Abstract
AIM: To determine the existence of a potential relationship between the methylation state of the Vimentin gene and its prognostic value in pancreatic cancer.
METHODS: Sixty-four primary tumor specimens and normal tissues were collected consecutively from pancreatic cancer patients during surgery at Hangzhou First People’s Hospital and Affiliated Hospital of the Logistics University of the Chinese People’s Armed Police Force. DNA was extracted from the samples and subsequently quantitative methylation-specific polymerase chain reaction was used to detect the Vimentin methylation status of the samples. All of the patients were followed up to December 2012. χ2 test, Kaplan-Meier survival and Cox regression statistical models were used.
RESULTS: Out of 64 pancreatic cancer tissues, 21 were marked as Vimentin methylation-positive, and 43 were marked as Vimentin methylation-negative. The location of pancreatic carcinoma was related to the Vimentin methylation state. The pathological T staging (P < 0.001), adjuvant chemotherapy (P = 0.003) and the Vimentin methylation state (P = 0.037) were independent prognostic factors.
CONCLUSION: In our study, Vimentin methylation status can predict the prognosis of pancreatic cancer patients. However, additional experiments and clinical trials are needed to accurately validate this observation.
Keywords: Vimentin, Methylation, Pancreatic carcinoma, Prognosis
Core tip: Vimentin is reported to be an important mesenchymal marker, and plays an important role in epithelial-mesenchymal transition in malignant tumors with regard to cellular adhesion, migration and signaling. In our study, we found that pathological T staging (P < 0.001), adjuvant chemotherapy (P = 0.003) and the Vimentin gene methylation state (P = 0.037) were independent prognostic factors. However, additional experiments and clinical trials are needed to accurately validate this observation.
INTRODUCTION
Pancreatic cancer is one of the most lethal malignancies in humans, wherein the 5-year survival rate is less than 5%[1,2]. Most patients are diagnosed at an advanced stage of disease because of the lack of easily observable symptoms[3]. As reported, only 20% of pancreatic cancer patients are determined as suitable candidates to be considered for surgical resection. Moreover, following surgery, the 5-year survival rate of pancreatic cancer patients is approximately only 15%-25% due to the high recurrence and metastatic rates[4,5]. In the advanced stage of disease, the prognosis for these patients remains poor, even after chemotherapy and radiotherapy. The median overall survival rate is 3-6 mo for patients with metastatic disease and 6-10 mo for patients with non-metastatic disease[6,7]. Thus, early detection of pancreatic cancer is clearly important in order to augment the potential benefits and rates of an operation, and the prognosis of the patients. Recently, CA19-9 has been commonly used as a biomarker of pancreatic cancer. However, the specificity and sensitivity of this biomarker have not been satisfactory[8-10].
Vimentin is reported as an important mesenchymal marker, and plays an important role in epithelial-mesenchymal transition in malignant tumors with regard to cellular adhesion, migration and signaling[11,12]. Several investigators have previously shown that Vimentin is an important marker for the early detection of cancer, such as bladder cancer, hepatocellular carcinoma and colorectal cancer[13,14]. In addition, methylation of the Vimentin gene is described as a marker in several malignant tumors, including gastric carcinoma, colorectal carcinoma, cervical cancer and bladder cancer[13,15-17]. In our current study, we have attempted to identify the relationship between the methylation state of Vimentin and pancreatic cancer.
MATERIALS AND METHODS
Sample collection and DNA preparation
Sixty-four primary tumor specimens and normal tissues were collected consecutively from pancreatic cancer patients undergoing surgery at Hangzhou First People’s Hospital and Affiliated Hospital of the Logistics University of the Chinese People’s Armed Police Force. All specimens were confirmed by histopathology. Written informed consent was obtained from all patients. All the collected samples were stored at -80°C. DNA from the samples was extracted by QIAamp DNA Mini Kit (Catalog number: 51306, Qiagen, Hilden, Germany). The clinicopathological characteristics of the patients who were enrolled in our study are shown in Table 1.
Table 1.
Clinicopathological features of pancreatic carcinoma n (%)
| Data | Vimentin methylated negative group (n = 43)2 | Vimentin methylated positive group (n = 21)3 | P value |
| Sex | 0.582 | ||
| Male | 27 (62.8) | 15 (71.4) | |
| Female | 16 (37.2) | 6 (28.6) | |
| Tumor position | 0.007 | ||
| Head | 11 (25.6) | 13 (61.9) | |
| Body and tail | 32 (74.4) | 8 (38.1) | |
| Preoperative CEA level | 0.294 | ||
| Normal | 20 (46.5) | 13 (61.9) | |
| Elevated | 23 (53.5) | 8 (38.1) | |
| Preoperative CA19-9 level | 0.600 | ||
| Normal | 24 (55.8) | 10 (47.6) | |
| Elevated | 19 (44.2) | 11 (52.4) | |
| Pathological N staging1 | 0.426 | ||
| N0 | 21(48.8) | 13 (61.9) | |
| N1 | 22 (51.2) | 8 (38.1) | |
| Pathological T staging1 | 0.753 | ||
| T1 | 14 (32.6) | 5 (23.8) | |
| T2 | 19 (44.2) | 11 (52.4) | |
| T3 | 10 (23.3) | 5 (23.8) | |
| Adjuvant chemotherapy | 0.791 | ||
| No | 20 (46.5) | 11 (52.4) | |
| Yes | 23 (53.5) | 10 (47.6) | |
The pathological T and N staging was based on the UICC staging systems for pancreatic cancer;
Median age 54 years, range: 36-71 years;
Median age 53 years, range: 41-68 years.
Sodium bisulfite modification
Genomic tumor DNA (1 μg) and the corresponding normal pancreatic tissue specimens were subjected to bisulfite treatment using an Epitect Bisulfite Kit (Catalog No. 59104, Qiagen, Hilden, Germany)[18].
Quantitative methylation-specific polymerase chain reaction
The bisulfite-treated DNA was amplified using a quantitative methylation-specific polymerase chain reaction and a Thermal Cycler Dice® Real-time System TP800 (Takara Bio Inc., Otsu, Japan). Thermo-cycling was carried out in a final volume of 25 μL containing 1.0 μL of the DNA sample, 100 nmol/L each of the Vimentin or β-actin primers (forward and reverse sequences), and 12.5 μL of SYBR Premix Ex Taq II (Takara Bio Inc., Otsu, Japan), which consists of Taq DNA polymerase, PCR reaction buffer and deoxynucleotide triphosphate mixture. The qPCR primer sequences for Vimentin were: Vimentin MS (sense), 5’-TCGTTTCGAGGTTTTCGCGTTAGAGAC-3’, and Vimentin MAS (antisense), 5’-CGACTAAAACTCGACCGACTCGCGA-3’. The PCR amplification consisted of 40 cycles (95°C for 5 s and 55°C for 30 s) after an initial denaturation step (95°C for 10 s). To correct for differences between the samples in terms of both the quality and quantity of the purified DNA, β-actin was used as an internal control. The targets were obtained from the same bisulfite treated DNA.
Vimentin methylation scores
The relative levels of methylated Vimentin DNA in pancreatic cancer and the corresponding normal pancreatic tissues were calculated by normalizing to the internal control of β-actin. The Vimentin methylation scores were calculated by comparing the relative levels of Vimentin in pancreatic carcinoma with the corresponding levels in normal pancreatic tissue. The methylation status was recorded as positive when the Vimentin methylation score was greater than 1.0.
Follow-up
After treatment, the patients were monitored every month for the first year, every 3 mo for the second year, and then every 6 mo thereafter. Telephone calls and letters were used to identify patients who could not attend regular follow-up assessments. Complete data were collected for all 64 patients through December 31, 2012. The follow-up period ranged from 6 to 38 mo (with a median of 17 mo).
Statistical analysis
The χ2 test was used to compare categorical variables between the palliatively operated group with the other groups. Student’s t-tests were used to compare paired continuous variables. Univariate survival analysis was performed by Kaplan-Meier methods. Survival curves were compared using the log-rank test. Statistical analyses were performed with SPSS software version 20.0 for Windows (SPSS, Inc., Chicago, IL, United States). Statistical significance was defined as an alpha value of P < 0.05.
RESULTS
Vimentin methylation in pancreatic cancer and corresponding pancreatic tissues
We detected Vimentin methylation in pancreatic cancer and corresponding pancreatic tissues. Of the 64 pancreatic cancer tissues, 21 of them had a high-level methylation status and 45 of the corresponding pancreatic tissues had a high level of methylation. There were 9 pancreatic cancer tissues and 5 normal corresponding pancreatic tissues without methylation of the Vimentin gene. In addition, Vimentin methylation scores were recorded and informed that 43 of them were marked as Vimentin methylation-negative, and the remaining 21 were Vimentin methylation-positive.
Vimentin methylation state was related to the age and the diameter of the tumor
The clinicopathological factors seen between these two groups are summarized in Table 1. Moreover, we found that the location of the pancreatic carcinoma was associated with the status of Vimentin methylation. However, patient gender, preoperative serum tumor markers, lymph node metastasis and pathological T-stage were found not to be associated with the Vimentin methylation state.
Vimentin methylation state was an independent prognostic factor in pancreatic cancer
Univariate analysis showed that tumor position (P = 0.002), pathological T-staging (P < 0.001), adjuvant chemotherapy (P = 0.015) and the Vimentin methylation state (P = 0.013) were prognostic factors in pancreatic carcinoma (Table 2 and Figure 1). Multivariate analysis showed that the pathological T-staging (P < 0.001), adjuvant chemotherapy (P = 0.003) and the Vimentin methylation state (P = 0.037) were independent prognostic factors (Table 3).
Table 2.
Univariate analysis of overall survival in pancreatic carcinoma
| Variables | n | Median survival (mo) | P value |
| Sex | 0.819 | ||
| Male | 42 | 13.45 | |
| Female | 22 | 10.84 | |
| Preoperative CEA | 0.260 | ||
| Normal | 33 | 15.15 | |
| Elevated | 31 | 13.61 | |
| Preoperative CA19-9 | 0.947 | ||
| Normal | 40 | 14.12 | |
| Elevated | 24 | 15.09 | |
| Tumor position | 0.007 | ||
| Body and tail | 40 | 16.20 | |
| Head | 24 | 11.25 | |
| Pathological T stage | < 0.001 | ||
| T1 | 19 | 21.01 | |
| T2 | 30 | 13.10 | |
| T3 | 15 | 8.00 | |
| Pathological N stage | 0.311 | ||
| N0 | 34 | 14.28 | |
| N1 | 30 | 16.20 | |
| Adjuvant chemotherapy | |||
| Yes | 33 | 16.12 | 0.015 |
| No | 31 | 13.07 | |
| Vimentin methylation state | 0.013 | ||
| Positive | 21 | 11.09 | |
| Negative | 43 | 16.03 | |
Figure 1.
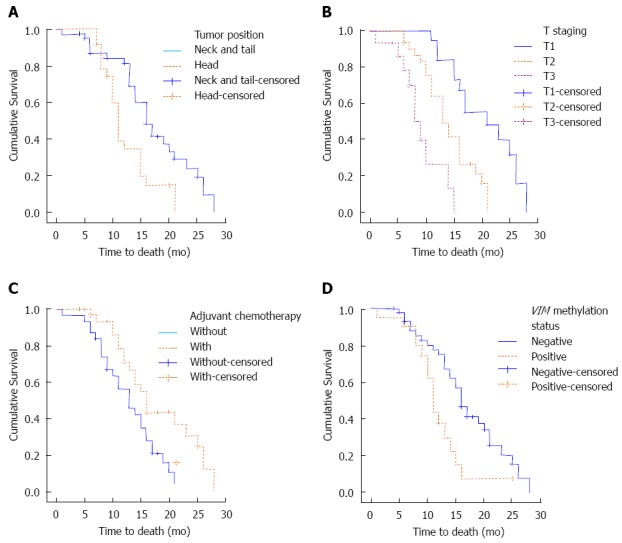
Univariate analysis: Tumor position, pathological T staging, adjuvant chemotherapy and vimentin methylation state were prognostic factors for the pancreatic carcinoma patients. A: Tumor position; B: T staging; C: Adjuvant chemotherapy; D: VIM methylation status.
Table 3.
Multivariate analyses of overall survival in pancreatic carcinoma (Cox’s regression model)
| Variable | HR | 95%CI | P value |
| OS in all pancreatic cancer patients | |||
| Tumor position | 1.789 | 0.873-3.666 | 0.112 |
| Pathological T staging | 4.026 | 2.283-7.101 | <0.001 |
| Adjuvant chemotherapy | 0.356 | 0.181-0.698 | 0.003 |
| Vimentin methylation state | 2.250 | 1.052-4.813 | 0.037 |
OS: Overall survival; HR: Hazard ratio.
DISCUSSION
An estimated 44030 people were diagnosed with pancreatic cancer and approximately 37660 people died of pancreatic cancer in the United States in 2011[19]. Although the technology of radiotherapy and chemotherapy has been developed, the incidence and mortality rates have remained approximately the same over the past two decades. A mutation in the CDKN2A (p16) gene has been reported in families with pancreatic cancer and melanoma[20,21]. An excess of pancreatic cancer is also seen in families harboring breast cancer susceptibility gene-2 mutations, and particular mutations in the PALB2 and MSH2 genes have recently been identified as possibly increasing pancreatic cancer susceptibility[22-25]. Vimentin is a key molecular marker of epithelial-mesenchymal transition, but it is not yet associated with either the occurrence or degree of malignancy of pancreatic carcinoma.
CpG methylation is a major epigenetic modification of genome DNA that is involved in the regulation of cell-specific gene expression[26]. It has been proposed that this modification may cause transcriptional repression by directly modulating transcription factor function or by triggering the formation of inactive chromatin[27,28]. We found that of 64 pancreatic cancer tissues, 21 of them displayed a high level of methylation status and 45 of the corresponding pancreatic tissues displayed a high level of methylation. Moreover, survival analysis showed that the Vimentin methylation status was an independent prognostic factor as well as a prognostic marker in T-staging and adjuvant chemotherapy. We are also aware that a low methylation status is always associated with high Vimentin expression levels. Additionally, Vimentin has been shown to be associated with several pathways, including cell adhesion, cytoplasmic microtubule assembly, and cytoskeleton remodeling. Higher Vimentin expression in pancreatic cancer cells may imply a higher state of malignancy of these cells, with an associated higher metastatic ability. The detailed mechanism of Vimentin and its gene methylation status requires further study.
Our observations only covered 64 pancreatic cancer patients, which is a small population sample. Although our results showed that the Vimentin methylation status could be used to predict prognosis in pancreatic cancer, more studies and clinical trials are needed to validate this result. In summary, our study showed that pancreatic cancer patients exhibiting a negative Vimentin methylation status displayed a poorer prognosis as compared with those with a positive status. The role of Vimentin methylation in pancreatic cancer warrants further empirical exploration.
COMMENTS
Background
Vimentin is reported as an important mesenchymal marker, and plays an important role in epithelial-mesenchymal transition in malignant tumors with regard to cellular adhesion, migration and signaling. In their current study, authors have attempted to identify the relationship between the methylation state of the Vimentin gene and pancreatic cancer.
Research frontiers
Several investigators have previously shown that Vimentin is an important marker for the early detection of cancer, such as bladder cancer, hepatocellular carcinoma and colorectal cancer. In addition, methylation of Vimentin is described as a marker in several malignant tumors, including gastric carcinoma, colorectal carcinoma, cervical cancer and bladder cancer.
Innovations and breakthroughs
The location of pancreatic carcinoma was related to the Vimentin methylation state. The pathological T staging (P < 0.001), adjuvant chemotherapy (P = 0.003) and the Vimentin methylation state (P = 0.037) were independent prognostic factors.
Applications
This result showed that the Vimentin methylation status could be used to predict prognosis in pancreatic cancer.
Peer review
The manuscript is very interesting. The authors try to determine the existence of a potential relationship between the methylation state of Vimentin and its prognostic value in pancreatic cancer. In total, 64 primary tumor specimens and normal tissues were collected in this study. The authors found that Vimentin methylation status can predict the prognosis of pancreatic cancer patients.
Footnotes
Supported by National Nature Science Foundation of China, No. 81001078
P- Reviewer: McHenry L, Schmidt S- Editor: Qi Y L- Editor: Logan S E- Editor: Du P
References
- 1.Michalski CW, Weitz J, Büchler MW. Surgery insight: surgical management of pancreatic cancer. Nat Clin Pract Oncol. 2007;4:526–535. doi: 10.1038/ncponc0925. [DOI] [PubMed] [Google Scholar]
- 2.Hirata K, Egawa S, Kimura Y, Nobuoka T, Oshima H, Katsuramaki T, Mizuguchi T, Furuhata T. Current status of surgery for pancreatic cancer. Dig Surg. 2007;24:137–147. doi: 10.1159/000102067. [DOI] [PubMed] [Google Scholar]
- 3.Cardenes HR, Chiorean EG, Dewitt J, Schmidt M, Loehrer P. Locally advanced pancreatic cancer: current therapeutic approach. Oncologist. 2006;11:612–623. doi: 10.1634/theoncologist.11-6-612. [DOI] [PubMed] [Google Scholar]
- 4.Carpelan-Holmström M, Nordling S, Pukkala E, Sankila R, Lüttges J, Klöppel G, Haglund C. Does anyone survive pancreatic ductal adenocarcinoma? A nationwide study re-evaluating the data of the Finnish Cancer Registry. Gut. 2005;54:385–387. doi: 10.1136/gut.2004.047191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 6.Sultana A, Smith CT, Cunningham D, Starling N, Neoptolemos JP, Ghaneh P. Meta-analyses of chemotherapy for locally advanced and metastatic pancreatic cancer. J Clin Oncol. 2007;25:2607–2615. doi: 10.1200/JCO.2006.09.2551. [DOI] [PubMed] [Google Scholar]
- 7.Hawes RH, Xiong Q, Waxman I, Chang KJ, Evans DB, Abbruzzese JL. A multispecialty approach to the diagnosis and management of pancreatic cancer. Am J Gastroenterol. 2000;95:17–31. doi: 10.1111/j.1572-0241.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 8.Wu X, Lu XH, Xu T, Qian JM, Zhao P, Guo XZ, Yang XO, Jiang WJ. Evaluation of the diagnostic value of serum tumor markers, and fecal k-ras and p53 gene mutations for pancreatic cancer. Chin J Dig Dis. 2006;7:170–174. doi: 10.1111/j.1443-9573.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- 9.Leung TK, Lee CM, Wang FC, Chen HC, Wang HJ. Difficulty with diagnosis of malignant pancreatic neoplasms coexisting with chronic pancreatitis. World J Gastroenterol. 2005;11:5075–5078. doi: 10.3748/wjg.v11.i32.5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou G, Chiu D, Qin D, Niu L, Cai J, He L, Huang W, Xu K. The efficacy evaluation of cryosurgery in pancreatic cancer patients with the expression of CD44v6, integrin-β1, CA199, and CEA. Mol Biotechnol. 2012;52:59–67. doi: 10.1007/s12033-011-9474-7. [DOI] [PubMed] [Google Scholar]
- 11.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 13.Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, Eknaes M, Skotheim RI, Rodrigues A, Magalhães JS, Oliveira J, Lothe RA, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16:5842–5851. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 14.Wong KF, Luk JM. Discovery of lamin B1 and vimentin as circulating biomarkers for early hepatocellular carcinoma. Methods Mol Biol. 2012;909:295–310. doi: 10.1007/978-1-61779-959-4_19. [DOI] [PubMed] [Google Scholar]
- 15.Shirahata A, Sakata M, Sakuraba K, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, et al. Vimentin methylation as a marker for advanced colorectal carcinoma. Anticancer Res. 2009;29:279–281. [PubMed] [Google Scholar]
- 16.Shirahata A, Sakuraba K, Kitamura Y, Yokomizo K, Gotou T, Saitou M, Kigawa G, Nemoto H, Sanada Y, Hibi K. Detection of vimentin methylation in the serum of patients with gastric cancer. Anticancer Res. 2012;32:791–794. [PubMed] [Google Scholar]
- 17.Jung S, Yi L, Kim J, Jeong D, Oh T, Kim CH, Kim CJ, Shin J, An S, Lee MS. The role of vimentin as a methylation biomarker for early diagnosis of cervical cancer. Mol Cells. 2011;31:405–411. doi: 10.1007/s10059-011-0229-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hibi K, Sakata M, Sakuraba K, Shirahata A, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, et al. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2008;28:1581–1584. [PubMed] [Google Scholar]
- 19.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 20.Ghiorzo P, Gargiulo S, Nasti S, Pastorino L, Battistuzzi L, Bruno W, Bonelli L, Taveggia P, Pugliese V, Borgonovo G, et al. Predicting the risk of pancreatic cancer: on CDKN2A mutations in the melanoma-pancreatic cancer syndrome in Italy. J Clin Oncol. 2007;25:5336–537; author reply 5336-537;. doi: 10.1200/JCO.2007.13.5624. [DOI] [PubMed] [Google Scholar]
- 21.Whelan AJ, Bartsch D, Goodfellow PJ. Brief report: a familial syndrome of pancreatic cancer and melanoma with a mutation in the CDKN2 tumor-suppressor gene. N Engl J Med. 1995;333:975–977. doi: 10.1056/NEJM199510123331505. [DOI] [PubMed] [Google Scholar]
- 22.Ferrone CR, Levine DA, Tang LH, Allen PJ, Jarnagin W, Brennan MF, Offit K, Robson ME. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J Clin Oncol. 2009;27:433–438. doi: 10.1200/JCO.2008.18.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 24.Slater EP, Langer P, Niemczyk E, Strauch K, Butler J, Habbe N, Neoptolemos JP, Greenhalf W, Bartsch DK. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78:490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 25.Lindor NM, Petersen GM, Spurdle AB, Thompson B, Goldgar DE, Thibodeau SN. Pancreatic cancer and a novel MSH2 germline alteration. Pancreas. 2011;40:1138–1140. doi: 10.1097/MPA.0b013e318220c217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 27.Shin TH, Paterson AJ, Grant JH, Meluch AA, Kudlow JE. 5-Azacytidine treatment of HA-A melanoma cells induces Sp1 activity and concomitant transforming growth factor alpha expression. Mol Cell Biol. 1992;12:3998–4006. doi: 10.1128/mcb.12.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammanamanchi S, Kim SJ, Sun LZ, Brattain MG. Induction of transforming growth factor-beta receptor type II expression in estrogen receptor-positive breast cancer cells through SP1 activation by 5-aza-2’-deoxycytidine. J Biol Chem. 1998;273:16527–16534. doi: 10.1074/jbc.273.26.16527. [DOI] [PubMed] [Google Scholar]


