Abstract
The recent discovery of short cis-acting RNA elements termed riboswitches has caused a paradigm shift in our understanding of genetic regulatory mechanisms. The three distinct superfamilies of S-adenosyl-L-methionine (SAM) riboswitches are the most commonly found riboswitch classes in nature. These RNAs represent three independent evolutionary solutions to achieve specific SAM recognition. This review summarizes research on 1) modes of gene regulatory mechanisms, 2) common themes and differences in ligand recognition, and 3) ligand-induced conformational dynamics among SAM riboswitch families. The body of work on the SAM riboswitch families constitutes a useful primer to the topic of gene regulatory RNAs as a whole.
1. Introduction
1.1 Riboswitches
RNA is capable of forming complicated structures. This complexity is achieved with a rather limited number of building blocks (four nucleotides). RNA lends itself well to situations necessitating multiple possible conformations via alternative base pairing and tertiary interactions, a concept elegantly illustrated by riboswitches. These structured regulatory RNA elements control gene expression by directly responding to cellular conditions without the direct involvement of protein factors. Canonical definition of a riboswitch refers to an RNA that binds and responses to specific small molecule metabolites or metal ions, though sometimes the term is used to refer regulatory RNAs sensing macromolecules (e.g. tRNA) and temperature as well.
cis-regulation by riboswitches is a distinct gene regulatory mechanism in prokaryotes. This theme is especially prevalent in Firmicutes, where 2-4% of all genes are under riboswitch control; it is less prevalent in other bacteria phyla[1]. Riboswitches almost exclusively function in cis, usually residing in the 5′ untranslated regions (5′-UTRs) of the host mRNAs. They regulate gene expression mainly by premature transcription termination or inhibition of translation initiation. Other regulatory mechanisms have been demonstrated, including the control of mRNA degradation or alternative splicing[2, 3]. Canonical riboswitches typically consist of two domains: an upstream aptamer domain responsible for ligand recognition and a downstream expression platform. The latter can adopt alternate “on” or “off” conformations to influence the decision of transcription or translation machinery (Figure 1). Ligand binding influences cross-talking between the two domains, which in turn triggers the expression platform conformation. Within riboswitch classes, the aptamer domains tend to be more highly conserved than expression platforms. Transcriptional riboswitches commonly form a rho-independent transcription terminator helix in the “off” state, leading to premature transcription termination (Figure 1). Conversely, the “on” state favors read-through by forming an alternate “antiterminator” structure. Translational riboswitches generally prevent initiation by sequestering the Shine-Dalgarno sequence in their “off” states. Many riboswitch-regulated genes are involved in transport or biosynthesis/breakdown of their target ligand, constituting classic feedback regulatory loops. The ligands for riboswitches range from general metabolic indicators (e.g. SAM, amino acids), signaling molecules (c-di-AMP and c-di-GMP)[4, 5], or toxic species (fluoride)[6]. To date, all known riboswitches respond to only one ligand. However, multiple riboswitches can be present in a single mRNA to integrate multiple inputs or regulate more than one process, producing a rather sophisticated logic output[7-9].
Figure 1. Schematics of riboswitch-mediated gene regulatory circuits.
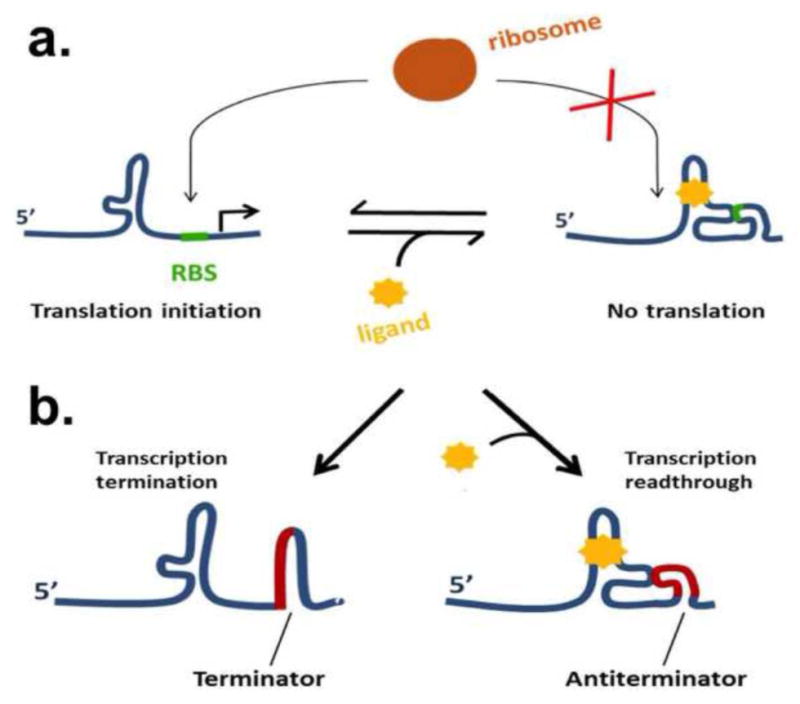
a.) Illustration of a typical translational riboswitch. Ligand binding favors a conformation in which the ribosome binding site (RBS) (green) is occluded, preventing translation initiation. b.) Illustration of a transcriptional riboswitch. Here, binding of the ligand to the aptamer domain causes the effector domain (red) to adopt an antiterminator conformation, preventing premature transcription termination.
1.2 SAM riboswitches
S-adenosyl-L-methionine (SAM) (Figure 2) is an important metabolite in all living organisms, synthesized from methionine and ATP by SAM synthetase. SAM is the universal methyl currency inside cells. The methyl group attached to the sulfonium ion in SAM is quite labile and can be easily transferred to substrates by methyltransferases. Perhaps due to its essential role in cell metabolism, SAM is the most common riboswitch effector known. Three evolutionarily distinct groups of SAM riboswitches have been identified: the SAM-I superfamily (referred to as clan in Rfam), consisting of the SAM-I (S-box), SAM-IV, and SAM-I/IV families; the SAM-II superfamily, consisting of SAM-II and SAM-V families; and the SAM-III (or SMK-box) family[10]. The body of work on the SAM riboswitch families constitutes a useful primer to the topic of riboswitches and, to an extent, to structured RNAs as a whole. This review summarizes research on 1) modes of gene regulatory mechanisms, 2) common themes and differences in ligand recognition, and 3) ligand-induced conformational dynamics among SAM riboswitch families.
Figure 2. S-Adenosyl-L-methionine (SAM).
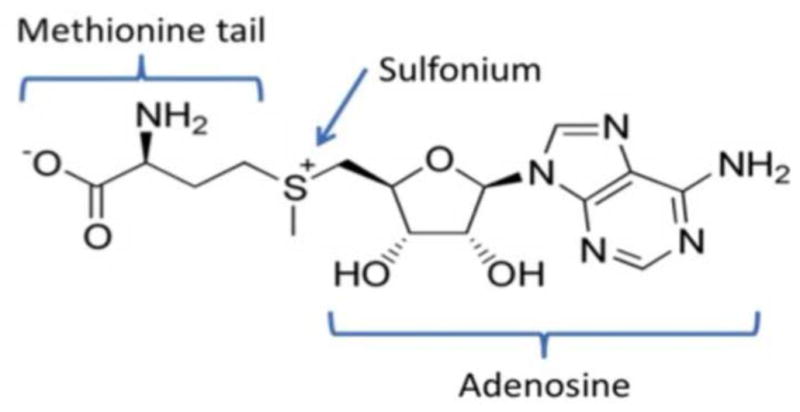
The three chemical “handles” by which riboswitches recognize SAM are the adenosine moiety, sulfonium ion, and methionine tail. The labile methyl attached to the sulfonium is transferred to the substrate in many methylation reactions. SAM is converted to S-Adenosyl-L-homocysteine as the result.
2. SAM riboswitch families and their gene regulatory mechanisms
[A note about terminology: As in Weinberg et al. 2008[11], we refer to each of the distinct groups of riboswitches that share clear evolutionary, structural and sequence relatedness as a family (e.g. SAM-I family, SAM-IV family, etc.). However, families of riboswitches that share similar structural features, but whose evolutionary relatedness is less well-defined, are grouped into superfamilies. For example, SAM-II and SAM-V are grouped into the SAM-II superfamily.]
2.1 The SAM-I superfamily
The Bacillus subtilis SAM-I (or S-box) riboswitch was the first SAM riboswitch family discovered and is one of the best studied. It was identified in the 5′ UTRs of sulfur metabolism genes that did not have any identifiable transcription regulator binding sites[12, 13]. The SAM-I riboswitch family is quite widespread and can be commonly found in low-GC Gram-positive bacteria[14]. Different isoforms of SAM-I, each tuned to respond optimally to different SAM concentration ranges, can be found within a single species. For example, B. subtilis contains at least eleven versions of SAM-I, each tuned to regulate different genes[13, 15].
The conserved secondary and crystal structures of the Thermoanaerobacter tengcongensis yitJ SAM-I are shown in Figure 3[16]. SAM-I controls transcription of its target genes at the level of transcription termination. When SAM levels are low, formation of a transcription antiterminator stem-loop is favored[13], while high SAM levels favor an alternative structure with one of the antiterminator strands instead forming part of P1, leading to terminator formation and transcription termination. One notable deviation from this mechanism is a branch of SAM-I riboswitches from Listeria monocytogenes that act in trans[17]. In this case, SAM-I aptamer binds to the 5′-UTR of the mRNA encoding virulence factor PrfA and inhibits expression, apparently in a SAM-independent manner.
Figure 3. SAM-I superfamily.
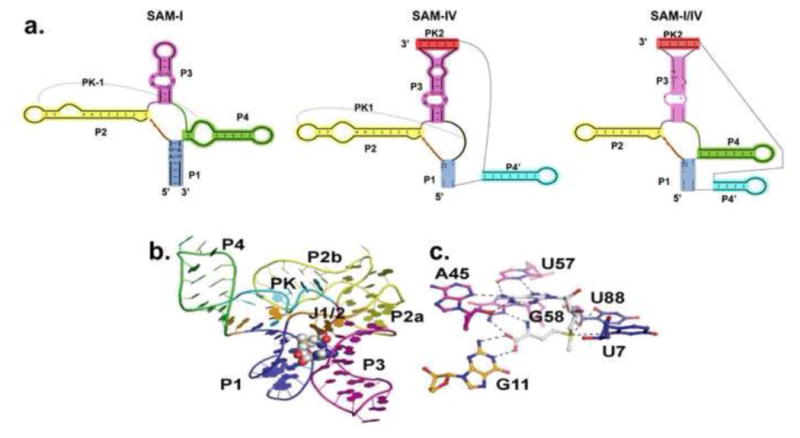
a.) Side-by-side comparison of the secondary structures of SAM-I, SAM-IV, and SAM-I/IV riboswitch families[10]. Equivalent helical elements among three families are drawn in the same color. Pseudoknot-1 (PK-1) structure is marked by a dashed line. b.) Representative SAM-I crystal structure from T. tengcongensis[25]. The RNA is in cartoon representation and colored according to panel a. SAM is in CPK representation. c.) Zoomed view of the SAM binding site in SAM-I riboswitch. SAM ( in white color) forms a base triple with A45 and U57. G58 and G11 interact with the methionine tail of SAM.
The SAM-IV family of riboswitches was identified in the 5′-UTRs of sulfur metabolism genes in Actinomycetales[18]. It shares a number of structural features with SAM-I and can therefore be included in the SAM-I superfamily. SAM-IV appears to bind SAM in a similar fashion to SAM-I using the same binding-site interactions. However, the scaffolding beneath the binding nucleotides differs (Figure 3) [11] . SAM-IV aptamers often appear near the translation start sites of genes, rather than near potential terminators, suggesting these instances likely regulate at the translational level[11]. This, however, remains to be experimentally verified. SAM-I riboswitches are more widely distributed than SAM-IV, suggesting that SAM-IV diverged from a SAM-I like ancestor, presumably by the loss of P4 and the addition of a new P4 and P5[11]. Due to its relatively recent discovery, many specific details of the SAM-IV family remain to be experimentally determined.
A distinct family in the SAM-I superfamily, “SAM-I/IV” was identified more recently from metagenome sequences [19] (Figure 3). Like SAM-IV, SAM-I/IV forms a pseudoknot between its 3′ end and the P3 stem-loop. However, it is missing a bulge in P2, which forms a conserved pseudoknot in both SAM-I and -IV [10]. The variation within the SAM-I family demonstrates RNA's ability to vary the peripheral scaffolding while preserving an intact ligand-binding core. It also nicely illustrates the modularity aspect of an aptamer domain – a common fold can be adapted to either transcription or translation regulation circuit.
2.2 The SAM-II superfamily
The first SAM-II riboswitches were identified as “metA” motifs in α-Proteobacteria [20]. The original discoveries, in Agrobacterium tumefaciens, were found near intrinsic transcription terminators; however, other examples (including the crystal structure from a Sargasso Sea metagenome sequence) appear to sequester the Shine-Dalgarno sequence (SD) [21]. These SAM-II riboswitches are typically short sequences, which allowed the full crystal structure, rather than just an aptamer domain, to be determined in complex with SAM [21]. The SAM-II structure forms an H-type pseudoknot when SAM is bound (Figure 4). The pseudoknot ends 2 nt upstream from the SD, but this appears to be sufficient to occlude ribosome binding in the “off” state.
Figure 4. SAM-II Superfamily.
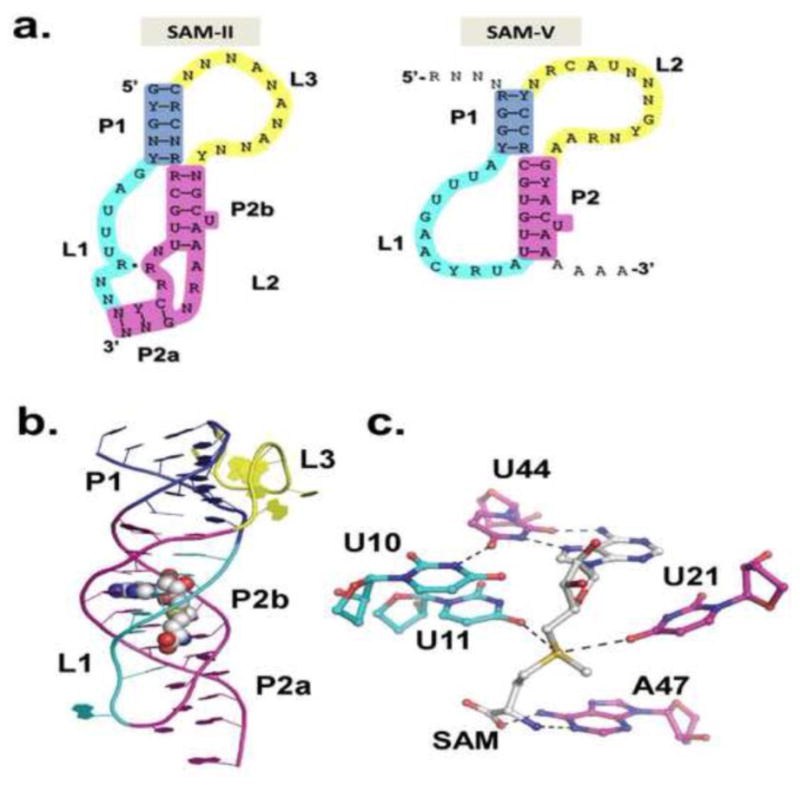
a) Consensus secondary structures of SAM-II and SAM-V riboswitch families[8]. The loops tolerate variability and long insertions. b) metX SAM-II crystal structure from Sargasso Sea metagenome[21]. Color scheme matches that in a. c.) SAM binding site in SAM-II. The Hoogsteen edge of SAM pairs with the Watson-Crick edge of U44 as part of a base triple. The SAM adenine stacks between G22 and A45. The sulfonium experiences favorable electrostatic interactions with U11 and U21 and the methionine tail hydrogen bonds with A47.
The structurally related SAM-V riboswitch is widespread in marine bacteria [8]. Like SAM-II, it appears to control expression mainly by SD sequestration. Its predicted binding site is very similar to that of SAM-II. However, as is the case for the SAM-I superfamily, SAM-V differs from SAM-II in the peripheral region outside the SAM binding site (Figure 4). Interestingly, examples have been found where SAM-II occurs in tandem with SAM-V to regulate a single gene. In these cases, SAM-II lies upstream of a putative transcriptional terminator, suggesting a deviant transcriptional rather than translational role [8]. This instance nicely exemplifies the common modularity of riboswitch domains. Many details regarding the distribution of SAM-II riboswitches and their regulatory mechanisms remain to be discovered.
2.3 The SAM-III family
Lastly, the SMK-box, or SAM-III riboswitch, is also a translational riboswitch. It was discovered in the 5′-UTR of metK (SAM synthetase) in Lactobacillales [22]. The Enterococcus faecalis SAM-III riboswitch SAM-bound “off” structure and its conserved secondary structure are shown in Figure 5[23]. It can be generally described as three helices, at the intersection of which is the SAM binding site. Like SAM-II, the SAM-III riboswitch also occludes ribosome binding to the SD. Its SD sequence is directly sequestered as part of the SAM-bound “off” state, actually making direct contacts with SAM in the binding site [23]. It is also possible that translational riboswitches like SAM-II and SAM-III indirectly affect RNA stability, since ribosomes play a protective role by physically blocking access to the RNA by nucleases.
Figure 5. SAM-III family.
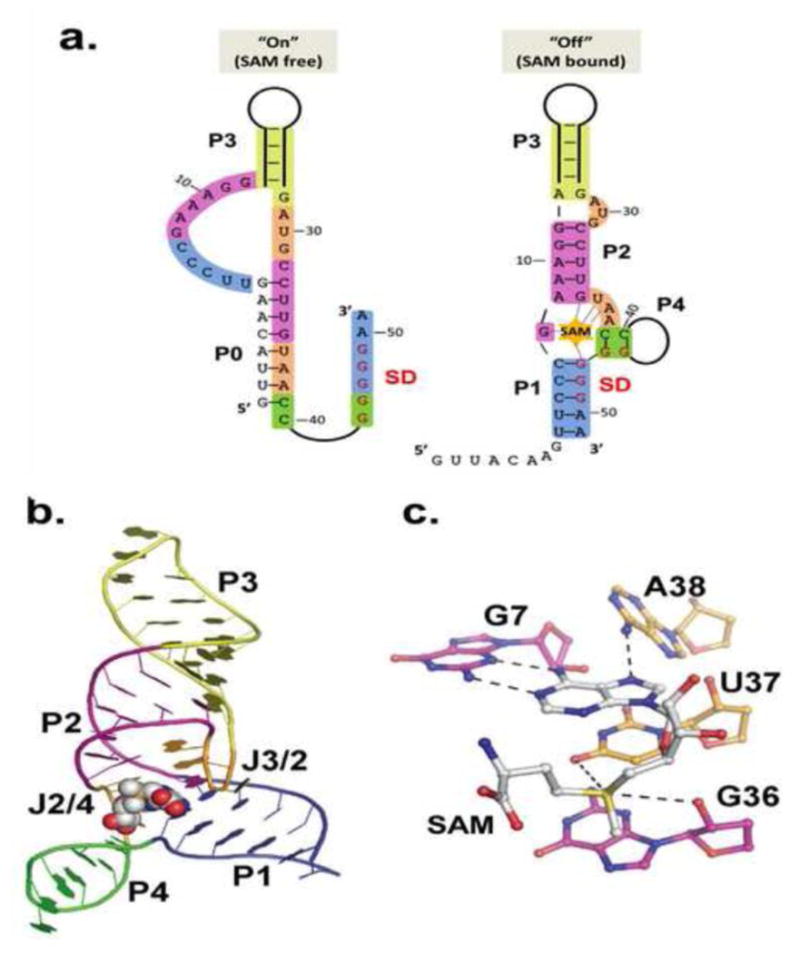
a.) Secondary structures of the on (SAM-free) and off (SAM-bound) states of the E. faecalis SAM-III riboswitch. Of the four secondary structure elements, only P3 is constant in both the on and off states. Bases involved in off state secondary structure elements are colored in both on and off state to emphasize changes. Coloring and numbering corresponds to off state crystal structure in B. b) Crystal structure of the SAM-bound E. faecalis SAM-III [23]. Coloring matches that in A. c.) SAM binding site in SAM-III. SAM stacks in the “binding pocket” between U37 (gold) and G48 (not shown). The adenosine pairs with G7 (purple) in the back of the pocket. Carbonyl oxygen atoms on U37 and G36 (in the SD) interact with the sulfonium.
2.4 Undiscovered SAM riboswitch families?
Many riboswitches, including SAM-II, -IV and -V were originally identified by bioinformatic searches [8, 18, 20]. With the availability of large amounts of genomic sequence data, covariance search models are a powerful technique for identifying conserved structured RNAs[19]. These methods use conservation and the co-variation of bases in predicted structures across multiple species to identify significant structured RNAs. It is likely that more SAM-binding and other riboswitches remain to be found. At this point, experimental verification and gene product characterization are often limiting factors in identifying riboswitches and characterizing their effectors[24]. For this reason, many conserved putative riboswitches remain “orphans”[19]. Interestingly, as research progresses on riboswitch classes for which some or all of the regulated genes are uncharacterized, riboswitches may in turn provide insight into the properties of their regulated genes’ products. One recent example is the SAM/SAH-binding element, a small putative riboswitch that was identified in a large comparative genomic screen and was subsequently shown to bind both SAM and its breakdown product, SAH with similar affinity [19]. Further structural and genetic study of this element will provide more definitive answers as to its biological function.
3. Common themes and differences in SAM recognition among SAM riboswitches
Recent X-ray crystal structures for representatives from SAM I-III superfamilies have provided significant insight into SAM binding by riboswitches [21, 23, 25]. For an excellent class-by-class review on the structural biology of SAM riboswitches, see Batey 2011 [10]. The three SAM riboswitch superfamilies are structurally distinct and their binding mechanisms can be differentiated by their interactions at three distinct “handles” on the SAM molecule: the adenosyl moiety, the positively charged sulfonium, and the methionine tail, including its amino and carboxyl groups. In addition to these chemically distinct handles, SAM itself binds in quite different conformations among the characterized classes. The adenosyl moiety mimics a regular adenosine residue in mediating stacking, base-pairing and base-triple interactions to the riboswitch. Not surprisingly, it is well anchored by all classes of SAM riboswitches; however, the specifics of the adenosine recognition differ significantly among the three superfamilies. The positively charged sulfonium ion in SAM is recognized by favorable electrostatic contacts, usually from uracil carbonyl oxygen atoms, in all SAM riboswitch superfamilies. This conserved recognition also forms the basis for ligand discrimination between SAM and its metabolized product S-adenosyl-L-homocysteine (SAH). Recognition of the third handle in SAM, the methionione tail, varies greatly, ranging from strong specification in SAM-I and II superfamilies, to little recognition in SAM-III.
3.1 SAM recognition by SAM-I
SAM-I is one of the most extensively characterized riboswitch structures. The X-ray crystal structure of the Thermoanaerobacter tengcongensis MetF-H2 SAM-I aptamer domain has been determined bound to SAM, SAH and sinefungin [25, 26]; in its ligand-free state[27]; as a scaffold for understanding the kink-turn motifs[28, 29]; and in complex with the kink-turn binding protein YbxF[30]. In addition, the Bacillus subtilis yitJ SAM-I riboswitch crystal structure was determined in complex with SAM [16]. The SAM-I aptamer domain consists of two sets of coaxially stacked helices (P1/P4 and P2/P3) connected by a 4-way junction, with SAM bound between the P1 and P3 helices. Within the binding pocket, SAM is in a compact cis conformation with the amino acid tail stacked underneath the adenine ring face[25]. The SAM adenosine and methionine tail moieties are both intimately recognized. The Watson-Crick (WC) and Hoogsteen (H) edges of the SAM adenosine are hydrogen (H)-bonded to the A45 sugar edge (S) and U57 WC edge, respectively, forming a base triple. The SAM adenosine ring is oriented to stack beneath C47, slightly offset. The sulfonium makes favorable electrostatic interactions with two uracil carbonyl oxygen atoms from two adjacent A-U pairs (U7 and U88). Finally, the amino acid tail forms multiple H-bonds with the G58 S edge and G11 WC edge (Figure 3C). These extensive contacts envelop the SAM molecule in the binding pocket, leaving minimal solvent exposure as compared to the other SAM-riboswitch classes.
3.2 SAM recognition by SAM-II
The much simpler SAM-II class forms a classic H-type pseudoknot and binds SAM in a pocket formed between P2 and L1[21]. The dissociation constant for SAM-II is significantly higher than SAM-I (weaker binding); however, despite its minimal topology, SAM-II is able to achieve similar levels of specificity as SAM-I in distinguishing SAM from its natural analogs[20]. SAM is bound in an extended, trans conformation in SAM-II. The adenine moiety of SAM is recognized in a base-triple, with its H edge similarly paired with the WC edge of U44, which in turn forms a H-WC pair with U10 (Figure 4C). In contrast to SAM-I, the adenine WC edge is solvent exposed. These interactions orient the SAM adenosine ring to optimally stack between G22 and A45. The sulfonium contacts also closely resemble those of its SAM-I counterpart, anchored by two highly conserved uracil carbonyl oxygen atoms (U11 and U21) from two adjacent A-U pairs. The amino acid tail is anchored by hydrogen bonds to the WC edge of A47.
3.3 SAM recognition by SAM-III
The SAM-III riboswitch represents yet another independently evolved RNA fold that specifically recognizes the SAM molecule. The SAM-binding pocket resides in a three-way junction with SAM bound in its syn conformation[23]. The floor of the binding pocket is formed by a base triple, C6-G48•A38 that ties P1 to J2/4, a contact that is important for sampling the intermediate “ready” state[23]. The ceiling of the pocket is formed by a sheared U37-A29 base pair, such that the SAM adenosine ring stacks slightly offset between U37 and G48. As in both previously discussed SAM riboswitch classes, the SAM adenosine forms part of a base triple with the S-edge of the absolutely conserved G7 and a hydrogen bond to A38 (Figure 5C). SAM-III differentiates itself from SAM-I and SAM-II in that the sulfonium is bound by a single carbonyl (U37) while the G36 O2′ provides the additional neutralizing bond. In stark contrast to the other classes, the amino acid tail in SAM-III is solvent exposed and poorly ordered in the electron density, suggesting it not required for recognition.
Additional SAM riboswitch families (SAM-IV, SAM-V and SAM-I/IV) currently lack a structurally determined representative[8, 11, 19]. However, despite the clear peripheral differences in these families, their SAM recognition modes are likely represented by examples within their superfamilies[8, 11]. SAM-III adopts a distinct fold from the other two superfamilies.
4. Mechanisms for fine-tuning in ligand recognition
Several important recent studies have clearly demonstrated that riboswitches are tuned across a gradient of activities to control gene expression. This has clearly been demonstrated for the guanine[31], guanine/adenine[32], TPP[33], c-di-GMP riboswitches[33] and PreQ riboswitches[34]. Tuning has even been demonstrated among multiple occurrences of the SAM-I riboswitch from a single organism[15]. Tomsic et al.[15] examined the 11 SAM-I riboswitches from Bacillus subtilis and determined that their in vitro SAM-binding affinity vary in a range of ∼250-fold. Notably, the range of affinities generally correlated with their in vivo sensitivities to SAM and induction levels, though additional factors, such as the strength of alternate structures, were proposed to be crucial to further tuning the systems’ responses[15]. For most riboswitches, the mechanism of tuning a response appears to be a complex combination of various levels, from the simple variance in affinity of a pre-organized binding pocket, to control of conformational selection[35]. The folding pathway and conformational dynamics of riboswitches is an area of intense study and is a prime means of tuning riboswitch response.
The SAM-I riboswitch samples bound-like conformations in the absence of ligand and ligand binding induces final structural compaction[16, 27, 36]. Switching between these conformations is a dynamic process and is influenced by peripheral tertiary contacts in the riboswitch, but also internal structures, such as the conserved kink-turn motif[37, 38]. Several lines of recent evidence demonstrate that the riboswitch undergoes conformational changes that resemble conformational capture, but upon ligand binding (and only in the presence of Mg2+), its core undergoes further compaction, resembling induced-fit binding [33, 39-42]. Clearly there are many factors at play in the dynamic SAM-I riboswitch and variance at any of these structural elements could lead to a grading of responses observed in the 11 B. subtilis SAM-I riboswitches[15].
The SAM-II and SAM-III riboswitches are also dynamic and tunable switches, but less detail exists for their conformational dynamics. SAM-II is stabilized in a near bound state by Mg2+, but like SAM-I, it only reaches its final stabilized form in the presence of SAM[43, 44]. SAM-III is also a dynamic structure that undergoes large conformational sampling between three discrete states: translation “on”, “ready”, and “off” states. Conformational entropy in peripheral regions critically influences the thermodynamic balance among three conformational states [13]. Using a simplified version of the E. faecalis metK switch, Wilson et al.[45] demonstrated that alternate pairing in P1 shifted the equilibrium of SAM-III riboswitches sampling the mutually exclusive “on” or “ready” states. Their work further highlighted the idea that unwinding and conformational selection, in this case could initiate by local unwinding of P1. These findings are in close agreement with recent experimental study[46] and theoretical modeling[47] of SAM-III that demonstrate melting of P1 and P4 precedes, P2 and suggested P3 remains stable throughout switching. These findings shed light on the physical reality of conformational switching and ligand binding, but the mechanisms of fine-tuning response to suit the required levels of gene expression remain unclear.
5. Conformational dynamics of SAM riboswitches
The ligand-bound states of many known riboswitches are structurally well characterized, but ligand-free states and the conformational dynamics governing the on/off switch are often poorly understood. Some of the recent challenges in understanding riboswitch dynamics include: 1) The structural differences between ligand-bound and free structures; 2) Whether ligand recognition occurs by conformation capture, induced-fit, or a combination of these two mechanisms; 3) If the switch is driven by kinetic or thermodynamic control; 4) The interplay with other conditions and processes in cells (e.g. transcription speed, translation). Historically, riboswitches have been classified into two broad types, I and II, referring to the degree of structural change in the aptamer between ligand-bound and -free states. Type I riboswitches have a preformed binding site in the absence of ligand, whereas Type II riboswitch aptamers undergo structural rearrangement when bound to ligand.
5.1 SAM-I conformational dynamics
A ligand-free crystal structure of the T. tencongensis SAM-I riboswitch aptamer domain was published by Stoddard et al. in 2010 [27]. This structure was nearly identical to the SAM-bound state (Figure 3), even crystallizing in the same crystal form. However, in the SAM-free structure, the SAM binding site was blocked by a base triple with a nearby adenosine (Figure 6). This suggests that SAM-I forms a binding-competent structure in solution, but that there was some flexibility in the binding site. Further experiments utilized Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) and Small-Angle X-ray Scattering (SAXS) to show that in the absence of SAM, SAM-I samples a number of both compact (bound-like) and less structured conformations in vitro [27, 40]. Particularly, P1 and P3 show increased structural heterogeneity in the absence of SAM. This conformational sampling eliminates the possibility of a pure “induced-fit” model for SAM-I. Furthermore, the simple “on”/”off” structural dichotomy is muddled by the fact that the “on” state is better explained as a population of multiple states, some of which are SAM binding-competent. Similarly, other riboswitches have also demonstrated conformational heterogeneity in the ligand-free state, suggesting it is a general principle of their function [48].
Figure 6. SAM-I binding site with and without SAM.
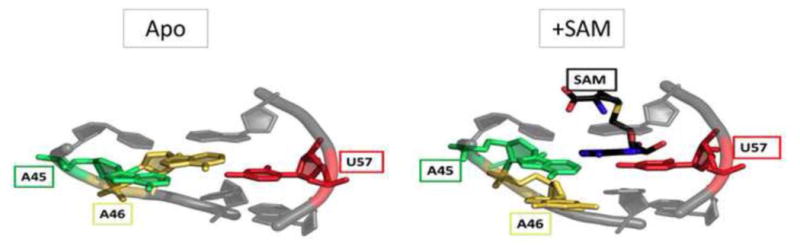
When SAM (black) binds, it forms a base triple with A45 and U57 (green and red). In the apo state, this interaction is blocked by A46 (yellow)[27].
The effect of “on” state conformational heterogeneity on SAM-I dynamics is further complicated by the finding that transcriptional riboswitches, like SAM-I, normally function under non-equilibrium conditions. The nascent RNA must make its decision co-transcriptionally, before RNA polymerase reaches a “decision point”, after which it has bypassed the terminator [49]. Consequently, kinetic, rather than thermodynamic, contributions are crucial to the SAM-I decision. For B. subtilis SAM-I, this is supported by the finding that its SAM dissociation constant (KD) of 4 nM is much lower than the ∼300 μM intracellular SAM concentration in B. subtilis [13][15]. Likewise, the half-life of the SAM-I/SAM complex is long, at over 7 min [15], much longer than the time it would take RNA polymerase to reach its decision point. Despite this, ∼10 μM SAM is sufficient to terminate transcription [50], though it is now known that multiple isoforms are tuned to respond differently to given SAM concentrations [45]. If SAM-I functioned under equilibrium conditions, it would likely never achieve a SAM-free state under physiological concentrations. Thus, the SAM-binding kinetics and folding pathway drive its function instead. Similar responses hold true for other transcriptional riboswitches, such as the FMN riboswitch [51] and computational predictions of folding rates show that transcription-attenuating riboswitches typically fold more quickly than those that sequester ribosome binding sites [52].
A number of studies have sought to dissect the determinants of SAM-I folding and SAM recognition [16, 27, 37, 41, 53]. Heppell et al. used single-molecule Fluorescence Resonance Energy Transfer (smFRET) to delineate at least 2 steps in the folding and SAM recognition pathway[41]. First, initial folding occurs upon Mg2+ binding to the SAM binding-competent state[41]. It was estimated that this folding would take on the order of 100 ms [16]. Compared to the rate of E. coli RNA polymerase, ∼40 nt/s, this allows a window of time after folding for ligand sensing. When SAM initially binds, small-scale rearrangements occur for an overall structural compaction. In particular, the B. subtilis yitJ crystal structure showed that J1/2 and J3/4 enclose the SAM binding site, via Mg2+ mediated tertiary contacts [16]. Furthermore, gel shift and smFRET assays showed that P1 twists upon SAM binding to coaxially stack with P4 [41, 42]. Together, these interactions may stabilize the SAM-bound state, making it essentially irreversible on a co-transcriptional timescale. Given the conformational heterogeneity of P1 and P3 before SAM binding and the induced twist in P1 after SAM binding, it can be argued that SAM-I incorporates both conformation selection and induced fit mechanisms. If SAM does not bind by the time the other half of the anti-terminator helix is transcribed, the folding of the anti-terminator (involving a strand of P1) occurs, and RNA polymerase passes the potential terminator site. It is important to note that other factors affecting the duration of the folding window, such as rate of transcription or the spacing and folding of the expression platform, also play key roles in transcriptional riboswitch function. Many of these roles await experimental validation.
5.2 SAM-II conformational dynamics
Ligand-sensing in SAM-II family of translational riboswitches may not rely on kinetics to the extent of the transcriptional riboswitches. This is often accompanied by higher KD values near the cellular concentration of its response. The A. tumefaciens SAM-II has a KD of ∼10 μM for SAM, similar to the SAM concentration that leads to transcription termination by SAM-I in vitro [20, 50]. The KD of the metX SAM-II that was crystallized (from a Sargasso sea metagenome sequence) was estimated at ∼140-200 nM by 2-AP fluorescence spectroscopy[44], though no intracellular SAM concentration data is available for the species.
Recent studies have provided deeper insight into the folding and switching of SAM-II. Using NMR and smFRET, Haller[44] demonstrated conformational heterogeneity of the SAM-free metX SAM-II, particularly at the 3′ end. These experiments showed that even in the absence of SAM, a stem-loop structure samples a pseudoknot (bound-like) state by making tertiary interactions between the 3′ strand and the loop. This state sampling is further supported by recent SAXS and NMR data[43]. Though the distribution of states depends on the concentration of Mg2+, the populations were roughly equal in the smFRET studies performed with 2 mM Mg2+ in the absence of SAM[44]. However, the half-life of the compacted (off) state was greatly increased in the presence of SAM, to about 1 s, resulting in the majority of molecules in the off state[44].These data suggest SAM-II functions by a conformational capture mechanism; however, 2-AP fluorescence experiments showed that further compaction of the riboswitch occurs after SAM binding, including in the binding site, in P2a, and further away in L3, like SAM-I, pointing to some combination of conformational selection and induced-fit binding[44].
5.3 SAM-III conformational dynamics
Like SAM-II, the SAM-III riboswitch family controls translation and is thus not constrained to act in a short time window. It was thus hypothesized that it could act as a true reversible “switch”[54], able to change its state over the mRNA half-life in response to fluctuating SAM concentrations. The SAM-III effective KD is ∼0.4-0.5 μM, as determined by filter binding assays and Isothermal Titration Calorimetry (ITC), significantly higher than that of SAM-I[23, 45]. Smith et al. used qRT-PCR to calculate the half-life of the SAM-III transcript in vivo at ∼3-4 min and demonstrated that the SAM off-rate is within the range of the mRNA half-life[54]. Further, using 2-aminopurine (2-AP) fluorescence studies, they showed reproducible conformational switching between “off” and “on” states upon repeated addition and removal of SAM within the half-life of the transcript.
Several experiments address the question of conformational selection vs. induced fit mechanism for SAM-III riboswitches. SHAPE[23], SAXS, and NMR[45] all show that the SAM-free SAM-III favors an “on” conformation, but samples an “off”-like or “ready” state. Upon addition of saturating SAM, the majority of RNAs exist in an “off” conformation. Constructs designed to destabilize the “on” state and stabilize the “ready” state have a much lower KD for SAM (∼60 nM), suggesting that the full-length construct's apparent KD of 0.4 μM is due to the relative stability of the “on” state over the “ready” state in the absence of SAM[45]. Since the SAM binding site is not pre-formed in the “on” state, this also supports the prediction that only the “ready” state is SAM-binding competent, clearly illustrating conformational selection. SHAPE experiments also demonstrate further protection of bases in the SAM binding pocket upon SAM binding, compared to the “ready” state, suggesting some degree of induced-fit mechanism as well[23, 46].
Given the potential for switching during its lifetime, the mechanism of switching is also of interest. As seen in Figure 5A, the P3 stem-loop is common to all the “on”, “off”, and ”ready” conformations. However, P1 and P4 in the “off” state are rearranged in the “on” state to free the SD and form a different helix, P0. At physiological temperatures, this transition happens quickly, in <1 min and temperature-dependent SHAPE suggests a 2-step folding/unfolding in SAM binding/release[42, 46]. Similar results were also found by force extension curve simulations for the “off” state[55]. The studies suggest a switching mechanism in which the off-state tertiary interactions break and SAM dissociates, followed by P1 and P4 unfolding, then P2 unfolding, allowing formation of P0 to stabilize the “on” state. The exact nature of the “on” state, the “ready” state, and any other folding intermediates, remain to be structurally determined. Though there is strong evidence for thermodynamic control, the effects of transient ribosome binding to the SD or transcription/translation coupling on SAM-III remain unclear.
6. Summary
As the largest known group of riboswitch, SAM riboswitches have highlighted an important facet of bacterial physiology. Knowledge accumulated from these studies exemplifies the variability of riboswitches in evolving distinct ligand binding pocket, maintaining proper conformational dynamics, and tuning its gene regulatory response. Many themes are shared by other riboswitch families. SAM riboswitches continue to attract strong interest. More details remain in understanding each class’ ligand-dependent gene regulatory mechanism. Additionally, given SAM's central metabolic role, its easily recognizable chemical “handles”, the abundance of genomic data, and improvements in bioinformatic identification, more SAM riboswitches are likely to be identified. Such cases will hopefully provide new modes of recognition and regulation. As research continues, the number of potential discoveries and applications may continue to grow.
Highlights.
SAM is the most common riboswitch effector known.
We review common themes and differences in SAM recognition by riboswitches.
Structural variation within each SAM riboswitch family fine-tunes its response to SAM.
Efforts are on-going to delineate the conformational switching landscape.
Acknowledgments
This work was supported by NIH operating grants GM-086766 to AK. JCG is supported by a CIHR Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Winkler WC. Metabolic monitoring by bacterial mRNAs. Archives of Microbiology. 2005;183:151–159. doi: 10.1007/s00203-005-0758-9. [DOI] [PubMed] [Google Scholar]
- 2.Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Masse E, Lafontaine DA. Dual-acting riboswitch control of translation initiation and mRNA decay. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3444–3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S, Breaker RR. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Research. 2013 doi: 10.1093/nar/gkt057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread Genetic Switches and Toxicity Resistance Proteins for Fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem Riboswitch Architectures Exhibit Complex Gene Control Functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 8.Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for common in marine bacteria. Rna. 2009;15:2046–2056. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson PY, Fedor MJ. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol. 2011;18:359–363. doi: 10.1038/nsmb.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batey RT. Recognition of S-adenosylmethionine by riboswitches, Wiley interdisciplinary reviews. RNA. 2011;2:299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. Rna. 2008;14:822–828. doi: 10.1261/rna.988608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Molecular Microbiology. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 13.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature structural biology. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 14.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, Bateman A. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Research. 2010 doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. Journal of molecular biology. 2010;404:803–818. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-Acting Riboswitch Controls Expression of the Virulence Regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 18.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Research. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome biology. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome biology. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 23.Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. Challenges of ligand identification for riboswitch candidates. RNA Biology. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 26.Montange RK, Mondragon E, van Tyne D, Garst AD, Ceres P, Batey RT. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. Journal of molecular biology. 2010;396:761–772. doi: 10.1016/j.jmb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder KT, Daldrop P, Lilley DM. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure. 2011;19:1233–1240. doi: 10.1016/j.str.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroeder KT, Daldrop P, McPhee SA, Lilley DM. Structure and folding of a rare, natural kink turn in RNA with an A*A pair at the 2b*2n position. Rna. 2012;18:1257–1266. doi: 10.1261/rna.032409.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baird NJ, Zhang J, Hamma T, Ferre-D′Amare AR. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. Rna. 2012;18:759–770. doi: 10.1261/rna.031518.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoddard CD, Widmann J, Trausch JJ, Marcano-Velazquez JG, Knight R, Batey RT. Nucleotides adjacent to the ligand-binding pocket are linked to activity tuning in the purine riboswitch. doi: 10.1016/j.jmb.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baird NJ, Kulshina N, Ferre-D′Amare AR. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 2010;7:328–332. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Souliere MF, Altman RB, Schwarz V, Haller A, Blanchard SC, Micura R. Tuning a riboswitch response through structural extension of a pseudoknot. doi: 10.1073/pnas.1304585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hennelly SP, Sanbonmatsu KY. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 2011;39:2416–2431. doi: 10.1093/nar/gkq1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 39.Baird NJ, Ferre-D′Amare AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hennelly SP, Novikova IV, Sanbonmatsu KY. The expression platform and the aptamer: cooperativity between Mg2+ and ligand in the SAM-I riboswitch. Nucleic Acids Res. 2013;41:1922–1935. doi: 10.1093/nar/gks978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7:384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 42.Eschbach S, St-Pierre P, Penedo JC, Lafontaine DA. Folding of the SAM-I riboswitch: A tale with a twist. RNA Biol. 2012;9 doi: 10.4161/rna.19648. [DOI] [PubMed] [Google Scholar]
- 43.Chen B, Zuo X, Wang YX, Dayie TK. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 45.Wilson RC, Smith AM, Fuchs RT, Kleckner IR, Henkin TM, Foster MP. Tuning riboswitch regulation through conformational selection. Journal of molecular biology. 2011;405:926–938. doi: 10.1016/j.jmb.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C, Smith AM, Ding F, Chowdhury A, Henkin TM, Ke A. Variable sequences outside the SAM-binding core critically influence the conformational dynamics of the SAM-III/SMK box riboswitch. Journal of molecular biology. 2011;409:786–799. doi: 10.1016/j.jmb.2011.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin JC, Thirumalai D. Kinetics of Allosteric Transitions in S-adenosylmethionine Riboswitch Are Accurately Predicted from the Folding Landscape. doi: 10.1021/ja408595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state, Wiley interdisciplinary reviews. RNA. 2012;3:369–384. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haller A, Souliere MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Accounts of chemical research. 2011;44:1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 50.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 52.Sauerwine B, Widom M. Folding Kinetics of Riboswitch Transcriptional Terminators and Sequesterers. Entropy. 2013;15:3088–3099. [Google Scholar]
- 53.Boyapati VK, Huang W, Spedale J, Aboul-ela F. Basis for ligand discrimination between ON and OFF state riboswitch conformations: The case of the SAM-I riboswitch. RNA. 2012:1230–1243. doi: 10.1261/rna.032177.111. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. The SAM-responsive S(MK) box is a reversible riboswitch. Mol Microbiol. 2010;78:1393–1402. doi: 10.1111/j.1365-2958.2010.07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin JC, Thirumalai D. Kinetics of Allosteric Transitions in S-adenosylmethionine Riboswitch Are Accurately Predicted from the Folding Landscape. Journal of the American Chemical Society. 2013;135:16641–16650. doi: 10.1021/ja408595e. [DOI] [PMC free article] [PubMed] [Google Scholar]


