Abstract
Many eukaryotic cells regulate their polarity and motility in response to external chemical cues. While we know many of the linear connections that link receptors with downstream actin polymerization events, we have a much murkier understanding of the higher order positive and negative feedback loops that organize these processes in space and time. Importantly, physical forces and actin polymerization events don't simply act downstream of chemotactic inputs but are rather involved in a web of reciprocal interactions with signaling components to generate self-organizing pseudopods and cell polarity. Here we focus on recent progress and open questions in the field, including the basic unit of actin organization, how cells regulate the number and speed of protrusions, and 2D vs. 3D migration.
The basic unit of actin assembly—reciprocal interactions between nucleators and the actin cytoskeleton generate oscillations and waves of polymerization
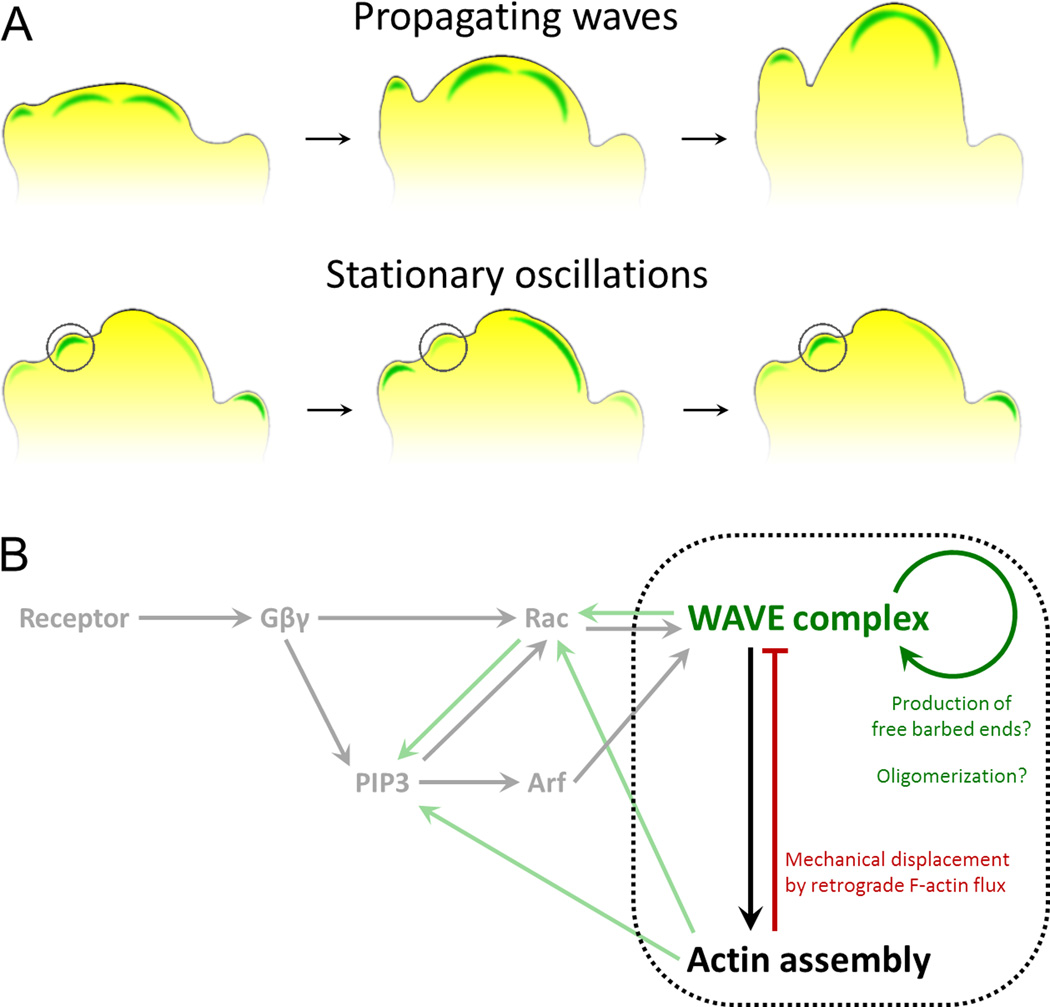
To move, cells need to control when and where they assemble actin polymer. What is the basic unit of actin assembly for eukaryotic chemotaxis? It has long been known that motile cells exhibit periodic changes in their morphology [1–3]; later work has observed that actin-binding proteins (e.g., WAVE regulatory complex (WRC), Arp2/3 complex, and coronin) exhibit oscillatory localization dynamics in Dictyostelium [4–10], neutrophils [11,12], and other mammalian cells [5,13–15], which take the form of either stationary oscillations or propagating waves at the plasma membrane (Figure 1A). A useful framework in which to conceptualize these waves has been to view them as an excitable system, similar to the action potentials which neurons use to transmit information. Among the advantages offered by an excitable system are the ability to ‘ignore’ stimuli below a particular threshold (i.e., filter out noise) and to amplify all signals above this threshold to the same (maximal) level of response [16]. This could enable subtle difference in the concentration of external chemoattractant or in the upstream regulators of cell polarity to be converted into large changes in the activation state and spatial distribution of actin assembly [8,17–19]. Additionally, since zones of inhibition exist directly behind propagating waves generated by an excitable system—preventing waves from reversing direction and making their survival dependent on their movement—this model can account for several of the behaviors displayed by migrating cells, including obstacle avoidance. Upon coming into contact with a barrier, waves of actin assembly-promoting factors are stalled, allowing inhibitory factors to ‘catch-up’ and terminate actin assembly [20]. Such an attribute is especially advantageous in confined environments, where cells would preferentially generate protrusions at locations free of obstructions. This and other potential roles for propagating waves of actin assembly/turnover have been reviewed elsewhere [21].
Figure 1. Actin polymerization as an excitable system.
A) Actin polymerization is organized as multiple wavelike (top) and oscillatory nucleation events (bottom, single oscillator denoted by circle) in motile cells, as visualized by the dynamics of the Wave Regulatory Complex (WRC) [9–11]. Waves that reach the cell boundary organize cellular protrusion. B) The behavior of these waves and oscillations are consistent with an excitable system, which typically has the topology of rapid positive feedback and delayed inhibition (components bound by black dotted line). Key components of the chemotactic signaling network and the flow of information to factors specifying actin assembly are depicted in gray. Candidates for the molecular basis of the positive and negative feedback loops (green and red arrows, respectively) are briefly summarized.
The upstream modulators of actin assembly within the chemotactic signaling network, such as PIP3 and Ras, exhibit properties of an excitable system, including refractory periods, all-or-nothing responses to input, and wave-like localization patterns [10,19,22]. The actin cytoskeleton in motile cells is also intrinsically excitable, but in a manner that may not be dependent on upstream signals such as Ras and PIP3 [10]. While multiple feedback loops do appear to exist both within and between the signaling network and cytoskeleton [20,23–27] (Figure 1B), the specific roles of each connection in modulating cell migration are not yet clear. In its most basic form, an excitable system comprises two components, one which produces fast-acting positive feedback and a second which produces slower negative feedback [28]. What components regulate positive feedback for actin waves/oscillations? Both WRC and Arp2/3 complex show wave-like localization dynamics in multiple systems, indicating that either might participate in wave generation, and both complexes play instrumental roles in driving cell motility throughout eukaryotes. Loss of WRC leads to defects in cell polarity, morphology, actin organization, and migration speed in a variety of systems [20,29–33] and disruption of Arp2/3 has produced similar defects in morphology and migration in many instances [34–37]. However, there is no consensus in the field on whether Arp2/3 is required for normal polarization and migration during chemotaxis [38,39].
Of these two complexes, WRC appears to be a stronger candidate for contributing to positive feedback (Figure 1B), given its biochemical activity in stimulating Arp2/3-dependent actin assembly [40] and genetic observations demonstrating its essential role in promoting chemotaxis in many cellular contexts [20,29–33]. Such positive feedback may operate though the recruitment of additional WAVE regulatory complexes to sites of Arp2/3-dependent barbed-end production (the WAVE2 component of WRC contains a WH2 domain, which has been shown in other contexts to associate with barbed-ends of actin filaments [41–43]). In addition, WRC may self-enrich through homo or hetero-oligomerization [44]. A recent study revealed such a mechanism at work for another Arp2/3 activator, N-WASP, which exhibits highly cooperative phase separation/multimerization with multivalent binding partners via interactions of their SH3 domains with proline-rich regions of N-WASP. These multivalent assemblies exhibit markedly more potent stimulation of Arp2/3-dependent actin assembly when compared to monomeric N-WASP [45]. One candidate for performing such a role in the context of WRC is IRSp53, a membrane-binding BAR domain protein, which directly interacts with WAVE2 via its SH3 domains to promote Arp2/3-dependent actin assembly [46,47], but the multitude of other WRC binding proteins could play a similar role.
What regulates the negative feedback necessary for actin oscillations? Actin filaments appear to play a prominent role, as treating motile cells with drugs blocking actin assembly increases the lifetimes and intensities of WRC waves, whereas treatment with drugs stabilizing filaments has the opposite effect [10,11,23]. This behavior is also observed for other actin nucleation-promoting factors: N-WASP recycles faster in the presence of Arp2/3 complex binding and actin polymerization [48]. While the specific mechanisms mediating negative feedback have yet to be identified, recent evidence using XTC cells suggests that actin filaments may mechanically remove WRC from the membrane while undergoing retrograde flow [23] (Figure 1B). Actin may also potentially play an indirect role via the recruitment of actin-binding proteins to the leading edge that promote dissociation of WRC from the plasma membrane.
Is cell migration in 3D qualitatively different than in 2D?
Amoeboid chemotaxis in two dimensions has been studied in great detail and has led to the general model (described above) in which the major pathway leading to motility relies on WRC binding and stimulation of Arp2/3-dependent actin assembly at the leading edge. While a body of work has identified additional modes for specifying actin assembly in migrating cells (e.g., formin- and Ena/VASP-dependent mechanisms; reviewed in [49]), the means by which these different factors contribute has proved challenging to unravel. WRC has long been thought to be required for chemotaxis; however, a recent study found that Dictyostelium lacking WRC could still efficiently undergo this process. In these cells, WASP, which normally localizes to clathrin patches to promote endocytosis, became enriched at sites that would typically contain WRC to stimulate Arp2/3-dependent actin assembly [9]. This is a profoundly significant result. WRC is typically localized to and thought to be responsible for building explosive integrated actin networks, such as lamellipodia, whereas N-WASP is typically localized to and thought to be responsible for building much more focal actin assemblies, such as filopodia and endocytic actin structures. The data in Dictyostelium suggest that the different actin structures aren’t intrinsic to the different actin nucleation-promoting factors, but rather that other elements (say lamellipodial or endocytic-specific scaffolds) are responsible for organizing the actin structures that are formed.
While the above study revealed only minor differences between the activities of WASP and WRC in driving 2D migration, later work demonstrated that N-WASP and WRC-dependent migration exhibited more distinct phenotypes in mammalian cells engaging in three-dimensional chemotaxis. Here, cells lacking WRC displayed impaired motility in 2D; but they were far more effective than wildtype cells at undergoing invasive migration in 3D [50]. Together both studies indicate that WASP-dependent migration may be subject to fewer regulatory inputs when compared to WRC-dependent migration, but it remains unclear to what degree (especially in three dimensions) that WASP proteins contribute to cell migration when WRC is present.
Comparatively little is known regarding the mechanisms mediating amoeboid migration in three dimensions. A report characterizing the movement of neutrophil-like HL-60 cells in microfluidics chambers mimicking a 3D environment found that cells created two distinct actin filament networks at the leading edge. One existed along the sides of the cell membrane in contact with the chamber wall and polymerized perpendicular to it, creating a wedge. A second network, which polymerized just behind the unobstructed edge of the cell, could use this wedge as a platform to funnel the force generated through its assembly into the free edge to drive the cell forward [51]. Like other studies that observed amoeboid migration in a confined environment [52–54], the model proposed here seems to suggest that cell-substrate adhesions, while instrumental for 2D migration, may not be strictly required for motility in a 3D environment. Surprisingly, these authors also found that Arp2/3—but not formin activity—was dispensable for this mode of migration [51]. Unlike Arp2/3, formins do not form branched networks; thus their contributions to the assembly of the highly-dendritic actin networks present at the leading edge was not initially apparent. While a handful of studies have implicated some formins (particularly those of the Dia subgroup) in mediating migration [55,56], the exact mechanisms by which they do so are not completely clear, even in 2D migration. One in vitro study has proposed a mechanism where formins associate with the free barbed ends of filaments nucleated by Arp2/3 to create some of the longer filaments seen at the leading edge of migrating cells [57]; and later work found that the formin Dia and Enabled (a member Ena/VASP family of actin assembly factors) directly interact to modulate the migration speed of Drosophila hematocytes [58]. It will thus be important to address how different mechanisms of actin assembly (e.g., WRC-, WASP-, and formin-mediated), which produce filament networks with distinct architectures, collaborate to drive cell migration under varying environmental conditions.
What controls the number and speed of cell protrusions?
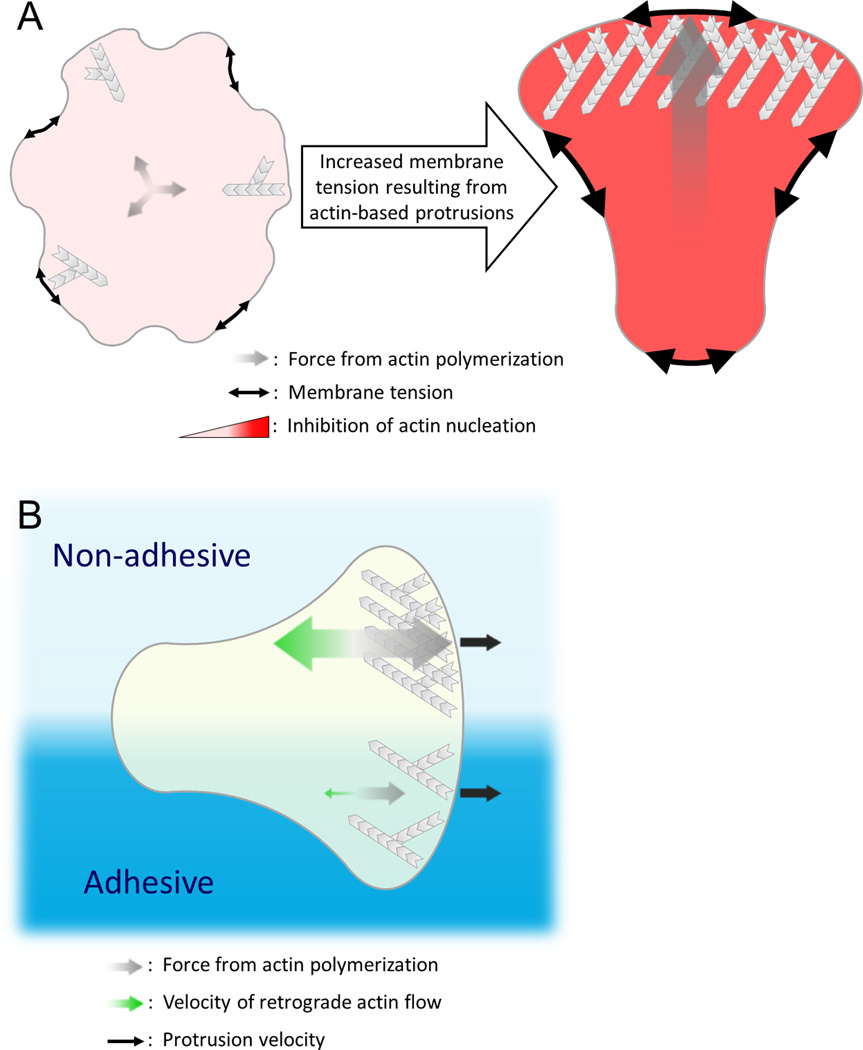
For efficient movement, cells need to limit their number of protrusions. How is this accomplished? A number of recent studies have implicated membrane tension as a global integrator of multiple cellular behaviors, including cell shape and movement [59–65]. In the context of chemotactic cells, neutrophils use tension within the plasma membrane to enable protrusions to compete with one another and establish a dominant front [66] (Figure 2A). The plasma membrane serves as a particularly useful conduit for transmitting information, as localized changes in tension can be propagated almost instantaneously throughout the entire membrane and act on a much faster timescale than alternative methods cells may use, such as reaction-diffusion systems [66].
Figure 2. Actin assembly reciprocally interacts with mechanical forces to regulate protrusion number and speed.
A) In the presence of uniform chemoattractant, cells form a single leading edge through competition between nascent protrusions. This competition occurs through long-range inhibition that is communicated by changes in membrane tension. Actin polymerization at each protrusion exerts force on the plasma membrane (grey arrows), rapidly increasing membrane tension (black double-arrows) throughout the cell. Increased membrane tension globally suppresses the formation of new sites of polarized actin assembly through inhibition of upstream signals such as Rac activation and WRC recruitment; however, assembly still occurs at the initial site since positive feedback components are already present at sufficiently high levels to overcome this inhibition. This system ensures that a single dominant leading edge emerges [66], but it is not yet clear how changes in tension are translated to changes in signaling. B) Dendritic cells in a confined environment maintain a steady rate of forward protrusion, independent of surface adhesiveness. To compensate for reduced traction on non-adhesive surfaces and the subsequent increase in the rate of retrograde flow of the actin network (green arrows), cells increase the rate of actin polymerization directed toward the leading edge (grey arrows) to produce the same protrusion velocity of the plasma membrane (black arrows). Even a single cell that finds itself overlapping surfaces with differing levels of adhesiveness is able to maintain distinct subcellular domains with different rates of actin assembly to produce the same net force across the leading edge and maintain persistence despite the adhesive differences. These data suggest that the speed of cell protrusion is not set by the amount of actin assembly but rather depends on how fast the plasma membrane is released into the protrusions [52]. How this is regulated is unknown.
How do cells ensure that tension remains within the proper range? In keratocytes, plasma membrane tension depends on actin-based based pushing forces from within the lamellipodium: Remarkably, when plasma membrane area is experimentally increased by ~30% (via fusion with giant unilamellar vesicles), cells rapidly increase total actin polymer levels within the lamellipodium to compensate for this enlarged area such that actin filament density (and thus force against the plasma membrane) remains constant [65]. Earlier work using keratocytes as an experimental system developed a mathematical model where the area, aspect ratio, and speed of motile cells could all be predicted through the interplay between actin dynamics and membrane tension, arguing that these are emergent self-organizing properties from the interaction between actin networks and physical forces transmitted through the plasma membrane [63].
Currently, the specific factors that sense and transmit the state of membrane tension to the signaling proteins that regulate actin cytoskeleton are unknown. Among the possibilities, proteins of the BAR domain family are particularly attractive candidates as they associate with membranes (by sensing/influencing their curvature), directly interact with Rho GTPases (which participate in many signal transduction pathways), and bind to—and modulate the activities of—factors promoting actin assembly (including N-WASP, WRC, and formins) in a diverse array of systems [67–71]. Since BAR proteins directly link the plasma membrane to both chemotactic signaling factors and actin assembly machinery, they seem ideally positioned to convey information among all three cellular currencies. Alternatively, the actin polymerization machinery itself could be the sensor. Blocking protrusion also results in an inhibition of WRC recruitment (and potentially its feedback to Rac activity [11,20]), and in vitro actin networks are known to change their activity in response to applied force [72].
Mechanical feedback also appears to set the speed of protrusions in amoeboid cells, as illustrated in an analysis of dendritic cell motility in confined environments as a function of substrate adhesivity. It was previously known that dendritic cell motility requires integrin-based adhesion in 2D, but integrins are dispensable in 3D [53,54]. Here, the authors used a confined chamber (basically two closely-spaced coverslips) to study the integrin-based versus adhesion-independent modes of motility. When placed in the chamber, cells migrated at the same velocity with the same morphology whether the surface was coated with PEG or with serum, preventing or promoting adhesion respectively [52]. Cells compensated for changes in the ‘stickiness’ of a surface by modulating their rates of actin polymerization. As ‘stickiness’ decreased, actin assembly increased to maintain a steady rate of forward protrusion, and this compensation could even be observed with half of the cell migrating in an adhesive mode and the other half migration without adhesions (Figure 2B) [52]. In contrast to other motile cells, such as fibroblasts, which are guided by changes in cell adhesion, dendritic cells appear to ignore adhesive differences during 3D migration. This could enable the cells to be guided by subtle changes in chemoattractant while ignoring large differences in substrate adhesiveness during their migration in vivo. The other surprising implication of this data is that actin is pushing as hard as it can at the leading edge, and what sets the rate of leading edge advance is not the amount of actin assembly but rather the speed with which the membrane can move forward. What sets this limit is a complete mystery, but could include friction in the membrane, release of membrane from other protrusions or the back through retraction or crushing of actin networks.
Future directions
Given the complexity of chemotaxis and the numerous connections among (and within) signaling pathways, actin dynamics, and the plasma membrane, it has been difficult to reconstitute these higher order behaviors in vitro to dissect the mechanisms by which oscillations of actin assembly/turnover contribute to cell migration. The field has recently succeeded in reconstituting the oscillatory dynamics of several self-organizing bacterial proteins in vitro (e.g., MinC/MinD/MinE [73] and FtsA/FtsZ [74]). WRC and N-WASP dependent actin nucleation have also been reconstituted in vitro [41,71,75,76], but they do not exhibit the same oscillatory dynamics as they do in vivo, and it is not clear whether this is due to missing components or a need to change the format in which these proteins are reconstituted (for example inside lipid bilayers instead of on the surface of vesicles). A complementary approach is to study the biochemistry of these components in vivo with more sophisticated tools to manipulate and measure signaling, actin organization, and physical forces. Tools such as atomic force microscopy have been employed to measure changes in membrane tension [77,78] and could potentially be used to interactively manipulate tension as well. Optogenetic [79–85] and chemical [86,87] tools may be used to rapidly modulate protein location/activity in living cells. Such approaches should provide further insight into the complex dance between signaling, forces, and the cytoskeleton that underlie the self-organization of protrusions and polarity.
Acknowledgments
We thank the Weiner lab for helpful discussion. This work was supported by NIH R01GM084040.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.De Bruyn PP. The amoeboid movement of the mammalian leukocyte in tissue culture. Anat Rec. 1946;95:177–191. doi: 10.1002/ar.1090950209. [DOI] [PubMed] [Google Scholar]
- 2.Hartman RS, Lau K, Chou W, Coates TD. The fundamental motor of the human neutrophil is not random: evidence for local non-Markov movement in neutrophils. Biophys J. 1994;67:2535–2545. doi: 10.1016/S0006-3495(94)80743-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Killich T, Plath PJ, Wei X, Bultmann H, Rensing L, Vicker MG. The locomotion, shape and pseudopodial dynamics of unstimulated Dictyostelium cells are not random. J Cell Sci. 1993;106(Pt 4):1005–1013. doi: 10.1242/jcs.106.4.1005. Through careful quantitative measure of cell shape and pseudopod dynamics, this paper provides evidence that the actin cytoskeleton is an oscillatory system more than a decade before fluorescence imaging enabled direct visualization of oscillatory and wavelike actin dynamics.
- 4.Vicker MG. Reaction-diffusion waves of actin filament polymerization/depolymerization in Dictyostelium pseudopodium extension and cell locomotion. Biophys Chem. 2000;84:87–98. doi: 10.1016/s0301-4622(99)00146-5. [DOI] [PubMed] [Google Scholar]
- 5.Vicker MG. Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp Cell Res. 2002;275:54–66. doi: 10.1006/excr.2001.5466. [DOI] [PubMed] [Google Scholar]
- 6. Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Muller-Taubenberger A, Kohler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. TIRF-based imaging of fluorescently-tagged Arp2/3 complex provided the first molecular handle on the excitable actin cytoskeleton in Dictyostelium.
- 7.Gerisch G, Bretschneider T, Muller-Taubenberger A, Simmeth E, Ecke M, Diez S, Anderson K. Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys J. 2004;87:3493–3503. doi: 10.1529/biophysj.104.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong Y, Huang CH, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci U S A. 2010;107:17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veltman DM, King JS, Machesky LM, Insall RH. SCAR knockouts in Dictyostelium: WASP assumes SCAR's position and upstream regulators in pseudopods. J Cell Biol. 2012;198:501–508. doi: 10.1083/jcb.201205058. Both WASP and SCAR/WAVE regulate actin nucleation, but these two proteins have very different upstream regulators and organize very different actin structures (endocytic sites for WASP, lamellipodia/pseudopodia for SCAR/WAVE). Astonishingly, in the absence of SCAR/WAVE, WASP localizes to and potentially supports pseudopod organization and dynamics. These data suggest that the different structures that WASP and WAVE build are not specified by these nucleation promoting factors but rather are likely to depend on other endocytic or lamellipod/pseudopod scaffolds.
- 10. Huang CH, Tang M, Shi C, Iglesias PA, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat Cell Biol. 2013;15:1307–1316. doi: 10.1038/ncb2859. First direct evidence that the polarity signaling network (Ras/PIP3) is an excitable system, with hallmark behaviors of maximal response to signals above a threshold as well as a refractory period between stimuli. Furthermore, the actin cytoskeleton was found to exhibit oscillatory localization patterns even in the absence of upstream inputs. Based on these observations, the authors develop a mathematical model with a slow excitable polarity system coupled to a rapid oscillatory actin network. The two networks reciprocally interact, but how they communicate with one another is not known.
- 11. Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. WAVE complex forms propagating waves in migrating neutrophils, and these waves are negatively regulated by actin assembly events. Waves of actin assembly may underlie some of the complex behaviors observed migrating cells, such as barrier avoidance.
- 12.Millius A, Dandekar SN, Houk AR, Weiner OD. Neutrophils establish rapid and robust WAVE complex polarity in an actin-dependent fashion. Curr Biol. 2009;19:253–259. doi: 10.1016/j.cub.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobereiner HG, Dubin-Thaler BJ, Hofman JM, Xenias HS, Sims TN, Giannone G, Dustin ML, Wiggins CH, Sheetz MP. Lateral membrane waves constitute a universal dynamic pattern of motile cells. Phys Rev Lett. 2006;97 doi: 10.1103/PhysRevLett.97.038102. 038102. [DOI] [PubMed] [Google Scholar]
- 14.Case LB, Waterman CM. Adhesive F-actin waves: a novel integrin-mediated adhesion complex coupled to ventral actin polymerization. PLoS One. 2011;6:e26631. doi: 10.1371/journal.pone.0026631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam Hui K, Kwak SI, Upadhyaya A. Adhesion-dependent modulation of actin dynamics in Jurkat T cells. Cytoskeleton (Hoboken) 2014;71:119–135. doi: 10.1002/cm.21156. [DOI] [PubMed] [Google Scholar]
- 16.Cross MC, Hohenberg PC. Pattern-Formation Outside of Equilibrium. Reviews of Modern Physics. 1993;65:851–1112. [Google Scholar]
- 17.Neilson MP, Veltman DM, van Haastert PJ, Webb SD, Mackenzie JA, Insall RH. Chemotaxis: a feedback-based computational model robustly predicts multiple aspects of real cell behaviour. PLoS Biol. 2011;9:e1000618. doi: 10.1371/journal.pbio.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibata T, Nishikawa M, Matsuoka S, Ueda M. Intracellular encoding of spatiotemporal guidance cues in a self-organizing signaling system for chemotaxis in Dictyostelium cells. Biophys J. 2013;105:2199–2209. doi: 10.1016/j.bpj.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishikawa M, Horning M, Ueda M, Shibata T. Excitable signal transduction induces both spontaneous and directional cell asymmetries in the phosphatidylinositol lipid signaling system for eukaryotic chemotaxis. Biophys J. 2014;106:723–734. doi: 10.1016/j.bpj.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4:e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allard J, Mogilner A. Traveling waves in actin dynamics and cell motility. Curr Opin Cell Biol. 2013;25:107–115. doi: 10.1016/j.ceb.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arai Y, Shibata T, Matsuoka S, Sato MJ, Yanagida T, Ueda M. Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc Natl Acad Sci U S A. 2010;107:12399–12404. doi: 10.1073/pnas.0908278107. First demonstration that the polarity signaling network (including PIP3 and PTEN) can self-organize even in the absence of actin polymer. Reciprocal interactions between PTEN and PI3K coupled through a reaction diffusion system could enable self-organization of polarity during chemotaxis.
- 23.Millius A, Watanabe N, Weiner OD. Diffusion, capture and recycling of SCAR/WAVE and Arp2/3 complexes observed in cells by single-molecule imaging. J Cell Sci. 2012;125:1165–1176. doi: 10.1242/jcs.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue T, Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS One. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, Cantley LC, Bourne HR. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunida K, Matsuda M, Aoki K. FRET imaging and statistical signal processing reveal positive and negative feedback loops regulating the morphology of randomly migrating HT-1080 cells. J Cell Sci. 2012;125:2381–2392. doi: 10.1242/jcs.096859. [DOI] [PubMed] [Google Scholar]
- 28.Cross MC, Hohenberg PC. Spatiotemporal chaos. Science. 1994;263:1569–1570. doi: 10.1126/science.263.5153.1569. [DOI] [PubMed] [Google Scholar]
- 29.Blagg SL, Stewart M, Sambles C, Insall RH. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr Biol. 2003;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- 30.Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, Yoshida N, Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- 32.Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, Fujiwara Y, Bronson R, Snapper SB, Kirschner MW, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Innocenti M, Gerboth S, Rottner K, Lai FP, Hertzog M, Stradal TE, Frittoli E, Didry D, Polo S, Disanza A, et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. 2008;65:904–922. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langridge PD, Kay RR. Mutants in the Dictyostelium Arp2/3 complex and chemoattractant-induced actin polymerization. Exp Cell Res. 2007;313:2563–2574. doi: 10.1016/j.yexcr.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 38.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Nardo A, Cicchetti G, Falet H, Hartwig JH, Stossel TP, Kwiatkowski DJ. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc Natl Acad Sci U S A. 2005;102:16263–16268. doi: 10.1073/pnas.0508228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu X, Kuhn JR. Actin filament attachments for sustained motility in vitro are maintained by filament bundling. PLoS One. 2012;7:e31385. doi: 10.1371/journal.pone.0031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montaville P, Jegou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier MF. Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biol. 2014;12:e1001795. doi: 10.1371/journal.pbio.1001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gohl C, Banovic D, Grevelhorster A, Bogdan S. WAVE forms hetero- and homo-oligomeric complexes at integrin junctions in Drosophila visualized by bimolecular fluorescence complementation. J Biol Chem. 2010;285:40171–40179. doi: 10.1074/jbc.M110.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. Using multivalent interactions, proteins form hetero-oligomers that phase-separate to create liquid protein droplets. Droplet formation allows proteins to dramatically increase their local concentration and might serve as a control point that cells could use to segregate processes to a specific location or for switch-like activation of signaling events.
- 46.Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- 48.Weisswange I, Newsome TP, Schleich S, Way M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature. 2009;458:87–91. doi: 10.1038/nature07773. [DOI] [PubMed] [Google Scholar]
- 49.Ydenberg CA, Smith BA, Breitsprecher D, Gelles J, Goode BL. Cease-fire at the leading edge: new perspectives on actin filament branching, debranching, and cross-linking. Cytoskeleton (Hoboken) 2011;68:596–602. doi: 10.1002/cm.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang H, Li A, Bi J, Veltman DM, Zech T, Spence HJ, Yu X, Timpson P, Insall RH, Frame MC, et al. Loss of Scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Curr Biol. 2013;23:107–117. doi: 10.1016/j.cub.2012.11.059. The roles played by WASP and SCAR/WAVE complex in driving cell motility depend on the mode of migration. WAVE complex is required for 2D migration but is dispensable for movement in 3D. In the absence of WAVE complex, WASP drives invasive migration specifically in 3D environments. Since SCAR/WAVE complex seems to inhibit this invasive migration, it appears, counterintuitively, to function as a tumor suppressor.
- 51. Wilson K, Lewalle A, Fritzsche M, Thorogate R, Duke T, Charras G. Mechanisms of leading edge protrusion in interstitial migration. Nat Commun. 2013;4:2896. doi: 10.1038/ncomms3896. When migrating in a confined environment, neutrophils appear to generate two types of actin networks that make distinct contributions to cell migration. The authors use small-molecule inhibitors to show, somewhat surprisingly, that formins are required for this mode of migration, while Arp2/3 complex is dispensable.
- 52. Renkawitz J, Schumann K, Weber M, Lammermann T, Pflicke H, Piel M, Polleux J, Spatz JP, Sixt M. Adaptive force transmission in amoeboid cell migration. Nat Cell Biol. 2009;11:1438–1443. doi: 10.1038/ncb1992. Migrating dendritic cells alter the rate of actin polymerization at the leading edge to maintain consistent protrusion velocity, even when the adhesive strength of their underlying substrate is not uniform. The surprising implication of this data is that actin is pushing as hard as it can at the leading edge, and what sets the rate of leading edge advance is not the amount of actin assembly but rather the speed with which the membrane can move forward. What sets this speed is unknown.
- 53.Malawista SE, de Boisfleury Chevance A, Boxer LA. Random locomotion and chemotaxis of human blood polymorphonuclear leukocytes from a patient with leukocyte adhesion deficiency-1: normal displacement in close quarters via chimneying. Cell Motil Cytoskeleton. 2000;46:183–189. doi: 10.1002/1097-0169(200007)46:3<183::AID-CM3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 54.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 55.Shi Y, Zhang J, Mullin M, Dong B, Alberts AS, Siminovitch KA. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol. 2009;182:3837–3845. doi: 10.4049/jimmunol.0803838. [DOI] [PubMed] [Google Scholar]
- 56.Kitzing TM, Wang Y, Pertz O, Copeland JW, Grosse R. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene. 2010;29:2441–2448. doi: 10.1038/onc.2009.515. [DOI] [PubMed] [Google Scholar]
- 57.Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bilancia CG, Winkelman JD, Tsygankov D, Nowotarski SH, Sees JA, Comber K, Evans I, Lakhani V, Wood W, Elston TC, et al. Enabled negatively regulates diaphanous-driven actin dynamics in vitro and in vivo. Dev Cell. 2014;28:394–408. doi: 10.1016/j.devcel.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masters TA, Pontes B, Viasnoff V, Li Y, Gauthier NC. Plasma membrane tension orchestrates membrane trafficking, cytoskeletal remodeling, and biochemical signaling during phagocytosis. Proc Natl Acad Sci U S A. 2013;110:11875–11880. doi: 10.1073/pnas.1301766110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gauthier NC, Fardin MA, Roca-Cusachs P, Sheetz MP. Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proc Natl Acad Sci U S A. 2011;108:14467–14472. doi: 10.1073/pnas.1105845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Batchelder EL, Hollopeter G, Campillo C, Mezanges X, Jorgensen EM, Nassoy P, Sens P, Plastino J. Membrane tension regulates motility by controlling lamellipodium organization. Proc Natl Acad Sci U S A. 2011;108:11429–11434. doi: 10.1073/pnas.1010481108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai J, Sheetz MP. Regulation of endocytosis, exocytosis, and shape by membrane tension. Cold Spring Harb Symp Quant Biol. 1995;60:567–571. doi: 10.1101/sqb.1995.060.01.060. [DOI] [PubMed] [Google Scholar]
- 65. Lieber AD, Yehudai-Resheff S, Barnhart EL, Theriot JA, Keren K. Membrane tension in rapidly moving cells is determined by cytoskeletal forces. Curr Biol. 2013;23:1409–1417. doi: 10.1016/j.cub.2013.05.063. Keratocytes modulate the amount of actin polymer within their lamellipodia in response to dramatic changes in plasma membrane area (elegantly generated through electrofusion of GUVs with cells), such that filament density stays constant. This property, which is suggested to operate independently of upstream regulatory inputs, ensures that membrane tension remains consistent in the face of significant changes in membrane area.
- 66. Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. For motile cells, protrusions compete with one another to ensure a dominant front, but the basis of this long-range inhibition was unknown. Through morphological perturbations and cell severing, the authors rule out passive diffusion as a mechanism of long-range inhibition. Tension in the plasma membrane is found to be the dominant source of long-range inhibition that constrain the number and size of cell protrusions.
- 67.Yan S, Lv Z, Winterhoff M, Wenzl C, Zobel T, Faix J, Bogdan S, Grosshans J. The F-BAR protein Cip4/Toca-1 antagonizes the formin Diaphanous in membrane stabilization and compartmentalization. J Cell Sci. 2013;126:1796–1805. doi: 10.1242/jcs.118422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Graziano BR, Yu HY, Alioto SL, Eskin JA, Ydenberg CA, Waterman DP, Garabedian M, Goode BL. The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol Biol Cell. 2014 doi: 10.1091/mbc.E14-03-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed S, Goh WI, Bu W. I-BAR domains, IRSp53 and filopodium formation. Semin Cell Dev Biol. 2010;21:350–356. doi: 10.1016/j.semcdb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 70.Fricke R, Gohl C, Dharmalingam E, Grevelhorster A, Zahedi B, Harden N, Kessels M, Qualmann B, Bogdan S. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr Biol. 2009;19:1429–1437. doi: 10.1016/j.cub.2009.07.058. [DOI] [PubMed] [Google Scholar]
- 71.Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 72.Chaudhuri O, Parekh SH, Fletcher DA. Reversible stress softening of actin networks. Nature. 2007;445:295–298. doi: 10.1038/nature05459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. Self-organizing pattern formation systems are thought to underlie a wide range of biological processes ranging from hydra body axis formation to stripes on a zebra. Self-organizing chemical reactions (Belousov-Zhabotinsky reaction) have previously been demonstrated, but this had never been achieved with biological components. This paper succeeds in reconstituting the first self-organizing protein based pattern formation system. This work is based on the bacterial proteins (MinD/E) that are used to find the center of dividing bacteria. It is the dream of many in the field to similarly reconstitute the signaling and cytoskeletal-based excitable systems that underlie eukaryotic cell movement.
- 74.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koronakis V, Hume PJ, Humphreys D, Liu T, Horning O, Jensen ON, McGhie EJ. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc Natl Acad Sci U S A. 2011;108:14449–14454. doi: 10.1073/pnas.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krieg M, Helenius J, Heisenberg CP, Muller DJ. A bond for a lifetime: employing membrane nanotubes from living cells to determine receptor-ligand kinetics. Angew Chem Int Ed Engl. 2008;47:9775–9777. doi: 10.1002/anie.200803552. [DOI] [PubMed] [Google Scholar]
- 78.Diz-Munoz A, Krieg M, Bergert M, Ibarlucea-Benitez I, Muller DJ, Paluch E, Heisenberg CP. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS Biol. 2010;8:e1000544. doi: 10.1371/journal.pbio.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang X, Jost AP, Weiner OD, Tang C. A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol Biol Cell. 2013;24:2419–2430. doi: 10.1091/mbc.E13-03-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 85.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 2005;435:365–369. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]