domain (ASC/PYCARD) into aggregates that are necessary for caspase-1 activation and cytokine production1 in a process that can be visualized by fluorescence microscopy36. We assessed spontaneous and stimulated ASC aggregation in NLRC4-MAS and control monocyte-derived macrophages. NLRC4-MAS macrophages displayed spontaneous ASC aggregation that was not substantially altered by LPS and/or IC-FLA stimulation, whereas control macrophages showed stimulation-dependent ASC aggregation (Fig. 4d,e). These results corroborate constitutive inflammasome activation in NLRC4-MAS macrophages.
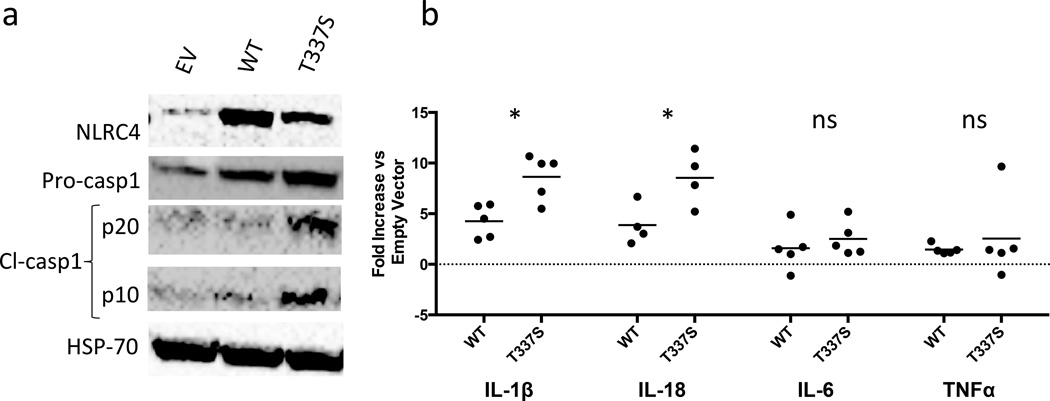
To investigate the activating effect of the NLRC4 mutation, we stably transduced WT or T337S mutant NLRC4 constructs, or empty vector (EV) virus into THP1 myelomonocytic cells and assessed inflammasome activity. THP1 cell expression of mutNLRC4 was consistently less than WT, despite infecting with up to 200 fold more mutant than WT virus, suggesting a proliferative disadvantage of mutant-transduced cells (Supplementary Fig. 8 & Fig. 5a). Despite lower NLRC4 expression, we measured increased spontaneous caspase-1 cleavage in mutNLRC4-transduced THP1 cells (Fig. 5a). Macrophages derived from mutNLRC4-transduced THP1 cells also showed enhanced secretion of IL-1β (mutNLRC4=5.5–10.6 fold over EV versus wtNLRC4=2.4–5.9 fold over EV, p=0.01) and IL-18 (mutNLRC4=5.2–11.4 fold over EV versus wtNLRC4=2.1–6.7 fold over EV, p=0.03) (Fig. 5b). These findings support gain-of-function of the NLRC4 mutation through constitutive caspase-1 activation.
Figure 5. Cells transduced with mutant NLRC4 exhibit spontaneous inflammasome activity.
THP1 monocytes were stably transduced with empty retrovirus or retrovirus carrying WT or T337S-mutant NLRC4 and differentiated into macrophages per Online Methods. (a) Immunoblots of whole cell lysates from transduced THP1 cells. (b) cytokines were measured by ELISA in culture supernatants from WT or mutant NLRC4-transduced THP1-cells differentiated into macrophages with low-dose Phorbol 12-myristate 13-acetate (PMA) in technical triplicate per online methods. The values expressed represent the average concentration of the technical replicates for WT- or T337S-transduced macrophages divided by the average concentration of technical replicates for EV-transduced macrophages within the same experiment. Each point represents an individual experiment. *p<0.05 by unpaired student’s T-test.
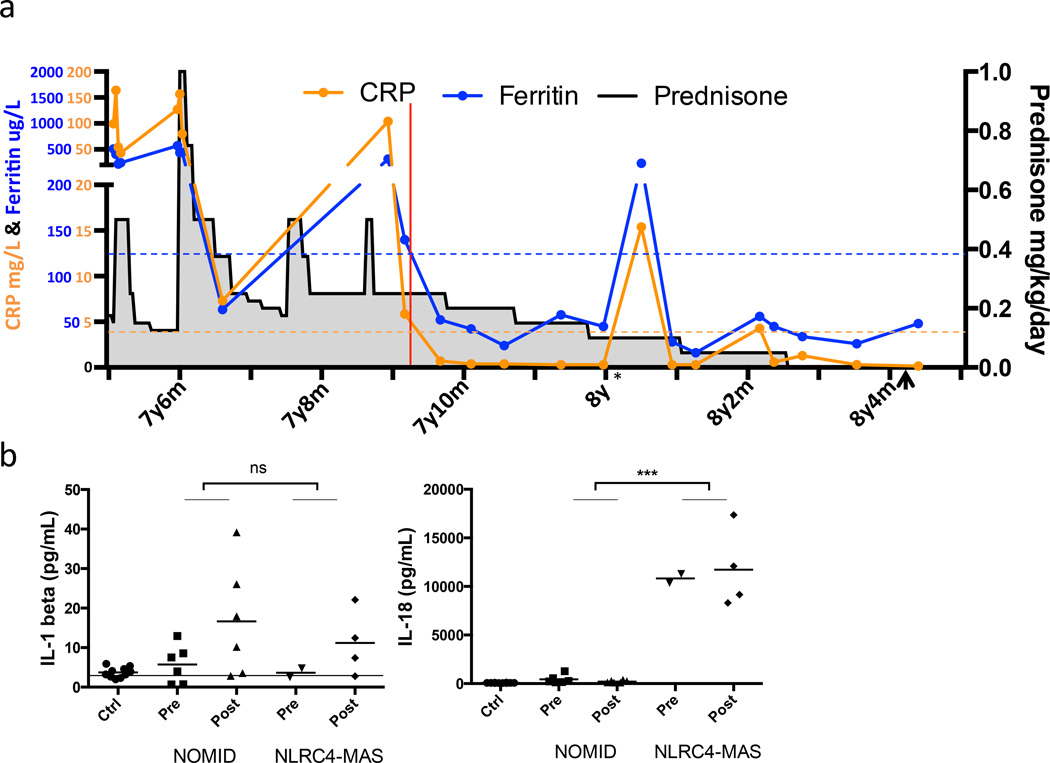
Given the successful use of IL-1 blocking strategies in NLRP3-mediated diseases8, the patient began treatment with recombinant IL-1 receptor antagonist (anakinra). The patient’s flare frequency, C-reactive protein, splenomegaly and prednisone dose were substantially reduced following IL-1 blocking therapy (Fig. 6a, Supplementary Fig. 1b,c, and Supplementary Note). After 7 months of treatment, the patient has had no fevers or clinical flares and has weaned off corticosteroids and colchicine (Fig. 6a). She has had no treatment-related adverse events, but has had transient transaminitis and elevated LDH and ferritin levels following a brief viral illness. Consistent with ongoing subclinical inflammation, her pattern of serum cytokinemia has not normalized following treatment with anakinra (Fig. 3, Supplementary Fig. 3), and cytokine production by her monocytes and macrophages did not change with treatment (Fig. 4, Supplementary Figs. 6 and 7).
Figure 6. IL-1 receptor antagonist treatment normalized markers of systemic inflammation and enabled cessation of steroids, while serum IL-18 remained elevated.
(a) Chronologic depiction of C-reactive protein (CRP), serum ferritin, and prednisone dose before and after IL-1 receptor antagonist (anakinra) therapy. Red line indicates the start of anakinra treatment. Dashed lines represent the upper limits of normal. * Indicates a transient viral illness with temporary elevation of CRP and ferritin. Arrowhead indicates cessation of colchicine therapy. (b) Serum cytokines were assayed in the NLRC4-MAS patient, healthy pediatric controls, the patient’s family, and matched samples from six NOMID patients pre and post anakinra treatment. Control samples are grouped together. Horizontal line represents lower limit of detection. ***p<0.0001 for unpaired two-tailed student’s T-test of all NOMID patient samples versus all samples from NLRC4-MAS patient.
Upon activation, NBD-mediated oligomerization of NLR proteins is critical for subsequent effector function4–6. Our study and the study by Romberg et al.23 link gain-of-function mutations in the NBD of NLRC4 to the clinical and immunologic phenotype of MAS. Mutations in the NBD of NLRP3 cause constitutive inflammasome activity and CAPS, and gain of function mutations in the NBD in CARD15/NOD2 cause Blau syndrome11. The presence of similar activating mutations in these NLRs, the strong species conservation of the NBD domain, and the role of the NBD in mediating autoinhibition6 support constitutive NLR oligomerization as an important mechanism in these three diseases. Unlike in CAPS/NOMID patients, the NLRC4 mutation in our patient causes very high circulating and stimulated levels of IL-18, similar to levels seen in other MAS-prone conditions26–28,37. The factors and molecular mechanisms that differentially regulate NLRC4 versus NLRP3 inflammasome activation are largely unknown and require further study. In addition, IL-18 production by non-hematopoietic cells (osteoblasts, adrenal cortex, intestinal epithelia, etc.38) may incrementally contribute to the constitutively high serum IL-18 levels in NLRC4-MAS versus CAPS patients.
Our functional data are consistent with a gain-of-function of the NLRC4 protein and support the mechanism of pathogenic macrophage activation with IL-1β and IL-18 overproduction and increased pyroptosis. Our data suggest for the first time that a macrophage-intrinsic defect can drive the MAS phenotype in the absence of a primary cytotoxic defect, thus providing a new paradigm for the pathogenesis of MAS. Activating mutations in NLRC4 cause a novel autoinflammatory disease presenting as periodic fever syndrome with MAS, distinct from CAPS and Blau Syndrome, and partially responsive to IL-1 inhibition. The persistence of high serum IL-18 levels and macrophage hyper-responsiveness even after IL-1 blocking treatment suggest a role for IL-18 in MAS and warrant an exploration of IL-18 as a therapeutic target.
Online Methods
Human Subjects
All research investigations were done as part of protocol 03-AR-0173 and as such were approved by the NIAMS/NIDDK Institutional Review Board. Written informed consent for conduct of research and publication of findings was obtained from the parents of all patients/healthy controls involved, and assent was obtained from the patients/healthy controls when possible.
Whole Exome Sequencing
Agilent SureSelect® Human 51 Mbp All Exon Kit (Agilent Technologies, Santa Clara, CA) was used for exome capture. Sequencing was performed on Illumina® HiSeq sequencers (Illumina, San Diego, CA) using 2×100 bp paired reads. The typical average on-target coverage is about 66×. A computational pipeline was developed to process the read data and perform tasks such as quality control (QC), variant discovery, annotation and filtering. Briefly, the sequencing reads in FASTQ format were aligned to the human reference genome (GRC B37) with the BWA (Burrows-Wheeler Aligner) mapping tool. The resulting BAM files (one per sample) were further processed to remove duplicate reads, refine alignment around indels and recalibrate base quality scores, according to the best practice guideline by the Genome Analysis Toolkit (GATK) from the Broad Institute. A variant caller (UnifiedGenotyper) from GATK was used to make joint variant calls across multiple samples, followed by a variant quality score recalibration step by the GATK VQSR tool. Besides quality scores, the variants were annotated with functional impact and allele frequency in public databases and local datasets. Sample relationship and gender were checked to identify potential sample mix-up and false family relationship. Likely disease-causing mutations were selected and prioritized based on quality score, allele frequency, functional impact, probable inheritance model (de novo mutation, autosomal recessive, autosomal dominant, or X-linked), and expert evaluation.
Sanger Sequencing
The reference sequence used for primer design and nucleotide numbering was NLRC4 NM_001199139. The exonic regions and flanking intronic sites of the gene were amplified by polymerase chain reaction (PCR) using specific primers (IDT, Coralville, IA) designed with Exon Primer (http://ihg.gsf.de/ihg/ExonPrimer). Supplementary Table 3 contains a list of all sequencing primers. The first PCR amplification reaction was performed in a final volume of 13 µL containing 10 picomoles of each primer, 6 µL of GoTaq® Hot Start Master Mix (Promega, Madison, WI) and 20–200 ng of template DNA. The reactions were carried out for 35 cycles: denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 1 minute. After the PCR assay, the DNA fragments were treated with Exo-Sap (GE Healthcare Bioscience, Pittsburgh, PA). PCR products underwent a second PCR reaction with ABI Prism® BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and directly sequenced in both directions using ABI 3100 Genetic Analyzer (Applied Biosystems). Results were analyzed using DNAStar Lasergene software. Targeted sequencing of the other 7 coding exons of NLRC4 was performed in the patient for the screening of single nucleotide polymorphisms (SNPs).
Gene-Expression Analysis by RNA Sequencing (RNA-Seq)
Total RNA was extracted from human blood samples collected in PaxGene tubes (Qiagen, Valencia, CA). Samples were obtained from five pediatric healthy controls, the NLRC4-MAS patient at seven different timepoints, seven NOMID patients with active disease prior to anakinra treatment, and the same seven NOMID patients with inactive disease following anakinra treatment. Complete blood counts were drawn on the same day for the NLRC4-MAS patient. RNA integrity was analyzed with Agilent 2100 Bioanalyzer. mRNA purification and fragmentation, complementary DNA synthesis and target amplification were performed using the Illumina® TruSeq RNA Sample Preparation Kit (Illumina). Pooled cDNA libraries were sequenced using HiSeq 2000 Illumina® platform (Illumina). Sequencing results were analyzed using Partek GS v6.6 software and are expressed in RPKM (reads per kilobase exon per million mapped). They are available via Gene Expression Omnibus accession number GSE57253. All RPKM values were offset by the addition of 0.5 prior to analysis to limit the effects of low-expressing samples40. Lists of differentially expressed genes (DEGs) between the NLRC4-MAS flare sample and controls were performed without statistical analysis. Lists of DEGs between NOMID patients and controls were generated by ANOVA analysis using Partek GS v6.6 software. Candidate gene lists for “Inflammatory Cytokines & Receptors,” “Apoptosis,” “Inflammasomes,” and “Type I Interferon Response,” were obtained from the human RT2 Profiler, while “Macrophage Activation” was derived from Biswas et al.41 (http://www.sabiosciences.com/NanoPreAMP_PCRArrays.php, and Supplementary Table 1). Overlapping genes between lists were eliminated. Differential expression of candidate genes was evaluated by generating fold changes between individual patient samples and the average RPKM across healthy controls. When multiple transcript IDs were present for a gene, the ID with the highest average expression in healthy controls was chosen.
Serum Cytokine Analysis
Sera were stored at −80°C. They were then analyzed using the Bio-Plex 21- and 27-plex Suspension Array System in duplicate per manufacturer’s specifications. All sera were analyzed simultaneously to avoid batch effects. Values below the limit of detection were reset at the limit of detection. Two-way hierarchical clustering was performed using Partek software.
Plasmids
A pCMV-SPORT6 vector containing the NLRC4 cDNA MGC: 35330 was obtained from Thermo Scientific. Subsequent cloning was performed by Bioinnovatise, Inc (Rockville, MD). Briefly, after DpnI digestion, the A to T mutation at position 1009 of NLRC4 was introduced into this plasmid using site-directed mutagenesis and complementary primers (see Supplementary Table 3) containing the patient’s mutation. An IRES-eGFP sequence was then cloned downstream of the WT and mutant NLRC4 open reading frames by seamless cloning. Finally, a 24 bp c-terminal FLAG tag was appended to each construct to generate pCMV-Sport6-NLRC4flag-GFP and pCMV-Sport6-T337SNLRC4flag-GFP. PCR fragments containing either WT or T337S mutant NLRC4flag were inserted into the multiple cloning site of the migR1 vector42 by seamless cloning to generate NLRC4flag-MigR1 and T337SNLRC4flag-MigR1. All constructs were completely sequenced to confirm position.
Transductions
THP1 monocytes (ATCC TIB-202) were maintained at 100,000–600,000 cells/mL in RPMI 1640 containing 10% fetal calf serum, 50µM β-mercaptoethanol, 50U/mL Streptomycin, and 20U/mL Penicillin (R10 medium). Equal amounts of empty MigR1, NLRC4flag-MigR1, and T337SNLRC4flag-MigR1 constructs were transfected into the Phoenix-Ampho cell line (ATCC CRL-3213) along with a pCl-Ampho helper plasmid. 1×106 THP1 cells were centrifuged with filtered, undiluted supernatants from transfected Phoenix cells. Transduced THP1 cells were then grown as above in R10 medium supplemented with 1× Non-essential Amino Acids (Gibco), 10mM HEPES, and 1mM Sodium Pyruvate (R10C). Transduced cells were sorted for viability and GFP positivity on a BD FACS Aria II about 1 week post transduction to >95% GFP positivity. Sorted cells were then passaged in R10C and stability of GFP expression assessed by flow cytometry at least weekly. Two different transduced cell lines were produced in the same manner and tested separately to ensure reproducibility. GFP positivity correlated with NLRC4 protein expression assessed by western blot (Fig. 5A).
Antibodies and immunoblotting
5×105 transduced THP1 cells were subjected to SDS-PAGE, transferred to nitrocellulose membranes and probed with appropriate antibodies. Antibodies used for protein blots were NLRC4 antibody (1:500 dilution; Millipore, 06-1125), pro-caspase1 antibody (1:1000 dilution; Cell Signaling Technology, 2225), cleaved caspase-1 antibody (1:500 dilution; Cell Signaling Technologies, 4199), HSPA8 antibody (1:2000 dilution; Cell Signaling Technologies, 8444). Horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technologies, 7074) and SuperSignal West Pico chemiluminescent substrate (Pierce) were used to detect primary antibodies. Equal protein loading was assessed by immunoblotting for HSPA8.
Primary monocyte purification & generation of monocyte-derived macrophages
Monocytes were isolated from Peripheral Blood Mononuclear Cells to >95% purity using CD14+ Magnetic Bead separation (Miltenyi) per manufacturer’s instructions. Monocytes were then stimulated directly. Monocyte-derived macrophages were generated by culturing purified monocytes in R10C medium supplemented with 10ng/mL recombinant human M-CSF. Culture medium was refreshed every 2 days for 7 days.
Generation of THP1 macrophages
1×106 stably-transduced THP1 cells were cultured in R10C containing low-dose (10ng/mL) phorbol 12-myristate 13-acetate (PMA) for 48 hours to generate THP1-derived macrophages. Adherent cells were then washed with and rested in OptiMEM (Gibco) serum-free medium at 37°C for 30 minutes prior to stimulation.
Monocyte and Macrophage Stimulation
After resting in medium, primary or transduced cells were then primed with LPS 1µg/mL for 4 hours before being stimulated with purified flagellin from S. typhimurium (FLA-ST, Invivogen) in DOTAP liposomes (IC-FLA, 6µg DOTAP/1µg FLA-ST, Roche) for an additional two hours. IC-FLA stimulation was performed at 5µg/mL unless otherwise indicated. For ATP stimulation, cells were primed in LPS for 5 hours and then stimulated for 1 hour with 1mM ATP. LPS priming was omitted for timecourse experiments.
Supernatant Cytokine Measurement
Cytokine production from primary human monocytes and monocyte-derived macrophages was performed in duplicate on undiluted culture supernatants using the Bio-plex Xpress suspension array system and read on a Bio-Rad Bio-Plex 200 HTF per manufacturer’s instructions. IL-1β, IL-6, TNFα, IL-10 (BD OptEIA), and IL-18 (MBL) from THP1 transduction experiments were measured by sandwich ELISA. For experiments comparing WT or Mutant samples to Empty Vector (EV), a nominal offset value near the limit of detection was added to each reading prior to normalization to minimize the effects of low values.
Macrophage Cell Death
Spontaneous and stimulated cell death was evaluated by measuring LDH release from 20,000 monocyte-derived macrophages using the Cytotox96 Non-radioactive cytotoxicity assay per manufacturer’s instructions (Promega). No LPS priming was used for these experiments.
Immunofluorescence and quantitation of ASC aggregates
Primary monocyte-derived macrophages were differentiated on glass coverslips as described. Adherent macrophages were washed twice in PBS, primed with 10ng/mL LPS for 6 hours, followed by stimulation with 5µg/mL IC-FLA as noted. Cells were incubated with 2 µM biotinyl-YVAD-CMK (Enzo) during the last 4 hours of stimulation. Cells were then rinsed with PBS, fixed and permeabilized with ice cold methanol for 7 minutes at −20°C, and blocked in PBS containing 1% fish skin gelatin (Sigma), 2% Bovine Serum Albumin, 5% Normal Goat Serum for 1 h at room temperature (RT). ASC was detected with rabbit anti-ASC antibodies (Adipogen) in blocking buffer (one hour at RT). A streptavidin-FITC conjugate (BD biosciences) and anti-rabbit AlexaFluor 568 (Life Technologies) were used for secondary detection. Nuclei were counterstained with Hoechst 33342 and coverslips were mounted in Vectashield. Macrophages were analyzed on a Zeiss 780 confocal microscope using a 63× or 40× oil immersion objective. Quantitation of ASC aggregates was performed visually at 63× in at least 100 cells adjacently positioned along an arbitrary line.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. SC was also supported by the Arthritis National Research Foundation. The authors are grateful to Drs. Oscar Navarro and Gino Somers for assistance with data acquisition, and Ms. Holly Convery and the patient and her family for assistance with research coordination.
Footnotes
Author Contributions: SWC and RGM conceived the study. SWC, JJO, RML and RGM directed the study. GAM, HK, DC, NP, YH, SB, and RML coordinated patient care and obtained clinical samples. AAJ performed Sanger sequencing. AAJ and TAF generated RNA-seq libraries. SWC, AAJ, SRB, AVV, and ZD analyzed genomic and transcriptional data. SWC and AB performed serum cytokine experiments and analysis. MAD performed structural analysis. SWC and SG performed all stimulation experiments, with assistance from BM and YL. SWC, SG, and KJMZ performed fluorescence microscopy, KJMZ performed the quantitation. JAD and AG provided reagents and critical direction. SWC, RML, and RGM guided clinical assessment and intervention. SWC and RGM wrote the manuscript with the assistance and final approval of all authors.
URLs:
Exome Variant Server: http://evs.gs.washington.edu/EVS/) [accessed 04/2014]
References
- 1.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ting JP, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. 2013;39:432–441. doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 7.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montealegre Sanchez GA, Almeida de Jesus A, Goldbach-Mansky R. Monogenic autoinflammatory diseases: disorders of amplified danger sensing and cytokine dysregulation. Rheum Dis Clin North Am. 2013;39:701–734. doi: 10.1016/j.rdc.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldbach-Mansky R, et al. Neonatal-onset multisystem inflammatory disease responsive to interleukin-1beta inhibition. N Engl J Med. 2006;355:581–592. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lachmann HJ, et al. Use of canakinumab in the cryopyrin-associated periodic syndrome. N Engl J Med. 2009;360:2416–2425. doi: 10.1056/NEJMoa0810787. [DOI] [PubMed] [Google Scholar]
- 11.Miceli-Richard C, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 12.Martin TM, et al. The NOD2 defect in Blau syndrome does not result in excess interleukin-1 activity. Arthritis Rheum. 2009;60:611–618. doi: 10.1002/art.24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoller EE, et al. Hemophagocytosis causes a consumptive anemia of inflammation. The Journal of experimental medicine. 2011;208:1203–1214. doi: 10.1084/jem.20102538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canna SW, Behrens EM. Not all hemophagocytes are created equally: appreciating the heterogeneity of the hemophagocytic syndromes. Current opinion in rheumatology. 2012;24:113–118. doi: 10.1097/BOR.0b013e32834dd37e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horneff G, Rhouma A, Weber C, Lohse P. Macrophage activation syndrome as the initial manifestation of tumour necrosis factor receptor 1-associated periodic syndrome (TRAPS) Clin Exp Rheumatol. 2013;31:99–102. [PubMed] [Google Scholar]
- 16.Pachlopnik Schmid J, et al. Inherited defects in lymphocyte cytotoxic activity. Immunological reviews. 2010;235:10–23. doi: 10.1111/j.0105-2896.2010.00890.x. [DOI] [PubMed] [Google Scholar]
- 17.Terrell CE, Jordan MB. Perforin deficiency impairs a critical immunoregulatory loop involving murine CD8+ T cells and dendritic cells. Blood. 2013;121:5184–5191. doi: 10.1182/blood-2013-04-495309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 19.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 20.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7(Unit7 20) doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: [Google Scholar]
- 23.Romberg N. Under Review. Nature Genetics. 2014 Under Review. [Google Scholar]
- 24.Sibley CH, et al. Sustained response and prevention of damage progression in patients with neonatal-onset multisystem inflammatory disease treated with anakinra: a cohort study to determine three- and five-year outcomes. Arthritis Rheum. 2012;64:2375–2386. doi: 10.1002/art.34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu M, et al. Distinct cytokine profiles of systemic-onset juvenile idiopathic arthritis-associated macrophage activation syndrome with particular emphasis on the role of interleukin-18 in its pathogenesis. Rheumatology. 2010;49:1645–1653. doi: 10.1093/rheumatology/keq133. [DOI] [PubMed] [Google Scholar]
- 26.Ichida H, et al. Clinical manifestations of adult-onset still's disease presenting with erosive arthritis: Association with low levels of ferritin and IL-18. Arthritis Care Res (Hoboken) 2013 doi: 10.1002/acr.22194. [DOI] [PubMed] [Google Scholar]
- 27.Mazodier K, et al. Severe imbalance of IL-18/IL-18BP in patients with secondary hemophagocytic syndrome. Blood. 2005;106:3483–3489. doi: 10.1182/blood-2005-05-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada T, et al. Sustained elevation of serum interleukin-18 and its association with hemophagocytic lymphohistiocytosis in XIAP deficiency. Cytokine. 2014;65:74–78. doi: 10.1016/j.cyto.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Russell TD, et al. IL-12 p40 homodimer-dependent macrophage chemotaxis and respiratory viral inflammation are mediated through IL-12 receptor beta 1. J Immunol. 2003;171:6866–6874. doi: 10.4049/jimmunol.171.12.6866. [DOI] [PubMed] [Google Scholar]
- 30.Fall N, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56:3793–3804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 31.Kessel C, Holzinger D, Foell D. Phagocyte-derived S100 proteins in autoinflammation: putative role in pathogenesis and usefulness as biomarkers. Clin Immunol. 2013;147:229–241. doi: 10.1016/j.clim.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Tang BM, Huang SJ, McLean AS. Genome-wide transcription profiling of human sepsis: a systematic review. Crit Care. 2010;14:R237. doi: 10.1186/cc9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wittkowski H, et al. S100A12 is a novel molecular marker differentiating systemic-onset juvenile idiopathic arthritis from other causes of fever of unknown origin. Arthritis Rheum. 2008;58:3924–3931. doi: 10.1002/art.24137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol Cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willingham SB, et al. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2007;2:147–159. doi: 10.1016/j.chom.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Man SM, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu M, Nakagishi Y, Yachie A. Distinct subsets of patients with systemic juvenile idiopathic arthritis based on their cytokine profiles. Cytokine. 2013;61:345–348. doi: 10.1016/j.cyto.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Novick D, Kim S, Kaplanski G, Dinarello CA. Interleukin-18, more than a Th1 cytokine. Semin Immunol. 2013;25:439–448. doi: 10.1016/j.smim.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 39.Megremis SD, Vlachonikolis IG, Tsilimigaki AM. Spleen length in childhood with US: normal values based on age, sex, and somatometric parameters. Radiology. 2004;231:129–134. doi: 10.1148/radiol.2311020963. [DOI] [PubMed] [Google Scholar]
- 40.Warden CD, Yate-Ching Y, Xiwei Wu. Optimal Calculation of RNA-Seq Fold-Change Values. International Journal of Computational Bioinformatics and In Silico Modeling. 2013;2:285–292. [Google Scholar]
- 41.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 42.Pear WS, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.