Abstract
Cyclin J (CycJ) is a poorly characterized member of the Cyclin superfamily of cyclin-dependent kinase regulators, many of which regulate the cell cycle or transcription. Although CycJ is conserved in metazoans its cellular function has not been identified and no mutant defects have been described. In Drosophila, CycJ transcript is present primarily in ovaries and very early embryos, suggesting a role in one or both of these tissues. The CycJ gene (CycJ) lies immediately downstream of armitage (armi), a gene involved in the Piwi-associated RNA (piRNA) pathways that are required for silencing transposons in the germline and adjacent somatic cells. Mutations in armi result in oogenesis defects but a role for CycJ in oogenesis has not been defined. Here we assessed oogenesis in CycJ mutants in the presence or absence of mutations in armi or other piRNA pathway genes. CycJ null ovaries appeared normal, indicating that CycJ is not essential for oogenesis under normal conditions. In contrast, armi null ovaries produced only two egg chambers per ovariole and the eggs had severe axis specification defects, as observed previously for armi and other piRNA pathway mutants. Surprisingly, the CycJ armi double mutant failed to produce any mature eggs. The double null ovaries generally had only one egg chamber per ovariole and the egg chambers frequently contained an overabundance of differentiated germline cells. Production of these compound egg chambers could be suppressed with CycJ transgenes but not with mutations in the checkpoint gene mnk, which suppress oogenesis defects in armi mutants. The CycJ null showed similar genetic interactions with the germline and somatic piRNA pathway gene piwi, and to a lesser extent with aubergine (aub), a member of the germline-specific piRNA pathway. The strong genetic interactions between CycJ and piRNA pathway genes reveal a role for CycJ in early oogenesis. Our results suggest that CycJ is required to regulate egg chamber production or maturation when piRNA pathways are compromised.
Background
Cyclin J (CycJ) is a poorly characterized member of the cyclin superfamily of proteins. Cyclins are eukaryotic proteins that contain a cyclin box, a domain that interacts with cyclin-dependent kinases (Cdks) (Hadwiger et al., 1989; Jeffrey et al., 1995). Many cyclins are known to have conserved roles in regulating the cell cycle. In metazoan species from Drosophila to human, for example, A and B cyclins regulate mitotic events, D cyclins regulate progression through G1, and E cyclins regulate entry into S phase (Minshull et al., 1989; Murray, 2004). Other cyclins have conserved roles in regulating transcription or other cellular processes (Lim and Kaldis, 2013). CycJ is conserved in all metazoans, yet it has only been studied in Drosophila were it was originally identified as a Cdk-interacting protein (Finley and Brent, 1994; Finley et al., 1996). The RNA expression pattern of CycJ is unique among Drosophila cyclins and suggests a possible role in oogenesis or embryogenesis. CycJ mRNA is present almost exclusively in ovaries and early embryos, whereas all other cyclins are expressed in multiple tissues and stages of development (Figure S1) (Arbeitman et al., 2002; Chintapalli et al., 2013; Finley et al., 1996; Graveley et al., 2011). A potential role for CycJ in embryogenesis was suggested in a study showing that injection of syncytial embryos with CycJ-inhibitory antibodies or peptide aptamers resulted in delays of the early nuclear division cycles (Kolonin and Finley, 2000). In another study, however, Althoff et al. examined embryos from CycJ null females and observed no obvious cell cycle defects (Althoff et al., 2009). That study also failed to detect CycJ protein expression in embryos using a genomic CycJ transgene fused to the green fluorescent protein gene, GFP. In contrast, the GFP-CycJ fusion could be detected in ovaries in all germline cells. In this study we set out to determine whether CycJ plays a role in ovaries where both the RNA and protein appear to be maximally expressed.
Oogenesis in Drosophila takes place in a series of parallel tubular structures called ovarioles, each of which is divided into an anterior region called the germarium and a posterior chain of developing egg chambers (Figure S2). Oogenesis is initiated when one of the two or three germline stem cells (GSCs) located at the anterior tip of a germarium undergoes mitotic division giving rise to a new stem cell and a differentiating daughter cell called a cystoblast (Schupbach et al., 1978; Spradling, 1993; Wieschaus and Szabad, 1979). The new stem cell remains in the stem cell niche at the anterior tip of the germarium where signaling from neighboring somatic terminal filament and cap cells leads to repression of differentiation factors (King and Lin, 1999; King et al., 2001; Song et al., 2004; Xie and Spradling, 1998, 2000). The cystoblast undergoes exactly four rounds of division with incomplete cytokinesis to give rise to 16 cystocytes interconnected by structural cell-cell connections known as fusomes and ring canals. The 16-cell cysts migrate toward the posterior region of the germarium where follicle cells encapsulate them to form egg chambers. One of the germline cells undergoes meiosis and becomes the oocyte while the other 15 undergo endoreduplication to become nurse cells that eventually donate their cytoplasm to the oocyte through the ring canals. Ovarioles consist of long chains of egg chambers that mature and increase in size during posterior migration culminating in the formation of a mature stage 14 oocyte.
Several aspects of oogenesis depend on gene silencing pathways that involve the ∼25-nucleotide small non-coding RNAs called PIWI-associated RNAs (piRNAs), which are synthesized from longer genome-encoded transcripts (Guzzardo et al., 2013; Khurana and Theurkauf, 2010; Thomson and Lin, 2009). piRNAs associate with the PIWI subfamily of argonaut proteins (Piwi, Aub, and Ago3) to silence transposons in the germline and adjacent somatic cells. The piRNA pathways silence transposons either by affecting chromatin structure or by targeting specific transposon RNAs for destruction (Brennecke et al., 2007; Peng and Lin, 2013; Vagin et al., 2006). Over 20 genes are known to be involved in the biogenesis and function of piRNAs, and many additional candidate piRNA pathway genes have been identified by large-scale screens (Czech et al., 2013; Handler et al., 2011; Muerdter et al., 2013). Mutations in several of the piRNA pathway genes (e.g., armi, and aub) result in transposon derepression accompanied by DNA damage accumulation (Haase et al., 2010; Khurana et al., 2010; Klattenhoff et al., 2007). The DNA damage activates checkpoint kinases that lead to disruption of the dorsal-ventral and anterior-posterior patterning of the oocyte (axis specification). The piRNA pathway also regulates germline stem cell maintenance both cell autonomously and from adjacent somatic cells by mechanisms that are not well understood (Juliano et al., 2011; Kirilly and Xie, 2007; Smulders-Srinivasan et al., 2010).
Here we set out to determine whether or not CycJ plays a role in oogenesis. We generated a deletion of the genomic region containing armi and CycJ, and by adding back individual transgenes, created null mutants for each gene. We show that while oogenesis is normal in the CycJ null, the armi null produces few egg chambers and mature eggs, all of which have axis specification defects. Surprisingly, in the armi-CycJ double null there was a further decrease in the number of egg chambers per ovariole, a drastic increase in the number of differentiated germline cells in each egg chamber, and no mature eggs. The armi null defects could be suppressed by mutation in the Chk2 checkpoint kinase gene as shown previously, but the armi-CycJ double null defects could not. We observed a similar genetic interaction between CycJ and two other piRNA pathway genes, piwi and aub, suggesting that CycJ plays a nonredundant role in oogenesis when the piRNA pathways are compromised.
Results
Construction of CycJ and armi null mutants
We set out to generate a CycJ null mutant by first creating a deletion of armi, CycJ and an uncharacterized gene, CG14971, and then replacing each of the three genes with genomic transgenes either individually or in combinations (Atikukke, 2009). We used flippase (FLP) induced recombination between two transposon insertion alleles, XPd07385 and RBe01160 that contain FLP recombination target (FRT) sites (Thibault et al., 2004). The resultant deletion strain, hereafter referred to as Df(3L)armi-J, eliminated the genomic region corresponding to all coding and noncoding exons of armi, CycJ and CG14971 (Figure 1). The deletion boundaries were confirmed by sequencing (Materials and Methods). The same three-gene deletion was created independently in other studies (Althoff et al., 2009; Olivieri et al., 2010), but oogenesis defects were not characterized in the individual and double mutants.
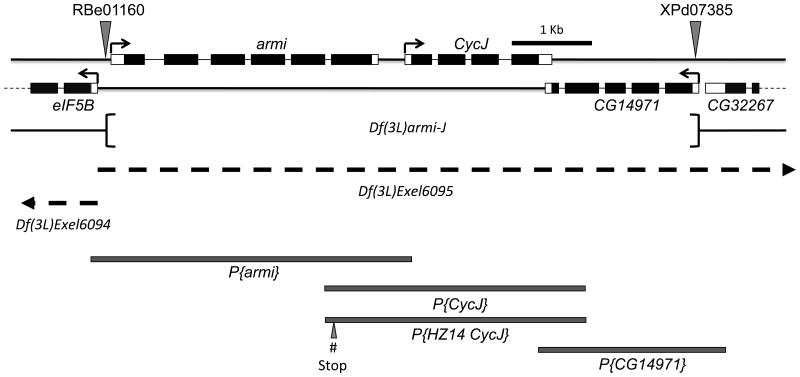
Figure 1.
Schematic representation of the genomic region corresponding to CycJ, armi, and the neighboring genes on chromosome 3L. The region shown corresponds to estimated cytological band 63E1. Black boxes represent coding regions and open boxes represent 5' and 3' untranslated regions. armi and CycJ are transcribed left to right while all other genes shown are transcribed right to left (arrows). FRT-bearing transposon insertions RBe00161 and XPd07385 (shown by triangles) were used to delete the intervening genomic region. The deleted genomic region is represented as Df(3L)armi-J. Rescue experiments were conducted by using independent transgenes, P{armi}, P{CycJ}, and P{CG14971} that were generated using the indicated genomic regions (shaded boxes). Rescue experiments also used a second CycJ genomic transgene, P{HZ14CycJ}, that introduced stop codons (# stop) in the armi coding region eliminating armi coding potential. Regions missing in the deficiency chromosomes Df(3L)Exel6094 and Df(3L)Exel6095 are indicated by dashed lines and extend beyond the region shown (arrowheads).
The homozygous Df(3L)armi-J mutant flies were viable indicating that armi, CycJ and CG14971 are not essential for viability or development to adulthood, a conclusion that was also reached in previous studies (Althoff et al., 2009; Olivieri et al., 2010). The deletion, however, resulted in complete male and female sterility, and the females did not lay any eggs (Table 1). As described in detail below, the three-gene deletion resulted in major defects in oogenesis. To analyze the contributions of individual genes to these phenotypes, we constructed transgenic lines containing genomic clones of each gene (Figure 1) individually and in different combinations and tested their ability to modify the Df(3L)armi-J phenotypes. Several lines of evidence suggest that all of the oogenesis defects that we observed with Df(3L)armi-J are due to loss of either armi, CycJ, or both genes and not to loss of CG14971 or disruption of the immediate upstream gene, eIF5B. First, Df(3L)armi-J complemented a lethal deficiency (Df(3L)Excel6094) (Figure 1) that removes eIF5B, which is an essential gene (Carrera et al., 2000), suggesting that eIF5B is not affected in the mutant that we generated. Transheterozygous Df(3L)armi-J / Df(3L)Excel6094 males and females are viable and fertile, as are animals with Df(3L)Exel6094 over a deficiency (Df(3L)Exel6095) that removes CycJ, armi, and several downstream genes (Figure 1). Furthermore, ovaries from Df(3L)6094/Df(3L)armi-J females or Df(3L)6094/Df(3L)6095 females showed show no signs of oogenesis defects (Figure S6). Second, we found that addition of the CG14971 transgene into Df(3L)armi-J females either in the presence or absence of the CycJ and armi transgenes had no effect on any of the phenotypes that we observed (Table 1). Finally, the combination of both armi and CycJ transgenes was sufficient to fully suppress all of the oogenesis defects described below, suggesting that CG14971 does not play a role in these phenotypes. Thus, we refer to either homozygous Df(3L)armi-J or transheterozygous Df(3L)armi-J/Df(3L)Exel6095 animals as armi-CycJ double null.
Table 1. Egg laying, morphology, and hatching rate defects in armi and CycJ mutants.
| Maternal Genotype | Genotype abbreviation | Eggs laida | Egg morphologyb (dorsal appendages) | Hatching Ratec | ||
|---|---|---|---|---|---|---|
| Two | One | None | ||||
| Df(3L)armi-J /+ | 100% | 100% | 0% | 0% | 96% | |
| Df(3L)armi-J or Df(3L)armi-J/Df(3L)Exel6095 | arminull CycJnull (CG14971null) | NONE | NA | NA | NA | NA |
| P{CG14971}; Df(3L)armi-J | arminull CycJnull | NONE | NA | NA | NA | NA |
| P{CycJ}; Df(3L)armi-J | arminull (CG14971null) | <1% | 0% | 0% | 100% | 0% |
| P{CG14971}; P{CycJ} ; Df(3L)armi-J | arminull | <1% | 0% | 0% | 100% | 0% |
| P{armi}; Df(3L)armi-J | CycJnull (CG14971null) | 65.3% | 92% | 7% | 1% | 30% |
| P{armi} / P{CG14971}; Df(3L)armi-J | CycJnull | 65.1% | 91% | 8% | 1% | 30% |
| P{armi}; P{CycJ}; Df(3L)armi-J | CG14971null | 66.7% | 94% | 6% | 0% | 73% |
| P{armi}; P{CycJ}; Df(3L)armi-J/Df(3L)Exel6095 | CG14971null | 79.2% | 90% | 7% | 3% | 76% |
| P{armi} / P{CG14971}; P{CycJ};Df(3L)armi-J | 67.4% | 93% | 7% | 0% | 75% | |
| Df(3L)armi-J / armi72.1 | armi72.1 (hypomorph) | 35.3% | 0% | 28% | 72% | 0% |
| aubHN / aubQC | aub | 75.0% | 8% | 68% | 24% | 0% |
| P{armi}; aubHN / aubQC; Df(3L)armi-J | CycJnull, aub | NONE | NA | NA | NA | NA |
| mnkP6; Df(3L)armi-J / armi72.1 | armi72.1, mnk | 89.4% | 85% | 8% | 7% | 0% |
| mnkP6; Df(3L)armi-J | arminull CycJnull, mnk | NONE | NA | NA | NA | NA |
| P{CycJ}; mnkP6;Df(3L)armi-J | arminull, mnk | 16.8% | 77% | 12% | 9% | 0% |
Percent of the number eggs laid by Df(3L)armi-J/+ (n=1118) over the same period of time. For the two armi null genotypes, repeated collections yielded only 4 or 5 eggs per collection. armi-CycJ null and CycJ null aub double mutants laid no eggs.
Number of dorsal appendages per egg (wild-type is two). Axis establishment defects lead to ventralized eggs, which have fewer than two dorsal appendages. NA, not applicable (no eggs laid).
Embryo hatching rates relative to wild-type (Materials and Methods).
Loss of both armi and CycJ results in severely disorganized ovarioles containing compound egg chambers
To look for oogenesis defects that may explain the lack of eggs from armi-CycJ double null females, we analyzed ovaries at various time points during the first week after eclosion. We determined that well-fed 3-day old flies were at the optimal age to allow observation of developmental abnormalities in the ovarioles before they became devoid of developing egg chambers. This analysis revealed that armi-CycJ double null females have oogenesis defects that were different from either single null fly (described below). Germaria from armi-CycJ double null animals were disorganized and the ovarioles generally contained only one egg chamber with many more than the normal number of germline cells (Figure 2B-C). The single egg chamber usually had a terminal filament (Figure 2C-D), suggesting that the germarium failed to package individual cysts and became the only egg chamber. The germline nuclei were of different sizes suggesting that they have undergone different numbers of endoreplicative cycles. Many egg chambers also showed an intense punctate DAPI staining pattern that increased as the females aged beyond 3 days, suggestive of increasing cell death (arrows in Figure 2E and F). We observed identical phenotypes in ovaries from animals homozygous for Df(3L)armi-J or transheterozygous for Df(3L)armi-J and the deficiency (Df(3L)Exel6095) that uncovers armi, CycJ, and a number of additional genes (Figure 2B-C). Ovaries from Df(3L)armi-J mutants that were over 1 week old contained ovarioles with disorganized germaria, no developing egg chambers, and few visible nuclei (Figure 2G). These observations suggest that as females age, further development ceases and the few egg chambers that initially developed are eliminated, leaving only disorganized germaria.
Figure 2.
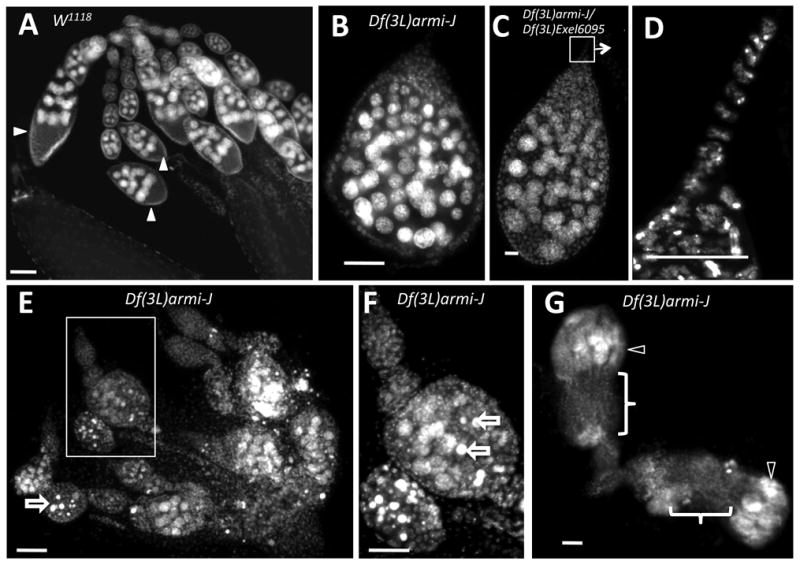
Deletion of both armi and CycJ results in accumulation of egg chambers with excess germline cells. Ovaries from wild type (w1118) and Df(3L)armi-J mutant females stained with DAPI to visualize nuclei. (A) Wild-type ovarioles from 3-day old or 8-day old females (not shown) consist of chains of developing egg chambers (arrows). Each egg chamber has 15 nurse cells undergoing endoreplication (large nuclei) and a single oocyte located at the posterior end (arrowheads). (B) A typical cyst from 3-day old Df(3L)armi-J females has many more than 16 nuclei undergoing endoreplicative cycles along with pycnotic nuclei. (C) An ovariole from a Df(3L)armi-J/Df(3L)Exel6095 transheterozygote, which have a phenotype identical to Df(3L)armi-J. (D) A higher magnification of the terminal filament region in C. (E) Ovaries from 3-day old Df(3L)armi-J females contain abnormal egg chambers with more than the normal number of endoreplicating nuclei. Some cells appear to be undergoing cell death as indicated by the characteristic pycnotic nurse cell nuclei (open arrows), which is evident at higher magnification in (F). (G) Ovaries from 8-day old Df(3L)armi-J females contain empty ovarioles (bracket) with disorganized germaria (open arrowheads). Anterior is generally toward the top of each figure; in (G) the anterior of the lower ovariole is toward the right. Size bar is 50 µm in all panels except E and G, in which it is 20 µm.
To determine whether the germline cells in homozygous Df(3L)armi-J were differentiated we stained ovaries with antibodies against Hts-RC, a component of the ring canals that connect cystocytes (Robinson et al., 1994), and Orb, an oocyte-specific marker (Lantz et al., 1994). In wild-type ovaries, each cystoblast undergoes four rounds of cell division with incomplete cytokinesis resulting in 16 cystocytes that are connected by 15 ring canals (Figure 3A-B) (Spradling, 1993). One of the two cystocytes with four ring canals eventually adopts the oocyte fate, as is evident from Orb staining, while the other 15 cystocytes undergo endoreduplication to become nurse cells (Figure 3E, H). In the Df(3L)armi-J mutant, ring canals appear to develop normally at least initially, though there are far more than the normal number in each egg chamber (Figure 3C-D). Orb staining revealed that the process of oocyte selection takes place, though again there were two or more Orb-staining cells in each egg chamber of the armi-CycJ double null, suggesting that multiple cysts are enclosed in a single follicular layer (Figure 3F-G). Combined, these results indicate that in the armi-CycJ double null, cystoblast division and differentiation occur, giving rise to several groups of germline cells consisting of nurse cells and an oocyte all contained in a single compound egg chamber.
Figure 3.
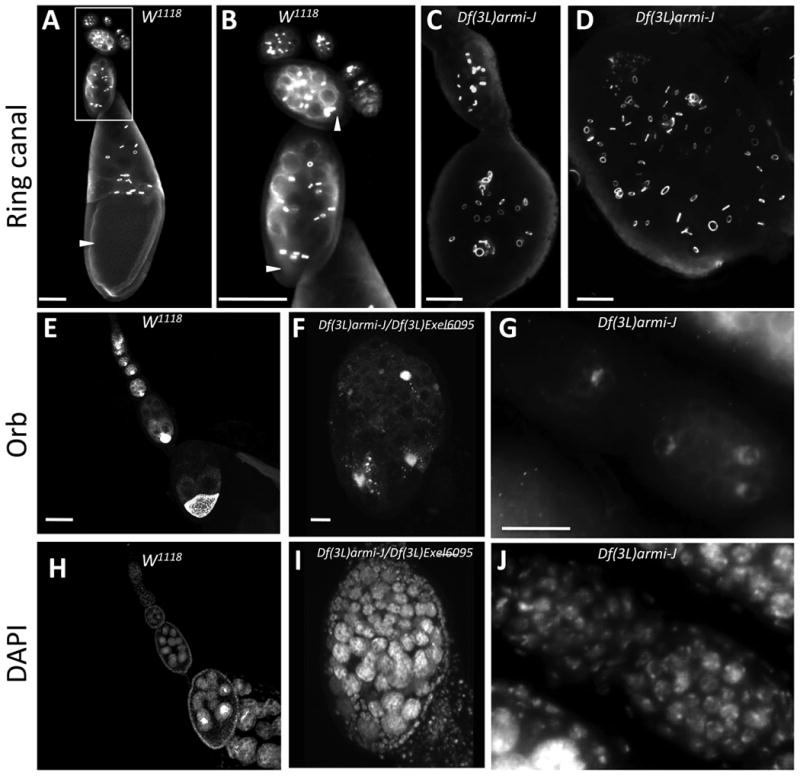
Multiple cystoblasts divide and differentiate within a single egg chamber in armi-CycJ mutants. Ovaries from wild type (w1118) (A, B) and Df(3L)armi-J (C,D) adults stained with anti-hts-RC antibody to visualize ring canals. In wild-type ovarioles each egg chamber contains a single posteriorly located oocyte (arrowheads) with four associated ring canals. (B) shows the boxed region in A at higher magnification. The Df(3L)armi-J mutant ovarioles show multiple abnormalities (C and D). Germarial patterning is disrupted and each egg chamber carries a giant cyst with multiple clusters of 16 cells interconnected by ring canals. (E-J) Ovarioles from wild-type (w1118) (E, H), Df(3L)armi-J/Df(3L)Exel6095 (F, I), and Df(3L)armi-J (G, J) stained with DAPI (H-J) and Orb (E-G) to visualize oocytes. In wild-type ovarioles each egg chamber contains one Orb-staining oocyte. In the mutant, each giant cyst contains multiple Orb-staining nuclei adjacent to endoreplicating nurse cells. Anterior is towards the top in A and upper left in the rest of the images. Size bars are 50 µm in A-E and H, and 20 µm in F, G, I, and J.
CycJ transgenes suppress oogenesis defects in the armi-CycJ double mutant
To determine whether the absence of CycJ contributes to defects observed in the armi-CycJ null, we introduced CycJ transgenes into Df(3L)armi-J homozygotes and Df(3L)armi-J/Df(3L)Exel6095 transheterozygotes. We used two different genomic CycJ transgenes and obtained similar results with each. One transgene contained CycJ and its upstream region including armi exon 9 and part of armi exon 8 (Figure 1). The other transgene included the same region but had stop codons introduced into armi exon 8 to guard against the possible expression of a C-terminal fragment of armi. We found that introduction of the CycJ transgenes significantly suppressed the armi-CycJ double null phenotypes (Figure 4C and D). Unlike Df(3L)armi-J, the Df(3L)armi-J females containing a CycJ transgene were capable of producing fully developed stage 14 egg chambers and of laying eggs, albeit at a dramatically reduced rate compared to heterozygotes (Table 1). Introduction of the CycJ transgenes also resulted in a dramatic decrease in the frequency of compound egg chambers (Figures 5 and S4). We observed similar results with the two different CycJ transgenes and with multiple independent insertion lines. Germline expression of a myc-tagged Cyclin J in the Df(3L)armi-J mutant (Figure S5) also reduced the frequency of compound egg chambers (Figure 5 and S4). This indicates that it is likely the coding region of CycJ that modifies the armi-CycJ phenotypes and suggests that CycJ can function in the germline. A CG14971 transgene did not modify the oogenesis phenotypes in any background. Combined, these results indicate that the oogenesis defects observed in Df(3L)armi-J are due to the absence of both armi and CycJ.
Figure 4.
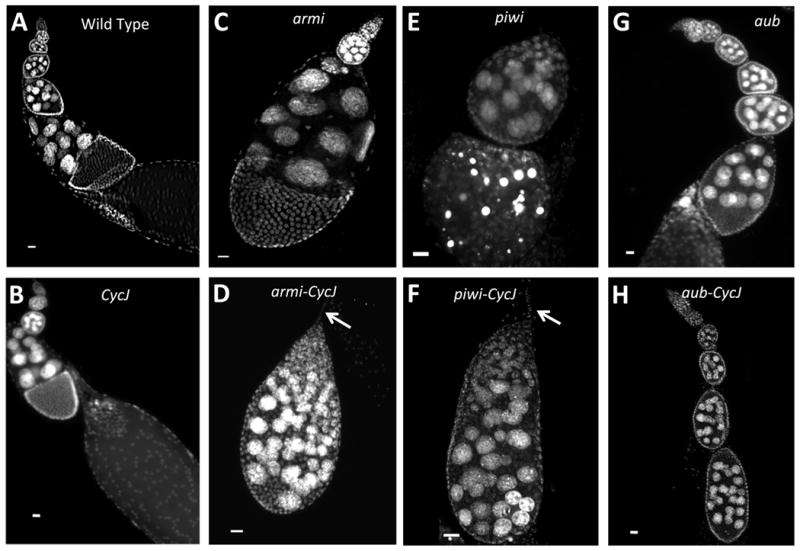
CycJ genetically interacts with armi, piwi, and aub. As in wild type (A), ovarioles from the CycJ null (Df(3L)armi-J with the P{armi} transgene) appear normal (B), with long chains of egg chambers each containing 15 nurse cell nuclei. Ovarioles from the armi null (C) (Df(3L)armi-J with a P{CycJ} transgene) frequently have just two egg chambers per ovariole, and egg chambers have 15 nurse cell nuclei. The armi-CycJ null (D) exhibits drastic oogenesis defects epitomized by production of ovarioles with one egg chamber that often has many more than 15 nurse cells. Like the armi null, piwi mutants (E) (piwi[06843]/Df(2L)BSC145) exhibit a decreased number of egg chambers per ovariole. piwi-CycJ double mutants (F) phenocopy the armi-CycJ, producing compound egg chambers with more than 15 nurse cells. The aub mutant (G) (aubHN/aubQC42) showed all stages of egg chamber development each with the normal number (15) of nurse cell nuclei. In aub-CycJ double mutants (H), oogenesis arrested prior to stage 8 with some egg chambers harboring many more than 15 nurse cells. Arrows in D and F point to the terminal filament. Anterior is towards the top and size bars are 20 µm in all panels.
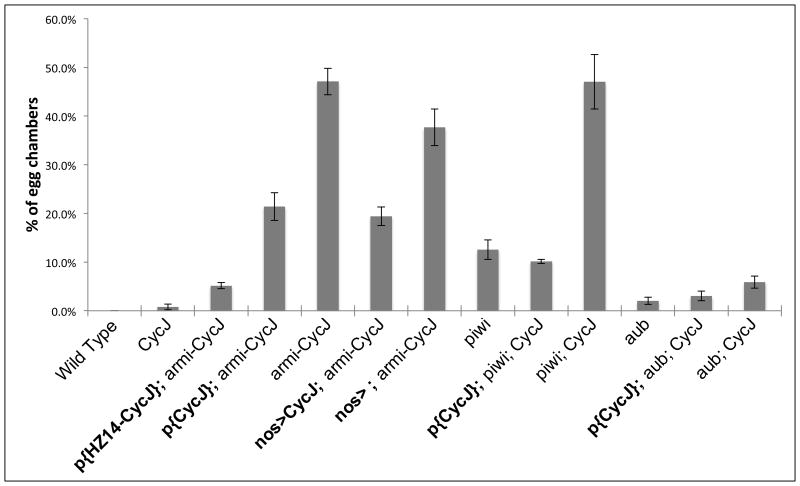
Figure 5.
CycJ expression suppresses compound egg chamber production in armi-CycJ, piwi-CycJ, and aub-CycJ. The percent of egg chambers with more than 15 nurse cell nuclei is shown, error bars = SEM (0.00% to 5.58%). Transgenes are in bold. CycJ null egg chambers are the same as wild type with respect to the number of nurse cell nuclei. In armi-CycJ null ovaries, 47.1% +/-2.7% of egg chambers have an excess of germline cells, which is indicative of a compound egg chamber (see examples in Fig. 2). CycJ genomic transgenes (P{CycJ} or P{HZ14_CycJ}) added to the armi-CycJ null significantly decreased the frequency of compound egg chambers. Germline specific expression of UAS-Myc-CycJ with the VP16::nos-Gal4 driver in armi-CycJ null also decreased compound egg chamber production. P{CycJ} added back to both piwi-CycJ and aub-CycJ was able to rescue compound egg chamber production back to the level of the piwi and aub single mutants, respectively. Note that armi-CycJ and piwi-CycJ exhibit the same high level of compound egg chamber production. The CycJ null is P{armi}, Df(3L)armi-J/Df(3L)Exel6095. The armi-CycJ double null in each case is Df(3L)armi-J/Df(3L)Exel6095. The piwi mutant is piwi[06843]/Df(2L)BSC145. The aub mutant is aubHN/aubQC42. P-values are shown in Figure S4. All ovaries are from females 2-4 days post eclosion except piwi mutants, which are 0-2 days.
These results also suggest that the defects observed in the Df(3L)armi-J animals that contain a CycJ transgene are due to the complete loss of armi; i.e., they represent the phenotype of an armi null. These armi null females produced mature eggs that were often collapsed and that had dorso-ventral axis establishment defects (Table 1), a phenotype previously observed for a strong armi loss-of-function allele, armi72.1 (Cook et al., 2004). The armi null ovaries had significantly fewer egg chambers per ovariole compared to wild-type (Figure S4). armi null ovaries also showed frequent mislocalization of oocytes within egg chambers (Figure S3). Rather than being positioned in the posterior end of each egg chamber as seen in wild-type ovarioles, oocytes were found located at random positions in a majority of the armi null egg chambers. This phenotype was originally identified in mutants of spindle C (spn-C) and was later observed for other spindle class genes including spn-A, spn-B, spn-D and spn-E (Gonzalez-Reyes et al., 1997; Gonzalez-Reyes and St Johnston, 1994). While this severe patterning defect was not reported for the previous armi loss of function mutant, it is consistent with the established role for armi in axis specification (Cook et al., 2004).
CycJ is not essential for oogenesis
The ability of P{CycJ} transgenes to rescue the production of egg chambers with normal numbers of germline cells shows that CycJ plays a role in egg chamber formation, packaging, or maturation, at least in an armi loss-of-function background. To assess whether CycJ is important in the presence of wild-type armi we introduced a genomic armi transgene (P{armi}) into the Df(3L)armi-J background to generate a CycJ null. Previously it was shown that an armi transgene was able to restore fertility to animals with the same three-gene deletion (Althoff et al., 2009). We further found that P{armi} completely suppressed all of the specific oogenesis defects associated with the Df(3L)armi-J deletion mutant or with the armi null (Figures 4B and S4). All oocyte developmental stages appeared normal in the P{armi}-complemented Df(3L)armi-J ovaries. Specifically, cysts developed with the appropriate number of nuclei and ring canals, the size of nurse cell nuclei suggested normal differentiation and endoreplication, and each ovariole had the normal number of egg chambers with a single oocyte located at the posterior end (Figures 4, 5, and S3). The P{armi}; Df(3L)armi-J/Df(3L)armi-J females were fertile and laid eggs with normal dorsal appendages, though only 30% of the eggs hatched and developed normally (Table 1). Providing a CycJ transgene along with the armi transgene in Df(3L)armi-J/Df(3L)armi-J or Df(3L)armi-J/Df(3L)Exel6095 significantly improved the hatching rate to 73 - 76% (Table 1). The introduction of a CG14971 transgene had no effect on any of the oogenesis phenotypes or hatching rates (Table 1). Thus, while the failure to rescue hatching rates to 100% of wild-type suggests that at least one of the three transgenes we introduced fails to fully complement for embryogenesis functions, all of the oogenesis defects that we describe are accounted for by loss of either armi or both armi and CycJ.
CycJ genetically interacts with multiple members of the piRNA pathways
The strong genetic interaction between CycJ and armi suggested that CycJ may be required when a piRNA pathway is compromised. To further test this possibility we examined the effect of removing CycJ in the background of mutants in two other piRNA pathway genes, piwi and aub (Figure 4). First, we examined a piwi loss-of-function mutant piwi[06843] over a deficiency Df(2L)BSC145 lacking piwi; we refer to piwi[06843]/ Df(2L)BSC145 as the piwi mutant in this study. piwi mutant ovaries were examined between zero and two days post eclosion because beyond two days they frequently contained agametic germaria and lacked egg chambers. Similar to the armi null, the piwi mutant produced only one to two egg chambers per ovariole and occasional compound egg chambers (Figures 4E and S4). Significantly, the piwi-CycJ double mutants exhibited a dramatic increase in the frequency of compound egg chambers to a level identical to that seen in the armi-CycJ double null (Figures 4F and 5). Addition of a CycJ transgene to the piwi-CycJ double mutant decreased the frequency of compound egg chambers to the levels seen in piwi mutants (Figure 5). These results define a strong genetic interaction between CycJ and a second piRNA pathway member.
Next we tested for a genetic interaction between CycJ and aub. aub is required for synthesis of piRNAs and for silencing of transposons specifically in germline cells (Aravin et al., 2004; Li et al., 2009; Malone et al., 2009; Vagin et al., 2006). aub mutants are similar to armi hypomorphs with respect to egg morphology, axis defects, accumulation of DNA double strand breaks, and checkpoint activation (Klattenhoff et al., 2007; Schupbach and Wieschaus, 1991). To test for a genetic interaction between aub and CycJ we generated females that were CycJ null and transheterozygous for the aub alleles, aubHN and aubQC42 (hereafter referred to as aub mutants). While the aub mutants exhibited a modest decrease in egg laying capacity (75% of wild-type), the aub-CycJ double mutants laid no eggs (Table 1). Ovaries from the aub mutants had fully developed ovarioles with egg chambers at all stages of development (Figure 4G). In contrast, oocyte development in the aub-CycJ double mutant was arrested, with egg chambers rarely advancing beyond stage 7 or 8 (Figure 4H). The aub-CycJ ovaries had a significant number of compound egg chambers with multiple oocytes and more than 15 nurse cell nuclei and ring canals (Figures 5 and S3). Unlike the armi-CycJ and piwi-CycJ double mutants, the compound egg chambers in aub-CycJ ovaries often included a disorganized layer of follicle cells nearly separating two fully developed cysts (Figures S3, arrow). The aub-CycJ double mutant phenotypes could be fully suppressed by introduction of a CycJ transgene (Figures 5 and S4). These observations define a genetic interaction between CycJ and aub, similar to the genetic interactions of CycJ with armi and piwi. The phenotypes of the respective double mutants have several similarities such that those of armi-CycJ and piwi-CycJ could be interpreted as more severe than those of aub-CycJ; e.g., armi-CycJ and piwi-CycJ egg chambers arrested at earlier stages of development and produced more compound egg chambers compared to the aub-CycJ mutants.
A checkpoint mutation partially suppresses loss of armi function but not the combined loss of armi and CycJ
Theurkauf and colleagues (Klattenhoff et al., 2007), have shown that strong loss-of-function armi alleles result in increased and persistent female germline DNA damage and activation of a DNA damage checkpoint dependent on the Chk2 kinase gene (known as loki or mnk in Drosophila) (Abdu et al., 2002; Masrouha et al., 2003). Activation of this checkpoint appears to be responsible for the axial patterning defect in eggs from armi mutants, since loss of mnk function significantly suppresses these armi mutant defects. To test if any of the phenotypes observed in the armi null or armi-CycJ double mutants are also due to checkpoint activation, we looked for suppression by a mnk mutant, using the mnkp6 allele (Brodsky et al., 2004; Takada et al., 2003). As previously demonstrated, the mnk mutation suppressed the patterning defects of the armi hypomorph, armi72.1 (Table 1) (Klattenhoff et al., 2007). Whereas none of the eggs laid by armi72.1/Df(3L)armi-J mothers had the normal number (two) of dorsal appendages, 85% of those from mnkP6 armi72.1/Df(3L)armi-J double mutant mothers were normal. The mnk mutation also suppressed the patterning defect in the armi null. None of the eggs from the armi null had normal dorsal appendages, while 77% of the eggs from the mnkP6 armi null had normal egg morphology (Table 1). Interestingly, the mnk mutation also partially suppressed the egg production defects of both the armi hypomorph and null mutants. Whereas the armi null produced only rare eggs (<1% of the number laid by heterozygous Df(3L)armi-J mothers), introduction of the mnk mutation resulted in a >17-fold increase in egg production (Table 1). The mnk mutant also rescued egg production in the armi hypomorph by >2-fold (Table 1). These results suggest that checkpoint activation contributes to not only the patterning defects but also the decrease in oocyte maturation associated with loss of armi function. In contrast, the mnk mutation did not suppress the developmental abnormalities of the armi-CycJ double null mutants. The mnkP6; Df(3L)armi-J females produced no eggs and had disorganized ovarioles essentially indistinguishable from those of Df(3L)armi-J (Table 1). Thus, while the checkpoint pathway clearly mediates the axis specification functions of armi, the armi-CycJ double mutant reveals functions for armi and CycJ that may be downstream or independent of the Chk2 checkpoint pathway.
Discussion
Cyclin J is one of a small handful of cyclins that remain poorly characterized despite their conservation in all metazoans. Cyclin J was originally identified in Drosophila where its mRNA was detected in adult females and embryos prior to cellularization but at no other stages (Finley et al., 1996). This pattern has been confirmed and further refined in several transcriptome studies, which have invariably shown that the CycJ message is highest in ovaries and early embryos and virtually absent in other tissues and developmental stages (Arbeitman et al., 2002; Chintapalli et al., 2013; Graveley et al., 2011), unlike all other cyclins that have broader expression patterns (Figure S1). Interestingly, the mosquito (Aedes aegypti) ortholog of CycJ is also expressed exclusively in ovaries and early embryos (Akbari et al., 2013). The conserved sequence and expression pattern suggested that Cyclin J may play an important role in oogenesis or early embryogenesis. Surprisingly, however, CycJ null Drosophila have no obvious defects and are fertile, albeit at a somewhat reduced rate compared to heterozygous controls. This could be explained if Cyclin J has an important function that is redundant with that of another protein, such as another cyclin. Althoff et al. explored this possibility by testing for genetic interactions between CycJ and CycE, CycA, CycB, and CycB3 (Althoff et al., 2009). They observed no genetic interactions, though their results could not exclude the possibility of redundancy with one of these or another cyclin. Instead of being fully redundant with another gene, it may be that CycJ is required only under specific conditions. Such condition-specific requirements have been observed for many genes in a number of organisms, most notably in yeast where only 17% of the genes are essential for growth in standard rich medium, whereas 97% of genes have been shown to be required under one or more specific environmental conditions (Giaever et al., 1999; Hillenmeyer et al., 2008; Winzeler et al., 1999). Our finding that a CycJ null mutant modifies the phenotypes of piRNA pathway mutants is consistent with a nonredundant role for Cyclin J in oogenesis under specific conditions; i.e., when some aspect of a piRNA pathway is compromised.
We have shown that mutation of CycJ alters the mutant phenotypes of armi, piwi, and aub. In the case of armi, the armi-CycJ double mutants have dramatic oogenesis defects, including only one egg chamber per ovariole, egg chambers with excess germline cells (compound egg chambers), and a complete failure to produce fully developed eggs. These defects can be partially rescued by introduction of CycJ transgenes, including a CycJ open reading frame driven by a germline promoter. These observations define a genetic interaction between CycJ and armi and indicate that CycJ plays some role in oogenesis. We observed a similar genetic interaction with piwi, where piwi-CycJ double mutants produced egg chambers with excess germline cells and this phenotype was rescued with CycJ transgenes back to levels observed in piwi. The genetic interaction with aub was also dramatic. While aub or CycJ mutant mothers produced eggs at 75% and 65% the rate of wild type, respectively, the double mutant produced no eggs.
The disruption of a piRNA pathway has a number of consequences, any of which could create a requirement for CycJ. piwi and armi are required for production of primary piRNAs in both the germline and associated somatic cells (Malone et al., 2009; Muerdter et al., 2013; Olivieri et al., 2010). Germline cells also possess a unique piRNA production mechanism known as ping-pong amplification that uses the germline-specific argonaut proteins Aub and Ago3 (Aravin et al., 2007; Li et al., 2009; Siomi et al., 2011). The requirement for CycJ may result from defects that these distinct pathways have in common since CycJ genetically interacts with members of each (e.g., armi and piwi, or aub). For example, loss of function mutations in members of either pathway result in increased transposon activity that can be accompanied by DNA double strand breaks in the germline (Klattenhoff et al., 2007). The DNA damage results in activation of checkpoint kinases such as Chk2 (encoded by loki/mnk) that in turn lead to defects in the localization of axis determinants. While any of these conditions may create a requirement for CycJ, our finding that a mnk mutant can suppress the armi null but not the armi-CycJ double mutant suggests that CycJ is not required specifically as a result of Chk2 activation. A similar analysis will be needed to test whether CycJ is required when other DNA damage responses are activated. Alternatively, it is possible that the genetic interactions we observe are a consequence of misregulation of genes other than transposon genes. The piRNA pathways, for example, have been shown to repress a small set of cellular genes (Sienski et al., 2012), any one of which could be responsible for uncovering a requirement for CycJ.
A common feature of the double mutants of CycJ and armi, piwi, or aub is the increased frequency of compound egg chambers. In both armi-CycJ and piwi-CycJ ovarioles there is often only one egg chamber and it has a terminal filament (Figure 4D and F), suggesting that packaging of individual egg chambers has completely failed and the germarium has itself become an egg chamber. These egg chambers contain an excess of differentiated germline cells including multiple oocytes, indicating that germline differentiation has occurred (Figure 3 and S3). Thus, they are unlike mutants that affect germline differentiation, which lead to germaria with an excess of undifferentiated GSC-like cells (Chen and McKearin, 2003; McKearin and Ohlstein, 1995). Proper egg chamber packaging requires several signaling pathways including the Notch and Hedgehog pathways. For example, a mutation in Delta (Bender et al., 1993), the main receptor for Notch signaling in the ovary, results in loss of stalk cells, which leads to fusion of adjacent egg chambers and formation of a compound egg chamber similar to those produced in aub-CycJ mutants. Mutation of Hedgehog (Forbes et al., 1996) results in failure of germline cyst encapsulation by follicle cells, which produces a single compound egg chamber located in the germarium, similar to those seen in armi-CycJ and piwi-CycJ. Previous studies have suggested that piRNA pathways may also be involved in proper egg chamber packaging, though the mechanisms are not known. For example, mutations of the piRNA pathway member maelstrom (Sato et al., 2011) or the piRNA-producing locus flamenco (Mevel-Ninio et al., 2007) were shown to produce compound egg chambers. We also observed a low frequency of compound egg chambers in the piwi mutants, aub mutants, and armi nulls (Figure 5). Thus, it appears that mutation of CycJ exacerbates a preexisting packaging defect seen in these piRNA pathway mutants. How the piRNA pathway and CycJ may affect the signaling pathways that are required for normal egg chamber packaging remains to be determined.
In conclusion, we have demonstrated strong genetic interactions between CycJ and three piRNA pathway members, armi, piwi, and aub. CycJ is not required for oogenesis under normal conditions but its role was uncovered in the armi, piwi, and aub mutants. The double mutants of CycJ and each of these genes have similar phenotypes that can be suppressed with CycJ transgenes. Taken together, these data suggest a nonredundant function for CycJ in regulating oogenesis when the piRNA pathways are compromised. The double mutant phenotypes suggest that CycJ may contribute to the role of the piRNA pathways in egg chamber production or maturation.
Methods
Drosophila strains
All animals were raised at 25°C on standard food. We carried out all crosses at 25°C unless otherwise stated. w1118 was used for wild type in all of the control experiments. FRT-bearing transposon insertion alleles, RBe01160 and XPd07385 (Thibault et al., 2004) were obtained from the Exelixis collection at Harvard Medical School (Artavanis-Tsakonas, 2004). The armi strong loss-of-function mutant, armi 72.1 (Cook et al., 2004), was kindly provided by William E. Theurkauf. An allele of the Drosophila Chk2 gene, mnkP6 (Brodsky et al., 2004; Takada et al., 2003), was kindly provided by Andrew Swan. Stocks for w1118, P{hsFLP}1, y1 w1118; DrMio/TM3, ry* Sb1, two of the aub mutants, aubHN cn1 bw1 and w1118; aubQC42 cn1 bw1 (Schupbach and Wieschaus, 1991), VP16::nos-Gal4, Df(3L)Exel6064, Df(3L)Exel6065, balancer strains, y[1] M{vas-int.Dm}ZH-2A w[*]; M{3xP3-RFP.attP'}ZH-51C, P{ry[+t7.2]=PZ}piwi[06843] cn[1]/CyO; ry[506], and w[1118] ; Df(2L)BSC145/CyO were obtained from the Bloomington Drosophila Stock Center. The P1 clone DS01105 and the BAC clone B22N9 were obtained from the Berkeley Drosophila Genome Project (BDGP) resource center.
Generation of CycJ and armi null alleles
We followed the procedure described by Parks et al., for generating a deletion (deficiency) mutant (Parks et al., 2004). We selected transposon elements, RB01160 and XP07385 with insertion sites flanking the region to be deleted (Figure 1). Males carrying RB01160 were mated with w1118, P{hsFLP}1, y1 w1118; DrMio/TM3, ry* Sb1 females that carry a heat shock inducible FLP recombinase transgene. Progeny males carrying both the RB01160 and FLP were then mated to females carrying XP07385 to generate progeny that contained FLP and the two FRT-bearing elements in trans. After 2 days, we subjected vials to a 1-hour heat shock by placing the vials into a 37°C water bath. After a total of 72 hours of egg-laying time, we removed the parents and subjected the vials to daily 1-hour heat shocks for 4 more days. We raised progeny to adulthood, collected virgin females and crossed them to males containing marked balancer chromosomes. Individual white-eyed progeny males having the putative deletion were then crossed pair-wise to virgin females to generate additional progeny for further confirmation by PCR and to make balanced stocks in an isogenic background. The deletion was confirmed by PCR from single flies using transposon-specific primer pairs and multiple pairs of gene-specific primers representing the genes deleted as well as the immediate upstream (eIF5B) and downstream (CG32267) genes. The resultant deletion mutant, referred to as Df(3L)armi-J, eliminated the genomic region corresponding to 670 bases upstream of the eIF5B start codon to 180 bases downstream of the stop codon of CG32267 (Figure 1). Animals bearing deficiencies were verified by PCR using the following primer pairs. Df(3L)Exel6094: Ex94L (5' CGA GGC CGG AAC CAA GGA GC) and TnLeft (5' TAC TAT TCC TTT CAC TCG CAC TTA TTG); Ex94R (5' CCG CAA TCC GGA AAG TTT TTC G) and TnRight (5' TTT ACT CCA GTC ACA GCT TTG); Df(3L)Exel6095: Ex95L (5' GCC AAG TTG GCA GGT GGG CA) and TnLeft; Ex95R (5' AGC GCC TAA GCA GTT GCA GCA) and TnRight. Df(3L)armi-J: PA_Df(3L)armiJ_R (5' TAA CTG TCG AGC GAA TGG AAG CGA) and RB (5' GCA TCA AAG AAC AAG CCG GCC AAG).
Transgenic constructs for rescue experiments
Independent transgenic lines carrying genomic copies of armi, CycJ, or CG14971 were generated by cloning the genomic region containing each individual gene along with the immediate upstream and downstream regions into the P-element vector, pCaSpeR (Thummel, 1992). To construct pCaSpeR-CycJ, we isolated a 4kb BglII fragment from genomic clone DS01105 and inserted it into the BamHI site of pCaSpeR1. This fragment contained from 1.1 kb upstream of the CycJ ATG to 1.9 kb downstream of the CycJ stop codon, and includes the last exon of armi, which encodes a part of the helicase domain but without regulatory elements, and 400 bp downstream of the CG14971 stop codon. pCaSpeR-HZ14-CycJ was constructed by cutting pCaSpeR-CycJ with SpeI, which uniquely digests in armi exon 8, filling in with Klenow (New England Biolabs), and religating. This introduced a frame shift 12 codons into the armi coding region in exon 8. To construct pCaSpeR-armi, a 6.3 kb genomic region containing armi and the neighboring region from 695 bp upstream of the armi ATG to 310 bp downstream of the armi stop codon, which excludes coding regions from the neighboring gene, was amplified from BAC clone B22N9 using the primers 5'GAATCTACGAGCGGCCGCCGATCACTAGGGTATTTATGG3' and 5'CCATGGACGCCTAGGCCATTGTATCGAAATTGAATGC3' (RA30 and RA39) that also introduced NotI and AvrII restriction sites, respectively. The PCR product was digested with NotI and AvrII and ligated to pCaSpeR2 cut with NotI and XbaI. To construct pCaSpeR-CG14971, a 4 kb genomic region corresponding to CG14971 from 820 bp upstream of the ATG to 897 bp downstream of the stop codon, excluding any of the neighboring gene coding regions, was amplified from P1 clone DS01105 using the primers, 5'CCATGGACGCTCTAGACGGCGTAGAAGAAAAAATGATCG3' and 5'GAATCTACGAGCGGCCGCCAAAGATTAGTAAAAGGG3' (RA09 and RA10) that introduced the restriction sites NotI and XbaI, respectively. The PCR product was digested with NotI and XbaI and ligated to pCaSpeR2 cut with NotI and XbaI to create pCaSpeR-CG14971. High fidelity polymerase, (Herculase, Stratagene Inc.) was used for all PCR and the resultant constructs were verified by DNA sequencing. Constructs were microinjected into w1118 Drosophila embryos to induce P-element mediated germline transformation as described (Rubin and Spradling, 1982). Injections were performed by the Model System Genomics facility at Duke University. Individual progeny bearing the transgenes as identified by eye color and verified by PCR were selected and mated with flies carrying marked balancer chromosomes to obtain balanced stocks. Chromosomes containing the transgenes are referred to as P{CycJ}, P{HZ14CycJ}, P{armi} and P{CG14971}. To express the CycJ open reading frame (ORF) from a UAS we used pHZ12 (Mairiang et al.), a vector derived from pUASattB (Bischof et al.) that contains an attB site, the mini-white gene, and a UAS driving expression of ORFs with an N-terminal 6His-3Myc tag. The CycJ ORF was amplified from a yeast two-hybrid clone (Stanyon et al., 2004) using recombination tag primers 5RT and 3RT (Parrish et al., 2004) and recombined into the 5RT and 3RT sites of vector pHZ12 as described (Parrish et al., 2004) to create pUAS-Myc-CycJ. The CycJ ORF in pUAS-Myc-CycJ was verified by sequencing and includes from the ATG to the stop codon of Cyclin J isoform A. pUAS-Myc-CycJ was inserted into the attP site at 51C in Drosophila line y[1] M{vas-int.Dm}ZH-2A w[*]; M{3xP3-RFP.attP'}ZH-51C (Bischof et al., 2007).
Immunohistochemistry
We followed the Cooley lab protocol (http://info.med.yale.edu/cooley/Protocol) (Verheyen and Cooley, 1994; Xue and Cooley, 1993) with slight modifications. We dissected ovaries from appropriately aged, well-fed, virgin females in EBR (Ringers solution; 100mM phosphate buffer pH6.8, 450mM KCl, 150mM NaCl, 20mM MgCl2) on ice, fixed in 100µl of devitellinizing buffer (6% formaldehyde, 16.7mM KH2PO4/K2HPO4 [pH 6.8], 75mM KCl, 25mM NaCl, 3.3mM MgCl2) and 600µl heptane for 10 minutes with gentle agitation, washed with PBS (150mM NaCl, 20mM K2HPO4/KH2PO4 [pH 7.4]) for 30 minutes and then with PBT (1X PBS, 0.3% Triton X-100 and 0.5% BSA) for an additional 10 minutes. After this, the ovaries were incubated with primary antibody for 1 hour at room temperature and then at 4°C overnight. Following this, the ovaries were washed with PBT for 1 hour, incubated with secondary antibody for 2 hours at room temperature, then washed with PBT for 1 hour, rinsed with PBS, and finally equilibrated with PBS: Glycerol (1:1) for at least 20 minutes. The following antibodies were used. Antibodies against hts-RC (Robinson et al., 1994), Orb 6H4 and 4H8 (Lantz et al., 1994), and c-myc 9E10 were obtained from the Developmental Studies Hybridoma Bank, University of Iowa. Primary antibodies were used at 1:50 dilution. Goat anti-mouse IgG FITC conjugate (Sigma) or DyLight 550 and Dylight 388 (Thermo Scientific), were used as the secondary antibodies at 1:200 dilution. For DAPI staining, fixed egg chambers were incubated with 1µg/ml DAPI (Sigma) in PBT for 30 minutes followed by several washes with PBT and then with PBS. Stained ovaries were mounted in 90% Glycerol containing 2.33% w/v DABCO (Sigma) for imaging. Fluorescence micrographs were obtained at room temperature using a Zeiss Axio imager upright microscope.
Quantification of phenotypes and rescue
To assess embryo hatching rates and morphology, equal numbers of newly emerged virgin females of each genotype were mated with w1118 males and the eggs were collected on apple juice plates and grape juice plates every day for 4 days. The number of eggs hatched was determined by counting the number of eggs that failed to hatch after aging for 24 hours at 25°C. Eggs were counted and categorized based on dorsal appendage morphology. At least 200 eggs were examined for each genotype except the armi null for which repeated collections provided 50 eggs to examine. To quantify oogenesis defects, females were ages for 2-4 days post eclosion and ovaries from at least 10 flies per genotype were extracted and stained with DAPI as described above. piwi mutant females were only aged for 0-2 days post eclosion due to rapid egg chamber loss. All ovarian material was transferred to a slide and ovarioles were mechanically separated from one another with forceps or insulin syringes for analysis. Number of mature oocytes per fly, number of egg chambers per ovariole, and percent of compound egg chambers per fly were quantified and averaged for each genotype. Statistical differences between single and double mutants for each phenotype were calculated using a type 3 student's T-test, which accounts for unequal sample sizes.
Supplementary Material
Figure S1. Cyclin J is expressed almost exclusively in Drosophila ovaries, a pattern that is unique among the superfamily of cyclins. This is a heatmap of cyclin expression in 55 stages and tissues of Drosophila as a percentage of each gene's maximal expression (% max). Expression data was acquired from two high throughput repositories, FlyAtlas for tissue expression (Chintapalli et al., Nat Genet. 2007 Jun;39(6):715-20) and modENCODE for stage expression (Graveley at al., Nature. 2011 Mar 24;471(7339):473-9). Raw values were obtained from each source, the highest value for each gene was determined, and that gene's expression was scaled as a percent of this maximal value as described (Murali and Finley, submitted). Percent max was determined for FlyAtlas and modENCODE data independently from one another. CycJ is maximally expressed only in early 0-2 hour old embryos and ovaries, and unlike all other cyclins, it is not expressed above ∼20% of its maximum at any other time or in any other tissue. ND = no data for Koko
Figure S2. Drosophila oogenesis begins in the germarium. A germline stem cell produces a daughter cell known as a cystoblast. This cell then undergoes exactly 4 rounds of division with incomplete cytokinesis, creating a group of 16 connected germline cells known as a cyst. One of these 16 cells will differentiate, undergo meiosis, and become the oocyte. The other 15 cells become nurse cells by undergoing specialized cell cycles termed endoreduplication consisting of extra rounds of DNA replication without cell division. Nurse cells ultimately provide their cytoplasm to the oocyte at the final stage of development (not shown). The 16 germline cells (1 oocyte and 15 nurse cells) come in contact with somatic stem cells, which give rise to the follicle cells that encapsulate the cyst to form an egg chamber (dotted lines). This process continuously produces egg chambers, which undergo posterior migration as they grow and develop into a mature oocyte. A, anterior; P, posterior.
Figure S3. Cystoblast division, differentiation, and packaging are relatively normal in CycJ null, armi null, and aub mutants, but not in aub-CycJ double mutants, which often have multiple cysts in one egg chamber. Egg chambers from CycJ null (A-C), armi null (D-F), and aub mutant (G-I) all have a single orb staining oocyte (B, E, H) and 15 ring canals (C, F, I). Note that the armi null (D, E, F) exhibits oocyte mislocalization to the middle of the egg chamber. aub-CycJ double mutants (J-L), on the other hand, have egg chambers containing multiple cysts as indicated by more than 15 nurse cell nuclei (J), two orb staining foci (K), and more than 15 ring canals (L). The arrow in J marks a group of somatic cells partially dividing two adjacent egg chambers that appear to have undergone incomplete formation or fusion resulting in one compound egg chamber. The CycJ null is P{armi}, Df(3L)armi-J/Df(3L)Exel6095. The armi null is P{CycJ}; Df(3L)armi-J/Df(3L)Exel6095. The aub mutant is aubHN/aubQC42. All size bars = 20 µm.
Figure S4. Quantification of oogenesis defects in armi-CycJ, piwi-CycJ, and aub-CycJ mutants. Three oogenesis phenotypes were affected in CycJ-piRNA pathway double mutants; number of mature oocytes per fly, number of egg chambers per ovariole, and percent of compound egg chambers per fly. armi-CycJ ovaries displayed significantly worse defects in all three phenotypes compared to armi null. Germline expression of UAS-myc-CycJ in an armi-CycJ background was able to increase egg chambers per ovariole and decrease compound egg chambers per fly, but did not affect mature oocyte production. aub-CycJ ovaries had more severe defects in mature oocyte and compound egg chamber production, but not in egg chambers per ovariole compared to aub mutants. piwi-CycJ ovaries had significantly more compound egg chambers than piwi mutants. CycJ transgenes were able to rescue all genetic interactions. All single and double null phenotypes are significantly worse than CycJ null and wild type. See Methods for details on quantification of these phenotypes and calculation of p values. For all genotypes excpet those noted, ovaries were scored at 2-4 days post eclosion.
Figure S5. Myc staining of VP16::nos-Gal4>UAS-CycJ.myc.cDNA in an armi-CycJ double null background. VP16::nos-Gal4 was able to drive the expression of UAS-CycJ.myc.cDNA in the germline of an armi-CycJ null, but not all germline cells exhibit myc staining, and some only show weak staining. This incomplete expression may be the reason there was only partial rescue of the armi-CycJ double null phenotypes. Egg chambers with myc-stained nuclei rarely had excess nurse cells, whereas egg chambers with myc-stained nuclei had excess nurse cells at a rate similar to the armi CycJ dobuble mutant (Figure S4). All size bars = 20 µm.
Figure S6. Transheterozygote females Df(3L)Exel6094 over Df(3L)Exel6095 or Df(3L)Exel6095 are viable and have normal oogenesis. (A) and (B) Diagnostic PCR showing that Df(3L)Exel6094/Df(3L)Exel6095 (A) and Df(3L)Exel6094/Df(3L)armi-J (B) are viable transheterozygotes containing both deficiencies and confirming the endpoints of these deficiencies. (C) and (D) DAPI stained ovarioles from Df(3L)Exel6094/Df(3L)Exel6095 (C) and Df(3L)Exel6094/Df(3L)armi-J (D) females. Oogenesis occurs without obvious defects in these transheterozygotes. Size bar = 20 μm. PCR was conducted on template DNA prepared from whole adult flies of the indicated genotypes. Gen=Genotype, Pri=primer pair (see methods). Ex94* are primer pairs specific for the *L left end and *R right end of Exel6094; Ex95* are primer pairs specific for the *L left end and *R right end of Exel6095. “armi-J” are primer pairs specific for Df(3L)armi-J. All primer pairs consisted of one transposon specific and one genome primer.
Cyclin J is a highly conserved cyclin without a described function.
Unlike all other cyclins, Drosophila Cyclin J is expressed specifically in ovaries
We characterized a Cyclin J null along with piRNA pathway mutants in oogenesis
The Cyclin J null is normal but modifies oogenesis defects in piwi and armi mutants
Cyclin J contributes to egg chamber development with piRNA pathway genes
Acknowledgments
We thank William E. Theurkauf and Andrew Swan for providing strains and reagents. We also thank Mark Van Berkum, Lori Pile, and members of the Finley laboratory for helpful discussions, and Stephen Guest and Dongmei Liu for comments on the manuscript. This work was supported in part by US National Institutes of Health grant HG001536.
Footnotes
Author's contributions: PA and GA performed all experiments and co-wrote the manuscript and are equal co-first authors. HZ constructed and tested CycJ transgenes and assisted with other experiments. RLF conceived the project, helped design and interpret the experiments, and co-wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Govindaraja Atikukke, Email: gatikukke@gmail.com.
Paul Albosta, Email: palbosta@med.wayne.edu.
Huamei Zhang, Email: heidizhang@yahoo.com.
Russell L. Finley, Jr., Email: rfinley@wayne.edu.
References
- Abdu U, Brodsky M, Schupbach T. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr Biol. 2002;12:1645–1651. doi: 10.1016/s0960-9822(02)01165-x. [DOI] [PubMed] [Google Scholar]
- Akbari OS, Antoshechkin I, Amrhein H, Williams B, Diloreto R, Sandler J, Hay BA. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013;3:1493–1509. doi: 10.1534/g3.113.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff F, Viktorinova I, Kastl J, Lehner CF. Drosophila Cyclin J is a mitotically stable Cdk1 partner without essential functions. Dev Biol. 2009;333:263–272. doi: 10.1016/j.ydbio.2009.06.042. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S. Accessing the Exelixis collection. Nat Genet. 2004;36:207. doi: 10.1038/ng1316. [DOI] [PubMed] [Google Scholar]
- Atikukke G. Cyclin J cooperates with the piRNA pathway to regulate early oocyte development in Drosophila (Doctoral Dissertation) Wayne State University; Detroit: 2009. p. 138. [Google Scholar]
- Bender LB, Kooh PJ, Muskavitch MA. Complex function and expression of Delta during Drosophila oogenesis. Genetics. 1993;133:967–978. doi: 10.1093/genetics/133.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Herzyk P, Davies SA, Dow JA. Data-mining the FlyAtlas online resource to identify core functional motifs across transporting epithelia. BMC Genomics. 2013;14:518. doi: 10.1186/1471-2164-14-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 2013;50:749–761. doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley RL, Jr, Brent R. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc Natl Acad Sci U S A. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley RL, Jr, Thomas BJ, Zipursky SL, Brent R. Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc Natl Acad Sci U S A. 1996;93:3011–3015. doi: 10.1073/pnas.93.7.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes AJ, Lin H, Ingham PW, Spradling AC. hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development. 1996;122:1125–1135. doi: 10.1242/dev.122.4.1125. [DOI] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Elliott H, Johnston D. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development. 1997;124:4927–4937. doi: 10.1242/dev.124.24.4927. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Johnston D. Role of oocyte position in establishment of anterior-posterior polarity in Drosophila. Science. 1994;266:639–642. doi: 10.1126/science.7939717. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzardo PM, Muerdter F, Hannon GJ. The piRNA pathway in flies: highlights and future directions. Current opinion in genetics & development. 2013;23:44–52. doi: 10.1016/j.gde.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499–2504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger JA, Wittenberg C, Richardson HE, de Barros Lopes M, Reed SI. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci U S A. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. The EMBO journal. 2011 doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King FJ, Lin H. Somatic signaling mediated by fs(1)Yb is essential for germline stem cell maintenance during Drosophila oogenesis. Development. 1999;126:1833–1844. doi: 10.1242/dev.126.9.1833. [DOI] [PubMed] [Google Scholar]
- King FJ, Szakmary A, Cox DN, Lin H. Yb modulates the divisions of both germline and somatic stem cells through piwi- and hh-mediated mechanisms in the Drosophila ovary. Mol Cell. 2001;7:497–508. doi: 10.1016/s1097-2765(01)00197-6. [DOI] [PubMed] [Google Scholar]
- Kirilly D, Xie T. The Drosophila ovary: an active stem cell community. Cell Res. 2007;17:15–25. doi: 10.1038/sj.cr.7310123. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Developmental cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kolonin MG, Finley RL., Jr A role for cyclin J in the rapid nuclear division cycles of early Drosophila embryogenesis. Dev Biol. 2000;227:661–672. doi: 10.1006/dbio.2000.9916. [DOI] [PubMed] [Google Scholar]
- Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes & Development. 1994;8:598–613. doi: 10.1101/gad.8.5.598. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- Mairiang D, Zhang H, Sodja A, Murali T, Suriyaphol P, Malasit P, Limjindaporn T, Finley RL., Jr Identification of new protein interactions between dengue fever virus and its hosts, human and mosquito. PLoS One. 2013;8:e53535. doi: 10.1371/journal.pone.0053535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrouha N, Yang L, Hijal S, Larochelle S, Suter B. The Drosophila chk2 gene loki is essential for embryonic DNA double-strand-break checkpoints induced in S phase or G2. Genetics. 2003;163:973–982. doi: 10.1093/genetics/163.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin D, Ohlstein B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 1995;121:2937–2947. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Pines J, Golsteyn R, Standart N, Mackie S, Colman A, Blow J, Ruderman JV, Wu M, Hunt T. The role of cyclin synthesis, modification and destruction in the control of cell division. Journal of cell science Supplement. 1989;12:77–97. doi: 10.1242/jcs.1989.supplement_12.8. [DOI] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell. 2013;50:736–748. doi: 10.1016/j.molcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Parrish JR, Limjindaporn T, Hines JA, Liu J, Liu G, Finley RL., Jr High-throughput cloning of Campylobacter jejuni ORfs by in vivo recombination in Escherichia coli. Journal of proteome research. 2004;3:582–586. doi: 10.1021/pr0341134. [DOI] [PubMed] [Google Scholar]
- Peng JC, Lin H. Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Current opinion in cell biology. 2013;25:190–194. doi: 10.1016/j.ceb.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cant K, Cooley L. Morphogenesis of Drosophila ovarian ring canals. Development. 1994;120:2015–2025. doi: 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sato K, Nishida KM, Shibuya A, Siomi MC, Siomi H. Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC. Genes Dev. 2011;25:2361–2373. doi: 10.1101/gad.174110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E, Nothiger R. Study of Female Germ Line in Mosaics of Drosophila. Roux Arch Dev Biol. 1978;184:41–56. doi: 10.1007/BF00848668. [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature reviews Molecular cell biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Smulders-Srinivasan TK, Szakmary A, Lin H. A Drosophila chromatin factor interacts with the Piwi-interacting RNA mechanism in niche cells to regulate germline stem cell self-renewal. Genetics. 2010;186:573–583. doi: 10.1534/genetics.110.119081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Spradling AC. Germline cysts: communes that work. Cell. 1993;72:649–651. doi: 10.1016/0092-8674(93)90393-5. [DOI] [PubMed] [Google Scholar]
- Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, Zhang H, Zhong J, Finley RL., Jr A Drosophila protein-interaction map centered on cell-cycle regulators. Genome biology. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Kelkar A, Theurkauf WE. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 2003;113:87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CSaP, V New pCaSpeR P-element vectors. Drosophila Information Service. 1992;150 [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Verheyen E, Cooley L. Looking at oogenesis. Methods in cell biology. 1994;44:545–561. [PubMed] [Google Scholar]
- Wieschaus E, Szabad J. The development and function of the female germ line in Drosophila melanogaster: a cell lineage study. Dev Biol. 1979;68:29–46. doi: 10.1016/0012-1606(79)90241-0. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290:328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cyclin J is expressed almost exclusively in Drosophila ovaries, a pattern that is unique among the superfamily of cyclins. This is a heatmap of cyclin expression in 55 stages and tissues of Drosophila as a percentage of each gene's maximal expression (% max). Expression data was acquired from two high throughput repositories, FlyAtlas for tissue expression (Chintapalli et al., Nat Genet. 2007 Jun;39(6):715-20) and modENCODE for stage expression (Graveley at al., Nature. 2011 Mar 24;471(7339):473-9). Raw values were obtained from each source, the highest value for each gene was determined, and that gene's expression was scaled as a percent of this maximal value as described (Murali and Finley, submitted). Percent max was determined for FlyAtlas and modENCODE data independently from one another. CycJ is maximally expressed only in early 0-2 hour old embryos and ovaries, and unlike all other cyclins, it is not expressed above ∼20% of its maximum at any other time or in any other tissue. ND = no data for Koko
Figure S2. Drosophila oogenesis begins in the germarium. A germline stem cell produces a daughter cell known as a cystoblast. This cell then undergoes exactly 4 rounds of division with incomplete cytokinesis, creating a group of 16 connected germline cells known as a cyst. One of these 16 cells will differentiate, undergo meiosis, and become the oocyte. The other 15 cells become nurse cells by undergoing specialized cell cycles termed endoreduplication consisting of extra rounds of DNA replication without cell division. Nurse cells ultimately provide their cytoplasm to the oocyte at the final stage of development (not shown). The 16 germline cells (1 oocyte and 15 nurse cells) come in contact with somatic stem cells, which give rise to the follicle cells that encapsulate the cyst to form an egg chamber (dotted lines). This process continuously produces egg chambers, which undergo posterior migration as they grow and develop into a mature oocyte. A, anterior; P, posterior.
Figure S3. Cystoblast division, differentiation, and packaging are relatively normal in CycJ null, armi null, and aub mutants, but not in aub-CycJ double mutants, which often have multiple cysts in one egg chamber. Egg chambers from CycJ null (A-C), armi null (D-F), and aub mutant (G-I) all have a single orb staining oocyte (B, E, H) and 15 ring canals (C, F, I). Note that the armi null (D, E, F) exhibits oocyte mislocalization to the middle of the egg chamber. aub-CycJ double mutants (J-L), on the other hand, have egg chambers containing multiple cysts as indicated by more than 15 nurse cell nuclei (J), two orb staining foci (K), and more than 15 ring canals (L). The arrow in J marks a group of somatic cells partially dividing two adjacent egg chambers that appear to have undergone incomplete formation or fusion resulting in one compound egg chamber. The CycJ null is P{armi}, Df(3L)armi-J/Df(3L)Exel6095. The armi null is P{CycJ}; Df(3L)armi-J/Df(3L)Exel6095. The aub mutant is aubHN/aubQC42. All size bars = 20 µm.
Figure S4. Quantification of oogenesis defects in armi-CycJ, piwi-CycJ, and aub-CycJ mutants. Three oogenesis phenotypes were affected in CycJ-piRNA pathway double mutants; number of mature oocytes per fly, number of egg chambers per ovariole, and percent of compound egg chambers per fly. armi-CycJ ovaries displayed significantly worse defects in all three phenotypes compared to armi null. Germline expression of UAS-myc-CycJ in an armi-CycJ background was able to increase egg chambers per ovariole and decrease compound egg chambers per fly, but did not affect mature oocyte production. aub-CycJ ovaries had more severe defects in mature oocyte and compound egg chamber production, but not in egg chambers per ovariole compared to aub mutants. piwi-CycJ ovaries had significantly more compound egg chambers than piwi mutants. CycJ transgenes were able to rescue all genetic interactions. All single and double null phenotypes are significantly worse than CycJ null and wild type. See Methods for details on quantification of these phenotypes and calculation of p values. For all genotypes excpet those noted, ovaries were scored at 2-4 days post eclosion.
Figure S5. Myc staining of VP16::nos-Gal4>UAS-CycJ.myc.cDNA in an armi-CycJ double null background. VP16::nos-Gal4 was able to drive the expression of UAS-CycJ.myc.cDNA in the germline of an armi-CycJ null, but not all germline cells exhibit myc staining, and some only show weak staining. This incomplete expression may be the reason there was only partial rescue of the armi-CycJ double null phenotypes. Egg chambers with myc-stained nuclei rarely had excess nurse cells, whereas egg chambers with myc-stained nuclei had excess nurse cells at a rate similar to the armi CycJ dobuble mutant (Figure S4). All size bars = 20 µm.
Figure S6. Transheterozygote females Df(3L)Exel6094 over Df(3L)Exel6095 or Df(3L)Exel6095 are viable and have normal oogenesis. (A) and (B) Diagnostic PCR showing that Df(3L)Exel6094/Df(3L)Exel6095 (A) and Df(3L)Exel6094/Df(3L)armi-J (B) are viable transheterozygotes containing both deficiencies and confirming the endpoints of these deficiencies. (C) and (D) DAPI stained ovarioles from Df(3L)Exel6094/Df(3L)Exel6095 (C) and Df(3L)Exel6094/Df(3L)armi-J (D) females. Oogenesis occurs without obvious defects in these transheterozygotes. Size bar = 20 μm. PCR was conducted on template DNA prepared from whole adult flies of the indicated genotypes. Gen=Genotype, Pri=primer pair (see methods). Ex94* are primer pairs specific for the *L left end and *R right end of Exel6094; Ex95* are primer pairs specific for the *L left end and *R right end of Exel6095. “armi-J” are primer pairs specific for Df(3L)armi-J. All primer pairs consisted of one transposon specific and one genome primer.