Abstract
PreQ1 riboswitches help regulate the biosynthesis and transport of PreQ1 (7-aminomethyl-7-deazaguanine), a precursor of the hypermodified guanine nucleotide queuosine (Q), in a number of Firmicutes, Proteobacteria, and Fusobacteria. Queuosine is almost universally found at the wobble position of the anticodon in asparaginyl, tyrosyl, histidyl and aspartyl tRNAs, where it contributes to translational fidelity. Two classes of PreQ1 riboswitches have been identified (PreQ1-I and PreQ1-II), and structures of examples from both classes have been determined. Both classes form H-type pseudoknots upon PreQ1 binding, each of which has distinct unusual features and modes of PreQ1 recognition. These features include an unusually long loop 2 in PreQ1-I pseudoknots and an embedded hairpin in loop 3 in PreQ1-II pseudoknots. PreQ1-I riboswitches are also notable for their unusually small aptamer domain, which has been extensively investigated by NMR, X-ray crystallography, FRET, and other biophysical methods. Here we review the discovery, structural biology, ligand specificity, cation interactions, folding, and dynamics, and applications to biotechnology of PreQ1 riboswitches.
Keywords: queuosine, tRNA modification, NMR, X-ray crystallography, queuine, preQ0
1. Introduction
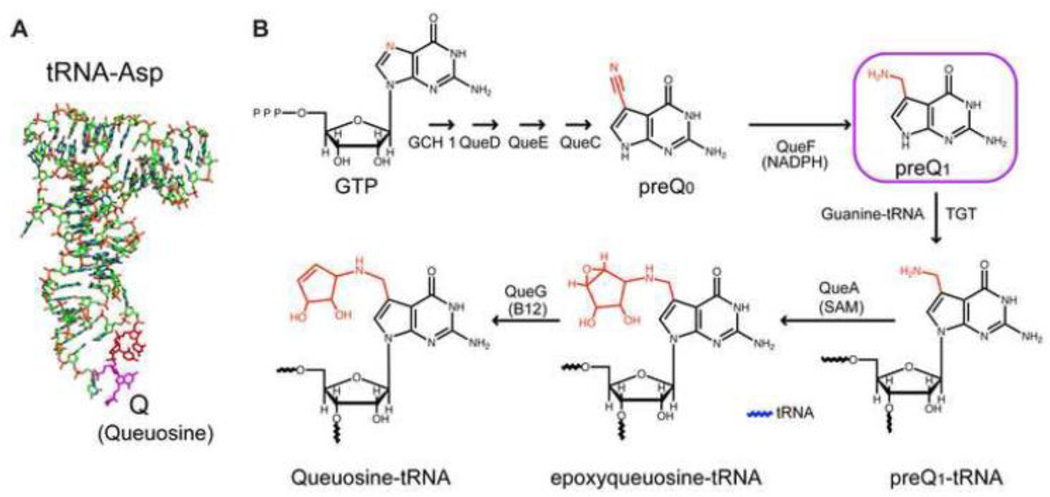
The prequeuosine 1 (PreQ1) riboswitch is a member of a large sub-set of riboswitches that are involved in regulation of purine synthesis and transport [1, 2]. These include riboswitches whose aptamer domain recognizes the bases adenine or guanine, the nucleoside 2’-deoxyguanosine, the second messenger cyclic-di-GMP, and the modified guanine PreQ1, as well as the purine moiety of the co-factors S-adenosylmethionine, S-adenosylhomocysteine, and adenosylcobalamin [1, 2]. PreQ1 riboswitches help regulate the biosynthesis and transport of PreQ1 (7-aminomethyl-7-deazaguanine), a precursor of the hypermodified guanine nucleotide queuosine (Q). Q is almost universally found in prokaryotes and eukaryotes (but not archeae, which utilize the Q homolog archaeosine [3]) in asparaginyl, tyrosyl, histidyl and aspartyl tRNAs at the anticodon wobble position [4–7] (Figure 1A).
Figure 1.
Structure and synthesis of queuosine in bacteria. (A) Structure of aspartyl tRNA with queuosine at the wobble position of the anticodon highlighted in magenta. (B) Pathway for synthesis of queuosine, highlighting the structures of preQ0 and PreQ1.
Modified nucleotides are found in all classes of RNA molecules and are essential for normal cellular function [8]. tRNAs are the most highly modified RNAs, containing on average 13 modified nucleotides per tRNA from over 100 unique RNA modifications [9]. These modifications contribute to tRNA stability [10], prevent ribosomal frameshifting [11], increase translation efficiency and fidelity [12], and improve anticodon-codon recognition [13]. Nearly all tRNAs contain a modification at the wobble position of the anticodon [14]. These modifications, including Q, are believed to influence the stability of the anticodon-codon interaction and promote translation accuracy. Q has been implicated to play a role in eukaryotic cellular development and proliferation [15–21], relieving hypoxic stress [22], neoplastic transformation [18, 23–27], tyrosine biosynthesis [28], translation frameshifting required in retroviral protein synthesis [29], and virulence of pathogenic bacteria such as Shigella flexneri [30–33] (reviewed in [8, 34]).
Prokaryotes synthesize PreQ1 de novo from GTP in a multienzyme pathway (Figure 1B) (reviewed in [35]. First, preQ0 is synthesized in a series of reactions successively involving GTP cyclohydrolase (GCH1), 6-carboxy-5,6,7,8-tetrahydropterin synthase (QueD), 7-carboxy-7-deazaguanine (CDG) synthase (QueE), and preQ0 synthase (QueC), which replaces the guanine N7 with an acetonitrile group, in a pathway that was recently elucidated [35]. PreQ0 is converted to PreQ1 by the preQ0 reductase (QueF), which reduces the nitrile group to an exocyclic aminomethyl substituent. The free base PreQ1 is then directly attached to the wobble position of the anticodon of cognate tRNAs by tRNA:guanine transglycosylase (TGT), which catalyzes the posttranscriptional exchange of guanine with PreQ1. The exocyclic amine at the 7 position is then modified by S-adenosylmethionine:tRNA ribosyltransferase-isomerase (QueA) to epoxyqueuosine-tRNA, and finally epoxyqueueosine (oQ) is converted to Q by oQ reductase (QueG). The cyclopentadiol substituent of Q is sometimes further modified in certain organisms with a glutamate, mannose, or galactosyl group [36, 37]. Because eukaryotes do not synthesize queuosine precursors de novo but rather scavenge Q from their diet or intestinal flora as the free base, queuine, PreQ1 riboswitches are only found in prokaryotes.
Two classes of PreQ1 riboswitches have been identified (PreQ1-I and PreQ1-II) with distinct aptamer secondary and tertiary structures. The PreQ1-I riboswitches have the smallest known aptamer domain discovered to date, which has facilitated numerous biophysical and computational modeling studies on structure and folding. The larger PreQ1-II riboswitch aptamer provides an interesting example of convergent evolution, utilizing a distinct mode of ligand recognition from the PreQ1-I riboswitch. Here we review X-ray crystal and NMR solution structures of PreQ1-I and PreQ1-II riboswitches and what has been learned from structural and biophysical studies about PreQ1 riboswitch folding, dynamics, cation interactions, and specificity of ligand binding, and potential applications to biotechnology.
2. Identification of PreQ1 riboswitches and sequence conservation
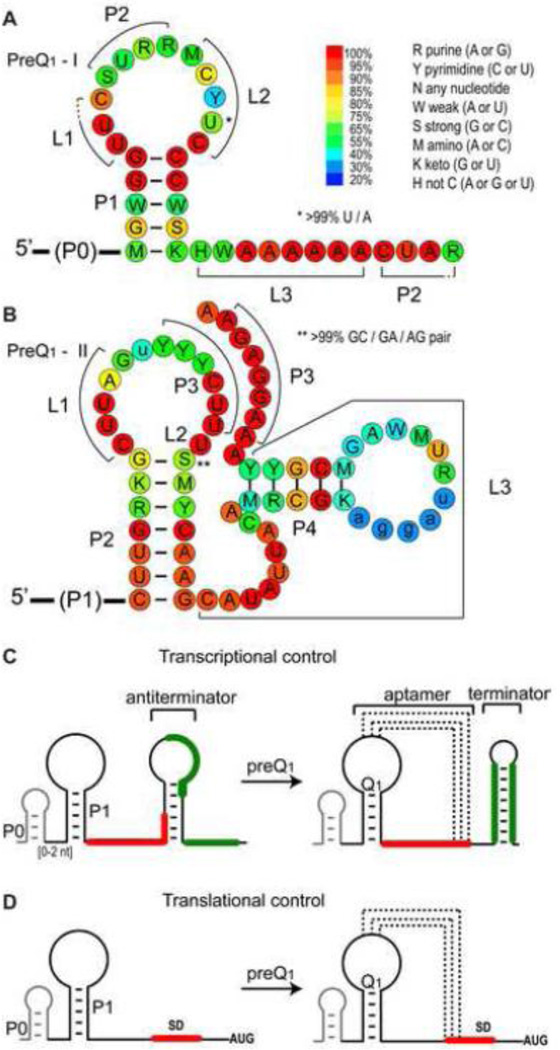
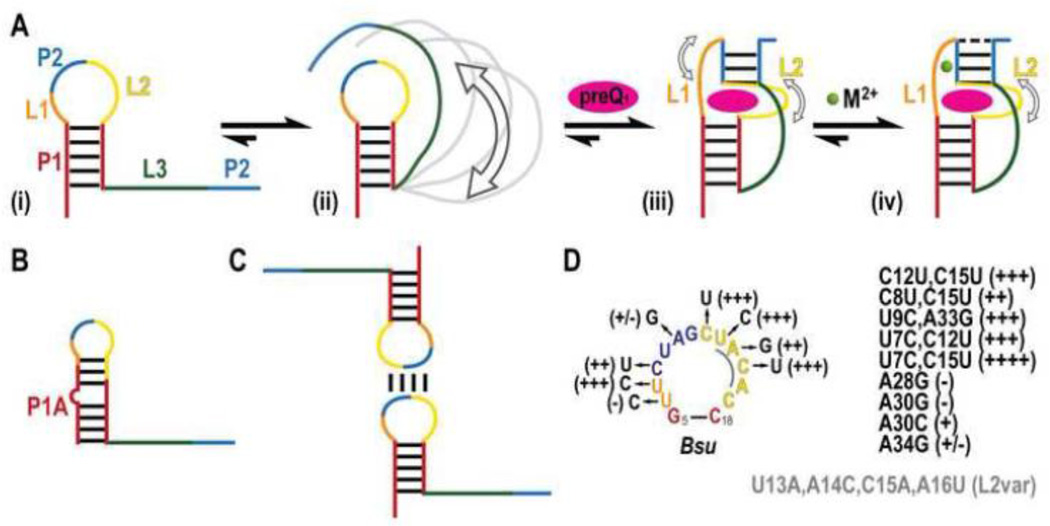
PreQ1-I riboswitches were first identified in a bioinformatics survey of noncoding DNA regions across 91 microbial genomes, and were commonly found in the 5’ UTR of the ykvJKLM operon encoding QueC, QueD, QueE, and QueF [38, 39]. The unusually small size of the aptamer (later determined to be minimally 34 nucleotides (nt) [39] posed a challenge in the initial survey, and the PreQ1-I motif was only observed in a few species in the orders Bacialles and Clostridia; upon revision of the initial search algorithm PreQ1 riboswitches were identified across the phyla Firmicutes, Proteobacteria, and Fusobacteria [39]. PreQ1-I riboswitches were initially sorted into two types, type 1 and 2. These two types are highly similar and differ mainly in the conserved apical loop sequence [39]. However, the solved PreQ1-bound structures of types 1 and 2 are nearly identical, and some riboswitches share type 1 and type 2 features. These types are combined in the Rfam database and fall under the generic PreQ1-I class; to date, nearly 900 sequences across 647 species have been identified for PreQ1-I riboswitches [40]. Phylogenetic analysis showed that the PreQ1-I riboswitches have a conserved secondary structure of a hairpin (P1) followed by an A-rich sequence (Figure 2A). The P1 hairpin is often preceded by a P0 hairpin (Figure 2C), which is not required for ligand binding and has only a small effect on Kd (~2-fold) [39].
Figure 2.
Predicted secondary structure, sequence conservation, and gene regulatory mechanism of PreQ1 riboswitches. (A,B) Sequence conservation of riboswitch aptamer domains of PreQ1-I (A) and PreQ1-II (B). Sequence conservation is from 894 sequences from 647 species for PreQ1-I and 429 sequences from 423 species for PreQ1-II from the Rfam database (1/31/2014) http://rfam.sanger.ac.uk/. (C,D) Schematic of gene regulation by PreQ1-I riboswitches for (C) transcription or (D) translation. The antiterminator in (C) and mechanism for regulation were revealed from structural studies [45, 46].
A second class of PreQ1 riboswitches (PreQ1-II) with a different conserved sequence, secondary structure, and apparently different mode of recognition was found associated with the COG4708 gene family, which are membrane proteins predicted to be involved in Q transport [41, 42]. This motif appears to be much more limited across phylogeny and has only been identified in the families Streptococcaceae and Lactobacillaceae within the order Lactobacillales; to date, 429 sequences across 423 species have been identified for PreQ1-II riboswitches in the Rfam database [40]. PreQ1-II riboswitches have a conserved secondary structure of three hairpins (P1, P2, and P4) and a potential pseudoknot hairpin (P3) (Figure 2B). Similar to the P0 hairpin in the PreQ1-I riboswitch, the P1 stem is dispensable for PreQ1 binding in vitro [42–44]. While PreQ1-I riboswitches have been identified that regulate transcription or translation (Figure 2C,D), PreQ1-II riboswitches identified to date are only involved in translation regulation (Figure 2D).
3. THE PreQ1-I RIBOSWITCH
3.1 PreQ1-I aptamer structures
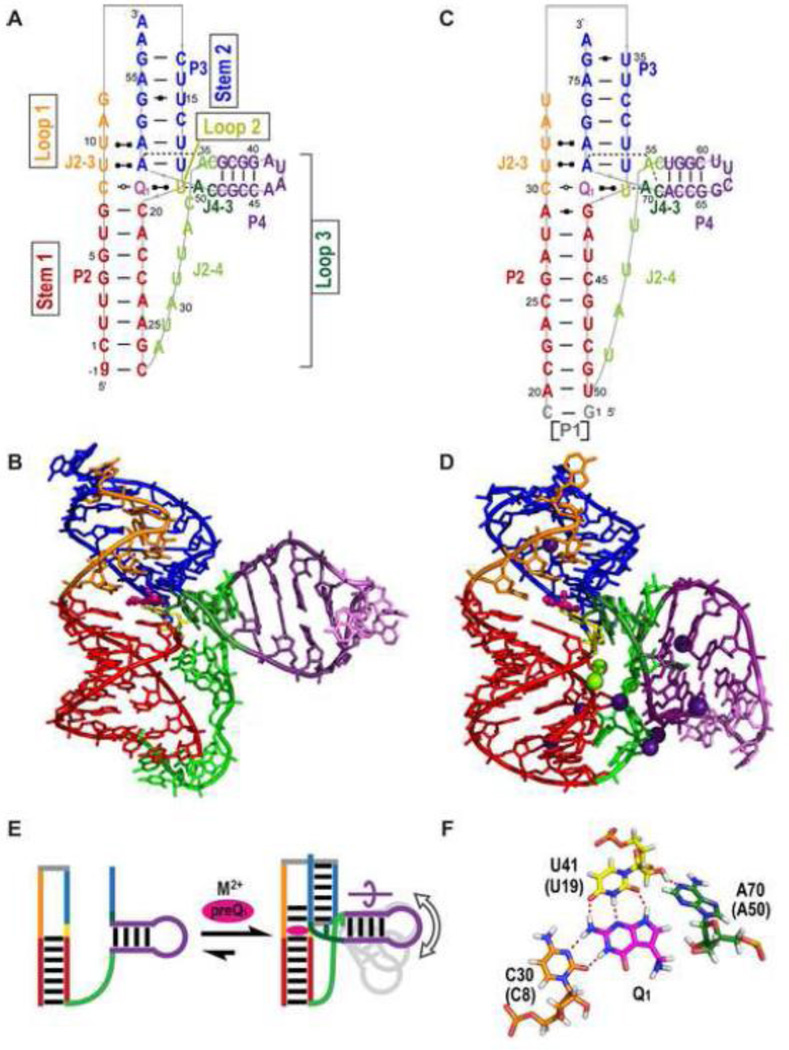
Structures of ligand-bound PreQ1-I riboswitch aptamers were reported from three laboratories at about the same time: the X-ray crystal and NMR solution structures of a transcription-regulating Bacillus subtilis (Bsu) riboswitch (type 2) bound to PreQ1 [45, 46] and the X-ray crystal structure of a translation-regulating Thermoanaerobacter tengcongensis (Tte) riboswitch (type 1) bound to preQ0 [47] (Table 1). A crystal structure of the Tte riboswitch bound to PreQ1 was later reported [48], and below we first compare the PreQ1-bound riboswitch structures (Figure 3). All of these structures were determined from minimal aptamer sequences, identified by in-line probing and deletions of the Bsu riboswitch [39], which did not include the putative P0 helix. The structures revealed that in the presence of PreQ1 the 3’ end of the A-rich tail of the PreQ1-I aptamer base pairs with the center of the P1 hairpin loop to form an H-type pseudoknot with two stems (stem 1 and stem 2) and three loops (loop 1, 2, and 3), and together provided detailed insight into the determinants of ligand recognition (Figure 3). Evidence for pseudoknot formation in the presence of PreQ1 was also shown for a Fusobacterium nucleatum (Fnu) PreQ1-I riboswitch (type 2) by imino proton NMR studies of site-specifically 15N-labeled RNA [49].
Table 1.
Structures of preQ1 riboswitches
| Class | Organism | Method | PDB | Ions in structures (Cations in buffer) | Ligand | Name (# nts) | Ref |
|---|---|---|---|---|---|---|---|
| I | Bsu | X-ray | 3FU2 | Ca2+ (Mg2+) | preQ1 | Bsu WT (34) | [46] |
| I | Bsu | X-ray | 3K1V | Ca2+ (Mg2+) | preQ1 | Bsu L2var L2:A13C14A15U16 (34) | [46] |
| I | Bsu | NMR | 2L1V | (K+) | preQ1 | Bsu C12, 15U (36, with 5’ GG) | [45, 50] |
| I | Tte | X-ray | 3Q50 | SO42− (Mg2+) | preQ1 | Tte WT (33) | [48] |
| I | Tte | X-ray | 3GCA | SO42−(Mg2+) | preQ0 | Tte WT (33) | [47] |
| I | Tte | X-ray | 3Q51 | SO42−, Mg2+ | none | Tte WT (33) | [48] |
| II | Lra | X-ray | 4JF2 | Mg2+, Cs+ | preQ1 | Lra WT’b(77, with 5’P1) | [43] |
| II | Spn | NMR | 2MIY | (Ca2+, K+) | preQ1 | Spn WT (58) | [44] |
L2var changes are A13C14A15U16
P4 loop changed from AAA to UUCG
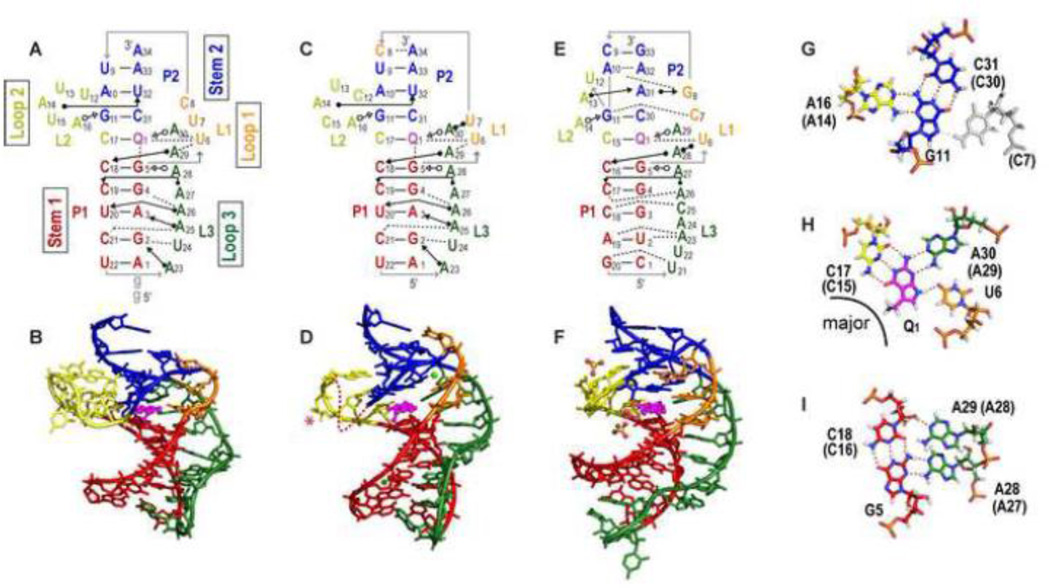
Figure 3.
Structures of PreQ1-I riboswitch aptamers. (A-B) Solution NMR structure of Bsu (C12,15U) [PDB ID 2L1V), (C-D) X-ray crystal structure of Bsu WT [PDB ID 3FU2] (E-F) X-ray crystal structure of Tte WT [PDB ID 3Q50]. (A,C,E) Schematics of secondary structures with base interactions illustrated using the symbols from Leontis and Westhof notation [87]. (B, D, F) Three-dimensional structures. Nucleotides C12 and U13 (shown as a dotted line) and the base of C15 (marked with an asterisk) are missing from the electron density in the Bsu WT crystal structure (D). Mg2+ and SO42− are depicted as green, and yellow and red spheres, respectively. (G, H, I) Stick representation of the three layers of the PreQ1-I binding pocket (from Bsu WT) showing (G) ‘ceiling’: loop 1-stem 2 A-G-C triple (A-G-C-C quartet for Tte, the loop 1 C shown in gray). (H) ‘binding core’: PreQ1-loop 2-loop 3-loop 1 PreQ1-C-A-U quartet, and (I) ‘floor’: loop 3-stem 1-loop 3 A-C-G-A quartet. The corresponding residue numbers for Tte WT are given in parentheses. For all structures, residues are colored as follows: P1 (red), P2 (dark blue), L1 (orange), L2 (gold), L3 (green), PreQ1 (magenta). For the PreQ1 binding core, hydrogen is white, nitrogen is blue, oxygen is red, phosphorus is orange, and carbon follows the main color scheme. For ease of comparison, structures are numbered starting with the first nucleotide in P1; note that this differs by two from the numbering used for the NMR structure of Bsu aptamer in references [45, 50].
Two crystal structures of the Bsu riboswitch aptamer were determined, one of the wild-type (WT) sequence (Bsu WT) (Figure 3C-D) and the other of a loop 2 sequence variant (Bsu L2var), where the central four of the six loop 2 nts are substituted, which diffracted to higher resolution (Table 1). The NMR structure has two nt substitutions in loop 2, C12U and C15U (Bsu C12,15U), and has two additional Gs at the 5’ end that precede the P1 stem in the WT sequence (Figure 3A-B). Only the last residue of loop 2 (C17) is highly conserved and these differences have little effect on the structures, but they do impact folding and apparent ligand affinity, as discussed in section 3.5. The crystal and solution structures of the Bsu riboswitch are nearly identical (Figure 3A-D), with small but significant differences in the positions of residues in the upper part of the pseudoknot, discussed below. The structure of the Tte riboswitch (Tte WT) is also highly similar to the Bsu riboswitch structures with a nearly identical binding pocket despite sequence differences, particularly in the residues above the binding pocket (Figure 3E,F).
In the solved pseudoknot structures, stem 1 is 5 bp, stem 2 is 3-4 bp, loop 1 is 2-3 nt, loop 2 is 4-6 nt, and loop 3 is 8-9 nt. Stems 1 and 2 form a continuous stack with PreQ1 inserted at the junction between them, where it forms a Watson-Crick (WC) base pair (bp) with the last base of loop 2 (C17 in Bsu, C15 in Tte) (Figure 3 A, C, E). As is usual for H-type pseudoknots loop 1 lies along the major groove and loop 3 lies along the minor groove, and loop 3 has extensive interactions with the bases in stem 1 (Figure 3A-F). Loop 2 connects stem 1 and stem 2, and is absent or short (0-2 nt) in most H-type pseudoknots. The unusually long loop 2 in the PreQ1-I riboswitch aptamer, which is largely looped out near the minor groove, likely explains why this pseudoknot fold was not predicted and is important for how the riboswitch functions in gene regulation, as discussed in section 3.5. Another important feature is the short stem 2, which in the Bsu riboswitch has only 3 WC bp possibly capped by a C-A bp and in the Tte riboswitch has two WC and two non-WC bps (Figure 3A-F). Thus, stem 2 would be intrinsically unstable in the absence of tertiary interactions from loop 1 and 2.
The first 8-9 residues of the A-rich tail that follows the P1 hairpin form loop 3, while the 3’ end of the tail pairs with the center of the P1 hairpin loop to form P2, with the residues on the 5’ and 3’ side of that hairpin loop comprising loop 1 and loop 2, respectively (Figure 2A). Loop 3 is highly conserved and A-rich (7 of 8 nts in Bsu, 6 of 9 nts in Tte) and interacts along the length of the minor groove of the 5 bp stem 1, notably through many adenine NH2 interactions with the sugar edge (N3 of A, O2 of C or U, 2’ OH) of bases in stem 1 (A amino-kissing motif) (Figure 3A,C,E,I). Loop 2 is 6 nts in the Bsu riboswitch and 4 nts in the Tte riboswitch. The only highly conserved residue in this loop, the last residue C17 (C15 in Tte), forms a WC bp with PreQ1 The two adenine residues in loop 2 interact in the minor groove of stem 2, forming a continuous stack with C17: A14 (A13 in Tte) has an A-minor interaction with U32 2’OH (U31 in Tte); A16 (A14 in Tte) forms an A-G-C triple, trans WC-sugar edge (Figure 3G). In the solution structure of Bsu (C12,15U) the bases of the remaining three loop 2 nts (12, 13, 15) are flipped out. In the crystal structure of Bsu WT, C12, U13, and the base of C15 are crystallographically disordered while for Bsu L2var all of loop 2 is crystallographically disordered except for C17. Thus, loop 2 is partially dynamic, as discussed in section 3.5. The shorter Tte loop lacks two of the pyrimidine residues but has a similar 3 nt stack with the remaining nt (U12) flipped out [47].
Loop 1 is short (2-3 residues), runs from the top of P1 to the top of P2, and forms a tight turn in the major groove of P2. The first residue, U6, interacts with PreQ1 and loop 3 residue A29 (A28 in Tte). In the crystal structures of the Bsu riboswitches, U7 hydrogen bonds to the Hoogsteen edge of A30 (the last residue of loop 3) and C8 pairs with A34 at the top of P2. In the solution structure of the Bsu C12,15U riboswitch, the positions of U7 and C8 are dynamic, the backbone turn is less sharp, and C8 is not within base pairing distance of A34. An NMR study in which the Bsu solution and crystal structures were rigorously compared using experimental and calculated residual dipolar couplings (RDCs), which provide information on the orientation of each bond vector, confirmed that these differences are largely due to Ca2+-induced structural changes [50], as discussed in sections 3.4 and 3.5. As this is the only example where the structure of the same riboswitch has been determined both by NMR and crystallography, it is worth noting that the RDC analysis also confirmed that the bottom half of the riboswitches, including P1 and loop 3, are virtually identical in solution and in the crystal. In the Tte riboswitch, the corresponding loop 1 residues C7 and G8 interact with the bottom half of stem 2 (Figure 3E,F).
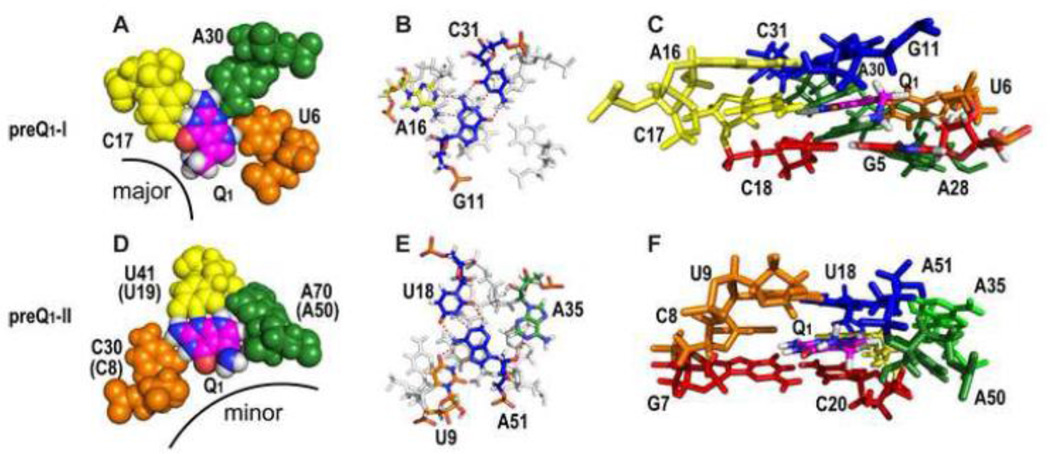
The PreQ1 binding pocket The PreQ1 binding pocket can be described as three layers (Figure 3G-I), the PreQ1 ‘binding core’ where PreQ1 hydrogen bonds to one residue from each of the three loops to form a PreQ1-C-A-U base quartet (Figure 3H), the ‘floor’ of the binding pocket where the C-G bp at the top of stem 1 forms an A-C-G-A base quartet with two successive As from loop 3 (Figure 3I), and the ‘ceiling’ where the G-C bp at the bottom of stem 2 forms an A-G-C triple (A-G-C-C quartet in Tte) above the binding core (Figure 3G). In the binding core, specificity for PreQ1 is achieved by hydrogen bonds to all of the proton donors and acceptors of the PreQ1 ring. The last C in loop 2 (C17 in Bsu, C15 in Tte) interacts via a standard WC bp, the last A in loop 3 (A30 in Bsu, A29 in Tte) and first U in loop 1 (U6) hydrogen bond to the sugar edge of PreQ1 (Figure 3H). In the Bsu riboswitch structures, the exocyclic amine group of PreQ1 points out of the major groove and down to the G-C bp below, at the ‘floor’, and has van der Waals contacts with guanine N7 and O6 (Figure 3B, D; see also Figure 7) [45, 51]. In the crystal structure of Tte WT the exocyclic amine is sterically occluded from interacting with G5 by a bound SO42− and is in the plane of the PreQ1 ring (Figure 3F). However, substitution of G5 for diaminopurine, which replaces the keto group at position 6 with an amine, decreased the binding affinity more than 450-fold for PreQ1 and 46-fold for preQ0, indicating that the interaction between the methyl amine of PreQ1 and the O6 of G5 is also important for binding affinity in the Tte riboswitch despite the absence of interaction in the crystal structure [48].
In addition to the interaction of the PreQ1 methylamine to G5 in the ‘floor) of the binding pocket, the base quartet and base triple below and above the binding core stabilize PreQ1 in the binding pocket by stacking interactions and also sterically occlude it from exiting except from the major groove side. The loop 2 adenine (A16 in Bsu, A14 in Tte) in the ‘ceiling’ A-G-C triple stacks on the loop 2 C that pairs with PreQ1 (C17 in Bsu, C15 in Tte). However, an A at this position is only 30% conserved, and is instead a U in 70% of the sequences (Figure 2A). A U-G-C triple would be nearly isosteric with the A-G-C triple (Figure 3G), and could in fact be more stable by adding an additional hydrogen bond between the 2’OH of G and O4 of U.
3.2 Mechanism of transcriptional and translational regulation
In the seminal study of the Bsu riboswitch the aptamer domain and a terminator hairpin were identified, but no obvious mechanism for ligand dependent control of terminator formation was found [39]. The solved structures of the Bsu riboswitch immediately suggested the classic terminator-antiterminator switching mechanism of riboswitch transcriptional control, and an antiterminator was identified whose stem would sequester the 3’-end of the A-rich tail required for formation of the pseudoknot (Figure 2C) [45, 46]. For the Bsu riboswitch, the antiterminator is predicted to be significantly less stable than the terminator. NMR studies of a 52 nt RNA construct containing the aptamer and anti-terminator sequence (but not the terminator) confirmed formation of the antiterminator in the absence of PreQ1, and showed that when the antiterminator is present PreQ1 addition did not induce the pseudoknot to form [45]. An 19F NMR study of a selectively labeled full-length transcriptional Fnu riboswitch found that in the absence of PreQ1 the expression platform had a 50:50 distribution between anti-terminator and terminator, which have comparable stabilities [52]. In this case, addition of PreQ1 shifted the equilibrium to 80% terminator. This is in contrast to several other transcriptional riboswitches, where addition of ligand to the full-length riboswitch does not cause a response in vitro [53, 54].
The structure of the Tte riboswitch bound to preQ0 [47] or PreQ1 [48] also suggested a translational control mechanism, where the 5’ end of the SD sequence is sequestered as part of P3 (stem 2 of H-type pseudoknot) of the folded aptamer. However, only the first two residues of the SD sequence are paired, at the top of P3 (Figure 3E).
3.3 Comparison to PreQ1-I riboswitch preQ0 bound and apo structures
Three structures of the Tte riboswitch have been determined: with preQ0 bound [49], with PreQ1 bound, and the “apo” form without ligand [48]. The preQ0 and PreQ1 bound aptamer structures are almost identical, with an RMSD of 0.578 Å, including the position of the ligands (Figure 3H, 4F). Improved affinity (5-10 fold) and selectivity for PreQ1 vs preQ0 is apparently conferred by the additional van der Waals or electrostatic interactions between the exocyclic amine of PreQ1 and the base pair below the PreQ1-quartet, at the floor of the binding pocket.
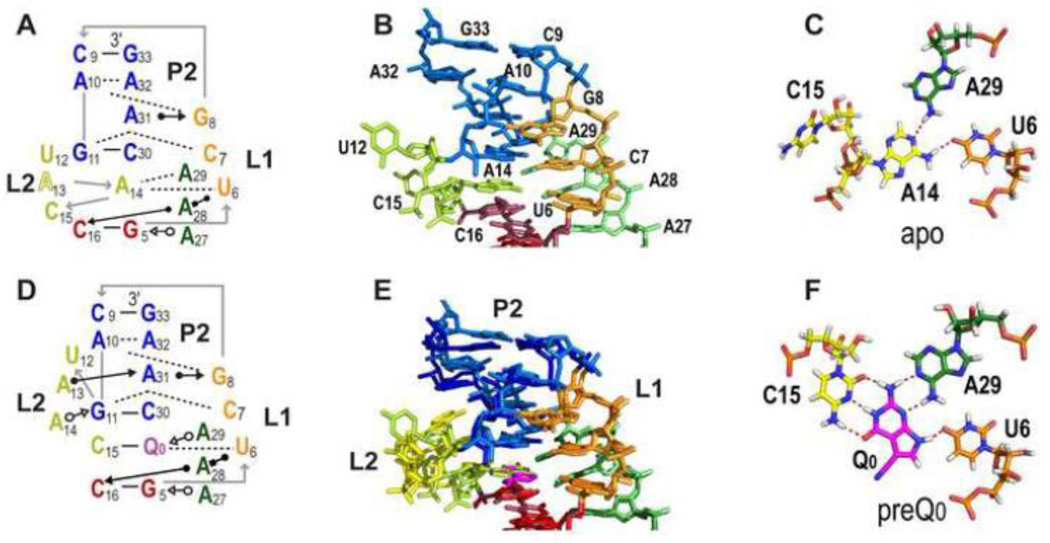
Figure 4.
X-ray crystal structures of the apo and preQ0 bound Tte PreQ1-I riboswitch. (A-C) apo Tte WT: (A) Schematic of secondary structures and base interactions at and above PreQ1 binding pocket. (B) Stick representation of structure of the regions shown in (A). (C) The loop 2-loop 3-loop 1 A14-A29-U6 base triple formed in the apo structure, where A14 replaces preQ0 or PreQ1 in the binding core. C15 is flipped out of the binding pocket. (D-F) Tte WT with preQ0: (D) Schematic of secondary structure at and above PreQ1 binding pocket. (E) Superposition of the apo and preQ0 bound structures, to highlight the similarities and differences (F) The preQ0-loop 2-loop 3-loop 1 preQ0-C15-A29-U6 quartet.
There are only a few structures of ligand-free riboswitch aptamers [55]. The structure of the Tte translation-regulating riboswitch in the absence of PreQ1 revealed a pre-organized binding pocket with high similarity to the bound conformation (1.7 Å overall RMSD) [48]. Intriguingly, loop 2 residue A14 occupies the position of PreQ1 in the PreQ1-bound structure where it interacts with A29 (loop 3) and U6 (loop 1), while loop 2 residue C15, which forms a WC bp with PreQ1 in the PreQ1 bound structure, is flipped out (Figure 4C). The position of A14 in the binding core disrupts the stacking of the loop 2 bases next to stem 2 (Figure 4B). The authors propose that this makes the last two residues of stem 2, A32G33, at the beginning of the Shine-Delgarno (SD) sequence more accessible to the 16S rRNA of the ribosome.
3.4 Cation interactions in PreQ1-I aptamers
Due to the close approach of phosphates from tertiary interactions in pseudoknots, they are in general stabilized by divalent cations. Many riboswitch pseudoknots also require divalent cations as a prerequisite for ligand binding [56–60]. The NMR structure of the Bsu PreQ1-I riboswitch was determined in 50 mM KCl, without added divalent cations, indicating that the aptamer can fold and bind PreQ1 tightly in the absence of divalent cations. The NMR study of an Fnu PreQ1-I riboswitch also showed evidence for pseudoknot formation with PreQ1 without Mg2+, but Mg2+ was apparently required for all molecules to fold [49, 61]. In the crystal structures of Bsu WT and Bsu L2var, two to five Ca2+ are present in different molecules of the asymmetric unit, but only one, located at the sharp turn in the backbone between U7 and C8 is shared in common among the crystal forms. A second Ca2+, visible only in the higher resolution Bsu L2var structure, interacts with the PreQ1 amino methyl group via a Ca2+ hydration water and was proposed to contribute to ligand binding [46]. To investigate the effect of Ca2+ on the PreQ1-I Bsu aptamer structure, Zhang et al [50] compared the solution and crystal structures of the PreQ1-I Bsu aptamer in the presence and absence of Ca2+ using NMR order tensor analysis of RDCs. This analysis indicated that in the presence of Ca2+ the solution structure of the PreQ1 bound Bsu C12,15U riboswitch becomes more similar to the crystal structure. Analysis of chemical shift changes during titration of Ca2+ revealed one higher affinity binding Ca2+, with an apparent Kd of 47 JLM located near the tight turn and a second weaker Ca2+ binding site likely corresponding to the one near the PreQ1 methylamine in the Bsu L2var crystal structure [50]. NMR relaxation studies also showed that loop 1 residues U7 and C8 are dynamic in solution without Ca2+, but that conformational exchange of C8 on the slow time scale (/is-ms) is completely quenched upon Ca2+ binding (Figure 5A) [50].
Figure 5.
Schematic of PreQ1-I riboswitch folding pathway (A) (i) The Bsu riboswitch initially folds into a hairpin followed by a flexible ssRNA tail. (ii) The ssRNA tail, in the presence of divalent cations, dynamically samples a variety of conformations (illustrated in gray), some of which bring L3 close to P1. (iii) On addition of PreQ1, the P2 stem forms and L3 docks into the P1 minor groove. L1 and L2 show dynamics on the µs-ms and ps-ns timescales, respectively (open arrows). (iv) Divalent cations bind to L1, quenching local flexibility while retaining L2 motions. (B) Alternate P1A hairpin observed in Bsu NMR construct as a result of additional 5’ GG residues. (C) A palindromic sequence commonly observed overlapping with the P2 site in the P1 loop gives rise to dimers. (D) Effect of Bsu loop mutagenesis on apparent PreQ1 binding. +++ better than WT, ++ equivalent to WT, + and +/− worse than WT, − no detectable binding. Multiple mutations or mutations to L3 are inset. Mutations in Bsu L2var are shown in gray.
The crystal structures of PreQ1- and preQ0-bound translation-regulating Tte riboswitches also did not contain any ordered divalent cations, although MgCl2 was present in crystallization buffer. Instead, they both contain 4 SO42− ions, two of which stabilize C16 and C18 in stem 1 by coordinating to the C amino group. In the apo Tte PreQ1-I riboswitch, one Mg2+ coordinates to N7 of G5 at the top of P1. Taken together, these results indicate that divalent cations stabilize but are not required for PreQ1 binding in PreQ1-I riboswitches.
3.5 PreQ1-I riboswitch folding and dynamics
SAXS and computational studies of folding of the translation-regulating Tte riboswitch Given that solution NMR data indicated that the PreQ1-I aptamer requires PreQ1 for pseudoknot formation [45, 49], the structure of the compact apo Tte riboswitch was somewhat surprising (Figure 4A-C). In the apo form, A14 rearranges to occupy the PreQ1 site, as discussed in section 3.3. This rearrangement collapses the “ceiling” of the binding pocket, increasing the solvent accessibility of the SD sequence nearly 50 Å2, coupled with a 7.6% increase in unit cell volume [48]. The increased exposure of the SD may explain how the Tte riboswitch can regulate translation in the absence of PreQ1. Wedekind and coworkers [48] also performed SAXS experiments, which provided evidence that the compact structure of the apo riboswitch seen in the crystal structure is also present in solution. However, deviations were observed when the SAXS scattering profile was compared to the crystallographic coordinates, indicating that in solution there likely exists an ensemble of ‘open’ and ‘closed’ states. Single molecule Förster Resonance Energy Transfer (smFRET) and computational studies by Suddala et al [62] on the Tte riboswitch showed that, consistent with the SAXS data, in the absence of PreQ1 the Tte riboswitch dynamically samples two states: a mid-FRET state, which is a diverse ensemble of partially folded conformations, and a high-FRET state, which is consistent with the folded apo Tte structure (Figure 4A). Molecular dynamics simulations by Banáš et al [63] of the Tte PreQ1-I riboswitch revealed that the relatively short P2 stem of the apo structure was highly unstable and irreversibly melted on the μs timescale while the P2 stem of the bound structure was stable. The stability of P2 appeared to be dependent upon stacking interactions between the terminal U10-A32 bp and L2 residue A13. In the absence of PreQ1, L2 showed conformational plasticity in contrast to the bound Tte structure, whose L2 conformation remained stable. This study concluded that loop 2 could act as a sensor for ligand by providing different stacking arrangements for P2 and modulating its stability [63].
The Tte apo conformation may allow for faster ligand recognition by weakly forming the binding pocket, A-minor interactions, and pseudoknot contacts, allowing PreQ1 to quickly enter the binding pocket and lock the pseudoknot in place. The ability of the Tte riboswitch to undergo exchange between a compact partially folded and bound-like conformation agrees with the notion that transcription-regulating riboswitches are kinetically controlled whereas translation-regulating riboswitches are thermodynamically controlled, in which the aptamer exists in equilibrium between open and closed states that is shifted to the bound state on binding ligand. It is worth noting that Tte is a thermophilic organism, ideally growing at 75 °C, and the equilibrium between open and closed states is likely to be shifted toward the former at that temperature. Suddala et al [62] tested the temperature dependence of PreQ1 binding for the Tte riboswitch, and found that the Kd was reduced ~60 fold from 25 °C to 60 °C, to 430 nM, in support of this hypothesis.
Biophysical studies of folding of transcription-regulating riboswitches Given the very small size of the aptamer, the PreQ1-I riboswitch posed a challenge in understanding how it could function within the timeframe of transcription. Early NMR studies and SAXS data [45, 48, 49] suggested that unlike the Tte translation-regulating riboswitch, the transcription-regulating Bsu and Fnu riboswitches do not form the pseudoknot in the absence of ligand. In the NMR construct of the Bsu riboswitch, the two additional G's at the 5’ end of the Bsu NMR result in the formation of a more stable alternate P1A helix in the absence of PreQ1 (Figure 5B). This hairpin occludes U6 and C17 that participate in the binding core, preventing formation of the ligand binding pocket. In the full-length riboswitch, P0, located 0-2 nt upstream from P1, may act to prevent incorrect (P1 A) hairpin formation during co-transcriptional folding (Figure 2C). For the Fnu riboswitch, Micura and coworkers [64] showed that the riboswitch aptamer forms a dimer at a palindromic sequence within the apical loop overlapping with the P2 site, preventing pseudoknot formation (Figure 5C). This dimer, which is unlikely to be relevant in vivo, would explain the relatively large radius of gyration observed in the SAXS data [48]. By using a construct with a base pair substitution in P2 to prevent dimer formation, the authors found NMR evidence that stem 2 of the pseudoknot could form in the absence of ligand in a temperature and/or Mg2+ dependent fashion [64]. This mutant Fnu riboswitch that reduced dimerization had a two-fold higher observed Kd [49], although it should be noted that this mutation swapped an A-U bp in P2 to a G-C bp thereby increasing the thermostability of P2, which may explain the observed prevalence of pseudoknot formed in the absence of PreQ1 (relative to WT) and the modest improvement in binding affinity. Additional fluorescence studies on the Fnu WT at low concentrations showed that the on-rate for PreQ1-binding in the Fnu riboswitch was extremely fast compared to other transcription-regulating riboswitches [52].
The WT Bsu riboswitch was also observed to form dimers by NMR and electrophoretic mobility shift assays, similar to the Fnu riboswitch [62, 65] (Figure 5C). The Bsu C12,15U construct used to solve the NMR solution structure, which disrupts the palindromic sequence and prevents dimer formation, had better NMR spectra and required less PreQ1 to saturate binding than the WT sequence [45] (Figure 5D). Kang et al made multiple substitutions to L2 for their NMR study (Figure 5D). Analysis of their results on PreQ1 binding reveals that mutations that would disrupt dimer formation improved the apparent binding affinity for PreQ1, as judged by NMR titrations (Figure 5D). The ability to modify L2 with no apparent loss in PreQ1 recognition is consistent with its low sequence conservation, indicating that the sequence identity is not important but rather its length and flexibility, as discussed below. Unlike the Fnu riboswitch, the NMR studies of the Bsu riboswitch showed no sign of pseudoknot formation in the absence of PreQ1 even in the Bsu C12,15U construct that does not form dimers [45, 65]. However, smFRET studies by Walter and co-workers [62] showed that in the presence of Mg2+, the Bsu WT riboswitch adopts a compact conformation in the absence of PreQ1. Although the absence of divalent cations could contribute partially to the unfolded conformation observed in the NMR study, it is also likely that the alternate P1A hairpin would prevent stable pseudoknot formation, as discussed above. The smFRET studies also provided evidence that the Bsu riboswitch transiently forms a compact conformation that brings the 3’ ssRNA tail close to the P1 apical loop at primarily <100 ms timescales. An additional explanation for the discrepancy between smFRET and NMR data is that the timescale of the transition between unfolded and partially folded conformations occurs outside the window of detection by NMR. For example, transient, lowly populated interactions of the 3’ tail with the P1 stem on the µs to low ms timescales would be invisible by NMR. A combined NMR and computational study on the Bsu PreQ1-I riboswitch 12 nt 3’ tail revealed that the A-rich tail contains a surprising degree of order, adopting an A-form-like conformation on average [65]. This partial ordering may facilitate rapid ligand recognition by reducing the conformational entropy of the single strand, docking the tail into the P1 minor groove on PreQ1 capture.
Comparing the smFRET studies of the Tte and Bsu riboswitch, Walter and coworkers [62] found that the Tte riboswitch exhibits much slower “switching” than the Bsu riboswitch. To obtain insights into the folding and ligand-binding pathway, Gō model molecular dynamics simulations were performed for both riboswitches. An earlier computational study posited that the Bsu aptamer folded in a concerted manner, with the P1 stem forming first, followed by A-minor interactions, PreQ1 recognition, and P2 formation occurring nearly concomitantly [66]. In contrast, the Tte riboswitch appears to have significantly different folding behavior: P1 forms first, quickly followed by PreQ1 binding, with A-minor interactions and P2 formation happening much later in the simulation [62]. It should be noted that Gō model simulations begin with an extended RNA chain and bias the simulation toward the fully folded state bound to ligand; therefore, it is unlikely that the experimentally determined apo structure will be observed under such conditions. Similar results were obtained from an independent computational study by Gong et al [67] using high-temperature unfolding simulations of the Tte and Bsu aptamers.
Zhang et al [50] investigated the flexibility of loops 1 and 2 of PreQ1-bound Bsu C12,15U using NMR 13C relaxation dispersion studies. U12, U13, and U15 are highly flexible, consistent with their extruded conformation, while A14, A16, and C17, which are stacked on each other in the minor groove of P2, have limited mobility. In the crystal structures of the Bsu riboswitch, L2 nts C12, U13 and the base of C15 in Bsu WT and most of L2 (U13-A16) in Bsu L2var had missing electron density, providing independent evidence for flexibility of L2 [46]. While L2 was found to have fast motions occurring on the ps-ns timescale, L1 residues U7, C8, and U9 have slower motions on the μs-ms timescale. Interestingly, C8 motions are quenched on addition of Ca2+ with concomitant changes to the L1 conformation, as described above in section 3.4. The authors conclude that L1, aided by divalent cations, effectively functions as a ‘lid’ to block PreQ1 exit upon ligand capture.
Together, these data paint a picture of the pathway of PreQ1 recognition (Figure 5A). By modulating variables peripheral to the binding core such as the length of L2, the thermostability of P2, and the degree of order in loop 3 to allow rapid docking on ligand recognition, the PreQ1-I riboswitch can tune its responsiveness to intracellular conditions. Variations in loop 2 sequence and length can modulate PreQ1 binding affinity and pseudoknot stability while the loop 3 sequence may influence kinetics of ligand recognition [62, 65]. Taken together, these studies suggest a general mechanism of ligand regulation wherein PreQ1-I riboswitches utilize a dynamic loop 2 and unstable P2 stem to prevent premature pseudoknot formation and loop 1 (with aid of divalent cations) to block PreQ1 exit on capture.
Computational model studies The small size of the minimal riboswitch aptamer (34 nt) lends itself well to computational studies as a model system for investigating the impact of small molecules on RNA stability. In one molecular dynamics study, Denning et al [68] found that in the presence of the osmolyte trimethylamine N-oxide (TMAO) the bound Bsu crystal structure becomes destabilized due to dehydration of the phosphate backbone caused by interactions between the backbone and a protonated form of TMAO. In another molecular dynamics study, Yoon et al [69] examined the mechanism of urea-induced denaturation of nucleic acids using the bound Bsu NMR structure and found that the two-step denaturation mechanism is the opposite of urea-induced unfolding of proteins.
4. THE PreQ1-II RIBOSWITCH
4.1 Structures of PreQ1-II riboswitches
The aptamer domain of PreQ1-II riboswitches is much larger (56 nt) than that of PreQ1-I riboswitches (34 nt). All PreQ1-II riboswitches identified to date regulate translation and contain a SD sequence at their 3’ end (Figure 2D) [40, 42]. Formation of a pseudoknot by this riboswitch was easily predicted based on the high conservation of pyrimidine residues complementary to the SD sequence on the 3’ side of the loop of the stem 1 (P2) hairpin (Figure 2B). The PreQ1-II riboswitch also contains a highly conserved unpaired C above the top of P1 that was hypothesized to form a WC bp to PreQ1 [42]. Although a C-to-U substitution reduced the binding affinity by two orders of magnitude, the ligand specificity did not change to favor adenine analogs as often occurs with purine riboswitches, suggesting that the PreQ1-II riboswitch does not form a canonical WC bp with ligand.
Two structures of PreQ1-II riboswitches have been determined, an X-ray crystal structure from Lactobacillus rhamnosus (Lra) containing 75 nt (Lra WT') [43] and subsequently an NMR solution structure from Streptococcus pneumoniae (Spn) containing 59 nt (Spn WT) [44]. The Lra WT' includes the hairpin preceding the folded aptamer domain, which by published convention is called P1, so for these riboswitches stem 1 is called P2 and stem 2 is called P3 (Figure 6). In the crystal structure, P1 stacks on the bottom of P2 but does not contribute to ligand binding. Excluding the single strand residues at the 5’ and/or 3’ ends, both aptamer domains are 56 nt long, but have different sequences.
Figure 6.
Structures of PreQ1-II riboswitches (A-B) Solution NMR structure of Spn WT [PDB ID 2MIY), (C-D) X-ray crystal structure of Lra WT' [PDB ID 4JF2]. (A, C) Schematics of secondary structures and base interactions. (B, D) Stick representation of three-dimensional structures. Mg2+, Cs+ are depicted as green and purple spheres, respectively. (E) Schematic illustrating the secondary structure of the Spn riboswitch in the absence of PreQ1 and the divalent cation and PreQ1 induced folding. The curved arrow above P4 indicates the rotation of P4 required to position the two As (dark and light green bars) in the binding pocket. The gray P4 hairpins illustrate that the position of P4 is dynamic. (F) Stick representations of interactions of PreQ1 in the binding core showing the PreQ1-loop 1-loop 2-loop 3 PreQ1-C-U-A quartet from Lra WT' For all structures, residues are colored as follows: P2 (red), P3 (dark blue), L1 (orange), L2 (gold), L3 (green, with P4 hairpin insert in purple), PreQ1 (magenta). Note that P2 is stem 1 and P3 is stem 2 in standard H-type pseudoknot nomenclature.
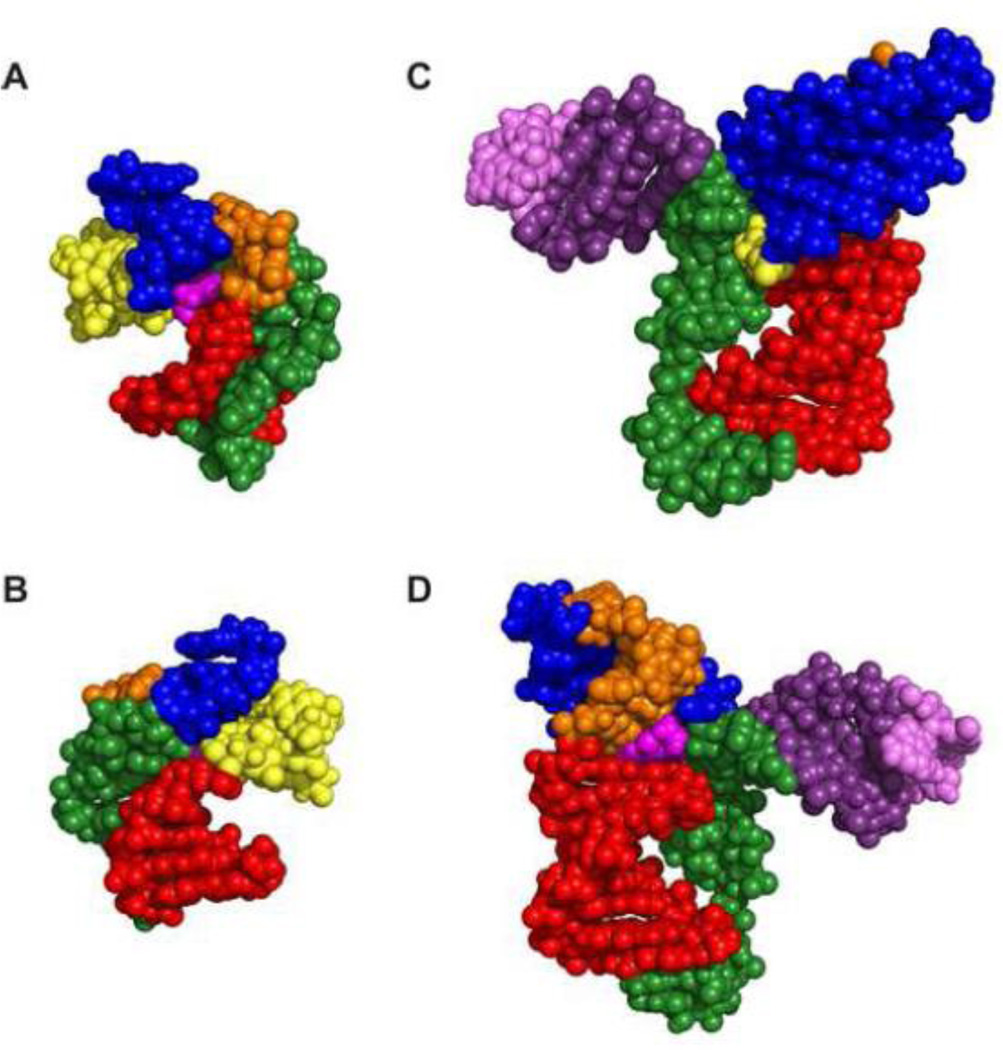
The PreQ1-II riboswitches form an unusual H-type pseudoknot with a hairpin embedded in loop 3, which is classified as HLout [70]. The pseudoknot has an 8-9 bp stem 1 (P2) and 6 bp stem 2 (P3). Loop 1 is 5 nt, loop 2 is a single nt, and loop 3 is 19-22 nt, including the embedded hairpin (P4) of 4 bp capped by a tetraloop (Figure 6A,C). Although both the PreQ1-I and PreQ1-II riboswitch aptamers form pseudoknots, their overall architecture and mode of sequence recognition are very different (Figure 6 and 7). Loop 1 (J2-3) interacts in the major groove of stem 1 (P2), with the first C forming a non-canonical bp with PreQ1 and the next two Us interacting with the two A-U bps at the bottom of stem 2 to form two U-A-U triples. This triple helical region is similar to that seen in the human telomerase RNA pseudoknot [71]. Since the SD sequence AAGGAGA forms one strand of stem 2 (P3), the two Us from loop 1 and two Us from stem 2 form a ‘U-trap’ [72] that both sequesters the SD sequence and stabilizes the PreQ1 binding pocket (Figure 6 A,C). This explains the highly conserved Us on either side of the stem 1 hairpin loop in the PreQ1-II riboswitch (Figure 2B). As in the PreQ1-I riboswitches, PreQ1 inserts at the junction between stem 1 and stem 2 and forms a base pair with loop 2, but in this case loop 2 is a single U nt, rather than the unusually long loop 2 in PreQ1-I riboswitches. The most unusual feature of the PreQ1-II pseudoknot is the long loop 3 with its embedded P4 hairpin. The single strand nucleotides 5’ to P4 in J2-4 (7 nts in Spn and 4 nts in Lra) do not interact with the minor groove as in the PreQ1-I and almost all other H-type pseudoknots, but rather span across the major groove without any contact to the bases of stem 1 (Figure 8 C-D). P4 is extended by two nts on each side (extended P4) and forms a three-way junction with stem 1 and stem 2 (Figure 6 A,C). Adenines on either side of the extended P4 form interactions in the binding pocket.
Figure 7.
The PreQ1 binding pocket for PreQ1-I and PreQ1-II riboswitch aptamers. (A-C) PreQ1-I structures of Bsu WT X-ray (A) and Bsu (C12,15U) NMR (B-C) and (D-F) PreQ1-II structures of Lra WT' crystal (D) and Spn WT NMR (E-F), comparing the PreQ1-1 and PreQ1-II binding cores (A,D), ceiling of the binding pockets (B,E), and binding pockets (C,F). (A,D) Sphere representation of Bsu WT PreQ1-loop 2-loop 3-loop 1 PreQ1-C-A-U quartet (A) and Lra WT' PreQ1-loop 1-loop 2-loop 3 PreQ1-C-U-A quartet (D), illustrating hydrogen bond and van der Waals interactions, especially between A50-PreQ1 and A50-U19. The corresponding residue numbers for Spn WT are given in parentheses. The exocyclic methylamine projects from the major groove in PreQ1-I and from the minor groove in PreQ1-II riboswitches. (B,E) Overlay of Bsu loop 2-stem 2 A-G-C triple on PreQ1-C-A-U quartet (B) and Spn loop 1-stem 2-loop 3 U-A-U-A quartet on PreQ1-C-U-A quartet (E). PreQ1 quartets are gray. (C,F) Stick representations of the three layers of binding pocket from Bsu (C) and Spn (F) NMR structures, with G-C bp below, PreQ1 binding core, and base triple or quartet above. The exocyclic amine protons point down toward the G-C pair below and have van der Waals contact with O6 and N7 of G5 (C), and point up toward the base quartet above, with possible interactions with O2 of U9 or phosphate backbone of A51 (F).
Figure 8.
Comparison of PreQ1-II vs PreQ1-I riboswitch structures. Sphere representation of (A,B) PreQ1-I Bsu (C12,15U) and (C,D) PreQ1-II Spn WT. (B, D) are the 180 degree rotation of (A, C), respectively. Loop 3 (green) lies along the minor groove in PreQ1-I aptamers (A) while shows loop 3 does not insert in a groove but spans the major groove in PreQ1-II aptamers (B). The exocyclic amine group of PreQ1 projects out from the major groove of PreQ1-I aptamers (A) and from the minor groove of PreQ1-II aptamers.
A major difference in the solved structures of the Spn WT and Lra WT' PreQ1-II riboswitches is the position of P4 (Figure 6 B,D). In the crystal structure of Lra WT', P4 is bent at a 98° angle relative to the rigid P2-P3 stem, while in the lowest energy NMR structure of Spn WT P4 is bent at a 77° angle relative to the P2-P3 stem. However, NMR dynamics measurements [44] and FRET studies [73] indicate that the position of P4 in the Spn riboswitch in solution is dynamic (Figure 6E), as discussed in section 4.4. Most PreQ1-II riboswitches have 7 bps in P2 and 9 or more nts in J2-4. The Lra riboswitches are an exception, with a longer 8-9 bp P2 and shorter 6-7 nt J2-4 [40]. The differences in the lengths of P2 and J2-4 between Lra WT' and Spn WT might result in the different position of P4 in the crystal and NMR structures, but it is also possible that in solution P4 of Lra WT' is dynamic. P4 has been proposed to play a major role in how the riboswitch regulates translation, preventing stable formation of the pseudoknot in the absence of PreQ1 [44, 73], as discussed in section 4.3.
The PreQ1-II binding pocket PreQ1 is stabilized in the binding pocket (Figure 7F) by hydrogen bond and van der Waals interactions that form a base quartet in the binding core (Figure 6F, 7D), a base quartet above (Figure 7E), and a base pair below PreQ1 (Figure 7F). In the binding core, specificity for PreQ1 is achieved from hydrogen bonding to a C residue from loop 1 (C8 in Spn, C30 in Lra) and the U residue that comprises loop 2 (U19 in Spn, U41 in Lra) and by van der Waals interactions to the last residue of loop 3 which is an A (A50 in Spn, A70 in Lra) (Figure 7D). Although PreQ1 is also recognized by C, A, and U residues from each of the three loops in the PreQ1-I riboswitches, the C and U are on different loops and the interactions of all three nucleotides differ between the two classes (Figure 7A, D). The C-PreQ1 interaction is an unusual trans WC/WC bp, in contrast to the cis WC/WC bp of the C-PreQ1 interaction in PreQ1-I riboswitch (Figure 7A,D). The loop 2 U has 3 hydrogen bonds to the ‘sugar’ edge of PreQ1 (N2H, N3, and N9H). The interaction of PreQ1 with the loop 3 A (A50 in Spn) is unusual in that specificity is achieved by van der Waals contacts rather than hydrogen bonds; the loop 3 A has a single hydrogen bond to the loop 2 U 2’OH, but its base and ribose C1’H1’ and 2’O pack tightly along the N9H-C8H-methylene edge of PreQ1 (Figure 6F, 7D). The van der Waals interactions between the PreQ1 exocyclic methylene group and A50 2’O explain the >100 fold lower binding affinity for 7-carboxamide-7-deazaguanine, which has a carbonyl group at that position [42]. In contrast, there is almost no decrease in binding affinity to the PreQ1-I riboswitch, where the PreQ1 methylene group does not have any specific interactions with the neighboring residues and there is enough room to accommodate a carbonyl group (Figure 7 A, C, D, F). The bottom of the binding pocket, below the PreQ1 binding core, is a G-C bp in Spn and an A-G bp in Lra located at the top of stem 1. Unlike the case for the PreQ1-I riboswitch, there is no specific interaction between the PreQ1 exocyclic amine group and the bottom of the binding pocket, perhaps explaining the observed sequence variability. Instead of pointing down toward the base pair below, the exocyclic methylamine points up, with possible interactions with O2 of U9 or the phosphate backbone of A51 (Figure 7C,F). Above the PreQ1 binding core, there are two loop 1-stem 2 U-A-U triples that stabilize the binding pocket. The bottom triple, which forms the top of the binding pocket, has an additional interaction with a loop 3 A (A35 in Spn, A55 in Lra) to form a loop 1-stem 2-loop 3 U-A-U-A base quartet (Figure 7E). This A is at the 5’ end of the extended P4 helix and forms an A-minor interaction with the stem 2 A 2’OH (A51 in Spn, A71 in Lra). The extended P4 positions the adenines at either end (A35 and A50 in Spn) to interact with the top and core of the binding pocket (Figure 7E,F) by a rotation of the P4 helix about its axis (Figure 6E). One of the most significant differences between the PreQ1-I and PreQ1-II binding pockets is that for the former PreQ1 enters from the major groove side, with only its exocyclic amine exposed to solvent, while for the latter PreQ1 enters from the minor groove side (Figure 7 and 8).
4.2 Cation interactions in the PreQ1-II aptamers
Unlike the PreQ1-I riboswitch, the PreQ1-II riboswitch requires divalent cations for high affinity binding [43, 44]. The NMR solution structure of Spn WT was solved in the presence of Ca2+ and the crystal structure of Lra WT contained both Mg2+ and Cs+. In the Lra WT crystal structure, two Mg2+ are bound to the backbone of the G at the top of P2 and the C in loop 1 that recognizes PreQ1, in the three-way junction between stems P2, P3, and P4. Several Cs+ and one Mg2+ are located in J2-4, the P4 stem, and at the bottom of P2, potentially stabilizing J2-4:P2 and J2-4:P4 interactions. ITC measurements indicated that addition of Mg2+ or Ca2+ increases the binding affinity of PreQ1 by 100-fold [44]. Based on chemical shift changes of residues near the binding pocket when Ca2+ was added in the presence of PreQ1, the authors propose that divalent cations stabilize the base triples and the position of A35 and A50 in the binding pocket. A smaller divalent cation effect for PreQ1 binding (4 fold) was observed for the Lra riboswitch [43], possibly due to its shorter J2-4.
The effect of divalent cations on the apo Spn riboswitch was also investigated using FRET, and the authors concluded that they stabilize formation of P3 in the pseudoknot [73]. NMR studies, which can give residue-specific interactions, indicate that in the absence of PreQ1 divalent cations only stabilize formation of the top half of P3 and not the base triples at and above the binding pocket [44].
4.3 PreQ1-II riboswitch folding and dynamics
The PreQ1-II riboswitch is less well-characterized than the PreQ1-I riboswitch, despite being discovered around the same time. Micura and coworkers [73] performed SHAPE experiments over a range of temperatures to determine residue-specific melting of the Spn PreQ1-II riboswitch in the free and PreQ1-bound states. The authors found that in all cases J2-4 had a high degree of reactivity relative to helical elements, indicating flexibility within this loop. NMR 13C spin relaxation measurements independently determined that this loop was highly dynamic [44]. smFRET studies showed that, similar to what was observed for the PreQ1-I riboswitch, formation of the pseudoknot in the absence of PreQ1 is Mg2+-dependent for the Spn riboswitch. Deletion of the P4 stem in loop 3 (residues 38-48) slightly increased the stability of stem 2 in the absence of ligand and decreased the binding affinity for PreQ1 ~10-fold. Interestingly, the lifetime of the high-FRET state (where stem 2 was formed) was significantly shorter than the WT construct, particularly in the presence of PreQ1, suggesting that although P4 deletion stabilizes the pseudoknot it reduces the ability of the riboswitch to stably bind PreQ1 [73]. Further, the P4 helix appears to have a high degree of flexibility even in the PreQ1-bound state. smFRET experiments with fluorophores placed at L1 and P4 in the WT construct showed that the PreQ1-bound structure falls approximately equally into two distributions: one in which the P4 helix is at an intermediate distance from L1 and one in which P4 is far from L1.
The NMR study of the Spn riboswitch by Kang et al [44] indicated that P2 and P4 form in the absence of PreQ1 and Ca2+ (Figure 6E). As discussed in section 4.2, in the presence of Ca2+, NMR spectra of apo Spn WT show imino proton resonances for the top part of P3 (3 bp), indicating that the pseudoknot is partially formed in agreement with smFRET data [73]. Deletion of the P4 stem (residues 36-49) (Spn AP4) results in a reduction in binding affinity as measured by ITC similar to that observed in the smFRET study. Examination of imino proton spectra of apo Spn AP4 with Ca2+ showed that the P3 stem is significantly stabilized in the absence of PreQ1, with five of the six base pairs observed. Analysis of NMR RDC measurements and 13C spin relaxation measurements are also consistent with the P4 dynamics observed in the smFRET study, revealing positional flexibility in P4 relative to a rigid P2-P3 (stem 1-stem 2) stem, corresponding to a 35° cone angle of motion about the P4 helical axis. This large motional amplitude would explain the ~20° difference observed in the P2-P4 inter-helical bend angle between Lra and Spn structures.
Taken together, the smFRET and NMR studies indicate that removing P4 reduces the range of motions accessible to loop 3, thereby increasing the likelihood to form the stem 2 of the pseudoknot. The P4 hairpin is a special feature of PreQ1-II riboswitch and was proposed to play a dual role by Kang et al [44] based on the experiments described above as well as NMR and ITC data on A35 and A50 substitutions. In the presence of PreQ1 and divalent cations (section 4.2), P4 positions the adenines at the 5’ and 3’ ends of P4 (A35 and A50 in Spn) to interact with the top and core of the binding pocket (Figure 6F, 7E,F). Thus, the extended P4 helix acts as a ‘screw’ cap, positioning A35 and A50 in the binding pocket and blocking off ligand exit from that side. In the absence of PreQ1, P4 destabilizes P3 therefore hindering pseudoknot formation.
5. Applications
Translational control via -1 frameshifting Riboswitches hold great potential in a number of biotechnology applications due to their ability to regulate gene expression in cis upon addition of a cognate ligand [74, 75]. To date, PreQ1 riboswitches have only been used in one such application. Yu et al [76] took advantage of the pseudoknot structure of the bound PreQ1-I aptamer to induce -1 ribosomal frameshifting in an in vitro translation assay. In the absence of PreQ1, the transcription-regulating Bsu and Fnu aptamers had near-zero -1 frameshifting whereas the Tte aptamer had 3.5% frameshifting at 28°. These results are consistent with SAXS and crystallography data supporting a pre-folded conformation for the apo Tte aptamer, and solution and NMR data that show that the apo Bsu and Fnu aptamers only transiently sample a partially pre-folded state. In addition, the Fnu aptamer was the most responsive to PreQ1, although the level of -1 frameshifting only reached a maximum of 20% even in the presence of 200 uM PreQ1. Replacement of the A-U bps in P1 with G-C bps in the Fnu aptamer increased -1 frameshifting to 39% when PreQ1 was present. Yu et al also found that the EC50 for this modified Fnu aptamer for PreQ1 was 180 nM. Although the Kd for this construct has not been measured, the most similar construct (Fnu WT aptamer) has a reported Kd of 283 nM [52], indicating that this riboswitch operates under thermodynamic control when regulating translation.
Antibacterial Targeting The prevalence of riboswitches in bacteria makes them attractive targets for antibacterial drug development [77–79]. Riboswitches have already been implemented with some success using ligand mimetics [80–82]. PreQ1 riboswitches are especially promising as antibiotic targets since PreQ1 is only synthesized in bacteria, PreQ1 riboswitches are therefore only present in bacteria, and Q plays an important role in biology and disease. For example, Q availability is a determinant of virulence for Shigella flexneri [30–33], and the PreQ1-II riboswitch is found in several virulent Streptococcal strains [42]. By developing 7-deazaguanine derivatives to act as PreQ1 mimetics, it may be possible to target these latter virulent bacteria.
Biofuels Engineering riboswitches for control of protein pathways is being pursued in applications for renewable energy and biofuels due to their ability to perform inducible gene expression. Riboswitches have also been proposed for use in biofuel cells as logic gates [83]. Cyanobacteria are an ideal system for manufacturing biofuels due to their ability to convert light into chemical energy and produce oxygen through oxygenic photosynthesis [84, 85]. The synthetic theophylline aptamer has already been used with some success to regulate protein expression in cyanobacteria [86]. PreQ1-I is advantageous in biofuel applications due to its small size and ability to regulate at either the transcription or translation level. By taking advantage of the principles governing PreQ1 recognition as well as pseudoknot folding and stability learned from the above studies, the PreQ1 riboswitches can be engineered to attenuate PreQ1 responsiveness in biofuel production.
6. Summary
The existence of multiple regulatory mechanisms for Q synthesis indicates its importance in cell fitness. The different structures and dynamics of the PreQ1-I and PreQ1-II riboswitches have provided new insights into how RNA can harness control of gene expression using differing modes of recognition of the same ligand. Despite the fact that both classes of PreQ1 riboswitch fold into H-type pseudoknots with PreQ1 at the junction between the two stems, the determinants of ligand specificity, orientation of PreQ1 in the binding pocket, and predicted pathways for ligand capture are very different. The wealth of structural and dynamic information within the two classes, described in this review, has provided insights into how ligand responsiveness can be fine-tuned by altering sequences peripheral to the binding pocket to modulate the binding kinetics and stability of the bound conformation. For example, in the PreQ1-I riboswitch, variations in the loop 2 sequence and length can modulate PreQ1 binding affinity and pseudoknot stability while the loop 3 sequence may influence kinetics of ligand recognition. In the PreQ1-I riboswitch the dynamic loop 2 and unstable P2 help to prevent premature pseudoknot formation and loop 1 (with aid of divalent cations) helps block PreQ1 exit, while in the PreQ1-I I riboswitch the embedded P4 in loop 3 helps to both prevent premature pseudoknot formation and function as a ‘screw’ cap to block ligand exit, although the mechanisms are likely more complex. Although folding and kinetics studies have been reported for the ligand recognition mechanism, no kinetics have yet been measured in the context of transcription or translation control, which will be important for a complete understanding of regulation of gene expression by these riboswitches.
Review Highlights.
There are two PreQ1 riboswitch classes with different folds and PreQ1 interactions
Structures of both classes have been solved by NMR and X-ray crystallography
PreQ1-I riboswitches have the smallest known aptamer and form an H-type pseudotknot
PreQ1-II riboswitches form an HL-out pseudoknot with an embedded hairpin.
Dynamics modulate aptamer folding and PreQ1 recognition
Acknowledgements
This work was supported by grants from the U.S. Department of Energy (DE-FC03-02ER63421) and National Institutes of Health (GM48123) to J.F. and a UCLA Tumor Biology USHHS Ruth L. Kirschstein NRSA T32 award (CA009056) to C.D.E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim JN, Breaker RR. Purine sensing by riboswitches. Biology of the cell / under the auspices of the European Cell Biology Organization. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 2.Batey RT. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edmonds CG, Crain PF, Gupta R, Hashizume T, Hocart CH, Kowalak JA, Pomerantz SC, Stetter KO, McCloskey JA. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria) Journal of bacteriology. 1991;173:3138–3148. doi: 10.1128/jb.173.10.3138-3148.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harada F, Nishimura S. Possible anticodon sequences of tRNA His, tRNA Asm, and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry. 1972;11:301–308. doi: 10.1021/bi00752a024. [DOI] [PubMed] [Google Scholar]
- 5.Morris RC, Elliott MS. Queuosine modification of tRNA: a case for convergent evolution. Mol Genet Metab. 2001;74:147–159. doi: 10.1006/mgme.2001.3216. [DOI] [PubMed] [Google Scholar]
- 6.Iwata-Reuyl D. An embarrassment of riches: the enzymology of RNA modification. Curr Opin Chem Biol. 2008;12:126–133. doi: 10.1016/j.cbpa.2008.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia GA, Kittendorf JD. Transglycosylation: a mechanism for RNA modification (and editing?) Bioorganic chemistry. 2005;33:229–251. doi: 10.1016/j.bioorg.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosjean H. DNA and RNA modification enzymes : structure, mechanism, function, and evolution. Landes Bioscience, Austin, Tex. 2009 [Google Scholar]
- 9.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground, Wiley interdisciplinary reviews. RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005;44:8078–8089. doi: 10.1021/bi050343f. [DOI] [PubMed] [Google Scholar]
- 11.Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. Aprimordial tRNA modification required for the evolution of life? The EMBO journal. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA. 2011;17:1846–1857. doi: 10.1261/rna.2628611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. Journal of molecular biology. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 14.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb2009: compilation of tRNA sequences and tRNA genes. Nucleic acids research. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langgut W, Kersten H. The deazaguanine-derivative, queuine, affects cell proliferation, protein phosphorylation and the expression of the proto oncogenes c-fos and c-myc in HeLa cells. FEBS letters. 1990;265:33–36. doi: 10.1016/0014-5793(90)80877-l. [DOI] [PubMed] [Google Scholar]
- 16.Langgut W, Reisser T, Nishimura S, Kersten H. Modulation of mammalian cell proliferation by a modified tRNA base of bacterial origin. FEBS letters. 1993;336:137–142. doi: 10.1016/0014-5793(93)81627-c. [DOI] [PubMed] [Google Scholar]
- 17.French BT, Patrick DE, Grever MR, Trewyn RW. Queuine, a Transfer-RNA Anticodon Wobble Base, Maintains the Proliferative and Pluripotent Potential of HI-60 Cells in the Presence of the Differentiating Agent 6-Thioguanine. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:370–374. doi: 10.1073/pnas.88.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada N, Shindo-Okada N, Sato S, Itoh YH, Oda K, Nishimura S. Detection of unique tRNA species in tumor tissues by Escherichia coli guanine insertion enzyme. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4247–4251. doi: 10.1073/pnas.75.9.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owenby RK, Stulberg MP, Jacobson KB. Alteration of the Q family of transfer RNAs in adult Drosophila melanogaster as a function of age, nutrition, and genotype. Mechanisms of ageing and development. 1979;11:91–103. doi: 10.1016/0047-6374(79)90027-7. [DOI] [PubMed] [Google Scholar]
- 20.Pathak C, Jaiswal YK, Vinayak M. Possible involvement of queuine in regulation of cell proliferation. BioFactors. 2007;29:159–173. doi: 10.1002/biof.5520290401. [DOI] [PubMed] [Google Scholar]
- 21.White BN, Tener GM. Activity of a transfer RNA modifying enzyme during the development of Drosophila and its relationship to the su(s) locus. Journal of molecular biology. 1973;74:635–651. doi: 10.1016/0022-2836(73)90054-5. [DOI] [PubMed] [Google Scholar]
- 22.Reisser T, Langgut W, Kersten H. The nutrient factor queuine protects HeLa cells from hypoxic stress and improves metabolic adaptation to oxygen availability. European journal of biochemistry/ FEBS. 1994;221:979–986. doi: 10.1111/j.1432-1033.1994.tb18814.x. [DOI] [PubMed] [Google Scholar]
- 23.Baranowski W, Dirheimer G, Jakowicki JA, Keith G. Deficiency of queuine, a highly modified purine base, in transfer RNAs from primary and metastatic ovarian malignant tumors in women. Cancer research. 1994;54:4468–4471. [PubMed] [Google Scholar]
- 24.Emmerich B, Zubrod E, Weber H, Maubach PA, Kersten H, Kersten W. Relationship of queuine-lacking transfer RNA to the grade of malignancy in human leukemias and lymphomas. Cancer research. 1985;45:4308–4314. [PubMed] [Google Scholar]
- 25.Huang BS, Wu RT, Chien KY. Relationship of the queuine content of transfer ribonucleic acids to histopathological grading and survival in human lung cancer. Cancer research. 1992;52:4696–4700. [PubMed] [Google Scholar]
- 26.Ishiwata S, Katayama J, Shindo H, Ozawa Y, Itoh K, Mizugaki M. Increased expression of queuosine synthesizing enzyme, tRNA-guanine transglycosylase, and queuosine levels in tRNA of leukemic cells. Journal of biochemistry. 2001;129:13–17. doi: 10.1093/oxfordjournals.jbchem.a002823. [DOI] [PubMed] [Google Scholar]
- 27.Ishiwata S, Ozawa Y, Katayama J, Kaneko S, Shindo H, Tomioka Y, Ishiwata T, Asano G, Ikegawa S, Mizugaki M. Elevated expression level of 60-kDa subunit of tRNA-guanine transglycosylase in colon cancer. Cancer letters. 2004;212:113–119. doi: 10.1016/j.canlet.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Marks T, Farkas WR. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochemical and biophysical research communications. 1997;230:233–237. doi: 10.1006/bbrc.1996.5768. [DOI] [PubMed] [Google Scholar]
- 29.Carlson BA, Kwon SY, Chamorro M, Oroszlan S, Hatfield DL, Lee BJ. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- 30.Durand JM, Dagberg B, Uhlin BE, Bjork GR. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Molecular microbiology. 2000;35:924–935. doi: 10.1046/j.1365-2958.2000.01767.x. [DOI] [PubMed] [Google Scholar]
- 31.Durand JM, Okada N, Tobe T, Watarai M, Fukuda I, Suzuki T, Nakata N, Komatsu K, Yoshikawa M, Sasakawa C. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. Journal of bacteriology. 1994;176:4627–4634. doi: 10.1128/jb.176.15.4627-4634.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurt JK, Olgen S, Garcia GA. Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro. Nucleic acids research. 2007;35:4905–4913. doi: 10.1093/nar/gkm473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stengl B, Meyer EA, Heine A, Brenk R, Diederich F, Klebe G. Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding. Journal of molecular biology. 2007;370:492–511. doi: 10.1016/j.jmb.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Iwata-Reuyl D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorganic chemistry. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- 35.McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorganic chemistry. 2012;43:15–25. doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasai H, Nakanishi K, Macfarlane RD, Torgerson DF, Ohashi Z, McCloskey JA, Gross HJ, Nishimura S. Letter: The structure of Q* nucleoside isolated from rabbit liver transfer ribonucleic acid. Journal of the American Chemical Society. 1976;98:5044–5046. doi: 10.1021/ja00432a071. [DOI] [PubMed] [Google Scholar]
- 37.Okada N, Shindo-Okada N, Nishimura S. Isolation of mammalian tRNAAsp and tRNATyr by lectin-Sepharose affinity column chromatography. Nucleic acids research. 1977;4:415–423. doi: 10.1093/nar/4.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, Iwata-Reuyl D, Breaker RR. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nature structural & molecular biology. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 40.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A. Rfam: updates to the RNA families database. Nucleic acids research. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic acids research. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14:685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberman JA, Salim M, Krucinska J, Wedekind JE. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nature chemical biology. 2013;9:353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang M, Eichhorn CD, Feigon J. Structural determinants for ligand capture by a class II preQ1 riboswitch. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1400126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. Erratum in: 2010, 2039, 2653-2655. [DOI] [PubMed] [Google Scholar]
- 46.Klein DJ, Edwards TE, Ferre-D'Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nature structural & molecular biology. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. The Journal of biological chemistry. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenkins JL, Krucinska J, McCarty RM, Bandarian V, Wedekind JE. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. The Journal of biological chemistry. 2011;286:24626–24637. doi: 10.1074/jbc.M111.230375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rieder U, Lang K, Kreutz C, Polacek N, Micura R. Evidence for pseudoknot formation of class I preQ1 riboswitch aptamers. Chembiochem. 2009;10:1141–1144. doi: 10.1002/cbic.200900155. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q, Kang M, Peterson RD, Feigon J. Comparison of solution and crystal structures of preQ1 riboswitch reveals calcium-induced changes in conformation and dynamics. Journal of the American Chemical Society. 2011;133:5190–5193. doi: 10.1021/ja111769g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein DJ, Ferre-D'Amare AR. Crystallization of the glmS ribozyme-riboswitch. Methods in molecular biology. 2009;540:129–139. doi: 10.1007/978-1-59745-558-9_10. [DOI] [PubMed] [Google Scholar]
- 52.Rieder U, Kreutz C, Micura R. Folding of a transcriptionally acting preQ1 riboswitch. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10804–10809. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 54.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 55.Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state, Wiley interdisciplinary reviews. RNA. 2012;3:369–384. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamauchi T, Miyoshi D, Kubodera T, Nishimura A, Nakai S, Sugimoto N. Roles of Mg2+ in TPP-dependent riboswitch. FEBS letters. 2005;579:2583–2588. doi: 10.1016/j.febslet.2005.03.074. [DOI] [PubMed] [Google Scholar]
- 57.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 58.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 60.Lipfert J, Sim AY, Herschlag D, Doniach S. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA. 2010;16:708–719. doi: 10.1261/rna.1985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 62.Suddala KC, Rinaldi AJ, Feng J, Mustoe AM, Eichhorn CD, Liberman JA, Wedekind JE, Al-Hashimi HM, Brooks CL, 3rd, Walter NG. Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic acids research. 2013 doi: 10.1093/nar/gkt798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banas P, Sklenovsky P, Wedekind JE, Sponer J, Otyepka M. Molecular mechanism of preQ1 riboswitch action: a molecular dynamics study. J Phys Chem B. 2012;116:12721–12734. doi: 10.1021/jp309230v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santner T, Rieder U, Kreutz C, Micura R. Pseudoknot preorganization of the preQ1 class I riboswitch. Journal of the American Chemical Society. 2012;134:11928–11931. doi: 10.1021/ja3049964. [DOI] [PubMed] [Google Scholar]
- 65.Eichhorn CD, Feng J, Suddala KC, Walter NG, Brooks CL, 3rd, Al-Hashimi HM. Unraveling the structural complexity in a single-stranded RNA tail: implications for efficient ligand binding in the prequeuosine riboswitch. Nucleic acids research. 2012;40:1345–1355. doi: 10.1093/nar/gkr833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng J, Walter NG, Brooks CL., 3rd Cooperative and directional folding of the preQ1 riboswitch aptamer domain. Journal of the American Chemical Society. 2011;133:4196–4199. doi: 10.1021/ja110411m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gong Z, Zhao Y, Chen C, Xiao Y. Computational study of unfolding and regulation mechanism of preQ1 riboswitches. PloS one. 2012;7:e45239. doi: 10.1371/journal.pone.0045239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denning EJ, Thirumalai D, MacKerell AD., Jr Profanation of trimethylamine N-oxide (TMAO) is required for stabilization of RNA tertiary structure. Biophys Chem. 2013;184:8–16. doi: 10.1016/j.bpc.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoon J, Thirumalai D, Hyeon C. Urea-induced denaturation of preQ1-riboswitch. Journal of the American Chemical Society. 2013;135:12112–12121. doi: 10.1021/ja406019s. [DOI] [PubMed] [Google Scholar]
- 70.Han K, Byun Y. PSEUDOVIEWER2: Visualization of RNA pseudoknots of any type. Nucleic acids research. 2003;31:3432–3440. doi: 10.1093/nar/gkg539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 72.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Souliere MF, Altman RB, Schwarz V, Haller A, Blanchard SC, Micura R. Tuning a riboswitch response through structural extension of a pseudoknot. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3256–E3264. doi: 10.1073/pnas.1304585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weigand JE, Suess B. Aptamers and riboswitches: perspectives in biotechnology. Applied microbiology and biotechnology. 2009;85:229–236. doi: 10.1007/s00253-009-2194-2. [DOI] [PubMed] [Google Scholar]
- 75.Wittmann A, Suess B. Engineered riboswitches: Expanding researchers' toolbox with synthetic RNA regulators. FEBS letters. 2012;586:2076–2083. doi: 10.1016/j.febslet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 76.Yu CH, Luo J, Iwata-Reuyl D, Olsthoorn RC. Exploiting preQ(1) riboswitches to regulate ribosomal frameshifting. ACS Chem Biol. 2013;8:733–740. doi: 10.1021/cb300629b. [DOI] [PubMed] [Google Scholar]
- 77.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 78.Deigan KE, Ferre-D'Amare AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Accounts of chemical research. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mulhbacher J, St-Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Current opinion in pharmacology. 2010;10:551–556. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 80.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nature chemical biology. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 81.Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS pathogens. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ster C, Allard M, Boulanger S, Lamontagne Boulet M, Mulhbacher J, Lafontaine DA, Marsault E, Lacasse P, Malouin F. Experimental treatment of Staphylococcus aureus bovine intramammary infection using a guanine riboswitch ligand analog. Journal of dairy science. 2013;96:1000–1008. doi: 10.3168/jds.2012-5890. [DOI] [PubMed] [Google Scholar]
- 83.Zhou M, Du Y, Chen C, Li B, Wen D, Dong S, Wang E. Aptamer-controlled biofuel cells in logic systems and used as self-powered and intelligent logic aptasensors. Journal of the American Chemical Society. 2010;132:2172–2174. doi: 10.1021/ja910634e. [DOI] [PubMed] [Google Scholar]
- 84.Gronenberg LS, Marcheschi RJ, Liao JC. Next generation biofuel engineering in prokaryotes. Curr Opin Chem Biol. 2013;17:462–471. doi: 10.1016/j.cbpa.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berla BM, Saha R, Immethun CM, Maranas CD, Moon TS, Pakrasi HB. Synthetic biology of cyanobacteria: unique challenges and opportunities. Front Microbiol. 2013;4:246. doi: 10.3389/fmicb.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakahira Y, Ogawa A, Asano H, Oyama T, Tozawa Y. Theophylline-dependent riboswitch as a novel genetic tool for strict regulation of protein expression in Cyanobacterium Synechococcus elongatus PCC 7942. Plant & cell physiology. 2013;54:1724–1735. doi: 10.1093/pcp/pct115. [DOI] [PubMed] [Google Scholar]
- 87.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]