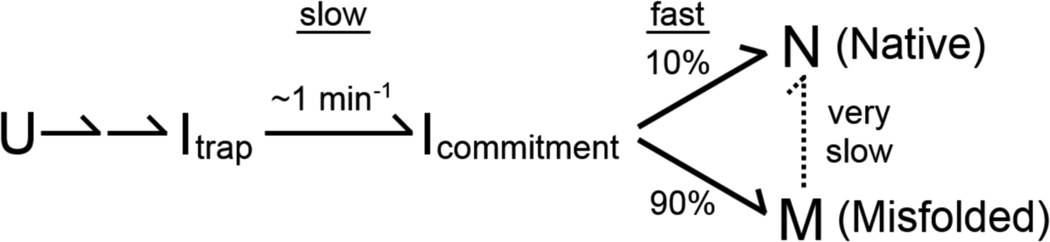Abstract
Like many structured RNAs, the Tetrahymena group I intron ribozyme folds through multiple pathways and intermediates. Under standard conditions in vitro, a small fraction reaches the native state (N) with kobs ≈ 0.6 min–1, while the remainder forms a long-lived misfolded conformation (M) thought to differ in topology. These alternative outcomes reflect a pathway that branches late in folding, after disruption of a trapped intermediate (Itrap). Here, we use catalytic activity to probe the folding transitions from Itrap to the native and misfolded states. We show that mutations predicted to weaken the core helix P3 do not increase the rate of folding from Itrap but they increase the fraction that reaches the native state rather than forming the misfolded state. Thus, P3 is disrupted during folding to the native state but not to the misfolded state, and P3 disruption occurs after the rate-limiting step. Interestingly, P3-strengthening mutants also increase native folding. Additional experiments show that these mutants are rapidly committed to folding to the native state, although they reach the native state with approximately the same rate constant as the wild-type ribozyme (~1 min–1). Thus, the P3-strengthening mutants populate a distinct pathway that includes at least one intermediate but avoids the M state, most likely because P3 and the correct topology are formed early. Our results highlight multiple pathways in RNA folding and illustrate how kinetic competitions between rapid events can have long-lasting effects because the ‘choice’ is enforced by energy barriers that grow larger as folding progresses.
Keywords: Structured RNA, group I intron, catalytic RNA, RNA folding, RNA topology
Introduction
Structured biopolymers face the difficult task of folding to a functional native conformation from the vast collection of non-functional conformations. For RNA, the energetics of specifying a native state may be simplified, at least conceptually, by the prevalence and independent stability of its local secondary structure.1 Local helices can form easily and persist even in the absence of tertiary structure, creating a hierarchy in which most of the secondary structure is determined locally and by relatively simple base-pairing rules, while the overall architecture is determined by the orientations of the helical elements and fixed by tertiary contacts between them.2, 3 On the other hand, the stability of local structure may amplify the challenge of finding the native structure rapidly.4 Non-native secondary structures can be long-lived, like their native counterparts,5–8 and even native contacts can slow folding if they form prematurely and must be disrupted to allow resolution of non-native structure elsewhere.9–15 In light of these properties, it is perhaps not surprising that RNA folding is found experimentally to be rife with kinetically trapped folding intermediates.16, 17 Still, relatively little is known about the folding pathways that lead to the formation and resolution of these intermediates or the non-native structural features of the intermediates.
The Tetrahymena thermophila group I intron and its ribozyme derivative have provided a powerful system to study a complex folding process (Fig. 1a). Early work using the ribozyme demonstrated multiple folding pathways, with the dominant pathways including two kinetically trapped intermediates (Scheme 1).18–22 Under standard conditions in vitro, essentially the entire population folds in seconds to the first intermediate, Itrap.9, 18, 23 From Itrap, a small fraction folds along a pathway to the native state, N, on the time scale of 1 min, while the remainder forms a long-lived misfolded intermediate termed M, from which refolding to N occurs on the timescale of hours.12, 13, 19
Figure 1.
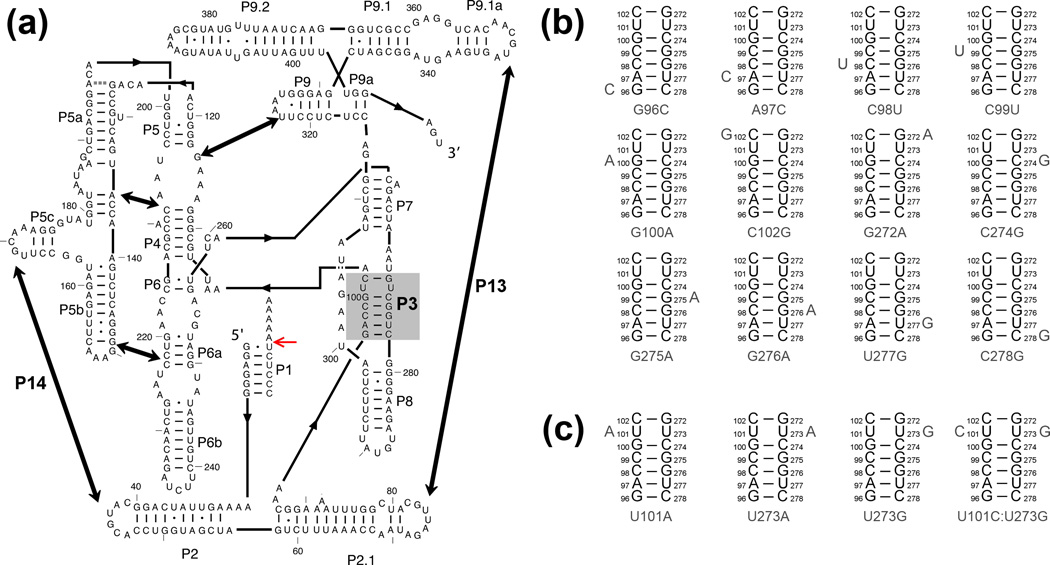
The Tetrahymena ribozyme and mutants used herein. (a) Ribozyme secondary structure. The ribozyme contains five long-range contacts, as indicated by thick arrows. The ribozyme is shown with substrate bound to form the P1 helix, and the red arrow indicates the substrate cleavage site. The P3 helix is shaded in gray. (b–c) Point mutations are designed to weaken the P3 helix by disrupting a single base pair (panel b) or to strengthen P3 by creating A–U, U-A, or U-G pairs or a C–G pair (panel c).
Scheme 1.
Further work has probed these two intermediates and their folding transitions to the native state. The misfolded state M closely resembles the native state,12, 24 yet extensive native structure is disrupted during its refolding to the native state, including all five long-range peripheral tertiary contacts and a native core helix named P3.11, 12 To explain why the M state must unfold to reach the native state even though it is structurally similar to the native state, the M state was proposed to differ in topology, such that extensive unfolding of the periphery and the P3 helix would be required for exchange to the native topology.11, 12 The Itrap intermediate is also compact24–27 and features extensive native secondary and tertiary structure,13, 22, 26 including the P3 helix.13 The folding transition from Itrap to the native state requires unfolding of peripheral structure,9, 13 but it is not known whether core unfolding is required in this transition and specific physical models have not been constructed.
Here we test whether the folding transitions from Itrap to the native and misfolded states require the transient disruption of the P3 helix. By using catalytic activity to monitor folding from Itrap for a series of P3 mutants, we show that P3 is indeed disrupted during folding to the native state, whereas P3 remains intact during the folding transition from Itrap to the M state. This result extends the known similarities of Itrap and M and suggests that the non-native topology is established early in folding along this pathway, exchanging to the native topology while P3 is disrupted during folding from Itrap to N. We also show that mutations that strengthen P3 increase the fraction of ribozyme that avoids the M state by promoting folding along a pathway that is rapidly committed to native folding, most likely by forming the native topology at the outset of folding. While the commitment to the pathway is made in seconds or faster, the native state is reached along this pathway on the timescale of minutes, with a rate constant comparable to that of the wild-type ribozyme. Our results underscore a competition between rapid processes of structure formation and disruption that, when mistimed relative to one another, can lead to alternative folded states that are separated by large energy barriers.
Results
We used catalytic activity to measure formation of the native state by the ribozyme, exploiting the fact that upon folding to the native state, the ribozyme cleaves its oligonucleotide substrate rapidly. Because substrate cleavage is fast, when Mg2+ is added to the unfolded ribozyme in the presence of the substrate, the progress of native state accumulation can be monitored over time from the amount of radiolabeled substrate that has been cleaved to the shorter product oligonucleotide (Fig. 2a; Fig. S1; see Methods).18, 28 For the wild-type ribozyme, the rate constant of an initial ‘burst’ phase of product accumulation reflects the rate constant for native state formation from Itrap and the amplitude of the burst reflects the probability of folding from Itrap to the native state rather than to the M state.18
Figure 2.
Native folding of P3-weakening mutants. (a) Reaction scheme. Folding and catalysis are initiated simultaneously with the addition of Mg2+, substrate and guanosine. Because substrate cleavage is faster than folding, the exponential burst of product formation in the rapid initial phase reveals both the rate constant for native state formation and, from its amplitude, the extent of native state formation. Release of the product oligonucleotide is slow, such that the data show a well-defined burst reflecting folding and the first round of substrate cleavage, followed by a slow linear accumulation of the product that reflects subsequent rounds of substrate cleavage. The concentration values indicate final concentrations after mixing. (b) Folding of the wild-type ribozyme with 10 mM Mg2+ at 25 °C (circles) or 37 °C (squares). (c) Folding of P3-weakening mutants with 10 mM Mg2+ at 25 °C (circles) or 37 °C (squares). Rate constants for the mutants are given in Table 1. The y-axis values represent the concentration of product relative to the concentration of active ribozyme, which was determined from the burst amplitude of a parallel reaction in which the ribozyme was prefolded to the native state.11 Folding of G272A at 37 °C is not shown because the results did not show a well-defined burst of product formation. (d) Rate constants for folding of the P3-weakening mutants, relative to that of the wild-type ribozyme, at 25 °C and 10 mM Mg2+. Error bars show standard errors from at least three independent measurements. (e) Fractions of the P3-weakening mutants that fold to the native state, relative to the value for the wild-type ribozyme at 25 °C and 10 mM Mg2+. Error bars show standard errors from at least three independent measurements.
To determine whether the P3 helix is transiently disrupted during folding from Itrap, we measured folding of sixteen ribozyme mutants that are predicted to weaken or strengthen P3 (Fig. 1b–c).11 The folding data were interpreted in the context of Scheme 1. From Itrap, a rate-limiting step involving partial unfolding generates an intermediate termed Icommitment, which then folds rapidly to the native and misfolded states.13, 29 If P3 is formed in Itrap, as suggested previously,13 and is disrupted transiently during the subsequent folding steps, modulating its stability by mutation is likely to affect the progress of native ribozyme accumulation. If P3 is disrupted in the rate-limiting transition for escape from Itrap, weakening P3 should increase the observed rate constant. If P3 is disrupted along only one of the two pathways after they diverge, weakening P3 should increase the fraction of the ribozyme population that folds along that pathway, changing the fraction that reaches the native state without changing the observed rate constant. If P3 is disrupted along both pathways after they diverge, weakening P3 would have no observed effect because it would increase the rates of the fast steps along both pathways equally, leaving the fraction of ribozyme that folds to the native state unchanged. For any effect of weakening P3, strengthening P3 would be most simply expected to have the opposite effect.
P3 disruption is required for folding from Itrap to N but not to M
We first measured folding of the twelve P3-weakening mutants and obtained rate constants for ten of them.* Most of these mutants folded slightly slower than the wild-type ribozyme, and none were more than 3-fold faster at either 25 °C or 37 °C (Fig. 2b–d and Table 1). There was no systematic dependence of Mg2+ concentration (10 – 200 mM Mg2+, Table S2), as observed previously for the wild-type ribozyme folding along this pathway.19 Overall, these results indicate that P3 is not disrupted during the rate-limiting transition from Itrap. If P3 is formed in the Itrap intermediate as suggested previously,13 it is apparently maintained in the rate-limiting transition state between Itrap and Icommitment (Scheme 1).
Table 1.
Rate constants for folding to the native state of wild-type and mutant ribozymes.
| Folding at 25 °C | Folding at 37 °C | |||||||
|---|---|---|---|---|---|---|---|---|
| Ribozyme | kfold (min–1) | Rela | % Native | Rela | kfold (min–1) | Rela | % Native | Rela |
| Wild Type | 0.29 ± 0.03 | (1) | 12 ± 3 | (1) | 0.61 ± 0.02 | (1) | 34 ± 2 | (1) |
| G96C | 0.07 ± 0.01 | 0.23 ± 0.04 | 100 | ≥ 8.3 | 0.24 ± 0.03 | 0.39 ± 0.04 | 100 | ≥ 2.9 |
| A97Cb | ― | ― | ― | ― | ― | ― | ― | ― |
| C98U | 0.33 ± 0.02 | 1.3 ± 0.1 | 30 ± 10 | 2.3 ± 0.3 | 0.69 ± 0.04 | 1.15 ± 0.09 | 55 ± 4 | 1.74 ± 0.06 |
| C99U | 0.19 ± 0.03 | 0.75 ± 0.08 | 31 ± 4 | 2.7 ± 0.3 | 0.57 ± 0.04 | 0.91 ± 0.08 | 60 ± 3 | 1.70 ± 0.07 |
| G100A | 0.21 ± 0.02 | 0.9 ± 0.1 | 18 ± 1 | 1.1 ± 0.1 | 0.54 ± 0.07 | 0.9 ± 0.1 | 39 ± 4 | 1.0 ± 0.1 |
| C102Gc | ― | ― | ― | ― | ― | ― | ― | ― |
| G272Ad | 0.17 ± 0.01 | 0.65 ± 0.07 | 15 ± 1 | 1.7 ± 0.3 | ― | ― | ― | ― |
| C274G | 0.78 ± 0.09 | 3.0 ± 0.3 | 29 ± 0 | 2.9 ± 0.3 | 0.82 ± 0.04 | 1.30 ± 0.05 | 72 ± 1 | 1.9 ± 0.1 |
| G275A | 0.20 ± 0.03 | 0.79 ± 0.09 | 40 ± 20 | 4.2 ± 0.6 | 0.51 ± 0.03 | 0.82 ± 0.05 | 62 ± 2 | 1.9 ± 0.1 |
| G276A | 0.10 ± 0.01 | 0.40 ± 0.06 | 30 ± 8 | 2.4 ± 0.3 | 0.37 ± 0.13 | 0.6 ± 0.2 | 26 ± 3 | 1.0 ± 0.1 |
| U277G | 0.13 ± 0.01 | 0.49 ± 0.04 | 50 ± 4 | 4 ± 1 | 0.34 ± 0.02 | 0.54 ± 0.04 | 70 ± 3 | 2.2e |
| C278G | 0.064 ± 0.007 | 0.22 ± 0.03 | 100 | ≥ 8.3 | 0.21 ± 0.02 | 0.34 ± 0.02 | 100 | ≥ 2.9 |
| U101A | 0.46 ± 0.05 | 1.6 ± 0.2 | 29 ± 0 | 2.4 ± 0 | 1.0 ± 0.1 | 1.6 ± 0.2 | 42 ± 3 | 1.2 ± 0.1 |
| U273A | 0.8 ± 0.2 | 3.4 ± 0.9 | 59 ± 7 | 6 ± 2 | 1.0 ± 0.2 | 1.8 ± 0.2 | 91 ± 9 | 3.4 ± 0.4 |
| U273G | 0.7 ± 0.2 | 3.0 ± 0.8 | 53 ± 8 | 5 ± 2 | 0.7 ± 0.1 | 1.3 ± 0.1 | 90 ± 10 | 3.8 ± 0.2 |
| U101C:U273G | 0.5 ± 0.2 | 2.0 ± 0.6 | 70 ± 10 | 7 ± 2 | 0.7 ± 0.1 | 1.1 ± 0.2 | 100 ± 8 | 4.1 ± 0.5 |
Errors in rate constants and fractions of native ribozyme are shown as standard error from at least two experiments.
Values were calculated as the folding rate of each mutant divided by that of the wild type from each individual experiment. The values shown are the averages and standard error from the individual experiments.
No burst of product was detected for A97C due to rapid product release, preventing measurement of the fraction of native ribozyme.
Slow substrate cleavage by C102G (0.52 min–1, ref. 11) prevented use of the continuous assay to measure folding.
The G272A mutant did not give a discrete burst phase in the continuous assay at 37 °C, preventing measurement of the folding rate constant and fraction of native ribozyme.
The value for the relative fraction of native ribozyme for U277G at 37 °C was obtained from a single experiment.
We also used these experiments to test whether P3 is disrupted along one of the pathways after they diverge. For all ten mutants whose folding could be measured, the bursts of product formation were significantly larger than for the wild-type ribozyme, indicating increased fractions of native ribozyme for the mutants (Fig. 2c, Fig 2e and Table 1). For eight of the ten mutants, flux through the M state remained detectable, allowing the effects of the mutations to be quantitated (Table 1). The other two mutants, G96C and C278G, which disrupt a terminal base pair in P3, folded to the native state predominantly in a single kinetic phase (Fig. S2a and Table 1), suggesting that these mutations give larger effects and that the M state is avoided within the limits of detection.**
The results that most or all of the P3-weakening mutations increase the fraction of the ribozyme that folds to the native state indicate that P3 is disrupted along this pathway specifically. The lack of a consistent effect on the observed rate constant indicates that P3 is not disrupted in the earlier, rate-limiting step from Itrap to Icommitment. Together, the results support the model in which P3 is formed in the intermediate Itrap,13 and they indicate that P3 remains formed during the rate-limiting transition to Icommitment. Subsequently, P3 is disrupted transiently during the folding transition to the native state, whereas it remains intact during the folding transition from Icommitment to the misfolded state (Scheme 1).
P3-strengthening mutants also increase folding to N
Based on the result that weakening P3 increases the fraction of native folding from Itrap, a simple prediction for the strengthening mutants would be that they would decrease the fraction of ribozyme that folds to the native state. However, previous work indicated that one strengthening mutant, U273A, dramatically increased native folding.19, 29–31 Thus, we used the set of four P3-strengthening mutants to test more broadly the effects of strengthening P3 on the folding outcome. We found that all four mutations increased the fraction of the ribozyme that folded to the native state rather than folding to the M state (Fig. 3 and Table 1). The P3-strengthening mutations also increased the rate constant for native folding modestly (<3-fold, Table 1). Analogous to the P3-weakening mutants, there was no systematic effect of increasing Mg2+ concentration on the rate constant (10 – 100 mM Mg2+, Table S2).
Figure 3.
Native folding of P3-strengthening mutants with 10 mM Mg2+ at 25 °C (circles) or 37 °C (squares). Rate constants for the mutants are given in Table 1. The y-axis values represent the concentration of product relative to the concentration of active ribozyme, as described in the Fig. 2 legend.11
P3-strengthening mutants increase flux along a pathway that avoids long-lived misfolding
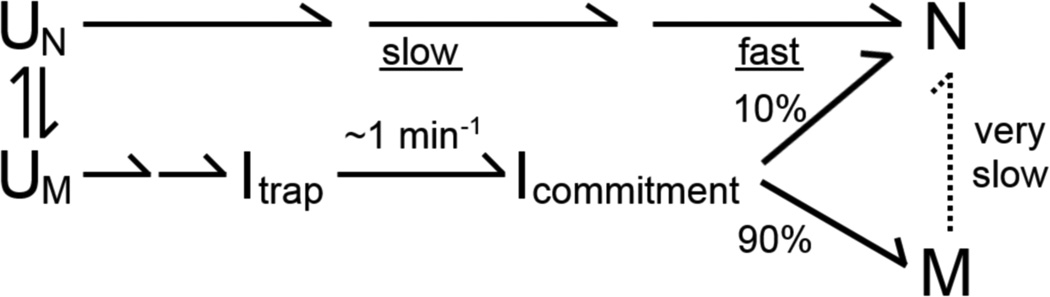
Both the P3-weakening and P3-strengthening mutations increase the fraction of ribozyme that avoids the M state and folds to the native state, a surprising result at face value. However, previous work hinted at a resolution by suggesting that early formation of P3, either by the U273A mutation or by increased Na+ concentration, favored folding to the native state along an alternative pathway in which Itrap and M are avoided.12, 29, 31 Thus, the effects of weakening P3 and strengthening P3 would be exerted at different points in folding, both resulting in increased native folding but via different mechanisms. We tested this model by using catalytic activity to probe when during folding the ribozyme mutants become committed to folding to the native or misfolded states. A prediction from this model is that the P3-weakening mutants would exhibit a ‘late’ commitment to native folding, at or after the rate-limiting transition state, as observed previously for the wild-type ribozyme (Scheme 2, lower pathway).19 In contrast, the P3-strengthening mutants would fold along the alternate pathway and thus commit ‘early’ to native folding (Scheme 2, upper pathway).
Scheme 2.
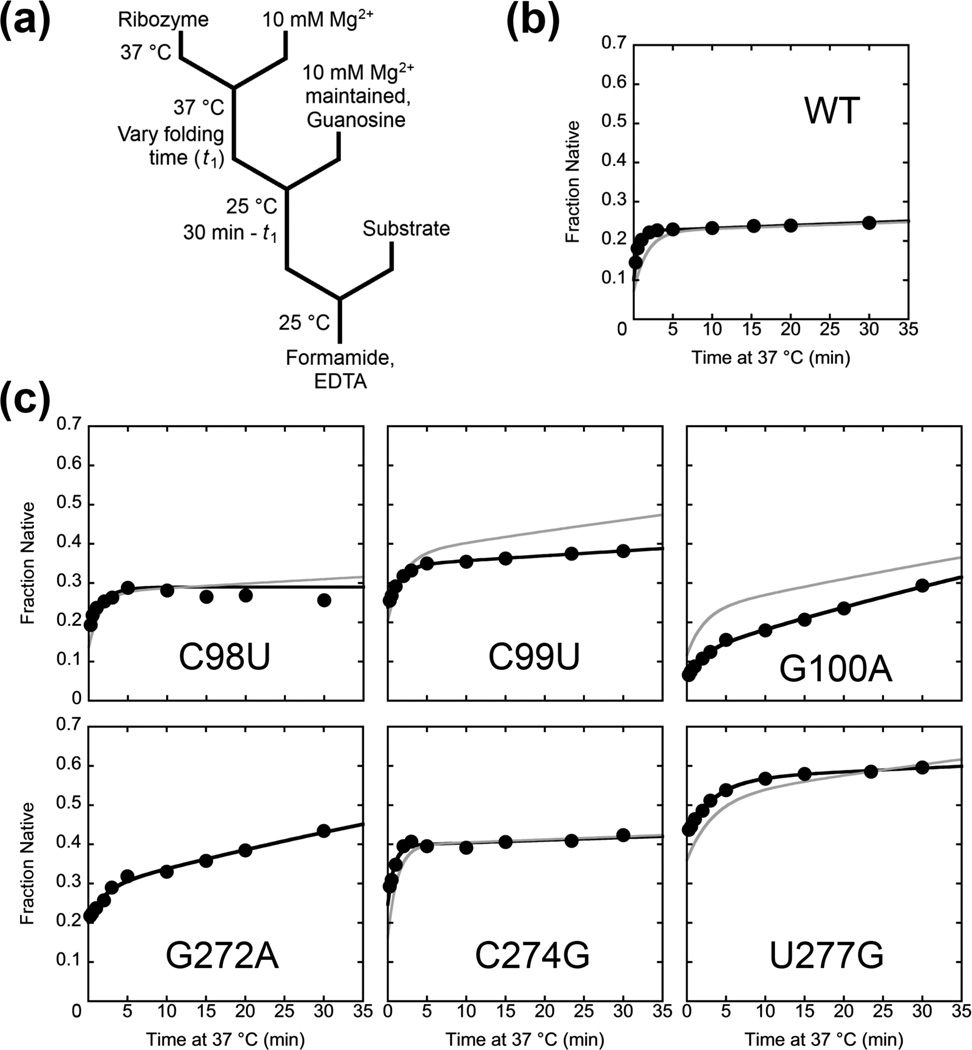
To measure the commitment point, we used a two-stage assay in which the ribozyme was initially folded in the presence of Mg2+ for varying amounts of time at 37 °C (Fig. 4a, t1).19 The temperature was then reduced to 25 °C and the ribozyme was incubated further for a total of 30 min, enough time to fold to the native and misfolded states (Table 1). The key point is that a smaller fraction of the ribozyme folds to the native state at 25 °C than at 37 °C (Fig. 2c and Table 1). Thus, the fraction of the ribozyme that had not yet committed to native folding at the time of the temperature shift would give the smaller fraction of native ribozyme associated with the lower temperature, allowing us to monitor the time dependence of the commitment to native folding at 37 °C.19
Figure 4.
Commitment to native folding or misfolding by P3-weakening mutants monitored by catalytic activity. (a) Reaction scheme. The ribozyme was incubated at 37 °C for various times t1, and then aliquots were transferred to solution at 25 °C and further incubated such that the total incubation time (37 °C + 25 °C) was 30 min. The fraction of native ribozyme was then determined by catalytic activity (see Methods). (b–c) Representative plots (black lines) and simulated folding curves (green lines) for folding of wild-type ribozyme (b) or P3-weakening mutants (c). The result for the wild-type ribozyme was consistent with earlier published results.19 Data from both the wild-type and mutant ribozymes showed two kinetic phases, and the simulations show that the first phase represents the rate constant for commitment to folding to the M or N states, and the second phase reflects refolding from the M state to the native state. A simulation could not be performed for G272A because there was not a distinct burst in the continuous folding assay for this mutant at 37 °C (Fig. 2 and Table 1).
We used this assay to test six of the twelve P3-weakening mutants, while properties of the other six mutants prevented their use in the assay (Figure S2).*** For the six mutants that could be measured, the rate constants for the commitment to native folding were similar to the overall rate constants for native folding at 37 °C, indicating that the commitment occurs late in folding (Fig. 4c and Table 2). For most of the mutants, the data were best described by two exponential curves, with the slower phase reflecting increased refolding from the M state with more time at 37 °C and correspondingly less time at 25 °C. This phase was also detectable for the wild-type ribozyme but was less prominent, presumably because refolding from the M state is slow enough that little additional native ribozyme would be expected within the 30 min of the experiment. To confirm the interpretations of the fast and slow phases, we simulated the reactions using folding parameters determined in independent experiments (Table 1).11 When the simulated results were scaled to account for the lower apparent fractions of native ribozyme in the experimental data, a difference that arises from the use of trace substrate in the experiment (see Methods),19 the simulated results were in good overall agreement with the experimental results (Fig. 4b, c and Fig. S4). Overall, our results show that the P3-weakening mutants fold along the same pathway as the wild-type ribozyme, with the commitment to fold to the native or misfolded states being made late in folding, after Itrap is formed and resolved (lower pathway in Scheme 2).
Table 2.
Rate constants for folding past the commitment point for P3-weakening ribozyme mutants.
| Ribozyme | kfold (min–1) | % Native (Initial) | % Native (End) |
|---|---|---|---|
| Wild Type | 1.4 ± 0.2 | 10.1 ± 0.5 | 21.0 ± 0.4 |
| C98U | 0.5 ± 0.1 | 17.2 ± 0.4 | 29 ± 1 |
| C99U | 0.5 ± 0.1 | 23 ± 1 | 38 ± 3 |
| G100A | 0.47 ± 0.09 | 9 ± 1 | 21 ± 6 |
| G272A | 0.45 ± 0.05 | 22 ± 2 | 32 ± 3 |
| C274G | 1.4 ± 0.4 | 23 ± 2 | 37 ± 1 |
| U277G | 0.4 ± 0.1 | 40 ± 3 | 53 ± 4 |
Uncertainties are shown as the standard errors from at least four experiments.
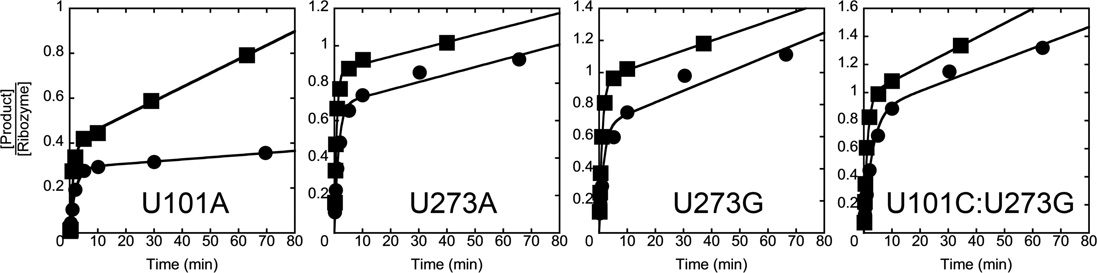
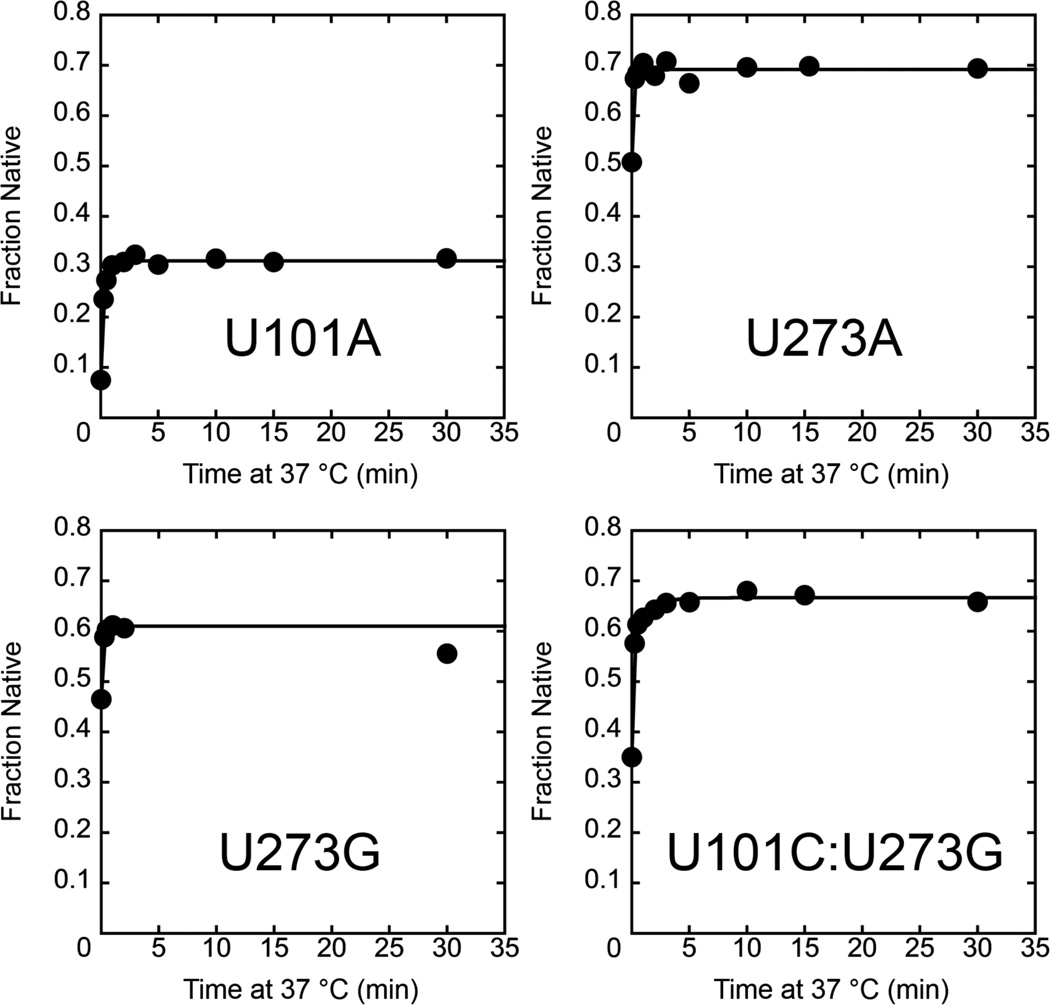
We used the same assay to monitor folding of the P3-strengthening mutants, again monitoring progression past the commitment point at 37 °C. Here we used a lower temperature of 15 °C for the temperature downshift to increase the difference in the amount of native folding between the high and low temperatures. Consistent with the model above, for all four mutants we observed a rapid increase in the fraction of native ribozyme with increased time at 37 °C, with rate constants of 4.1 min–1 to >7 min–1 (Fig. 5). These rate constants are much larger than those for the wild-type ribozyme and the P3-weakening mutants, and they are also much larger than the rate constants for overall folding of the P3-stengthening mutants (Table 1). Thus, these mutants are committed to fold primarily to the native state early in their folding process at 37 °C (upper pathway in Scheme 2). For the U101C:U273G mutant, there was also a slower phase, suggesting that a small fraction of this ribozyme folds via a pathway with a late commitment point, perhaps the pathway populated by the wild-type ribozyme and P3-weakening mutants. Taken together, the results in this section indicate that the P3-weakening mutants fold predominantly through the same pathway as the wild-type ribozyme but with a larger fraction that reaches the native state from the Itrap intermediate rather than forming the M state, whereas the P3-strengthening mutants fold predominantly along a pathway that avoids both Itrap and the M state and is rapidly committed to native folding.
Figure 5.
Commitment to point for native folding of P3-strengthening mutants. The ribozyme was incubated at 37 °C for various times, and then aliquots were transferred to 15 °C and further incubated such that the total incubation time (37 °C + 15 °C) was 30 min. The fraction of native ribozyme was then determined by catalytic activity. The rate constants are U101A, 4.1 min–1; U273A, U273G, and U101C:U273G, >7 min–1. Data from the U101C:U273G ribozyme included a second phase with a rate constant of 0.86 min–1.
Discussion
While structured RNAs are known to misfold in vitro and in vivo, the molecular origins of misfolding and the pathways that generate and resolve misfolded conformations remain largely unknown. Here we aimed to examine how modulating the stability of the long-range helix P3 impacts the lifetime of a kinetically-trapped folding intermediate and the subsequent flux through different folding pathways for the Tetrahymena ribozyme. As described below, the results increase our understanding of key folding steps and the connections between folding pathways (Fig. 6). A general theme is that the choices of folding pathways are governed by kinetic competitions that can occur rapidly but then be enforced by the formation of additional structure, such that the choice has a long-lasting impact on the folding process.
Figure 6.
Two dominant folding pathways of the Tetrahymena ribozyme. The wild-type ribozyme and P3-weakening mutants fold primarily through the lower pathway (red) and the P3-strengthening mutants fold primarily through the upper pathway (green). All of the intermediates on the lower pathway are postulated to include the non-native topology, while those on the upper pathway have the native topology (indicated for intermediates with a superscript N). On the red pathway, the transition from Itrap to Icommitment (abbreviated Icommit) is shown as occurring in two substeps, with native tertiary structure being disrupted in the first substep and then non-native structure being resolved in the second substep. From Icommitment, folding can continue to the misfolded state (M) with the reformation of tertiary contacts, or P3 can be disrupted to allow an exchange of topology, and reformation of P3 gives the intermediate IP3N, which folds rapidly to the native state (N). The rate constants for the transitions from Icommitment are not known but a lower limit of X ≥ 5 was established from earlier work.19, 29 For the P3-strengthening mutants, incubation in buffer solution allows formation of P3, which is proposed to be coupled to formation of the native topology, giving the intermediate I1N (dashed box). Upon Mg2+ addition, folding proceeds by tertiary structure formation to an intermediate that has the non-native structural feature of Itrap but the native topology (ItrapN). This intermediate is resolved in two substeps as described above, and reformation of tertiary contacts leads to formation of the native state.
P3 is disrupted transiently during the transition from Itrap to the native structure
The results of the P3-weakening mutants extend our understanding of how the structured intermediate Itrap is resolved to the native and misfolded states. First, there is a rate-limiting transition from Itrap to another intermediate, Icommitment.13 Weakening P3 does not accelerate the transition, indicating that P3 does not break as Itrap rearranges to the transition state for the step. Because prior footprinting experiments suggested that P3 is formed in Itrap intermediate,13 we infer that P3 likely remains formed throughout the transition and is retained in Icommitment. Mutations of the long-range tertiary contacts accelerate the transition from Itrap,13 leading to a model with two substeps (Fig. 6). Disruption of tertiary contacts occurs first, most likely reversibly, facilitating resolution of the non-native structure in Itrap in the second substep. The identity of this non-native structure in Itrap remains unknown, but when it is present it apparently allows formation of the peripheral tertiary contacts but limits the overall stability of Itrap, relative to the M state, perhaps by weakening some contacts or by limiting cooperativity. During the two substeps of this transition, the rate-limiting transition state is reached for overall folding to both the native and misfolded states (Fig. S5).
From the resulting intermediate Icommitment, folding continues rapidly to the native or misfolded structures. Our finding that weakening P3 increases the flux to the native state indicates that P3 is disrupted during the transition to the native state but not to the misfolded state. Because it is a long-range helix, disruption of P3 allows the core domains to move relative to each other and to exchange topologies between the native topology and the putative misfolded topology.11 Thus, the requirement for P3 disruption to reach the native state suggests that a change in topology is required, while the lack of this requirement in the transition to the misfolded state indicates that the misfolded topology is present in Icommitment, and most likely also in Itrap. This view is consistent with the structural similarity of Itrap and the M state, as indicated by footprinting and small angle X-ray scattering experiments.13, 24 Further, it establishes a strong parallel between the productive folding transitions from Itrap and from the M state, suggesting that these two transitions may converge to a common pathway. We suggest that the convergence may occur at the intermediate Icommitment, which has the misfolded topology and P3 formed but some or all of the tertiary contacts broken (Fig. 6). From this intermediate, the ribozyme can fold toward the N state by breaking P3 or it can fold to M by reforming tertiary contacts while maintaining P3.11 This decision represents a kinetic competition during folding (see below).
P3-strengthening mutants fold along a distinct pathway
Instead of reducing the fraction of native ribozyme, as would be expected from the model above, strengthening P3 increases the ribozyme fraction that avoids the M state to reach the native state. Previous work demonstrated that one of these mutants, U273A, largely avoids long-lived misfolding and instead folds predominantly to the native state.30, 31 Our results strongly support the interpretation that the increase in native folding results from increased stability of P3,30, 31 as all four of the P3-strengthening mutants tested here displayed increased native folding. Preincubation of the wild-type ribozyme in high Na+ concentration also increases native folding upon Mg2+ addition, and footprinting experiments suggested that the Na+ incubation promotes formation of P3 as well as other contacts.29 Thus, we infer that the P3 strengthening mutations allow P3 to form prior to Mg2+ addition (giving the intermediate I1N in Fig. 6), which biases folding by promoting a native arrangement of the core domains and their connections to favor a native topology. Our finding that these mutants display an early commitment to native folding indicates that the dominant pathway for these mutants is separated by large energy barriers from the pathway that results in misfolding and dominates folding of the wild-type ribozyme. Nevertheless, small fractions of these P3-strengthened mutants do fold to M, perhaps because P3 formation is incomplete, allowing formation of the misfolded topology, or because formation of the native topology is incomplete even with P3 formed. For these mutants, any ribozyme that folds through the ‘standard’ pathway is presumably strongly biased to form the M state because of the increased difficulty in disrupting P3 to allow topological exchange (Fig. S5).
Although the P3-strengthening mutants fold predominantly along the pathway that avoids Itrap and M and is committed to native folding, they still reach the native state relatively slowly, with rate constants of approximately 1 min–1. This rate constant is much lower than would be expected in the absence of kinetic barriers, suggesting formation of at least one folding intermediate. The results that these ribozyme mutants become committed to fold to the native state much faster than they reach the native state support this interpretation and indicate that this intermediate is committed to native folding. Although a previous study suggested that the U273A mutant folds up to 50-fold faster than the wild-type ribozyme and without populating intermediates, this conclusion relied on time-resolved footprinting and native gel shift analysis, which may not distinguish the native state from highly structured intermediates, and a catalytic activity assay with limited time resolution.31 In the current work, the higher time resolution and the additional commitment point assay demonstrate the intermediate and the moderate folding rate. Both the value of the folding rate constant and the lack of Mg2+ dependence on this value parallel the behavior of the wild type ribozyme (see Fig. 3, Table 1, and Table S2), suggesting that the intermediate that limits folding of these mutants may possess the non-native structural feature of Itrap, although it presumably has the native topology (ItrapN in Fig. 6). When the non-native structure in this intermediate is resolved, the bulk of the population apparently folds to the native state rather than reversing the topology and misfolding.
Competition between rapid events influences RNA folding decisions
Our results highlight two points at which the duration and outcomes of ribozyme folding can be strongly influenced by alternative events that happen rapidly. The first point is early in folding, where two pathways arise and remain separate. If P3 is formed, folding is biased to the native pathway. If it is not formed, global compaction and the formation of tertiary structure rapidly lock in a non-native structural feature, most likely the non-native topology, to give Itrap with a lifetime >1 min. For the wild-type ribozyme and the P3-weakening mutants, this competition strongly favors the pathway that includes Itrap, and for the P3-strengthening mutants the competition is avoided in our experiments because the mutants form P3 prior to Mg2+ addition. It will be interesting to determine whether the P3-strengthening mutants can fold along the native pathway when folding is begun from conditions that destabilize secondary structure, such as high temperature or urea concentration.
After partial unfolding of Itrap, there is a competition from the intermediate Icommitment, between P3 disruption, which allows folding to N, and alternative folding steps that result in formation of the M state. The time scale for this choice is not known, but it occurs after the rate-limiting step for folding from Itrap, i.e. in seconds or faster. Once formed, the M state persists for hours under standard in vitro conditions, or for minutes under conditions that more closely mimic physiological conditions,12, 19 and the native state is even more stable. Thus, a kinetic competition that takes place in seconds leads to divergent pathways that are separated by very large energy barriers.
This behavior is most likely a consequence of the high stability of RNA structure, including local structure. The kinetic competitions involve alternative events that happen fast, and then the outcome of the competition is enforced by the formation of additional stable structure, some or all of which must be disrupted to allow exchange of the structural differences. Approximately half of the Azoarcus group I intron ribozyme misfolds, while the remainder folds rapidly to the native state.32 Similar behavior was observed for pre-tRNAIle, which harbors the intron, although it is not clear whether the origin of misfolding is the same.33 The glmS riboswitch also partitions between pathways to native and non-native structures.34 In general, observations of misfolding by only a portion of an RNA population suggest a kinetic competition and a bifurcation of pathways to the native and misfolded states.35
Implications of P3 mutations for folding in vivo
Remarkably, both weakening P3 and strengthening P3 increase the amount of ribozyme that folds to the native state while avoiding the M state. Although it is possible that the relative flux through folding pathways is altered in vivo, it is also possible that the M state is formed to some extent in nature and that sequence changes that strengthen or weaken P3 could decrease the amount of M formed. However, each of these changes would have risks. First, strengthening P3 would not necessarily decrease the amount of misfolded ribozyme, because P3 formation would be in kinetic competition with events favoring formation of Itrap. Further, because it is a long-range contact, P3 would presumably be disfavored in co-transcriptional folding relative to the formation of local contacts. In addition, even if strengthening P3 did decrease the amount of misfolded ribozyme formed, it would extend the lifetime of the misfolded RNA that does form. Weakening P3 would decrease the lifetime of the M state by decreasing its stability relative to the largely unfolded transition state, but it might also decrease the stability of the structurally similar native state. Therefore, nature may have struck a balance between folding efficiency and structural stability, perhaps relying on cellular factors to ensure efficient resolution of the misfolded structure.
Materials and Methods
Materials
The L-21/ScaI Tetrahymena ribozyme was purified using Qiagen RNeasy columns as described.36 The oligonucleotide substrate (CCCUCUA5, rSA5) (Dharmacon, Lafayette, CO) was 5´-end-labeled with [γ-32P] ATP by using T4 polynucleotide kinase and purified by non-denaturing polyacrylamide gel electrophoresis as previously described.37 RNA concentrations were determined spectrophotometrically using the following extinction coefficients: wild-type and mutant ribozymes, 3.9 × 106 M–1 cm–1; rSA5, 1.09 × 105 M–1 cm–1.
Measuring formation of the native state
RNA folding reactions using the continuous assay were initiated by the simultaneous addition of substrate (300 nM, including a trace amount of 5´-32P-labeled substrate) and MgCl2 to wild-type and mutant ribozymes (100 nM in 50 mM Na-MOPS, pH 7.0, and 1 mM guanosine). The Mg2+ concentration was varied from 10 mM to 200 mM and the temperature was 25 °C or 37 °C. At various times, aliquots were removed and quenched by addition of formamide and ethylenediaminetetraacetic acid (EDTA). Radiolabeled product (CCCUCU) was separated from uncleaved substrate on a 7 M urea/20% acrylamide gel. Progress curves displayed a rapid burst of product formation, reflecting a single ribozyme turnover that is rate limited by folding to the native state, followed by a slower linear phase of product formation that is rate limited by release of the product.19 The burst amplitudes were normalized by amplitudes from parallel reactions in which the ribozymes were prefolded to the native state. Wild-type and P3-weakening mutants were prefolded by incubation at 50 °C with 10 mM Mg2+ for 30 – 45 min.18, 38 P3-strengthening mutants were prefolded by incubation at 50 °C with 10 mM Mg2+ and 50 mM Na+ for 60 min, followed by dilution into reaction conditions.11
For P3-weakening mutants G96C, A97C, C102G, and C278G, folding was also monitored by using a discontinuous assay in which folding was initiated by addition of 10 mM or 50 mM MgCl2 to 100 nM ribozyme at 25 °C (50 mM Na-MOPS, pH 7.0, and 1 mM guanosine). At various times, aliquots were removed and shifted to 0 °C to inhibit further folding from Itrap.39 A trace amount of radiolabeled substrate was added and the fraction of native ribozyme was determined by catalytic activity as described previously.11
Measuring commitment points to native folding
The rate constant for commitment to folding to the misfolded vs. native states was measured by catalytic activity.19 Folding of wild-type and mutant ribozymes (50 mM Na-MOPS, pH 7.0) was initiated by adding 10 mM Mg2+ at 37 °C. At various times, aliquots were shifted to 25 °C by dilution into a solution containing 10 mM Mg2+ and 1 mM guanosine and further incubated at 25 °C for a total of 30 min (time at 37 °C + time at 25 °C = 30 min). A trace amount of rSA5 was added while maintaining buffer, Mg2+, and guanosine concentrations, and the fraction of native ribozyme was determined from the fraction of the substrate cleaved in the burst phase. For P3-strengthening mutants, aliquots were transferred to 15 °C instead of 25 °C to increase the difference in the fraction of native ribozyme between the temperatures.
Kinetic simulation of commitment to native folding
Kinetic simulations that modeled the commitment to native folding of wild-type and mutant ribozymes were generated using Kinetic Explorer.40 The values for the rate constants for folding from Itrap, the fraction of ribozyme folding to the native state, and the rate constants for refolding of the M state were measured in independent experiments at 10 mM Mg2+ and 25 °C or 37 °C.11 Rate constants for refolding from the M state at 100 mM Mg2+ and 25 °C or 37 °C, used in separate simulations, were calculated from the Mg2+ dependences at lower Mg2+ concentrations. The simulations used the simplified folding pathway shown in Scheme 1. Two ‘mixing’ steps were used, with the first mixing step to initiate folding and the second mixing step to simulate the temperature shift. The fraction of native ribozyme after a total folding time of 30 min was plotted as a function of time at 37 °C, in correspondence with the experiment. To compare the simulations with the experiments (Fig. 4 and Fig. S4), the simulated data were normalized to account for small differences in the burst amplitudes between the simulations, which used parameters obtained from experiments with excess substrate relative to ribozyme, and the commitment point experiments, which used trace amounts of substrate. Smaller bursts are typically observed with trace substrate, most likely because the M state binds the substrate slightly faster than the N state does, so that the M state is slightly overrepresented when using trace substrate.19
Supplementary Material
Acknowledgements
We thank members of the Russell laboratory for helpful comments on the manuscript. This work was supported by grants to R.R. from NIGMS (GM070456) and the Welch Foundation (F-1563). D.M was supported by National Institutes of Health Award F31GM084692.
Footnotes
Two of the mutations, A97C and C102G, were shown previously to decrease the substrate cleavage rate, such that folding from Itrap would not be rate limiting for substrate cleavage and the observed rate constant would not reflect native state formation.11 By measuring the dissociation rate constant for the oligonucleotide product, we found that these mutants are compromised for tertiary docking of the P1 helix, which is required for substrate cleavage (Table S1). This effect presumably accounts at least in part for the reduced rates of substrate cleavage. We attempted to use a discontinuous assay in which folding from Itrap was quenched at 0 °C prior to substrate addition, as this assay does not require substrate cleavage to be faster than folding under the tested set of conditions.28, 39 However, both mutants failed to cleave the substrate detectably at 0 °C (data not shown), preventing the use of this assay to measure their folding.
A discontinuous folding assay also indicated essentially complete native folding for C278G with a rate constant that did not depend on Mg2+ concentration. The absence of a decrease in rate constant with increasing Mg2+ concentration indicates that the experiment monitored folding from Itrap rather than from the M state (Fig. S2b–d). Thus, C278G either avoided forming the M state or it refolded rapidly, such that the M state did not accumulate. Although reduced catalytic activity at low temperature prevented use of the discontinuous assay for G96C, because the same base pair is disrupted in both mutants, the basic features of folding of the two mutants are likely to be the same.
The mutants G96C, A97C, C102G, and C278G were excluded for the reasons noted in the previous sections. G275A was excluded because it showed complex behavior in control assays used to set up this experiment (Fig. S3). We attempted to use G276A but there was no detectable change in the fraction of native ribozyme, presumably because this mutant folds to the same fraction of native ribozyme within error at 25 °C and 37 °C (Table 1).
References
- 1.Sigler PB. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4:477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 2.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 3.Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 4.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 5.Pichler A, Schroeder R. Folding problems of the 5' splice site containing the P1 stem of the group I thymidylate synthase intron: substrate binding inhibition in vitro and mis-splicing in vivo. J Biol Chem. 2002;277:17987–17993. doi: 10.1074/jbc.M111798200. [DOI] [PubMed] [Google Scholar]
- 6.Chadalavada DM, Knudsen SM, Nakano S, Bevilacqua PC. A role for upstream RNA structure in facilitating the catalytic fold of the genomic hepatitis delta virus ribozyme. J Mol Biol. 2000;301:349–367. doi: 10.1006/jmbi.2000.3953. [DOI] [PubMed] [Google Scholar]
- 7.Chadalavada DM, Senchak SE, Bevilacqua PC. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J Mol Biol. 2002;317:559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- 8.Woodson SA, Cech TR. Alternative secondary structures in the 5' exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991;30:2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- 9.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 10.Thirumalai D, Woodson SA. Maximizing RNA folding rates: a balancing act. RNA. 2000;6:790–794. doi: 10.1017/s1355838200000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell D, 3rd, Jarmoskaite I, Seval N, Seifert S, Russell R. The Long-Range P3 Helix of the Tetrahymena Ribozyme Is Disrupted during Folding between the Native and Misfolded Conformations. J Mol Biol. 2013;425:2670–2686. doi: 10.1016/j.jmb.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Wan Y, Suh H, Russell R, Herschlag D. Multiple unfolding events during native folding of the Tetrahymena group I ribozyme. J Mol Biol. 2010;400:1067–1077. doi: 10.1016/j.jmb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 15.Shcherbakova I, Brenowitz M. Perturbation of the hierarchical folding of a large RNA by the destabilization of its scaffold's tertiary structure. J Mol Biol. 2005;354:483–496. doi: 10.1016/j.jmb.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 16.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 18.Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 19.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 20.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 21.Zarrinkar PP, Williamson JR. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat Struct Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]
- 22.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 23.Pan T, Sosnick TR. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat Struct Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 24.Russell R, Millett IS, Doniach S, Herschlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 25.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SG, Thiyagarajan P, Doniach S, Pollack L, Herschlag D. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 26.Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, Smith H, Brenowitz M, Pollack L. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J Mol Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- 27.Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SG, Pande V, Doniach S, Herschlag D, Pollack L. Rapid compaction during RNA folding. Proc Natl Acad Sci U S A. 2002;99:4266–4271. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan Y, Mitchell D, 3rd, Russell R. Catalytic activity as a probe of native RNA folding. Methods Enzymol. 2009;468:195–218. doi: 10.1016/S0076-6879(09)68010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci U S A. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 31.Pan J, Deras ML, Woodson SA. Fast folding of a ribozyme by stabilizing core interactions: evidence for multiple folding pathways in RNA. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 32.Sinan S, Yuan X, Russell R. The Azoarcus group I intron ribozyme misfolds and is accelerated for refolding by ATP-dependent RNA chaperone proteins. J Biol Chem. 2011;286:37304–37312. doi: 10.1074/jbc.M111.287706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangan P, Masquida B, Westhof E, Woodson SA. Architecture and folding mechanism of the Azoarcus group I pre-tRNA. J Mol Biol. 2004;339:41–51. doi: 10.1016/j.jmb.2004.03.059. [DOI] [PubMed] [Google Scholar]
- 34.Brooks KM, Hampel KJ. A rate-limiting conformational step in the catalytic pathway of the glmS ribozyme. Biochemistry. 2009;48:5669–5678. doi: 10.1021/bi900183r. [DOI] [PubMed] [Google Scholar]
- 35.Pan J, Thirumalai D, Woodson SA. Folding of RNA involves parallel pathways. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 36.Russell R, Herschlag D. Specificity from steric restrictions in the guanosine binding pocket of a group I ribozyme. RNA. 1999;5:158–166. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 38.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 39.Russell R, Tijerina P, Chadee AB, Bhaskaran H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.