Abstract
The folding dynamics of riboswitches are central to their ability to modulate gene expression in response to environmental cues. In most cases, a structural competition between the formation of a ligand-binding aptamer and an expression platform (or some other competing off-state) determines the regulatory outcome. Here, we review single-molecule studies of riboswitch folding and function, predominantly carried out using single-molecule FRET or optical trapping approaches. Recent results have supplied new insights into riboswitch folding energy landscapes, the mechanisms of ligand binding, the roles played by divalent ions, the applicability of hierarchical folding models, and kinetic vs. thermodynamic control schemes. We anticipate that future work, based on improved data sets and potentially combining multiple experimental techniques, will enable the development of more complete models for complex RNA folding processes.
Keywords: single molecule, optical trap, optical tweezers, gene regulation, regulatory mechanism
1. Introduction
A foundational principle of molecular biology is that the explanations of living processes are bound up in the relationship between structure and function in biomolecules. A grand challenge of biology has been to build predictive models of organisms by leveraging our understanding of the structure-function relationship. One milestone in this pursuit would be the ability to predict the three-dimensional structures of proteins and nucleic acids directly from their genetic sequences [1–6]. But static structural information alone is insufficient: biomolecules are also highly dynamic. The interplay between structural dynamics and function is therefore a key to understanding the mechanisms underlying biological activity at the molecular level. Because RNA molecules can form comparatively simple structures that are capable of gene regulation and catalysis, in addition to storing and conveying genetic information, they have become model systems for studying the relationship between molecular structure and function in cellular processes. The riboswitch, a regulatory element that acts in cis on one or more genes encoded by its mRNA [7, 8], is an exquisite example. The dynamics of riboswitch folding determine the regulatory fate of the gene(s) under its control. Any quantitative model of riboswitch function must therefore, of necessity, involve a detailed description of its folding dynamics. Folding behavior may be modeled as a diffusive process over a multi-dimensional energy landscape (section 2). The physical mechanisms underpinning riboswitch function can then be understood in terms of this folding landscape and its response to a target ligand in the context of the regulatory mechanism employed by the switch (for example, the regulation of transcription, translation, or alternative splicing).
Accurately reconstructing folding energy landscapes requires the compilation of molecular trajectories in sufficient numbers, and with sufficient observation time, to ensure that all relevant subpopulations have been sampled and identified. Single-molecule approaches afford a means to observe directly individual folding trajectories in a manner free of issues that arise from population averaging and lack of synchronization, which otherwise hamper the interpretations of ensemble-based measurements. In particular, single-molecule Förster resonance energy transfer (smFRET) [9] or fluorescence quenching [10] can measure conformational changes subtending distances of 2–10 nm on as many as hundreds of molecules simultaneously, with a typical time resolution ranging from ~1 to 100 ms (section 3.2). Optical trapping (OT) [11], atomic force microscopy (AFM) [12], and magnetic tweezers (MT) [13, 14] facilitate observations of folding trajectories via the measurement of end-to-end changes in molecular extension. These techniques have achieved spatial resolution down to ~0.3 nm [15] and temporal resolution below 100 µs [16] and allow for applied forces ranging from ~0.1 to ~100 pN (section 3.3). The various available single-molecule methods are versatile and, in many respects, complementary.
2. Riboswitch Folding: Models and Mechanisms
Folding studies of riboswitches are organized around one or more key concepts: these include free energy landscapes, the effects of ligand binding, aptamer-expression platform competition, kinetic versus thermodynamic control, the role of counterions, the nature of intermediates and misfolded states, and the interplay between secondary and tertiary structures. Riboswitches generally employ a modular architecture consisting of an aptamer domain and expression platform. To date, most folding investigations have been confined to studies of the isolated riboswitch aptamer. The ligand-bound form of the aptamer will either interfere with, or promote, the formation of the expression platform and thereby, depending upon the context, up- or down-regulate gene expression. Folding experiments typically measure the lifetimes of various conformational states and the responses of those lifetimes to ligands, cations, forces, or chemical denaturants, in order to ascertain the physical mechanisms responsible for riboswitch behavior.
Folding free energy landscapes [17, 18] provide a unifying framework for interpreting results from different experimental techniques and disparate riboswitches. A ribonucleotide polymer chain has a great many conformational degrees of freedom, and the folding energy associated with any given molecular configuration can therefore be represented on a hyper-dimensional “surface,” or folding energy landscape. For simplicity, this hyper-surface is generally flattened (projected) onto just a one- or two-dimensional surface, where the multiple conformational degrees of freedom become represented by one or two well-chosen reaction coordinates [19] (Figure 1). As RNA folds, it samples, in principle, from a vast conformational space—the ensemble of all possible structures—to settle into its functional, native state (or, in some cases, one of several native states [20–22]), which is often the minimum free-energy state. The fact that biological polymers manage to fold into native conformations on biological timescales, despite the vastness of the conformational space, has been dubbed the “Levinthal Paradox” [23]. The development of a “folding funnel” picture of the folding free energy landscape offered one resolution to the paradox. In this picture, the conformational search takes place on a funnel-like energy landscape leading to the native state(s), thereby biasing the entire folding process and constraining the space that gets explored [24]. There are often a series of locally accessible, non-native intermediate states found on the funnel landscape between the unfolded ensemble and the native state (Figure 1; section 5.1). Prior work has suggested that RNA folding landscapes tend to be fairly rugged, with substantial activation energy barriers separating multiple intermediate [25, 26] and native [20–22] states. Occasionally, the intermediate states compete with longer-lived, misfolded states [27], which—in the case of riboswitches—can act to delay or prevent aptamer formation (section 4.2).
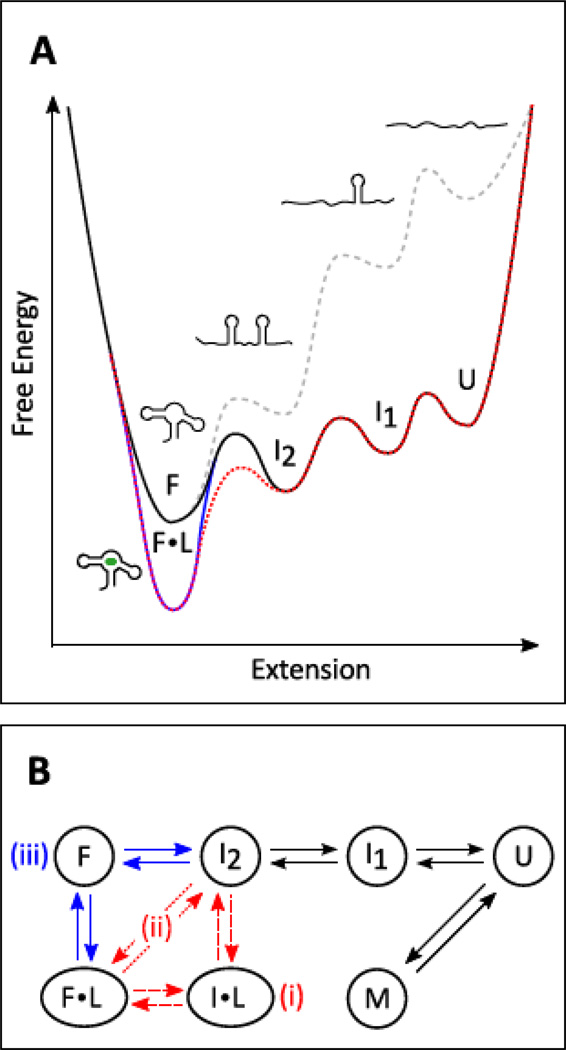
Figure 1. Riboswitch aptamer folding.
(A) Notional free energy landscape for the folding and ligand binding of a generic riboswitch aptamerdomain. Energy is plotted vs. end-to-end extension, the reaction coordinate used in optical trappingexperiments. Example conformational states (U: unfolded, I1,2: intermediates; F: folded, F•L: folded andbound to ligand) are shown for each energy well (bound ligand in green). Ligand-free energy landscapesat zero applied load (gray dashed curve) and under external tension (black curve) are plotted againstextension. Also shown are folding landscapes in the presence of ligand, following either a conformational selection (blue curve) or an induced fit (red dotted curve) model for binding, both under applied tension.
(B) Kinetic scheme matching the energy landscape in (A), with additional potential states represented(M: a misfolded state, I•L: an induced-fit intermediate). Roman numerals indicate two possible induced-fit pathways (i, ii) and the conformational selection pathway (iii).
Generally speaking, RNA folding tends to be hierarchical, with secondary structures (which tend to have lower energy than tertiary structures) typically folding first, followed by tertiary contact formation [28–31]. A hierarchical folding model has been invoked to describe the folding of many RNAs fairly well (section 4.4). However, numerous counterexamples have been identified that violate a strict hierarchy (for example, ribonucleoprotein assembly [32]), demonstrating a more complex interplay between secondary and tertiary structure formation (sections 4.1 and 4.2). In recognition of the limitations and approximations implicit in the hierarchical model, an alternative “quasi- hierarchical” view of RNA folding has been proposed [33]. The hierarchical framework may nevertheless serve as a starting point for RNA structure prediction, without necessarily capturing the detailed RNA folding kinetics [29].
RNA is a polyanion at physiological pH, and the formation of compact structures requires cations to screen its negatively-charged phosphate backbone [30, 34–38]. Divalent magnesium ions, in particular, facilitate tertiary interactions through both non-specific “screening” and specific “binding” interactions, and are sometimes necessary for riboswitch ligand binding (sections 4.4 and 4.5). The significant role played by counterions in structured RNAs compels most folding studies to explore the effect of monovalent and divalent salts, even when no specific interactions are anticipated.
Single-molecule experiments can probe both the effects of ligand binding on folding dynamics and the mechanisms that dictate how such effects propagate through the riboswitch to produce genetic regulation. Riboswitch aptamers are able to bind their cognate ligands with high specificity and affinity, and studies of mutant aptamer folding dynamics can reveal the origin of these characteristics (sections 4.2, 4.5, and 4.8). Ligand binding perturbs the free energy landscape—at a minimum, decreasing the energy of the fully folded state—and can potentially influence folding rates, as well. The effect of ligand binding on RNA folding has been conceptualized in terms of two models: (1) induced fit and (2) conformational selection (also called conformational capture).
To define these models in the context of riboswitch folding, we consider the case of an aptamer folding from an unfolded state (U) via a series of intermediates to its fully folded RNA state (F, or F•L with ligand bound), with a notional landscape shown in Figure 1A. We then focus on the transition from the final intermediate, I2, to the fully folded aptamer, F, where F is the “ON” switch state promoted by ligand. [In principle, an induced-fit mechanism could affect multiple transitions along the folding trajectory, but here we consider I2→F for simplicity.] In an induced fit model, the shape of the activation energy barrier from I2 to F or F•L must change in the presence of the ligand, because the metabolite changes the dominant pathway itself to the folded conformation: this is the meaning of ‘induced’. In particular, the ligand can catalyze riboswitch aptamer folding by lowering the barrier height and changing its position. Changes in the shape of the folding energy barrier are the hallmark of induced fit. [The folded state is also lower in energy in presence of ligand due to the binding energy, but this is not a specific characteristic of induced fit.] In the kinetic scheme shown in Figure 1B, i and ii represent induced-fit folding pathways, where F•L is reached via the interaction of ligand with I2, either through a bound intermediate (path i) or via a ligand-induced transition state (path ii). Paths i and ii are often hard to distinguish experimentally. Alternatively, in the conformational selection model for ligand-promoted folding, the energy landscapes in the presence or absence of ligand differ predominantly in the depth of the folded-state energy well (Figure 1A), and not so much in the barrier shape. This is because in the conformational selection model, the ligand does not interact with the riboswitch in an appreciable way until the riboswitch spontaneously fluctuates into its folded state (F), whereupon the ligand ‘captures’ this conformation, stabilizing it through binding energy (F•L). In the kinetic scheme of Figure 1B, pathway iii represents conformational selection. We depict both induced-fit and conformational-selection pathways in the same diagram (Figure 1B) to emphasize that these schemes are not mutually exclusive, and may, in principle, coexist for a single riboswitch. Indeed, kinetic models incorporating both mechanisms have been proposed previously [39]. When both pathways are present, the dominant pathway may depend upon environmental conditions.
The modular architecture of many riboswitches comes with a twist: the ligand-binding aptamer and regulatory expression platform correspond to mutually exclusive secondary structures that involve shared RNA sequences. The folding competition between these structures shifts upon ligand binding, and this coupling forms the physical basis for gene regulation [40, 41]. If an associated gene’s regulatory fate is sensitive to the exact timing of folding events, for example, through co-transcriptional folding or via transcription-translation coupling [42, 43], then the riboswitch is kinetically controlled (section 4.1). If regulation depends upon the increased structural stability of the ligand-bound aptamer, and the aptamer is given sufficient time to out-compete the expression platform (that is, the system has time to reach thermal equilibrium), the riboswitch is thermodynamically controlled (section 4.2). Depending upon the expression platform, the regulatory context, and the environmental conditions, the same riboswitch can exhibit either kinetic or thermodynamic control [44].
3. Single-Molecule Methods
3.1 Introduction
Most biological macromolecules fold independently of their neighbors. Moreover, even identically prepared molecules will tend to fold along different trajectories on the landscape. The population averaging that takes place in bulk biochemical measurements obscures such aspects of folding and makes any determination of the true folding pathway nearly impossible. Various synchronization techniques (e.g. stopped-flow [45], T-jump [46]) have been developed to reduce ensemble heterogeneity by preparing every molecule in the same initial state. However, because each molecule evolves structure independently, any initial synchronization is quickly lost after the first few transitions in any dynamic process.
Direct observations of the time-resolved trajectories of individual molecules can avoid such synchronization and heterogeneity issues altogether. A single-molecule record reports on individual molecular characteristics, but data acquired from multiple records can also permit the ensemble reconstruction of the parent distributions for kinetic processes (for example, building up the exponential distribution of lifetimes from a Poisson process). FRET (smFRET) and optical trapping (OT) have emerged as the favored techniques for single-molecule folding studies, along with AFM, MT, and other fluorescence approaches. The following sections delve into how energy landscape parameters may be extracted from data acquired by smFRET or OT, although many of the analytical approaches described carry over to other single-molecule techniques. In all cases, a key challenge is to assign features in the collected dataset to specific conformational states.
3.2 Single-molecule FRET
In FRET, energy is transferred nonradiatively from an excited donor fluorophore to an acceptor fluorophore through the coupling of donor and acceptor transition dipoles. The excited acceptor fluorophore can subsequently relax via radiative emission. The FRET efficiency is computed from the relative emission strengths of the donor and the acceptor fluorophores. This efficiency is highly sensitive to donor-acceptor separation, depending on the inverse sixth power of the distance. The efficiency also depends on the fluorescence quantum yield of the isolated donor, the relative orientations of the donor and acceptor transition dipoles, and the spectral overlap between donor emission and acceptor excitation. Donor quantum yield, relative orientation, and spectral overlap can be combined into the Förster length parameter, R0, which specifies the characteristic distance for signaling between a given donor and acceptor dye, typically assuming free relative rotation of these fluorophores (this corresponds to setting the orientational parameter, κ2, to be 2/3) [47–49].
FRET can be used to monitor the relative distances between (and, occasionally, the relative orientations of) two domains of a given macromolecule via the attachment of donor and acceptor fluorophores to specific sites. In particular, measurements of FRET efficiency on a labeled RNA can be used to discriminate among different folding states. Typical fluorophores used in biomolecular assays exhibit R0~4–5 nm, leading to a useful range of ~2–10 nm between labeled sites [50]. Given the considerations above (section 3.1) single-molecule FRET is the preferred approach when one wishes to follow folding dynamics at high resolution. To reduce background fluorescence signals, smFRET experiments are commonly performed using total internal reflection fluorescence (TIRF) microscopy [9]. Typically, a riboswitch aptamer domain is site-specifically labeled with both donor and acceptor fluorophores for FRET, then attached to a glass (or quartz) surface via linker chemistry (Figure 2) [51]. To date, lysine [52], SAM class I [53], SAM class II [54], guanine [55], adenine [56–58], TPP [59], c-di-GMP class I [60], and preQ1 class I & II riboswitches [61, 62] have been explored using smFRET methodologies (section 4).
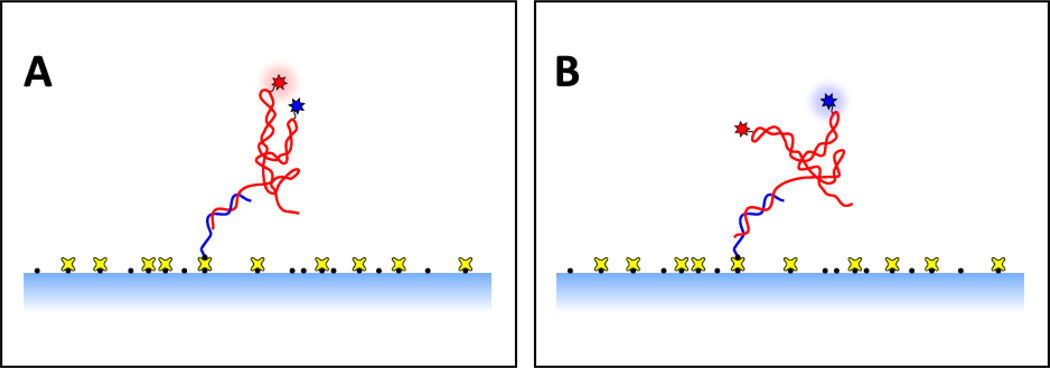
Figure 2. Typical experimental geometry for a surface-based smFRET assay of riboswitch folding.
The riboswitch (red) is anchored to a treated glass surface (light blue shading) via a single-stranded DNA handle (blue), which is specifically attached to the treated surface via chemical linkers (black dots, yellow crosses). Conformational changes alter the proximity of the donor and acceptor fluorophores (blue and red stars), producing a change in the FRET efficiency. Panel (A) shows close proximity of the donor and acceptor, which results in a high FRET state. Panel (B) shows a molecular conformation in which the donor and acceptor are separated, resulting in a low FRET state.
Fluorophore labeling sites must be very carefully chosen to be sensitive to relevant folding transitions, which may coincide with ligand binding. However, it is equally essential that such labeling does not (significantly) perturb either the folding kinetics or the energetics of the riboswitch. In practice, it can be difficult to assess the extent to which the labeling affects such characteristics. Label-independent methods, such as isothermal titration calorimetry (ITC), can be used to compare some properties of labeled and unlabeled constructs, and crystallographic and chemical reactivity data, as well as sequence gazing, can inform the rational selection of labeling sites (see for instance [61]). The choice of dyes is critical here because donor and acceptor dye photophysics are a central constraint in smFRET experiments. Emission brightness, which is influenced by both donor and acceptor quantum yield, must be sufficient to supply a robust smFRET signal [49]. Dye photostability is another critical parameter, because blinking, long-lived on/off switching, and bleaching photochemistry can all limit the useful single-molecule observation time (section 5.2) [47, 49]. Improved photostability can be achieved by removing oxygen from solution through the use of scavenger systems (using enzymes such as protocatechuate 2,3-dioxygenase and glucose oxidase) and/or by introducing protective agents that work through a variety of chemical mechanisms, e.g., Trolox, ascorbic acid, cyclooctatetraene (COT), and 4-nitrobenzyl alcohol (NBA) [63]. The conjugation of certain of these protective agents (e.g., Trolox and COT) directly to carbocyanine dyes has also resulted in improved photostability, but without the need to introduce additional agents, thereby avoiding potential issues of biocompatibility or toxicity [64].
In principle, intramolecular distances can be extracted quantitatively from smFRET records. Whenever FRET is used as a “molecular ruler,” the chemical nature of the linkers used for dye attachment to RNA can become a limiting factor. Direct covalent fluorophore attachment, or use of excessively short linker moieties, can impede the rotational freedom of dyes, making it necessary to consider the dye orientation (which may be unknown) explicitly in any FRET-distance calculations. Conversely, the use of longer, flexible linkers can support free dye rotation (and thereby improve the validity of the κ2 = 2/3 approximation for random dipole orientation), but the increased lengths of such linkers also lead to greater uncertainties in determinations of intermolecular distances in structured RNAs. Dye chemistry can also affect quantitative measurements of distance. For example, Cy3 and Cy5 dyes are known to stack with some nucleic acid bases, thereby reducing their orientational freedom. [49]
Perhaps owing to these complications, most smFRET studies of riboswitches have not tended to focus on quantitative measurements of distance, but have instead used FRET data for largely qualitative inferences about transitions among conformational states. Such determinations can nevertheless supply quantitative information about the dwell times and equilibrium occupancies of these states. The extraction of FRET states, and estimates of their kinetic and energetic parameters, may be performed using a number of analytical tools. In the simplest instance, the histograms of FRET efficiency for a given donor-acceptor pair will typically exhibit one or more peaks that correspond to different stable or metastable inter-fluorophore distances (and/or orientations), and by implication, to different conformational states. Fitting the histogram peaks to sums of Gaussians allows estimation of the relative occupancies of the states. Once assigned, the histograms of dwell times in particular FRET states can be used to extract kinetic parameters, for example, the exponential docking and undocking rate constants for the add adenine riboswitch aptamer (section 4.2). More complex kinetic analysis is also possible: entire kinetic schemes with multiple transitions among states with characteristic FRET efficiencies and dwell times can be reconstructed from single-molecule trajectories using Hidden Markov Models (HMMs) and advancements thereof [65, 66]. Notably, HMM approaches can allow for discrimination among states with degenerate FRET efficiencies but different kinetic properties (section 5.2).
3.3 Optical Trapping
Significant optical forces can be generated when a small dielectric object interacts with the light of an intense, diffraction-limited focus produced by a high-intensity laser [67, 68], and such forces can be exploited to trap the object. In a single-beam optical trap, the steep gradient in intensity in the immediate vicinity of such a focus produces a restoring force that tends to pull the particle towards the region of brighter light. Lasers in the near infrared region are typically used to minimize any absorption of light by biomolecules that might lead to unwanted heating. The spring-like physical properties of an optical trap can be accurately characterized in all three dimensions, allowing the trap to be used to apply controlled forces on much smaller, individual biomolecules by means of an attached particle. Generally, a transparent glass or polystyrene microsphere (bead) is used for this purpose. In a typical experimental geometry, micron-scale polystyrene beads are chemically functionalized with linkers that bind to a specific partner conjugated to a long DNA molecule (typically, ~1 kb), which in turn serves as a convenient ‘handle’ on the bead. An RNA molecule of interest can then be hybridized to a short, single-stranded overhang at the distal end of the DNA handle. In a dual optical trap arrangement, two beads, each carrying different handle overhang sequences, can be used to capture the opposite ends of a single RNA molecule. This geometry allows the system to exert pN-scale forces and measure nm-scale motions as the RNA folds or unfolds. Two arrangements for optically trapping riboswitches are illustrated in Figure 3.
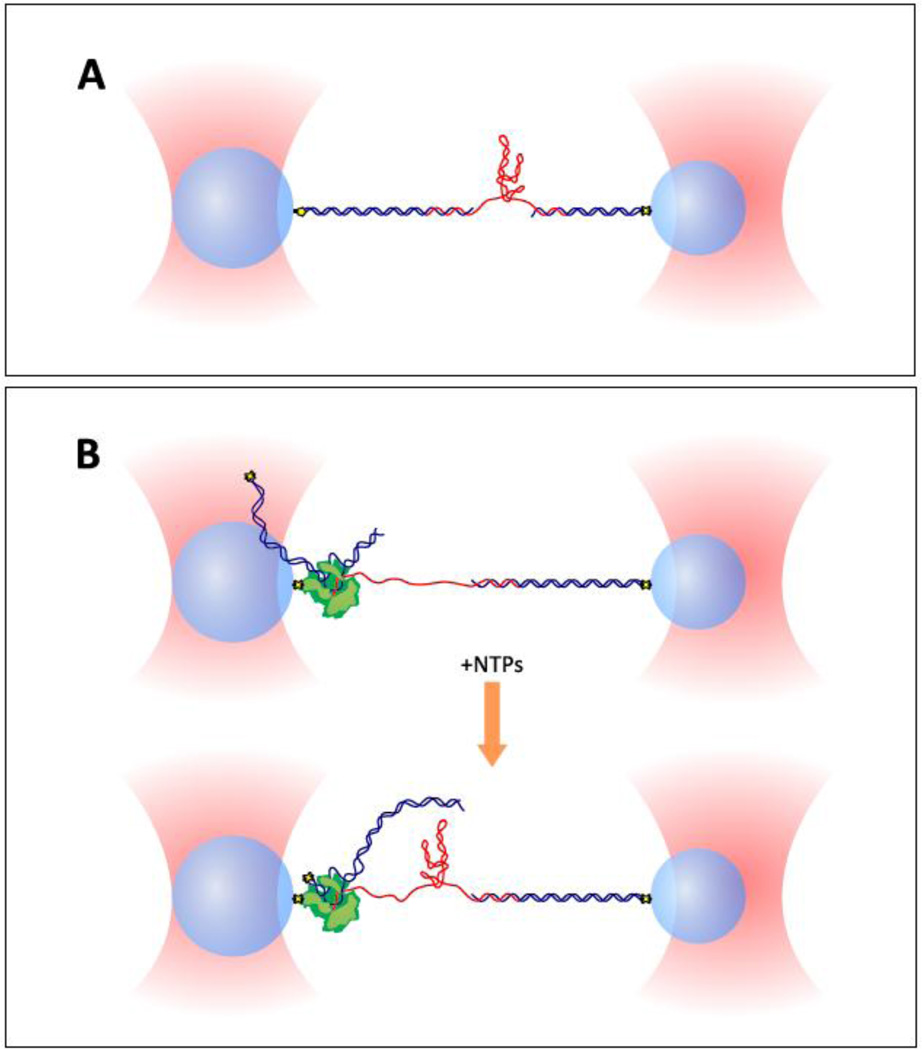
Figure 3. Two dual-beam optical trap assays for riboswitch folding.
Dual optical traps (salmon) hold functionalized polystyrene beads (light blue spheres) which are each attached to a DNA handle (blue) or RNA polymerase (green) via specific attachment chemistries (yellow and black shapes). The riboswitch RNA (red) is hybridized to single-stranded overhang regions at the ends of double-stranded DNA handles. The end-to-end extension of the riboswitch construct can be measured and controlled with nanometer-level precision. Panel (A) shows a geometry for studying riboswitch constructs synthesized in vitro. Panel (B) describes a setup for studying riboswitches transcribed in situ by RNA polymerase from a DNA template capped with a “roadblock” (yellow and black shapes); addition of NTPs triggers elongation of the nascent RNA (upper image), producing the riboswitch construct (lower image). The geometry in (B) allows the study of co-transcriptional as well as post-transcriptional RNA folding.
Single-Molecule Force Spectroscopy (SMFS) uses force to characterize molecular folding landscapes, and has been used successfully to study DNA [69], RNA [11], and proteins [70]. In effect, the application of an external force tilts the folding energy landscape for a given biomolecule (Fig 1A), biasing it towards a more unfolded conformation. This perturbation can be used to supply information about molecular energetics, kinetics, and folded conformations. In an unbranched polymer such as RNA, the measured end-to-end molecular extension supplies a natural reaction coordinate for tracking the folding trajectory [19]. By systematically exploring the molecular extension in response to a range of applied forces, one can directly observe metastable folding intermediates and measure their corresponding lifetimes. Transitions between intermediates that produce changes in extension must then be assigned to the folding, or unfolding, of known structural elements. Folding intermediates often exhibit different extensions, which facilitates the assignment of a given extension signal to a particular conformational state. In some cases, however, two (or more) conformations may coexist at the same extension. This degeneracy can sometimes be disambiguated through the differential sensitivity of these two conformations to the applied force, or through differences in their lifetimes. For example, the ligand-bound form of an aptamer may exhibit a greater resistance to opening force than the apo-form (Figure 1A, states F•L and F), yet have very similar structure. Similarly, certain tertiary conformational transitions may not result in an extension change, in which case their presence must be inferred in this manner (section 4.4).
Broadly speaking, SMFS employs two different classes of experiment to extract landscape features from data records: (1) non-equilibrium measurements (e.g. force ramps and force jumps), and (2) near-equilibrium measurements (e.g. force and extension clamps and constant-position measurements.) In force ramp experiments, the force is increased (or decreased) monotonically over time as the corresponding extension is recorded. The resulting force-extension curve (FEC) displays both molecular stretching and structural transitions. At various forces, structural elements become destabilized, and abrupt discontinuities (“rips”) appear in the FEC that correspond to unfolding events, where structural elements give way to the next (quasi-stable) intermediate. The smooth segments of the FEC situated on either side of such rips may be fit to a worm-like chain (WLC) or similar polymer model of RNA elasticity, and thereby used to extract the opening distance associated with the rip itself, which can be converted into an equivalent number of base pairs that open up [71–73]. In many cases, knowledge of this base-opening value is sufficient to make a structural assignment. The activation energy and the position of the energy barrier for a specific transition can be extracted from its rip-force distribution, compiled from hundreds, or even thousands, of FECs [74]. In addition, the free energy for folding can be calculated from the distribution of the mechanical work required to unfold and fold the molecule using non-equilibrium analysis methods [75].
Constant-force folding experiments use a force clamp, which may be passive [76] or active [77], to maintain a given force on RNA irrespective of its molecular extension: this has the effect of tilting the folding-energy landscape with respect to this reaction coordinate (Figure 1A). With sufficient bias, two (or more) given conformational states may be made approximately isoenergetic. Under such conditions, thermal fluctuations are sufficient to drive repeated transitions between these states, with typical lifetimes ranging from milliseconds to minutes, depending upon the energy barrier height. The relative free energies and activation energy barriers can then be estimated from the relative occupancies and apparent lifetimes for the observed states, using Boltzmann and Arrhenius relationships, respectively (for example, [69]). To map the entire landscape, the clamping force may be stepped down in discrete increments from some maximum value (typically, ≥20 pN) while observing conformational transitions for minutes at a time (depending upon the observed state lifetimes), until all observable intermediates have been scored. In some instances, using the latest generation of ultra-stable instruments, it has proved possible to extract more information about the folding energy landscape than just the locations and relative energies of the wells and barriers. High-resolution data from SMFS can be transformed into a complete, fine-grained landscape by using deconvolution methods. In such cases, actual landscape shapes have been reconstructed for DNA hairpins [69] and RNA aptamers [78] (section 4.2).
4. Results
4.1 The pbuE adenine riboswitch
The pbuE riboswitch of B. subtilis modulates the transcription of a gene encoding an adenine efflux pump. The pbuE aptamer domain consists of a three-helix central junction (Figure 4A) that specifically binds adenine through Watson-Crick pairing to a uridine residue. In the adenine-bound aptamer, the P2 and P3 stems are observed to dock with one another via a kissing-loop interaction between L2 and L3 [79]. A portion of the sequence on the 3′ side of aptamer stem P1 can alternatively participate in the formation of a terminator hairpin. In the absence of adenine, which stabilizes the aptamer domain, terminator formation outcompetes aptamer folding, ending transcription with high probability.
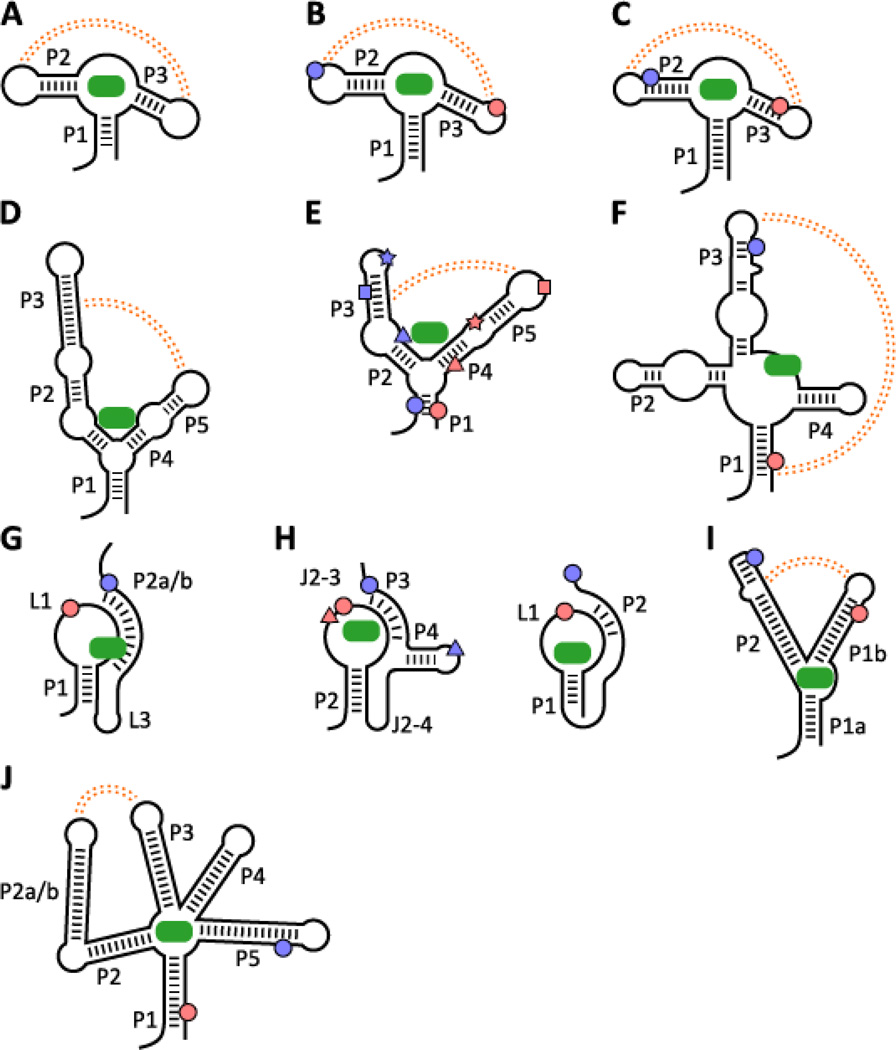
Figure 4. Riboswitches studied by single-molecule techniques.
Secondary structures are represented, with ligand (green oblong) binding represented schematically and key tertiary interactions in the ligand-bound folded state indicated notionally in orange or as pseudoknot basepairs (G, H). (A) pbuE adenine riboswitch aptamer; (B) add adenine riboswitch aptamer; (C) xpt guanine riboswitch aptamer; (D) thiC TPP riboswitch aptamer; (E) thiM TPP riboswitch aptamer;(F) metI SAM class I riboswitch aptamer; (G) metX SAM class II riboswitch; (H) left: preQ1 class IIriboswitch, right: preQ1 class I riboswitch; (I) tfoX c-di-GMP class I riboswitch aptamer; (J) lysC lysineriboswitch aptamer. FRET labeling sites are indicated with blue and salmon dots (triangles, squares andstars indicate additional possible labeling sites) for the donor and acceptor locations, respectively.
Greenleaf and colleagues performed force ramp and constant-force measurements on an in situ transcribed pbuE aptamer (Figure 3B) to reconstruct a five-state energy landscape for aptamer folding [80]. They demonstrated that the P2 and P3 stems (but not P1) fold prior to binding pocket formation, and that an intermediate folding state competent to bind the ligand (A-comp), involving tertiary contacts, forms just prior to full aptamer folding (state F). The existence of A-comp confirmed earlier suggestions about a likely pre-organization of the ligand binding pocket [57, 79]. Perhaps surprisingly, the tertiary contacts associated with this intermediate contributed 2.7 ± 0.3 kcal/mol of energetic stabilization, an amount comparable to the folding energy of stem P1. The formation of a tertiary structure (as observed in A-comp) prior to a secondary structure (the folding of P1 in F) provides a clear counterexample to the strict hierarchical folding model for RNA (section 5.4). Despite a significant stabilization by adenine of 4 ± 1 kcal/mol in F•L, the weak P1 switch helix remained “brittle:” once the lone GC pair in P1 gets disrupted, the entire three-helix junction falls apart, prompting the proposal that this GC may function more broadly as a “structural keystone” in all purine riboswitches, where it is a common element [80] (section 4.2). The reconstructed landscape also showed that the presence of adenine shifted the location (as well as changed the height) of the final energy barrier to folding, although the observed shift may lack statistical significance. If borne out by further work, such a shift would imply that an adenine-induced fit occurs in the aptamer after binding-pocket formation.
In general, the full-length pbuE riboswitch will not bind ligand because terminator hairpin formation easily outcompetes aptamer folding at equilibrium: it is favored energetically by a Boltzmann factor of ~1010. This led to the assumption that any regulation must therefore occur co-transcriptionally, out of equilibrium [57]. Frieda and coworkers [81] demonstrated directly how regulation arises through a kinetic competition between the aptamer and expression platform by observing co-transcriptional behavior of the full-length riboswitch in real time (section 2). A constant force was applied during in situ transcription of the nascent mRNA, as transcription and folding were followed simultaneously. At lower forces, P2 and P3 folded sequentially, regardless of the adenine concentration (in agreement with [80]), but the further folding of the aptamer was adenine-dependent. When the aptamer had successfully folded and bound ligand, the probability of termination dropped from 85% to 5%. The adenine-bound aptamer delayed terminator formation only transiently—typically, for just a few seconds—but sufficiently long for the associated RNA polymerase to get past the critical U-tract of the terminator element [82]. Similar reductions in termination efficiency could be achieved by increasing the applied force to 10.4 pN, which mechanically delayed formation of the terminator. In these experiments, the adenine-bound aptamer lifetime dropped to ~2 s from ~10 s in aptamer-only studies, suggesting that some mechanism, e.g. branch migration [83, 84], may facilitate terminator hairpin invasion of stem P1 [81]. This effect highlights the importance of understanding the behavior of the aptamer in the context of the full-length riboswitch. Together, these experiments [80, 81] demonstrate how adenine promotes and stabilizes pbuE aptamer folding, forming a kinetic trap that prevents the terminator from folding in sufficient time to abort transcription.
4.2 The add adenine riboswitch
Like the pbuE aptamer from B. subtilis, the add riboswitch aptamer from V. vulnificus consists of an adenine-binding three-helix junction with P2 and P3 stems that dock via L2-L3 interactions (Figure 4B). In V. vulnificus, ligand binding promotes translation initiation by preventing the sequestration of the Shine-Delgarno (SD) sequence and start codon [85]. When the P1 helix is unfolded, the expression platform locks the SD sequence and start codon into a stable “sequesterer” hairpin (SH). When stabilized by adenine, the aptamer (including P1) is fully folded, SH formation is precluded, and the start codon becomes available to the ribosome.
Neupane and colleagues performed OT experiments on both the add aptamer and the full-length add riboswitch [86]. Constant-force and force-ramp experiments revealed a 5-state folding free energy landscape for the add aptamer, with a transient intermediate, P1_U, located between the P2P3 state (P2 and P3 folded) and the fully folded state, F. Similarly to the pbuE state A-comp (section 4.1), this intermediate was interpreted as a conformation with the central junction pre-organized and P1 unfolded [86]. Tertiary contacts contributed ~1.5 kcal/mol of additional free energy to P1_U, consistent with the docking of P2 and P3 in this state. Contrary to the hierarchical folding model, comparatively weak tertiary interactions seem to nucleate the long-range conformational changes required to fold the P1 stem in this riboswitch (section 5.4). The transition state position for P1_U → F suggested that the “structural keystone” of the add riboswitch may be a UA-U triplex, rather than the GC pair inferred in prior work on pbuE (section 4.1). In the absence of adenine, P1 stem unfolding was measured to be four orders of magnitude slower (koff ≈ 10−5 s−1) than its counterpart in pbuE (section 4.1). The addition of saturating adenine produced 8 kcal/mol stabilization of F, compared to 4 kcal/mol in pbuE [80], and also increased the lifetime of F dramatically.
Lemay and coworkers performed smFRET experiments on a P1-stabilized add aptamer variant with FRET labels on the L2 and L3 loops (Figure 4B). Their measurements revealed a novel docking intermediate, I, evident at low Mg2+ concentration ([Mg2+] < 0.05 mM) [57]. Raising [Mg2+] increased the population of the I state at the expense of the undocked state, U, and further increasing the [Mg2+] stabilized a long-lived docked conformation, D, consistent with the coordinated Mg2+ ions observed in the adenine-bound crystal structure [79]. To test if I could bind ligand, adenine was introduced under extremely low Mg2+ concentrations (2 nM). Under such conditions, the D state was unstable (without Mg2+ to facilitate any loop-loop interaction), but the persistence of a significant I population was interpreted to indicate an adenine-dependent stabilization. Such behavior is consistent with an induced-fit kinetic model, where I is an adenine-bound undocked, or partially docked, state “U•L” (Figure 1B, pathway i).
But what relationship exists, if any, between state I, as observed by smFRET [57], and state P1_U, as observed by SMFS [86]? Because the occupancies of I and D are both sensitive to [Mg2+], OT experiments performed at various magnesium concentrations may help to decipher how the docking states are related to P1_U. Absent any additional information, direct comparisons are difficult because stem P1 is open in P1_U, but presumably closed in the U, I, and D states, and the end-to-end extension is therefore insensitive to any further transitions that occur once P1 folds. In principle, simultaneous observation of stem P1 folding and P2-P3 docking using multi-color FRET [87, 88] or the combined force-FRET technique [89, 90] (section 6) could be used to reconstruct a two-dimensional folding energy landscape and thereby resolve this question [91].
In a follow-up smFRET study, Dalgarno et al. further explored the add aptamer L2-L3 docking interaction by using urea to selectively disrupt tertiary contacts [58]. Urea is known to destabilize secondary, as well as tertiary, structures in RNA by mechanisms that remain unclear [92], but the authors used sufficiently low urea concentrations that the overall secondary structure content appeared to remain intact, based on circular dichroism measurements. Two subpopulations with distinct dynamics were observed in the presence of various amounts of urea, Mg2+, and adenine. In one subpopulation, RNA molecules rapidly fluctuated between closed (“docked”) and open (“undocked”) arm states, DLF and U, while another subpopulation exhibited a long-lived docked state, DLB, with only occasional visits to the undocked state, U. The occupancy of each subpopulation was strongly influenced by the concentration of adenine present: DLB was interpreted as ligand-bound, and DLF as ligand-free [32]. With 4 mM Mg2+ and 5 M urea present, adenine stabilized DLB by ~50-fold relative to DLF. These findings indicate that an additional stabilization energy ΔΔG ≈ 2.3 kcal/mol is conferred upon ligand binding, which is the of the same order as the ~8 kcal/mol measured in OT experiments [86]. However, the presence of urea in these experiments confounds more direct comparisons: specifically, the urea dependence of the rates kdock(LF), kdock(LB), and kundock in the presence of adenine was not determined, preventing any extrapolation of these results to no-urea conditions.
In the add riboswitch, transcription-translation coupling is not necessary for regulation, and the full riboswitch can bind and release the ligand on a timescale of seconds [85]. Both properties suggest thermodynamic control (section 2). Refolding time experiments, in which the full-length add riboswitch was subjected to unfolding force-ramps repeated at various intervals, during which it had an opportunity to refold [86], demonstrated how ligand binding modulates a competition between aptamer and SH expression platform folding through a thermodynamic control scheme. Thermodynamic control requires the minimum free energy conformation of the switch to change in response to bound ligand. In support of this, the SH-folded state was found to be 1.4 kcal/mol more stable than the folded aptamer state in the absence of adenine, whereas the ligand-bound aptamer is more stable than the SH by 6.6 kcal/mol [86]. Interestingly, an AFM experiment by Heus and colleagues indicated that the “OFF” state (with SH folded) is further stabilized by a previously unidentified pseudoknot [12], although the comparatively large measurement uncertainties in this study could be improved upon in future work, for example using laser-based cantilever stabilization [93]. For thermodynamic control, the full riboswitch must have sufficient time to achieve equilibrium before the regulatory decision is made. Based on refolding-time experiments [86], the probability of SH formation was fit to an exponential model, yielding an equilibration time τ ≈ 0.2 s. During refolding, the aptamer folded first (t ≈ 0.033 s), but after ~3τ, the riboswitch reached folding equilibrium. Given a transcription rate of 40–50 nt/s in vivo, the riboswitch has sufficient time (1.5τ) to achieve a near-equilibrium state before the earliest possible start of translation [86].
High-resolution OT techniques are now capable of discerning fairly intricate details of energy landscapes. Constant-force experiments by Neupane et al. revealed a rare misfolded state, M, that formed from the unfolded state, U, in just 0.3% of records [86]. The properties of M were consistent with it being a 32-nt hairpin that transiently assumes additional misfolded states (M2 and M3) [86]. However, the low probabilities of formation and short lifetimes of these misfolded states make it unlikely that they exert any substantial effect on riboswitch function.
As discussed, folding can be considered as a diffusive process over a conformational energy landscape (section 2). In another study by Neupane and coworkers, the authors determined the effective diffusion constant and the transition path time over the net folding barrier for the add riboswitch aptamer [16]. Gupta et al. demonstrated the practicability of non-equilibrium OT experiments in reconstructing finegrained folding-energy landscapes for complex RNAs, using the add aptamer as a working example [78]. An effective two-state folding landscape was recovered, although certain previously-observed aptamer configurations were not well-resolved using this approach [86]. With further improvements, such finegrained landscape determinations may offer a promising direction for riboswitch studies.
Data from several experiments support an induced fit mechanism for aptamer ligand binding, as proposed by Dalgarno et al. [57, 58, 86]. Their model features two alternative folding pathways, with different degrees of conformational rearrangement associated with the induced fit. In one pathway, U proceeds to DLB via DLF, whereas in the other, the transition to DLB proceeds directly from U [58]. Presumably, the docking intermediate(s) detected in earlier work [57, 86] also fall(s) along one or both of these pathways. Data from both Lemay et al. and Dalgarno et al. indicate that the folding rate, kfold, increases roughly 2-fold in the presence of saturating adenine and magnesium (versus magnesium alone) [57, 58]. Hence, from these data, the activation energy barrier for folding is reduced in the presence of ligand (ΔΔG‡ ≈ −0.4 kcal/mol), matching our criteria for an induced-fit mechanism (Figure 1). However, the reduction in the activation barrier height is only moderate. Results from Neupane and colleagues, on the other hand, point to a more substantial barrier reduction (ΔΔG‡ ≈ −3.7 kcal/mol for the final transition of P1_U to F vs. F•L) [86]. In addition, the FRET results indicated only minor stabilization of the folded state by adenine (ΔΔG ≈ −0.65 kcal/mol [57]), compared to a value of −8 kcal/mol estimated from OT experiments [86].
This large discrepancy remains to be resolved, but there are several candidate explanations. The OT experiments were performed in the presence of physiological levels of magnesium (4 mM) [86], whereas the FRET measurements were carried out in 0.5 mM [57] or 0.1 mM [58] magnesium. Also, the “undocked” state for the FRET calculations included both U and I states, due to rapid interconversion between these states at the magnesium concentration used [57]. Finally, the ligand-free and ligand-bound folded states could not be distinguished in the FRET experiments. When urea was used to differentiate DLB and DLF (at 4 mM [Mg2+]), improved agreement with the OT results was obtained, with ΔΔG‡ ≈ ΔΔG ≈ −2.3 kcal/mol [58]. However, based on the available data, the use of urea apparently changed the mechanism from induced fit to conformational selection (Figure 1; with urea present, kdock was roughly independent of [adenine]).
4.3 The xpt guanine riboswitch
Single-molecule investigations of purine riboswitch folding have not been limited to adenine riboswitches. Brenner et al. studied the xpt guanine riboswitch aptamer from B. subtilis, using both bulk FRET and TIRF-smFRET approaches [55]. Just as in the case of the adenine riboswitches (sections 4.1, 4.2), the xpt riboswitch contains a P1-P2-P3 three-way junction aptamer, with P2-P3 docking in the ligand-bound aptamer state (Figure 4C) [94]. The guanine ligand is bound specifically via Watson-Crick base-pairing to a cytosine residue in this junction. The xpt riboswitch regulates the expression of downstream genes in the xpt-pbuX operon, coding for proteins involved in purine metabolism and transport. The expression platform consists of an anti-terminator hairpin that shares nucleotides with the alternatively-formed P1 hairpin of the aptamer [95]. Guanine binding promotes folding of the aptamer domain over the anti-terminator a leading to transcription termination via an intrinsic terminator hairpin.
Labeling each of the three possible pairs of the P1, P2, and P3 elements in separate FRET experiments [55] allowed a more comprehensive exploration of aptamer folding dynamics than could be obtained by labeling the P2 and P3 arms alone, as in other work. smFRET with the (P1, P2) element pair revealed three interconverting FRET states, with an intermediate-efficiency FRET state that was promoted by both magnesium and guanine. These states were interpreted as corresponding to unfolded (undocked), intermediate, and folded (docked) configurations. Similarly, the (P2, P3) element pair revealed two dominant FRET states at low [Mg2+], which were subsumed into a single state with a slightly different FRET efficiency at higher [Mg2+]. Together, the (P1, P2) and (P2, P3) data suggest a folding intermediate, I, where the P2 and P3 elements interact in a docked-like state. FRET data at higher magnesium levels revealed compaction beyond that in the docked state, with the base of P1 moving closer to P2 and P3. The combined data from the three FRET pairs clarified the relative motions of all three elements, allowing the authors to propose a folding model in which the P1 and P3 elements form a static platform, to which the dynamic P2 stem-loop is anchored. Their folding model consists of three states: (1) an ensemble “unfolded” state, U, with P2 remaining dynamic between two preferred orientations, corresponding to the undocked and docking intermediate (I) states; (2). A folded native state, FNS, with stabilized L2-L3 contacts, yielding P2-P3 docking (promoted by magnesium and guanine); and (3) a further compactified state, FC, induced by high magnesium (>5 mM), and perhaps less likely to be biologically relevant [55].
The study by Brenner and colleagues provides some support for a conformational selection mechanism for folding of the xpt riboswitch. When (P2, P3) smFRET experiments were performed in the presence of 0.1 mM or greater [Mg2+], the undocking rate constant kunfold decreased roughly 2-fold in the presence of guanine ligand vs. the guanine-free case, whereas the docking rate constant, kfold, did not change appreciably (changes were within the errors of the measurement). Hence, the unfolding energy barrier is raised only slightly (by ~0.4 kcal/mol), and the folding energy barrier is not significantly affected by presence of the metabolite. As a result, the docked state with guanine is in a lower-energy well, but the undocked-to-docked energy barrier is unchanged by guanine. This situation is consistent with a conformational selection mechanism (Figure 1). Hence, despite the high degree of structural similarity between the xpt and add aptamers [79], the xpt riboswitch seems to bind its cognate ligand somewhat differently from the add riboswitch (section 4.2). This contrast underscores the importance of performing functional biophysical studies in addition to structural investigations.
4.4 The thiC TPP riboswitch
Versions of the thiamine pyrophosphate (TPP), or “thi-box,” riboswitch have been found in all three domains of life (prokayotes, archaea, and eukaryotes) [96], leading to speculation about its ancient origin in the RNA world [97]. Depending upon the organism and context, this element can regulate genes by modulating transcription, translation, or even (in eukaryotes) alternative RNA splicing [96, 98]. Its aptamer domain includes two “sensor” helices, P2/P3 and P4/P5 (Figures 4D, 4E), each containing unpaired internal loop regions that form highly specific binding pockets for the pyrimidine and pyrophosphate groups of the ligand, respectively [99, 100]. These helices are connected through a central junction to P1. The P1 element can act as a “switch helix” in transcriptional or translational contexts, where P1 nucleotides can alternatively participate in expression platform structures (section 4.5). The eukaryotic TPP riboswitches regulate gene expression by promoting alternative splicing outcomes that modulate the mRNA lifetime or efficacy in the cell [96, 101, 102]. Crystal structures of the ligand-bound aptamer [99, 100] reveal a tertiary interaction between L5 and P3, suggesting that tertiary dynamics may play an important role in ligand binding.
The thiC riboswitch (Figure 4D) regulates alternative splicing of the thiamine biosynthesis gene thiC in A. thaliana in response to TPP levels. Anthony and colleagues used OT to reconstruct a 7-state folding landscape of the thiC aptamer, from the fully unfolded ssRNA to the stable native conformation [103]. In this instance, all the observed structural transitions could be explained by secondary structure formation; any formation of tertiary contacts presumably occurred during, or after, the final P1 folding event, and therefore did not produce any significant change in the end-to-end extension. Ligand binding was observed indirectly, when P1 folded irreversibly at low force in the presence of saturating levels of TPP.
A central junction point mutation can confer bacterial resistance to the antimicrobial compound pyrithiamine pyrophosphate (PTPP), a TPP analog that targets TPP riboswitches [104], suggesting that the junction sequence is functionally important. In OT experiments, the PTPP-resistant central junction mutant A105G displayed reduced tertiary stabilization of the folded state, due to the P1 region pairing “out of register” (i.e., mismatched by slippage), preventing both the formation of native contacts in the junction and the proper alignment of the helix arms. The central junction thus seems to serve as a structural scaffold for the correct orientation of the sensor helices and their binding domains. Taken together, the wild type and A105G mutant results suggest that all secondary structures and the central junction may need to fold before the aptamer can bind ligand.
Force-ramp experiments performed on the TPP-bound aptamer revealed two populations with distinct rupture-force distributions [103]. The “strong-binding” state at high rupture force was 30% more stable than the “weak-binding” state. The weak-binding state was the only bound state that formed in the presence of thiamine or thiamine monophosphate (the absence of the pyrophosphate moiety is expected to reduce the affinity of the aptamer for these substrates). Upon mutation of a base implicated in stem-loop docking of P3/L5 (A90G), the strong-binding state was abolished, leaving only weak binding for all ligands, showing that the P3/L5 interaction is necessary for strong TPP binding. Because the weak-to-strong binding transition was the only one to depend upon ligand identity, the authors proposed that P4/P5 (and not P1, by contrast with prokaryotic TPP riboswitches and purine riboswitches) may operate as the “switching” element that regulates splicing in thiC, corroborating conclusions from previous biochemical and in vivo studies [102]. In addition, the kinetic model that emerges from this study, which incorporates two-step ligand binding, is consistent with an induced fit mechanism (pathway i in Figure 1B), where the P1-P2-P3 folded state corresponds to I2, the weak-binding state to I•L, and the strongly bound state (which is never observed in the absence of ligand) to F•L.
4.5 The thiM TPP riboswitch
Haller et al. investigated the thiM TPP riboswitch aptamer domain from E. coli (shown in Figure 4E) using TIRF-smFRET [59]. In this riboswitch, the short P1 element functions as a switch helix: the 3′ end of the P1 stem contains a nucleotide sequence that can alternatively basepair with an anti-Shine-Delgarno sequence, thereby releasing the SD sequence from a translational repressor stem. Hence, TPP represses thiM gene expression by binding to, and stabilizing, the folded aptamer domain.
The thiM aptamer was investigated via multiple FRET pairs, which reported on the relative conformations of the P3 and P5 “forearms” (which dock through a P3-L5 interaction), the P2 and P4 “upper arms”, and the two strands of the P1 switch helix (Figure 4E). The authors observed a folding intermediate in the opening/closing dynamics of the P3 and P5 forearms [59]. This was interpreted as a state where the P2/P1/P4 junction is pre-organized, but the P3 and P5 forearms remain open in a “Y-shaped” configuration. Multiple, distinct dynamical behaviors were observed for P3-L5 docking in the presence of magnesium and TPP. The dominant behaviors were: (1) a relatively stable docked state, interchanging with an undocked state, and (2) a dominant undocked state that fluctuated transiently to a docked state. In the absence of magnesium and TPP, docked states were not observed, but the addition of 2 mM Mg2+ permitted a transient docked state to form. TPP-dependent stabilization of the docked conformation (with 2 mM Mg2+ present) was demonstrated, although these (presumably ligand-bound) states were still surprisingly labile: in thiM, the lifetimes were <1 s in the “stable” docked state, and docked state occupancy was at most 30%. These values are to be compared with the corresponding values of >60 s and ~90% observed for thiC [103]).
As with the pbuE and add riboswitches (sections 4.1, 4.2), the P1 switch helix of the thiM riboswitch did not fold in accordance with a strictly hierarchical model of RNA folding. In the absence of magnesium and TPP, a condition in which the tertiary P3-L5 docking interaction was not observed, P1 folding was highly dynamic (4:5 ratio of unfolded:folded stem). When magnesium was introduced, the folded state dominated, but the P1 helix remained dynamic, fluctuating between folded and unfolded states. Even with both TPP and magnesium present, heterogeneous dynamics were observed, where P1 would transition between a stably folded state and a dynamic state that fluctuated.
In contrast to the heterogeneous dynamics observed for P1 and P3-P5 folding, Haller and colleagues reported that P2 and P4 remained predominantly in the same stable, compact state regardless of the presence of Mg2+ or TPP ligand, based on smFRET between labels on the P2 and P4 elements. The extent to which P2 is dynamic relative to P4 is not entirely clear, however, because single-molecule trajectories and associated dwell times were not reported for those experiments. Nonetheless, the results suggest that the P2-P4 junction may, on average, be pre-folded even in the absence of magnesium or TPP. Curiously however, the addition of 2 mM Mg2+ slightly shifted the peak efficiency and promoted occupancy of the (apparent) low-FRET unfolded state of P2-P4. The addition of TPP together with Mg2+ reversed the shift. Based on the slowly interchanging behaviors of P1 and P3-P5, combined with the relative stability of the P2-P4 junction, the authors proposed a folding model that invoked a remote coupling of the dynamics of the P1 element and the P3-L5 docking interaction [59].
Consistent with the notion of some type of coupling between P1 and P3-L5, the probability of P3-L5 being in a docked conformation increased 15% when the P1 stem was stabilized by two additional GC pairs, supporting the view that P1 kinetics are somehow able to influence P3/L5 kinetics despite the reportedly “rigid” and “pre-organized” intervening P2/P4 junction. Understanding the physical basis for this influence, which is central to the mechanism underlying ligand-induced regulation, represents a challenge for future biophysical studies. Because the kinetic rates derived from hidden Markov modeling were not available (possibly due to large uncertainties arising from a limited average observation time, ~3.5 s), more work may be required to establish a rigorous correspondence between FRET states and conformational states in this system, and to clarify the extent and nature of any coupling. Simultaneous measurements of P1 folding and P3-L5 docking (section 6) could, in principle, assist in developing a more quantitative understanding of how these elements interact.
The A69G mutation in thiM, which is homologous to A90G in thiC and therefore thought to disrupt P3-L5 docking, markedly increased the dynamics of P3 relative to P5. Paradoxically, however, it also increased the occupancy of a high-FRET state that the authors had previously associated with the ligand-bound, native conformation. As anticipated, the A69G mutation decreased the lifetime of the P3-L5 docked conformation (by ~2.5 fold), but surprisingly, the lifetime of the low-FRET (open) conformation also decreased (by ~3 fold), biasing the aptamer towards its folded state. To explain this finding, the authors postulated that allosteric effects might globally alter the folding landscape. However, an alternative possibility is that the mutation may have promoted an intermediate state with rapid kinetics (with EFRET~0.65, and perhaps misfolded) that occasionally transitions to the less-populated, native conformation (with EFRET~0.8). We note that these two high-FRET peaks seem to have been combined for the HMM analysis of the mutant, whereas for the wild-type aptamer, a single FRET peak was observed (with EFRET~0.75).
4.6 The SAM class I riboswitch
The structurally diverse and broadly distributed S-adenosylmethionine (SAM) riboswitches regulate the expression of proteins involved in the synthesis and transport of several related metabolites, including SAM. Based on sequence comparisons, the known instances of this switch may be categorized into six classes [105]. The class I SAM riboswitch aptamer consists of a 4-helix junction (Figure 4F), and portions of the P1 and P3 elements form a SAM-binding pocket [105]. When the P1 helix of the aptamer is formed, a transcription terminator hairpin can fold downstream, and termination occurs. However, when P1 is unfolded, some P1 nucleotides can alternatively participate in an antiterminator hairpin that promotes transcriptional read-through.
Heppell et al. studied the B. subtilis metI SAM-I riboswitch using an ensemble of techniques including single-round in vitro transcription, electrophoretic mobility shift assays (EMSA), selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), bulk 2-aminopurine (2-AP) fluorescence, bulk FRET, and TIRF-smFRET [53]. smFRET experiments on the aptamer domain with labels on the P1 and P3 stems revealed three FRET states, and all three were accessible simultaneously under appropriate magnesium concentrations. These states were interpreted as an open “unfolded” state, U (but with secondary structure presumed formed), a magnesium-induced folding intermediate, FMg, and a magnesium- and SAM-induced native state, FNS [53]. In the absence of magnesium, all molecules appeared to be in state U. Whereas magnesium was required to access FMg, the presence of ligand was not strictly required to enter FNS: in 3 mM magnesium, most molecules were in FMg, but FNS had ~32% occupancy. The introduction of 100 µM SAM increased the population in FNS at the expense of FMg.
EMSA experiments performed on a mutant aptamer construct containing a 4-A bulge that bends P1 axially suggested that the conformational change between FMg and FNS likely involves some rotation of the P1 helix, based on the altered electrophoretic mobility of the bent construct in the presence of SAM [53, 106]. This aspect of SAM-I riboswitch folding might be particularly interesting to study further using single-molecule fluorescence polarization anisotropy (smFPA) [107], which may permit a more direct measurement of rotational dynamics within the metI riboswitch aptamer.
4.7 The SAM class II riboswitch
The metX SAM-II riboswitch, first identified in the Sargasso Sea metagenome, is thought to regulate gene expression at the translational level by sequestering the SD sequence of its associated gene in an H-type pseudoknot (Figure 4G), with pseudoknot formation being promoted by ligand binding. Here, the presence of SAM down-regulates the target gene. Haller and colleagues performed a study of the metX riboswitch using NMR, bulk 2-AP fluorescence, bulk FRET, and TIRF-smFRET [54]. The folding of the full-length riboswitch was directly interrogated by smFRET experiments.
Monitoring pseudoknot formation via FRET labels placed on the P2a and L1 elements, the authors observed transitions between two discrete FRET values in the presence of 2 mM Mg2+. However, based on the clustered timing of the transitions, which varied from record to record and did not correspond to simple, exponential distributions (“heterogeneous dynamics”), the authors concluded that more than one distinct conformational state was likely present in each of the two FRET levels, which therefore represented “ensemble states” with potentially silent transitions. In this instance, it was not possible to assign unambiguously unique configurations to each of the FRET levels from the available data [54]. Any conformational changes presumed to be associated with folding within a given FRET level appeared to be slow with respect to the comparatively short single-molecule observation time (limited to τFRET~3.5 s), because changes in dynamic switching behavior were observed in only 1–5% of individual records (most of the observed variation was record-to-record). Here, further experiments using selected FRET pairs that supply additional conformational coordinates may be necessary to disambiguate the conformational states inferred from any dynamic analysis. Such an strategy was employed recently in a smFRET study of the TPP riboswitch [59] (section 4.5).
In the folding model for the metX riboswitch presented by Haller and coworkers, an “unfolded” state (open pseudoknot, but P1 stem folded) exhibits only transient access to a high-FRET, closed-pseudoknot state in the absence of magnesium. This state subsequently transitions to a pair of partly-folded, intermediate states (an open- and a closed-pseudoknot state) in a magnesium-dependent manner. These two intermediates were proposed to be a SAM binding-incompetent state with some triplex interactions formed (but an open pseudoknot) and a SAM binding-competent, partial-pseudoknot state (with a closed pseudoknot). SAM binding to the latter intermediate generates the native state with a stable pseudoknot [54]. Ligand binding was proposed to proceed by a conformational selection mechanism, with SAM-binding capturing the transient intermediate state, followed by adaptive rearrangements that complete the pseudoknot structure.
The proposal for a conformational selection mechanism was presumably influenced by the ready accessibility of the closed-pseudoknot conformation—and in particular, a SAM binding-competent state—even in the absence of ligand. However, we note that access to a fully folded state, F, in absence of ligand does not rule out the alternative induced fit mechanism, because the presence of the ligand may alter the dominant folding reaction pathway from type iii to either type i or ii (Figure 1B). The rate constant for the pseudoknot folding transition in the presence of SAM was not determined, precluding a quantitative comparison of the barrier heights for folding in the presence and absence of ligand (Figure 1A). Absent additional information, one cannot discriminate between induced fit and conformational selection models in this case.
4.8 The preQ1 class II riboswitch
In addition to adenine and guanine (sections 4.1–3), the purine riboswitch family includes members that respond to 7-aminomethyl-7-deazaguanine (preQ1), a precursor for the biosynthesis of the modified purine queuosine [94, 95]. Soulière et al. studied the class II preQ1 riboswitch from S. pneumonia R6 using TIRF-smFRET and SHAPE [61]. Much like the metX SAM-II riboswitch (section 4.7), the preQ1 riboswitch folds into a pseudoknot that sequesters the SD sequence of a downstream target gene, preventing translation initiation when the riboswitch is fully folded. Pseudoknot folding, and hence down-regulation of the target gene, is promoted by the preQ1 ligand. The preQ1 class II riboswitch is a variation on the standard H-type pseudoknot architecture, but with a nested helical element added: the P4 stem-loop (Figure 4H, left).
Two different FRET pairs were used in smFRET experiments that led to a kinetic model for riboswitch folding and preQ1 ligand binding [61]. In the absence of magnesium and preQ1 ligand, FRET labels placed on the J2–3 and SD sequences revealed a dominant open state (low-FRET) that frequently transitioned to a closed state (high-FRET). In 2 mM Mg2+, however, the riboswitch instead remained predominantly in the closed state, with transient visits to a slightly more compact open state. With both 2 mM Mg2+ and saturating ligand (100 µM) present, the closed state was stabilized even further. The authors developed a three-state model for folding and ligand binding, consisting of (1) an unfolded-pseudoknot state, (2) a partial-pseudoknot intermediate state promoted by magnesium, and (3) a fully folded pseudoknot state with preQ1 bound. A second FRET vector with labels on J2–3 and the P4 hairpin loop was also investigated. Based on a comparison of results from both FRET vectors, the P4 helix was reported to move flexibly relative to the stacking axis of the P2-P3 pseudoknot in all three folding states, but fluctuate more slowly as the pseudoknot became stabilized in states (2) and (3).
The FRET data from the SD–J2–3 pair shed light on the mechanism of ligand-promoted folding. In the presence of Mg2+ and saturating preQ1 ligand, kfold did not change, whereas kunfold was reduced by ~50% compared to experiments conducted in Mg2+ alone. This outcome is most consistent with a conformational selection mechanism: the folding energy barrier does not depend on [preQ1], but the folded state free energy in the presence of preQ1 is ~0.4 kcal/mol lower than in its absence (Figure 1A).
The P4 helix insert of the preQ1-II pseudoknot is not found in either the SAM-II (section 4.7) or preQ1-I pseudoknot riboswitches [61]. A P4 deletion mutant of the preQ1-II riboswitch was therefore constructed to explore the role of this additional element. In the presence of 2 mM Mg2+, the P4 deletion mutant exhibited reduced lifetimes for the partial-pseudoknot intermediate state and, when saturating preQ1 was introduced, for the fully folded pseudoknot state. On the other hand, the deletion of P4 increased the lifetime of the closed-pseudoknot state accessed in the absence of Mg2+ and preQ1. Hence, one role of P4 may be to destabilize the pseudoknot under conditions of low magnesium and ligand, reducing leaky down-regulation of downstream gene expression. However, the mutation-induced changes in the relative stability of the riboswitch folding states did not correspond very well with bulk measurements of ligand affinity in the deletion mutant [61]. This discrepancy may indicate that the identification of the fully folded state in the FRET data was incorrect or incomplete. It is also possible that (unreported) changes in the folded structure of the mutant reduced its ligand binding affinity. In this regard, the crystal structure of the ligand-bound class II preQ1 riboswitch from Lactobacillus rhamnosus revealed a binding site formed by a 3-way junction of P2, P3, and P4 [108]. This finding raises the possibility that the ΔP4 mutant of Soulière et al. [61], where the P4 helix was replaced by a single guanosine nucleotide, may compromise the preQ1 binding pocket, together with any binding interactions that it may form, even if the fold of the overall pseudoknot is preserved. Changes in these binding interactions may also explain the observed discrepancy between the folded state dwell times and the ligand affinity.
4.9 The c-di-GMP class I riboswitch
Cyclic diguanylate (c-di-GMP) is an important second messenger in a variety of bacterial species, where it globally regulates cellular activities that include motility, virulence gene expression, and biofilm formation [109]. The tfoX class I c-di-GMP riboswitch from Vibrio cholerae has a three-helix aptamer domain, with a three-way junction between the P1a, P1b, and P2 elements (Figure 4I). In the folded aptamer, the P1b and P2 arms dock, with a tetraloop-tetraloop receptor interaction formed between them (the tetraloop is at the end of P1b, and the receptor is in P2) [110, 111]. The ligand c-di-GMP binds to the aptamer domain in the three-helix junction, interacting with P1a, P1b, and the helix-helix junctions, and hence stabilizes the folded aptamer.
Wood et al. studied the tfoX riboswitch by TIRF-smFRET, with additional experimental support from isothermal titration calorimetry (ITC) [60]. Measurements were carried out on the tfoX aptamer with fluorescent labels situated on the P1b and P2 arms. Reminiscent of the data reported in the case of the metX-SAM-II riboswitch (section 4.7), heterogeneous dynamics were observed between two FRET states (high and low), with clustered timing for transitions that could not be modeled by simple exponential distributions, and which often varied from record to record. The presence or absence of such clustered transitions led the authors to propose the existence of both “static” and “dynamic” versions for each of the two FRET states, high (docked) and low (undocked). These four possibilities were modeled. The dynamic, docked state was postulated to correspond to a configurational intermediate in which the arms fluctuate between open and closed states. The authors conjectured that this intermediate is the c-di-GMP binding-competent state [60]. In their model, an undocked state can transition to this binding-competent, but unstable docked state, promoted by Mg2+. Indeed, ~96% of molecules were found in the static-undocked state in the absence of Mg2+, and ~95% remained trapped in that state under saturating c-di-GMP in a magnesium-free background. In the presence of Mg2+, the binding-competent intermediate state can transition to the static-docked state—presumably, the stable, active signaling state—promoted by c-di-GMP. However, even at high Mg2+, only ~10% of molecules were static-docked in the absence of c-di-GMP [60]. Hence the c-di-GMP ligand, in addition to Mg2+, was required for appreciable occupancy of the static-docked state. Because neither the ligand nor Mg2+ alone was sufficient for complete folding, it was proposed that Mg2+ directly mediates c-di-GMP binding [60]. Although the smFRET data may be interpreted in several other ways, that proposal is consistent with structural evidence for the position of bound Mg2+ in the riboswitch [112].
However, it is not entirely clear whether the dynamic, docked state is the sole binding-competent state. The available smFRET data in the presence of Mg2+ and c-di-GMP seem to indicate that the two undocked states (static and dynamic) might also be able to transition directly to the regulatory-active state (static, docked), corresponding to a complex free energy landscape with multiple pathways. Alternatively, additional transient intermediate states may exist that were missed altogether. Especially because of the apparent degeneracy in FRET values observed for different proposed conformations, it will be important in future experiments to explore additional FRET pairs that might supply information capable of disambiguating states whose existence can only be indirectly inferred from a kinetic analysis.
4.10 The lysine riboswitch
The known lysine riboswitches share an aptamer domain architecture consisting of a five-way helical junction (Figure 4J) which folds into a lysine-bound conformation dominated by three coaxial stacks (P1/P2/P2a, P2b/P3, and P4/P5) [113–116]. In the lysC lysine riboswitch from B. subtilis, nucleotides from the P1 hairpin of the aptamer can alternatively participate in the formation of an antiterminator hairpin. When the aptamer folds, the terminator hairpin can form and terminate transcription before the downstream lysC gene is reached. Lysine binding promotes aptamer folding, and hence transcription termination: the riboswitch thereby down-regulates gene expression in response to ligand. The lysC riboswitch controls expression of the gene for lysine-sensitive aspartokinase (AKIII), which catalyzes the first step in lysine biosynthesis [117].
Fiegland and colleagues studied the lysC riboswitch by bulk FRET in combination with three different smFRET approaches: TIRF-smFRET with tethered aptamers, raster-scanned smFRET of tethered aptamers using alternating-laser excitation (ALEX), and single-molecule burst FRET of free aptamers in solution with ALEX [52]. The use of multiple approaches facilitates measurements across a broad range of time scales. ALEX collects separate signals arising from the excitation of the donor or acceptor dyes, and thereby makes it possible to exclude data from any molecules carrying just a single active fluorophore, arising, for example, from dye bleaching or incomplete labeling.
smFRET was performed with labels placed at the junction-distal ends of the P1 and P5 elements. Fiegland et al. observed that over the 2–500 s observation time, ~30% of molecules remained in a persistent low-FRET state, regardless of the lysine concentration [52]. In raster-scanned ALEX-smFRET of surface-attached aptamers, and depending upon the lysine concentration, this low-FRET state coexisted with (1) a more compact (higher-FRET) state, interpreted as a lysine-binding competent “open” state, and (2) an even higher-FRET state, interpreted as the lysine-bound “closed” state. The persistent low-FRET state was interpreted as corresponding to molecular conformations with inactive binding pockets. In principle, a variety of different inactive forms could contribute to this unresponsive, low-FRET population, which was evidently long-lived (no switching between active and inactive forms was reported). On the one hand, unresponsive molecules could arise from misfolded states with biological relevance. On the other, such molecules could simply be a byproduct of partial purification or incomplete annealing procedures used to prepare them. Single-molecule urea denaturation, followed by renaturation [58], might supply further insights into the identity of the unresponsive population. The ~70% of molecules that were not in a static, low-FRET state exhibited rapid hopping between open and closed states.
Fiegland et al. did not address the issue of induced fit vs. conformational selection. However, the kinetic scheme they proposed may be mapped to an induced-fit model of the type shown in pathway ii in Figure 1B, namely, a 3-state model with an open state interconverting with a lysine-bound open state, which, in turn, exchanges with a lysine-bound closed state. The rate for the overall open-to-closed transition, kclose, increased dramatically with increasing ligand concentration. In fact, in the absence of lysine, the closed state was not observed. Over the range of lysine concentrations tested (25–2000 µM), kclose increased 20-fold, corresponding to an activation energy reduction of ~1.7 kcal/mol. The activation energy barrier for aptamer folding thus is substantially lowered in the presence of ligand (Figure 1). On the other hand, the opening rate, kopen, was not found to depend appreciably on lysine concentration, suggesting that lysine may lower the closed state free energy about as much as it does the open-to-closed activation energy.
5. Discussion
5.1 Identification of riboswitch folding states
In a growing number of cases, novel transient folding states have been identified and measured in single-molecule experiments, and these states help to inform our understanding of both folding and gene regulation. Such identifications are facilitated by the excellent spatiotemporal resolution achievable through single-molecule techniques, as well as by the absence of population averaging, which can confound interpretations of bulk measurements. In smFRET, intermediate folding states can sometimes be detected directly, as transient states with their own characteristic FRET efficiencies. Other times, intermediate states can only be inferred indirectly, based on a kinetic analysis of the transitions among various FRET states, which may be degenerate with respect to some molecular configurations (section 5.2). In either case, the fundamental experimental challenge is first to identify the detectable states, then to assign candidate molecular configurations to each of these states, and thereby to build up a picture of the molecular trajectories through which riboswitch folding occurs. In many cases, a single, strategically placed FRET dye pair will not suffice to resolve all the relevant RNA conformations. In such situations, additional dye pair labels, used in separate experiments or simultaneously (section 6), may become necessary.
In OT experiments, transient intermediate folding states are most often detected directly, as characteristic changes in the end-to-end extensions of individual molecules that have been subjected to controlled forces by an optical trap. Such changes may either be recorded near thermal equilibrium (e.g., using force clamps) or well away from equilibrium (e.g., using force ramps): a variety of theoretical analysis methods has been developed to handle each case. Here again, the fundamental experimental challenge is to identify all detectable states and to assign molecular configurations to each of these, thereby building up a picture of folding. However, not all the molecular rearrangements of interest may generate changes in the end-to-end extension of a given riboswitch. Conversely, certain molecular rearrangements may generate similar changes in extension, and therefore require further means of disambiguation. In such cases, additional experimental strategies may be called for. These include attaching force probes to different parts of the riboswitch (section 6), using mutant constructs, assessing the effects of ligand (or ligand-analog) binding on folding energetics and kinetics, or subdividing the molecule and performing measurements on its constituent parts. Optical traps may also be usefully combined with FRET or other single-molecule approaches (section 6).
We note that even when most of the relevant folding states can be identified and scored from transitions in single-molecule records (either directly or indirectly), it is nevertheless a challenge to map these identified states to specific conformations of the riboswitch. The final, ligand-bound form of the aptamer may be known from independent structural studies (crystallography, NMR), but all the other folding forms—particularly any transient intermediates—tend to be largely conjectural. Evidence for their assignment is therefore indirect, of necessity, and a certain amount of informed guesswork is usually involved. Strong clues generally come from the transitions themselves, which produce specific changes in molecular length or FRET distance. Further clues may be gained from computational or structural modeling methods, but these approaches are insufficiently mature to be fully trustworthy, and they are more predictive of secondary than tertiary structure. Useful information may be developed in additional experiments, for example, by studying the sensitivity of any intermediate structures to chaotropic agents, metal ions, cross-linking reagents, small reactive species, and the like. In selected cases, the addition of short blocking oligonucleotide sequences, which pair with targeted structural elements and destabilize these, can help to confirm structural assignments. Similarly, studies of partial sequences or engineered mutant constructs can build confidence by providing ways to test candidate structures. That said, the assignment of experimentally-observed folding states to structural intermediates remains part art, part science.
Notwithstanding such caveats, the identification of metastable states in the folding landscape, including intermediate and misfolded states, is critical to our understanding of riboswitching at a mechanistic level: namely, how the riboswitch regulates its gene, and what role the target ligand plays in that process. Using smFRET, a misfolded (or inactive) state was detected in the lysC riboswitch aptamer (section 4.10), and folding intermediates have been directly observed in the add, xpt, thiM, and metI aptamers (sections 4.2, 4.3, 4.5, 4.6). OT studies have revealed misfolded and intermediate states in the add riboswitch, and intermediate states were also reported for the pbuE and thiC riboswitches (sections 4.1, 4.4).
The identification of transient states also represents an important step towards developing a finer-grained view of the energy landscapes upon which riboswitches fold. This, in turn, is critical for gaining an understanding of how such states interact with the target ligand and trigger switching activity. Intermediate states expose details of the landscape that are particularly important for understanding those riboswitches that act co-transcriptionally. Because the competition between aptamer-and expression platform in this context is highly sensitive to the presence of kinetic traps, certain folding intermediates can play an outsized role in the regulatory decision, in a manner which cannot be handled satisfactorily by existing theoretical or computational approaches (however, recent work involving the prediction of different possible secondary structures during simulated elongation of nascent riboswitch RNAs shows promise [118, 119]). Studies of folding intermediates can also reveal the roles played by divalent cations (section 5.5) and the interplay between secondary and tertiary structure formation (section 5.4).
5.2 Insights from conformational state dwell times
Measurements of the dwell times for riboswitch folding states have proved to be especially informative. In many cases, where abrupt transitions among distinct states can be scored in records of smFRET or OT experiments, the associated distributions of dwell time can be reconstructed directly. In other cases, for example, when folding states share similar FRET efficiency or molecular extension, the identification of folding states and the reconstruction of their dwell times can nevertheless be carried out indirectly, using appropriate modeling (e.g., HMM methods) and kinetic analysis. The observation of “heterogeneous dynamics” in transitions between otherwise similar FRET levels has led to the inference of additional folding states in the add, thiM, and tfoX riboswitch aptamers (sections 4.2, 4.5, 4.9). Slowly interchanging dynamic behaviors were also observed for the metX riboswitch aptamer, but further work may be needed before the inferred states can be assigned to candidate configurations (section 4.7).
Heterogeneities observed in the average dwell times from one single-molecule record to the next may, in principle, reflect a complex energy landscape: one containing multiple folded or docked states, producing more than one dominant folding trajectory. [Alternatively, folding heterogeneity may simply reflect residual subpopulations of malformed or inactive molecules in the experimental preparation. It is not trivial to distinguish these possibilities, in practice.] For example, two different docked states, whose relative populations depended upon the adenine concentration, were reported for the add riboswitch by Dalgarno et al. [58] (section 4.2); these states were revealed in the presence of urea. By contrast, different docked states were not apparent from smFRET dwell time distributions in several other purine riboswitch studies [55–57].
In the case of the add adenine riboswitch (under urea-free conditions), the single-record average dwell times for both the docked and the undocked states ranged over a factor of 100 in the absence of adenine, but this variation was reportedly reduced 10-fold in the presence of adenine (50 µM) [57]. This finding raises the possibility that one role of adenine may be to restrict the states available for folding. Supporting this interpretation, a constitutively “on” mutant of the add riboswitch also exhibited a lower variation in average dwell times in the absence of ligand, although the reduction was only 2-fold in that case [56]. However, for a different purine riboswitch (xpt guanine, section 4.3), the variation in dwell times for the docked and undocked states was also a factor of ~100 [55], but the addition of ligand had no substantial effect on that variation. This divergent behavior might reflect different ligand-binding mechanisms used by the xpt and add riboswitches, as suggested by single-molecule experiments (sections 4.2, 4.3, 5.3). However, one technical concern that we raise for dwell time estimates is that these can be very sensitive to the durations of the individual records collected, particularly when such records are limited by comparatively short dye lifetimes, and therefore not significantly longer than some of the transition times being estimated. Another concern relates to how any “variation” is actually determined (including the type of metric employed, which would seem to be the range and not the variance), and its level of statistical significance.
5.3 Ligand roles in riboswitch folding: induced fit vs. conformational selection
Several single-molecule investigations have provided evidence regarding the question of whether the activation of a riboswitch by its cognate ligand is best understood in terms of an induced fit or a conformational selection model. As discussed, these competing models differ critically in the role played by the ligand in the folding pathway (section 2). In practice, distinguishing these possibilities by experiment can be difficult, and fraught with ambiguity.
Some riboswitches appear to follow the induced fit mechanism. Evidence from single-molecule experiments has provided support for induced fit in the cases of the add adenine riboswitch (section 4.2), the thiC TPP riboswitch (section 4.4), and the lysC lysine riboswitch (section 4.10). Conformational selection also appears to obtain for some riboswitches: single-molecule results consistent with that mechanism have been reported for the xpt guanine riboswitch (section 4.3) and the preQ1-II riboswitch (section 4.8). Caution should be exercised in considering the heights of energy barriers and well depths derived from smFRET data, however, because the reported energies would not seem to match up particularly well, in several instances, with estimates from other techniques. For example, one bulk study of adenine binding to the add aptamer using 2-AP fluorescence reported a binding energy of ~8 kcal/mol [120], which agreed closely with the ligand-induced stabilization of the folded aptamer derived from single-molecule OT data (ΔΔG ≈ −8 kcal/mol) [86]. However, a different value was estimated from smFRET data (ΔΔG ≈ −0.65 kcal/mol) [57]. On the other hand, smFRET results for the add riboswitch in the presence of urea [58] were more consistent with those derived from OT data (section 4.2), as well as with 2-AP fluorescence measurements. Possible explanations for such discrepancies were discussed (section 4.2). In part, discrepancies in measurements of ΔΔG (the stabilization energy of binding) and ΔΔG‡ (the barrier height change for binding) may be associated with difficulties in discriminating ligand-free from ligand-bound states using smFRET alone. This contrasts with the situation in OT, where such states are readily distinguished by their differential response to unfolding force. In future work, more definitive experimental support for one binding mechanism or the other might be obtained by measuring two (or more) different reaction coordinates simultaneously (section 6).
5.4 The hierarchical folding model and riboswitches
Typically, smFRET investigations have not probed the folding dynamics of riboswitch secondary structure. Most of the duplex elements found in riboswitches, particularly the sensor arms, are sufficiently stable that the unfolded state has negligible occupancy in physiological buffers at room temperature. For example, a computation of the stability of the P2 hairpin from the add aptamer using mfold [121] predicts just a 0.1% chance of occupying the unfolded state. Nevertheless, even for stem-loop elements with high stability, transient openings (fraying) near helix bases or bulges may be critical for riboswitch folding or switching. Ensemble measurements of secondary structure formation are also problematic, as these may miss folding states, owing to the averaging over fast dynamics across many molecules (typically ~10–100 µs [122, 123]). The ability of optical trapping to bias the folding energy landscape using force, combined with its comparatively good time resolution (typically ~1 msec) puts it in a fairly unique position to provide an in-depth examination of secondary structure formation, along with its relationship to tertiary structure formation. Success has already been achieved in reconstructing, in a stepwise fashion, folding-energy landscapes that span the range from the completely unfolded form to the fully-folded, ligand bound form for the pbuE and add (adenine) [80, 86] and thiC (TPP) [103] riboswitch aptamers (sections 4.1, 4.2, and 4.4).
The thiC TPP riboswitch aptamer (Figure 4D) appears to follow a hierarchical folding scheme, with mechanical renaturation experiments showing that half of P3 folds first, followed by P5, P4, and then the other half of P3, followed by P2, and finally P1. In the presence of TPP, ligand binding and the formation of tertiary interactions after P1 folding were inferred from the mechanical stabilization of the P1-folded form, although tertiary interactions were not directly observed along the extension coordinate.
Several riboswitch studies have now shown that, contrary to the hierarchical picture of folding, the sequence of secondary and tertiary folding events can interleave in the “nearly-folded” regime, particularly when the structural elements in question have comparable folding energies (section 4.1). For the add and pbuE riboswitches, the P1 helix folds concomitant with tertiary structure formation via a structural intermediate with P1 unfolded, but with some tertiary contacts formed: this intermediate is thought to reflect a pre-organization of the ligand-binding pocket [80, 86] (sections 4.1 and 4.2). When smFRET was used to investigate the folding of the P1 “switch helix” directly in the thiM TPP riboswitch aptamer (section 4.5), the results suggested linked dynamics for P1 duplex folding and sensor arm docking [59]. This relationship is central to how aptamer folding and ligand binding jointly stabilize the switch helix.
A grand challenge for future single-molecule riboswitch studies will be to go beyond the folding of isolated aptamer regions to study the dynamic competition between aptamers and expression platforms in a functional (i.e., in situ) context. A start along these lines has already been made with an investigation of cotranscriptional folding in the complete pbuE riboswitch [81] (section 4.1), whose expression platform regulates gene expression via a transcriptional terminator. Owing to their greater complexity, still greater experimental challenges await with riboswitches whose expression platforms regulate protein translation or alternative RNA splicing.
5.5 The role of metal ions in riboswitch folding
Magnesium ions are critical for the stabilization of complex, structured RNAs [124], and even simple hairpins are strongly stabilized by divalent cations [28]. In riboswitches, magnesium seems to promote folding intermediates and/or fully folded states that are competent to bind ligand. With experimental methods that are sensitive to the formation of transient intermediates and can follow individual molecular folding trajectories, it becomes possible to determine, in certain cases, specific states that may be stabilized by magnesium, in addition to observing gradual compaction as a function of [Mg2+], which is something that can also be measured by ensemble methods (such as bulk FRET).
In the case of the xpt guanine, metI SAM-I, metX SAM-II, and tfoX c-di-GMP riboswitches (sections 4.3, 4.6, 4.7, 4.9), magnesium appears to be required for the formation of a stable folded or docked state. For example, the metX riboswitch is found predominantly in an unfolded state in the absence of magnesium and SAM, but makes frequent and short-lived (~30 ms) fluctuations to a closed pseudoknot form (section 4.7). The addition of 2 mM Mg2+ strongly modulates metX folding behavior, eliciting a ~50% occupancy of a compact, low-FRET intermediate state (open pseudoknot) and ~50% occupancy of a high-FRET state (consisting of two closed pseudoknot forms) [54].
In other cases, such as the preQ1-II riboswitch (section 4.8) and a mutant add adenine riboswitch (section 5.2), magnesium effects a modest increase in folded state occupancy, but folded states are readily accessible even in its absence [56, 61]. For the add mutant in NaCl alone (50 mM), 38% of molecules were already in a docked state, in stark contrast to the wild-type aptamer in the absence of magnesium (section 4.2) [57]. The addition of 2 mM magnesium to the mutant aptamer produced an 83% docked-state occupancy. The mutant therefore appears to fold by a different pathway than the wild-type, with a dramatically reduced magnesium dependence: this interpretation would be consistent with the lack of an observable folding intermediate in the mutant [56], in contrast to the wild-type [57].
Limited insights into possible roles for specifically bound magnesium ions have been obtained from some single molecule studies, including investigations of the add and tfoX riboswitches (sections 4.2, 4.9). Analysis of add dwell-time histograms for the docked and undocked states showed that, with adenine present, the addition of 2 M urea reduced the docking rate by ~2-fold in the presence of 4 mM Mg2+, but by ~8-fold in the presence of 0.1 mM Mg2+ [58]. According to a mechanism proposed by Sosnick and Pan [125], the enhanced effect of urea at low magnesium concentration is consistent with a folding pathway involving a rate-limiting step that includes the formation of a metal-ion binding site around one (or more) of the five specifically bound magnesium ions identified by crystallography [79]. In future work, characterization of the effects of magnesium on high-resolution folding landscapes may supply more mechanistic insights into the roles played by any specifically bound, as well as “screening,” cations.
6. Conclusion and Future Directions
Without doubt, single-molecule approaches have substantially improved our working understanding of riboswitches. In particular, single-molecule studies have revealed sequences of folding events for the various elements comprising individual riboswitch aptamers, and in selected cases, have permitted reconstructions of entire folding energy landscapes. Novel intermediate states (and ensembles of such states) have been successfully identified in single molecule records. So, too, have individual folding trajectories. Fresh insights have been gleaned into the roles played by both ligand and metal-ion binding in the aptamer folding process, including increasingly detailed views of transient structures, which are notoriously hard to obtain by any other means. In particular, the specific effects of ligand binding on energy landscapes have helped to clarify the fundamental mechanisms for ligand-promoted folding (i.e., induced fit vs. conformational selection). Recently, single-molecule work has moved beyond studies of isolated aptamers to explore the actual riboswitching mechanism, that is, the competition between the aptamer region and the expression platform that leads to gene regulation. Such work makes it possible not only to distinguish between kinetic and thermodynamic control mechanisms, but also to improve our understanding of how RNA folds co-transcriptionally inside cells.
On the theoretical side, advanced analytical methods, based on non-equilibrium thermodynamics and statistical mechanics, have been expressly developed for extracting useful information from single-molecule records. Such methods are increasingly being brought to bear on work with structured RNAs. At the same time, experimental findings from single-molecule techniques also provide a convenient “proving ground” for the validation of structural predictions, including dynamics, generated by increasingly sophisticated computational methods.
Experimental difficulties persist for the twin challenges faced in single-molecule work on riboswitches (section 5.1), namely: (1) the identification of all relevant intermediate folding states, and (2) the assignment of such folding states to candidate structures. Typically, structural assignments cannot be based on single-molecule data alone, and therefore often involve an inspired fusion of structural, computational, biochemical, genetic, and single-molecule information. With current techniques, it is often ambiguous whether an observed transition corresponds to a change in secondary or tertiary structure. A single experimental coordinate for folding—such as the end-to-end molecular extension, or the FRET distance between strategically placed dyes—may not be able to discriminate among relevant conformations [19, 74]. However well-chosen, any one reaction coordinate, which projects a high-dimensional folding space into a single dimension, cannot be expected to fully recapitulate the folding process except under exceptional circumstances.
In principle, this limitation could be relieved by simultaneously recording single-molecule data along separate, approximately “orthogonal” reaction coordinates that are sensitive to different aspects of folding. A number of smFRET studies have already compared—albeit, in separate experiments—constructs carrying different FRET pairs, to obtain a more complete picture of the folding process [55, 59, 61]. In a similar vein, ‘click’ chemistry [126] (or similar) could be used to bioconjugate handles to internal sites of a riboswitch, rather than hybridizing these to the 3′ and 5′ ends, and thereby permit a different pulling coordinate to be used in OT assays. But true simultaneous measurement along more than one reaction coordinate could provide still greater insight, helping to address such issues as secondary vs. tertiary structure formation, coupling between the folding of different structural elements, and discrimination between ligand-bound and ligand-free states [59, 80, 86, 103]. New methodologies to record more than one reaction coordinate, which are being vigorously pursued but are still in their infancy, include combined OT and smFRET (“Force-FRET”) [89, 127–130], multicolor smFRET [87, 88] (which can also be combined with OT [131]), and smFRET combined with AFM [132, 133] or magnetic tweezers [134]. Each single-molecule approach individually poses its own unique set of technical challenges, and these tend to get compounded when techniques are combined. Nevertheless, success on any of these fronts has the potential to improve our understanding of gene regulatory mechanisms.
Highlights.
Single-molecule studies of riboswitch folding are reviewed
Insights gained into general themes of riboswitch folding are discussed
Unanswered questions, limitations, and future directions for the field are considered
Acknowledgements
The authors thank Irena Hwang-Fisher for able assistance with graphics. A.S. was supported by an NSF predoctoral fellowship. This work was supported by grant GM57035 to S.M.B. from the NIH.
Abbreviations used
- OT
Optical trapping
- smFRET
single-molecule FRET
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anfinsen CB. Principles that govern folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Anfinsen CB, Haber E, Sela M, White FH. Kinetics of formation of native ribonuclease during oxidation of reduced polypeptide chain. Proc Natl Acad Sci U S A. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rother K, Rother M, Boniecki M, Puton T, Bujnicki JM. RNA and protein 3D structure modeling: similarities and differences. J Mol Model. 2011;17:2325–2336. doi: 10.1007/s00894-010-0951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laing C, Schlick T. Computational approaches to RNA structure prediction, analysis, and design. Curr Opin Struct Biol. 2011;21:306–318. doi: 10.1016/j.sbi.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris ER, Searle MS. Overview of protein folding mechanisms: experimental and theoretical approaches to probing energy landscapes. Curr Protoc Protein Sci, Chapter. 2012;28:1–22. doi: 10.1002/0471140864.ps2802s68. [DOI] [PubMed] [Google Scholar]
- 7.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 8.Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Gene Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang XW, Ha T, Kim HD, Centner T, Labeit S, Chu S. Fluorescence quenching: A tool for single-molecule protein-folding study. Proc Natl Acad Sci U S A. 2000;97:14241–14244. doi: 10.1073/pnas.97.26.14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodside MT, Garcia-Garcia C, Block SM. Folding and unfolding single RNA molecules undertension. Curr Opin Chem Biol. 2008;12:640–646. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heus HA, Puchner EM, Vugt-Jonker AJvan, Zimmermann JL, Gaub HE. Atomic force microscope-based single-molecule force spectroscopy of RNA unfolding. Anal Biochem. 2011;414:1–6. doi: 10.1016/j.ab.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Wen JD, Manosas M, Li PTX, Smith SB, Bustamante C, Ritort F, Tinoco I. Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys J. 2007;92:2996–3009. doi: 10.1529/biophysj.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlaminck IDe, Dekker C. Recent advances in magnetic tweezers. Annu Rev Biophys. 2012;41:453–472. doi: 10.1146/annurev-biophys-122311-100544. [DOI] [PubMed] [Google Scholar]
- 15.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neupane K, Ritchie DB, Yu H, Foster DA, Wang F, Woodside MT. Transition path times for nucleic acid folding determined from energy-landscape analysis of single-molecule trajectories. Phys Rev Lett. 2012;109:068102. doi: 10.1103/PhysRevLett.109.068102. [DOI] [PubMed] [Google Scholar]
- 17.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 18.Woodside MT, Block SM. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu Rev Biophys. doi: 10.1146/annurev-biophys-051013-022754. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best RB, Paci E, Hummer G, Dudko OK. Pulling direction as a reaction coordinate for the mechanical unfolding of single molecules. J Phys Chem B. 2008;112:5968–5976. doi: 10.1021/jp075955j. [DOI] [PubMed] [Google Scholar]
- 20.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 22.Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DMJ, Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci U S A. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levinthal C. How to fold graciously. In: Marcus RA, editor. Mossbaun Spectroscopy in Biological Systems Proceedings. Vol. 67. Urbana, IL: University of Illinois; 1969. pp. 22–24. 61801, 1969. [Google Scholar]
- 24.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 25.Pan J, Thirumalai D, Woodson SA. Folding of RNA involves parallel pathways. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 26.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struc Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 27.Russell R, Das R, Suh H, Traver KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Bioph Biom. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 29.Tinoco I, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 30.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J Mol Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 31.Pereira MJB, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. Single VS ribozyme molecules reveal dynamic and hierarchical folding toward catalysis. J Mol Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duncan CDS, Weeks KM. Nonhierarchical ribonucleoprotein assembly suggests a strain-propagation model for protein-facilitated RNA folding. Biochemistry. 2010;49:5418–5425. doi: 10.1021/bi100267g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Misra VK, Draper DE. Mg(2+) binding to tRNA revisited: the nonlinear Poisson-Boltzmann model. J Mol Biol. 2000;299:813–825. doi: 10.1006/jmbi.2000.3769. [DOI] [PubMed] [Google Scholar]
- 35.Misra VK, Draper DE. The interpretation of Mg2+ binding isotherms for nucleic acids using Poisson-Boltzmann theory. J Mol Biol. 1999;294:1135–1147. doi: 10.1006/jmbi.1999.3334. [DOI] [PubMed] [Google Scholar]
- 36.Misra VK, Draper DE. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 37.Misra VK, Shiman R, Draper DE. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- 38.Anthony PC, Sim AYL, Chu VB, Doniach S, Block SM, Herschlag D. Electrostatics of nucleic acid folding under conformational constraint. J Am Chem Soc. 2012;134:4607–4614. doi: 10.1021/ja208466h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: A flux description of reaction mechanism. Proc Natl Acad Sci U S A. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176–181. doi: 10.1016/j.mib.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Lau MW, Ferre-D’Amare AR. Ribozymes and Riboswitches: Modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira MJB, Behera V, Walter NG. Nondenaturing purification of co-transcriptionally folded RNA avoids common folding heterogeneity. PLOS One. 2010;5 doi: 10.1371/journal.pone.0012953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer IM, Miklos I. Co-transcriptional folding is encoded within RNA genes. Bmc Mol Biol. 2004;5 doi: 10.1186/1471-2199-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 45.Fabian H, Naumann D. Methods to study protein folding by stopped-flow FT-IR. Methods. 2004;34:28–40. doi: 10.1016/j.ymeth.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Kubelka J. Time-resolved methods in biophysics. 9. Laser temperature-jump methods for investigating biomolecular dynamics. Photoch Photobio Sci. 2009;8:499–512. doi: 10.1039/b819929a. [DOI] [PubMed] [Google Scholar]
- 47.Preus S, Wilhelmsson LM. Advances in quantitative FRET-based methods for studying nucleic acids. Chembiochem. 2012;13:1990–2001. doi: 10.1002/cbic.201200400. [DOI] [PubMed] [Google Scholar]
- 48.Wu PG, Brand L. Resonance energy-transfer - methods and applications. Anal Biochem. 1994;218:1–13. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 49.Lilley DMJ. The structure and folding of branched RNA analyzed by fluorescence resonance energy transfer. Methods Enzymol. 2009;469:159–187. doi: 10.1016/S0076-6879(09)69008-X. [DOI] [PubMed] [Google Scholar]
- 50.Rheenen Jvan, Langeslag M, Jalink K. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys J. 2004;86:2517–2529. doi: 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemay JF, Penedo JC, Mulhbacher J, Lafontaine DA. Molecular basis of RNA-mediated gene regulation on the adenine riboswitch by single-molecule approaches. Methods Mol Biol. 2009;540:65–76. doi: 10.1007/978-1-59745-558-9_6. [DOI] [PubMed] [Google Scholar]
- 52.Fiegland LR, Garst AD, Batey RT, Nesbitt DJ. Single-molecule studies of the lysine riboswitch reveal effector-dependent conformational dynamics of the aptamer domain. Biochemistry. 2012;51:9223–9233. doi: 10.1021/bi3007753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7:384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 54.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 55.Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49:1596–1605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tremblay R, Lemay JF, Blouin S, Mulhbacher J, Bonneau E, Legault P, Dupont P, Penedo JC, Lafontaine DA. Constitutive regulatory activity of an evolutionarily excluded riboswitch variant. J Biol Chem. 2011;286:27406–27415. doi: 10.1074/jbc.M111.229047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 58.Dalgarno PA, Bordello J, Morris R, St-Pierre P, Dube A, Samuel ID, Lafontaine DA, Penedo JC. Single-molecule chemical denaturation of riboswitches. Nucleic Acids Res. 2013;41:4253–4265. doi: 10.1093/nar/gkt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haller A, Altman RB, Souliere MF, Blanchard SC, Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci U S A. 2013;110:4188–4193. doi: 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood S, Ferre-D’Amare AR, Rueda D. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem Biol. 2012;7:920–927. doi: 10.1021/cb300014u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Souliere MF, Altman RB, Schwarz V, Haller A, Blanchard SC, Micura R. Tuning a riboswitch response through structural extension of a pseudoknot. Proc Natl Acad Sci U S A. 2013;110:E3256–E3264. doi: 10.1073/pnas.1304585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suddala KC, Rinaldi AJ, Feng J, Mustoe AM, Eichhorn CD, Liberman JA, Wedekind JE, Al-Hashimi HM, Brooks CL, 3rd, Walter NG. Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res. 2013;41:10462–10475. doi: 10.1093/nar/gkt798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Q, Juette MF, Jockusch S, Wasserman MR, Zhou Z, Altman RB, Blanchard SC. Ultra-stable organic fluorophores for single-molecule research. Chem Soc Rev. 2014;43:1044–1056. doi: 10.1039/c3cs60237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altman RB, Terry DS, Zhou Z, Zheng Q, Geggier P, Kolster RA, Zhao Y, Javitch JA, Warren JD, Blanchard SC. Cyanine fluorophore derivatives with enhanced photostability. Nat Methods. 2012;9:68–71. doi: 10.1038/nmeth.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blanco M, Walter NG. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010;472:153–178. doi: 10.1016/S0076-6879(10)72011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Svoboda K, Block SM. Biological applications of optical forces. Annu Rev Biophys Biomol Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 68.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gebhardt JCM, Bornschlogla T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc Natl Acad Sci U S A. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of Lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 72.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 73.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophys J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dudko OK, Hummer G, Szabo A, Theory analysis. and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci U S A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collin D, Ritort F, Jarzynski C, Smith SB, Tinoco I, Bustamante C. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature. 2005;437:231–234. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive all-optical force clamp for high-resolution laser trapping. Phys Rev Lett. 2005;95:208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta AN, Vincent A, Neupane K, Yu H, Wang F, Woodside MT. Experimental validation of free-energy-landscape reconstruction from non-equilibrium single-molecule force spectroscopy measurements. Nat Phys. 2011;7:631–634. [Google Scholar]
- 79.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem & Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLOS Biol. 2010;8 doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xayaphoummine A, Viasnoff V, Harlepp S, Isambert H. Encoding folding paths of RNA switches. Nucleic Acids Res. 2007;35:614–622. doi: 10.1093/nar/gkl1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLOS Genet. 2011;7 doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neupane K, Yu H, Foster DA, Wang F, Woodside MT. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39:7677–7687. doi: 10.1093/nar/gkr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S. Single-molecule four-color FRET. Angew Chem Int Edit. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lang MJ, Fordyce PM, Block SM. Combined optical trapping and single-molecule fluorescence. J Biol. 2003;2:6. doi: 10.1186/1475-4924-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou RB, Schlierf M, Ha T. Force-fluorescence spectroscopy at the single-molecule level. Methods Enzymol. 2010;475:405–426. doi: 10.1016/S0076-6879(10)75016-3. [DOI] [PubMed] [Google Scholar]
- 91.Hohng S, Zhou RB, Nahas MK, Yu J, Schulten K, Lilley DMJ, Ha TJ. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.England JL, Haran G. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annu Rev Phys Chem. 2011;62:257–277. doi: 10.1146/annurev-physchem-032210-103531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sullan RMA, Churnside AB, Nguyen DM, Bull MS, Perkins TT. Atomic force microscopy with sub-picoNewton force stability for biological applications. Methods. 2013;60:131–141. doi: 10.1016/j.ymeth.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Batey RT. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim JN, Breaker RR. Purine sensing by riboswitches. Biol Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 96.Li SS, Breaker RR. Eukaryotic TPP riboswitch regulation of alternative splicing involving longdistance base pairing. Nucleic Acids Res. 2013;41:3022–3031. doi: 10.1093/nar/gkt057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Breaker RR. Riboswitches and the RNA World. Csh Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 99.Thore S, Leibundgut M, Ban NN. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 100.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2010;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 102.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3’ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anthony PC, Perez CF, Garcia-Garcia C, Block SM. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci U S A. 2012;109:1485–1489. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem & Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 105.Batey RT. Recognition of S-adenosylmethionine by riboswitches. WIREs RNA. 2011;2:299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eschbach SH, St-Pierre P, Penedo JC, Lafontaine DA. Folding of the SAM-I riboswitch: a tale with a twist. RNA Biology. 2012;9:535–541. doi: 10.4161/rna.19648. [DOI] [PubMed] [Google Scholar]
- 107.Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat Struct Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 108.Liberman JA, Salim M, Krucinska J, Wedekind JE. Structure of a class II preQ(1) riboswitch reveals ligand recognition by a new fold. Nat Chem Biol. 2013;9:353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith KD, Lipchock SV, Livinston AL, Shanahan CA, Strobel SA. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry. 2010;49:7351–7359. doi: 10.1021/bi100671e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Serganov A, Huang LL, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci U S A. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Gene Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cassan M, Ronceray J, Patte JC. Nucleotide sequence of the promoter region of the E. coli lysC gene. Nucleic Acids Res. 1983;11:6157–6166. doi: 10.1093/nar/11.18.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Quarta G, Sin K, Schlick T. Dynamic energy landscapes of riboswitches help interpret conformational rearrangements and function. PLOS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quarta G, Kim N, Izzo JA, Schlick T. Analysis of riboswitch structure and function by an energy landscape framework. J Mol Biol. 2009;393:993–1003. doi: 10.1016/j.jmb.2009.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 121.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Draper DE. Parallel worlds. Nat Struct Biol. 1996;3:397–400. doi: 10.1038/nsb0596-397. [DOI] [PubMed] [Google Scholar]
- 123.Thirumalai D, Woodson SA. Maximizing RNA folding rates: A balancing act. RNA. 2000;6:790–794. doi: 10.1017/s1355838200000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sosnick TR, Pan T. RNA folding: models and perspectives. Curr Opin Struc Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 126.Yu ZB, Koirala D, Cui YX, Easterling LF, Zhao Y, Mao HB. Click chemistry assisted single-molecule fingerprinting reveals a 3D biomolecular folding funnel. J Am Chem Soc. 2012;134:12338–12341. doi: 10.1021/ja303218s. [DOI] [PubMed] [Google Scholar]
- 127.Gross P, Farge G, Peterman EJ, Wuite GJ, tweezers Combining optical, microscopy single-molecule fluorescence. and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 2010;475:427–453. doi: 10.1016/S0076-6879(10)75017-5. [DOI] [PubMed] [Google Scholar]
- 128.Sirinakis G, Ren Y, Gao Y, Xi Z, Zhang Y. Combined versatile high-resolution optical tweezers and single-molecule fluorescence microscopy. Rev Sci Instrum. 2012;83:093708. doi: 10.1063/1.4752190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tarsa PB, Brau RR, Barch M, Ferrer JM, Freyzon Y, Matsudaira P, Lang MJ. Detecting force-induced molecular transitions with fluorescence resonant energy transfer. Angew Chem Int Edit. 2007;46:1999–2001. doi: 10.1002/anie.200604546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat Methods. 2011;8:335–U382. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee S, Hohng S. An optical trap combined with three-color FRET. J Am Chem Soc. 2013;135:18260–18263. doi: 10.1021/ja408767p. [DOI] [PubMed] [Google Scholar]
- 132.He Y, Lu M, Cao J, Lu HP. Manipulating protein conformations by single-molecule AFM-FRET nanoscopy. ACS Nano. 2012;6:1221–1229. doi: 10.1021/nn2038669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gumpp H, Stahl SW, Strackharn M, Puchner EM, Gaub HE. Ultrastable combined atomic force and total internal reflection fluorescence microscope [corrected] Rev Sci Instrum. 2009;80:063704. doi: 10.1063/1.3148224. [DOI] [PubMed] [Google Scholar]
- 134.Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A, Liphardt J. Biocompatible force sensor with optical readout and dimensions of 6 nm(3) Nano Lett. 2005;5:1509–1514. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]