Abstract
Regulators of differentiated cell fate can offer targets for managing cancer development and progression. Here we identify Runx2 as a new regulator of epithelial cell fate in mammary gland development and breast cancer. Runx2 is expressed in the epithelium of pregnant mice in a strict temporally and hormonally-regulated manner. During pregnancy, Runx2 genetic deletion impaired alveolar differentiation in a manner that disrupted alveolar progenitor cell populations. Conversely, exogenous transgenic expression of Runx2 in mammary epithelial cells blocked milk production, suggesting that the decrease in endogenous Runx2 observed late in pregnancy is necessary for full differentiation. In addition, overexpression of Runx2 drove EMT-like changes in normal mammary epithelial cells, while Runx2 deletion in basal breast cancer cells inhibited cellular phenotypes associated with tumorigenesis. Notably, loss of Runx2 expression increased tumor latency and enhanced overall survival in a mouse model of breast cancer, with Runx2-deficient tumors exhibiting reduced cell proliferation. Together, our results establish a novel function for Runx2 in breast cancer that may offer a novel generalized route for therapeutic interventions.
Keywords: Runx2, mammary gland, differentiation, breast cancer, epithelial progenitors
Introduction
Identifying new regulators of breast cancer progression is critical to enable the discovery of novel therapies that will improve patient survival. To this end, how cell fate is controlled in the breast is an area of intense interest. The mammary gland is a hierarchically organized epithelial tissue within a stromal fat pad consisting of adipocytes, fibroblasts and endothelium (1). The epithelial compartment contains stem cells capable of reconstituting an entire functional gland, with the two main epithelial lineages (luminal and basal) then arising through a series of progenitors (2-4). The complexity of the mammary epithelial hierarchy is continuing to expand with much still to be determined (5, 6).
As direct regulators of gene expression, transcription factors are commonly involved in cell fate decisions. Transcription factors, such as Gata3, Elf5 and Notch have established roles in controlling mammary epithelial cell fate during development (7-11). Significantly, many of the transcription factors that regulate normal organogenesis also contribute to breast cancer phenotype. For example, Gata3, Elf5 and Notch influence tumor aggressiveness, subtype and cancer stem cells (12-15). Thus, understanding the mechanisms that control cell fate during normal development will also provide insight into breast tumorigenesis, where these signaling networks are disrupted.
Runx transcription factors are evolutionarily conserved regulators of cell fate, with 3 family members present in mammals (Runx1-3). The Runx proteins have well-established roles in hematopoietic (Runx1), bone (Runx2) and gastrointestinal/neuronal (Runx3) development, although their expression is not restricted to these tissues (16). All 3 family members are associated with cancer progression, acting as tumour suppressors or oncogenes in a context dependent manner (17). Runx2 is essential for bone development and homeostasis in the mouse, with Runx2−/− mice dying perinatally due to failed osteoblast differentiation (18, 19). Referred to as the master regulator of bone development, relatively few studies have examined Runx2 function in other developmental contexts. In the mammary gland, Runx2 is expressed in embryonic mammary buds, and both basal and luminal cell lineages in the adult (19-21), where it directly regulates the expression of a number of genes associated with mammary gland development (22). Runx2 is also highly expressed in breast cancer cell lines, compared to normal epithelial cells (23), with Runx2 target gene expression inversely correlating with estrogen receptor expression (24). In vitro studies suggest Runx2 functions in epithelial tumorigenesis by inducing aberrant proliferation and inhibiting apoptosis (25). Moreover, Runx2 may contribute to aspects of breast cancer metastasis, such as epithelial-to-mesenchymal transition (EMT), disruption of acini morphology, colonisation of distal sites or bone osteolysis (24, 26), potentially through the regulation of genes such as estrogen receptor, matrix metalloproteinase 9 & 13, transforming growth factor beta receptor and vascular endothelial growth factor, that are associated with a metastatic phenotype (27, 28). Despite the evidence that Runx2 can influence mammary cell function in vitro, the role of Runx2 in normal mammary gland development and breast cancer has not been examined in vivo.
We used two genetic methods of Runx2-deletion to test Runx2 function in normal mammary gland development and breast tumor progression. Conditional Runx2 deletion in mammary epithelial cells using MMTV-cre (Runx2f/f;MMTV-cre) impaired ductal outgrowth during puberty and disrupted progenitor cell differentiation in pregnancy. Runx2−/− mammary epithelium transplanted into fat pads of immunodeficient recipients also exhibited a marked developmental defect during pregnancy, with the same alterations in the proportion of progenitor cells observed in Runx2f/f;MMTV-cre mice. Exogenous Runx2 expression impaired differentiation in HC11 mammary epithelial cells, with concurrent phenotypic changes consistent with EMT. In a mouse model of luminal breast cancer, Runx2 deletion significantly increased animal survival and was associated with reduced proliferation and cyclin D1 expression. Together our data demonstrate that Runx2 is a novel regulator of mammary progenitors and breast tumorigenesis.
Methods
Mice were housed in Bosch Rodent Facility and Walter and Eliza Hall Institute Animal Facility in accordance with ethics approvals K00/6-11/3/5545 and Walter and Eliza Hall Institute Animal Ethics Committee 2011.002. Mammary glands of Balb/c mice were collected at virgin, pregnancy, lactation and involution time points (V, P, L and I, respectively), snap frozen and homogenised in Trizol, followed by extraction of RNA and protein. Lymph nodes were removed from mammary glands prior to freezing. RT-PCR was performed with SuperscriptIII (Life Technologies) and qPCR performed on ABI7900HT with taqman probes (Applied Biosystems).
Mammary placodes dissected from Runx2+/+ and Runx2−/− (19) E14.5 embryos were transplanted into cleared 4th inguinal mammary fat pads of 3-week-old Rag1−/−recipients (Animal Resource Centre, Perth, Australia) as previously described (8). Time matings were performed overnight with the following day termed day 0.5 of pregnancy. The floxed Runx-2 construct and mice were generated by OzGene (Perth, Australia). Targeted mice were crossed with MMTV-cre (line A) mice (a kind gift from K.-U. Wagner, Omaha, NE (29)) to generate Runx2f/f;MMTV-cre animals. Longitudinal and cross-sectional analysis of Runx2 deletion in the Polyoma Middle T transgenic (PyMT) breast cancer model (30) was performed by crossing Runx2+/− mice with MMTV-PyMTtg/+ (PyMT) mice (30) and back crossing to Runx2+/− mice, to produce Runx2+/+;PyMT and Runx2−/−;PyMT embryos that were transplanted to 3-week-old Rag1−/− mice as previously described. The cross-sectional analyses of Runx2+/+;PyMT and Runx2−/−;PyMT transplants were performed 13 weeks post-transplantation. Mammary glands were fixed in 10% NBF overnight and stained with carmine. Following imaging of whole mounts, mammary glands were paraffin embedded and 5 μm sections cut for hematoxylin and eosin (H&E) and immunostaining analyses (SMA (Sigma), ZO1 (Life Technologies), cytokeratin-8 (Progen), p63 (Abcam), Ki67 (Thermo Scientific), Cyclin D1 (Abcam), Runx2 – D130.3 (MBL) β-casein (generous gift from Professor Charles Streuli, University of Manchester)). Flow cytometry analyses of progenitor cell populations were performed as previously described (31). Briefly, single cell suspensions of inguinal and thoracic mammary glands were incubated with antibody (CD31, CD45, TER119 (BD Biosciences), CD24, CD29 (Biolegend), CD14 (eBioscience) and ckit (in-house)) and analysed in the following combinations: lineage negative (lin−: CD31−CD45−TER119−), MaSC-enriched population (lin-CD29hiCD24+), luminal population (lin-CD29loCD24+), luminal progenitor population (lin−CD29loCD24+CD14+ckit+) and alveolar progenitor population (lin−CD29loCD24+CD14+ckit−/lo). In the PyMT studies, tumor-free survival is the age when tumors were first detected and overall survival was determined to be when the tumor reached 10% of body weight (ethical end point). Cell lines were validated by the Tissue Culture Facility, Garvan Institute of Medical Research. Culture media were as follows; MDA-MB-231 (RPMI, 10% FBS), MCF10A-EcoR (DMEM/F12, 5% Horse Serum, 0.5 μg/ml hydrocortisone, 20 ng/ml EGF, 10 μg/ml insulin, 100 ng/ml Cholera toxin), HC11 (RPMI, 10% FBS, 10 ng/ml EGF, 5 μg/ml insulin). HC11 cells were differentiated by addition of 100 nM dexamethasone, 5 ug/ml insulin and 5 ug/ml prolactin (DIP), in the absence of EGF. Runx2 cDNA (NM_001024630; Origene) was subcloned into pMIG-IRES-EGFP and retrovirus created by transfecting PLAT-E cells with pMIG or pMIGRunx2. Viral-containing supernatant was added to cultures of HC11 and MCF10A-EcoR cells and stably transfected cells isolated by FACS. MDA-MB-231 cells were transfected with 10 nM Runx2 or non-target control siRNA (Ambion) using Lipofectamine 2000 and migration assay performed 48 hours post-transfection. Real-time migration analyses were performed on xCelligence (Roche) by measuring increasing electrical impedance as cells migrated across electrodes. 10% FBS was used as the chemoattractant. Cell growth was analysed on Incucyte Zoom using associated software (Essen Bioscience). For western blotting, cells were lysed in RIPA buffer supplemented with protease inhibitor cocktail (Roche). Antibodies (not already described) were Notch 1, Cleaved Notch 1 (NICD), pStat5 (Tyr694), Stat5a, Runx2 (Cell Signalling), pFak (Y397) (Biosource), E-Cadherin and N-Cadherin (BD Transduction Laboratories) and β actin (Sigma).
For IHC analysis, antigen retrieval was performed in citrate buffer (pH 6.0). Cyclin D1 IHC was performed without antigen retrieval. Metamorph software was used to quantify Ki67 positive cells, with equal image analysis parameters applied to at least 10 epithelial regions of interest/gland. Cyclin D1 staining was quantified manually with ImageJ particle counter of at least 4000 cells/gland.
Results
Developmentally-regulated expression of Runx2 in the mammary gland
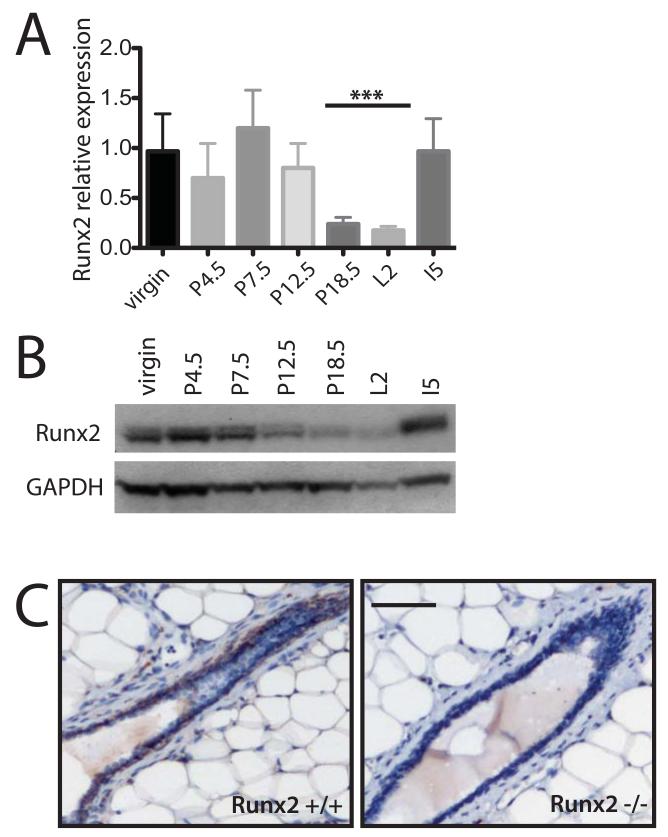
Runx2 is expressed in both embryonic and adult mammary epithelium in the basal and luminal cell linages (19-21), but its developmental profile is unknown. To examine the developmental regulation of Runx2 expression during mammopoiesis, we examined Runx2 mRNA and protein levels at different stages of mouse mammary gland development. Quantitative PCR analysis revealed that Runx2 expression remained relatively unchanged between adult virgins and mice in either early or mid-pregnancy, but fell significantly towards the end of pregnancy and remained low in lactation (Figure 1A). Analyses of Runx2 protein levels also exhibited a similar trend with Runx2 levels lowest at late pregnancy and early lactation (Figure 1B). Runx2 immunostaining confirmed previous microarray data (20, 21), with Runx2 expressed in both the ducts and terminal end buds of virgin mice (Figure 1C, Figure S2D). Together these data suggest that Runx2 plays a role during the early to mid-phases of pregnancy but may not be required in late pregnancy or lactation.
Figure 1.
Runx2 expression is developmentally regulated in the mammary gland.
A) Quantitative PCR of Runx2 expression across a developmental time course (n=3 mice at each time point) P-pregnancy; L-lacation; I-involution (*** p<0.01).
B) Representative western blot of Runx2 expression of protein extracted from same tissue as mRNA used in (A).
C) Runx2 immunohistochemistry of Runx2+/+ and Runx2−/− mammary glands generated by transplantation of Runx2+/+ and Runx2−/− embryonic mammary tissue to Rag1−/− hosts. Glands collected 16 weeks post-transplantation, from virgin Rag1−/− recipient mice with cross-section of duct shown (Scale bar 50 μm).
Runx2 regulates epithelial progenitors during pregnancy
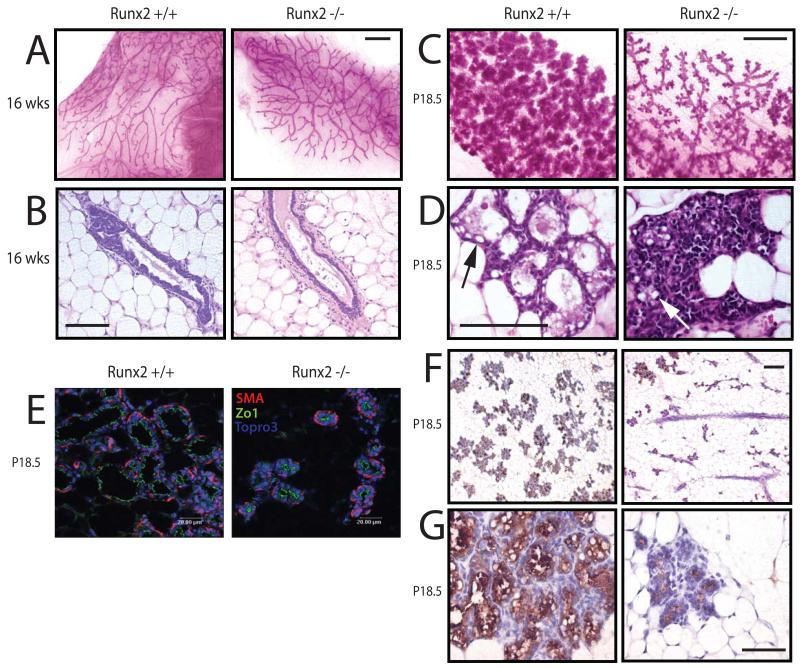
To determine the developmental role of Runx2 in the breast, we analysed the mammary glands of Runx2 knockout (Runx2−/−) mice. As Runx2−/− mice die soon after birth due to failed skeletal development, we transplanted embryonic mammary buds from Runx2−/− and control (Runx2+/+) mice into the cleared fat pads of 3-4 week old immunocompromised Rag1−/− recipients. This assay addresses phenotypes intrinsic to the transplanted mammary epithelium. Runx2 deletion was confirmed by IHC (Figure 1C) and in virgin mice at 16 weeks post-transplantation, no differences were observed between the Runx2+/+ and Runx2−/− mammary glands, indicating that Runx2 may not be required for branching morphogenesis (Figure 2A-B). No developmental differences were observed at days 7 or 12 of pregnancy (Figure S1A, data not shown). However, it was apparent by day 18.5 of pregnancy (P18.5) that Runx2−/− mammary glands were under-developed, with a lower density of alveoli compared with Runx2+/+ glands (Figure 2C). The same phenotype was observed on the first day postpartum (Figure S1B).
Figure 2.
Failed lobuloalveolar development in Runx2−/− mammary glands.
A) Whole mounts of transplanted Runx2+/+ and Runx2−/− mammary glands isolated from virgin Rag1−/− recipients at 16 weeks post-transplantation (Scale bar 1 mm).
B) H&E stained section through epithelial duct of glands shown in (A) (Scale bar 12.5 μm).
C-D) Same as in (A-B), but transplanted mammary glands collected at day 18.5 of pregnancy (P18.5) Arrows indicate lipid globules.
E) Sections of P18.5 Runx2+/+ and Runx2−/− mammary glands were analysed by immunofluorescence for polarity markers ZO1 (apical) and smooth muscle actin (SMA: basal).
F) Immunohistochemical analysis of β casein expression in Runx2+/+ and Runx2−/− mammary glands at P18.5 (Scale bar 25 μm). Higher magnification image shown in (G).
The alveoli in Runx2−/− glands did not have a discernable lumen, although immunofluorescence staining for the polarity markers ZO1 and SMA indicated that the alveoli were correctly polarized (Figure 2D-E & S1C). Thus, the lack of engorgement of the Runx2−/− alveoli suggested a potential secretory differentiation defect. While some epithelial areas showed no signs of secretory activation, other regions of the Runx2−/− gland displayed alveoli containing fat globules, indicating that secretory activation had occurred. Immunohistochemistry showed that milk proteins were detected in the Runx2−/− gland at both P18.5 and L1 (Figures 2F and S1D). Quantitation of mRNA expression of several milk protein genes in the P18.5 glands indicated that WAP and β casein levels were moderately downregulated in Runx2−/− glands (Figure S1E). These data demonstrate that Runx2 is required for alveolar development during late pregnancy, but that alveolar polarisation and lactational differentiation still occur in the absence of Runx2.
To further investigate the function of Runx2 during mammary gland development, we created mice in which loxP sites were engineered into the endogenous Runx2 locus to enable conditional silencing of Runx2 expression. Mouse mammary tumor virus (MMTV) promoter-driven Cre line A (29) expression facilitated tissue-specific deletion of Runx2 in the mammary gland (Figure S2). In the mammary glands of Runx2f/f;MMTV-cre mice, we observed a transient delay in ductal morphogenesis during puberty, with a mild reduction in the percentage of fat pad filling compared to control littermates at 6 weeks of age (Figure S3A). By 8 weeks the glands of Runx2f/f;MMTV-cre and control mice were indistinguishable, suggesting that Runx2 contributes to ductal morphogenesis in puberty but is not essential at this time. Moreover, the ducts of Runx2f/f;MMTV-cre mammary glands appeared normal, containing both the outer myoepithelial (p63-positive) and inner luminal epithelial (keratin-8 positive) cells (Figure S3B).
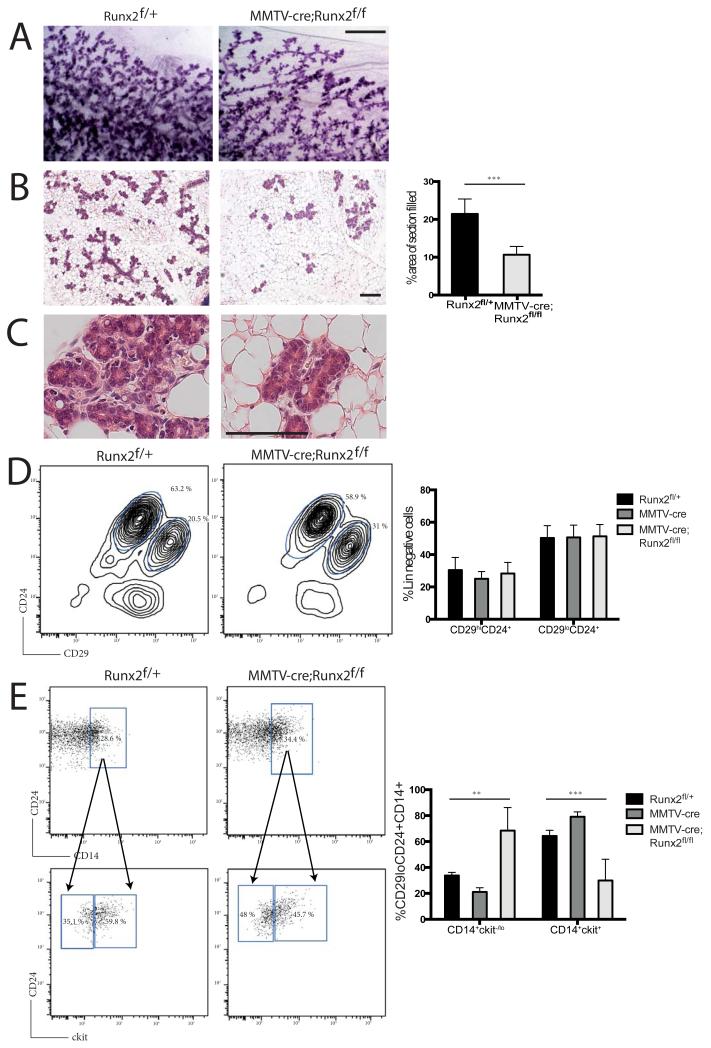
During pregnancy, Runx2f/f;MMTV-cre mammary glands exhibited developmental defects consistent with Runx2−/− transplanted mammary glands. Significantly less of the Runx2f/f;MMTV-cre fat pad was filled with alveoli at day 14.5 of pregnancy, compared with glands from Runx2f/+ mice (Figure 3A-C). Lineage analysis of different mammary cell populations has provided mechanistic insight into the function of a number of transcriptional regulators of mammary gland development (6-11). To further investigate cellular changes in the Runx2-deficient model, we employed flow cytometry to isolate mammary epithelial subpopulations (25). The overall proportions of basal (CD29hiCD24+) and luminal (CD29loCD24+) cell populations were unchanged at mid-pregnancy between Runx2f/+ and Runx2f/f;MMTV-cre mammary glands (Figure 3D). Next, CD14 and ckit were used to discriminate the luminal progenitor and alveolar progenitor populations (25). We observed an increase in the CD14+ckit−/lo alveolar progenitor population in Runx2f/f;MMTV-cre mammary glands at 14.5 days of pregnancy compared to controls at the same time point (Figure 3E). Similarly, we found that Runx2−/− luminal cells at 18.5 days of pregnancy in the Runx2 transplant model demonstrated the same shift from luminal (CD14+ckit+) to alveolar (CD14+ckit−/lo) progenitors compared to Runx2+/+ controls (Figure S4). Therefore, these data demonstrate that loss of Runx2 results in deregulation of the luminal lineage during alveolar maturation in mid to late pregnancy and suggest that Runx2 is involved in the specification of alveolar cell maturation.
Figure 3.
Runx2 regulates alveolar progenitor populations.
A) Carmine stained mammary gland whole mounts from Runx2f/+ and MMTV-cre;Runx2f/f at day 14.5 of pregnancy (P14.5) (Scale bar 1 mm).
B) Representative H&E stained sections of tissues shown in (A) (Scale bar 25 μm). Quantification of area of fat pad occupied by epithelium is shown in graph alongside images (p<0.001). Higher magnification images shown in (C).
D) Flow cytometry analysis of Runx2f/+ and MMTV-cre;Runx2f/f mammary epithelial populations. MaSC-enriched (lin-CD29hiCD24+) and luminal (lin-CD29loCD24+) populations were analysed at P14.5.
E) Flow cytometry analysis of the luminal population (CD29loCD24+) by fractionating progenitor CD14+ cells and then analysing ckit expression. Representative FACS plots are shown. Data for MMTV-cre control mammary glands are included in the graph quantifying luminal progenitor populations (** p<0.01 & *** p<0.001).
Aberrant Runx2 expression blocks differentiation and induces EMT-like changes in mammary epithelial cells
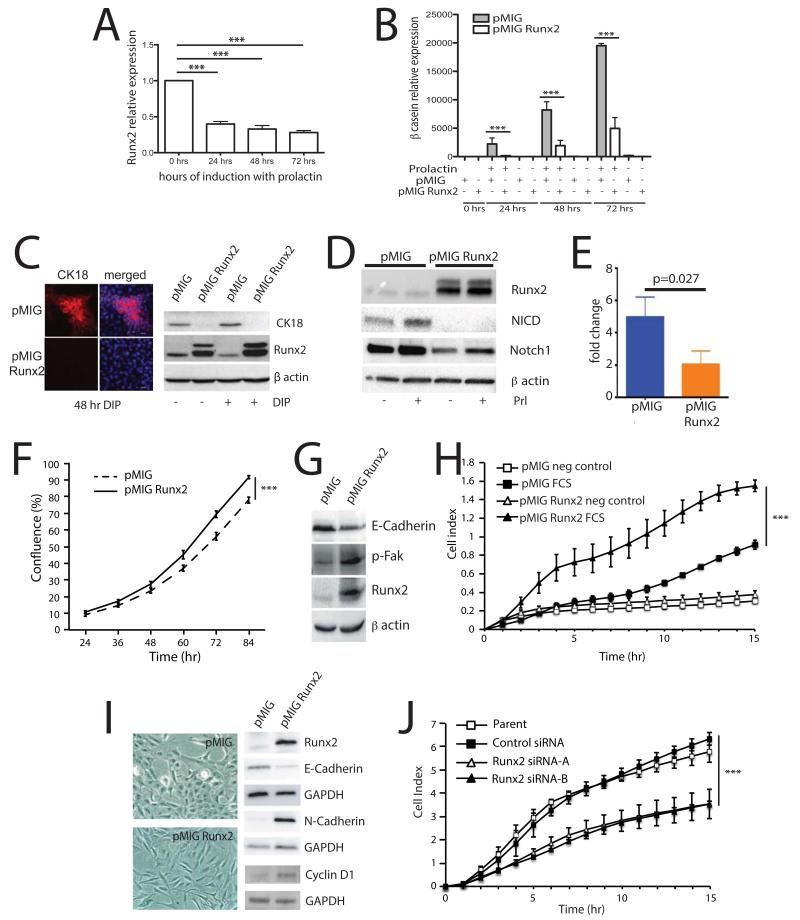
To further investigate the role of Runx2 in mammary epithelial cell fate control, we examined the functional significance of the Runx2 inhibition during late pregnancy. To this end, we utilised the mammary epithelial cell line HC11, which was originally derived from mammary glands of mid-pregnant mice, as a model of differentiation (32). To demonstrate their suitability as a model to study Runx2 function during differentiation we examined Runx2 expression in response to prolactin stimulation and found that Runx2 mRNA expression was significantly downregulated to less than half its regular expression at 24, 48 and 72 hrs following prolactin treatment (Figure 4A). This change in Runx2 level is similar to that observed in vivo towards the end of pregnancy (Figure 1A) and demonstrated that Runx2 expression levels decreased concurrent with the induction of lactational differentiation.
Figure 4.
Maintaining Runx2 expression blocks differentiation and induces cancer-associated changes in mammary epithelial cells
A) Runx2 expression by qPCR in HC11 cells, left untreated or treated with differentiation media (dexamethasone, insulin and prolactin) for 24, 48 and 72 hrs (n=3, error bars represent S.E.M.).
B) HC11 cells stably over-expressing Runx2 or EGFP were treated with dexamethasone and insulin, with and without prolactin to induce differentiation as indicated for the times shown and β-casein expression was assessed by qPCR (n=3, error bars represent S.E.M.).
C) Cytokeratin-18 expression (CK18) in HC11 stable lines treated with and without differentiation media (DIP) for 48 hrs was examined by immunofluorescence (left panel) and western blotting (right panel). Merged = CK18+DAPI.
D) Western blot of whole cell lysates from pMIG and pMIGRunx2 HC11 cells in differentiation media for 24 hours with or without prolactin as indicated.
E) Quantitative PCR of Hey1 expression from pMIG and pMIGRunx2 HC11 cells in differentiation media with prolactin for 72 hours (n=3, error bars represent S.E.M. p=0.027).
F) Growth rates of pMIG and pMIGRunx2 HC11 cells was determined using live-cell imaging and quantifying cell confluence using phase-contrast image mask (n=3, performed in triplicate, 4 image positions/well, error bars represent S.E.M.).
G) Western blot of whole cell lysates from pMIG and pMIGRunx2 HC11 cells in growth media.
H) Migration of pMIG and pMIGRunx2 HC11 cells was analysed in real-time by xCelligence, using 10% FCS as chemoattractant or blank media as a control (n=3, error bars represent S.E.M.).
I) MCF10A cells expressing pMIG and pMIGRunx2 were selected by FACS. Phase-contrast images show a more fibroblastic morphology in pMIGRunx2-expressing cells (left). Expression of EMT-markers was determined by western blotting (right).
J) MDA-MB-231 cells were treated with Runx2 shRNA or non-target control shRNA and 48 hours later subjected to migration analysis on Excelligence using 10 % FCS as chemoattractant.
In all experiments *** represents p<0.01.
To test the hypothesis that this decrease in Runx2 expression is required for differentiation to occur, we generated HC11 cells stably overexpressing Runx2 and examined β-casein expression in response to prolactin. In control HC11 cells, β-casein expression was induced after 24 hours treatment with prolactin and continued to increase up to 72 hours (Figure 4B). However, β-casein expression was significantly reduced in HC11-Runx2 cells at all time points. Analysis of Stat5 activation following prolactin stimulation in the same cells (Figure S5), showed no difference in the level of pStat5 between control and Runx2 overexpressing cells, suggesting that the Runx2-mediated impairment of differentiation occurs independently of Stat5. To examine whether the block in differentiation was due to altered cell type specification or just inhibition of milk production, we examined expression of the luminal epithelial cell marker cytokeratin-18 (CK18). In control HC11 cells, we found populations of CK18-positive cells, although the majority of cells were CK18-negative (Figure 4C). This is consistent with HC11 cells being a heterogeneous population. In Runx2 overexpressing cells, CK18 expression was not detected, either by immunofluorescence or western blotting (Figure 4C). Notch1 regulates the specification of luminal progenitors, while expression of activated Notch1 (NICD) induces luminal progenitor differentiation (9, 33), therefore we next investigated whether Notch1 signaling is regulated by Runx2. Forced expression of Runx2 in HC11 cells suppressed Notch1 activation, inhibiting both Notch1 and NICD protein levels (Figure 4D) and reducing expression of hey1 (Figure 4E) a downstream target of Notch signaling. Together, these data demonstrate that sustaining Runx2 expression in mammary epithelial cells perturbs differentiation and maintains HC11 cells in a less differentiated state, potentially through inhibition of Notch1 activation. Given that previous studies have indicated a potential role for Runx2 in breast cancer progression, we examined whether Runx2 overexpression in HC11 cells induced a more cancer-like phenotype. There was a small but significant increase in the growth rate of HC11-Runx2 cells compared with controls (Figure 4F). Consistent with Runx2 inducing EMT-like changes, HC11-Runx2 cells showed decreased E-cadherin expression, increased levels of phospho-FAK and significantly increased migration rates compared with controls (Figure 4G-H). In agreement with previous work, Runx2 overexpression induced morphological changes in normal human breast (MCF10A) cells and Runx2 siRNA reduced migration of human breast cancer (MDA-MD-231) cells (Figure 4I-J; (34)). Runx2 induced upregulation of cyclin D1 in MCF10A cells, as well as altered cadherin expression expected in cells undergoing EMT (i.e. decreased E-Cadherin and increased N-Cadherin; Figure 4I). Thus, manipulating Runx2 expression drove cell phenotypes consistent with Runx2 promoting tumorigenesis in normal mammary cells.
Runx2 deletion delays breast cancer development and prolongs survival
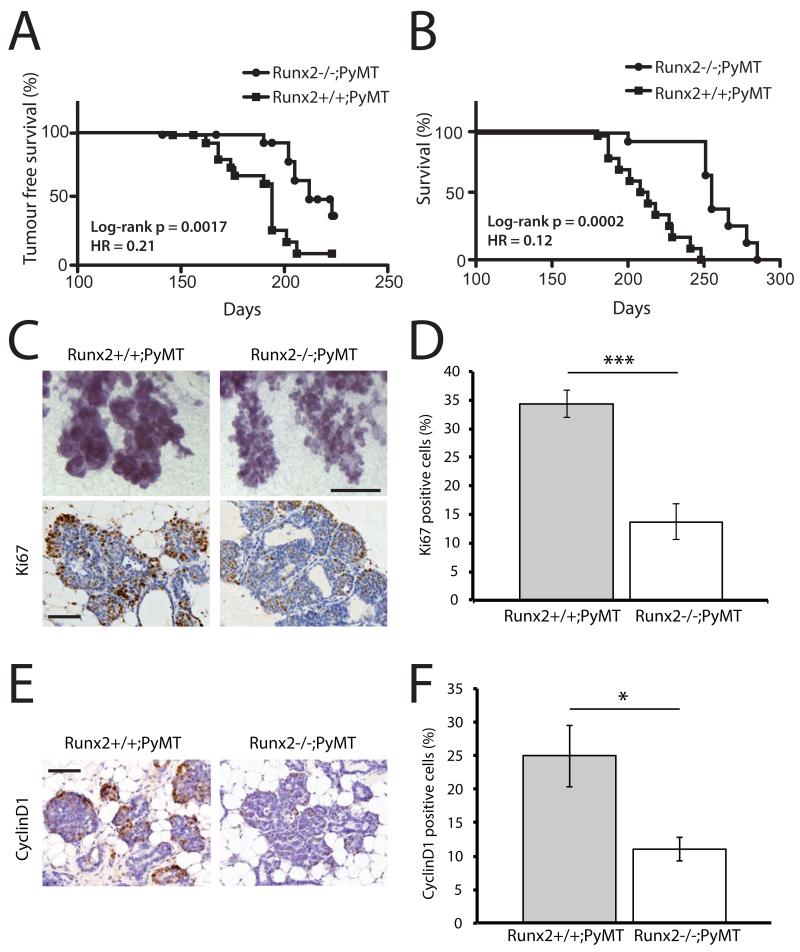
Runx2 has an established role in determining the osteolytic activity of breast cancer cells, however, its role in the initiation of mammary tumourigenesis is largely unknown. To address this question, we deleted Runx2 in the MMTV-PyMT mouse model by crossing Runx2+/− and PyMT mice together to generate either PyMT;Runx2−/− or PyMT;Runx2+/+ embryos. The dissected embryonic mammary buds were then transplanted to Rag1−/− hosts and tumor incidence and survival analyzed in recipient mice. There was a significant increase in time to tumor detection in Rag1−/− recipients of PyMT;Runx2−/− mammary epithelium compared to recipients of PyMT;Runx2+/+ epithelium (HR=0.21; p=0.0017, Figure 5A). There was an even more significant increase in overall survival in the absence of Runx2 (HR=0.12; p=0.0002, Figure 5B). Metastases were not detected in Rag1−/− immunocompromised mice transplanted with either PyMT;Runx2+/+ or PyMT;Runx2−/− epithelium, probably because of the established role the immune system has in mediating metastasis, thus precluding our evaluation of a potential role for Runx2 in metastasis within this system.
Figure 5.
Runx2 promotes tumorigenesis in pre-clinical breast cancer model
A-B) Kaplan-Meier survival plots of tumor-free survival (A) and overall survival (B) of Rag1−/− recipient mice carrying either Runx2+/+;PyMT or Runx2−/−;PyMT transplanted mammary glands.
C) Representative images of whole-mounts (top panels) (Scale bar 0.5 mm) and Ki67 IHC (bottom panels) of Runx2+/+;PyMT and Runx2−/−;PyMT mammary glands 13 weeks post-transplant (Scale bar 12.5 μm).
D) Quantification of Ki67 IHC described in (C) (n=3 mice, error bars represent S.E.M.).
E) Representative images of cyclin D1 IHC of Runx2+/+;PyMT and Runx2−/−;PyMT mammary glands 13 weeks post-transplant (Scale bar 12.5 μm).
F) Quantification of Ki67 IHC. (n=3 mice, error bars represent S.E.M.).
In all experiments * represents p<0.05 and *** represents p<0.01.
To examine the mechanism of prolonged survival in the absence of Runx2 we performed a cross-sectional analysis at 13 weeks post-transplantation, prior to detection of palpable tumours. Morphologically, the PyMT;Runx2−/− glands were consistently less neoplastic than the PyMT;Runx2+/+ tissues (Figure 5C). Indeed, proliferation levels were significantly lower in the hyperplastic regions of PyMT;Runx2−/− epithelium (Figure 5C-D). Furthermore, cyclin D1 expression was also reduced in the absence of Runx2 (Figure 5E-F). Together, these data demonstrate that Runx2 functions to promote tumour progression in vivo by facilitating increased proliferation rates, leading to decreased survival.
Discussion
This is the first study to use genetic models to demonstrate a role for Runx2 in breast development and tumorigenesis in vivo. Runx2 expression exhibits tight temporal regulation during pregnancy, which corresponds to the time at which expansion of the luminal progenitor population occurs (31). Runx2 protein and mRNA levels then fall significantly at late pregnancy for the final stages of differentiation to occur. The importance of developmental stage-specific regulation of Runx2 is also evident in osteoblasts where Runx2 deficiency causes a failure in osteoblast development (18). However, maintaining Runx2 expression in osteoblasts disrupts their final maturation, similar to our finding in HC11 cells, where differentiation is blocked by forced expression of Runx2 (35).
The mammary epithelial cell hierarchy is currently an area of intense interest due to providing insight into the cells of origin in breast cancer. We show here that Runx2 is necessary for the specification of luminal progenitor cells. Without Runx2, the proportion of progenitors shifts towards a more alveolar-committed population that have a more differentiated phenotype (31). Interestingly, expression of Notch1 intracellular domain in the developing mammary gland increases the mature luminal cell population, suggesting that Notch1 promotes luminal cell differentiation (33). We observed decreased Notch1 activation in HC11 cells overexpressing Runx2, raising the possibility that inhibitory cross-talk between Runx2 and Notch1 controls luminal cell lineage specification, similar to the interaction between Runx2 and Notch1 during osteoblast differentiation (36, 37). Disrupting progenitor populations in the primary setting perturbs normal breast development and while the precise contributions of the different luminal progenitor subsets to development are not currently known, lineage tracing studies, preferably with a doxycycline –inducible system, as recently described (38) will help define the role of different cell lineages during mammopoiesis.
The Runx2−/− and Runx2f/f;MMTV-cre mice result in deletion of Runx2 in all and the majority of mammary epithelial cells, respectively. We observe no difference in the proportions of basal and luminal cells during development, although microarrays have identified Runx2 mRNA in both basal (CD29hiCD24+) and luminal (CD29+CD24+) cell populations (21). Runx2 expression in ER negative luminal cells may act in a cell-autonomous manner to control progenitor populations during pregnancy. The use of lineage-specific or temporally restricted Cre-recombinase driven by promoters, such as p63 (basal epithelial cells) or WAP (luminal cells during pregnancy) to delete Runx2 in the Runx2f/f mice will enable the requirement of Runx2 in specific cell-types to be characterized.
In the PyMT breast cancer model, we demonstrate a reduction in proliferative cells and cyclin D1 levels in the absence of Runx2. The percent and distribution of cyclin D1 cells in Runx2+/+;PyMT mice at the early stage of tumorigenesis is consistent with prior observations (30). There is much debate on the role of Runx2 has in proliferation, with cell-type, temporal and levels of expression all expected to play a role (39). How removal of Runx2 reduces cyclin D1 expression is not known, in fact cyclin D1 has been shown to induce downregulation of Runx2 in other systems (40), potentially indicating a negative feedback mechanism or Runx2 might be required for paracrine signals to induce cyclin D1. For instance, in MDA-MB-231 cells, TGFβ-induced cyclin D1 expression is attenuated upon Runx2 knockdown by siRNA. (34) Cyclin D1 is an established oncogene in a number of tissues, however correlation of cyclin D1 expression with patient prognosis provides varying results (41-43). Well known for its requirement for cells to transition from G1 to S-phase of the cell cycle, cyclin D1 is also required for activation of many estrogen-responsive genes (44). Furthermore, the kinase activity of cyclin D1 is necessary for maintenance of pregnancy-induced mammary epithelial cell progenitors (PI-MECs) (45). Recently, Notch3-induced mouse breast tumors were attributed to cyclin D1-dependent expansion of luminal progenitors (46). Thus, in addition to regulating cell proliferation, Runx2 regulation of cyclin D1 may also control other aspects of tumour phenotype, and we aim to examine this further in the future.
Epithelial-to-mesenchymal transition is associated with cancer cells acquiring characteristics that support metastasis, such as increased migration and invasion. Our findings of the effects of Runx2 expression on EMT are consistent with previously reported data. Runx2 over-expression in MCF7 breast cancer cells induced EMT and promoted cancer cell-invasion through matrigel (24). Conversely, blocking Runx2 function by inhibiting Runx2 sub-nuclear trafficking reduced MDA-MB-231 cell invasion in culture assays (28). Runx2 drives EMT through downstream targets such as TGFβ, Wnt and Snai2, which are also associated with driving breast cancer metastasis.
Metastases were not observed in the Rag1−/− recipients of PyMT epithelium, precluding us from examining more advanced stages of tumorigenesis. However, studies suggest that in addition to EMT, Runx2 may contribute to several further stages of the metastatic process. Upregulation of VEGF by Runx2 could stimulate the growth of tumor vasculature, while Runx2-mediated expression of the anti-apoptotic protein Bcl-2 may aid tumor cell survival as they pass through the circulatory system to distant sites around the body (25, 28). Breast cancer commonly metastasizes to bone and xenotransplantation studies support a role for Runx2 in metastatic bone disease. For example, knockdown of Runx2 in MDA-MB-231 cells reduced osteolysis when the cancer cells were injected into the tibia of recipient mice (34). Recently, Runx2 was demonstrated to be upregulated by serotonin, which is associated with breast cancer progression and bone demineralization and may indicate a mechanism by which Runx2-dependent osteolysis is activated (47). Ultimately the delineation of Runx2 function in breast cancer metastasis will require the development of models in which Runx2 expression in tumour cells can be manipulated in an immune-competent host.
Progenitor cells are prime candidates for cells of origin in several breast tumor subtypes. Parity-identified mammary epithelial cell (PI-MEC) progenitors are thought to be the cell of origin in MMTV-ErbB2/neu/HER2 driven tumours (45, 48). Although, subtyped as basal, Brca1-mutant basal-like breast cancers are derived from luminal progenitors (49). Furthermore, luminal progenitors have also recently been shown to contribute to tumor progression in the MMTV-PyMT breast cancer model (50). Interestingly, a Runx2 metagene dataset was enriched in the basal-like tumor subtype, as well as some of the HER2 subtype (17). Using our conditional Runx2 knockout mice crossed onto basal (SV40) and Neu (MMTV-Neu) tumor models will help delineate the role of Runx2 in these aggressive tumor subtypes.
In summary, we have identified a new regulator of normal and transformed mammary epithelial cells in vivo. It will now be of significant interest to delineate relationships between Runx2 and other established cancer-driving transcription factors and examine the potential of Runx2 as a therapeutic target in breast cancer.
Supplementary Material
Acknowledgements
The authors acknowledge the support received from the Bosch Institute’s Molecular Biology, Advanced Microscopy and Rodent Facilities, and the expert help of Facility staff, especially Donna Lai and Louise Cole. We also thank Gillian Lehrbach at the Tissue Culture Facility, Garvan Institute of Medical Research. We would also like to thank the Animal, FACS and Histology facilities at WEHI. This work was supported by the Australian National Health and Medical Research Council (NHMRC); Cancer Council NSW, Australian Research Council, the Victorian State Government through the Victorian Breast Cancer Research Consortium and Operational Infrastructure Support; the Australian Cancer Research Foundation. M.J.N & A.S were supported by NHMRC Career Development Fellowship’s, M.J.N & P.T.S by National Breast Cancer Foundation of Australia Fellowships, S.B. by a NHMRC Postgraduate Scholarship (1017256), C.J.O by a NHMRC Research Fellowship and J.E.V. by a NHMRC Australia Fellowship.
References
- 1.Hennighausen L, Robinson GW. Signaling Pathways in Mammary Gland Development. Dev Cell. 2001;1:467–75. doi: 10.1016/s1534-5807(01)00064-8. [DOI] [PubMed] [Google Scholar]
- 2.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 3.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Smith GH. Murine Mammary Epithelial Stem Cells: Discovery, Function, and Current Status. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Šale S, Lafkas D, Artavanis-Tsakonas S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nat Cell Biol. 2013;15:451–60. doi: 10.1038/ncb2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheta M, Teschendorff A, Sharp G, Novcic N, Russell A, Avril S, et al. Phenotypic and functional characterisation of the luminal cell hierachy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 8.Oakes SR, Naylor MJ, Asselin-Labat ML, Blazek KD, Gardiner-Garden M, Hilton HN, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–6. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–41. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Buono KD, Robinson GW, Martin C, Shi S, Stanley P, Tanigaki K, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–80. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 11.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 Maintains the Differentiation of the Luminal Cell Fate in the Mammary Gland. Cell. 2006;127:1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–52. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–18. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalyuga M, Gallego-Ortega D, Lee HJ, Roden DL, Cowley MJ, Caldon CE, et al. ELF5 Suppresses Estrogen Sensitivity and Underpins the Acquisition of Antiestrogen Resistance in Luminal Breast Cancer. PLoS Biol. 2012;10:e1001461. doi: 10.1371/journal.pbio.1001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owens TW, Naylor MJ. Breast cancer stem cells. Front Physiol. 2013;4 doi: 10.3389/fphys.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen MM., Jr Perspectives on RUNX genes: an update. American journal of medical genetics Part A. 2009;149A:2629–46. doi: 10.1002/ajmg.a.33021. [DOI] [PubMed] [Google Scholar]
- 17.Chimge NO, Frenkel B. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2013;32:2121–30. doi: 10.1038/onc.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 19.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 20.Kouros-Mehr H, Werb Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev Dynamics. 2006;235:3404–12. doi: 10.1002/dvdy.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendrick H, Regan JL, Magnay FA, Grigoriadis A, Mitsopoulos C, Zvelebil M, et al. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genomics. 2008;9:591. doi: 10.1186/1471-2164-9-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inman CK, Shore P. The osteoblast transcription factor Runx2 is expressed in mammary epithelial cells and mediates osteopontin expression. J Biol Chem. 2003;278:48684–9. doi: 10.1074/jbc.M308001200. [DOI] [PubMed] [Google Scholar]
- 23.Barnes GL, Javed A, Waller SM, Kamal MH, Hebert KE, Hassan MQ, et al. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 2003;63:2631–7. [PubMed] [Google Scholar]
- 24.Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, et al. Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2. Breast Cancer Res. 2011;13:R127. doi: 10.1186/bcr3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratap J, Imbalzano KM, Underwood JM, Cohet N, Gokul K, Akech J, et al. Ectopic runx2 expression in mammary epithelial cells disrupts formation of normal acini structure: implications for breast cancer progression. Cancer Res. 2009;69:6807–14. doi: 10.1158/0008-5472.CAN-09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendoza-Villanueva D, Zeef L, Shore P. Metastatic breast cancer cells inhibit osteoblast differentiation through the Runx2/CBFbeta-dependent expression of the Wnt antagonist, sclerostin. Breast Cancer Res. 2011;13:R106. doi: 10.1186/bcr3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, et al. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–91. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, et al. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci U S A. 2005;102:1454–9. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner K-U, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–30. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asselin-Labat ML, Sutherland KD, Vaillant F, Gyorki DE, Wu D, Holroyd S, et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol Cell Biol. 2011;31:4609–22. doi: 10.1128/MCB.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 1988;7:2089–95. doi: 10.1002/j.1460-2075.1988.tb03048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons MJ, Serra R, Hermance N, Kelliher MA. NOTCH1 inhibition in vivo results in mammary tumor regression and reduced mammary tumorsphere-forming activity in vitro. Breast Cancer Res. 2012;14:R126. doi: 10.1186/bcr3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, et al. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W, Toyosawa S, Furuichi T, Kanatani N, Yoshida C, Liu Y, et al. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155:157–66. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ann EJ, Kim HY, Choi YH, Kim MY, Mo JS, Jung J, et al. Inhibition of Notch1 signaling by Runx2 during osteoblast differentiation. J Bone Miner Res. 2011;26:317–30. doi: 10.1002/jbmr.227. [DOI] [PubMed] [Google Scholar]
- 37.Xu N, Liu H, Qu F, Fan J, Mao K, Yin Y, et al. Hypoxia inhibits the differentiation of mesenchymal stem cells into osteoblasts by activation of Notch signaling. Exp Mol Path. 2013;94:33–9. doi: 10.1016/j.yexmp.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014 doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 39.Lucero CM, Vega OA, Osorio MM, Tapia JC, Antonelli M, Stein GS, et al. The cancer-related transcription factor Runx2 modulates cell proliferation in human osteosarcoma cell lines. J Cell Physiol. 2013;228:714–23. doi: 10.1002/jcp.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen R, Wang X, Drissi H, Liu F, O’Keefe RJ, Chen D. Cyclin D1-cdk4 induce runx2 ubiquitination and degradation. J Biol Chem. 2006;281:16347–53. doi: 10.1074/jbc.M603439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 42.Peurala E, Koivunen P, Haapasaari KM, Bloigu R, Jukkola-Vuorinen A. The prognostic significance and value of cyclin D1, CDK4 and p16 in human breast cancer. Breast Cancer Res. 2013;15:R5. doi: 10.1186/bcr3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu XL, Chen SZ, Chen W, Zheng WH, Xia XH, Yang HJ, et al. The impact of cyclin D1 overexpression on the prognosis of ER-positive breast cancers: a meta-analysis. Breast Cancer Res Treat. 2013;139:329–39. doi: 10.1007/s10549-013-2563-5. [DOI] [PubMed] [Google Scholar]
- 44.Casimiro MC, Wang C, Li Z, Di Sante G, Willmart NE, Addya S, et al. Cyclin D1 determines estrogen signaling in the mammary gland in vivo. Mol Endocrinol. 2013;27:1415–28. doi: 10.1210/me.2013-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, et al. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling H, Sylvestre JR, Jolicoeur P. Cyclin D1-dependent induction of luminal inflammatory breast tumors by activated notch3. Cancer Res. 2013;73:5963–73. doi: 10.1158/0008-5472.CAN-13-0409. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez LL, Gregerson KA, Horseman ND. Mammary gland serotonin regulates parathyroid hormone-related protein and other bone-related signals. Am J Physiology Endo Metab. 2012;302:E1009–15. doi: 10.1152/ajpendo.00666.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner KU, Booth BW, Boulanger CA, Smith GH. Multipotent PI-MECs are the true targets of MMTV-neu tumorigenesis. Oncogene. 2013;32:1338. doi: 10.1038/onc.2012.452. [DOI] [PubMed] [Google Scholar]
- 49.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 50.Luo M, Zhao X, Chen S, Liu S, Wicha MS, Guan JL. Distinct FAK activities determine progenitor and mammary stem cell characteristics. Cancer Res. 2013;73:5591–602. doi: 10.1158/0008-5472.CAN-13-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.