Abstract
Microtubules (MTs) are pivotal for numerous eukaryotic processes ranging from cellular morphogenesis, chromosome segregation to intracellular transport. Execution of these tasks requires intricate regulation of MT dynamics. Here, we identify a new regulator of the Schizosaccharomyces pombe MT cytoskeleton: Asp1, a member of the highly conserved Vip1 inositol polyphosphate kinase family. Inositol pyrophosphates generated by Asp1 modulate MT dynamic parameters independent of the central +TIP EB1 and in a dose-dependent and cellular-context-dependent manner. Importantly, our analysis of the in vitro kinase activities of various S. pombe Asp1 variants demonstrated that the C-terminal phosphatase-like domain of the dual domain Vip1 protein negatively affects the inositol pyrophosphate output of the N-terminal kinase domain. These data suggest that the former domain has phosphatase activity. Remarkably, Vip1 regulation of the MT cytoskeleton is a conserved feature, as Vip1-like proteins of the filamentous ascomycete Aspergillus nidulans and the distantly related pathogenic basidiomycete Ustilago maydis also affect the MT cytoskeleton in these organisms. Consistent with the role of interphase MTs in growth zone selection/maintenance, all 3 fungal systems show aspects of aberrant cell morphogenesis. Thus, for the first time we have identified a conserved biological process for inositol pyrophosphates.
Author Summary
Fungi are an extremely successful and diverse group of organisms ranging from the small single-celled yeasts to the indefinitely growing filamentous fungi. Polarized growth, where growth is restricted to defined regions, leads to the specific cell shape of yeast cells, as well as the very long hyphae of filamentous fungi. Fungal polar growth is controlled by an internal regulatory circuit of which the microtubule cytoskeleton comprises the transport road for numerous cargos needed for polarized growth. However, the microtubule cytoskeleton is not static, but a dynamic structure, which is modulated by microtubule-associated proteins and the interaction with other cellular structures. Our present analysis has identified a new regulator of the microtubule cytoskeleton in the fission yeast S. pombe: a member of the highly conserved Vip1 inositol polyphosphate kinase family. Vip1 proteins have a dual domain structure consisting of an N-terminal kinase domain which synthesizes inositol pyrophosphates and a C-terminal domain, which we show to negatively regulate the kinase output. Our results suggest that modulation of microtubule dynamics is correlated to Vip1 kinase activity. Importantly, polarized growth and microtubule dynamics were also modulated by Vip1 family members in A. nidulans and U. maydis thus uncovering a conserved biological role for inositol pyrophosphates.
Introduction
Cell polarization can be viewed as the generation and upkeep of a defined cellular organization. The readout of cell polarization in fungal systems is polarized growth resulting in a specific cell shape and size. This ranges from the 14 µm long cylindrical Schizosaccharomyces pombe fission yeast cell, which maintains its shape by restricting growth zones in a cell cycle dependent manner to the extremely polarized growth of filamentous fungi such as Aspergillus nidulans where hyphal extension can occur in a continuous and infinite manner [1]–[3]. Fungi are capable of morphological transitions in response to external signals and this represents an important virulence trait of pathogenic fungi such as the corn smut fungus Ustilago maydis. Here, the transition from a non-pathogenic haploid yeast-like form to a dikaryotic filament is required for the fungus to enter the host tissue [4]. Such an alteration in growth form is also present in non-pathogenic model yeasts such as S. cerevisiae and S. pombe where it acts as a foraging response [5]–[7]. Polarized growth in fungi depends on the interplay between the MT and actin cytoskeletons and in some systems septins [8]. In S. pombe, where growth occurs at the cell tips which contain oscillating Cdc42, actin cables are used for the transport of growth vesicles. On the other hand, MT plus-end dependent transport of the landmark complex Tea1-4 via the kinesin Tea2 is required for marking potential zones of growth [1], [9]–[13]. Correct delivery of Tea1-4 requires alignment of antiparallel interphase MTs along the long axis of the fission yeast cell. The dynamic MT plus-ends are oriented and polymerize towards the cell end; upon contact with the tip MT dynamics are modified, the landmark complex unloaded and anchored at the cell tip [14]–[16]. MT dynamics are regulated mainly by the diverse group of proteins at the MT plus-end. Here, the central component is the conserved EB1 family, which is essential for plus-end association of numerous +TIPs [17]. Interestingly, the Tea1-4 complex is also present in filamentous fungi where a recent publication has uncovered additional functions namely regulating MT dynamics and MT guidance at the hyphal tip. Loss of the S. pombe Tea1 homologue TeaA in A. nidulans results in an inability to maintain the direction of growth and thus results in meander-like growing hyphae [3], [18]. TeaA present at the hyphal tip is responsible for focusing of MTs at a single point and the regulation of MT plus-end dynamics via negative modulation of the XMAP214 family member AlpA [19]. If this negative regulatory function on MT dynamics is a common feature of Tea1-like proteins remains to be determined but the MT phenotype of S. pombe tea1Δ (deletion) cells supports such a scenario [14].
Although core mechanisms of growth zone definition and maintenance are conserved in fungi, the consistently growing hyphae of filamentous fungi require a much more sophisticated system of MT-based transport than is necessary for yeast cell growth [3], [20]–[27]. For example in U. maydis the MT cytoskeleton is required for long distance endosomal transport via plus- and minus-end -directed motor proteins such as kinesin and dynein, respectively [24], [28]–[32]. This transport process has been shown to be crucial for efficient secretion [33], [34]. Important molecular cargos of these endosomes are septins, mRNAs and ribosomes [35]–[37]. Interestingly, local translation of septin mRNA on shuttling endosomes loads these membranous carriers with newly synthesized septin protein for transport towards the hyphal tip [35].
In this work we describe a new core element of fungal growth zone selection and MT cytoskeleton regulation: the conserved Vip1 family which synthesizes diphospho-myo-inositol polyphosphates (inositol pyrophosphates). These high energy molecules are mainly made from inositol hexakiphosphate (IP6) and are generated by two classes of enzymes: IP6Ks and the PPIP5Ks (called Vip1 family throughout this work) (recently reviewed in: [38]–[41]. The Vip1 family, which was discovered in S. pombe and S. cerevisiae, was shown to have enzymatic activity by using an S. cerevisiae strain where the genes coding for the IP6K Kcs1 and the nudix hydrolase Ddp1 had been deleted [42], [43]. In humans two homologues exist named PPIP5K1 and PPIP5K2 [44], [45]. All members of this enzyme class have a dual domain structure consisting of an N-terminal “rimK”/ATP-grasp superfamily domain which phosphorylates position 1 on the fully phosphorylated inositol ring and a C-terminal domain with homology to histidine-acid-phosphatases [44]–[47]. The function of the latter domain remains a matter of debate. Key histidine residues are conserved in this domain, but unusually Vip1-like proteins do not have an aspartate residue next to the second histidine [45]. Furthermore detailed analysis of the human Vip1 phosphatase-like domain demonstrated that this domain is catalytically inactive [48]. However phenotypic analysis of S. pombe strains expressing Asp1 variants (fission yeast Vip1 member) with mutations of conserved C-terminal domain histidine residues suggested that these residues are required for negative regulation of the kinase activity [7]. In addition, truncated S. cerevisiae and human Vip1 variants which only contained the N-terminal kinase domain generated more inositol pyrophosphates than the full-length versions pointing to a kinase antagonizing activity of the C-terminal domain [45], [49]. Now, we provide evidence that Vip1-like proteins harbor two enzymatic activities.
Inositol pyrophosphates regulate cellular processes by two different modes of action: (i) modulation of protein function by reversible binding of these high energy molecules and (ii) protein pyrophosphorylation [50], [51]. An example for the first type of regulation is the Akt kinase which is involved in insulin signaling. Here, specific inositol pyrophosphates were shown to bind to the Akt PH domain thus blocking activation of this kinase [52]. An example for the second type of action is the regulation of the antiviral response via activation of the interferon transcription factor IRF3. In a cell free system IRF3 was phosphorylated by specific inositol pyrophosphates and this process required the transfer of the β-phosphate of the pyrophosphate group [49].
The cellular processes regulated by inositol pyrophosphates are wide-ranging and diverse. These include the phosphate availability response in S. cerevisiae, the chemotactic response in Dictyostelium, the antiviral response and insulin signaling in mammals and the dimorphic switch in S. pombe [7], [49], [50], [52], [53]. We have now extended our analysis of Vip1 biological function and have found that inositol pyrophosphates have a conserved role in fungal morphogenesis.
Results
The Vip1 family member Asp1 generates IP7 in vitro
We had previously generated S. pombe strains that expressed the endogenous Asp1 variants Asp1D333A and Asp1H397A [7]. The former Asp1 variant has a single amino acid change at position 333, a key catalytic residue for kinase activity, while H397 is a highly conserved histidine residue of the C-terminal acid phosphatase-like domain (Figure 1A) [43]. Phenotypic analysis of these Asp1 variant expressing strains suggested that Asp1D333A and Asp1H397A have an altered enzymatic activity compared to the wild-type Asp1 protein [7]. We therefore assayed the ability of Asp1D333A and Asp1H397A to generate inositol pyrophosphates.
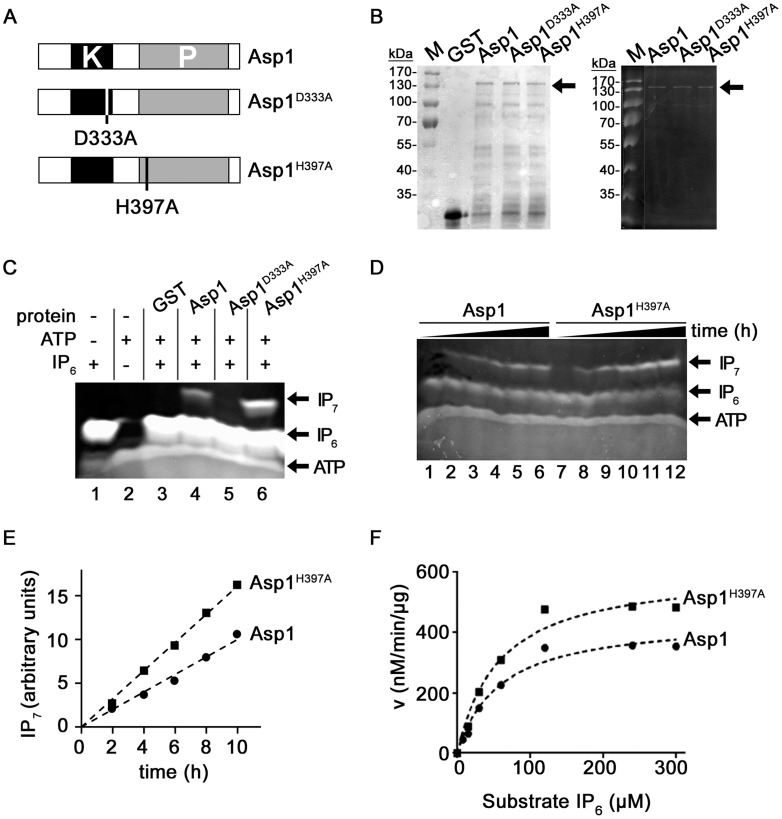
Figure 1. Generation of IP7 by Asp1 variants.
(A) Diagrammatic representation of the Asp1 variants used. (B) Western blot analysis using a GST antibody of GST-tagged wild-type Asp1, GST-Asp1D333A and GST-Asp1H397A. Proteins were purified from E.coli, quantified (Coomassie stained gel is shown in left panel) and equal amounts loaded on a 10% PAGE. The tagged Asp1 variants (arrow) run at the expected size of 139 kDa. (C) Generation of IP7 by GST-Asp1 variants. 4 µg of the indicated proteins were used in an ATP-dependent enzymatic reaction (16 hrs) and the resulting inositol pyrophosphates were resolved on a 35.5% PAGE and stained with Toluidine Blue [54]. −, component not present; +, component present. (D) Time-dependent (0–10 hrs in 2 hr. steps) generation of IP7 by Asp1 and Asp1H397A. Assay conditions were as in 1C. (E) Quantification and diagrammatic representation of the IP7 bands obtained in the assay shown in (D). (F) Determination of Km and Vmax values. 2 µg GST-Asp1 and GST-Asp1H397A were incubated with varying amounts of substrate for 6 hrs. The IP7 was detected as in 1C, and quantified using an IP6 calibration curve (Materials and Methods). v, reaction rate. Vmax Asp1: 450.5 nM/min/µg; Vmax Asp1H397A: 611.2 nM/min/µg.
As it had not been demonstrated previously that the S. pombe Asp1 protein could generate inositol pyrophosphates, we first tested with an in vitro assay if this was the case. Asp1 was expressed in bacteria as a glutathione-S-transferase (GST) fusion protein and purified. Using a recently published method that allows analysis of inositol pyrophosphates by PAGE, we found that purified GST-Asp1 generated inositol pyrophosphates (from now on called IP7) in an ATP-dependent manner using IP6 as a substrate (Figure S1A, right panel) [54]. This activity was dose-dependent, as the amount of IP7 generated increased with increasing amounts of protein used (Figure S1B). We next tested if the GST-tagged Asp1D333A and Asp1H397A proteins (Figure 1B) also generated IP7 and found that Asp1D333A was unable to convert IP6 to IP7 (Figure 1C, lane 5). Interestingly, comparing equal amounts of protein, the Asp1H397A variant generated more IP7 than the wild-type Asp1 protein (Figure 1C, lane 6 and 4, respectively). Analysis of IP7 production by Asp1 and Asp1H397A proteins over a time period of 0 to 10 hrs revealed that IP7 production increased with time and that Asp1H397A could produce up to 100% more IP7 than the wild-type Asp1 protein (Figure 1D, left panel, lanes 7–12 and 1–6, respectively; quantification shown in 1E). Similar results were obtained when comparing IP7 production of the wild-type Asp1 and the Asp11-364 variant (contains only the kinase domain) (Figure S12). These data point to a negative role of the C-terminal acid phosphatase-like domain. To analyze this further we determined the Km and Vmax values for Asp1 and Asp1H397A (Figure 1F). The Km values for Asp1 and Asp1H397A were 58.18 µM and 57.32 µM respectively implying a similar affinity for the substrate. However the Vmax for Asp1H397A was 36% higher than that of Asp1 (Figure 1F).
To learn more about the negative impact of the Asp1 phosphatase-like domain on IP7 production, we (i) tested if addition of Asp1365-920 reduced the inositol pyrophosphate out-put in an Asp1 in vitro kinase assay and (ii) determined the in vivo read-out of Asp1 variants with mutations in conserved residues of the phosphatase-like domain (Figure 2A).
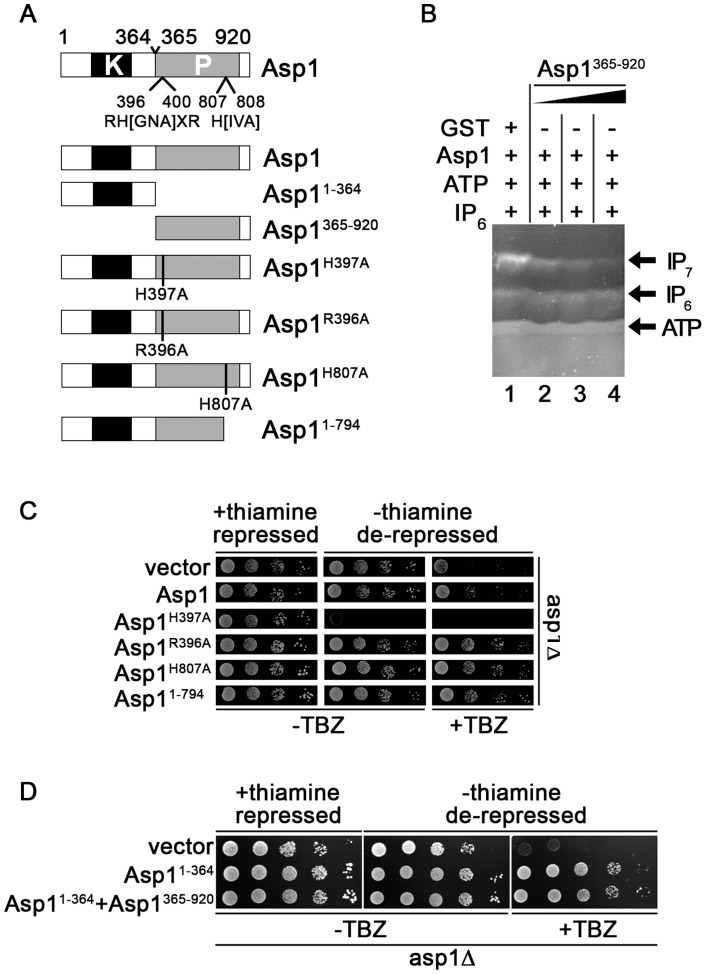
Figure 2. Function of the Asp1 phosphatase-like domain.
(A) Diagrammatic representation of the Asp1 protein with the conserved phosphatase signature motif (motif: RH(GNA)XR-HD in Asp1 RHADR-HI)(top) and the Asp1 variants used. Top: (B) Generation of IP7 by GST-Asp1 with varying amounts (2,4,8 µg) of Asp1365-920. Enzymatic reaction was carried out as for 1C. −, component not present; +, component present. (C) Serial dilution patch tests (104–101 cells) of the asp1Δ (deletion) strain expressing the indicated asp1 variants via the nmt1+ promoter. This promoter is repressed in the presence of thiamine and de-repressed in its absence. Cells were grown for 6 days at 25°C on plasmid selective minimal medium without (−) or with (+) TBZ. (D) Serial dilution patch tests (105–101 cells) of the asp1Δ strain expressing either asp11-364, asp1365-920 or asp11-364 and asp1365-920. Cells were grown for 7 days at 25°C on plasmid selective minimal medium without (−) or with (+) TBZ.
The presence of purified bacterially expressed GST-tagged Asp1365-920 in the IP7 in vitro assay together with full length Asp1 reduced the amount of IP7 in a dose-dependent manner (Figure 2B). IP6 amounts were unaffected by Asp1365-920 as shown by the incubation of only this Asp1 variant with IP6 in the in vitro assay (Figure S13). Thus, the negative effect was only seen for the IP7 output. We therefore propose that the Asp1 C-terminal phosphatase-like domain has phosphatase activity and its substrate is inositol pyrophosphate generated by the Asp1 N-terminal kinase domain (see discussion).
We had shown previously that the asp1H397A strain was more resistant to microtubule poisons such as thiabendazole (TBZ) while the asp1D333A strain was more sensitive to TBZ compared to the wild-type strain [7]. A deletion of asp1+ (asp1Δ strain) also led to TBZ hypersensitivity (Figure S2A). Furthermore a strain where the wild-type asp1+ had been replaced by the asp1D333A, H397A variant also showed the same increased TBZ sensitivity as the asp1D333A and asp1Δ strains (Figure S2A). These data strongly suggest that the TBZ resistance/sensitivity of these strains is solely dependent on the function of the Asp1 kinase. Absence of Asp1 kinase activity results in TBZ hypersensitivity (asp1Δ, asp1D333A and asp1D333A, H397A strains) while increased Asp1 kinase function (asp1H397A strain) results in TBZ resistance. The Asp1 C-terminal phosphatase-like-domain appears to modulate only the function of the Asp1 N-terminal kinase domain as the asp1D333A, H397A strain has the same TBZ phenotype as asp1Δ and asp1D333A strains (Figure S2A).
These results demonstrate that increased TBZ resistance can be used as an in vivo read-out for a non-functional Asp1 phosphatase domain. We expressed wild-type asp1+ and the mutant versions asp1H397A, asp1R396A, asp1H807A and asp11-794 on a plasmid from the thiamine-repressible nmt1+ promoter [55] in the asp1Δ strain. Western blot analysis revealed that expression of full length Asp1 variants was similar (Figure S14). Expression of these asp1 variants except asp1H397A did not affect cell growth (Figure 2C, growth on thiamine (nmt1+ promoter repressed) versus growth on thiamine-less (nmt1+ promoter de-repressed) plates. Plasmid-borne high expression of asp1H397A is lethal as has been shown previously [43].
As shown in Figure 2C plasmid-borne expression of full length asp1+ allowed partial growth of the asp1Δ strain on TBZ containing plates. However expression of Asp1R396A, Asp1H807A and Asp11-794 resulted in better growth of the asp1Δ strain on TBZ medium. We conclude that the conserved phosphatase signature motif is required for the function of the C-terminal domain.
To test if the Asp1 C-terminal domain is also able to regulate Asp1 kinase activity in trans in vivo, we constructed a plasmid, which expressed Asp11-364 and Asp1365-920 from two separate nmt1+ promoters on the same plasmid (Figure S3A). Expression of this plasmid in the asp1Δ strain resulted in a similar phenotype as expression of a plasmid expressing only Asp11-364 (Figure 2D, protein levels shown in Figure S3B–C). Thus in this in vivo situation, it appears that both Asp1 domains need to be on the same molecule for the negative impact of the C-terminal domain to be exerted.
Asp1 affects interphase MT organization
Our in vitro kinase assay demonstrated that the Asp1D333A variant has no enzymatic activity, while that of Asp1H397A is higher than that of the wild-type Asp1 protein. Thus it is very likely that the resistance/sensitivity to microtubule poisons is a result of different intracellular inositol pyrophosphate levels.
We had previously identified a truncated Asp1 variant (Asp11-794) as a multi-copy suppressor of the TBZ-hypersensitivity of a mal3 mutant strain [7]. Mal3 is the fission yeast member of the EB1 family of MT associated proteins [56]. We therefore determined if Asp1 function modulated the MT cytoskeleton by analyzing the interphase MT cytoskeleton of the various asp1 strains via expression of GFP-α-tubulin (using the endogenous nmt81::gfp-atb2+ construct) [57]. asp1 variant strains with or without the presence of GFP-α-tubulin had a similar growth phenotype (Figure S4A).
In S. pombe, interphase MTs are polymerized in the vicinity of the nucleus, align along the long axis of the cell and grow with their plus-ends towards the cell end, where they pause prior to de-polymerization [58]. The organization of interphase MTs of the gfp-atb2 asp1H397A strain was comparable to the wild-type strain while those of the fainter fluorescent gfp-atb2 asp1D333A and gfp-atb2 asp1Δ MTs appeared to be more disorganized (Figure 3A). In particular, the number of interphase MTs that were not oriented along the long axis of the cell was increased in asp1D333A (Figure S15) and asp1Δ cells although this was not statistically significant. However the number of interphase MTs that depolymerized at the lateral cortex/cytoplasm and not at the cell tip was increased significantly in asp1D333A and asp1Δ cells compared to wild-type cells (Figure 3B). An example is shown for an asp1D333A MT that touched the lateral cortex and became depolymerized instead of being deflected as seen for such MTs in asp1+ cells (Figure S4B).
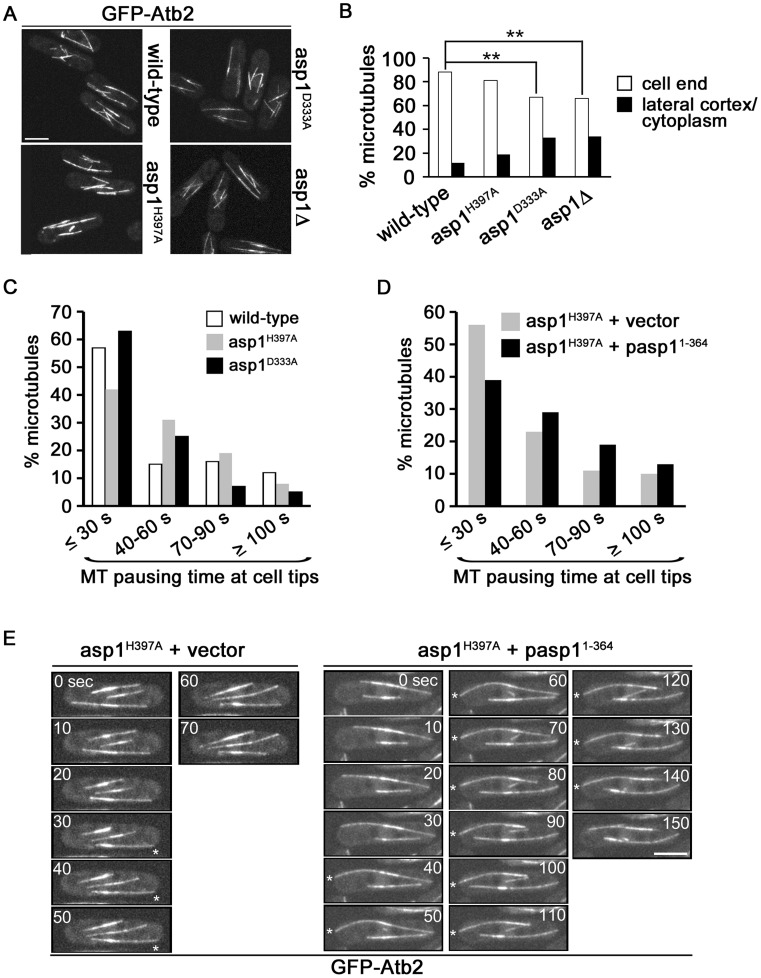
Figure 3. Asp1 kinase function affects MT organization.
(A) Live cell images of the indicated strains expressing nmt81::gfp-atb2+. The same imaging and image-processing conditions were used for all strains. Bar, 5 µm. (B) Percentage of MTs depolymerising at a cell end or at the lateral cortex/in the cytoplasm. Wild-type: n = 102, asp1H397A: n = 218, asp1D333A: n = 166, asp1Δ: n = 131. ** P<0.005 for asp1D333A or asp1Δ compared to wild-type as determined using χ2-test. (C) MT pausing time (sec) at cell ends in the indicated strains. Overall MT pausing time of these strains is shown in table 1. We arbitrarily defined the 4 categories to show the variability within this system. Wild-type: n = 100, asp1H397A: n = 67, asp1D333A: n = 75. (D) MT pausing time (sec) at cell ends in the asp1H397A strain transformed with a vector control or expressing pasp11-364. Overall MT pausing time of these strains is shown in table 1. Cells were grown in plasmid-selective minimal medium. asp1H397A+vector: n = 105, asp1H397A+pasp11-364: n = 110. pasp11-364 denotes plasmid-borne expression of Asp11-364 via the nmt1+ promoter under promoter de-repressing conditions. (E) Live cell images of the nmt81::gfp-atb2 + expressing asp1H397A strain transformed with the vector control or expressing pasp11-364. Images shown are 10 sec intervals. Asterisks (*) denote MTs touching the cell end. Bar, 5 µm.
Inositol pyrophosphates regulate interphase MT dynamics
Measurement of MT parameters in the 3 asp1 variant strains revealed that MT dynamics were altered. asp1D333A cells showed an increased MT growth rate while the rate of MT shrinkage was decreased in asp1H397A cells (Table 1). Interphase MTs of asp1D333A cells had an increased number of catastrophe events while those of asp1H397A cells were reduced compared to wild-type cells (Table 1). The average MT length for both asp1 mutant strains was increased compared to the MTs of the wild-type strain. Thus, all measured MT parameters were affected by intracellular inositol pyrophosphate levels. asp1D333A MTs are more dynamic, whereas asp1H397A MTs have the opposite phenotype.
Table 1. Interphase MT dynamics in asp1 variant strains.
| strain | growth (nm/sec) | rate of shrinkage (nm/sec) | growth before catastrophe (sec) | length (µm) | pausing at tips (sec) |
| wild-type | 56.8±26.5 (n = 90) | 154.8±66.5 (n = 72) | 81.2±32.3 (n = 100) | 6.3±1.2 (n = 100) | 47.3±38.0 (n = 100) |
| asp1D333A | 66.7±27.2* ↑ (n = 90) | 145.6±67.5 (n = 45) | 68.3±29.4* ↓ (n = 126) | 6.9±1.1* ↑ (n = 86) | 34.5±29.1* ↓ (n = 75) |
| asp1H397A | 58.2±30.9 (n = 90) | 126.7±50.2* ↓ (n = 92) | 99.0±39.2* ↑ (n = 155) | 7.3±1.4* ↑ (n = 100) | 50.7±35.7 (n = 67) |
Parameters of MT dynamics were measured for the indicated strains expressing nmt81::gfp-atb2+. Cells were grown in non-selective minimal medium. n, number of MTs measured; asterisks denote significance between wild-type and mutant; ↓ or ↑, parameter is decreased or increased compared to wild-type. growth: * p<0.01 for asp1D333A vs. wild-type (t-test). Rate of shrinkage: * p<0.005 for asp1H397A vs. wild-type (Welch-test). Growth before catastrophe: * p<0.005 for asp1D333A vs. wild-type (t-test), * p<0.0005 for asp1H397A vs. wild-type (t-test). Length: * p<0.0005 for asp1D333A vs. wild-type (t-test), * p<0.0005 for asp1H397A vs. wild-type (Welch-test). Pausing at tips: * p<0.01 for asp1D333A vs. wild-type (Welch-test).
Interestingly, we found that the residence time of the MT plus-end at the cell tip was dependent on the asp1 variant expressed in the cell. Measurement of the time that a MT stays at the cell tip showed that the residence time of a MT plus-end at the cell tip is variable. Nevertheless, when we compared this MT parameter for wild-type and asp1D333A cells we found that the latter MTs had on average a significantly reduced pausing time at the cell tip before depolymerization (Table 1). For example, only 12% of asp1D333A MTs but 28% of wild-type MTs paused at the cell tip for more than 60 seconds (Figure 3C). In contrast, the residence time of asp1H397A MT plus-ends at the cell tip appeared to be increased compared to wild-type MTs but this was not statistically significant (Table 1). We therefore increased Asp1 generated inositol pyrophosphate levels even further by plasmid-borne expression of the Asp1 variant Asp11-364 (kinase only) in the asp1H397A strain. Under these conditions the average MT pausing time was increased by 30% in cells expressing Asp11-364 (asp1H397A strain plus vector: 42.2±36.8 seconds; n = 105; asp1H397A strain plus pasp11-364: 53.6±38.4 seconds; n = 110; p<0.025 (t-test)). A detailed depiction of MT pausing time in this assay is shown in Figure 3D and an example of the increased MT residence at the cell tip is shown in Figure 3E.
Thus, inositol pyrophosphate levels appear to regulate the residence time of a MT plus-end at the cell tip. Increasing the levels of Asp1 generated inositol pyrophosphates increases pausing at the tip prior to a catastrophe event, while lowering the amount of Asp1 generated inositol pyrophosphates has the opposite effect.
Asp1 regulated MT dynamics occur independently of the +TIP protein Mal3
Proteins associated with MT plus-ends play a leading role in regulating MT dynamics [59]. Of particular importance is the EB1 family, which is central to the association of other +TIPs with the MT plus-end. To determine if Asp1 affects MT dynamics via the EB1 family member Mal3, double mutant strains between mal3Δ (mal3 deletion) and the asp1 alleles asp1H397A, asp1D333A and asp1Δ were constructed. The asp1H397A mal3Δ strain showed a reduced TBZ sensitivity compared to the single mutant mal3Δ strain, demonstrating that increased Asp1 kinase function rescues the mal3Δ mutant TBZ phenotype (Figure 4A). Loss of Asp1 kinase activity increased the TBZ hypersensitivity of mal3Δ strains as shown for the asp1D333A mal3Δ and asp1Δ mal3Δ strains (Figure 4A). Similar results were obtained when these strains grew on medium containing the MT drug methyl-benzimidazol-2-yl-carbamate (MBC) (Figure 4B).
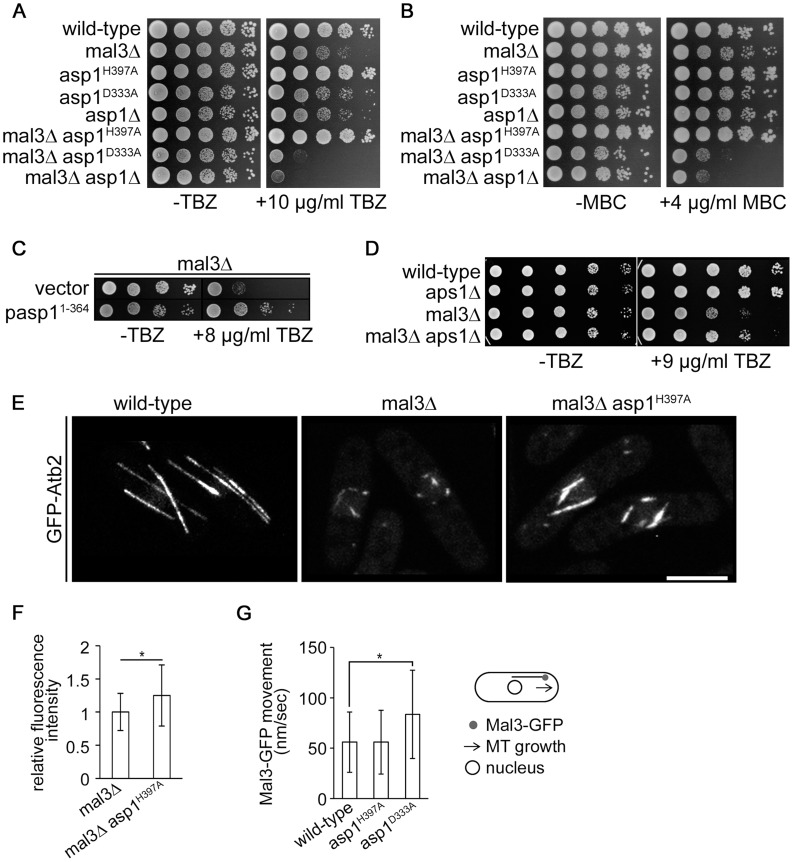
Figure 4. Asp1 MT regulation functions independently of Mal3.
(A) Serial dilution patch test (104–101 cells) of the indicated strains grown for 5 days at 25°C on YE5S without (−) or with (+) TBZ. (B) Serial dilution patch test (104–101 cells) of the indicated strains grown for 5 days at 25°C on YE5S without (−) or with (+) MBC. (C) Serial dilution patch test of the mal3Δ transformants grown under plasmid selective conditions at 25°C for 5 or 9 days without (−) or with (+) TBZ, respectively. (D) Serial dilution patch tests (105–101 cells) of the indicated strains grown at 25°C on MM without (−) or with (+) TBZ. (E) Photomicrographs of living wild-type, mal3Δ and mal3Δ asp1H397A cells grown at 30°C expressing nmt81::gfp-atb2 +. Bar, 5 µm. (F) MT relative fluorescent intensity (mal3Δ strain, 1+/−0.28, n = 26; mal3Δ asp1H397A strain, 1.25+/−0.46, n = 16; arbitrary units; *, P<0.05 as determined using Welch-Test). (G) Movement of outmost outbound Mal3-GFP comets (see diagram). Speed of comets (nm/sec): wild-type, 56±30, n = 93; asp1H397A, 56±31.6, n = 69; asp1D333A, 83.7±43,8, n = 75. * p<0.0005 for asp1D333A versus wild-type (Welch-test).
We next assayed if plasmid borne overexpression of the Asp1 variant Asp11-364 (Asp1 kinase domain only), rescued the mal3Δ TBZ hypersensitivity, and found this to be the case (Figure 4C). Furthermore increasing intracellular IP7 levels by other means than asp1 + manipulation, namely by using a strain where the ORF coding for the nudix hydrolase Aps1 was deleted (aps1Δ), also decreased the TBZ hypersensitivity of the mal3Δ strain (Figure 4D). Nudix hydrolases degrade inositol pyrophosphates and disruption of the nudix hydrolase encoding gene increases the intracellular concentration of inositol pyrophosphates 3-fold [60], [61].
As Mal3 stabilizes MTs, mal3Δ cells do not have a normal interphase MT-cytoskeleton, where MTs are aligned in antiparallel bundles along the cell axis to the cell ends. Instead such interphase cells have very short MT stubs present around the nucleus as MT catastrophe events are increased (compare wild-type MTs to mal3Δ MTs) (Figure 4E) [56], [62], [63].
This short interphase MT phenotype was rescued partially in the mal3Δ asp1H397A strain (Figure 4E).We determined MT parameters in the mal3Δ and mal3Δ asp1H397A strains and observed no difference in the number of MTs/cell (Figure S16). An analysis of the interphase MTs of a mal3Δ asp1D333A strain was not possible as interphase MTs of this strain were extremely short and unstable.
Interestingly, MT length, MT growth time and the relative MT intensity were all increased significantly in the double mutant mal3Δ asp1H397A strain compared to the single mutant mal3Δ strain (Table 2 and Figure 4F). MTs grew longer before a catastrophe event in the mal3Δ asp1H397A strain compared to the mal3Δ strain and MT length was increased in the former compared to the latter strain (Table 2). Furthermore the relative MT fluorescence intensity was increased 1.25 fold in the mal3Δ asp1H397A strain compared to mal3Δ strain (Figure 4F). Thus MT dynamics regulation by inositol pyrophosphates does not require the EB1 protein.
Table 2. Interphase MT dynamics in mal3Δ variant strains.
| strain | growth (nm/sec) | length (µm) |
| mal3Δ | 37±23 (n = 90) | 2.1±0.3 (n = 54) |
| mal3Δ asp1H397A | 54±35* ↑ (n = 91) | 2.7±0.6* ↑ (n = 53) |
Parameters of MT dynamics were measured for the indicated strains expressing nmt81::gfp-atb2+. Cells were grown in non-selective minimal medium. n, number of MTs measured; asterisks denote significance between mal3Δ and mal3Δ asp1H397A; ↑, parameter is increased compared to mal3Δ. Growth: * p<0.0005 for mal3Δ asp1H397A vs. mal3Δ (t-test). Length: * p<0.0005 for mal3Δ asp1H397A vs. mal3Δ (t-test).
Next, we analyzed Mal3-GFP particle movement in the various asp1 strains. The EB1 family decorates MTs and forms the comet-shaped structures at the MT plus-end characteristic of plus-end tracking proteins [59], [63]. The Mal3-GFP distribution on MTs was similar in all asp1 strains and was as described [63]. We determined the speed of the outmost outbound Mal3-GFP comets moving towards the cell end. As shown in Figure 4G movement of such Mal3-GFP particles in the wild-type and asp1H397A strain was similar, while asp1D333A Mal3-GFP comets were faster. The speed of movement of outmost outbound Mal3-GFP was directly correlated to the MT growth rate of the particular asp1 strain (Table 1 and Figure 4G). We also assayed movement of the kinesin Tea2-GFP in the 3 asp1 variant strains and found that the speed of Tea2-GFP signals at the end of MTs was comparable to Mal3-GFP comets (Figure S17).
Asp1 kinase function is required for growth zone selection in S. pombe
Interphase MTs in S. pombe control proper polarized growth by delivering the Tea1-4 landmark protein complex to potential sites of growth at the cell tip [10], [12], [64], [65]. Consequently, an aberrant interphase MT cytoskeleton can result in an altered positioning of the growth zones and in cells with a branched or bent morphology.
In wild-type fission yeast cells growth at a specific cell end is cell cycle controlled. After cytokinesis, cells will first grow in a monopolar manner selecting the old end (the end present before the previous cell division) as the first growth zone. The attainment of a critical cell size and completion of S-phase allow a switch to bipolar growth (NETO transition) at both cell ends in the G2 cell cycle phase [66], [67]. We had shown previously that Asp1 kinase function is essential for NETO, as 84% of asp1D333A cells grow exclusively monopolar on an agar surface using the old end as the site of growth [7]. However, the cylindrical cell shape was maintained in most asp1D333A cells demonstrating that the growth zone was still at the cell end. Abnormal growth zone positioning i.e. the selection of a growth zone not at the cell tip was observed in less than 5% of asp1D333A cells [7].
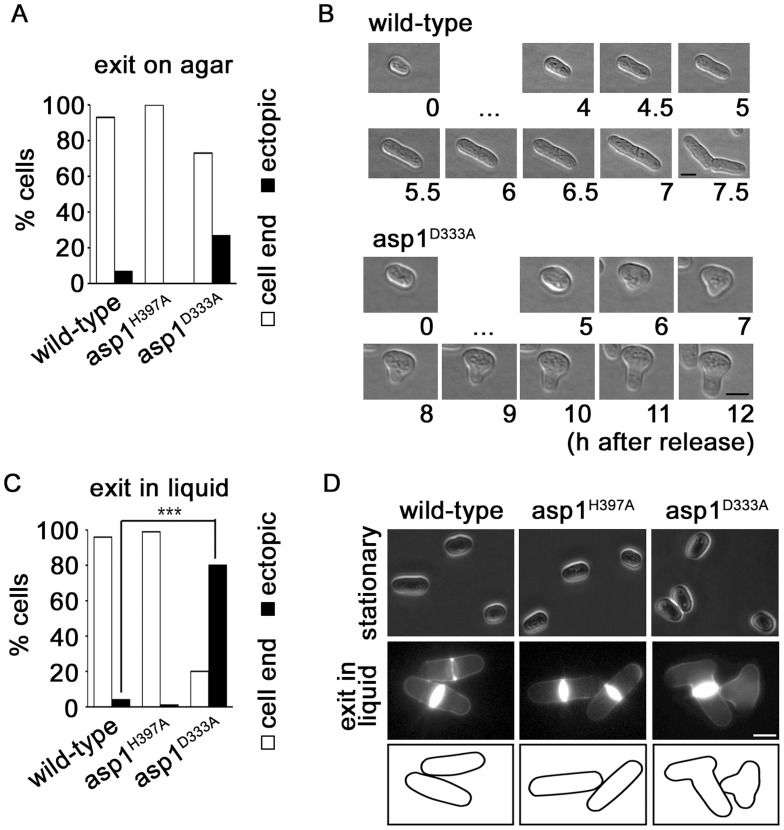
Next we asked, if proper polarized growth could also be re-established in asp1 mutants that were re-entering the vegetative cell cycle after nutrient starvation. Re-entry of cells into the cell cycle from G0 requires a de novo definition of the growth zones. [13], [16]. We thus examined the morphology of asp1+ and asp1 mutant cells after exit from stationary phase: on agar 93% of asp1+, 100% of asp1H397A but only 73% of asp1D333A cells grew as normal cylindrically shaped cells (Figure 5A). The remaining 27% of growing cells had an abnormal morphology, indicating that proper polarized growth was not re-established (Figure 5A–B). Incubation of stationary asp1D333A cells into fresh liquid medium massively aggravated the ectopic growth phenotype: under these conditions 80% of the cells had an aberrant, branched or lemon-shaped appearance indicating that Asp1 kinase activity was required for polarized growth and growth zone selection under these circumstances (Figure 5C–D). We also determined if cells when exiting from G0 state on solid medium showed the monopolar to bipolar growth pattern of exponentially growing cells. However we found a wide variety of growth patterns even for the asp1+ cells indicating that cells need to undergo a number of cell divisions before the normal growth pattern is stably re-established. It was thus not possible to determine if asp1D333A cells deviate from the norm.
Figure 5. Asp1 is required for growth zone selection.
(A) The indicated strains were released from stationary phase by streaking cells on solid YE5S at 25°C and analyzing the cell shape microscopically. Wild-type strain, n = 16; asp1H397A, n = 19; asp1D333A strain, n = 15. (B) Photomicrograph of a wild-type and asp1D333A cell that exits stationary phase on solid agar. Bars, 5 µm. […] denotes no change in cell morphology at these time points. (C) The indicated strains were released from stationary phase by inoculating an aliquot with YE5S liquid medium. Phenotype was scored after 7 hrs at 25°C. Wild-type strain, n = 142; asp1H397A, n = 110; asp1D333A strain, n = 122 ***p<0.001 asp1D333A compared to wild-type as determined using χ2-test. (D) Photomicrographs of cells in stationary phase (top panels) and after release into YE5S liquid medium (bottom panels). Cells were stained with Calcofluor white. Bar, 5 m.
A. nidulans Vip1-like protein is required for polarized growth
As the Vip1 family is conserved from yeast to man, we determined if Vip1 members also played a role in cell morphogenesis in other organisms. We therefore analyzed the function of Asp1-homologues in the filamentous ascomycete Aspergillus nidulans and the dimorphic basidiomycete Ustilago maydis. In both fungi, the importance of the MT cytoskeleton for fungal growth has been investigated extensively [18], [21]–[26], [31], [68], [69]. We decided to generate and characterize strains where the genes coding for the Asp1-homologues had been deleted as we have shown for S. pombe that the asp1Δ strain behaved identical to the asp1D333A strain under all conditions tested (Figure S2A–B; [7]).
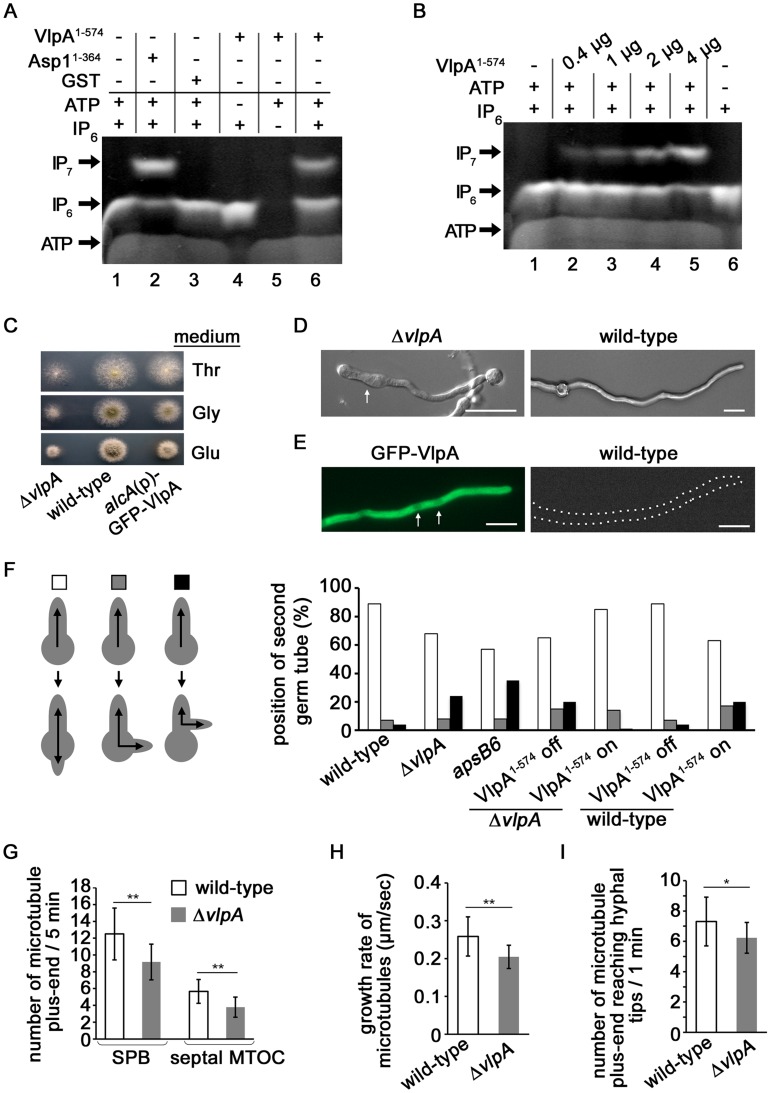
The A. nidulans Asp1 orthologue AN5797.2 has the characteristic Vip1 family dual domain structure (Figure S5) and was named vlpA (Vip1-like protein). To test if VlpA generates inositol pyrophosphates, bacterially expressed and purified GST-VlpA1-574, which contains the putative kinase domain was used in the in vitro kinase assay [54]. VlpA1-574 generated IP7 in an ATP dependent manner using IP6 as a substrate (Figure 6A, lanes 6, 4 and 5, respectively). This activity increased with increasing amounts of VlpA1-574 (Figure 6B).
Figure 6. Function of Vip1-like protein A. nidulans VlpA.
(A) VlpA has kinase activity. 2 µg GST-VlpA1-574 or GST-Asp11-364 (kinase domain only) were used in an enzymatic reaction as described [54]. −, component not present in assay; +, component present in assay. (B) Correlation between GST-VlpA1-574 protein amount/assay and the amount of IP7 generated. (C) Colonies of the vlpA-deletion strain ΔvlpA (left), wild-type (middle) and a strain in which GFP-VlpA was expressed under the control of alcA promoter (right). The strains were grown on minimal medium agar with threonine (upper), glycerol (middle) or glucose (bottom) for 2 days at 37°C. (D) The ΔvlpA strain (SCoS94, left) and wild-type strain (right) were grown in minimal medium with glucose overnight at 28°C. Some hyphae showed swelling in the ΔvlpA strain (arrow). Bars, 10 µm. (E) Growth conditions of the GFP-VlpA strain (Scos176) as in (D). GFP-VlpA expressed under native promoter localized predominantly in the cytoplasm with weak nuclear staining (arrows). The wild-type strain photographed under the same condition is shown on the right. Bars, 10 µm. (F) Left panel: position of the second germtube or branch: second germtube opposite first germtube (white), random position (grey) or second hypha branching out of the first hypha (black). Right panel: quantification of the number of germlings with a second germtube in the indicated strains. Spores were grown in minimal medium with glucose overnight. To induce expression of VlpA1-574, minimal medium with glycerol was used. N germlings/strain = 100. (G) Number of MT plus-ends appearing from the SPB or septal MTOC during 5 minutes in the wild-type (SSK92, white) and the vlpA-deletion strain (SDO2, grey). At SPB, wild-type: n = 20, ΔvlpA: n = 12, **p<0.01 as determined using t-test. At MTOC, wild-type: n = 10, ΔvlpA: n = 9, **p<0.01 as determined using t-test. (H) Growth rate of MT in the wild-type (SSK92) and the ΔvlpA strain (SDO2, grey). N = 10 cells for each of the strains, **p<0.01 as determined using t-test. (I) Number of MT plus-ends reaching hyphal tips during 1 minute in the wild-type (SSK92) and ΔvlpA strain (SDO2). N = 13 cells for each of the strains, *p<0.05 as determined using t-test.
We next deleted the endogenous vlpA gene and found that the vlpA-deletion strain (ΔvlpA) showed a growth delay and smaller colonies than the wild-type strain (approximate 50% diameter of colony on glucose medium) (Figure 6C). The majority of hyphae in the ΔvlpA strain displayed a normal morphology however swelling of hyphae was observed in some instances (Figure 6D, left). This phenotype could be caused by mis-positioning of the growth zone.
We constructed a strain expressing N-terminally GFP-tagged VlpA fusion protein under the control of the inducible alcA promoter instead of native VlpA. Under repressed conditions with glucose as carbon source, the strain exhibited a growth delay (Figure 6C, bottom right most panel). Under de-repressed conditions with glycerol or induced conditions with threonine, the slow growth phenotype was alleviated implying that GFP-VlpA can complement the growth defect of the vlpA deletion. We constructed a strain expressing GFP-VlpA under the native promoter and found that GFP-VlpA fluorescence was observed predominantly in the cytoplasm (Figure 6E, left).
Interestingly, the A. nidulans VlpA is needed for correct growth zone selection as it is required for the correct positioning of the second germtube. Once the first hypha reaches a determinate length, a second germ tube appears on the spore after the first septum at the base of the first hypha was formed [70]. This second germination site normally lies opposite of the first hypha (Figure 6F). In A. nidulans, MTs are formed from spindle pole bodies (SPB) and from septum-associated MT-organizing centers (septal-MTOCs) [71], [72]. MTs emanating from the septum of the first hypha grow towards the first germtube as well as into the direction of the spore. The MTs from the septa towards the spore are required for the positioning of the second germtube [70]. In the vlpA deletion strain, 24% of the spores did not produce a second germtube from the spore but produced a second hypha by branching out of the first hypha situated between septum and spore (Figure 6F). This aberrant phenotype was rescued by expressing a VlpA variant GFP-VlpA1-574 (contains the kinase domain) from the alcA promoter in the vlpA deletion strain (Figure 6F). Interestingly, expression of this VlpA variant in the wild-type strain altered growth zone selection (Figure 6F). These results demonstrate that (i) VlpA kinase activity is required for growth zone selection and (ii) physiological levels of VlpA kinase are required for proper growth zone selection. Thus, the Vip1-like proteins from A. nidulans and S. pombe are both required for growth zone selection.
A. nidulans VlpA modulates the MT cytoskeleton
A comparable phenotype of aberrant growth zone selection had been observed previously for the apsB6 mutant (Figure 6F) [70]. The apsB gene was identified by mutant screening. Anucleate primary sterigmata (aps) mutants are partially blocked in conidiation due to failure of the organized migration of nuclei into the conidiophore metulae. The mutants also show irregular distribution of nuclei in vegetative hyphae [73]. ApsB is a MTOC component that interacts with gamma-tubulin [74]. The apsB6 mutant shows an altered MT organization as it forms fewer MTs out of SPBs, compared to the wild-type and substantially fewer MTs from septa [72]. We therefore analyzed such parameters in the vlpA-deletion strain.
GFP tagged KipA, which is a kinesin localizing at growing MT plus-end, was used as plus-end marker to determine MT parameters [71]. Comparing wild-type to the vlpA-deletion strain during a five minute time period, we observed a reduction of newly emanating GFP-KipA signals in the vlpA-deletion strain at SPBs (27%) and at septal-MTOC (33%) (Figure 6G, Figure S6, Movie S1 and S2). The growth rate of the MT plus-ends was slightly reduced in the vlpA-deletion strain (21%) (Figure 6H). Pausing of MT plus-ends at hyphal tips was analyzed by using GFP-α-tubulin. Since the pausing time at hyphal tips was too short to determine if differences existed between the wild-type and the vlpA deletion strains, we scored the number of MT plus-ends reaching hyphal tips during a 1 minute time period. We counted fewer MT plus-ends in the vlpA deletion strain compared to the wild-type strain indicating that MT dynamics at the hyphal tip was altered in the absence of VlpA (Figure 6I).
The Asp1-like protein UmAsp1 is important for proliferation and polar growth in U. maydis
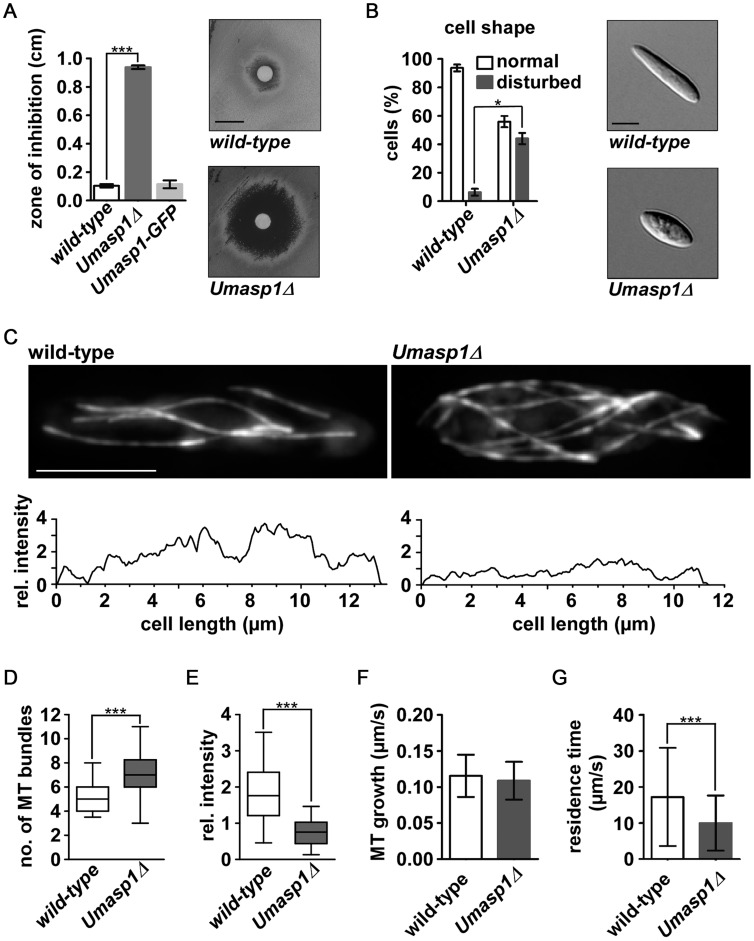
Finally, we studied the function of an Asp1 homologue in a distantly related fungus, the basidiomycete U. maydis. Sequence comparison revealed a protein designated UmAsp1(um06407 in MUMDB; MIPS Ustilago maydis database [75], with 922 amino acids and 49% sequence identity to S. pombe Asp1 over its entire length (Figure S5). To study its function we generated deletion strains in laboratory strain AB33. This strain is a derivative of wild-type strain FB2 that contains an active bW2/bE1 heterodimeric transcription factor under control of the nitrate-inducible promoter Pnar1. Thereby, b-dependent filamentation can be elicited by changing the nitrogen source in the medium [76]. We observed that a corresponding deletion strain of Umasp1Δ exhibited reduced proliferation during yeast-like growth in comparison to wild-type (Figure S7). Assaying TBZ sensitivity revealed that Umasp1Δ strains were hypersensitive to this MT inhibitor (Figure 7A; Figure S7B–C). For microscopic analysis we compared wild-type and Umasp1Δ strains expressing GFP-Tub1 (GFP fused to α-tubulin). The Umasp1Δ strain showed an increased number of cells that were clearly different from the cigar-shaped wild-type cells. Cells exhibited an increased diameter in the central region and/or were rounded-up at the poles (Figure 7B; Figure S8). Such cells were classified as having a disturbed shape and quantification revealed that about 40% of Umasp1Δ cells had an abnormal cell morphology (Figure 7B). Analysis of the MT cytoskeleton showed specific deviations from wild-type MTs. In wild-type cells 4 to 5 microtubular bundles are observed that are facing with their plus ends towards the poles [23], [25], [77]. We observed that the MT organization was altered in Umasp1Δ cells: a conservative quantification scoring only cells with drastic changes revealed that in comparison to the wild-type MT organization was altered (Figure S9). The most profound differences observed were (i) Umasp1Δ cells with large buds exhibited depolymerized MTs (Figure S9B, bottom panels); a phenotype rarely observed for wild-type cells. (ii) Umasp1Δ cells mostly with no bud or a small bud (early G2 phase) [23] had significantly more MT bundles. Instead of the 4 to 5 bundles present in wild-type cells, we observed 6 to 8 (Figure 7C–D; Movies S3 and S4). The fluorescence intensity of the GFP-Tub1 signal was drastically reduced in these bundles (Figure 7C, E) suggesting that loss of UmAsp1 results in an increased number of MT bundles with fewer MTs within such a bundle.
Figure 7. Loss of Umasp1 causes aberrant morphology of and altered MT organization in U. maydis yeast cells.
(A) Diagrammatic representation of growth inhibition test. The radius of growth zone inhibition was determined for the indicated strains on plates with 10 µl TBZ (concentration: 10 mg/ml) at the centre (experiments, n = 3. Error bars show SEM. ***, p<0.001; t-test). Representative examples are shown on the right and in Figure S7C (size bar, 1 cm). (B) Percentage of cells with disturbed cell shape. Bars show the mean of three independent experiments with n>100 cells (error bar shows SEM. *p<0.03, t-test). Representative examples are shown on the right (size bar, 5 µm). (C) Top: Deconvolved fluorescence photomicrographs depicting MT morphology (via expression of GFP-Tub1 (GFP fused to α-tubulin)) of wild-type and Umasp1Δ cells (size bar, 5 µm). Note that due to deconvolution fluorescence for the Umasp1Δ cell appears brighter. Bottom: Corresponding intensity profile showing longitudinal maximum intensity of background subtracted raw images. (D and E) Whisker diagrams showing the number of wild-type and Umasp1Δ MT bundles (D) and their relative intensity (E). Whiskers indicate 90%/10% percentiles (n>49 cells in (D) and n = 10 cells in (E); *** p<0.001 Mann-Whitney test for (D) and (E)). (F) MT growth parameters. Growth of MTs was determined by analyzing the comet-like movement of Pep1-GFP (error bars indicate standard deviation). Only MTs that grew >2 µm were analyzed (n = 225 and 125 for wild-type and Umasp1Δ, respectively). (G) The residence time of dynamic MTs was determined in GFP-Tub1 strains. n = 79 and 96 for wild-type and UMasp1Δ respectively. Error bar indicates standard deviation (unpaired t-test, *** p<0.001).
Studying the subset of intact MTs indicated that MT growth rate, which was analyzed by determining the velocity of the GFP-tagged U. maydis EB1 protein Pep1 [77], was not significantly different compared to wild-type (Figure 7F), but the residence time of MTs pausing at the cell end was significantly reduced (Figure 7G). In summary, UmAsp1 is needed for correct morphology and MT organization during proliferation of yeast-like cells.
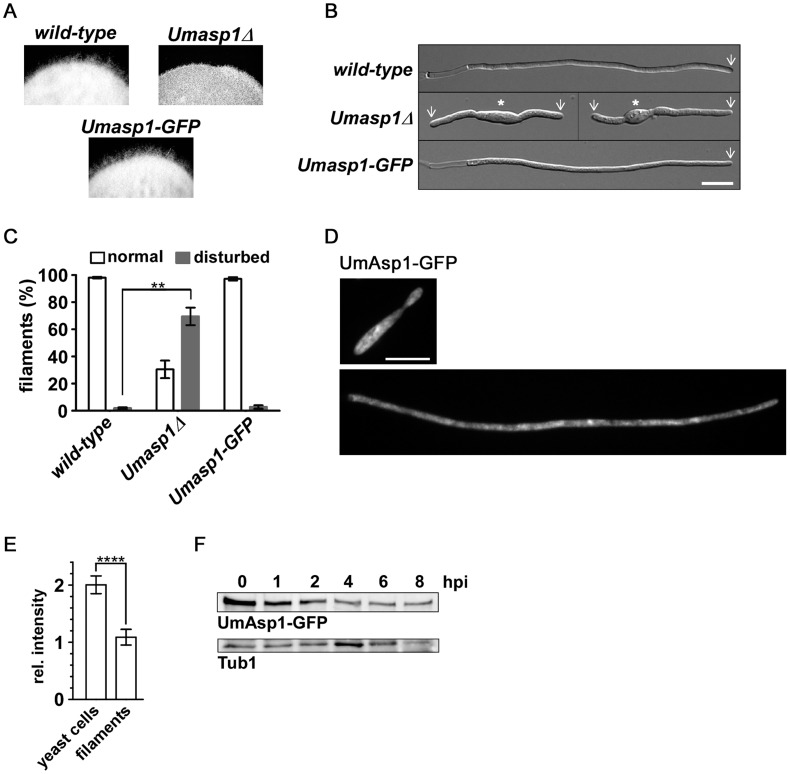
To investigate the function of UmAsp1 during hyphal growth, AB33 filamentation was induced on plates and in liquid medium. Wild-type forms a fuzzy colony indicative for efficient hyphal growth (Figure 8A, top, left panel). This was disturbed in Umasp1Δ strains (Figure 8A, top, right panel). Filaments were shorter, often bipolar and the amount of abnormal filaments was clearly increased in Umasp1Δ strains (Figure 8B–C, Figure S10). Thus, as in hyphae of A. nidulans, UmAsp1 is important for filamentous growth.
Figure 8. Loss of UmAsp1 causes defects in filamentous growth.
(A) Edges of colonies of the indicated AB33 derived strains grown on charcoal plates. Aerial hyphae are emanating from the colony. (B) Photomicrographs (DIC) of the indicated strains grown for 8 hrs under filament inducing conditions. Wild-type and UmAsp1-GFP filaments form characteristic empty sections at the basal pole. White arrow: growth zone; white star: yeast cell (bar, 10 µm). (C) Bar diagram showing percentage of filaments exhibiting normal or disturbed growth. Bars show the mean of three independent experiments with n>100 cells (error bar, SEM; ** p = 0.0108). (D) Examples of UmAsp1-GFP yeast cells and filaments (8 hours post induction) (bar, 10 µm). (E) Bar chart showing mean average fluorescence intensity of UmAsp1-GFP in yeast and hyphae (yeast, n = 10 cells and hyphae, n = 7 cells; see Figure S11 for example photomicrographs). Error bars indicate standard deviation (*** p<0.001 unpaired t-test). (F) Western blot analysis of protein extracts of strain AB33 Umasp1-GFP after induction (0–8 hrs) of filamentous growth. Tub1 served as a loading control (hpi, hours post induction).
To study the subcellular localization we generated strains expressing UmAsp1-GFP (C-terminal fusion to GFP). The resulting strain was phenotypically indistinguishable from wild-type (Figures 7A, 8A–C) demonstrating that the fusion protein is fully functional. Studying the subcellular localization in yeast or hyphal cells did not reveal any pronounced subcellular accumulation of the protein as has been shown for other Vip1-like proteins (Figure 8D). However, UmAsp1-GFP fluorescence was reduced in hyphae, suggesting that the protein amount decreases after filament induction (Figure 8E, Figure S11). Indeed, western blot analysis demonstrated that UmAsp1-GFP protein amounts decreased over time (Figure 8F). Thus, UmAsp1 protein amounts decrease and hence presumably intracellular inositol pyrophosphate levels appear to be down-regulated during the switch to hyphal growth.
Discussion
In this work we have defined the function of the C-terminal domain of the Vip1 family member Asp1 from S. pombe and have identified a new role for inositol pyrophosphates in fungal polarized growth and the modulation of MTs. In all three fungal model systems analyzed transport-based processes along the MT cytoskeleton are essential for proper polarized growth. However the long hyphal compartments of the filamentous fungi require a more sophisticated system of localized delivery [3], [18], [24]. Thus although Vip1-like proteins play a role in polarized growth in S. pombe, A. nidulans and U. maydis their specific roles are not expected to be identical.
Function of the Asp1 C-terminal histidine acid phosphatase domain
All Vip1 family members have a dual domain structure consisting of an N-terminal kinase domain and a C-terminal histidine acid phosphatase-like domain. Generation of inositol pyrophosphates has been shown for the budding yeast and human Vip1 family members [43]–[45], [48]. In this work we have extended the analysis to two further fungal Vip1-like proteins: the S. pombe Asp1 and the A. nidulans VlpA. Both proteins generated inositol pyrophosphates in vitro. The use of Asp1 and VlpA N-terminal- only-variants mapped the kinase activity to the N-terminal part of the respective protein.
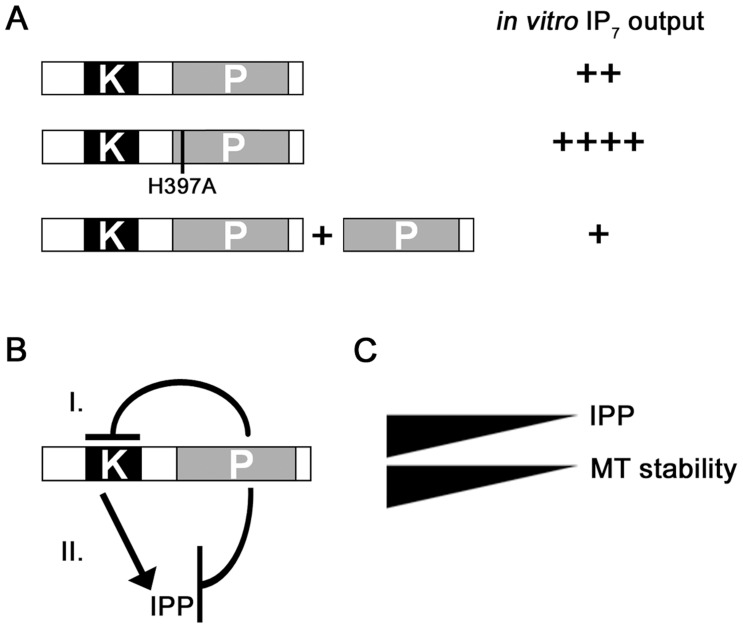
The precise function of the C-terminal phosphatase-like domain of Vip1-like proteins has been elusive. The histidine acid phosphatase signature motif is in principle present in Vip1-like proteins but the conserved “HD” motif has been replaced by H(I,V,A) [45]. A recent publication has shown that the phosphatase-like domains of the human Vip1 members are catalytically inactive. Instead the authors show that this domain plays a role in inositol lipid binding [48]. On the other hand, a comparison of the amounts of inositol pyrophosphates generated by human and the S. cerevisiae full-length Vip1 proteins versus N-terminal kinase-domain-only-variants, showed that the latter variants exhibited more specific activity [43], [45]. This implied a negative impact of the phosphatase-like domain on inositol pyrophosphate production. However it was unclear, if this effect was due to the large size differences between the full length and the kinase-domain-only-variants [45]. In this paper we demonstrate that the phosphatase-like domain has a regulatory function: (i) the Asp1H397A variant generated significantly more inositol pyrophosphates in vitro than the equally sized wild-type Asp1 protein (Figure 9A). The Km values for these two proteins were similar, but Vmax for the mutant Asp1 variant Asp1 H397A was higher. (ii) Addition of the phosphatase-only variant Asp1365-920 to an Asp1 protein containing in vitro kinase assay massively reduced the IP7 output (Figure 9A). However the presence of a mutated phosphatase variant, Asp1365-920 H397A in the assay did not have this effect (Figure S18). Thus our results suggest that the C-terminal phosphatase-like domain of Asp1 has enzymatic activity and its substrates are the inositol pyrophosphates produced by the N-terminal kinase domain of the protein (Figure 9B, model II). However as we have not formally proven that the C-terminal domain has phosphatase activity other modes of regulation are possible as shown in model I (Figure 9B).
Figure 9. Model for the regulation of Asp1 kinase function by the C-terminal phosphatase domain.
(A) In vitro IP7 output of wild-type Asp1 (top), Asp1H397A (middle) and Asp1 plus Asp1365-920 (phosphatase domain only). ++++ - +, high to low IP7 output. (B) Two possible modes of action are shown. (I) The phosphatase domain could modulate the function of the kinase domain directly leading to reduced inositol pyrophosphate generation. (II) The phosphatase domain has enzymatic activity using the inositol pyrophosphate generated by the kinase domain as a substrate. IPP = inositol pyrophosphate. (C) MT stability correlates directly with intracellular inositol pyrophosphate levels.
We have shown previously that specific extrinsic signals appear to up-regulate Asp1 kinase activity via the cAMP PKA pathway [7]. We speculate that such an up-regulation might occur by modification and result in down-regulation of the Asp1 C-terminal domain function. Such a scenario could also be envisaged for other external signal induced processes regulated by Vip1 family members, such as the antiviral response [49].
Inositol pyrophosphate signaling is an important modulator of fungal growth
The present work has defined a new role for inositol pyrophosphates generated by the Vip1 family: the modulation of fungal growth and the MT cytoskeleton. In S. pombe, interphase MT organization and MT dynamics were strongly altered in the asp1 mutant strains. In A. nidulans MT arrays from the SPB and the septal MTOC were affected in the vlpA deletion strain while in U. maydis loss of the Vip1-like protein resulted in increased TBZ sensitivity and an increase of cells with aberrant MT organization.
How then do inositol pyrophosphates modulate the MT cytoskeleton? In all systems tested to date and shown for U. maydis and A. nidulans in this work, Vip1 proteins are predominantly cytoplasmic without a specific subcellular localization [42], [48]. However as inositol pyrophosphates appear to modulate processes by binding to proteins or by pyrophosphorylation of proteins, direct association of Vip1 proteins with their targets might not be necessary. Our analysis in S. pombe demonstrated that in the absence of the +TIPs EB1 family member Mal3 the MT cytoskeleton can still be modulated by inositol pyrophosphates. Furthermore MT localization of EB1 proteins appeared unaffected in the S. pombe asp1D333A and the Umasp1Δ strain. As the EB1 protein family is at the center of the +TIP network of MT plus-ends and required for the recruitment of the majority of +TIPs [17], [59], we reason that such MT proteins are unlikely targets of inositol pyrophosphates. We have started to search for MT relevant inositol pyrophosphate targets by expressing either asp11-364 (kinase domain only) or asp1365-920 (phosphatase domain only) in various S. pombe mutants with an altered MT cytoskeleton. Our rationale is that the mutant phenotype of a strain with a deletion of a direct Vip1 target should not be affected by varying inositol pyrophosphate levels. We found that inositol pyrophosphates show a “genetic interaction” with the MT plus-end components that can associate with MTs independently of EB1 (our unpublished observations). However other MT structures might also be modulated by inositol pyrophosphates: MTs emanating from SPBs and septal MTOCs are reduced in the A. nidulans vlpA-deletion strain as has been shown for the apsB mutant strain [72]. ApsB is a conserved MTOC associated protein that interacts with γ-tubulin [74].
Of particular interest is the observed direct correlation between intracellular inositol pyrophosphate levels and the time that S. pombe MT plus-ends stay at the cell tip before a catastrophe event. Components of a fungal growth zone can regulate MT plus-end dynamics as has been shown for A. nidulans Tea1 family member TeaA, which negatively regulates the activity of the XMAP215 protein AlpA [19]. Thus it is feasible that Asp1 enzymatic activity regulates MT dynamics at the cell tip. Although immunofluorescence analysis of S. pombe Asp1-GFP did not show a specific cytoplasmic localization [42], localization of the human Vip1 member PPIP5K1 was slightly enhanced at the plasma membrane [48]. Plasma membrane targeting of PPIP5K1 in NIH3T3 cells was increased dramatically following PtdIns3 kinase activation [48].
Inositol pyrophosphates and growth zone selection
In fission yeast the switch from mono- to bipolar growth (NETO) is a complicated process that is regulated by a number of interwoven processes [10], [12]. These range from the correct positioning of landmark proteins by the MT cytoskeleton to the successful completion of S-phase and cytokinesis [64]–[66], [78]. We have shown previously that asp1D333A cells are able to correctly initiate growth at the old end after cytokinesis but cannot undergo NETO [7]. Correct selection of the first growth zone was also observed for the positioning of the first germtube of spores of an A. nidulans vlpA-deletion strain. However, similar to the S. pombe NETO event the positioning of the second growth zone (second germtube) was aberrant. Interestingly, plasmid-borne expression of A. nidulans VlpA1-574 (kinase domain) in a wild-type background also led to an alteration in the positioning of the second germ tube. We presume that VlpA1-574 expression increases intracellular inositol pyrophosphate levels and thus propose that fine-tuning of inositol pyrophosphate levels is required for the correct positioning of the second germ tube in A. nidulans.
In accordance with this hypothesis we observed that hyphal growth was also disturbed in U. maydis. Loss of UmAsp1 caused reduced and aberrant filamentous growth, including an increase of bipolar filaments. This is reminiscent of strains treated with MT-inhibitors or carrying mutations in MT-dependent motors such as kinesin-3 type Kin3, dynein Dyn1/2 or missing the RNA-binding protein Rrm4 involved in endosomal mRNA transport [35]–[37].
Interestingly, UmAsp1 levels decreased after the switch to hyphal growth indicating that alternative growth forms require a modulation of intracellular inositol pyrophosphate levels. Noteworthy, these filaments are arrested in the G2 cell cycle [79] suggesting a connection to cell cycle control.
A change in inositol pyrophosphate levels also regulates the environmentally controlled switch to an alternative growth form of S. pombe namely pseudohyphal invasive growth [7]. Here, Asp1 generated inositol pyrophosphates were essential for the switch to occur and increasing intracellular levels of these high energy molecules increased the cellular response. A similar scenario has been described recently for the regulation of the antiviral response by human Vip1 generated inositol pyrophosphates [49]. Ectopic expression of human Vip1 family members strongly increased the interferon response. Thus, modulation of the kinase activity of Vip1-like proteins might be a general mechanism of eukaryotic cells to react to extrinsic signals.
Materials and Methods
In vitro enzymatic activity of Vip1-like proteins
PCR-generated DNA fragments containing the S. pombe asp1+, asp1D333A, asp1H397A asp1364-920 ORFs, the S. cerevisiae VIP1-535 and the A. nidulans vlpA1-574 were cloned into E. coli expression vector pKM36 (a gift from Dr. K. Mölleken, Heinrich-Heine-Universität, Düsseldorf, Germany) to generate GST-tagged proteins. These proteins were expressed and purified from E.coli strain Rosetta (DEB) according to protocol (Sigma Aldrich). Protein concentration was determined using Bradford. Defined quantities of the Vip1-like proteins were used in an enzymatic reaction followed by PAGE analysis [54]. Intensity of IP7 bands was determined with ImageJ 1.44 (NIH). Determination of Km and Vmax: Enzymatic reactions with 2 µg of protein were carried out for 6 hrs using 0–300 µM IP6 substrate. The amount of IP7 generated per reaction was determined by quantifying the relevant IP7 band and converting this number using an IP6 calibration curve. IP6 was obtained from Sigma-Aldrich. Michaelis-Menten enzyme kinetics were calculated with GraphPad Prism6 (GraphPad Software, Inc.).
Strains and media
All strains used are listed in Table 3. S. pombe strains were grown and new strains were obtained as described [7]. A. nidulans was grown in supplemented minimal medium including 2% glucose, 2% glycerol or 2% threonine [80]. A. nidulans strain constructions were as described [81].To generate a N-terminal GFP fusion construct of VlpA a 900 bp fragment of vlpA (starting from ATG) was amplified from genomic A. nidulans DNA with appropriate primers. This AscI-PacI-digested PCR fragment was cloned into the corresponding sites of pCMB17apx (for N-terminal GFP fusion proteins of interest expressed under the control of alcA promoter, containing Neurospora crassa pyr4 as a selective marker) [82], generating pCoS105. The 1.5-kb promoter of vlpA was amplified from genomic DNA with appropriate primers and cloned into the corresponding sites of pCoS105, generating pCoS228. They were transformed into wild-type strain TN02A3. To express VlpA variant GFP-VlpA1-574 (contains the kinase domain) from the alcA promoter, the fragment of vlpA was amplified from genomic A. nidulans DNA with appropriate primers. This AscI-PacI-digested PCR fragment was cloned into the corresponding sites of pCMB17-pyroA (pyr-4 was replaced with pyroA in pCMB17apx), generating pCoS197, which was transformed into the wild-type strain TN02A3 and vlpA-deletion strain. Integration event was confirmed by PCR. vlpA was deleted via transformation of a deletion cassette (Program Project grant GM068087) into TN02A3 and the deletion confirmed by southern blotting. U. maydis strain constructions and growth of yeast like cells was performed according to published protocols [76]. Filamentous growth of AB33 and variants was induced by shifting 20 or 50 ml of exponentially growing cells (OD600 = 0.4–0.5) from complete medium (CM) to nitrate minimal medium each supplemented with 1% glucose. Cells were incubated at 28°C shaking at 200 rpm for 4 to 8 h prior to microscopy. For serial dilution patch tests, cells were pre-grown to OD600 = 0.5 before plating. For quantitative inhibition studies, cells were grown to OD600 = 0.5 and 300 µl were streaked out on a CM-plate. The filter paper present at the plate centre contained either 10 µl DMSO (solvent control) or 10 µl TBZ (10 mg/ml). After three days of growth at 28°C the radius of growth inhibition was measured.
Table 3. Strains used in this study.
| S. pombe | ||
| name | genotype | source |
| UFY1156 | h− asp1Δ::kanR his3-D1 ade6-M216 leu1-32 ura4-D18 | U. Fleig |
| UFY605 | h− his3-D1 ade6-M210 leu1-32 ura4-D18 | K. Gould |
| UFY1579 | h+ asp1H397A::kanR his3-D1 ade6-M210 leu1-32 ura4-D18 | U. Fleig |
| UFY1511 | h+ asp1D333A::kanR his3-D1 ade6-M210 leu1-32 ura4-D18 | U. Fleig |
| UFY857 | h− kanR::nmt81::gfp-atb2+ leu1-32 | T. Toda |
| UFY963 | h+ kanR::nmt81::gfp-atb2+ ade6-M216 his3D1 leu1-32 | This study |
| UFY1318 | h+ kanR::nmt81::gfp-atb2+ ade6-M210 his3D1 leu1-32 ura4-D18 | This study |
| UFY1763 | h+ asp1H397A::kanR kanR::nmt81::gfp-atb2+ leu1-32 ura4-D18 | This study |
| UFY1529 | h+ asp1D333A::kanR kanR::nmt81::gfp-atb2+ leu1-32 ura4-D18 | This study |
| UFY1407 | h+ asp1Δ::kanR kanR::nmt81::GFP-atb2+ leu1-32 his3-D1 ade6-M216 | This study |
| UFY135 | h+ mal3Δ::his3+ his3Δ ade6-M210 leu1-32 ura4-D18 | U. Fleig |
| UFY561 | h+ mal3Δ::ura4+ ade6-M210 his3Δ leu1-32 ura4-D18 | U. Fleig |
| UFY1641 | h− asp1H397A::kanR mal3Δ::his3+ his3− ade6-M210 leu1-32 ura4-D18 | U. Fleig |
| UFY1528 | h− asp1D333A::kanR mal3Δ::his3+ his3− ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1322 | h− asp1Δ::kanR mal3Δ::his3+ his3− ade6-M216 leu1-32 ura4-D18, | This study |
| UFY1729 | h− aps1Δ::his3+ his3-D1 ade6-M210 leu1-32 ura4-D18 | U. Fleig |
| UFY2164 | h+ aps1Δ::his3+ mal3Δ::ura4+ his3− ade6-M210 leu1-32 ura4-D18 | This study |
| UFY880 | h− mal3Δ::ura4+ kanR::nmt81::gfp-atb2+ his3Δ ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1762 | h− asp1H397A::kanR mal3Δ::ura4+ kanR::nmt81::gfp-atb2+ his3− ade6-M210 leu1-32 ura4-D18 | This study |
| UFY596 | h− mal3-pk-GFP::ura4+ ade6-M210 his3D1 leu1-32 ura4-D18 | H. Browning |
| UFY2015 | h− asp1H397A::kanR mal3-pk-GFP::ura4+ ade6-M210 his3D1 leu1-32 ura4-D18 | This study |
| UFY2014 | h+ asp1D333A::kanR mal3-pk-GFP::ura4+ ade6-M210 his3D1 leu1-32 ura4-D18 | This study |
| UFY1582 | h− asp1D333A H397A::kanR ade6-M210 leu1-32 ura4-D18 his3-D1 | This study |
Generation of asp1 variant containing plasmids and western blot analysis
asp1+, asp11-364 (plasmid p672), asp1H397A plasmids are derivatives of pJR2-3XL and have been described previously [7]. For the asp11-364+asp1365-920 containing plasmid, p672 was cut with SapI and a PCR generated DNA fragment containing the nmt1+ promoter followed by the DNA sequence encoding asp1365-920 inserted via homologous recombination in S. cerevisiae [83]. asp1R397A and asp1H807 were generated by directed mutagenesis using the QuikChangeII Site-Directed Mutagenesis Kit (Stragene) and after verification of sequence by sequence analysis cloned into pJR-3XL via S. cerevisiae homologous recombination. To determine expression of plasmid-borne asp1 variants, the appropriate asp1 containing DNA sequences were fused to gfp and expression of the fusion protein was determined by western blot analysis as has been described [7]. U. maydis Vlp1G expression was determined via western blot analysis as has been described [36].
Microscopy
For imaging of living S. pombe cells, cells were pre-grown in minimal medium at 25°C or 30°C and slides were prepared by mounting cells on agarose pads as described in [84]. Images were obtained at room temperature using a Zeiss Spinning Disc confocal microscope, equipped with a Yokogawa CSU-X1 unit and a MRm Camera. Slides were imaged using AxioVision software. Images shown are maximum intensity projections of 10–25 z-slices of 0.24–0.5 µm distance. For measurement of MT dynamics, strains expressing GFP-Atb2 [57] under control of the nmt81 promoter were pre-grown under promoter-derepressing conditions for at least 48 hrs. For technical reasons, we used the nmt81::gfp-atb2+ construct, as this facilitated the measurement of the sometimes faint MTs of the asp1D333A strain. Time-lapse images were acquired in 5–10 sec intervals.
For live-cell imaging of A.nidulans germlings and young hyphae, cells were grown on coverslips in 0.5 ml of Supplemented minimal media with 2% glycerol (de-repression of the alcA promoter, moderate induction). Cells were incubated at 30°C overnight/1 day. Coverslips were mounted on slide glass. Tempcontrol mini (Pepcon) was used for a constant temperature of the slide glass during microscopy. Images were captured using an Axiophot microscope using a Planapochromatic 63 times oil immersion objective lens, the Zeiss AxioCam MRM camera and the HBO103 mercury arc lamp (Osram) or HXP 120 (Zeiss, Jena, Germany). Images were collected and analyzed with the AxioVision system (Zeiss). Signal intensity was quantified with ImageJ software.
Live cell imaging of U. maydis was performed according to published protocols [36]. Microscope and camera were controlled by MetaMorph (Version 7.7.0.0, Molecular Devices, Seattle, IL, USA). The same software was used for measurements and image processing including the adjustment of brightness and contrast. MT bundles were visualized with a 63× Planapochromat (NA 1.4, Zeiss) or 100× α-Planapochromat (NA 1.46, Zeiss) in combination with a HXP lamp or laser illumination (488 nm), respectively. Z- stacks were composed of 38 planes with 270 nm spacing (63×) and 66 planes with 240 nm spacing (100×). Exposure time was 100 ms. Deconvolution was performed with Fiji. A theoretical PSF was determined with the diffraction PSF 3D plugin and images were generated using the Deconvolve 3D plugin [85], [86]. 3D movies were generated with MetaMorph. To determine the number of MT bundles z-stacks were collapsed to a maximum projection and after cytoplasmic background subtraction the number of bundles was determined. For determination of MT bundle intensity the maximum values of a longitudinal line scan (Fig. 7C) were plotted over distance. Each value from the x-axes was included in a whisker diagram (Fig. 7E) showing the median and range of fluorescent MT bundles (n = 10 cells for wild-type and Umasp1Δ, respectively). Fluorescence micrographs of Umasp1-GFP were acquired with 500 ms exposure time in a single plane. Before determining average cytoplasmic fluorescence images were background subtracted. For measurement of MT growth (Fig. 7F) strains expressing GFP-Tub1 were used. Z- stacks were composed three planes with 1 µm spacing (100× objective). Exposure time was 100 ms. For measurement of MT residence time (Fig. 7G) strains expressing Peb1-GFP were used. Z- stacks were composed of 5 planes with 800 nm spacing (100× objective). Exposure time was 100 ms. Statistical analysis was done with Prism5 (Graphpad).
Supporting Information
Asp1 converts IP6 to IP7 in vitro. (A) Left panel: S. cerevisiae Vip1 for which enzymatic activity had been demonstrated was used as a positive control for IP7 generation [43]. 1 µg bacterially expressed and purified GST-Vip11-535 (contains kinase domain) was used in an enzymatic reaction as described [54] followed by resolution of the products via PAGE and staining of the gel with Toluidine Blue. −, component not present in assay; +, component present in assay. Right panel: Asp1 generates IP7 from IP6 in an ATP-dependent reaction. 1 µg bacterially expressed and purified GST-Asp1 was used in the above mentioned in vitro assay. (B) Correlation between Asp1 protein input and the amount of IP7 generated. Left panel: Toluidine Blue stained PAGE showing IP7 produced by varying amounts of GST-Asp1 protein. Incubation time: 16 hrs. Right panel: Diagrammatic representation of the quantification of the IP7 bands shown in the left panel.
(TIF)
(A) asp1D333A, asp1Δ and asp1D333A, H397A strains show TBZ hypersensitivity. Serial dilution patch tests (105–101 cells) of the indicated strains on YE5S plates with (+) or (−) without TBZ. Plates were incubated for 5 days at 25°C. (B) asp1D333A and asp1Δ strains are sensitive to NaCl and caspofungin and resistant to treatment by the cell wall enzyme zymolyase. Serial dilution patch tests (105–101 cells) on YE5S plates with (+) or without (−) 50 mM NaCl or 1.5 µg/ml caspofungin, respectively. Plates were incubated for 4 days at 25°C. For zymolyase experiments cells were incubated with zymolyase and OD600 determined at the indicated time intervals. Reduction in OD600 is due to cell lysis.
(TIF)
Expression of plasmid-borne asp1 variants in the asp1Δ strain. (A) Diagrammatic representation of the S. pombe LEU2 plasmids used in B–C. P, nmt1+ promoter. (B) Western blot analysis of the asp1Δ strain expressing the indicated Asp1-GFP variants. Similar amounts of protein were resolved by SDS-PAGE and probed with an anti-GFP antibody or an anti-γ-tubulin antibody (loading control). (C) Quantification and diagrammatic representation of the Asp1-GFP signals obtained in (B).
(TIF)
(A) Serial dilution patch tests (105–101 cells) of the indicated strains grown on minimal medium without thiamine (promoter on conditions) for 5 or 4 days at 25°C or 30°C, respectively. Incubation on TBZ containing plates was for 9 days at 25°C. (B) Live cell images of the indicated strains expressing gfp-atb2+. Time between the images is 10 seconds. In each case the arrow indicates a short MT that polymerizes from the cell middle but is not oriented along the long axis of the cell. In the wild-type strain this MT reaches the cell cortex (80 seconds image), becomes deflected and continues to grow. In the asp1D333A strain, such a MT touches the cell cortex (100 second image) and then depolymerizes. Bars, 5 µm.
(TIF)
(A) Sequence comparison of the Vip1 family members from S. pombe, S. cerevisiae, A. nidulans (AN5797.2) and U. maydis (UM06407.1). Multiple sequence alignment was performed with MultAlin using BLOSUM62 matrix [87]. (B) The respective kinase and phosphatase domains are indicated in green and grey, respectively.
(TIF)
GFP-KipA, a marker of growing MT plus-ends, in the wild-type (SSK92) and the vlpA-deletion strain (SDO2). (A) Diagrammatic representation of the components shown in (B) and (C). (B) and (C) We compared newly emanating GFP-KipA signals in the wild-type (B) and the ΔvlpA strain (C) during a 5 minute time period at SPBs (asterisks) and at septal-MTOC (white arrows). Bar, 10 µm. Kymographs at septa during a 5 minute time period are shown. GFP signals coming from the septum are shown by blue arrows. GFP signals arriving at the septum are shown by red arrows. Bar, 1 µm.
(TIF)
Loss of UmAsp1 causes defects in proliferation and leads to TBZ sensitivity. (A) Growth of the indicated yeast strains over time. (B) Serial dilution patch test (107 to 105 cells) of the indicated strains grown with/without 10 µg/ml TBZ. (C) Filter paper with/without 10 µg/ml TBZ was placed on a lawn of U. maydis cells (strains indicated above). The region indicated by a white bar was measured to determine the zone of inhibition (radius in cm) given in Figure 7A. Note, plates of Umasp1Δ cells appear slightly darker due to secretion of an unknown pigment.
(TIF)
Loss of UmAsp1 causes alterations in cell morphology. Representative DIC images of wild-type (A) and Umasp1Δ (B) cells, quantified in Figure 7B are shown (Bars, 10 µm).
(TIF)
Loss of UmAsp1 causes defects in MT organization. Representative fluorescence images of wild-type (A) and Umasp1Δ (B) cells are shown. (C) The indicated MT categories were determined in wild-type and Umasp1Δ strains. Bars show the mean of three independent experiments with n>100 cells (error bars show SEM, p<0.0001; two-way ANOVA test).
(TIF)
Loss of UmAsp1 causes defects in filamentous growth. Representative DIC images of wild-type (A) and Umasp1Δ (B) hyphae 8 hours after filament inducing conditions (Bars, 10 µm). Quantification is shown in Figure 8C.
(TIF)
UmAsp1-GFP signal decreases during switch to filamentous growth. Fluorescence micrographs of mixed cultures expressing either UmAsp1-GFP (*) or Rrm4-mCherry (#) [35] are shown. Micrographs detecting either green or red fluorescence were taken subsequently from the same region of interest: (A) yeast, (B) filaments. Thereby, the degree of green auto-fluorescence (seen in the Rrm4-mCherry control) can be judged.
(TIF)
(A) Time dependent generation of IP7 by GST-Asp1 variants. 4 µg of the indicated proteins were used in an ATP-dependent enzymatic reaction and the resulting inositol pyrophosphates were resolved on a 35,5% PAGE and stained with Toluidine Blue. −, component not present; +, component present. (B) Quantification and diagrammatic representation of the IP7 bands obtained in the assay shown in (A).
(TIF)
IP6 amounts in the presence (+) or absence (−) of 9 µg Asp1365-920. Assay conditions and detection of IP6 were as described for the in vitro kinase assay.
(TIF)
Western blot analysis of the asp1Δ strain expressing the indicated Asp1-GFP (arrow shows full length fusion protein) variants. Similar amounts of protein were resolved by SDS-PAGE and probed with an anti-GFP antibody or an anti-γ-tubulin antibody (left and right panels, respectively).
(TIF)
Percentage of MTs polymerizing towards the lateral cortex (black bars) or towards a cell end (white bars). Wild-type: n = 77, asp1H397A: n = 73, asp1D333A n = 83.
(TIF)
Diagrammatic representation of the number of interphase MTs in the indicated strains (mal3Δ strain, n = 95; mal3Δ asp1H397A strain, n = 99).
(TIF)
Movement of outmost outbound Tea2-GFP comets (see diagram). Speed of comets (nm/sec): wild-type, 60±30, n = 89; asp1H397A, 60±26,7, n = 64; asp1D333A, 90±43,3, n = 71. * p<0.0005 for asp1D333A vs. wild-type (Welch-test).
(TIF)
Generation of IP7 by GST-Asp1 with varying amounts (2,4,8 µg) of Asp1365-920 H397A. Enzymatic reaction was carried out as described in Figure 1C. −, component not present; +, component present.
(TIF)
GFP-KipA, a marker of growing MT plus-ends in the wild-type strain (SSK92). 2 seconds intervals, total 5 minutes. Scale bar, 10 µm.
(AVI)
GFP-KipA in the vlpA-deletion strain (SDO2). 2 seconds intervals, total 5 minutes. Scale bar, 10 µm.
(AVI)
3D reconstruction of a wild-type cell expressing GFP-Tub1. The underlying z-stack is depicted in Figure 7. Size of angle images of the z-stack was doubled and pixels resampled. Ratio of xy-distance and xz-distance was chosen 1∶1 to obtain cubic voxels. Movie comprises 14 frames in 12 seconds.
(MOV)
Acknowledgments
We thank Kathleen Gould (Vanderbilt University, Nashville, USA) for insightful comments on the manuscript and Kathleen Gould, Takashi Toda (Cancer Research UK) and Katja Mölleken (Heinrich-Heine-University, Düsseldorf) for reagents. We are grateful to Marina Pascual Ortiz for help with Figures 2B, 4A and S2 and Natascha Künzel for help with Figures 4B and S2 (Heinrich-Heine-University, Düsseldorf). We thank the Center for Advanced Imaging (CAi) at the Heinrich-Heine-University, Düsseldorf.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (http://www.dfg.de/): project SFB590 (UF) and project FOR1334 (UF, MF, RF, NT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Piel M, Tran PT (2009) Cell shape and cell division in fission yeast. Curr Biol 19: R823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin SG, Arkowitz RA (2014) Cell polarization in budding and fission yeasts. FEMS Microbiol Rev 38: 228–253. [DOI] [PubMed] [Google Scholar]
- 3. Fischer R, Zekert N, Takeshita N (2008) Polarized growth in fungi–interplay between the cytoskeleton, positional markers and membrane domains. Mol Microbiol 68: 813–826. [DOI] [PubMed] [Google Scholar]
- 4. Vollmeister E, Schipper K, Baumann S, Haag C, Pohlmann T, et al. (2012) Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol Rev 36: 59–77. [DOI] [PubMed] [Google Scholar]
- 5. Gancedo JM (2001) Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol Rev 25: 107–123. [DOI] [PubMed] [Google Scholar]
- 6. Amoah-Buahin E, Bone N, Armstrong J (2005) Hyphal Growth in the Fission Yeast Schizosaccharomyces pombe. Eukaryot Cell 4: 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pöhlmann J, Fleig U (2010) Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol Cell Biol 30: 4535–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lichius A, Berepiki A, Read ND (2011) Form follows function – the versatile fungal cytoskeleton. Fungal Biol 115: 518–540. [DOI] [PubMed] [Google Scholar]
- 9. Lo Presti L, Martin SG (2011) Shaping fission yeast cells by rerouting actin-based transport on microtubules. Curr Biol 21: 2064–2069. [DOI] [PubMed] [Google Scholar]
- 10. Huisman SM, Brunner D (2011) Cell polarity in fission yeast: a matter of confining, positioning, and switching growth zones. Semin Cell Dev Biol 22: 799–805. [DOI] [PubMed] [Google Scholar]
- 11. Das M, Drake T, Wiley DJ, Buchwald P, Vavylonis D, et al. (2012) Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth. Science 337: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin SG (2009) Microtubule-dependent cell morphogenesis in the fission yeast. Trends Cell Biol 19: 447–454. [DOI] [PubMed] [Google Scholar]
- 13. Browning H, Hayles J, Mata J, Aveline L, Nurse P, et al. (2000) Tea2p Is a Kinesin-like Protein Required to Generate Polarized Growth in Fission Yeast. J Cell Biol 151: 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mata J, Nurse P (1997) tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89: 939–949. [DOI] [PubMed] [Google Scholar]
- 15. Drummond DR, Cross RA (2000) Dynamics of interphase microtubules in Schizosaccharomyces pombe. Current Biology 10: 766–775. [DOI] [PubMed] [Google Scholar]
- 16. Snaith HA, Sawin KE (2003) Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423: 647–651. [DOI] [PubMed] [Google Scholar]
- 17. Akhmanova A, Steinmetz MO (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9: 309–322. [DOI] [PubMed] [Google Scholar]
- 18. Takeshita N, Higashitsuji Y, Konzack S, Fischer R (2008) Apical sterol-rich membranes are essential for localizing cell end markers that determine growth directionality in the filamentous fungus Aspergillus nidulans. Mol Biol Cell 19: 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takeshita N, Mania D, Herrero S, Ishitsuka Y, Nienhaus GU, et al. (2013) The cell-end marker TeaA and the microtubule polymerase AlpA contribute to microtubule guidance at the hyphal tip cortex of Aspergillus nidulans to provide polarity maintenance. J Cell Sci 126: 5400–5411. [DOI] [PubMed] [Google Scholar]
- 20. Egan MJ, McClintock MA, Reck-Peterson SL (2012) Microtubule-based transport in filamentous fungi. Curr Opin Microbiol 15: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horio T, Oakley BR (2005) The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol Biol Cell 16: 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egan MJ, Tan K, Reck-Peterson SL (2012) Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol 197: 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steinberg G, Wedlich-Soldner R, Brill M, Schulz I (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci 114: 609–622. [DOI] [PubMed] [Google Scholar]
- 24. Steinberg G (2014) Endocytosis and early endosome motility in filamentous fungi. Curr Opin Microbiol 20C: 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banuett F, Herskowitz I (2002) Bud morphogenesis and the actin and microtubule cytoskeletons during budding in the corn smut fungus, Ustilago maydis . Fungal Genet Biol 37: 149–170. [DOI] [PubMed] [Google Scholar]
- 26. Fuchs U, Manns I, Steinberg G (2005) Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol Biol Cell 16: 2746–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Penalva MA (2009) Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic 10: 57–75. [DOI] [PubMed] [Google Scholar]
- 28. Göhre V, Vollmeister E, Bölker M, Feldbrügge M (2012) Microtubule-dependent membrane dynamics of Ustilago maydis: trafficking and function of Rab5a-positive endosomes. Comm Integra Biol 5: 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schuster M, Kilaru S, Fink G, Collemare J, Roger Y, et al. (2011) Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol Biol Cell 22: 3645–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wedlich-Söldner R, Straube A, Friedrich MW, Steinberg G (2002) A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis . EMBO J 21: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wedlich-Söldner R, Bolker M, Kahmann R, Steinberg G (2000) A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. Embo J 19: 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lenz JH, Schuchardt I, Straube A, Steinberg G (2006) A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J 25: 2275–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koepke J, Kaffarnik F, Haag C, Zarnack K, Luscombe NM, et al. (2011) The RNA-binding protein Rrm4 is essential for efficient secretion of endochitinase Cts1. Mol Cell Proteom 10 M111.011213 011211-011215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schuchardt I, Assmann D, Thines E, Schuberth C, Steinberg G (2005) Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis . Mol Biol Cell 16: 5191–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baumann S, König J, Koepke J, Feldbrügge M (2014) Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep 15: 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrügge M (2012) Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci 125: 2740–2752. [DOI] [PubMed] [Google Scholar]
- 37. Higuchi Y, Ashwin P, Roger Y, Steinberg G (2014) Early endosome motility spatially organizes polysome distribution. J Cell Biol 204: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson MS, Livermore TM, Saiardi A (2013) Inositol pyrophosphates: between signalling and metabolism. Biochem J 452: 369–379. [DOI] [PubMed] [Google Scholar]
- 39. Tsui MM, York JD (2010) Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv Enzyme Regul 50: 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shears SB, Weaver JD, Wang H (2013) Structural insight into inositol pyrophosphate turnover. Adv Biol Regul 53: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wundenberg T, Mayr GW (2012) Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol Chem 393: 979–998. [DOI] [PubMed] [Google Scholar]
- 42. Feoktistova A, McCollum D, Ohi R, Gould KL (1999) Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 152: 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, et al. (2007) A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316: 106–109. [DOI] [PubMed] [Google Scholar]
- 44. Choi JH, Williams J, Cho J, Falck JR, Shears SB (2007) Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem 282: 30763–30775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fridy PC, Otto JC, Dollins DE, York JD (2007) Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem 282: 30754–30762. [DOI] [PubMed] [Google Scholar]
- 46. Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, et al. (2009) Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J Biol Chem 284: 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang H, Falck JR, Hall TM, Shears SB (2011) Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol 8: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gokhale NA, Zaremba A, Shears SB (2011) Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem J 434: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pulloor NK, Nair S, Kostic AD, Bist P, Weaver JD, et al. (2014) Human genome-wide RNAi screen identifies an essential role for inositol pyrophosphates in Type-I interferon response. PLoS Pathog 10: e1003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee YS, Mulugu S, York JD, O'Shea EK (2007) Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH (2004) Phosphorylation of proteins by inositol pyrophosphates. Science 306: 2101–2105. [DOI] [PubMed] [Google Scholar]
- 52. Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, et al. (2010) Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 143: 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, et al. (2003) Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell 114: 559–572. [DOI] [PubMed] [Google Scholar]
- 54. Loss O, Azevedo C, Szijgyarto Z, Bosch D, Saiardi A (2011) Preparation of quality inositol pyrophosphates. J Vis Exp e3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130. [DOI] [PubMed] [Google Scholar]
- 56. Beinhauer JD, Hagan IM, Hegemann JH, Fleig U (1997) Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol 139: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garcia MA, Vardy L, Koonrugsa N, Toda T (2001) Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. Embo J 20: 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sawin KE, Tran PT (2006) Cytoplasmic microtubule organization in fission yeast. Yeast 23: 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kumar P, Wittmann T (2012) +TIPs: SxIPping along microtubule ends. Trends Cell Biol 22: 418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ingram SW, Safrany ST, Barnes LD (2003) Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene, and effects on growth rate, morphology and intracellular diadenosine 5′,5″′-P1,P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem J 369: 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, et al. (1998) A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. Embo J 17: 6599–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Katsuki M, Drummond DR, Osei M, Cross RA (2009) Mal3 masks catastrophe events in Schizosaccharomyces pombe microtubules by inhibiting shrinkage and promoting rescue. J Biol Chem 284: 29246–29250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Busch KE, Brunner D (2004) The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr Biol 14: 548–559. [DOI] [PubMed] [Google Scholar]
- 64. Sawin KE, Nurse P (1998) Regulation of cell polarity by microtubules in fission yeast. J Cell Biol 142: 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Verde F, Mata J, Nurse P (1995) Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol 131: 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kume K, Koyano T, Kanai M, Toda T, Hirata D (2011) Calcineurin ensures a link between the DNA replication checkpoint and microtubule-dependent polarized growth. Nat Cell Biol 13: 234–242. [DOI] [PubMed] [Google Scholar]
- 67. Mitchison JM, Nurse P (1985) Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci 75: 357–376. [DOI] [PubMed] [Google Scholar]
- 68. Oakley BR (2004) Tubulins in Aspergillus nidulans. Fungal Genet Biol 41: 420–427. [DOI] [PubMed] [Google Scholar]
- 69. Takeshita N, Manck R, Grun N, de Vega SH, Fischer R (2014) Interdependence of the actin and the microtubule cytoskeleton during fungal growth. Curr Opin Microbiol 20C: 34–41. [DOI] [PubMed] [Google Scholar]
- 70. Takeshita N, Fischer R (2011) On the role of microtubules, cell end markers, and septal microtubule organizing centres on site selection for polar growth in Aspergillus nidulans. Fungal Biol 115: 506–517. [DOI] [PubMed] [Google Scholar]
- 71. Konzack S, Rischitor PE, Enke C, Fischer R (2005) The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol Biol Cell 16: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Veith D, Scherr N, Efimov VP, Fischer R (2005) Role of the spindle-pole-body protein ApsB and the cortex protein ApsA in microtubule organization and nuclear migration in Aspergillus nidulans. J Cell Sci 118: 3705–3716. [DOI] [PubMed] [Google Scholar]
- 73. Clutterbuck AJ (1994) Mutants of Aspergillus nidulans deficient in nuclear migration during hyphal growth and conidiation. Microbiology 140 (Pt 5): 1169–1174. [DOI] [PubMed] [Google Scholar]
- 74. Zekert N, Veith D, Fischer R (2010) Interaction of the Aspergillus nidulans microtubule-organizing center (MTOC) component ApsB with gamma-tubulin and evidence for a role of a subclass of peroxisomes in the formation of septal MTOCs. Eukaryot Cell 9: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kämper J, Kahmann R, Bölker M, Ma LJ, Brefort T, et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature 444: 97–101. [DOI] [PubMed] [Google Scholar]
- 76. Brachmann A, Weinzierl G, Kämper J, Kahmann R (2001) Identification of genes in the bW/bE regulatory cascade in Ustilago maydis . Mol Microbiol 42: 1047–1063. [DOI] [PubMed] [Google Scholar]
- 77. Straube A, Brill M, Oakley BR, Horio T, Steinberg G (2003) Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis . Mol Biol Cell 14: 642–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thadani R, Huang D, Oliferenko S (2011) Robust polarity specification operates above a threshold of microtubule dynamicity. Cytoskeleton (Hoboken) 68: 290–299. [DOI] [PubMed] [Google Scholar]
- 79. Mielnichuk N, Sgarlata C, Perez-Martin J (2009) A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J Cell Sci 122: 4130–4140. [DOI] [PubMed] [Google Scholar]
- 80. Hill TW, Kafer E (2001) Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genetics Newsletter 48: 20–21. [Google Scholar]
- 81. Yelton MM, Hamer JE, Timberlake WE (1984) Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A 81: 1470–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Efimov VP, Zhang J, Xiang X (2006) CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans. Mol Biol Cell 17: 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jakopec V, Walla E, Fleig U (2011) Versatile use of Schizosaccharomyces pombe plasmids in Saccharomyces cerevisiae. FEMS Yeast Res 11: 653–655. [DOI] [PubMed] [Google Scholar]
- 84. Tran PT, Paoletti A, Chang F (2004) Imaging green fluorescent protein fusions in living fission yeast cells. Methods 33: 220–225. [DOI] [PubMed] [Google Scholar]
- 85. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, et al. (2006) A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Asp1 converts IP6 to IP7 in vitro. (A) Left panel: S. cerevisiae Vip1 for which enzymatic activity had been demonstrated was used as a positive control for IP7 generation [43]. 1 µg bacterially expressed and purified GST-Vip11-535 (contains kinase domain) was used in an enzymatic reaction as described [54] followed by resolution of the products via PAGE and staining of the gel with Toluidine Blue. −, component not present in assay; +, component present in assay. Right panel: Asp1 generates IP7 from IP6 in an ATP-dependent reaction. 1 µg bacterially expressed and purified GST-Asp1 was used in the above mentioned in vitro assay. (B) Correlation between Asp1 protein input and the amount of IP7 generated. Left panel: Toluidine Blue stained PAGE showing IP7 produced by varying amounts of GST-Asp1 protein. Incubation time: 16 hrs. Right panel: Diagrammatic representation of the quantification of the IP7 bands shown in the left panel.
(TIF)
(A) asp1D333A, asp1Δ and asp1D333A, H397A strains show TBZ hypersensitivity. Serial dilution patch tests (105–101 cells) of the indicated strains on YE5S plates with (+) or (−) without TBZ. Plates were incubated for 5 days at 25°C. (B) asp1D333A and asp1Δ strains are sensitive to NaCl and caspofungin and resistant to treatment by the cell wall enzyme zymolyase. Serial dilution patch tests (105–101 cells) on YE5S plates with (+) or without (−) 50 mM NaCl or 1.5 µg/ml caspofungin, respectively. Plates were incubated for 4 days at 25°C. For zymolyase experiments cells were incubated with zymolyase and OD600 determined at the indicated time intervals. Reduction in OD600 is due to cell lysis.
(TIF)
Expression of plasmid-borne asp1 variants in the asp1Δ strain. (A) Diagrammatic representation of the S. pombe LEU2 plasmids used in B–C. P, nmt1+ promoter. (B) Western blot analysis of the asp1Δ strain expressing the indicated Asp1-GFP variants. Similar amounts of protein were resolved by SDS-PAGE and probed with an anti-GFP antibody or an anti-γ-tubulin antibody (loading control). (C) Quantification and diagrammatic representation of the Asp1-GFP signals obtained in (B).
(TIF)
(A) Serial dilution patch tests (105–101 cells) of the indicated strains grown on minimal medium without thiamine (promoter on conditions) for 5 or 4 days at 25°C or 30°C, respectively. Incubation on TBZ containing plates was for 9 days at 25°C. (B) Live cell images of the indicated strains expressing gfp-atb2+. Time between the images is 10 seconds. In each case the arrow indicates a short MT that polymerizes from the cell middle but is not oriented along the long axis of the cell. In the wild-type strain this MT reaches the cell cortex (80 seconds image), becomes deflected and continues to grow. In the asp1D333A strain, such a MT touches the cell cortex (100 second image) and then depolymerizes. Bars, 5 µm.
(TIF)
(A) Sequence comparison of the Vip1 family members from S. pombe, S. cerevisiae, A. nidulans (AN5797.2) and U. maydis (UM06407.1). Multiple sequence alignment was performed with MultAlin using BLOSUM62 matrix [87]. (B) The respective kinase and phosphatase domains are indicated in green and grey, respectively.
(TIF)
GFP-KipA, a marker of growing MT plus-ends, in the wild-type (SSK92) and the vlpA-deletion strain (SDO2). (A) Diagrammatic representation of the components shown in (B) and (C). (B) and (C) We compared newly emanating GFP-KipA signals in the wild-type (B) and the ΔvlpA strain (C) during a 5 minute time period at SPBs (asterisks) and at septal-MTOC (white arrows). Bar, 10 µm. Kymographs at septa during a 5 minute time period are shown. GFP signals coming from the septum are shown by blue arrows. GFP signals arriving at the septum are shown by red arrows. Bar, 1 µm.
(TIF)
Loss of UmAsp1 causes defects in proliferation and leads to TBZ sensitivity. (A) Growth of the indicated yeast strains over time. (B) Serial dilution patch test (107 to 105 cells) of the indicated strains grown with/without 10 µg/ml TBZ. (C) Filter paper with/without 10 µg/ml TBZ was placed on a lawn of U. maydis cells (strains indicated above). The region indicated by a white bar was measured to determine the zone of inhibition (radius in cm) given in Figure 7A. Note, plates of Umasp1Δ cells appear slightly darker due to secretion of an unknown pigment.
(TIF)
Loss of UmAsp1 causes alterations in cell morphology. Representative DIC images of wild-type (A) and Umasp1Δ (B) cells, quantified in Figure 7B are shown (Bars, 10 µm).
(TIF)
Loss of UmAsp1 causes defects in MT organization. Representative fluorescence images of wild-type (A) and Umasp1Δ (B) cells are shown. (C) The indicated MT categories were determined in wild-type and Umasp1Δ strains. Bars show the mean of three independent experiments with n>100 cells (error bars show SEM, p<0.0001; two-way ANOVA test).
(TIF)
Loss of UmAsp1 causes defects in filamentous growth. Representative DIC images of wild-type (A) and Umasp1Δ (B) hyphae 8 hours after filament inducing conditions (Bars, 10 µm). Quantification is shown in Figure 8C.
(TIF)
UmAsp1-GFP signal decreases during switch to filamentous growth. Fluorescence micrographs of mixed cultures expressing either UmAsp1-GFP (*) or Rrm4-mCherry (#) [35] are shown. Micrographs detecting either green or red fluorescence were taken subsequently from the same region of interest: (A) yeast, (B) filaments. Thereby, the degree of green auto-fluorescence (seen in the Rrm4-mCherry control) can be judged.
(TIF)
(A) Time dependent generation of IP7 by GST-Asp1 variants. 4 µg of the indicated proteins were used in an ATP-dependent enzymatic reaction and the resulting inositol pyrophosphates were resolved on a 35,5% PAGE and stained with Toluidine Blue. −, component not present; +, component present. (B) Quantification and diagrammatic representation of the IP7 bands obtained in the assay shown in (A).
(TIF)
IP6 amounts in the presence (+) or absence (−) of 9 µg Asp1365-920. Assay conditions and detection of IP6 were as described for the in vitro kinase assay.
(TIF)
Western blot analysis of the asp1Δ strain expressing the indicated Asp1-GFP (arrow shows full length fusion protein) variants. Similar amounts of protein were resolved by SDS-PAGE and probed with an anti-GFP antibody or an anti-γ-tubulin antibody (left and right panels, respectively).
(TIF)
Percentage of MTs polymerizing towards the lateral cortex (black bars) or towards a cell end (white bars). Wild-type: n = 77, asp1H397A: n = 73, asp1D333A n = 83.
(TIF)
Diagrammatic representation of the number of interphase MTs in the indicated strains (mal3Δ strain, n = 95; mal3Δ asp1H397A strain, n = 99).
(TIF)
Movement of outmost outbound Tea2-GFP comets (see diagram). Speed of comets (nm/sec): wild-type, 60±30, n = 89; asp1H397A, 60±26,7, n = 64; asp1D333A, 90±43,3, n = 71. * p<0.0005 for asp1D333A vs. wild-type (Welch-test).
(TIF)
Generation of IP7 by GST-Asp1 with varying amounts (2,4,8 µg) of Asp1365-920 H397A. Enzymatic reaction was carried out as described in Figure 1C. −, component not present; +, component present.
(TIF)
GFP-KipA, a marker of growing MT plus-ends in the wild-type strain (SSK92). 2 seconds intervals, total 5 minutes. Scale bar, 10 µm.
(AVI)
GFP-KipA in the vlpA-deletion strain (SDO2). 2 seconds intervals, total 5 minutes. Scale bar, 10 µm.
(AVI)
3D reconstruction of a wild-type cell expressing GFP-Tub1. The underlying z-stack is depicted in Figure 7. Size of angle images of the z-stack was doubled and pixels resampled. Ratio of xy-distance and xz-distance was chosen 1∶1 to obtain cubic voxels. Movie comprises 14 frames in 12 seconds.
(MOV)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.