Summary
Messenger RNA decay in Bacillus subtilis is accomplished by a combination of exoribonucleases and endoribonucleases. Intermediates in the decay process have not been readily detectable, and previous studies on mRNA decay have used a handful of highly expressed transcripts as models. Here, we use RNA-Seq analysis to probe mRNA turnover globally. A significant fraction of messages showed differential accumulation of RNA fragments that mapped near the 5’ or 3’ end of the coding sequence, consistent with initiation of decay from either the 5’ end or from an internal cleavage site. Patterns of mRNA decay in the wild type were compared with patterns in a mutant strain lacking polynucleotide phosphorylase (PNPase), which is considered the major 3’ exonuclease activity in mRNA decay and which is one of four known 3’ exonucleases in B. subtilis. The results showed a striking dependence on PNPase for mRNA turnover in many cases, suggesting specificity in the ability of 3’ exonucleases to degrade from 3’-hydroxyl termini. RNA-Seq data demonstrated a sharp decrease in expression of Sigma D in the PNPase-deletion strain. Reduction in sigD regulon expression explained the chain growth phenotype of the PNPase mutant and also predicted a defect in swarming motility.
Keywords: Bacillus subtilis, mRNA decay, polynucleotide phosphorylase
Introduction
Bacterial messenger RNAs have relatively short half-lives. RNA decay, which is necessary for recycling of nucleotides and for rapid changes in the gene expression program, is achieved by the activities of endoribonucleases and exoribonucleases, and this can occur in several ways. In one mechanism, mRNA decay initiates with an endonuclease cleavage, followed by 3’ exonucleolytic degradation of the upstream fragment. The downstream fragment is either subject to further endonuclease cleavages and 3’-to-5’ decay, or is degraded, in the organisms that have one, by a 5’-to-3’ exoribonuclease activity (Bechhofer, 2009, Lehnik-Habrink et al., 2012). An alternative mRNA decay mechanism initiates with conversion of the protective triphosphorylated 5’ end to a susceptible 5’-monophosphorylated end, by the action of an RNA pyrophosphohydrolase, and the mRNA is then a substrate for 5’-to-3’ exonucleolytic decay (Celesnik et al., 2007, Richards et al., 2011). It is likely that individual mRNAs are degraded by a combination of these mechanisms, as we have shown in Bacillus subtilis for the model mRNA △ermC (Yao et al., 2011). In some organisms, degradation from the 3’ end of mRNAs occurs by a polyadenylation-dependent mechanism (Dreyfus & Regnier, 2002); however, it is not known whether this latter mechanism is a significant one in B. subtilis (Bechhofer, 2009).
Four 3’-to-5’ exoribonucleases are known to exist in B. subtilis: polynucleotide phosphorylase (PNPase), RNase R, RNase PH, and YhaM (Luttinger et al., 1996, Mitra et al., 1996, Oussenko & Bechhofer, 2000, Craven et al., 1992, Oussenko et al., 2002). PNPase, encoded by the pnpA gene, releases nucleotides processively from the 3’ end by phosphorolysis, as does RNase PH; RNase R and YhaM are hydrolytic enzymes. Biochemical studies have suggested that PNPase is the major RNA turnover enzyme in B. subtilis (Deutscher & Reuven, 1991, Wang & Bechhofer, 1996). It has been proposed that phosphorolytic nuclease activity is important for organisms such as B. subtilis, which dwells in a nutrient-poor soil environment and needs to conserve phosphate bond energy (Deutscher & Reuven, 1991). We and others have shown that a pnpA deletion strain grows at a slightly slower rate than wild type and has several interesting phenotypes: cold sensitivity, chain growth in culture, competence deficiency, and extreme tetracycline sensitivity (Luttinger et al., 1996, Wang & Bechhofer, 1996). The basis of the cold sensitivity is likely to be similar to that found in E. coli, where the exonuclease activity of PNPase is required for adaptation to low temperature (Awano et al., 2008). The basis of the other pnpA strain phenotypes has not been clarified.
In accordance with the hypothesis that PNPase is the major mRNA decay enzyme, we have shown that the absence of PNPase results in an accumulation of decay intermediates for several small, monocistronic mRNAs (Bechhofer & Wang, 1998, Oussenko et al., 2005). In addition, dysregulation of the trp operon is observed in a strain lacking PNPase, due to impeded turnover of the regulatory trp leader RNA (Deikus et al., 2004). In vitro studies of purified B. subtilis PNPase on trp leader RNA demonstrated that PNPase requires a single-stranded 3’ extension for binding and is incapable of degrading past strong secondary structure (Deikus & Bechhofer, 2007). It is therefore expected that PNPase-mediated mRNA decay does not initiate at the native 3’ end, which abuts the transcription terminator structure, but from internal 3’ ends generated by endonuclease cleavage. RNase Y is believed to be the major endonuclease in the initiation of decay in B. subtilis (Shahbabian et al., 2009, Lehnik-Habrink et al., 2011, Durand et al., 2012).
In addition to PNPase, the three other B. subtilis 3’-to-5’ exoribonucleases may be involved in mRNA decay (Oussenko et al., 2005), but this has not been investigated in detail. B. subtilis contains a 5’-to-3’ exonuclease activity, RNase J1 (Even et al., 2005, Britton et al., 2007, Li de la Sierra-Gallay et al., 2008), and RNase J1 is involved in decay of full-length mRNA from the native 5’ end (Daou-Chabo et al., 2009, Richards et al., 2011, Yao et al., 2011) and in decay of mRNA fragments that are generated by endonuclease cleavage (Deikus et al., 2008, Yao & Bechhofer, 2010).
It has been thought for some time that bacterial mRNA decay occurs by an “all-or-nothing” mechanism (Kaberdin et al., 2011), i.e., the decay rate is dependent on an initial enzymatic reaction, such as endonuclease cleavage or removal of the 5’ pyrophosphate. After the initial reaction, the mRNA is degraded so rapidly by a combination of endonucleolytic and exonucleolytic activities that intermediates in the decay process do not accumulate and are difficult to detect. Indeed, on Northern blot analysis of individual mRNAs, one often observes full-length mRNA but not mRNA processing intermediates. For the current study, we anticipated that RNA-Seq analysis might give a more detailed picture of decay intermediates in the mRNA decay process. An earlier study on bacterial mRNA decay (Kristoffersen et al., 2012), which employed RNA-Seq methodology to determine mRNA half-lives in the related Gram-positive organism, B. cereus, motivated the current study. Importantly, RNA-Seq data obtained from B. subtilis wild-type and pnpA knockout strains could be used to address the role of PNPase in global mRNA decay.
Results
The B. subtilis transcriptome was probed by RNA-Seq analysis performed in triplicate on steady-state RNA isolated from wild-type and pnpA strains grown to mid-logarithmic phase in Luria-Bertani broth (see Experimental Procedures for details). The NCBI database reference genome (http://www.ncbi.nlm.nih.gov/genome/665) was used to map reads to coding sequences (CDSs), and 93%-98% reads from each RNA-Seq sample were unambiguously mapped to the B. subtilis genome (Fig. S1). Because of the uncertainty of transcription start and end sites, and the use of more than one promoter for a CDS in many cases (Nicolas et al., 2012), we used CDSs rather than transcription units to assign reads. Reads from antisense genes that could affect interpretation of results were excluded (see below). To differentiate background transcription from true expression, a cutoff of 1 read per base in at least one of the strains was used. Pearson coefficients of triplicate samples of the same strain were >0.95 (Fig. S2).
Of 4,176 genes in the B. subtilis database, 2,276 (~55%) were expressed above the cutoff level in one or both strains (Table S1). This is similar to earlier studies that found about half of B. subtilis genes were expressed in cells grown in rich medium (Rasmussen et al., 2009, Durand et al., 2012). In Table S2, CDSs are grouped into 20 functional categories, as defined by the SubtiWiki website (Michna et al., 2014). As expected for cells growing in exponential phase in rich medium, fewer genes involved in transport, metabolism, and sporulation were significantly expressed (29%-55%) compared to genes involved in cell division, ATP synthesis, nucleotide metabolism, and protein synthesis (63-81%). The differential gene expression data for wild-type and pnpA mutant strains shown in Table S2 is addressed later.
RNA processing in the wild-type strain
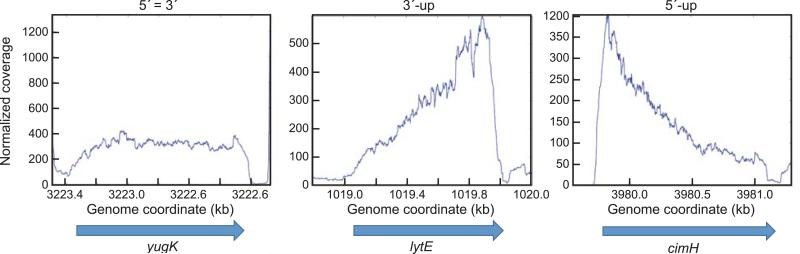
We analyzed RNA-Seq data from the wild-type strain, with an eye toward detecting distributions of reads that might reflect processing or turnover mechanisms. The level of reads from the 5’ one-third and the 3’ one-third of each gene was compared, to determine whether there was significant differential accumulation at either end. The data for all expressed genes is in Table S3, and the genes were divided into three categories: (1) those with a relatively equal number of reads in the 5’ one-third vs. the 3’ one-third of the CDS, i.e., the 5’/3’ ratio being less than 1.5 (referred to as the “5’=3’ category”); (2) those with ≥1.5-fold increased reads in the 3’ one-third relative to the 5’ one-third (referred to as the “3’-up category”); and (3) those with ≥1.5-fold increased reads in the 5’ one-third relative to the 3’ one-third (referred to as the “5’-up category”). Examples of RNA-Seq histograms for a gene in each of these three categories is shown in Fig. 1 (discussed below). Since the RNA-Seq data was not strand-specific, we took into account reads that might arise from antisense RNAs. Ten mRNAs whose 5’ or 3’ ends are located in genomic regions that are also the site of antisense transcription (Nicolas et al., 2012), and that could therefore affect the grouping into 5’-3’ categories, were not included in the analysis.
Fig. 1.
(A) RNA-Seq analysis of genes in three 5’-3’ ratio categories in the wild-type strain. Genome coordinates on the X-axis; normalized reads on the Y-axis. Extent of CDSs indicated by the rightward facing arrows.
Most of the mRNAs (67.5%) expressed in the wild-type strain above the cutoff level were in the 5’=3’ category, with no significant difference in read numbers from either end of the CDS. An example of mRNAs in this class is yugK (Fig. 1), which showed a uniform read distribution and for which the 5’/3’ ratio was 1.1.
We found that 26.9% of mRNAs above the cutoff were in the 3’-up category, with a ≥1.5-fold greater number of reads in the 3’ region compared to the 5’ region. The example of the lytE gene is shown in Fig. 1, for which there were increasing reads in the 5’-to-3’ direction, with a 3’/5’ ratio of 3.1. Other genes in the 3’-up category had a more complex pattern (cf. wild-type curves for pgcA and yabE in Fig. 2B) that likely reflected endonuclease cleavage(s) followed by degradation upstream of the cleavage site(s) by 3’ exonucleases and downstream of the cleavage site(s) by RNase J1.
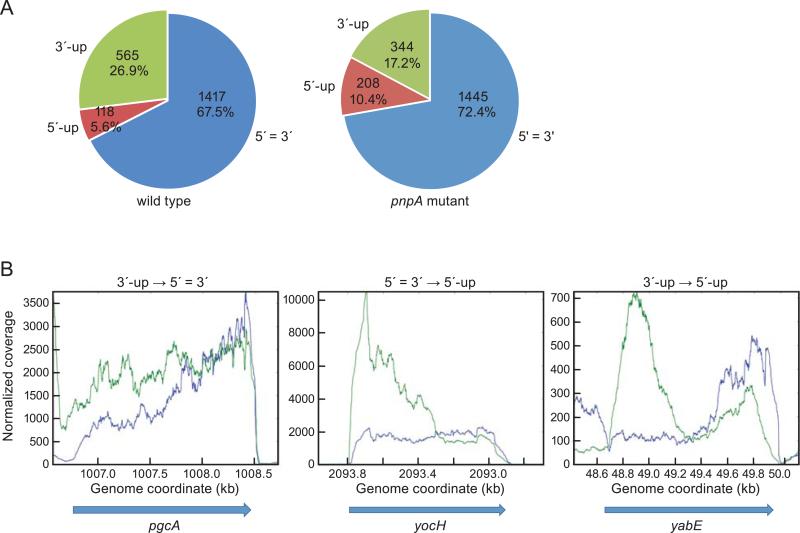
Fig. 2.
(A) Percent of genes in three 5’-3’ ratio categories in the wild-type and pnpA mutant strains. The categories are indicated outside the segments, with number of genes and percent of total inside each segment. (B) Change in 5’/3’ ratios upon loss of PNPase. Read data from wild-type (blue) and pnpA mutant (green) strains are shown, with the shift in category from wild type to mutant indicated above each graph.
A gradual increase in reads from the 5’ end toward the 3’ end, as seen for lytE (Fig. 1), might be expected for mRNAs that are degraded from the 5’ terminus by the processive 5’ exonuclease activity of RNase J1, as 5’-proximal sequences may be degraded earlier than 3’-proximal sequences. If so, we would expect this pattern to be unaffected by the loss of 3’ exonuclease activity. Indeed, the pattern of lytE RNA processing is unchanged in the pnpA mutant strain, although the read level is 3-fold lower (see below). We further analyzed mRNAs that might be degraded exclusively from the 5’ end by RNase J1, as we hypothesized for those with a lytE-like pattern, by comparing our data with the tiling array data of Durand et al. (2012), which was used to identify those mRNAs that are affected by depletion of RNase J1. Of 1206 genes reported by Durand et al. to show ≥1.5-fold increased expression in a strain depleted for RNase J1, 619 had read levels above the cutoff in the current study. (A similar microarray study of Putzer and colleagues (Mader et al., 2008) had only 183 CDSs above the cutoff in our studies, so we chose to compare our results with those of Durand et al.) To limit the number of RNA-Seq patterns that needed to be checked by eye for a lytE-like pattern, we restricted this analysis to monocistronic genes. Of 471 monocistronic genes in our RNA-Seq analysis, 144 had ≥1.5-fold increased mRNA in the RNase J1 depletion strain. 50 of the 471 monocistronic genes showed a lytE-like pattern, and 27 of these (54%) overlapped with genes that were significantly up when RNase J1 was depleted. Of the 421 monocistronic genes that did not show a lytE-like pattern in our study, only 117 (27.7%) overlapped with genes that were up when RNase J1 was depleted. In other words, genes with a lytE-like pattern correlated to a much higher degree with genes that had increased signal when RNase J1 was depleted. It should be noted that the Durand et al. study was done before it was discovered that RNase J1 is not essential and can be knocked out completely (Figaro et al., 2013), and was therefore done on strains that still contained RNase J1 but at a lower level than wild type. It is likely that merely depleting RNase J1 will not have a measurable direct effect on mRNAs that are good substrates for the enzyme activity. We suggest that mRNAs in the 3’-up category with a lytE-like RNA-Seq pattern have a high likelihood of being degraded in the 5’-to-3’ direction by RNase J1.
Lastly, we found that 5.6% of mRNAs were in the 5’-up category, with a ≥1.5-fold greater number of reads in the 5’ region compared to the 3’ region. An example of a gene in this category is cimH (Fig. 1), for which the 5’/3’ ratio was 2.3. The RNA-Seq reads histogram shows a 5’-proximal peak, followed by a gradual decrease in read coverage to the end of the CDS. We hypothesized that initiation of cimH mRNA is by a decay-initiating cleavage some distance from the 5’ end, and decay of 5’-proximal sequences is delayed relative to rapid decay of 3’-proximal sequences, perhaps due to particular structures or RNA-protein binding in the region upstream of the endonuclease cleavage site.
RNA processing in the pnpA strain
RNA-Seq analysis of the pnpA mutant strain was able to reveal an effect of the loss of PNPase on accumulation of decay intermediates, as represented by the three 5’-3’ categories described above. Whereas 67.5% of mRNAs were in the 5’=3’ category in the wild type, this number went up to 72.4% in the pnpA mutant (Fig. 2A). The increase in genes in the 5’=3’ category was mostly accounted for by the 228 genes that were in the 3’-up category in the wild type strain and shifted to the 5’=3’ category in the pnpA strain (Table 1). An example of this shift is shown for the pgcA gene in Fig. 2B, which had a higher number of 3’-proximal reads in the wild-type strain but a more uniform distribution in the pnpA strain. In other words, for these genes there is an accumulation in the pnpA strain of 5’-proximal RNA such that 3’ reads are no longer in greater abundance than 5’ reads. Similarly, the percent of genes in the 5’-up category almost doubled from 5.6% in the wild type to 10.4% in the pnpA mutant (Fig. 2A). Most of this increase in the 5’-up category was due to the 103 genes that were in the 5’=3’ category in the wild type and were shifted to the 5’-up category in the pnpA mutant (Table 1). An example of this is the yocH gene shown in Fig. 2B, for which there is an even distribution in the wild-type strain but a marked increase in 5’-proximal reads in the pnpA strain. There were even three genes (yabE, ywmB, rapF) that shifted from the 3’-up category in the wild type to the 5’-up category in the pnpA mutant (Table 1). The read distribution pattern for yabE is shown in Fig. 2B, where the high 3’/5’ ratio in the wild type becomes a high 5’/3’ ratio in the pnpA strain. Finally, the percent of genes in the 3’-up category went from 26.9% in the wild type down to 17.2% in the pnpA mutant (Fig. 2A). Thus, the absence of PNPase caused a significant increase in the number of mRNAs that accumulate 5’-proximal sequence. This is consistent with the hypothesis that PNPase is required for degradation in the 3’-to-5’ direction from a site of endonucleolytic cleavage. The loss of PNPase would make upstream products of endonuclease cleavage less susceptible to rapid turnover, and 5’-proximal decay intermediates would be in greater abundance than in the wild-type strain. The downstream products, however, would not be affected by the lack of PNPase, as they are likely degraded by RNase J1.
Table 1.
Altered distribution of genes in three 5′-3′ categories in wild-type and pnpA mutant strains.
| Wild-type | pnpA | Number |
|---|---|---|
| 5′=3′ | 5′=3′ | 1079 |
| 5′=3′ | 5′-up | 103 |
| 5′=3′ | 3′-up | 46 |
| 3′-up | 3′-up | 267 |
| 3′-up | 5′=3′ | 228 |
| 3′-up | 5′-up | 3 |
| 5′-up | 5′-up | 80 |
| 5′-up | 5′=3′ | 25 |
| 5′-up | 3′-up | 0 |
Interestingly, for a small percentage of genes (~4%), the ratio of 3’/5’ reads increased in the pnpA strain (Table 1), i.e., 46 genes in the 5’=3’ category in the wild type shifted to the 3’-up category in the pnpA strain, and 25 genes in the 5’-up category in the wild type shifted to the 5’=3’ category in the pnpA strain. We cannot rule out complex transcriptional and post-transcriptional effects in the pnpA strain.
Effect of PNPase deletion on rnjA expression
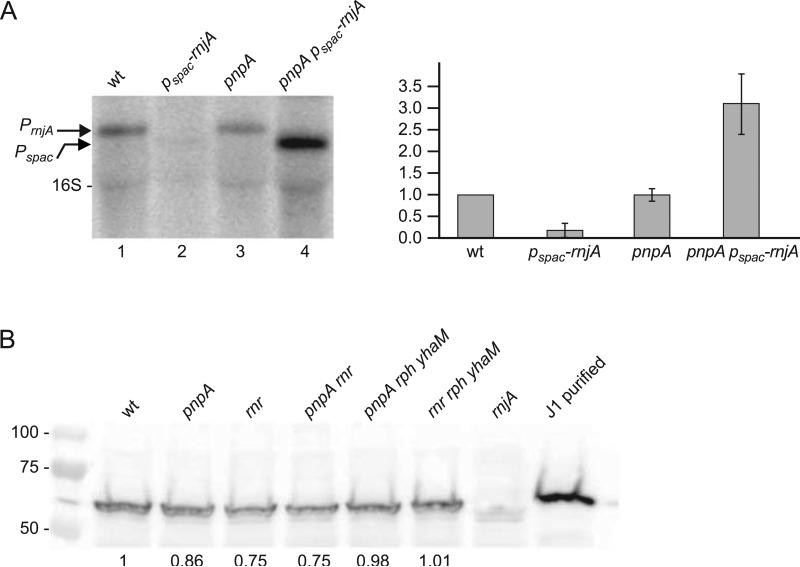
Before analyzing further the impact of PNPase on global RNA processing, we addressed the issue of possible effects of deleting PNPase on the expression level of RNase J1, which is believed to be the other major exoribonuclease in mRNA decay (Condon, 2010). Inefficient turnover of mRNA due to the lack of PNPase might cause a depletion of the free nucleotide pool and impede redirection of gene expression programs, which could be compensated by increased expression of the 5’ exonuclease RNase J1. In fact, preliminary experiments with a strain in which the RNase J1-encoding rnjA gene was under control of the IPTG-inducible Pspac promoter showed that there is selective pressure to elevate rnjA expression in the pnpA background. As seen in Fig. 3A, rnjA mRNA levels were assessed by Northern blot analysis in strains in which the gene was expressed either from its native promoter (lanes 1 and 3) or from the Pspac promoter (lanes 2 and 4) in the wild-type background (lanes 1 and 2) and the pnpA background (lanes 3 and 4). All strains carried the pMAP65 plasmid, which provides a high level of lac repressor (Petit et al., 1998). In the wild-type background, IPTG-induced transcription of rnjA from the Pspac promoter was about five-fold lower than expression from its native promoter, as expected from earlier results of Western blot analysis (Durand et al., 2012). In the pnpA background, the level of rnjA transcription from its native promoter was unchanged but the level of transcription from the Pspac promoter increased 17-fold. Sequencing of the lac operator in this strain identified several point mutations that likely eliminated lac repressor binding and resulted in elevated rnjA expression, conferring a growth advantage. Thus, the absence of PNPase selects for cells that have increased RNase J1 expression. This is consistent with the failure to construct a strain that is deleted for both pnpA and rnjA genes (Figaro et al., 2013).
Fig. 3.
Analysis of RNase J1 expression in response to loss of PNPase. (A) Northern blot analysis of rnjA mRNA using a 5’-end-labeled oligonucleotide probe complementary to an internal sequence in the RNase J1 CDS. Arrows on left indicate migration of rnjA mRNA transcribed from its native promoter (PrnjA) or from an IPTG-inducible promoter (Pspac). Migration of 16S rRNA is also indicated. Quantitative data on right are average of three experiments, with rnjA mRNA level in the wild-type strain set to 1. (B) Western blot analysis of RNase J1 in ribonuclease mutant strains. Numbers below the blot are relative to RNase J1 detected in the wild-type strain. 50 ng of purified RNase J1 was loaded in the control lane on the right. Numbers on the left indicate migration of protein molecular weight markers (kilodaltons).
The RNA-Seq data for transcript levels of rnjA and a number of genes involved in RNA decay showed that these were not greatly affected by the loss of PNPase (Table 2). There is a significant increase in the read level for the gene encoding RNase III and a small increase for the gene encoding RNase PH. (The small increase for RNase R is less than 1.5-fold and barely reaches significance.) We do not know whether these changes are biologically relevant. We nevertheless thought it prudent to use Western blot analysis to determine the relative level of RNase J1 in ribonuclease mutant strains, in order to eliminate the possibility that the loss of PNPase caused changes at the translational level (Jamalli et al., 2014). The results (Fig. 3B) show that no significant difference in RNase J1 protein level was observed in the pnpA strain or in any of several single or multiple exoribonuclease mutant strains that were tested.
Table 2.
Read levels of genes involved in mRNA decay in wild-type and pnpA strains.
| Gene Name | Enzyme name (activity) | wild type | pnpA | pnpA/wt Ratio | P-value |
|---|---|---|---|---|---|
| rnr | RNase R (3′ exo) | 6.15 | 8.78 | 1.43 | 0.04 |
| rph | RNase PH (3′ exo) | 3.33 | 5.34 | 1.60 | 0.01 |
| yhaM | YhaM (3′ exo) | 6.02 | 3.81 | 0.63 | 0.00 |
| rnjA | RNase J1 (5′ exo/endo) | 16.05 | 18.60 | 1.16 | 0.04 |
| rnjB | RNase J2 (5′ exo/endo) | 13.54 | 17.30 | 1.28 | 0.00 |
| rny | RNase Y (endo) | 38.36 | 37.59 | 0.98 | 0.85 |
| rnc | RNase III (endo) | 9.92 | 22.42 | 2.26 | 0.01 |
| nrnA | nanoRNase | 4.50 | 2.85 | 0.63 | 0.00 |
| rppH | RNA pyrophosphohydrolase | 2.34 | 1.49 | 0.64 | 0.01 |
Differential gene expression in the pnpA strain – direct effects
RNA-Seq results from the pnpA and wild-type strains revealed a surprisingly widespread effect of the pnpA deletion on levels of gene expression. Of the 2,276 genes included in the analysis (at least 1 read per base in either strain), ~66% showed significant differential expression in the wild-type vs. pnpA strains (P-values <0.05; Table S1). Of these, fully three quarters had differential expression (increase or decrease) of at least 1.5-fold, which we used as a cut-off for further analysis. No single functional category had a significantly greater percentage of differentially expressed genes (Table S2). While it might be expected that the absence of PNPase would mean less efficient degradation of mRNA intermediates and overall increases in the number of RNA reads, we found that, among genes with a ≥1.5-fold difference in gene expression, 18.1% (412 genes) had increased expression while 31.6% (720 genes) had decreased expression (Table S2). Significant increases in read number in the pnpA strain could be due to a direct effect of lower degradative activity, or could be due to indirect effects (e.g., increased transcription due to increased expression of a sigma factor). Significant decreases in read number in the pnpA strain are likely due to indirect effects: either the absence of PNPase could allow other ribonucleases to degrade an mRNA more efficiently, or the absence of PNPase could lead to decreased transcription (e.g., decreased expression of a sigma factor).
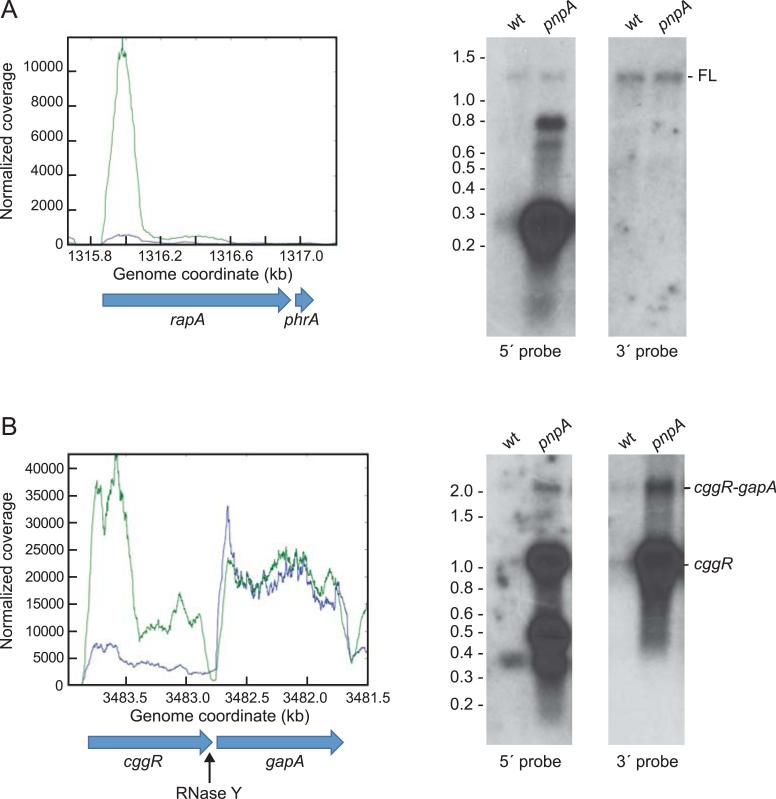
We hypothesized that mRNAs that were directly affected by the absence of PNPase are those whose initiation of decay begins with an endonuclease cleavage, generating a 3’ hydroxyl that is then acted upon by PNPase. To identify such mRNAs, we calculated the ratio of reads in the 5’-proximal one-third vs. the 3’-proximal one-third of each CDS in both pnpA and wild-type strains, to give a “ratio of ratios” or “RR.” This is a sensitive measure of the direct effect of PNPase on mRNA turnover, as it is independent of relative expression levels. Genes with an RR of >1.5 would likely be those whose decay from a 3’ end generated by endonuclease cleavage requires the 3’ exonucleolytic activity of PNPase. A list of genes with RR >1.5 is in Table S4. The gene with the highest RR value was rapA (RR = 10.5) for which RNA-Seq data and Northern blot analysis are shown in Fig. 4A. The RapA CDS, which encodes a response regulator aspartate phosphatase, is transcribed together with that of its inhibitor, PhrA (Diaz et al., 2012), to give a ~1.3 kb transcript. In the wild-type strain, the 5’ rapA probe detected full-length mRNA and an additional RNA of ~250 nts. This decay intermediate is seen in the RNA-Seq histogram as a small, 5’-proximal peak. The 5’/3’ ratio of rapA in the wild-type strain is 3.2, placing it in the 5’-up category. In the pnpA strain, the 5’ probe detected the same amount of full-length transcript but about a 20-fold higher level of the 250-nt 5’-proximal fragment, as reflected in the RNA-Seq histogram. The 5’ probe also detected additional 600-800-nt decay intermediates, which are represented in the histogram by an extended shoulder of the 5’-proximal peak that reaches out to ~800 nts. Using the 3’ probe, only the full-length mRNA was detected, at equal amounts in both strains.
Fig. 4.
Analysis of mRNAs with high ratio of ratios (RR). (A) RNA-Seq and Northern blot analysis of rapA mRNA. RNA-Seq data from wild type in blue and from pnpA mutant in green. 5’-and 3’-directed oligonucleotide probes were used in Northern blots, as indicated below the blots. FL = full-length rapA-phrA mRNA. Migration of RNA molecular weight markers indicated on the left of the blot. (B) RNA-Seq and Northern blot analysis of cggR-gapA mRNA. Location of RNase Y cleavage near the end of the cggR CDS is indicated below the CDS schematic.
Another noteworthy example of an mRNA with an RR>1.5 is cggR mRNA, which is a well-characterized substrate for RNase Y, the putative major decay-initiating endonuclease in B. subtilis (Lehnik-Habrink et al., 2012). cggR, which encodes a transcriptional repressor protein, is the first gene in the hexacistronic gapA operon that contains several glycolytic genes. The level of CggR protein in the cell is ~100-fold lower than the level of GapA protein encoded by the second cistron of the gapA operon, and this has been shown to be due to differential instability of the cggR-encoding mRNA. The mechanism involves RNase Y cleavage at a site several nucleotides upstream of the cggR stop codon (Ludwig et al., 2001, Commichau et al., 2009), which is presumably followed by 3’-to-5’ exonucleolytic decay of cggR mRNA. In Fig. 4B are shown RNA-Seq reads for cggR-gapA in the wild-type and pnpA mutant strains, and a Northern blot analysis using cggR 5’ and 3’ probes. The RNA-Seq data indicated a large increase (~6-fold) in cggR 5’-proximal reads in the pnpA strain, as well as a smaller increase in reads that are closer to the cggR 3’ end. gapA mRNA reads were the same in both strains. In the Northern blot analysis of the wild-type strain using the 5’ cggR probe, the 2.2 kb cggR-gapA discistronic transcript and the 1.2 kb cggR RNA were barely detected. (The cggR-gapA transcript is the result of transcription termination downstream of the gapA CDS, while the cggR RNA is due to RNase Y cleavage.) These species were detected also by the cggR 3’ probe. The 5’ probe also detected a ~360-nt 5’-proximal intermediate, which is reflected in the RNA-Seq pattern by the small 5’-proximal peak. In the pnpA strain, cggR-gapA RNA was easily detected by either probe, as well as a large increase in cggR RNA. The 5’ probe also detected a species of ~500 nts as well as smaller RNAs, which correlate with the 5’-proximal peaks in the histogram. Accumulation of cggR RNA in the pnpA mutant strain suggests that PNPase is absolutely required for decay of the upstream fragment generated by RNase Y cleavage.
The overall RR data showed that out of 1831 genes above the cutoff in both strains, 178 gave an RR of >1.5 and a P value of <0.05 (Table S4). In other words, the decay of about 10% of mRNAs is highly dependent on PNPase activity, and none of the other 3’ exonucleases can substitute efficiently in the decay process.
Location of genes affected by loss of PNPase
We hypothesized that the location of a gene relative to the transcription start site (TSS) would have an effect on its decay pattern. Genes located closer to the TSS would be more likely to house the 5’-most endonuclease cleavage target site, and so would be most affected by the absence of the 3’ exonuclease (PNPase) that is required for subsequent turnover. Genes that are located more distally from the TSS would likely have an endonuclease target site located upstream and would be subject to RNase J1 turnover and less affected by the absence of PNPase. The operonic context of 123 of the 178 genes with an RR >1.5 for which the operon location is known was analyzed (Table 3). Genes were annotated as being either monocistronic, the first cistron in a polycistronic operon, the second cistron in a polycistronic operon, or the third cistron and later. Of genes with an RR >1.5 and known operonic location, 51.1% were either monocistronic or the first gene in an operon, compared to 33.1% of all genes with >1 reads/base whose location is known. Thus, genes that show a 5’ accumulation that is directly due to the loss of PNPase show a position bias near a TSS. This is consistent with initiation of decay by endonuclease cleavage, followed by PNPase 3’ exonuclease activity.
Table 3.
Operon position of genes with RR > 1.5.
| All CDSs | Above cutoff in both strains | RR>1.5 | ||||
|---|---|---|---|---|---|---|
| Number of genes | Percent | Number of genes | Percent | Number of genes | Percent | |
| Total | 4176 | 1831 | 178 | |||
| unknown operon position | 1645 | 39.4 | 685 | 37.4 | 55 | 30.9 |
| Monocistronic genes | 965 | 23.1 | 397 | 21.7 | 62 | 34.8 |
| First gene in operon | 457 | 10.9 | 208 | 11.4 | 29 | 16.3 |
| Second gene in operon | 459 | 11.0 | 230 | 12.6 | 20 | 11.2 |
| Third or later gene in operon | 650 | 15.6 | 311 | 17.0 | 12 | 6.7 |
Differential gene expression in the pnpA strain – indirect effects
To address at least some of the indirect effects of loss of PNPase on gene expression, changes in sigma factor mRNA levels were analyzed. For nine sigma factor RNAs, the read level was greater than the cutoff of 1 read/base in at least one of the strains (Table 4), and for five of these the difference in read number did not reach significance (P >0.05). Expression of Sigma A, the major vegetative sigma factor was only slightly affected by loss of PNPase (30% decrease). Sigma B gene expression was 2-fold higher in the pnpA strain. Of the 128 genes in the sigB regulon with a read abundance above the cutoff, 33 genes (25.8%) had >1.5-fold increase in expression (Table 4, lower portion). Thus, the 2-fold increase in sigB expression could account for some but not most of the changes in sigma B-controlled genes. It is likely that the small increase in Sigma B levels is only sensed by high-affinity promoters in the sigB regulon. Sigma Y gene expression was decreased by 60%, perhaps explaining why six of the seven genes in the sigY regulon showed a decrease of >1.5-fold. Sigma D expression was reduced more than 3-fold. Correspondingly, of the 72 sigD regulon genes with a read abundance above the cutoff, 62 had decreased RNA levels of at least 1.5-fold in the pnpA strain. The one gene that showed a significant increase, gdpP, is actually controlled by an anti-sense transcript that is expressed from a sigma D promoter (Luo & Helmann, 2012). Thus, a large percentage (87.5%) of differentially expressed sigD regulon genes in the pnpA strain could be explained by an indirect effect of the loss of PNPase on sigD expression.
Table 4.
Read abundance for sigma factors in wild-type and pnpA mutant strains
| Gene Name | wild type | pnpA mutant | pnpA/WT | P-value |
|---|---|---|---|---|
| sigA | 18.66 | 12.84 | 0.69 | 0.00 |
| sigB | 35.74 | 72.67 | 2.03 | 0.00 |
| sigD | 4.69 | 1.28 | 0.27 | 0.00 |
| sigH | 6.54 | 7.87 | 1.20 | 0.28 |
| sigL | 4.27 | 4.24 | 0.99 | 0.90 |
| sigM | 1.77 | 1.39 | 0.79 | 0.06 |
| sigW | 35.17 | 33.48 | 0.95 | 0.66 |
| sigX | 3.44 | 4.47 | 1.30 | 0.10 |
| sigY | 1.03 | 0.44 | 0.43 | 0.00 |
| Number of genes | Above cutoff | Increase >1.5 | Decrease >1.5 | |
|---|---|---|---|---|
| sigB regulon | 164 | 128 | 33 | 21 |
| sigY regulon | 7 | 7 | 0 | 6 |
| sigD regulon | 82 | 72 | 1 | 62 |
Analysis of fla/che operon RNA
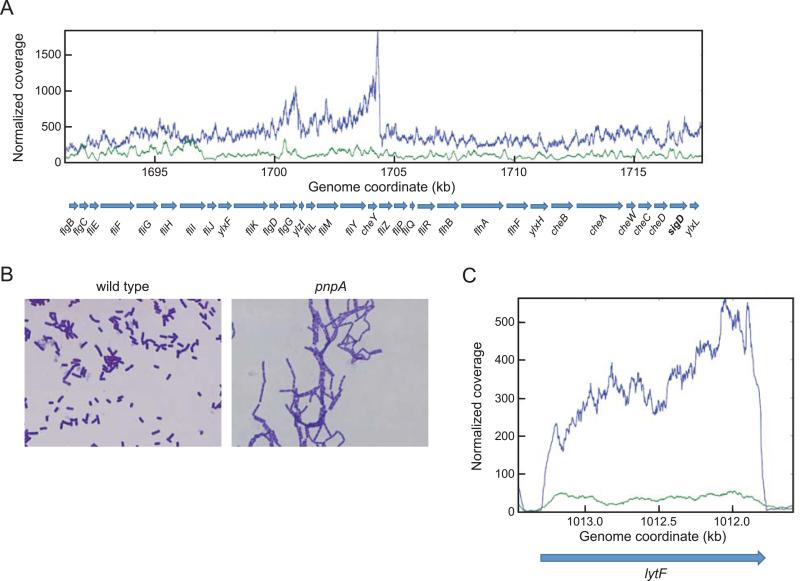
The sharp decrease in sigD expression led us to examine the transcription pattern of the fla/che operon, a 32-gene operon of which sigD is the penultimate gene (Fig. 5A). Transcription of the fla/che operon is controlled for the most part by the sigma A-dependent fla/che promoter. There is an additional promoter located in the fliJ gene (the 8th gene of the operon), which is designated the PylxF3 promoter (the ylxF gene follows fliJ in the operon). An earlier study of fla/che transcription by qRT-PCR noted that transcript levels increase from the promoter-proximal region to about the first third of the operon, and then decrease with distance from the fla/che promoter such that sigD transcription is relatively low (Cozy & Kearns, 2010). As shown in the RNA-Seq analysis in Fig. 5A, in the wild-type strain fla/che read levels rose gradually until reaching a maximum in the cheY-fliZ region, and then dropped off abruptly and remained at a relatively constant level until the end of the operon. In the pnpA mutant strain there was no rise in read levels moving downstream in the operon, and the levels throughout the operon remained 2-4-fold lower than the wild type (Table 5). This explains the 3-fold decrease in sigD expression (Table 4). The differential pattern of fla/che read levels in the two strains suggests some form of post-transcriptional processing that likely involves PNPase.
Fig. 5.
(A) Read data from RNA-Seq analysis of the fla/che operon in wild-type and pnpA mutant strains. Gene names are indicated below the graph, with the penultimate sigD gene in bold. (B) Photomicrographs of overnight wild-type and pnpA mutant cultures. (C) RNA-Seq analysis of lytF in wild-type and pnpA mutant strains; wild type in blue, pnpA mutant in green.
Table 5.
Read abundance of genes involved in motility and cell separation
| Wild-type | pnpA mutant | pnpA/WT | P-value | |
|---|---|---|---|---|
| fla-che operon | ||||
| flgB | 2.07 | 1.39 | 0.67 | 0.04 |
| flgc | 2.61 | 2.10 | 0.80 | 0.02 |
| fliE | 3.39 | 1.61 | 0.47 | 0.00 |
| fliF | 3.51 | 1.58 | 0.45 | 0.00 |
| fliG | 4.75 | 1.98 | 0.42 | 0.00 |
| fliH | 5.52 | 2.24 | 0.40 | 0.00 |
| flil | 4.26 | 1.85 | 0.43 | 0.00 |
| fliJ | 4.43 | 1.23 | 0.28 | 0.00 |
| ylxF | 4.44 | 1.35 | 0.31 | 0.00 |
| fliK | 4.74 | 1.25 | 0.26 | 0.00 |
| flgD | 5.65 | 1.39 | 0.25 | 0.00 |
| flgG | 6.94 | 1.64 | 0.24 | 0.00 |
| ylzI | 6.15 | 1.57 | 0.26 | 0.00 |
| fliL | 5.83 | 1.41 | 0.24 | 0.00 |
| fliM | 6.81 | 1.61 | 0.24 | 0.00 |
| fliY | 7.55 | 1.78 | 0.24 | 0.00 |
| cheY | 11.73 | 2.13 | 0.18 | 0.00 |
| fliZ | 7.02 | 1.61 | 0.23 | 0.00 |
| fliP | 4.15 | 1.33 | 0.32 | 0.00 |
| fliQ | 4.53 | 1.37 | 0.30 | 0.00 |
| fliR | 3.71 | 1.07 | 0.29 | 0.00 |
| flhB | 3.80 | 1.05 | 0.28 | 0.00 |
| flhA | 3.44 | 1.05 | 0.30 | 0.00 |
| flhF | 3.27 | 0.84 | 0.26 | 0.00 |
| ylxH | 3.09 | 0.93 | 0.30 | 0.00 |
| cheB | 3.16 | 0.86 | 0.27 | 0.00 |
| cheA | 4.33 | 1.16 | 0.27 | 0.00 |
| cheW | 5.25 | 1.39 | 0.27 | 0.00 |
| cheC | 4.56 | 1.13 | 0.25 | 0.00 |
| cheD | 4.39 | 1.18 | 0.27 | 0.00 |
| sigD | 4.69 | 1.28 | 0.27 | 0.00 |
| ylxL | 5.19 | 1.37 | 0.27 | 0.00 |
| Autolysis genes | ||||
| lytA | 2.92 | 1.59 | 0.54 | 0.02 |
| lytB | 1.79 | 0.86 | 0.48 | 0.00 |
| lytC | 3.56 | 0.89 | 0.25 | 0.00 |
| lytD | 1.35 | 0.32 | 0.23 | 0.00 |
| lytE | 3.12 | 0.98 | 0.32 | 0.00 |
| lytF | 3.42 | 0.38 | 0.11 | 0.00 |
| Flagellin | ||||
| hag | 205.50 | 11.63 | 0.06 | 0.00 |
Chain growth and swarming phenotypes of pnpA mutant strains
The effect of PNPase loss on sigD expression led us to examine the basis for the chaining phenotype of the pnpA strain, which is shown in Fig. 5B. Separation of dividing cells is controlled by the major autolysis genes (lytABC, lytD, and lytF), and these are under control of sigma D promoters (Marquez et al., 1990). Indeed, there was a significant reduction of expression of these genes in the pnpA strain (Table 5), especially for lytF, which encodes the major enzyme for cell separation (Chen et al., 2009) (Fig. 5C). These data immediately explain why the pnpA strain grows as chains.
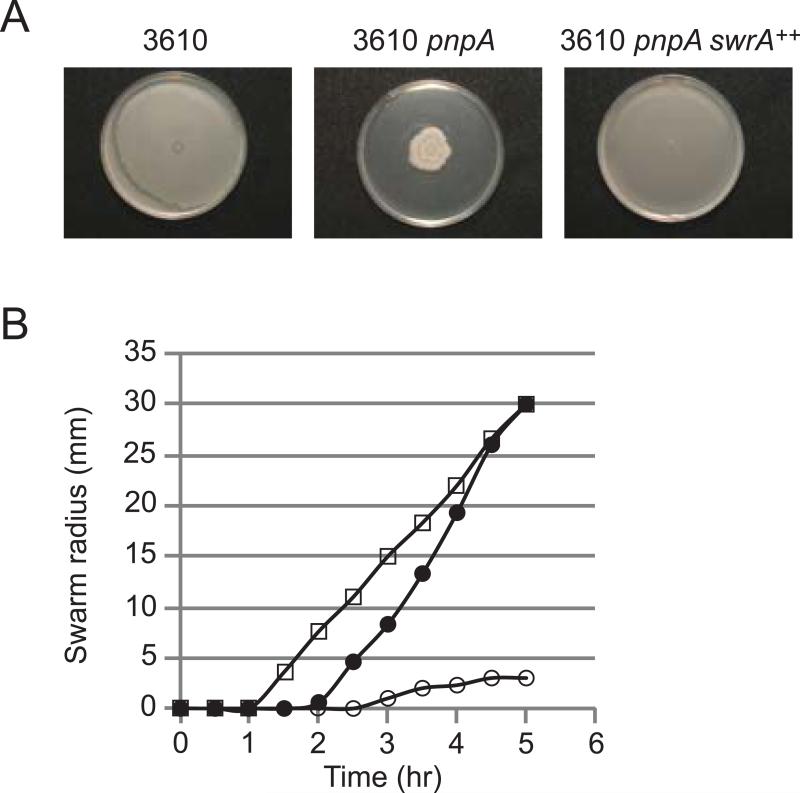
Another sigD regulon gene that was affected in the pnpA strain was the flagellin-encoding hag gene, which showed a 17-fold decrease in RNA reads (Table 5). Since B. subtilis swarming motility is dependent on a high level of flagellar synthesis (Kearns, 2010), we hypothesized that disruption of pnpA might also affect swarming motility. The B. subtilis 168 derivatives that were used to analyze mRNA decay are domesticated strains that do not show swarming motility on agar plates (Patrick & Kearns, 2009). Thus, to determine whether the loss of PNPase affected swarming, the pnpA mutation was transferred via SPP1 phage transduction into the undomesticated and swarming-competent B. subtilis 3610 strain. Qualitative and quantitative assays of swarming shown in Fig. 6 demonstrated that loss of PNPase indeed resulted in a severe swarming defect.
Fig. 6.
Swarming motility assay. (A) Wild-type swarming phenotype of 3610 strain compared with swarming-deficient 3610 pnpA strain and complementation in strain overexpressing swrA. (B) Quantitative swarming assay of three strains: 3610, open squares; 3610 pnpA, open circles; 3610 pnpA swrA++, closed circles.
The swarming defect in the 3610 pnpA strain could be a result of the decrease in fla/che operon expression (notably sigD), or could be due to an effect of the loss of PNPase on a different locus. To probe this, a derivative of the 3610 pnpA strain was made that had increased fla/che transcription. SwrA acts positively to increase fla/che transcription (Kearns & Losick, 2005), likely in partnership with DegU at the major fla/che promoter (Mordini et al., 2013), and IPTG-induced overexpression of swrA has been shown to complement motility defects (Kearns & Losick, 2005). As shown in Fig. 6, transduction of the 3610 pnpA strain to swrA++ resulted in restoration of the swarming phenotype. This result suggests that the severe swarming defect caused by the pnpA deletion is due to effects on fla/che operon expression.
Discussion
We found that RNA-Seq analysis of B. subtilis mRNA is useful in revealing patterns of mRNA decay/processing that show an accumulation of mRNA fragments at steady state. In cases where Northern blot analysis was used to confirm RNA-Seq results, we found excellent correspondence between RNA-Seq data and the pattern detected by Northern blotting using probes from different ends of the gene (cf. Fig. 4 for examples). This validates the use of RNA-Seq to probe RNA processing. A previous study done on wild-type B. cereus strains gave similar results, and was able to show differential decay patterns for different segments of the same transcript (Kristoffersen et al., 2012).
In accordance with the “all-or-nothing” mechanism of mRNA decay (Kaberdin et al., 2011), two-thirds of mRNAs showed equal amounts of RNA reads across the CDS (the 5’=3’ category). For such genes, only full-length mRNA would be detected by probes that could be complementary to any part of the CDS. For genes in the 5’-up or 3’-up categories, characterization of mRNA decay by Northern blotting will very much depend on probe design. For example, the data for rapA mRNA in Fig. 4A show that use of a 3’ probe would indicate no difference between wild-type and pnpA mutant strains, with only full-length mRNA being detected, while the use of a 5’ probe revealed a remarkable difference in RNA accumulation between the two strains.
In the wild-type strain, one third of mRNAs showed a skewed distribution of reads, with accumulation of either 5’-proximal or 3’-proximal RNA fragments (Fig. 1). The RNA-Seq pattern for genes in the 3’-up category, where there is a gradient of read levels from the 5’ to the 3’ end (e.g., lytE; Fig. 1), is consistent with decay from a monophosphorylated 5’ end by RNase J1 5’ exonuclease activity. Such a pattern could reflect a differential rate of RNase J1 processivity that might be affected by translating ribosomes, RNA secondary structure, or bound proteins. We found a significant overlap of mRNAs that had a lytE-like pattern with mRNAs shown to be increased by RNase J1 depletion in earlier study (Durand et al., 2012), suggesting that many mRNAs with a lytE-like pattern are candidates for decay by RNase J1 from the 5’ end. The lytE-like pattern could also be due multiple endonuclease cleavages that occur in a 5’-to-3’ direction. In fact, this latter mechanism is needed to explain the observation (not shown) that a third of the lytE-like genes shift from the 3’-up category in the wild-type strain to the 5’=3’ category in the pnpA strain. Degradation of mRNAs strictly by processive RNase J1 activity in the 5’-to-3’ direction would likely not be affected by loss of PNPase.
On the other hand, the pattern for genes in the 5’-up category is consistent with endonuclease cleavage, followed by degradation of the upstream fragment by a 3’ exonuclease and of the downstream fragment by RNase J1 or by additional endonuclease cleavages. Various 3’ exonucleases differ in their ability to degrade past secondary structure. We have shown using purified enzymes that PNPase processivity in the 3’-to-5’ direction is sensitive to mRNA secondary structure (Deikus & Bechhofer, 2007), while the mechanism of RNase R 3’ exonucleolytic activity allows it degrade structured RNA efficiently (Vincent & Deutscher, 2009). Thus, the pattern observed for genes in the 5’-up category, where 5’-proximal sequences accumulate relative to downstream sequences, may result from structured RNA regions and their effect on processivity, depending on which 3’ exonuclease accessed the 3’ end generated by endonuclease cleavage.
Accumulation of 5’-proximal fragments at steady state was even more apparent when PNPase was absent (Figs. 2 and 4; Table 1). An important conclusion from these data is that the loss of PNPase is not compensated for by the activity of RNase R, RNase PH or YhaM. In other words, there appears to be specificity in the exonuclease activity that degrades upstream cleavage fragments, such that only PNPase can do so efficiently in some cases. Based on the ratio of ratios measure used to identify mRNAs that are directly affected by the loss of PNPase, we estimate that ~10% of mRNAs are dependent specifically on PNPase for degradation. Since biochemical studies of phosphate-dependent 3’ exonuclease activity indicated that PNPase was the major such enzyme in B. subtilis (Deutscher & Reuven, 1991, Wang & Bechhofer, 1996), it is likely that, when present, PNPase is indeed responsible for the bulk of mRNA turnover. When PNPase is not present, other 3’ exonucleases, such as RNase R, can efficiently substitute for PNPase in many cases but not in the 10% of cases that appear to depend on PNPase for rapid turnover. The basis of this specificity will require further investigation of individual mRNAs that we have shown here are most affected by the absence of PNPase, e.g., rapA and cggR mRNA (Fig. 4).
RNA-Seq data identified a significant decrease in sigD expression, which could explain the chain growth phenotype of the pnpA mutant, an observation that was made when the pnpA mutant was first constructed (Wang & Bechhofer, 1996). The effect on sigD expression also predicted a pnpA-related swarming motility defect, which was confirmed in the undomesticated 3610 strain (Fig 6). The pnpA-mediated swarming defect was complemented by swrA overexpression, which is likely to be occurring at the level of increased fla/che operon expression. Further experiments are needed to determine whether RNA processing mechanisms dictate the complex fla/che read pattern (Fig. 5A), and whether the lack of PNPase has a direct effect on processing of the fla/che transcript. For example, although swrA is a pseudogene and non-functional in the 168 strain, and therefore does not appear in the list of read data in Table S1, the RNA-Seq data for the swrA locus in the pnpA strain showed a 4-5-fold decrease in read levels from wild type. This is not surprising, since swrA transcription is itself controlled by sigD in an autoregulatory loop with the fla/che operon (Kearns & Losick, 2005, Calvio et al., 2008, Mordini et al., 2013). The severe swarming defect of the 3610 pnpA strain (Fig. 6) provides a convenient platform to screen for genetic alterations that restore swarming, which should be useful in determining the role that post-transcriptional processing by PNPase plays in complex regulatory pathways such as swarming motility and biofilm production.
Experimental Procedures
Bacterial Strains
The wild-type strain, BG1, is a trpC2 thr-5 derivative of B. subtilis strain 168. Construction of a disruption of the pnpA gene with a neomycin-resistance cassette was described previously (Wang & Bechhofer, 1996). The pnpA strain used in the current study was BG546, which is a neomycin-resistant transformant of BG1 using the original BG119 pnpA chromosomal DNA. BG119 had one or more additional mutations that affected nitrogen metabolism (unpublished results). The NCIB3610 strain (“3610”) was generously provided by Dr. Roberto Kolter. The neomycin-resistant pnpA deletion derivative of the 3610 strain was constructed by transduction with SPP1 phage carrying chromosomal DNA from BG546. Note that the pnpA gene is located at 1740 kb on the B. subtilis chromosome, far from the two genes that are mutated and cause lack of swarming in the domesticated 168 strain (sfp at 407.6 kb and swrA at 3622 kb). Since SPP1 phage packages around 40 kb of DNA, there was no concern that these mutant genes would be transduced into the 3610 strain along with the pnpA locus.
The strain with rnjA transcription under control of the Pspac promoter was described previously (Britton et al., 2007). The Pspac-rnjA strain was grown in the presence of 1 mM IPTG, which results in a level of rnjA expression that is approximately five-fold lower than in the wild-type strain (Durand et al., 2012).
RNA preparation and sequencing
For RNA extraction, 1 ml of an overnight culture of wild-type (BG1) or pnpA (BG546) strains was inoculated into 50 ml of LB media. Cells from three independent cultures were harvested at mid-logarithmic stage (OD600 0.4), and total RNA was extracted by the hot phenol method. RNA concentrations were determined using a Qubit Fluorometer (Invitrogen) and RNA integrity was checked on a 1% agarose formaldehyde-MOPS gel. Ribosomal RNA was depleted with the Ribo-Zero rRNA removal kit (Epicentre). The quality of rRNA-depleted RNA samples was checked on a Bioanalyzer (Agilent Technologies). RNA-seq libraries for single-end 100-nucleotide sequencing were constructed using TruSeq RNA Sample Prep Kit v2 (Illumina) following the manufacturer's instructions exactly. Sequencing of the libraries was performed on a HiSeq 2000 sequencing system (Illumina).
Data analysis
Reads from sequencing were mapped to the B. subtilis strain 168 reference genome (NCBI database; NC_000964.3) using Bowtie2 (2.1.0) (Langmead & Salzberg, 2012) with “local” alignment (“--local”). Coding sequence information obtained from the NCBI database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/Bacillus_subtilis_168_uid57675/) was used in BEDtools to map reads to specific coding sequences. Custom Python scripts were developed to perform data normalization, data analysis and to generate histograms. We adopted the normalization method in the DESeq Bioconductor package (Anders & Huber, 2010). Briefly, for each RNA sample a ratio was calculated for each gene as read counts/geometric mean of read counts across all RNA samples. The median of this ratio was set as the normalization factor for each sample. Raw read counts were normalized by dividing the normalization factor of each sample. To quantify gene expression with RNA-seq data, the levels of gene expression were set as the normalized number of reads mapped to the CDS divided by the length of the CDS (read/base). The mean of triplicate samples was calculated.
Northern blotting
RNA isolated from Bacillus subtilis strains grown to mid-logarithmic phase in LB medium was fractionated either on a 1% denaturing agarose MOPS gel and blotted by wicking. 5’ end-labeled oligonucleotides were used as probes. The rapA 5’ and 3’ probes were complementary to nts 5-37 and 943-976 of the rapA CDS, respectively. The cggR 5’ and 3’ probes were complementary to nts 37-66 and 681-711 of the cggR CDS, respectively. To control for RNA loading in Northern blot analyses, membranes were stripped and probed for 5S rRNA as described (Sharp & Bechhofer, 2003).
Western blotting
Western blot analysis of RNase J1 was performed essentially as described (Durand et al., 2012) except that the secondary antibody was linked to horseradish peroxidase and revealed by chemiluminesce (GE; ECL Select). Quantification was done using a Chemidoc apparatus (Biorad).
Growth and motility phenotypes
Photos of wild-type and pnpA overnight cultures were taken on a Zeiss Axioplan2 camera after staining with 0.5% crystal violet. Qualitative and quantitative swarming motility assays were performed as described (Patrick & Kearns, 2009). SwrA expression in the 3610 pnpA swrA++ strain was induced with 1 mM IPTG.
Supplementary Material
Acknowledgements
This work was supported by Public Health Service grants GM-100137 (to D.H.B.) and GM-093030 (to D.B.K.) from the National Institutes of Health. We thank Sylvan Wallenstein for helpful discussion on statistical analysis, Hyung Min Cho for assistance in Python programming, and Hera Vlamakis of the Kolter lab for strains and advice on phage transduction. This work was supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano N, Inouye M, Phadtare S. RNase activity of polynucleotide phosphorylase is critical at low temperature in Escherichia coli and is complemented by RNase II. J Bacteriol. 2008;190:5924–5933. doi: 10.1128/JB.00500-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer DH. Chapter 6 Messenger RNA Decay and Maturation in Bacillus subtilis. Prog Nucleic Acid Res Mol Biol. 2009;85:231–273. doi: 10.1016/S0079-6603(08)00806-4. [DOI] [PubMed] [Google Scholar]
- Bechhofer DH, Wang W. Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J Bacteriol. 1998;180:5968–5977. doi: 10.1128/jb.180.22.5968-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- Calvio C, Osera C, Amati G, Galizzi A. Autoregulation of swrAA and motility in Bacillus subtilis. J Bacteriol. 2008;190:5720–5728. doi: 10.1128/JB.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Guttenplan SB, Blair KM, Kearns DB. Role of the sigmaD-dependent autolysins in Bacillus subtilis population heterogeneity. J Bacteriol. 2009;191:5775–5784. doi: 10.1128/JB.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010;7:316–321. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- Cozy LM, Kearns DB. Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol. 2010;76:273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven MG, Henner DJ, Alessi D, Schauer AT, Ost KA, Deutscher MP, Friedman DI. Identification of the rph (RNase PH) gene of Bacillus subtilis: evidence for suppression of cold-sensitive mutations in Escherichia coli. J Bacteriol. 1992;174:4727–4735. doi: 10.1128/jb.174.14.4727-4735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou-Chabo R, Mathy N, Benard L, Condon C. Ribosomes initiating translation of the hbs mRNA protect it from 5′-to-3′ exoribonucleolytic degradation by RNase J1. Mol Microbiol. 2009;71:1538–1550. doi: 10.1111/j.1365-2958.2009.06620.x. [DOI] [PubMed] [Google Scholar]
- Deikus G, Babitzke P, Bechhofer DH. Recycling of a regulatory protein by degradation of the RNA to which it binds. Proc Natl Acad Sci U S A. 2004;101:2747–2751. doi: 10.1073/pnas.0307343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikus G, Bechhofer DH. Initiation of decay of Bacillus subtilis trp leader RNA. J Biol Chem. 2007;282:20238–20244. doi: 10.1074/jbc.M702747200. [DOI] [PubMed] [Google Scholar]
- Deikus G, Condon C, Bechhofer DH. Role of Bacillus subtilis RNase J1 Endonuclease and 5′- Exonuclease Activities in trp Leader RNA Turnover. J Biol Chem. 2008;283:17158–17167. doi: 10.1074/jbc.M801461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Reuven NB. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz AR, Core LJ, Jiang M, Morelli M, Chiang CH, Szurmant H, Perego M. Bacillus subtilis RapA phosphatase domain interaction with its substrate, phosphorylated Spo0F, and its inhibitor, the PhrA peptide. J Bacteriol. 2012;194:1378–1388. doi: 10.1128/JB.06747-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M, Regnier P. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Durand S, Gilet L, Bessieres P, Nicolas P, Condon C. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 2012;8:e1002520. doi: 10.1371/journal.pgen.1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J Bacteriol. 2013;195:2340–2348. doi: 10.1128/JB.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamalli A, Hebert A, Zig L, Putzer H. Control of expression of the RNases J1 and J2 in Bacillus subtilis. J Bacteriol. 2014;196:318–324. doi: 10.1128/JB.01053-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdin VR, Singh D, Lin-Chao S. Composition and conservation of the mRNA-degrading machinery in bacteria. J Biomed Sci. 2011;18:23. doi: 10.1186/1423-0127-18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen SM, Haase C, Weil MR, Passalacqua KD, Niazi F, Hutchison SK, Desany B, Kolsto AB, Tourasse NJ, Read TD, Okstad OA. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biol. 2012;13:R30. doi: 10.1186/gb-2012-13-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Lewis RJ, Mader U, Stulke J. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol. 2012;84:1005–1017. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Schaffer M, Mader U, Diethmaier C, Herzberg C, Stulke J. RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011;81:1459–1473. doi: 10.1111/j.1365-2958.2011.07777.x. [DOI] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- Ludwig H, Homuth G, Schmalisch M, Dyka FM, Hecker M, Stulke J. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol Microbiol. 2001;41:409–422. doi: 10.1046/j.1365-2958.2001.02523.x. [DOI] [PubMed] [Google Scholar]
- Luo Y, Helmann JD. A sigmaD-dependent antisense transcript modulates expression of the cyclic-di-AMP hydrolase GdpP in Bacillus subtilis. Microbiology. 2012;158:2732–2741. doi: 10.1099/mic.0.062174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- Mader U, Zig L, Kretschmer J, Homuth G, Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Marquez LM, Helmann JD, Ferrari E, Parker HM, Ordal GW, Chamberlin MJ. Studies of sigma D-dependent functions in Bacillus subtilis. J Bacteriol. 1990;172:3435–3443. doi: 10.1128/jb.172.6.3435-3443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michna RH, Commichau FM, Todter D, Zschiedrich CP, Stulke J. SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res. 2014;42:D692–698. doi: 10.1093/nar/gkt1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Hue K, Bechhofer DH. In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase. Mol Microbiol. 1996;19:329–342. doi: 10.1046/j.1365-2958.1996.378906.x. [DOI] [PubMed] [Google Scholar]
- Mordini S, Osera C, Marini S, Scavone F, Bellazzi R, Galizzi A, Calvio C. The role of SwrA, DegU and P(D3) in fla/che expression in B. subtilis. PLoS One. 2013;8:e85065. doi: 10.1371/journal.pone.0085065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Hartig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stulke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Volker U, Bessieres P, Noirot P. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oussenko IA, Bechhofer DH. The yvaJ gene of Bacillus subtilis encodes a 3′-to-5′ exoribonuclease and is not essential in a strain lacking polynucleotide phosphorylase. J Bacteriol. 2000;182:2639–2642. doi: 10.1128/jb.182.9.2639-2642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oussenko IA, Sanchez R, Bechhofer DH. Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in gram-positive bacteria. J Bacteriol. 2002;184:6250–6259. doi: 10.1128/JB.184.22.6250-6259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J Bacteriol. 2009;191:7129–7133. doi: 10.1128/JB.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J, Liu Q, Pellegrini O, Celesnik H, Yao S, Bechhofer DH, Condon C, Belasco JG. An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol Cell. 2011;43:940–949. doi: 10.1016/j.molcel.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JS, Bechhofer DH. Effect of translational signals on mRNA decay in Bacillus subtilis. J Bacteriol. 2003;185:5372–5379. doi: 10.1128/JB.185.18.5372-5379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J Mol Biol. 2009;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bechhofer DH. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Bechhofer DH. Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y. J Bacteriol. 2010;192:3279–3286. doi: 10.1128/JB.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Richards J, Belasco JG, Bechhofer DH. Decay of a model mRNA in Bacillus subtilis by a combination of RNase J1 5′ exonuclease and RNase Y endonuclease activities. J Bacteriol. 2011;193:6384–6386. doi: 10.1128/JB.05939-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.