Abstract
Medical image retrieval and classification have been extremely active research topics over the past 15 years. With the ImageCLEF benchmark in medical image retrieval and classification a standard test bed was created that allows researchers to compare their approaches and ideas on increasingly large and varied data sets including generated ground truth. This article describes the lessons learned in ten evaluations campaigns. A detailed analysis of the data also highlights the value of the resources created.
Introduction
While development of image retrieval approaches and systems began as a research field over two decades ago [5,34,38], progress has been slow for a variety of reasons. One problem is the inability of image processing algorithms to automatically identify the content of images in the manner that information retrieval and extraction systems have been able to do so with text [4,38]. A second problem is the lack of robust test collections and in particular, realistic query tasks with ground truth that allow comparison of system performance [4,18,25,32]. In general, the limits of systematic comparisons have been analyzed in several publications [44], but also an important impact could be shown when evaluating the results of such benchmarks [42,40], particularly economic value but also scholarly impact in terms of citations.
The lack of realistic test collections is one of the motivations for the ImageCLEF initiative, which is a part of the Cross-Language Evaluation Forum (CLEF), a challenge evaluation for information retrieval from diverse languages [24]. The goals of CLEF are to build realistic test collections that simulate real world retrieval tasks, enable researchers to assess the performance of their systems, and compare their results with others. The goal of test collection construction is to assemble a large collection of content (text, images, structured data, etc.) that resemble collections used in the real world. Builders of test collections also seek a sample of realistic tasks to serve as topics that can be submitted to systems as queries to retrieve content [18,33]. The final component of test collections is relevance judgments that determine which content is considered relevant to each topic.
Biomedical information retrieval systems are complex, comprising many key components. These include image modality classification [29], visual image similarity computation, multimodal image and text information retrieval, and others that may be specific to individual systems. Performance evaluation needs to be conducted on these components to determine the overall system performance. With the exponential increase in available biomedical data repositories it is important for the evaluation to be close to real world in its size and scope. The ImageCLEF1 medical retrieval tasks have provided such an evaluation forum and framework for evaluating the state of the art in biomedical image information retrieval [4,18,24,26,28,29,30,41].
Motivation
An important goal is to develop systems that can support the management of the large amounts of data, including images, that are routinely created and stored in hospital information systems [17]. The use of image retrieval systems for medical and other applications is growing, yet we know little about the motivations and tasks for which they are used [19]. In order to set realistic goals for ImageCLEF over the years, a number of user-studies were conducted to understand the needs and motivation of users of clinical image retrieval systems [19,25]. Such user studies were performed at Oregon Health and Science University (OHSU) [19] and in University Hospitals, Geneva in 2005–2006 [25] then again in 2009 at OHSU and in 2011 in Geneva [22]. Analyses of log files were also performed to derive tasks that are representative for medical information search [32]. In 2005, Hersh et al. performed a qualitative study to analyze real users’ tasks for which image retrieval can be used [19]. They examined the information needs of 13 biomedical professionals with the following roles: clinician, researcher, educator, librarian and student. The results of this study showed that medical image needs of biomedical professionals could be categorized into four groups: research-related, patient care-related, education-related and other.
Müller et al. also performed a qualitative study that year to learn about the image use and search behavior in the medical domain [25]. They conducted a survey with 18 users at the University Hospitals of Geneva. The participants were asked questions in order to explain how they would use and search for images in their roles as clinicians, educators, researchers, librarians, and/or students. The obtained results showed that the tasks performed with images and the ways to search for images vary strongly depending on the role of the person and the department they belong to. As reported earlier [19], students, researchers and lecturers search for images for use in presentations and reports. Many clinicians create their personal image archives from clinical routine for further use. They usually add clinical information to illustrate interesting cases, particularly for teaching and research. Results also showed that image search was not restricted to the hospital archive or teaching file; many users searched for images using Google and specialized websites. But in these cases, participants said that quality was a problem and was hard to judge. The participants in this study identified some key needs of an image retrieval system including the ability to search pathology and anatomical location. Searching by visual similarity was highlighted as desired but not implemented feature of search engines. Many participants had expressed the desire to be able to index and search their PACS systems. In [23] these results were validated and an evolution could also be shown in that radiologists knew about visual search and desired to be able to search by image regions and for entire medical cases.
In 2008, Sedghi et al., performed a user study with 26 participants in order to investigate the relevance criteria used by health care professionals to make relevance judgments when searching for medical images [36]. In total, 26 criteria were identified; amongst them, visual relevancy, background information and image quality were the most frequent criteria used by participants. The study showed that the importance of each criterion was dependent upon the user specialty and their information needs, and that users apply different criteria in different situations when evaluating the relevancy of medical images.
In 2009, OHSU recruited a sample of clinicians, educators, and researchers from the local health sciences university. OHSU provided demonstrations of state-of-the-art image retrieval systems to prompt real information needs from the participants and then observed them search with six different systems. Participants were asked in an open-ended manner to collate the data for themes and trends. Participants were also asked to provide data about search tasks they might undertake. In particular, they were asked to describe tasks where medical images are helpful, and to provide information about the types of images they find most useful. In general, image supported tasks fell into five broad groups: Education, Publication, Diagnosis, Research, and Other. Some examples of how the participants self-reported using image retrieval systems included:
Self-Education: use of images to review instructions for specific procedures, to review how to interpret images, to learn about a new clinical topic.
Professional Education: use of images to educate medical students and residents in the clinic or the classroom (e.g., to demonstrate concepts that are necessary to perform an exam, teaching what to look for in an image in a given condition).
Patient Education: mostly, in this task images are used as visual aid in patient education (e.g., give clinical explanation about their conditions). Participants mentioned other cases where images are used such as giving an illustration to providers.
Diagnosis: images are used in difficult diagnosis situations (e.g., uncommon or nonspecific rashes) to confirm a diagnosis or generate a differential diagnosis.
Publications: images are used to prepare medical presentations or to give a talk.
Research: images are used to develop a research idea, to write grants, to make a presentation at research conferences.
These and other user studies have helped define ImageCLEF over the years by creating a basis for realistic topics for the image retrieval task as well as expanding beyond simple ad-hoc retrieval tasks into automatic annotation, case-based retrieval [12,35], and modality classification.
Methods
The ImageCLEF annual image retrieval challenge started in 2003 as a task of the CLEF campaign, which had its beginnings in the Text REtrieval Challenge (TREC) and has been an independent workshop since 2000. Thus, the organization of ImageCLEF has been modeled after TREC [15] and follows a similar set of stages as described below.
The organizers distribute a “collection”, which in the case of ImageCLEF consists of images and annotations for modality classification; images and associated text for ad-hoc retrieval; and case descriptions with images for case-based retrieval. A set of 25–100 search “topics” is then distributed. These are typically realistic domain specific information needs. Participants then apply their tools and techniques to produce ordered sets of images or “runs” that are responding to each search topic; automatic modality classification results; or ranked lists of cases similar to a topic in the case-based retrieval task. The next step in the evaluation campaign is to have human experts assess the relevance of the returned images or case descriptions for the information need presented in the search topic. Subject matter experts or “judges” are recruited to help with the assessment. However, due to practicality constraints, only a subset of all images can be assessed in a reasonable timeframe. “Pooling” is used to identify candidate images to be assessed [37]. Typically, the top 30–60 results from each run for each topic are combined to create pools of about 1000 images or cases that are assessed manually. The detailed criteria for assessing relevance by the judges are prescribed as part of the protocol. ImageCLEF uses a ternary judgment scheme wherein each image in each pool was judged to be “relevant”, “partly relevant”, or “non relevant”. Images clearly corresponding to all criteria specified in the protocol were judged as “relevant”; images were marked as “partly relevant” for when the relevance could not be accurately confirmed; and, images for which one or more criteria of the topic were not met were marked as “non relevant”. The results were manually verified during the judgment process. Judges were clinicians and many topics were judged by two or more judges to add robustness to the rankings of the systems and analyze inter-rater disagreement. This also allowed excluding a few judges for whom the interpretation of the judgment rules was not respected but this remained a rare case.
Results and Discussions
Evolution of ImageCLEF over the years
ImageCLEF was first organized as a track of the Cross Language Evaluation Forum (CLEF) in 2003 [9]. The first year was a pilot experiment with the following objective “given a multilingual statement describing a user need, find as many relevant images as possible” [9]. This pilot project used historic photographs from St. Andrews University and 50 search topics were provided in a language other than that of the collection. The participants employed purely text-based image retrieval approaches for this task and only four groups participated.
2004 was the beginning of the medical image retrieval and classification tasks at ImageCLEF [8]. The collection used for this task was a subset of the CasImage collection, a dataset of anonymized medical images and associated notes from the University Hospitals of Geneva. These textual annotations, in English or French, consisted of a number of fields including diagnosis, clinical presentation, keywords, title and unstructured description and were associated with a case that can include multiple images. Not all fields were populated for all cases and the annotations that were present may have had issues typical of real-life clinical notes such as abbreviations, spelling errors, and other linguistic problems as well as challenges with multilingual collections such as incorrect French accents. The query tasks were selected by a radiologist and were made available to participants in the form of a sample image. Thus, this was a query by example task and the goal was to retrieve similar images, where similarity was based on modality, anatomical locations and imaging protocols. Participants could use purely visual techniques (content-based image retrieval or CBIR) as well as textual retrieval techniques based on the notes associated with the sample image. A radiologist, a medical doctor and a medical computer scientist performed the relevance assessments on pools created from the submissions. Images were judged using a ternary scale as relevant, partially relevant or not relevant. Based on these assessments, relevance sets used for the judging were created in a number of ways. These include deeming an image to be relevant only if all 3 agree (most strict), relevant if all three judges said that the images were relevant or partially relevant, relevant if at least 2 judges say that the image is relevant, relevant if any of judges say that the image is at least partially relevant (most lenient).
The size of the dataset was greatly increased for the 2005 medical retrieval task from the 6’000 images in 2004. In addition to the CasImage collection, images from the Pathology Education Instructional Resource (PEIR), images from the Mallinckrodt Institute of Radiology (MIR) and the PathoPic collection were also made available. The PEIR collection of about 33,000 pathology images included annotations in English associated at the image level, the MIR dataset consisted of about 2000 nuclear medicine images and had English annotations at the case level and the Pathopic collection consisted of about 9000 images with extensive German annotations and incomplete English translations. Thus, this large and diverse collection of over 50,000 images contained images from radiology, nuclear medicine and pathology with annotations in English, French and German that were associated with the images at either the images level or the case level where a single annotation could apply to multiple images. Twenty-five search topics were defined based on a user survey conducted at OHSU and developed along the following axes: anatomy, modality, pathology or disease and abnormal visual observation. Twelve of the 25 topics were thought to be best suited for visual systems, eleven for mixed systems while a couple were semantic topics where visual features were not expected to improve performance [7]. Relevance assessments were performed by 9 judges, most of who were clinicians while one was an image-processing specialist. Pools were created using the top 40 results from each run resulting in pools of approximately 900 images. A ternary scale was used again and relevance sets were created in a few different ways from most strict to most lenient.
The same dataset was used again in 2006. However, the topics were selected based on search logs of a medical media search engine created by the Health on the Net (HON) foundation [23]. Thirty search topics were generated with ten each expected to be amenable to visual, textual and mixed search methods. Seven clinicians from OHSU performed the relevance assessments.
In 2007, in addition to the dataset used in 2005 and 2006, two more datasets were added. These included the myPACS dataset of about 15,000 primarily radiology images annotated in English at the case level and about 1500 images from the Clinical Outcomes Research Institute (CORI) dataset of endoscopic images annotated in English at the image level. This combined dataset of more than 66,000 images had annotations in English, French and German and images of a variety of modalities. Thirty topics from PubMed® log files were selected that sought to cover at least two of the axes (modality, anatomy, pathology and visual observation) and again 30 search topics were created. The top thirty images from each run were combined to create the pools with an average pool size of about 900. Judges were clinicians that were also students in the OHSU biomedical informatics graduate program.
A new database was used in 2008 but the task remained essentially the same as in 2007. The Radiological Society of North America (RSNA) had made available a set of about 66,000 images published in two radiology journals (Radiology and Radiographics). These images were a subset of the images used by the Goldminer search engine [21]. The high quality annotations associated with the images were the figure captions published in the journal. However, the images were primarily radiology focused unlike in previous years where pathology and endoscopic images were also represented. The query topics were selected from the topics previously used between 2005 and 2007. Training data was also made available. This consisted of the images and annotations as well as the topics, sample images and relevance judgments (“qrels”). The judges again were clinicians who were students in OHSU’s biomedical informatics training program.
In 2009, the size of the image dataset increased to over 74,000. These images again were provided by RSNA (similar to 2008) and were part of the Goldminer database. The 2009 search topics were selected from a set of queries created by clinicians participating in a user study of medical search engines. In addition to ‘ad-hoc’ search topics, case-based topics were introduced for the first time. These case-based topics are meant to more closely resemble the information needs of a clinician in a diagnostic role. Teaching files in CasImage were used to create five topics. A textual description and a set of images were provided for each case but the diagnosis was withheld and only given to the judges for assessment.
The RSNA dataset of about 77,500 images was used in 2010. The 16 image-based search topics were selected, as in 2009, from topics that had been searched for in the above-mentioned user study. Additionally, fourteen case-based topics were provided. Based on research that had demonstrated the improvements in early precision obtained in filtering out images of non-relevant modalities [2], a modality classification sub-task was added in 2010. The goal of this subtask was to classify an image into one of 8 classes (computed tomography, magnetic resonance imaging, nuclear medicine, positron emission tomography, ultrasound, x-ray, optical and graphics). A training dataset of 2390 images was provided and the test set had 2620 images.
The size of the database was increased significantly in 2011 to about 231,000 images. These images were part of the articles published in open access journals and available in PubMed Central. The subset chosen were from journals that allow for the redistribution of data. Since the images were published in a wide range of journals, there was substantially more diversity in the kinds of images available and the number of potentially non-clinically oriented images as a large part of the journals was from BioMed Central and thus more biology oriented. A substantial portion of the images included charts and graphs and other similar non-clinical image types, thus highlighting the need for efficient filtering. The image-based and case-based topics were subsets of those used in previous years. The modality classification task, started in 2010, was continued in 2011. However, the number of classes was increased from 8 to a hierarchical scheme of 18 classes. 1000 training and 1000 test images were provided.
In 2013, the medical task of ImageCLEF was organized as a workshop at the annual meeting of the American Medical Informatics Association (AMIA). The same dataset was used as in 2012 [26]. The ad-hoc and case-based tasks were continued with 35 topics each. The modality classification hierarchy was further refined to now include 38 classes, of which 31 were present in the dataset [29]. A new compound figure separation task was added in 2013. As approximately half of the images in the database used in 2012 and 2013 were compound figures, the separation of these into the component sub-figures is an important first step in the classification and retrieval tasks. A training set of 1538 and a test set of 1429 images were made available.
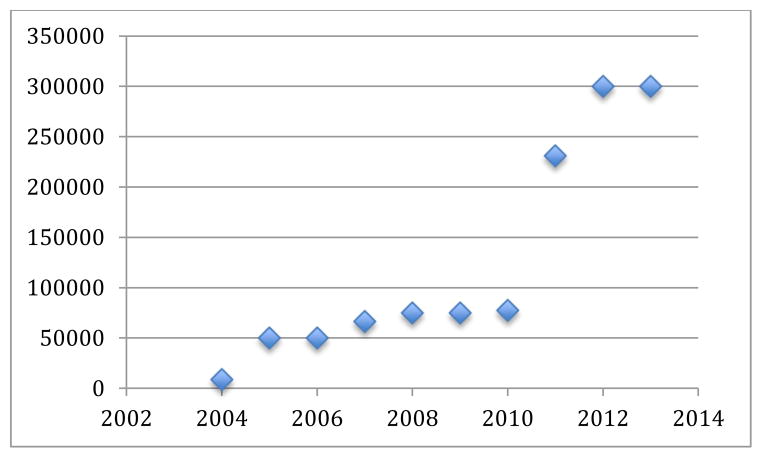
As seen in Figure 1, the number of images in the collections has grown from 6,000 to over 300,000 over 10 years.
Figure 1.
Number of images in the collection
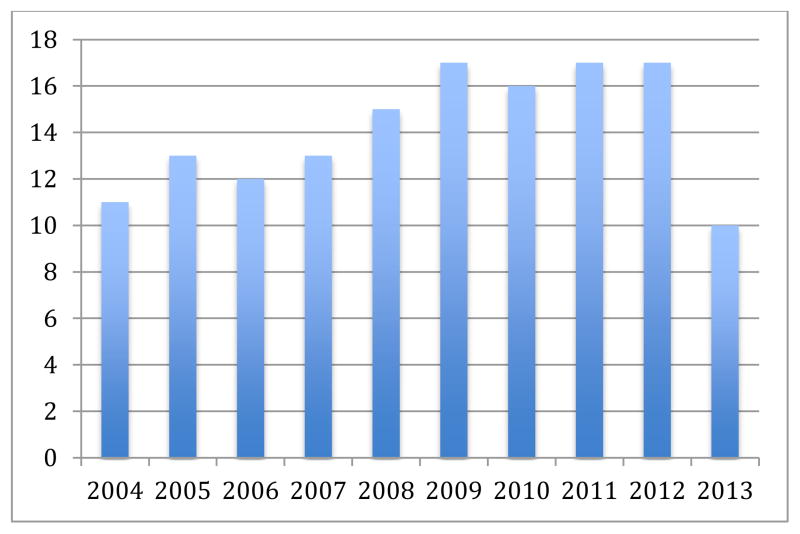
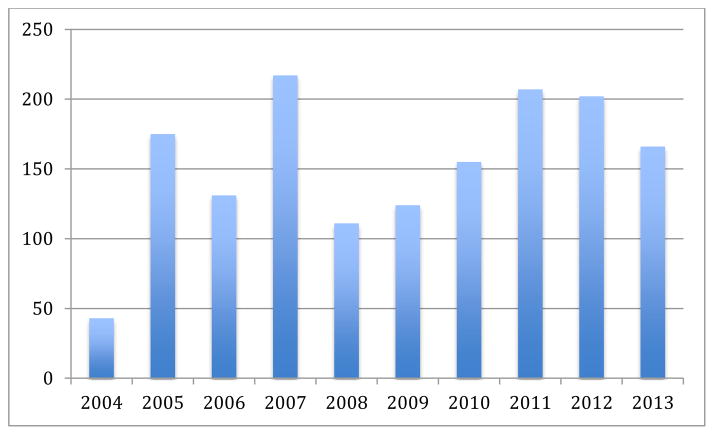
As seen in Figure 2, the number of groups submitting runs generally increased from ten during the first year to about 17. The total number of runs submitted fluctuated over the years as seen in Figure 3 depending on the number of sub-tasks being organized. The number of registrations has increased strongly from about 10 in 2004 to around 70 in 2012. Many groups then do not feel confident about the results but often continue working and publishing on the data well after the collections. Participation in tasks is often seen as good when sure to obtain good results even though the workshop highlighted talks from interesting techniques and not necessarily the best performing techniques.
Figure 2.
Number of groups submitting runs for the medical task
Figure 3.
Total runs submitted for the medical task
Overview of participant methods
Textual retrieval
Text retrieval has often been used to search on the biomedical literature [10].
The title, abstract, figure caption, figure citation in the articles have been used by the ImageCLEFmed participants to find key textual features for retrieval. Since 2004, participants in the medical task explored a vast variety of textual information retrieval techniques. Stop word and special character removal, tokenization and stemming (e.g. Porter stemmer) have been broadly applied [6].
Additional resources were also used to support participants’ retrieval approaches. The open source Lucene2 tool has been used since 2005 by the participants. The Lucene search engines provide a simple and useful framework for text search [16]. The Terrier IR3 platform, an open source search engine for large-scale data, has been explored for indexing documents and query expansion since 2011. In the last two ImageCLEF campaigns the Essie tool [20] was also included in the experiments and obtained best results in 2013. Essie is a phrase-based search engine with term and concept query expansion and probabilistic relevancy ranking, which serves several web sites at the National Library of Medicine. Lemur4 software was used in 2005 and it was reused again in 2010. And some systems such as SMART (System for the Mechanical Analysis and Retrieval of Text) [3], easyIR [27] or Morphosaurus5 were used in only one of the benchmarks.
Although text retrieval does not require the use of specialized or annotated corpora, manually annotated collections have been used to utilize medical knowledge in connection with the original text. The use of Medical Subject Headings (MeSH) has been exploited since 2005. Tools such as MetaMap6 were commonly used to map biomedical text to the Unified Medical Language System Metathesaurus (UMLS). WordNet7, an on-line English thesaurus and lexical database, and EuroWordNet, similar to WordNet, for other languages of Europe, [43] have been used for term expansion. Even Wikipedia8, Google Search API9and the translator Babelfish10 have been exploited to extract external terms.
Since 2010, some participants have included modality filters (using either text-based or image-based modality detection) in their retrieval approaches [2]. Modality filtration was found to be useful by some participants while others found only minimal benefit using modality.
Similar techniques were applied for the retrieval and modality classification tasks. On the other hand, only one group applied textual techniques (in combination with visual techniques) for the compound figure separation task. This group determined the number of image panels comprising a compound figure by identifying textual panel labels in the figure’s caption [1].
Visual Retrieval
Content-based image retrieval has been an active area of research for the last decade. Image features used for retrieval have traditionally been low level features such as those based on image intensity, color and texture.
Over the years, participants in ImageCLEF explored different color spaces including RGB, YCbCr and CIE L*a*b*. Image histograms provided means to compare the distributions of color in the images. Participants utilized a wide range of local and global texture features. These included Tamura features of coarseness, contrast and directionality; Gabor filters at different scales and directions, Haar filters, Haralick’s co-occurrence matrix based features, fractal dimensions and others.
A simple, yet surprisingly effective feature set was based on generating thumbnails and using these down-sampled images as features. Visual words or similar feature modeling techniques have obtained best results in the past years of the challenges, similar to good result of methods using patches in the past [13].
A range of distance metrics used to compute similarity between image features were also studied. In addition to the commonly used Euclidean distance, participants used Earth Mover’s Distance (EMD) and Jeffrey divergence and Jensen-Shannon divergence for histogram comparisons, as well as statistical distance measures. Participants also used cross-correlation functions and image distortion models to calculate distances between images. Some participants evaluated segmenting the images and extraction of shape features or using “blob” based features.
Teams also were successful in “learning” semantic terms, or connections of visual features and text terms.
Many groups used the popular medGIFT (Gnu Image Finding Tool) search engine [39]; as the team from the University of Geneva had made available the baseline results using this tool to all participants.
Combined multimodal retrieval
Participants explored the use of early and late fusion as well as a variety of schemes for filtering images based to modality as a way to combine the results from text-based and image-based search engines. The effect of weights used in combining the results was also studied. An overview of all fusion techniques used in ImageCLEF from 2004–2009 can be found in [11]. A task on information fusion organized at the ICPR conference only on fusing some of the participants’ runs also delivered more insight to information fusion on such very uneven data, with text retrieval obtaining very good results and visual retrieval often quite low results [31]. Lessons learned from the fusion evaluation were that rank-based methods often performed better than score-based methods as the score curves for visual and text retrieval differ strongly. The importance of the ranks could be adapted based on the probability of finding relevant images. In the fusion competition much better results were obtained than any participant obtained in the competition. This underlines that good fusion techniques can massively improve the results, likely much more than tweaking of parameters in either visual or text retrieval could.
Conclusions and lessons learned after ten years
In ten years, the ImageCLEFmed campaign on medical image classification and retrieval has evolved strongly to adapt to current challenges in the domain. Many systems and techniques have been explored and tested over the years to identify promising techniques and directions. The databases grew from 6,500 to over 300,000 images and now contain a large noise component requiring more complex filtering but being representative of the literature that stores most medical knowledge. Also the tasks increased in complexity from simple visual image retrieval in 2004 to a task consisting of image-based retrieval, case-based retrieval, modality classification and compound figure separation. ImageCLEF has had an important scholarly impact [42] and many groups have worked with the data, allowing PhD students to concentrate on research instead of preparing data sets. The user surveys and analyses of log files have created insight into the changes in visual information search behavior and create the basis for system testing. Several clear lessons have been learned over the years:
Visual retrieval alone has low performance unless used for very precise tasks such as modality classification and a limited number of classes;
Visual retrieval can obtain high early precision, particularly for tasks that can be considered visual retrieval tasks;
A variety of features need to be used for good visual retrieval and these features should be modeled as in the visual words paradigm; whereas global features such as thumbnails can deliver basic information local feature such as patches have a potential to increase results;
Text retrieval works well and has by far the best performance in terms of Mean Average Precision (MAP) compared with visual retrieval;
Text retrieval has generally a very good recall but sometimes not the optimal early precision that most users might be interested in;
Fusion of visual and text information can improve the results as the two retrieval paradigms are complementary but fusion needs to be done with care as the characteristics of the two are not the same and many poor approaches for fusion actually decrease the text retrieval results;
Mapping of free text to semantics can improve results over using text, only;
Using modality information of images can improve performance of image retrieval where one modality is the query objective
For the modality classification results the main limiting factor was the training data that did not cover the diversity of the test data; the best techniques all used automatic or manual techniques for the extension of the training data set;
Early vs. late fusion each have scenarios where they perform best and it is hard to indicate which techniques would be best;
Compound figure separation is an important step to focus search on single figures but keep their context that is often important.
These lessons learned show the importance of such benchmarks and of systematic evaluation. Research can now be focused on promising techniques and allows concentrating on real research challenges and reproducible approaches, which is clearly not the case when small, private databases are used.
Having a forum such as a workshop where participants can compare their experiences with those of other researchers who worked on the same data is another important part. These discussions frequently lead to new, improved research ideas and also collaborations between participants. Research lives off these exchanges and cannot be done alone anymore. Sharing work to create recourses and evaluation platforms creates an added value for everyone involved and has many advantages in terms of research organization.
Future work
In 2014 the general medical retrieval task was not held and rather a semantic annotation challenge of the liver is organized. This has also as a goal to concentrate on a reflection of what had been achieved in ten years. This reflection can then lead to new challenges that can be addressed in future evaluation campaigns.
One of the already identified challenges is the retrieval from extremely large collections of data that can potentially not be shared by simple download anymore. This would require a different architecture, for example to store data centrally in the cloud and move the algorithms to the data by executing them in a central virtual machine in the cloud [14]. Such an infrastructure can potentially also lead to a possibility to work on copyrighted or confidential data as participants only need to work on a small training data set and the full analysis of the tools does not require data access and the virtual machines can be isolated in this phase.
Such architecture could also lead to more collaboration between participants who could easily share components when working on the same data and the same infrastructure. This can lead to a more detailed comparison of components and combinations of really the best performing techniques of all participants. It can also reduce the effort of each participant as researchers can concentrate on specific aspects and the combine with the other existing tools. A more systematic combination of techniques can also lead to new insights.
Then, the final goal is obviously to obtain tools that have real clinical impact and thus pave the way for digital medicine where data on the patient are understood, interpreted and mapped to semantics and then linked to current knowledge finding outliers, defining risk factors and giving a global picture to the physicians. Images are a part of this system but need to be integrated with a large variety of other data.
Acknowledgments
JKC is funded in part by the NIH grants U01CA154602 and R00LM009889.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Apostolova E, You D, Xue Z, Antani S, Demner-Fushman D, Thoma GR. Image retrieval from scientic publications: text and image content processing to separate multi-panel Figures. Journal of the American Society for Information Science. 2013 [Google Scholar]
- 2.Bedrick S, Radhouani S, Kalpathy Cramer J. ImageCLEF experimental evaluation in image retrieval. Springer; 2010. Improving Early Precision in the ImageCLEF Medical Retrieval Task. [Google Scholar]
- 3.Buckley C. Implementation of the SMART information retrieval system. Cornell University; 1985. [Google Scholar]
- 4.Caputo B, Müller H, Thomee B, Villegas M, Paredes R, Zellhofer D, Goeau H, Joly A, Bonnet P, Martinez Gomez J, Garcia Varea I, Cazorla C. Working Notes of CLEF 2013 (Cross Language Evaluation Forum) 2013. ImageCLEF 2013: the vision, the data and the open challenges. [Google Scholar]
- 5.Chang SK, Kunii T. Pictorial data base applications. IEEE Computer. 1981;14(11):13–21. [Google Scholar]
- 6.Clough P, Müller H. ImageCLEF – Experimental evaluation of visual information retrieval. Springer; 2010. Seven Years of ImageCLEF; pp. 3–18. [Google Scholar]
- 7.Clough P, Müller H, Deselaers T, Grubinger M, Lehmann T, Jensen J, Hersh W. The CLEF 2005 Cross-Language Image Retrieval Track, Springer Lecture Notes in Computer Science LNCS 4022. 2006:535–557. [Google Scholar]
- 8.Clough P, Müller H, Sanderson M. The CLEF Cross Language Image Retrieval Track (ImageCLEF) 2004. CLEF Proceedings – Springer Lecture Notes in Computer Science. 2005:597–613. volume LNCS 3491. [Google Scholar]
- 9.Clough P, Sanderson M. The CLEF 2003 Cross Language Image Retrieval Task, working notes of CLEF. 2013 [Google Scholar]
- 10.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Briefings in bioinformatics. 2005 doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 11.Depeursinge A, Müller H. Fusion techniques for combining textual and visual information retrieval. In: Müller H, Clough P, Deselaers T, Caputo B, editors. ImageCLEF, The Springer International Series On Information Retrieval. Vol. 32. Springer; Berlin Heidelberg: 2010. pp. 95–114. [Google Scholar]
- 12.Depeursinge A, Vargas A, Platon A, Geissbuhler A, Poletti PA, Müller H. 3D Case-Based Retrieval for Interstitial Lung Diseases. MICCAI workshop on Medical Content-Based Retrieval for Clinical Decision Support, Springer Lecture Notes in Computer Science LNCS; London, UK. 2010. pp. 39–48. [Google Scholar]
- 13.Deselaers T, Weyand T, Keysers D, Macherey W, Ney H. FIRE in ImageCLEF 2005: Combining content-based image retrieval with textual information retrieval. Accessing Multilingual Information Repositories; Vienna, Austria. 2006; pp. 688–698. Volume 4022 of LNCS. [Google Scholar]
- 14.Hanbury A, Müller H, Langs G, Weber MA, Menze BH, Salas Fernandez T. Bringing the algorithms to the data: cloud based benchmarking for medical image analysis. CLEF conference, Springer Lecture Notes in Computer Science; 2012. [Google Scholar]
- 15.Harman D. Overview of the First Text REtrieval Conference (TREC-1) TREC. 1992;1992:1–20. [Google Scholar]
- 16.Hatcher E, Gospodnetic O, McCandless M. Lucene in action. (2004). structured biomedical text. Journal of the American Medical Informatics Association. 2007;14(3):253–263. doi: 10.1197/jamia.M2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haux R. Hospital information systems – past present future. International Journal of Medical Informatics. 2005;75:268–281. doi: 10.1016/j.ijmedinf.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Hersh W, Müller H, Kalpathy-Cramer J, Kim E, Zhou X. The consolidated ImageCLEFmed medical image retrieval task test collection. Journal of Digital Imaging. 2009;22(6):648–655. doi: 10.1007/s10278-008-9154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hersh W, Müller H, Gorman P, Jensen J. Task analysis for evaluating image retrieval systems in the ImageCLEF biomedical image retrieval task. Slice of Life conference on Multimedia in Medical Education (SOL 2005); Portland, OR, USA. June 2005. [Google Scholar]
- 20.Ide NC, Russell FL, Demner-Fushman D. Essie: a concept-based search engine for. doi: 10.1197/jamia.M2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahn CE, Jr, Thao C. GoldMiner. A Radiology Image Search Engine. Amercial Journal of Roetgenology. 2007;188(6) doi: 10.2214/AJR.06.1740. [DOI] [PubMed] [Google Scholar]
- 22.Markonis D, Holzer M, Dungs S, Vargas A, Langs G, Kriewel S, Müller H. A survey on visual information search behavior and requirements of radiologists. Methods of information in Medicine. 2012;51(6):539–548. doi: 10.3414/ME11-02-0025. [DOI] [PubMed] [Google Scholar]
- 23.Müller H, Boyer C, Gaudinat A, Hersh W, Geissbuhler A. Analyzing Web Log Files of the Health On the Net HONmedia Search Engine to Define Typical Image Search Tasks for Image Retrieval Evaluation. medinfo 2007; Brisbane, Australia. 2007; [PubMed] [Google Scholar]
- 24.Müller H, Clough P, Deselaers T, Caputo B, editors. Volume 32 of The Springer International Series On Information Retrieval. Springer; Berlin Heidelberg: 2010. ImageCLEF – Experimental Evaluation in Visual Information Retrieval. [Google Scholar]
- 25.Müller H, Despont-Gros C, Hersh W, Jensen J, Lovis C, Geissbuhler A. Health care professionals’ image use and search behaviour. Studies in Health Technology and Informatics; Proceedings of the Medical Informatics Europe Conference (MIE 2006); Maastricht, The Netherlands: IOS Press; Aug, 2006. pp. 24–32. [Google Scholar]
- 26.Müller H, Garcia Seco de Herrera A, Kalpathy-Cramer J, Demner Fushman D, Antani S, Eggel I. Overview of the ImageCLEF 2012 medical image retrieval and classification tasks. CLEF 2012 working notes; Rome, Italy. 2012. [Google Scholar]
- 27.Müller H, Geissbühler A, Marty J, Lovis C, Ruch P. Using medGIFT and easyIR for the ImageCLEF 2005 evaluation tasks. CLEF 2005 working notes; Vienna. September 2005. [Google Scholar]
- 28.Müller H, Kalpathy-Cramer J, Eggel I, Bedrick S, Radhouani S, Bakke B, Kahn JCE, Hersh W. Overview of the CLEF 2009 medical image retrieval track. Proceedings of the 10th international conference on Cross-language. [Google Scholar]
- 29.Müller H, Kalpathy-Cramer J, Demner-Fushman D, Antani S. Creating a classification of image types in the medical literature for visual categorization. SPIE medical imaging; San Diego, USA. 2012. [Google Scholar]
- 30.Müller H, Kalpathy-Cramer J, Jr, CEK, Hatt W, Bedrick S, Hersh W. Overview of the ImageCLEFmed 2008 medical image retrieval task. Evaluating Systems for Multilingual and Multimodal Information Access - 9th Workshop of the Cross-Language Evaluation Forum. Volume 5706 of Lecture Notes in Computer Science (LNCS); Aarhus, Denmark. 2009.pp. 500–510. [Google Scholar]
- 31.Müller H, Kalpathy-Cramer J. The ImageCLEF Medical Retrieval Task at ICPR 2010 Information Fusion to Combine Visual and Textual Information. ICPR 2010 contest proceedings; Istanbul, Turkey. Springer; 2010. pp. 99–108. LNCS 6388. [Google Scholar]
- 32.Müller H, Kalpathy-Cramer J, Hersh W, Geissbuhler A. Using Medline queries to generate image retrieval tasks for benchmarking. Medical Informatics Europe (MIE2008); Gothenburg, Sweden. May 2008; [PubMed] [Google Scholar]
- 33.Müller H, Rosset A, Vallée JP, Terrier F, Geissbuhler A. A reference data set for the evaluation of medical image retrieval systems. Computerized Medical Imaging and Graphics. 2004;28(6):295–305. doi: 10.1016/j.compmedimag.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Niblack W, Barber R, Equitz W, Flickner MD, Glasman EH, Petkovic D, Yanker P, Faloutsos C, Taubin G. QBIC project: querying images by content, using color, texture, and shape. In: Niblack W, editor. Storage and Retrieval for Image and Video Databases, volume 1908 of SPIE Proceedings. Apr, 1993. pp. 173–187. [Google Scholar]
- 35.Quellec G, Lamard M, Bekri L, Cazuguel G, Roux C, Cochener B. Medical case retrieval from a committee of decision trees. IEEE Transactions on Information Technology in Biomedicine. 2010;14(5):1227–1235. doi: 10.1109/TITB.2010.2053716. [DOI] [PubMed] [Google Scholar]
- 36.Sedghi S, Sanderson M, Clough P. A study on the relevance criteria for medical images. Pattern Recognition Letters. 2009 [Google Scholar]
- 37.Sparck Jones K, van Rijisbergen C. Report on the Need for and Provision of an “ideal” Information Retrieval Test Collection. 1975 [Google Scholar]
- 38.Smeulders AWM, Worring M, Santini S, Gupta A, Jain R. Content based image retrieval at the end of the early years. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2000;22 (12):1349–1380. [Google Scholar]
- 39.Squire DMG, Müller W, Müller H, Pun T. Content-based query of image databases: inspirations from text retrieval. Pattern Recognition Letters. 2000;21(13–14):1193–1198. Special Issue of SCIA 1999. [Google Scholar]
- 40.Thornley CV, Johnson AC, Smeaton AF, Lee H. The scholarly impact of TRECVid (2003–2009) Journal of the American Society for Information Science and Technology. 2011 Apr;62(4):613–627. [Google Scholar]
- 41.Tsikrika T, Müller H, Kahn CE., Jr Log Analysis to Understand Medical Professionals’ Image Searching Behaviour. Medical Informatics Europe; Pisa, Italy. 2012; [PubMed] [Google Scholar]
- 42.Tsikrika T, Larsen B, Müller H, Endrullis S, Rahm E. The Scholarly Impact of CLEF (2000–2009). CLEF 2013; Valencia, Spain. Springer; 2013. pp. 1–12. LNCS 8138. [Google Scholar]
- 43.Vossen P. EuroWordNet: a multilingual database with lexical semantic networks. Boston: Kluwer Academic; 1998. [Google Scholar]
- 44.Zobel J. How reliable are the results of large scale information retrieval experiments?. 21st Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; Melbourne, Australia. August 1998.pp. 307–314. [Google Scholar]