Abstract
The pathogenesis of the Brucella-induced inflammatory response in the bovine placenta is not completely understood. In this study we evaluated the role of the B. abortus Type IV secretion system and the anti-inflammatory factor BtpB in early interactions with bovine placental tissues. Transcription profiles of chorioallantoic membrane (CAM) explants inoculated with wild type (strain 2308), ΔvirB2 or ΔbtpB Brucella abortus were compared by microarray analysis at 4 hours post infection. Transcripts with significant variation (>2 fold change; P<0.05) were functionally classified, and transcripts related to defense and inflammation were assessed by quantitative real time RT-PCR. Infection with wild type B. abortus resulted in slightly more genes with decreased than increased transcription levels. Conversely, infection of trophoblastic cells with the ΔvirB2 or the ΔbtpB mutant strains, that lack a functional T4SS or that has impaired inhibition of TLR signaling, respectively, induced more upregulated than downregulated genes. Wild type Brucella abortus impaired transcription of host genes related to immune response when compared to ΔvirB and ΔbtpB mutants. Our findings suggest that proinflammatory genes are negatively modulated in bovine trophoblastic cells at early stages of infection. The virB operon and btpB are directly or indirectly related to modulation of these host genes. These results shed light on the early interactions between B. abortus and placental tissue that ultimately culminate in inflammatory pathology and abortion.
Introduction
Brucellosis is an important zoonotic disease with worldwide distribution, caused by bacteria of the genus Brucella. It causes significant economic losses due to abortions and culling of infected cattle, whereas in humans it is associated with a febrile illness with variable symptoms, and it may occasionally be fatal [1]–[3].
Most cases of bovine brucellosis are due to Brucella abortus infection, which is transmitted by contact with contaminated aborted fetuses, fetal membranes, and uterine secretions after abortion or during the postpartum period [4], [5]. Aborted fetuses resulting from B. abortus infection often exhibit signs of fibrinous pleuritis, which may be associated with suppurative bronchopneumonia and fibrinous pericarditis [6], [7]. During pregnancy, after the initial infection of the erythrophagocytic trophoblasts located at the base of the chorionic villi, the bacteria spread throughout the placenta following a periplacentomal pattern, infecting trophoblastic cells of the intercotyledonary region, mostly at the end of gestation (180 to 240 days) [8]–[11]. Thus, B. abortus triggers an intense acute inflammatory response in the placenta, which is associated with abortion [7]. While this inflammatory pathology is well-described, very little is known about the initial interactions between B. abortus and placental cells that ultimately result in placentitis and abortion, two processes that are key components of disease transmission. Because of the difficulty of studying these early interactions in pregnant animals, ex vivo infection of cultured chorioallantoic membrane (CAM) explants, which results in localization of B. abortus to trophoblasts, has been used to study the initial phases of placental infection [12], [13].
Virulence factors of B. abortus have been studied in the context of persistent infection of the mononuclear phagocyte system, however few studies have been performed in the context of placental infection in the natural host. The type IV secretion system (T4SS) is considered to be a key virulence factor of Brucella spp., and it is responsible for secretion of effector proteins across the bacterial cell envelope [14]–[16]. The T4SS has been shown to be involved in abortion in goats [17], raising the question of its contribution to early interactions with the placenta.
A second set of virulence factors, shown to be involved in immune evasion, are the TIR domain proteins, BtpA and BtpB [18]–[20]. BtpA is present in B. abortus, B. melitensis, and B. suis ATCC 23445 (biovar 2), but it is absent in B. suis 1330 (biovar 1). BtpA binds directly to MyD88, preventing signaling via TLR2 and TLR4, impairing innate immune response and inhibiting maturation of in vitro infected dendritic cells by blocking the TLR2 signaling pathway [18], [20]. BtpB is present in all species of Brucella. It inhibits TLR signaling, preventing activation of dentritic cells, and together with BtpA can modulate the host inflammatory responses during Brucella sp. infection [19]. Interestingly, translocation of BtpB into host macrophages was shown to depend on the VirB T4SS [19].
In this study, the CAM explant model was used as an ex vivo model to study the pathogenesis of Brucella infection during the initial phase of the bacteria-host interaction [12]. Carvalho Neta et al [13], using this model, demonstrated that B. abortus modulates the innate immune response by trophoblastic cells by inhibiting the transcription of proinflammatory mediators at early stages of infection. The aim of this study was to interrogate the role of the VirB T4SS and BtpB in early suppression of inflammatory responses in the placenta.
Materials and Methods
Bacterial strains
The inocula were prepared by growth for 12–15 hours under agitation at 37°C of the three bacterial strains: B abortus 2308 was cultivated in Brucella broth (Difco, Lawrence, KS, USA) and the ΔvirB2 B. abortus and ΔbtpB B. abortus strains were cultivated in Tryptic Soy Broth (Difco) supplemented with kanamycin (100 µg/µL). After incubation, the optical density of bacterial suspensions was determined by spectrophotometry (OD600) and adjusted to 1.0×108 CFU/mL. To confirm the concentration of bacteria, the inocula were serially diluted in PBS (pH 7.4), and 100 µL of each dilution were plated on Tryptic Soy Agar (Difco), in duplicate. After 48 h of incubation at 37°C with 5% CO2, colonies counted and the number of colony forming units (CFU) was obtained by the average of duplicates. The handling of agent and infected material was performed under biosafety level 3 containment.
Generation of mutant strains
The ΔvirB2 B. abortus strain used in this study was obtained by allelic exchange of the virB2 gene (BAB2_0067), inserting a kanamycin cassette. The plasmid used to construct the mutant strains was the pAV2.2, a kanamycin resistant vector, which has been described previously [21]. The ΔbtpB B. abortus strain was obtained by allelic exchange of the btpB gene (BAB1_0279) for a kanamycin cassette. The plasmid used for btpB mutagenesis was generated in this study. For this purpose a plasmid called pUKD/btpB was made by using a previously described three-step cloning strategy [22]. Briefly, a fragment of the 5′ region of BAB1_0279 with engineered SmaI site was amplified from B. abortus genomic DNA using primers BMEI1674UP-F (TGAATGTGGCAAGCCCTCGAC) and BMEI1674UP-R (ACCCGGGCTTGTTTCTCTTTAGAC). A fragment of the 3′ region of BAB1_0279 with engineered SmaI and PstI sites was amplified using primers BMEI1674DN-F (ACCCGGGCAGATGCAAATATGGCCGTAAG) and BMEI1674DN-R (TCTGCAGCCGGAGGAATGGCATCAC). Both amplicons were TOPO cloned into pCR2.1 to yield plasmids pUP/btpB and pDN/btpB respectively. The orientation of the inserted 5′ fragment of BAB1_0279 was determined by PCR and restriction analysis to make sure the unique SmaI site was next to the T7 promoter in pCR2.1 vector. The 3′ fragment of BAB1_0279 was then excised by SmaI/PstI double digestion and cloned into the same sites of pUP/btpB to generate the pUD/btpB. The resulting plasmid was selected for ampicillin resistance as double digestion of SmaI/PstI truncates the original kanamycin resistance gene in pCR2.1. In the third cloning step, a 1.3-kb SmaI fragment of pUC4-KIXX (Pharmacia) containing the Tn5 kanamycin resistance gene was cloned into the SmaI site of pUD/btpB to give rise topUKD/btpB, which was selected for both kanamycin and ampicillin resistance. These plasmids were transformed into electrocompetent B. abortus cells by electroporation as previously described [23]. Colonies that were kanamycin resistant and ampicillin sensitive were selected as mutant candidates. Deletion of the virB2 and btpB genes was confirmed by PCR using primers flanking the deleted region.
Chorioallantoic membrane explant culture and infection
Snapwell plates (Transwell Cell Culture Permeable Supports – Snapwell Inserts - Corning Incorp., NY, EUA) were used for culturing CAM explants [12], [13]. Seven intact pregnant bovine uteruses at the final third of gestation were obtained at local slaughterhouses. Gestational age was estimated by cephalococcygeal length (CR - Crown-rump length) [24]. All fetuses were serologically negative for Brucella spp. by using the Acidified Antigen Buffered test with fetal amniotic fluid. Prior to obtaining the CAM, the perimetrium was thoroughly decontaminated with iodinated alcohol, the uterus was then opened and CAM removed and placed in RPMI 1640 sterile medium (Invitrogen, São Paulo, Brazil) containing antibiotics (100 U/mL penicillin and 100 µg/mL streptomycin) for 20 minutes. CAM explants were then washed twice in RPMI (Invitrogen) at 37°C for complete removal of the antibiotic. Sterile rings and detachable supports were positioned over the intercotyledonary portion of the CAM explants. Excess tissue was removed from CAM explants, which were placed in 6 well culture plates (Corning, NY, USA) containing sterile culture medium RPMI 1640 supplemented with 4 mM glutamine, 1 mM pyruvate, 1 mM nonessential amino acids, 2.9 mM sodium bicarbonate and 15% fetal bovine serum (Invitrogen), and incubated at 37°C in a humidified atmosphere with 5% CO2. The experimental protocol was approved by the Ethics Committee on Animal Experimentation of UFMG (CETEA – Protocol 183/2010).
The trophoblastic surface of the CAM explants was inoculated with 200 µL of culture medium (RPMI 1640) containing 2.0×107 CFU, which corresponded to a multiplicity of infection of approximately 1000 (MOI 1000∶1) as previously used by Carvalho Neta et al. [13]. CAM explants were inoculated in triplicate with wild type, ΔvirB2 or ΔbtpB strains of B. abortus 2308. Plates were incubated at 37°C with 5% CO2 for 4 h, the medium was then replaced with RPMI 1640 medium supplemented with 50 µg/mL gentamicin (Invitrogen, São Paulo, Brazil) to inactivate extracellular bacteria. Plates were maintained at 37°C with an atmosphere containing 5% CO2 for 1 h followed by washing three times with PBS (phosphate buffered saline - pH 7.4) to eliminate the antibiotic. Uninfected CAM controls were inoculated with sterile RPMI 1640 medium and kept under the same conditions.
Determining the number of internalized bacteria
To determine the number of internalized bacteria, three explants inoculated with wild type, ΔvirB2 or ΔbtpB B. abortus 2308 were incubated for 4 h, followed by 1 h incubation with RPMI 1640 medium supplemented with 50 µg gentamicin/mL (Invitrogen, São Paulo, Brazil) to inactivate extracellular bacteria, washed three times with PBS (pH 7.4), and then lysed with 200 µL of sterile 0.1% Triton X-100 (Roche, Mannheim, Germany). Serial dilutions of the lysates were prepared in PBS (pH 7.4), and 100 µL of each dilution were plated on tryptose agar (Difco) in duplicate. After 48 h of incubation at 37°C with 5% CO2, the number of colony forming units (CFU) was counted in each plate.
RNA extraction and preparation of cDNA
After removal of RPMI culture medium supplemented with gentamicin (n = 4 for microarray analysis; n = 3 for qRT-PCR), TRIzol Reagent (Invitrogen) was added to the trophoblastic surface of the CAM explants of uninfected controls or explants infected with wild type, ΔvirB2 or ΔbtpB B. abortus 2308, for total RNA extraction, according to the manufacturer's instructions. Purity and concentration of RNA samples were assessed by spectrophotometry, and RNA quality evaluated by agarose/formaldehyde gel electrophoresis. RNA samples were stored at −80°C. Synthesis of cDNA was performed using Superscript III First Strand S (Invitrogen), following the manufacturer's specifications using a RNA concentration of 1,500 ng in a reaction with a final volume of 20 µL, and cDNA was stored at −20°C.
Microarray analysis
Gene expression profiles were evaluated using the Agilent two color microarray-based gene expression platform according to manufacturer's instructions. Briefly, RNA (500 ng) was amplified and labeled using the two-color Quick Amp labeling kit (Agilent Technologies, CA, USA). Complementary RNA (cRNA) was synthesized from triplicates of CAM explants of four independent experiments. cRNA from uninfected control explants labeled with Cy3 and cRNA from explants infected with either wild type, ΔvirB2 or ΔbtpB B abortus 2308, labeled with Cy5 were hybridized on the same slide at 65°C, for 17 hours and 10 RPM in a high-density microarray containing 4×44,000 genes representing fully sequenced bovine genome (#G2519F Agilent Technologies, Palo Alto, CA, USA). After hybridization, slides were washed in Gene Expression wash buffers 1 and 2 as per instructions, scanned with an Agilent DNA microarray scanner (Agilent Technologies) and the hybridization signals were extracted using the Agilent Feature Extraction software version 11.0.
Quantitative real-time PCR (qRT-PCR)
After functional classification using the Funcat (Functional Classification) platform (http://mips.helmholtzmuenchen.de/genre/proj/mfungd/Search/Catalogs/searchCatfirstFun.html) genes of interest, i.e. related to inflammation and immune response that had at least a 2-fold change in transcription levels and that were statistically significant (P<0.05) were selected for confirmation based on qRT-PCR. Levels of transcripts were normalized based on β-actin transcript level. qRT-PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems, NY, USA) and the StepOnePlus thermal cycler (Applied Biosystems, NY, USA). Primers used in this study are described in Table 1. The data were analyzed using the comparative Cycle threshold (Ct) method [25].
Table 1. Primers used in this study.
| Gene | Primers (5′-3′) | Product size (nt) |
| IFN-alpha G | TCAAGCCATCTCTGTGCTCC | 72 |
| ACGGCTGAACCCTCTACACT | ||
| Chemokine (C-X-C motif) ligand 12 | GATGCCAAGGTCTTCGTCGT | 104 |
| TCAAAGAATCGGCAAGGGCA | ||
| Interleukin 15 | TGGGCTGTATCAGTGCAAGT | 148 |
| ACTTTGCAATTGGGATGAGCA | ||
| Interleukin 1 family, member 6 (epsilon)-like | GCCGGAGCTTTGTCTCTTCT | 136 |
| CCTGCCATTCTGGTCATGGT | ||
| Transmembrane 4 L six family member 19 | CCCTGCCGAAGGATGCTTAT | 85 |
| GCAAATCAAGGCTCCAAGCA | ||
| Tumor necrosis factor receptor superfamily, member 9 | ACATGGCATCTGTCGACCTT | 82 |
| ACGTCACTTTCCTTCGTCCC | ||
| Apolipoprotein L, 3 | CAGAGACACACGAAAGGCG | 108 |
| GGCTGGAAGAAGGTGTCGTT | ||
| Heat shock 70 kDa protein 1- like | AAACTGGATCGAAGGCGGC | 53 |
| GCTGCAGCCATGATTTTCCT | ||
| Platelet/endothelial cell adhesion molecule | GCTGACCCTTCTGCTCTGTT | 116 |
| GTGTCAGGTTCTCCCCGTTT | ||
| Pellino homolog 2 | CCCAATAAGGAGCCCGTGAA | 136 |
| TGGGTTTGACTCCGTTAGCC | ||
| β-actina | ACTTGCGCAGAAAACGAGAT | 84 |
| CACCTTCACCGTTCCAGTTT |
Statistical analysis
Normalization and statistical analysis of the microarray data were performed using the GeneSpring software (Agilent Technologies, CA, USA). Analysis of variance (ANOVA) and the Student's t-test were performed with significance level of P<0.05. Analysis of variance (ANOVA) was performed after logarithmic transformation of CFU values and means were compared by the Tukey's Multiple Comparison Test (P<0.05).
Microarray data accession number
The microarray data set has been submitted to the Gene Expression Omnibus database at NCBI (http://www.ncbi.nlm.nih.gov/geo/) and assigned accession number GSE58216.
Results
Internalization of wild type, ΔvirB2 and ΔbtpB Brucella abortus strains by bovine trophoblastic cells
In order to evaluate whether the Brucella strains used in this study had comparable levels of internalization in trophoblastic cells of bovine CAM explants, CFU numbers at 4 h post inoculation followed by 1 h of incubation with gentamicin. There was no difference between the number of internalized wild type and ΔbtpB B. abortus 2308. It has been demonstrated that in infected CAM, B. abortus is found intracellularly in trophoblasts [13]. In contrast, the number of internalized ΔvirB2 mutant B. abortus was significantly lower than the other two strains (Figure 1).
Figure 1. Internalization of wild type, ΔvirB2 or ΔbtpB Brucella abortus by bovine trophoblastic cells.
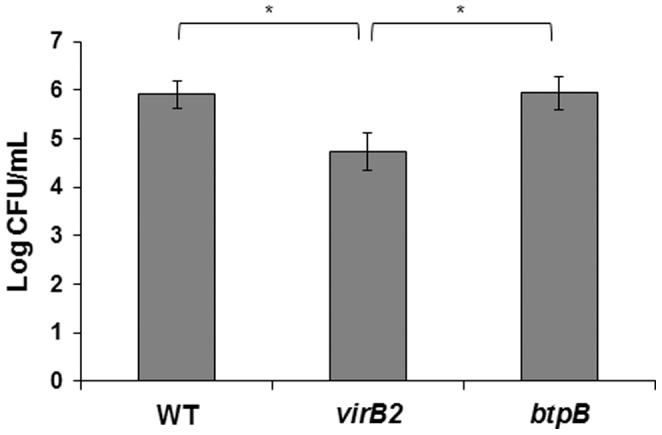
Chorioallantoic membrane (CAM) explants were inoculated, incubated for 4 h, followed by 1 h incubation with gentamicin, and then lysed for intracellular CFU counting. Data represents the average log of CFU numbers from CAM explants, from three independent experiments performed in triplicates. Data underwent logarithmic transformation followed by analysis of variance (ANOVA) and the Tukey's multiple comparison test with significance level of P<0.05.
Transcription profile of bovine trophoblastic cells during infection with wild type, ΔvirB2 or ΔbtpB Brucella abortus 2308 strains
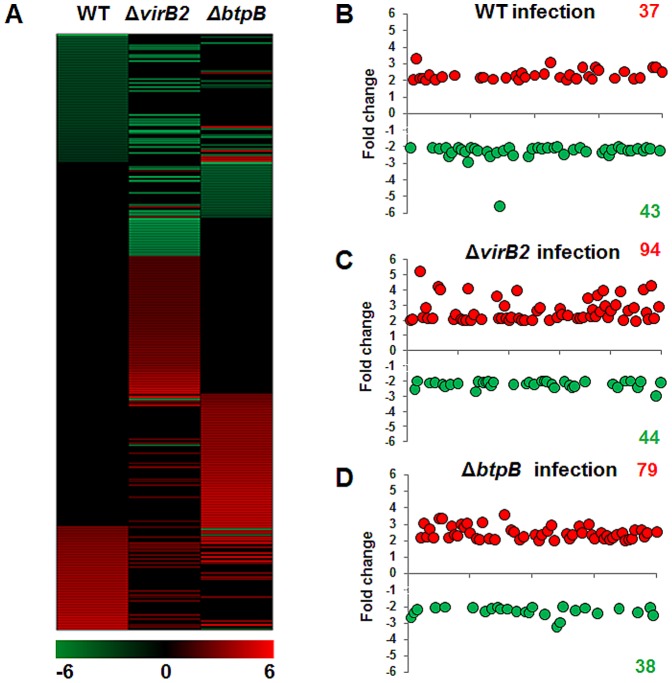
Considering that B. abortus modulates the innate immune response of bovine trophoblastic cells [13], and that TIR domain-containing Brucella proteins, such as BtpB have been shown to impair the host innate immune response [19], whereas the virB-encoded T4SS is required for Brucella survival within host cells [26], a comparison of the transcription profile of bovine trophoblastic cells infected with wild type B. abortus, or the isogenic ΔvirB2 or ΔbtpB strains was performed in this study. A heat map was generated to analyze transcripts with >2 fold change that had statistically significant differences in expression (P<0.05) (Figure 2). Infection of bovine trophoblastic cells with wild type B. abortus 2308 resulted in 80 transcripts with differential levels of expression (i.e. at least a 2-fold change). Among those transcripts, 37 were upregulated and 43 were downregulated. In contrast, infection with ΔvirB2 or ΔbtpB B. abortus mutant strains resulted in a higher number of differentially expressed transcripts (138 and 117, respectively). While the number of downregulated genes was similar between wild type B. abortus and the mutant strains, remarkably, we observed an increased number of upregulated genes in CAM explants infected with both the ΔvirB2 and the ΔbtpB mutants (Figure 2 and 3, Table 2).
Figure 2. Gene transcription profiling of the host response to B. abortus strains at 4 hours after infection of bovine trophoblastic cells.
(A) A heat map of gene transcription changes in bovine trophoblastic cells infected with wild type, ΔvirB2 and ΔbtpB B. abortus– strains, compared to mock-infected controls. (B) Fold changes in gene transcription for genes that were significantly (P<0.05) up or downregulated during wild type, ΔvirB2 and ΔbtpB B. abortus infection compared to mock-infected controls. These data represent results from pools of total RNA obtained from triplicates of CAM explants obtained from four independent experiments. Increase or decrease in mRNA levels are indicated in red or green, respectively.
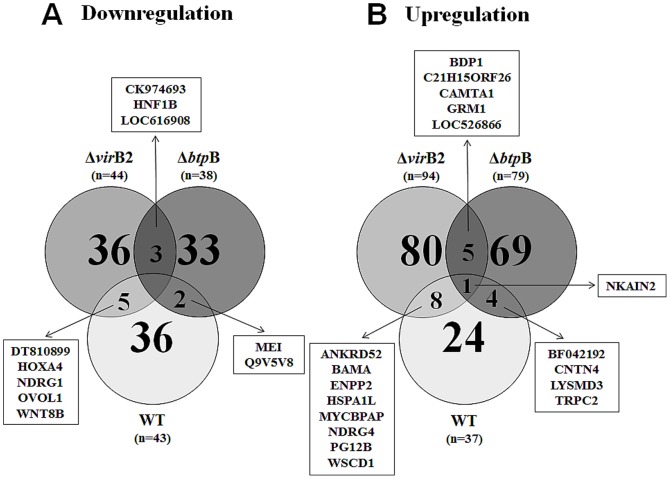
Figure 3. Venn diagram indicating the number of genes with significant changes in mRNA levels assessed by microarray analysis in bovine trophoblastic cells from CAM explants obtained from 4 placentas at the last trimester of pregnancy (n = 4) infected with wild type, ΔvirB2 and ΔbtpB B. abortus compared to mock-infected controls.
Changes in transcription higher than 2-fold and values of P<0.05 were considered significant. (A) Downregulated genes and (B) upregulated genes. Abbreviations: 4105740 BARC 9BOV cDNA clone 9BOV30_O11 5′ (CK974693), HNF1 homeobox B (HNF1B), Hypothetical protein LOC616908 (LOC616908), LB01613.CR_H15 GC_BGC-16 cDNA clone IMAGE:8082593 (DT810899), Homeobox A4 (HOXA4), N-myc downstream regulated 1 (NDRG1), Ovo-like 1 (Drosophila) (OVOL1), PREDICTED: wingless-type MMTV integration site family, member 8B (WNT8B), Hypothetical LOC516011 (MEI), Q9V5V8_DROME CG13214-PA, isoform A (Q9V5V8), BDP1 protein Fragment (BDP1), Chromosome 15 open reading frame 26 ortholog (C21H15ORF26), Calmodulin binding transcription activator 1 (CAMTA1), GRM1 protein Fragment (GRM1), hCG1653800-like (LOC526866), Na+/K+ transporting ATPase interacting 2 (NKAIN2), Ankyrin repeat domain 52 (ANKRD52), 001128BAMA005012HT BAMA cDNA (BAMA), Ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2), Heat shock 70 kDa protein 1-like (HSPA1L), Hypothetical LOC505551 (MYCBPAP), NDRG family member 4 (NDRG4), PG12B_HUMAN (Q9BX93) Group XIIB secretory phospholipase A2-like protein precursor [TC318659] (PG12B), WSC domain containing 1 (WSCD1), BP250013A20E1 Soares normalized bovine placenta cDNA (BF042192), Contactin 4, isoform c precursor (CNTN4), LysM, putative peptidoglycan-binding, domain containing 3 (LYSMD3), Transient receptor potential channel 2 (TRPC2).
Table 2. Genes with significant decrease in transcription level in bovine trophoblastic cells (chorioallantoic membrane explants) infected with wild type (strain 2308), ΔvirB2, or ΔbtpB in comparison to uninfected controls at 4 hours post infection.
| Function* and GenBank identification | Strain** | Fold Change | P value |
| Cell biogenesis | |||
| NOMO3-like protein Fragment [ENSBTAT00000046252] | ΔbtpB | 2.180 | 0.0420 |
| tubulin polymerization promoting protein [NM_173976] | ΔbtpB | 2.353 | 0.0004 |
| LMNB2_HUMAN (Q03252) Lamin-B2 [TC345309] | ΔvirB2 | 2.338 | 0.0467 |
| microfibrillar associated protein 5 [NM_174386] | ΔvirB2 | 2.651 | 0.0089 |
| keratin 10 [NM_174377] | WT | 2.554 | 0.0319 |
| Cell cycle/DNA processing | |||
| AT rich interactive domain 1A, transcript variant 1 [XM_592084] | ΔbtpB | 2.361 | 0.0151 |
| hypothetical LOC516011 [XM_594137] | ΔbtpB | 2.031 | 0.0048 |
| nuclear receptor subfamily 2, group E, member 3 [NM_001167900] | ΔbtpB | 3.237 | 0.0254 |
| SWI/SNF related, matrix associated [NM_001172224] | ΔbtpB | 2.116 | 0.0012 |
| centrosomal protein 97 kDa [NM_001192424] | ΔvirB2 | 2.201 | 0.0321 |
| ephrin-A5 [NM_001076432] | ΔvirB2 | 2.205 | 0.0113 |
| hepatocyte nuclear factor 4, alpha [NM_001015557] | ΔvirB2 | 2.367 | 0.0496 |
| misc_RNA (ETV4), miscRNA [ENSBTAT00000010973] | ΔvirB2 | 2.057 | 0.0492 |
| ovo-like 1 [NM_001081521] | ΔvirB2 | 2.112 | 0.0196 |
| polymerase I and transcript release factor [NM_001081731] | ΔvirB2 | 2.908 | 0.0010 |
| zinc finger, ZZ-type with EF-hand domain 1 [XM_864249] | ΔvirB2 | 2.435 | 0.0063 |
| hypothetical LOC516011 [XM_594137] | WT | 2.109 | 0.0476 |
| ovo-like 1 [NM_001081521] | WT | 2.082 | 0.0145 |
| Q6P1W9_HUMAN Histone deacetylase 7 A protein [TC363092] | WT | 2.041 | 0.0057 |
| Cell fate | |||
| triple functional domain (PTPRF interacting) [ENSBTAT00000007252] | ΔvirB2 | 2.306 | 0.0159 |
| Defense and inflammation | |||
| CD200 molecule [NM_001034620] | ΔbtpB | 2.073 | 0.0250 |
| platelet/endothelial cell adhesion molecule [NM_174571] | ΔbtpB | 2.240 | 0.0460 |
| N-myc downstream regulated 1 [NM_001035009] | ΔvirB2 | 2.189 | 0.0192 |
| polymeric immunoglobulin receptor [NM_174143] | ΔvirB2 | 2.211 | 0.0280 |
| chemokine (C-X-C motif) ligand 12 mRNA [NM_001113174] | WT | 5.527 | 0.0436 |
| N-myc downstream regulated 1 [NM_001035009] | WT | 2.222 | 0.0349 |
| pellino homolog 2 [XM_612354] | WT | 2.040 | 0.0366 |
| serine peptidase inhibitor. Kazal type 5 [NM_001102102] | WT | 2.118 | 0.0015 |
| TRAF2 and NCK interacting kinase [ENSBTAT00000015600] | WT | 2.211 | 0.0097 |
| Signal transduction | |||
| oculocerebrorenal syndrome of [NM_001102191] | ΔbtpB | 2.956 | 0.0060 |
| odorant receptor MOR10-like [XM_591864] | ΔbtpB | 4.476 | 0.0062 |
| neurogranin (protein kinase C substrate. RC3) [NM_001113313] | ΔvirB2 | 2.414 | 0.0222 |
| olfactory receptor Olfr399-like [XM_002695734] | ΔvirB2 | 2.214 | 0.0109 |
| olfactory receptor. family 13. subfamily C. member 3-like [XM_001256440] | ΔvirB2 | 2.003 | 0.0077 |
| wingless-type MMTV integration site family. member 8B [XM_582222] | ΔvirB2 | 2.017 | 0.0492 |
| olfactory receptor Olr136-like [XM_002693219] | WT | 2.106 | 0.0471 |
| olfactory receptor Olr1654-like [XM_001255032] | WT | 2.001 | 0.0205 |
| olfactory receptor Olr1654 [XM_001255418] | WT | 2.025 | 0.0407 |
| olfactory receptor. family 4. subfamily K. member 13-like [XM_001255280] | WT | 2.048 | 0.0460 |
| phosphodiesterase 7A [ENSBTAT00000015427] | WT | 2.025 | 0.0092 |
| wingless-type MMTV integration site family. member 5B [XM_584724] | WT | 2.434 | 0.0412 |
| wingless-type MMTV integration site family. member 8B [XM_582222] | WT | 2.283 | 0.0412 |
| Transport | |||
| 1254442 MARC 7BOV cDNA 5′ [DN516021] | ΔbtpB | 2.659 | 0.0189 |
| ELMO/CED-12 domain containing 1 [NM_001078108] | ΔbtpB | 2.135 | 0.0376 |
| ArfGAP with SH3 domain. ankyrin repeat and PH domain 2 [ENSBTAT00000003007] | ΔvirB2 | 2.003 | 0.0184 |
| sodium channel. voltage-gated. type V. alpha subunit [NM_174458] | ΔvirB2 | 2.216 | 0.0095 |
| syntaxin-1B (Syntaxin-1B2) (Synaptocanalin I) [ENSBTAT00000044107] | ΔvirB2 | 2.020 | 0.0409 |
| discs. large homolog [NM_001191307] | WT | 2.132 | 0.0067 |
| N-ethylmaleimide-sensitive factor attachment protein. beta [NM_001046233] | WT | 2.122 | 0.0228 |
| potassium voltage-gated channel. shaker-related subfamily. beta member 1 [NM_001025336] | WT | 2.870 | 0.0002 |
| RAB6A. member RAS oncogene family (RAB6A). mRNA [NM_001193115] | WT | 2.477 | 0.0284 |
| Systemic development | |||
| Ellis van Creveld syndrome 2 [NM_173927] | ΔbtpB | 2.030 | 0.0344 |
| HNF1 homeobox B [NM_001192855] | ΔbtpB | 2.286 | 0.0035 |
| HUMHRX {Homo sapiens} (exp = −1; wgp = 0; cg = 0) [TC345413] | ΔbtpB | 2.170 | 0.0209 |
| myeloid/lymphoid or mixed-lineage leukemia [NM_001192549] | ΔbtpB | 2.464 | 0.0004 |
| HNF1 homeobox [NM_001192855] | ΔvirB2 | 2.426 | 0.0167 |
| homeobox A4 [NM_001076134] | ΔvirB2 | 2.315 | 8 E-05 |
| mitofusin 2 (MFN2). transcript variant 1 [NM_001190269] | ΔvirB2 | 2.371 | 0.0020 |
| myeloid/lymphoid or mixed-lineage leukemia [NM_001192025] | ΔvirB2 | 2.031 | 0.0124 |
| homeobox A4 [NM_001076134] | WT | 2.005 | 0.0010 |
| Interaction with environment | |||
| contactin-4-like [XM_600040] | ΔbtpB | 2.485 | 0.0004 |
| extracellular matrix protein 2. female organ and adipocyte specific [NM_001034597] | ΔbtpB | 2.150 | 0.0377 |
| neurocan (NCAN). mRNA [NM_001193082] | ΔvirB2 | 2.201 | 0.0117 |
| Neuronal cell adhesion molecule Fragment [ENSBTAT00000008864] | ΔvirB2 | 2.047 | 0.0035 |
| binder of sperm 1[NM_001001145] | WT | 2.185 | 0.0221 |
| neuroligin 2 [NM_001191242] | WT | 2.001 | 0.0026 |
| SCO-spondin homolog mRNA [NM_174706] | WT | 2.143 | 0.0118 |
| Metabolism | |||
| 4-hydroxyphenylpyruvate dioxygenase-like [NM_001099371] | ΔbtpB | 2.038 | 0.0057 |
| acyl-CoA dehydrogenase. long chain (ACADL) [NM_001076936] | ΔbtpB | 2.022 | 0.0375 |
| inter-alpha (globulin) inhibitor H3 [NM_001101898] | ΔvirB2 | 2.112 | 0.0272 |
| phospholipase A2. group [NM_001193052] | ΔvirB2 | 2.127 | 0.0342 |
| pipecolic acid oxidase [NM_001014878] | ΔvirB2 | 2.206 | 0.0239 |
| steryl-sulfatase-like [XM_001789191] | ΔvirB2 | 2.039 | 0.0008 |
| adenosine deaminase. RNA-specific [ENSBTAT00000009896] | WT | 2.342 | 0.0200 |
| ADP-ribosyltransferase 5 [NM_001076515] | WT | 2.204 | 0.0048 |
| phosphoinositide-3-kinase. catalytic. delta polypeptide [XM_580673] | WT | 2.074 | 0.0283 |
| retinol dehydrogenase 12 (all-trans/9-cis/11-cis) [NM_183363] | WT | 2.189 | 0.0146 |
| Protein fate | |||
| leucine rich repeat containing 41 (LRRC41) [NM_001045873] | ΔbtpB | 2.372 | 0.0161 |
| peptidylprolyl isomerase (cyclophilin)-like 2 [NM_001038081] | ΔbtpB | 2.014 | 0.0093 |
| ribosomal protein S6 kinase. 90 kDa. polypeptide 4 [NM_001191400] | ΔbtpB | 2.390 | 0.0029 |
| F-box protein 32 [NM_001046155] | ΔvirB2 | 2.341 | 0.0159 |
| Protein synthesis | |||
| eukaryotic elongation factor-2 kinase (EEF2K). mRNA [NM_001192542] | ΔvirB2 | 2.056 | 0.0075 |
| alanyl-tRNA synthetase 2. mitochondrial (putative) [NM_001191211] | WT | 2.066 | 0.0296 |
| Binding function | |||
| EH domain binding protein 1-like 1 [NM_001191243] | ΔbtpB | 2.120 | 0.0095 |
| PR domain containing 13 [NM_001193010] | ΔbtpB | 2.078 | 0.0181 |
| suppressor of sable-like [XM_582657] | ΔbtpB | 2.582 | 0.0487 |
| syndecan binding protein (syntenin) 2 [ENSBTAT00000039310] | ΔvirB2 | 2.171 | 0.0358 |
| Regulation of metabolism | |||
| cystatin SC [NM_001038122] | ΔbtpB | 2.276 | 0.0247 |
| Serine protease inhibitor Kazal-type 6 Precursor [ENSBTAT00000047606] | ΔbtpB | 2.212 | 0.0325 |
| dedicator of cytokinesis 4 [XM_613918] | ΔvirB2 | 2.006 | 0.0180 |
| LB00228.CR_H15 GC_BGC-02 cDNA clone IMAGE:7950401 [DT823428] | WT | 2.067 | 0.0031 |
| Transcription | |||
| Q5TAY6_HUMAN (Q5TAY6) Zinc finger protein 31 [TC342188] | ΔbtpB | 2.108 | 0.0298 |
| zinc finger and BTB domain containing 9 [NM_001191462] | ΔbtpB | 2.045 | 0.0121 |
| zinc finger protein 296 [NM_001192262] | ΔbtpB | 2.538 | 0.0158 |
| zinc finger protein 30 homolog [XR_083922] | ΔbtpB | 2.295 | 0.0294 |
| LB02718.CR_O08 GC_BGC-27 cDNA clone IMAGE:8313058 [DV891009] | ΔvirB2 | 2.998 | 0.0025 |
| FtsJ methyltransferase domain containing 2 [NM_001082430] | WT | 2.043 | 0.0280 |
| hypothetical LOC618701 [NM_001099723] | WT | 2.316 | 0.0041 |
| lin-28 homolog B [XM_612469] | WT | 2.024 | 0.0152 |
| sterol regulatory element Binding Protein family member (sbp-1)-like [XM_583656] | WT | 2.087 | 0.0268 |
| zinc finger protein 213 [ENSBTAT00000037526] | WT | 2.506 | 0.0012 |
| zinc finger protein 42. transcript variant 7 [XM_872583] | WT | 2.302 | 0.0396 |
| Unclassified | |||
| 4105740 BARC 9BOV cDNA clone 9BOV30_O11 5′ [CK974693] | ΔbtpB | 2.381 | 0.0330 |
| 4121344 BARC 8BOV cDNA clone 8BOV_34C06 5′ [CN787257] | ΔbtpB | 2.189 | 0.0149 |
| chromosome 16 open reading frame 7 [XM_592406] | ΔbtpB | 2.099 | 0.0197 |
| hypothetical protein LOC616908 [NM_001076437] | ΔbtpB | 2.311 | 0.0208 |
| LRRN4 C-terminal like [NM_001195069] | ΔbtpB | 2.024 | 0.0208 |
| Q9V5V8_DROME (Q9V5V8) CG13214-PA [TC333751] | ΔbtpB | 2.704 | 0.0059 |
| 1429623 MARC 7BOV cDNA 3′ [DR113203] | ΔvirB2 | 2.193 | 0.0128 |
| 4105740 BARC 9BOV cDNA clone 9BOV30_O11 5′ [CK974693] | ΔvirB2 | 2.706 | 0.0170 |
| AL450489 match: proteins: Q8VCL4 Q9CUK0 Q9Y4E5 [TC317712] | ΔvirB2 | 2.001 | 0.0179 |
| family with sequence similarity 3. member D. transcript variant 2 [XM_865037] | ΔvirB2 | 2.144 | 0.0251 |
| hypothetical protein LOC616908 [NM_001076437] | ΔvirB2 | 2.558 | 0.0058 |
| LB01613.CR_H15 GC_BGC-16 cDNA clone IMAGE:8082593 [DT810899] | ΔvirB2 | 2.035 | 0.0297 |
| LB01652.CR_A20 GC_BGC-16 cDNA clone IMAGE:8385118 5′ [EH174578] | ΔvirB2 | 2.094 | 0.0009 |
| misc_RNA (LOC785410). miscRNA [ENSBTAT00000025153] | ΔvirB2 | 2.041 | 0.0060 |
| similar to Myeloid-associated differentiation marker [NM_001101279] | ΔvirB2 | 2.177 | 0.0396 |
| chromosome 10 open reading frame 120 ortholog [NM_001076476] | WT | 2.237 | 0.0331 |
| hypothetical LOC619120 [XM_871440] | WT | 2.208 | 0.0093 |
| hypothetical LOC785309 [XM_001253376] | WT | 2.028 | 0.0256 |
| LB01613.CR_H15 GC_BGC-16 cDNA clone IMAGE:8082593 [DT810899] | WT | 2.243 | 0.0250 |
| MIPOL1 protein [ENSBTAT00000000857] | WT | 2.571 | 0.0281 |
| misc_RNA (LOC524074). miscRNA [ENSBTAT00000000329] | WT | 2.040 | 0.0279 |
| Q9V5V8_DROME (Q9V5V8) CG13214-PA. isoform A [TC333751] | WT | 2.541 | 0.0239 |
*Functional classification generated by Funcat (http://mips.helmholtz-muenchen.de/genre/proj/mfungd/Search/Catalogs/searchCatfirstFun.html);
**WT = wild type Brucella abortus 2308.
These results indicated that downregulation of host trophoblastic cell transcripts at early stages of infection is directly or indirectly associated with the virB Type IV secretion system and BtpB.
Wild type B. abortus impairs host transcription of genes related to immune response when compared to ΔvirB and ΔbtpB mutants
Differentially expressed transcripts of trophoblastic cells in response to infection with wild type B. abortus or the ΔvirB2 and ΔbtpB mutant strains were functionally classified (Tables 2 and 3). Explants infected with wild type B. abortus had the highest number of downregulated genes that are related to signal transduction (7/43; 16.2%), transcription (6/43; 13.9%), and defense and inflammation (5/43; 11.6%). Upregulated transcripts were related to metabolism (6/37; 16.2%), transport (5/37; 16.2%), and signal transduction (4/37; 10.8%). Explants infected with the ΔvirB2 mutant strain had downregulated transcripts mostly in the following categories: cell cycle/DNA processing (7/44; 15.9%), metabolism (4/44; 9.1%), and signal transduction (4/44; 9.1%), and upregulated genes related to defense and inflammation (17/94; 18.0%), metabolism (14/94; 14.8%), and signal transduction (8/94; 8.5%). Explants infected with the ΔbtpB strain had larger numbers of downregulated transcripts associated with cell cycle/DNA processing (4/38; 10.5%), transcription (4/38; 10.5%), and systemic development (4/38; 10.5%). Genes upregulated were classified as transport-related (10/79; 12.6%), signal transduction (9/79; 11.3%), and cell cycle/DNA processing (7/79; 8.8%) (Figure 4, Table 3). Interestingly, while in wild type B. abortus-infected explants most of the differentially expressed transcripts that were classified as associated with defense and inflammation were downregulated (5/7; 71.4%), explants infected with either the ΔvirB2 or ΔbtpB strains had mostly upregulated transcripts in that category (17/19; 89.4%), supporting the idea that infection by B. abortus 2308 causes a downregulation of the immune response at early stages of infection in order to prevent a robust inflammatory response. Therefore, deletion of the virulence genes virB2 and btpB results in strains that trigger increased transcription of several genes related to the immune response.
Table 3. Genes with significant increase in transcription level in bovine trophoblastic cells (chorioallantoic membrane explants) infected with wild type (strain 2308), ΔvirB2, or ΔbtpB in comparison to uninfected controls at 4 hours post infection.
| Function* and GenBank identification | B. abortus infection** | Fold Change | P value |
| Cell biogenesis | |||
| actin. gamma 2. smooth muscle. enteric [NM_001013592] | ΔbtpB | 3.388 | 0.0139 |
| calponin 1. basic. smooth muscle [NM_001046379] | ΔbtpB | 3.047 | 0.0061 |
| LysM. putative peptidoglycan-binding. domain containing 3 [NM_001192982] | ΔbtpB | 2.404 | 0.0076 |
| myosin. heavy chain 11. smooth muscle [NM_001102127] | ΔbtpB | 2.986 | 0.0016 |
| tropomyosin 1 (alpha) [NM_001013590] | ΔbtpB | 2.048 | 0.0243 |
| tropomyosin 2 (beta) [NM_001010995] | ΔbtpB | 2.710 | 0.0047 |
| keratin 6A [NM_001083510] | ΔvirB2 | 3.474 | 0.0202 |
| matrilin 3 [XM_591137] | ΔvirB2 | 5.058 | 6 E-05 |
| myosin light chain 2a-like (LOC789339) [XM_001256122] | ΔvirB2 | 2.817 | 0.0170 |
| LysM. putative peptidoglycan-binding. domain containing 3 [NM_001192982] | WT | 2.204 | 0.0357 |
| Cell cycle/DNA processing | |||
| esophageal cancer related gene 4 protein [NM_001038113] | ΔbtpB | 3.599 | 0.0450 |
| establishment of cohesion 1 homolog 2-like (LOC786089 [XM_001253877] | ΔbtpB | 2.120 | 0.0201 |
| HUMRECQ DNA helicase [TC350379] | ΔbtpB | 2.148 | 0.0214 |
| NDC80 homolog. kinetochore complex component [XM_582722] | ΔbtpB | 2.182 | 0.0184 |
| PC4 and SFRS1-interacting protein [ENSBTAT00000036721] | ΔbtpB | 2.024 | 0.0350 |
| polycomb group ring finger 5 [NM_001077980] | ΔbtpB | 2.910 | 0.0040 |
| SRY (sex determining region Y)-box 7 [XM_615317] | ΔbtpB | 2.122 | 0.0205 |
| cAMP responsive element modulator [NM_001034710] | ΔvirB2 | 2.059 | 0.0380 |
| histone cluster 1. H1d (HIST1H1D) [NM_001101066] | ΔvirB2 | 2.292 | 0.0155 |
| nuclear receptor coactivator 7 [ENSBTAT00000049978] | ΔvirB2 | 2.345 | 0.0433 |
| centromere-binding factor 5 like PUA domain containing protein with a type I pseudouridine synthase domain [TC372398] | ΔvirB2 | 6.112 | 0.0379 |
| SAMD9_HUMAN Sterile alpha motif domain-containing protein 9 [TC347659] | ΔvirB2 | 2.119 | 0.0026 |
| T-box 19 mRNA [NM_001075663] | ΔvirB2 | 2.135 | 0.0244 |
| Defense and inflammation | |||
| CCAAT/enhancer binding protein (C/EBP). epsilon [NM_001192808] | ΔbtpB | 2.494 | 0.0084 |
| complement factor H (CFH) [NM_001033936] | ΔbtpB | 3.172 | 0.0291 |
| tumor necrosis factor (ligand) superfamily. member 10-like [XM_583785] | ΔbtpB | 3.290 | 0.0267 |
| tumor necrosis factor receptor superfamily. member 13C [NM_001193192] | ΔbtpB | 2.692 | 0.0317 |
| apolipoprotein L. 3 [NM_001100351] | ΔvirB2 | 3.191 | 0.0168 |
| chemokine (C-C motif) ligand 5 mRNA [NM_175827] | ΔvirB2 | 2.121 | 0.0331 |
| chemokine (C-C motif) receptor-like 2 [NM_001075732] | ΔvirB2 | 2.165 | 0.0295 |
| chemokine (C-X-C motif) receptor 5 [NM_001011675] | ΔvirB2 | 2.076 | 0.0285 |
| chemokine binding protein 2 [NM_001015581] | ΔvirB2 | 2.068 | 0.0105 |
| complement component 3a receptor 1 [NM_001083752] | ΔvirB2 | 2.851 | 0.0072 |
| Fc fragment of IgG binding protein [XM_614095] | ΔvirB2 | 2.072 | 0.0406 |
| fms-related tyrosine kinase 3-like [XM_590263] | ΔvirB2 | 2.652 | 0.0419 |
| heat shock 70 kDa protein 1-like [NM_001167895] | ΔvirB2 | 2.150 | 0.0015 |
| interleukin 1 family. member 6 (epsilon)-like (IL1F6) [XM_601728] | ΔvirB2 | 4.025 | 0.0148 |
| interleukin 15 [NM_174090] | ΔvirB2 | 5.250 | 0.0004 |
| interleukin 2 receptor. alpha [NM_174358] | ΔvirB2 | 2.669 | 0.0156 |
| MPV17 mitochondrial membrane protein-like [NM_001046602] | ΔvirB2 | 2.134 | 0.0018 |
| radical S-adenosyl methionine domain containing 2 [NM_001045941] | ΔvirB2 | 2.824 | 0.0480 |
| rCG28728-like (LOC533818) Protein Gbp4 [XM_001789771] | ΔvirB2 | 2.302 | 0.0076 |
| transmembrane 4 L six family member 19 [ENSBTAT00000057570] | ΔvirB2 | 4.074 | 0.0023 |
| tumor necrosis factor receptor superfamily. member 9 [NM_001035336] | ΔvirB2 | 4.246 | 0.0294 |
| heat shock 70 kDa protein 1-like [NM_001167895] | WT | 2.017 | 0.0055 |
| toll-like receptor 6 [NM_001001159] | WT | 2.111 | 0.0407 |
| Cell differentiation | |||
| neuropilin 2 [NM_001193237] | ΔvirB2 | 2.681 | 0.0464 |
| Signal transduction | |||
| A kinase (PRKA) anchor protein 12 [XM_591518] | ΔbtpB | 2.152 | 0.0478 |
| ADAM metallopeptidase with thrombospondin type 1 motif. 6 [NM_001193016] | ΔbtpB | 2.194 | 0.0198 |
| GRM1 protein Fragment [ENSBTAT00000057021] | ΔbtpB | 2.779 | 0.0282 |
| olfactory receptor Olr1242-like (LOC788607) [XM_001255616] | ΔbtpB | 2.001 | 0.0184 |
| olfactory receptor. family 11. subfamily G. member 2-like [XM_002690673] | ΔbtpB | 2.122 | 0.0021 |
| olfactory receptor. family 2. subfamily A. member 4-like [XM_001790464] | ΔbtpB | 2.813 | 0.0035 |
| Q2ABB1_BOVIN (Q2ABB1) Bitter taste receptor. partial (12%) [TC320439] | ΔbtpB | 2.066 | 0.0299 |
| Ras-related associated with diabetes [NM_001045913] | ΔbtpB | 2.394 | 0.0208 |
| spermatogenesis associated 7 [NM_001098928] | ΔbtpB | 2.411 | 0.0332 |
| brain and acute leukemia. cytoplasmic [NM_001083508] | ΔvirB2 | 2.854 | 0.0281 |
| cerebellin 3 precursor [NM_001079603] | ΔvirB2 | 2.967 | 0.0233 |
| draxin [NM_001195012] | ΔvirB2 | 2.111 | 0.0437 |
| EDAR-associated death domain [ENSBTAT00000061420] | ΔvirB2 | 4.127 | 0.0499 |
| G protein-coupled receptor 115 [NM_001143875] | ΔvirB2 | 3.983 | 0.0298 |
| GRM1 protein Fragment [ENSBTAT00000057021] | ΔvirB2 | 2.180 | 0.0058 |
| prostaglandin E receptor 2 (subtype EP2) [NM_174588] | ΔvirB2 | 2.055 | 0.0338 |
| thromboxane A2 receptor [NM_001167919] | ΔvirB2 | 2.225 | 0.0365 |
| olfactory receptor. family 5. subfamily I. member 1-like [XM_002693698] | WT | 2.388 | 0.0441 |
| olfactory receptor. family 8. subfamily A. member 1-like [XM_581848] | WT | 2.131 | 0.0329 |
| PIGLHHCGB luteinizing hormone receptor precursor variant B [TC374819] | WT | 2.041 | 0.0479 |
| similar to obscurin. cytoskeletal calmodulin and titin-interacting RhoGEF [NM_001102196] | WT | 2.094 | 0.0228 |
| Transport | |||
| BC015727 solute carrier organic anion transporter family member 4A1 [TC352480] | ΔbtpB | 2.283 | 0.0079 |
| ceruloplasmin-like [XM_002685007] | ΔbtpB | 2.025 | 0.0133 |
| cytochrome P450. family 4. subfamily V. polypeptide 2 [NM_001034373] | ΔbtpB | 2.185 | 0.0105 |
| hephaestin-like [XM_587920] | ΔbtpB | 2.225 | 0.0456 |
| Na+/K+ transporting ATPase interacting 2 [NM_001102345] | ΔbtpB | 2.037 | 0.0496 |
| phosphatidylinositol binding clathrin assembly protein [NM_001101977] | ΔbtpB | 2.132 | 0.0410 |
| potassium inwardly-rectifying channel. subfamily J. member 10 [NM_001081601] | ΔbtpB | 2.526 | 0.0063 |
| RAB GTPase activating protein 1-like [NM_001103263] | ΔbtpB | 3.033 | 0.0415 |
| reticulon 1 [ENSBTAT00000015218] | ΔbtpB | 2.264 | 0.0315 |
| ryanodine receptor 3 [XM_590220] | ΔbtpB | 3.020 | 0.0285 |
| AB051866 RIM long form [TC358572] | ΔvirB2 | 2.041 | 0.0086 |
| ATPase. H+ transporting V0 subunit e2 [NM_001097574] | ΔvirB2 | 2.007 | 0.0134 |
| Na+/K+ transporting ATPase interacting 2 [NM_001102345] | ΔvirB2 | 2.137 | 0.0129 |
| potassium inwardly-rectifying channel. subfamily J. member 13 [NM_001193254] | ΔvirB2 | 2.176 | 0.0023 |
| potassium voltage-gated channel. subfamily H (eag-related) [NM_001191394] | ΔvirB2 | 2.429 | 0.0015 |
| small calcium-binding mitochondrial carrier 1-like [XM_609165] | ΔvirB2 | 2.154 | 0.0417 |
| solute carrier organic anion transporter family [XM_002689417] | ΔvirB2 | 2.026 | 0.0265 |
| ATPase. Ca++ transporting. plasma membrane 2 [NM_001191245] | WT | 2.251 | 0.0153 |
| component of oligomeric golgi complex 5 [BC149439] | WT | 2.143 | 0.0081 |
| Na+/K+ transporting ATPase interacting 2 (NKAIN2). mRNA [NM_001102345] | WT | 3.087 | 0.0001 |
| sodium-dependent noradrenaline transporter [ENSBTAT00000046433] | WT | 2.188 | 0.0049 |
| unc-13 homolog C [XM_001249446] | WT | 2.520 | 0.0306 |
| Systemic development | |||
| shisa homolog 2 [NM_001101265] | ΔbtpB | 2.197 | 0.0304 |
| NDRG family member 4 [NM_001075695] | ΔvirB2 | 2.054 | 0.0350 |
| Q69Z70_MOUSE (Q69Z70) MKIAA1910 protein (Fragment) [TC349493] | ΔvirB2 | 2.288 | 0.0273 |
| NDRG family member 4 [NM_001075695] | WT | 2.533 | 0.0276 |
| Interaction with environment | |||
| BC026119 contactin 4. isoform c precursor [TC348690] | ΔbtpB | 2.747 | 0.0276 |
| SCO-spondin homolog [NM_174706] | ΔbtpB | 2.103 | 0.0497 |
| AF311284 erythroid membrane-associated protein [TC330508] | ΔvirB2 | 2.943 | 0.0056 |
| ectonucleotide pyrophosphatase/phosphodiesterase 2 [NM_001080293] | ΔvirB2 | 2.208 | 0.0225 |
| BC026119 contactin 4. isoform c precursor [TC348690] | WT | 2.777 | 3 E-05 |
| ectonucleotide pyrophosphatase/phosphodiesterase 2 [NM_001080293] | WT | 2.193 | 0.0290 |
| kinesin family member 3B-like. transcript variant 3 [XM_863957] | WT | 2.778 | 0.0242 |
| Metabolism | |||
| Acp1 protein-like [XM_868782] | ΔbtpB | 3.137 | 0.0429 |
| ATPase. Na+/K+ transporting. alpha 2 polypeptide [NM_001081524] | ΔbtpB | 2.419 | 0.0372 |
| glycerol kinase [NM_001075236] | ΔbtpB | 2.55 | 0.0022 |
| mitochondrial carnitine palmitoyltransferase 1A [FJ415874] | ΔbtpB | 2.069 | 0.0432 |
| phospholipid scramblase 4 [NM_001081732] | ΔbtpB | 2.3899958 | 0.0197 |
| transient receptor potential channel 2 [NM_174477] | ΔbtpB | 2.50079 | 0.0329 |
| acyl-CoA synthetase short-chain family member 2 [NM_001105339] | ΔvirB2 | 2.183 | 0.0024 |
| acylglycerol kinase [NM_001098969] | ΔvirB2 | 2.256 | 0.0417 |
| ankyrin repeat domain 52 [NM_001192530] | ΔvirB2 | 2.319 | 0.0195 |
| cDNA clone IMAGE:8166104 [BC114144] | ΔvirB2 | 3.904 | 0.0002 |
| cDNA clone IMAGE:8233560 [BC153862] | ΔvirB2 | 2.413 | 0.0455 |
| cytochrome P450. family 2. subfamily J. polypeptide 2-like [XM_587546] | ΔvirB2 | 2.419 | 0.0104 |
| glycine-N-acyltransferase [NM_177513] | ΔvirB2 | 2.811 | 0.0005 |
| LOC781710 protein Fragment [ENSBTAT00000056121] | ΔvirB2 | 3.699 | 0.0107 |
| nicotinamide nucleotide adenylyltransferase 2 [NM_001075486] | ΔvirB2 | 2.162 | 0.0180 |
| PG12B_HUMAN Group XIIB secretory phospholipase A2-like protein precursor [TC318659] | ΔvirB2 | 2.030 | 0.0275 |
| Q6UWU2_HUMAN (Q6UWU2) APKK229 [TC386487] | ΔvirB2 | 2.204 | 0.0345 |
| UDP glucuronosyltransferase 1 family. polypeptide A1 [NM_001105636] | ΔvirB2 | 3.970 | 0.0026 |
| WNK lysine deficient protein kinase 2 [XM_582977] | ΔvirB2 | 2.059 | 0.0433 |
| WSC domain containing 1 [XM_617236] | ΔvirB2 | 2.211 | 0.0212 |
| ankyrin repeat domain 52 [NM_001192530] | WT | 2.178 | 0.0400 |
| glutathione S-transferase. theta 3-like [XM_001256131] | WT | 2.345 | 0.0440 |
| PG12B_HUMAN Group XIIB secretory phospholipase A2-like protein precursor [TC318659] | WT | 2.260 | 0.0337 |
| polo-like kinase 3 [NM_001075153] | WT | 2.799 | 0.0023 |
| transient receptor potential channel 2 [NM_174477] | WT | 2.067 | 0.0262 |
| WSC domain containing 1 [XM_617236] | WT | 2.024 | 0.0180 |
| Protein fate | |||
| ADAMTS-like 3 [Source:HGNC Symbol;Acc:14633] [ENSBTAT00000006101] | ΔbtpB | 3.114 | 0.0331 |
| carboxypeptidase E [NM_173903] | ΔbtpB | 3.077 | 0.0390 |
| F-box protein 16 [NM_001078119] | ΔbtpB | 2.710 | 0.0311 |
| ubiquitin specific protease 16-like. transcript variant 2 [XM_866110] | ΔbtpB | 2.180 | 0.0066 |
| von Hippel-Lindau tumor supressor [NM_001110019] | ΔbtpB | 2.279 | 0.0439 |
| YOD1 OTU deubiquinating enzyme 1 homolog [NM_001080309] | ΔbtpB | 2.509 | 0.0374 |
| ADAM metallopeptidase with thrombospondin type 1 motif. 3 [NM_001192797] | ΔvirB2 | 2.659 | 0.0124 |
| heat shock 22 kDa protein 8 [NM_001014955] | ΔvirB2 | 2.337 | 0.0349 |
| misc_RNA (LOC613597). miscRNA [ENSBTAT00000019285] | WT | 2.315 | 0.0422 |
| Binding function | |||
| ELAV (embryonic lethal. abnormal vision. Drosophila)-like 4 | ΔbtpB | 2.126 | 0.0388 |
| multiple C2 domains. transmembrane 2 [BC118493] | ΔbtpB | 2.366 | 0.0429 |
| PHD finger protein 14 [BC148049] | ΔbtpB | 2.471 | 0.0011 |
| phosphodiesterase 4D interacting protein [ENSBTAT00000061284] | ΔbtpB | 3.121 | 0.0365 |
| RNA binding motif protein 20 [NM_001192613] | ΔbtpB | 2.526 | 0.0167 |
| RUN and FYVE domain containing 4 [NM_001102081] | ΔbtpB | 2.320 | 0.0400 |
| calcineurin-like phosphoesterase domain containing 1 [NM_001031771] | ΔvirB2 | 2.020 | 0.0497 |
| hypothetical LOC505551 [XM_581851] | ΔvirB2 | 2.433 | 0.0181 |
| ribosomal protein SA-like [XR_083612] | ΔvirB2 | 2.037 | 0.0153 |
| RNA binding motif protein 43 [NM_001099168] | ΔvirB2 | 2.186 | 0.0282 |
| TRIM6-TRIM34 readthrough [NM_001046461] | ΔvirB2 | 2.223 | 0.0417 |
| tripartite motif-containing 55. transcript variant 1 [XM_001789975] | ΔvirB2 | 4.279 | 0.0149 |
| hypothetical LOC505551 [XM_581851] | WT | 2.456 | 0.0446 |
| Regulation of metabolism | |||
| ArfGAP with coiled-coil. ankyrin repeat and PH domains 2 [NM_001105337] | ΔbtpB | 2.909 | 0.0049 |
| RPTOR independent companion of MTOR. complex 2 [NM_001144096] | ΔbtpB | 2.147 | 0.0249 |
| low density lipoprotein receptor-related protein 8. apolipoprotein e receptor [NM_001097565] | ΔvirB2 | 2.630 | 0.0135 |
| regulator of G-protein signaling 16 [NM_174450] | ΔvirB2 | 2.047 | 0.0101 |
| family with sequence similarity 129. member A [NM_001191282] | WT | 2.301 | 0.0063 |
| Transcription | |||
| BDP1 protein Fragment [ENSBTAT00000000471] | ΔbtpB | 2.226 | 0.0048 |
| calmodulin binding transcription activator 1 [NM_001100294] | ΔbtpB | 2.336 | 0.0111 |
| pseudouridylate synthase 7 homolog (S. cerevisiae)-like | ΔbtpB | 2.052 | 0.0460 |
| regulation of nuclear pre-mRNA domain-containing protein 2 (RPRD2) | ΔbtpB | 2.139 | 0.0382 |
| regulatory factor X 7 [NM_001192820] | ΔbtpB | 2.136 | 0.0104 |
| TBP-associated factor 11 [XM_867002] | ΔbtpB | 2.747 | 0.0340 |
| zinc finger protein 667 [NM_001102078] | ΔbtpB | 2.560 | 0.0113 |
| adenosine deaminase. RNA-specific. B2 [NM_001192588] | ΔvirB2 | 2.043 | 0.0150 |
| BDP1 protein Fragment [ENSBTAT00000000471] | ΔvirB2 | 2.320 | 0.0031 |
| calmodulin binding transcription activator 1 [NM_001100294] | ΔvirB2 | 2.064 | 0.0012 |
| EP300 interacting inhibitor of differentiation 3 [NM_001100312] | ΔvirB2 | 2.181 | 0.0017 |
| zinc finger protein 565 [NM_001102216] | ΔvirB2 | 2.141 | 0.0036 |
| POU class 6 homeobox 2 [NM_001102046] | WT | 3.300 | 0.0430 |
| TOX high mobility group box family member 3 [XM_880075] | WT | 2.346 | 0.0435 |
| Elements of external origin | |||
| Q52KE6_MOUSE (Q52KE6) Pgbd5 protein [TC306097] | WT | 2.120 | 0.0246 |
| Unclassified | |||
| A17601A FFB cDNA clone A1760 3′ [EE903511] | ΔbtpB | 2.243 | 0.0274 |
| BP250013A20E1 Soares normalized bovine placenta cDNA [BF042192] | ΔbtpB | 3.374 | 0.0263 |
| chromosome 15 open reading frame 26 ortholog [NM_001075536] | ΔbtpB | 2.145 | 0.0133 |
| chromosome 9 open reading frame 30 ortholog [NM_001076019] | ΔbtpB | 2.111 | 0.0187 |
| hCG1653800-like [XM_002695883] | ΔbtpB | 2.853 | 0.0319 |
| hypothetical protein LOC100125939 [NM_001105502] | ΔbtpB | 2.131 | 0.0483 |
| hypothetical protein LOC534992 [NM_001079608] | ΔbtpB | 2.290 | 0.0212 |
| LB029104.CR.1_D17 GC_BGC-29 cDNA clone IMAGE:8491939 [EE363922] | ΔbtpB | 2.740 | 0.0270 |
| LB03019.CR_A20 GC_BGC-30 cDNA clone IMAGE:8139166 [DV930234] | ΔbtpB | 2.218 | 0.0201 |
| misc_RNA (LOC784747). miscRNA [ENSBTAT00000002161] | ΔbtpB | 2.416 | 0.0075 |
| myeloid-associated differentiation marker-like [XM_608387] | ΔbtpB | 2.139 | 0.0025 |
| Q4SMS2_TETNG (Q4SMS2) Chromosome 8 SCAF14545 [TC378905] | ΔbtpB | 2.117 | 0.0304 |
| trophoblast Kunitz domain protein 5-like [XR_083836] | ΔbtpB | 3.024 | 0.0220 |
| 001128BAMA005012HT BAMA cDNA [DY090836] | ΔvirB2 | 2.889 | 3 E-05 |
| 596479 MARC 6BOV cDNA 3′ [CB423273] | ΔvirB2 | 2.106 | 0.0237 |
| cDNA clone IMAGE:8190996 [BC119997] | ΔvirB2 | 2.108 | 0.0029 |
| chromosome 15 open reading frame 26 ortholog [NM_001075536] | ΔvirB2 | 2.043 | 0.0057 |
| hCG1653800-like [XM_002695883] | ΔvirB2 | 2.780 | 0.0446 |
| Hw_FAT_14_050513_F05 CF-24-HW fat cDNA library cDNA[DV775976] | ΔvirB2 | 2.113 | 0.0218 |
| hypothetical LOC100140997 [XM_001787456] | ΔvirB2 | 2.523 | 0.0305 |
| hypothetical LOC514143 [XM_591946] | ΔvirB2 | 2.760 | 0.0153 |
| hypothetical LOC524181 [XM_002702390] | ΔvirB2 | 2.224 | 0.0180 |
| hypothetical LOC614176 [XM_865557] | ΔvirB2 | 2.209 | 0.0181 |
| LB004140.C21_N19 GC_BGC-04 cDNA clone IMAGE:9059349 3′ [EV693857] | ΔvirB2 | 2.007 | 0.0298 |
| LB02816.CR_B07 GC_BGC-28 cDNA clone IMAGE:8225577 [DV909965] | ΔvirB2 | 2.004 | 0.0107 |
| LB02963.CR_E02 GC_BGC-29 cDNA clone IMAGE:8476204 [EE364260] | ΔvirB2 | 2.854 | 0.0497 |
| misc_RNA (LOC100140452). miscRNA [ENSBTAT00000053419] | ΔvirB2 | 2.851 | 0.0383 |
| P-glycoprotein Fragment [ENSBTAT00000047541] | ΔvirB2 | 3.031 | 0.0386 |
| putative uncharacterized protein FLJ41210 Fragment [ENSBTAT00000010299] | ΔvirB2 | 2.177 | 0.0388 |
| Q61107_MOUSE (Q61107) Purine nucleotide binding protein [TC353960] | ΔvirB2 | 2.383 | 0.0108 |
| similar to LOC129881 protein [XM_868516] | ΔvirB2 | 2.988 | 0.0424 |
| uncharacterized protein ENSP00000334415 homolog [NM_001110092] | ΔvirB2 | 2.042 | 0.0338 |
| 001128BAMA005012HT BAMA cDNA [DY090836] | WT | 2.148 | 0.0013 |
| 693743 MARC 6BOV cDNA 3′ [CB442901] | WT | 2.629 | 0.0263 |
| cDNA clone IMAGE:8031759 [BC149666] | WT | 2.018 | 0.0354 |
| coiled-coil domain containing 54 [NM_001105490] | WT | 2.043 | 0.0249 |
| hypothetical protein LOC785476 [NM_001099194] | WT | 2.152 | 0.0139 |
| Q4SID5_TETNG (Q4SID5) Chromosome 5 SCAF14581 [TC371777] | WT | 2.299 | 0.0367 |
| Q5JS37_HUMAN (Q5JS37) OTTHUMP00000018294 [TC345240] | WT | 2.127 | 0.0460 |
| uMC-bcl_0B02-014-e08 Day 14 CL from a pregnant animal bcl cDNA 3′ [CV976853] | WT | 2.775 | 0.0030 |
*Functional classification generated by Funcat (http://mips.helmholtz-muenchen.de/genre/proj/mfungd/Search/Catalogs/searchCatfirstFun.html);
**WT = wild type Brucella abortus 2308.
Figure 4. Functional classification of genes with significant changes in transcription levels in bovine trophoblastic cells at 4 hours after infection with wild type, ΔvirB2, and ΔbtpB B. abortus.
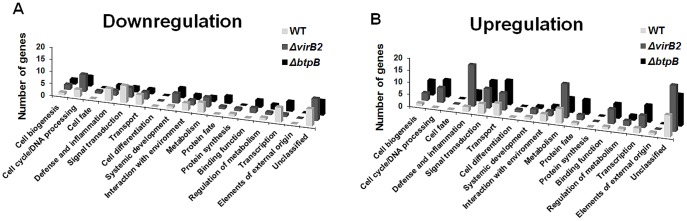
(A) Downregulated genes, and (B) upregulated genes in comparison to mock-infected controls.
Transcription of genes related to immune response during the early stages of infection of bovine trophoblasts with wild type, ΔvirB2 or ΔbtpB B. abortus strains
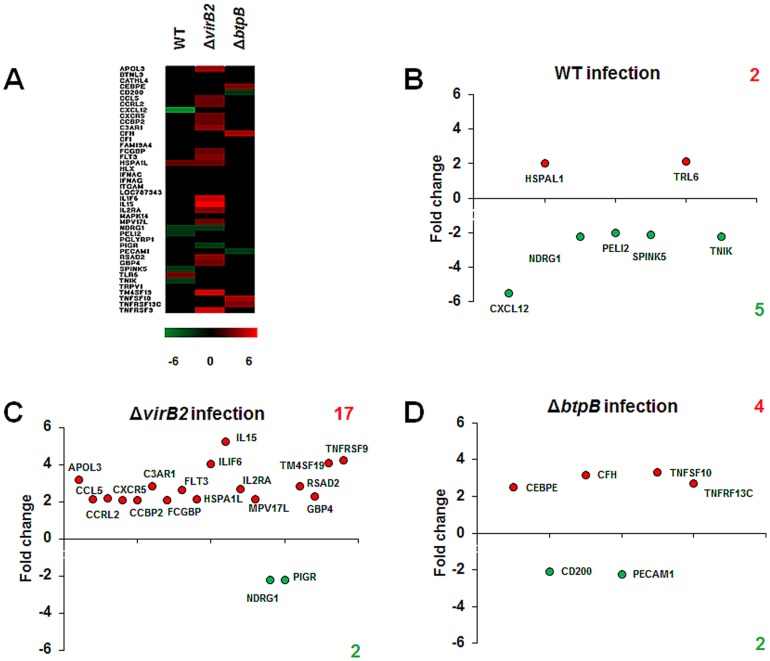
Considering that acute inflammation in the placenta is a hallmark of B. abortus infection in cattle [7] and that B. abortus influences expression of proinflammatory transcripts by bovine trophoblastic cells [13], here we focused the analysis of differentially expressed transcripts associated with defense and inflammation. The microarray data presented above demonstrated downregulation of transcripts in wild type B. abortus-infected explants that included chemokines, genes involved in signaling pathways by TLR, in regulation of proliferation and cellular differentiation, cellular response to stress and anti-inflammatory responses. Only two genes in these categories had significantly increased transcription, namely: Toll-like receptor 6 (TLR6) and heat shock 70 kDa protein 1-like (HSPA1L) (Figure 5B). In contrast, trophoblastic cells infected with either the ΔvirB2 mutant strain or the ΔbtpB strain had a significant increase of transcripts of cytokines and chemokines as well as genes associated with the complement cascade (Figure 5C). Transcription of only two genes related to defense and inflammation were significantly decreased, namely platelet/endothelial cell adhesion molecule (PECAM1) and CD200 molecule (C200) (Figure 5D).
Figure 5. Changes in transcript abundance of defense and inflammation genes during infection of bovine trophoblastic cells with wild type, ΔvirB2 and ΔbtpB B. abortus, compared to mock-infected controls.
(A) Heat map of transcription changes during infections with wild type, ΔvirB2 and ΔbtpB B. abortus. (B–D) Fold changes in gene transcription of genes classified as defense and inflammation that were significantly (P<0.05) up or downregulated during wild type (B), ΔvirB2 (C), and ΔbtpB (D) B. abortus, compared to mock-infected controls. Fold changes >2 with P<0.05 were considered significant. Abbreviations: apolipoprotein L, 3 (APOL3), butyrophilin-like 9 (BTNL9), cathelicidin 4 (CATHL4), CCAAT/enhancer binding protein (C/EBP), epsilon (CEBPE), CD200 molecule (CD200), chemokine (C-C motif) ligand 5 (CCL5), chemokine (C-C motif) receptor-like 2 (CCRL2), chemokine (C-X-C motif) ligand 12 (CXCL12), chemokine (C-X-C motif) receptor 5 (CXCR5), chemokine binding protein 2 (CCBP2), complement component 3a receptor 1 (C3AR1), complement factor H (CFH), complement factor I (CFI), family with sequence similarity 19 (chemokine (C-C motif)-like), member A4 (FAM19A4), Fc fragment of IgG binding protein (FCGBP), fms-related tyrosine kinase 3-like (FLT3), heat shock 70 kDa protein 1-like (HSPA1L), HLX H2.0-like homeobox (HLX), IFN-alpha C (IFNAC), integrin, alpha M (complement component 3 receptor 3 subunit) (ITGAM), interferon alpha G (IFNAG), interferon-alphaomega-like (LOC787343), interleukin 1 family, member 6 (epsilon)-like (IL1F6), interleukin (IL15), interleukin 2 receptor, alpha (IL2RA), mitogen-activated protein kinase 14 (MAPK14),MPV17 mitochondrial membrane protein-like, nuclear gene encoding mitochondrial protein (MPV17L), N-myc downstream regulated 1 (NDRG1), pellino homolog 2 (PELI2), peptidoglycan recognition protein 1 (PGLYRP1), PIGR polymeric immunoglobulin receptor (PIGR), platelet/endothelial cell adhesion molecule (PECAM1), radical S-adenosyl methionine domain containing 2 (RSAD2), rCG28728-like (GBP4), serine peptidase inhibitor, Kazal type 5 (SPINK5), toll-like receptor 6 (TLR6), TRAF2 and NCK interacting kinase (TNIK), transient receptor potential cation channel subfamily V member 1-like, transcript, variant 2 (TRPV1), transmembrane 4 L six family member 19 (TM4SF19), tumor necrosis factor (ligand) superfamily, member 10-like (TNFSF10), tumor necrosis factor receptor superfamily, member 13C (TNFRSF13C), tumor necrosis factor receptor superfamily, member 9 (TNFRSF9).
Validation of the microarray data with qRT-PCR
In order to validate the results obtained with the microarrays analysis, selected differentially expressed transcripts were evaluated by qRT-PCR. Genes encoding IL15, HSPA1L, TNFRSF9, APOL3, PECAM1, PELI2, IL1F6, and TM4SF19 were amplified. Six of the eight selected genes had results that were parallel to those observed by microarray analysis (Figure 6).
Figure 6. Validation of the microarray results by real-time qRT-PCR.
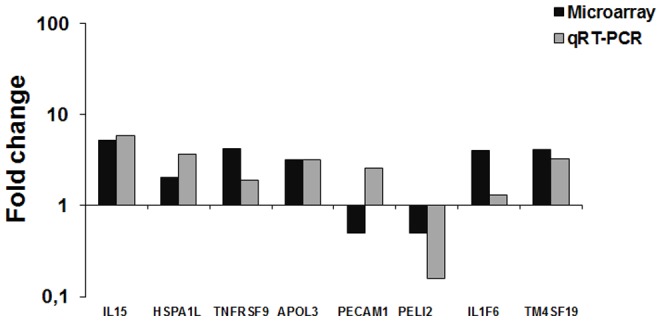
Bars represent the average of fold change values from three independent experiments. Chorioallantoic membranes were infected with wild type B. abortus for evaluation of HSPA1L and PELI2 expression; with the ΔvirB2 Brucella abortus strain for evaluation of IL15, TNFRSF9, IL1F6, and TM4SF19; and with ΔbtpB Brucella abortus strain for evaluation of PECAM1. Abbreviations: interleukin 15 (IL15), heat shock 70 kDa protein 1-like (HSPA1L), tumor necrosis factor receptor superfamily, member 9 (TNFRSF9), apolipoprotein L, 3 (APOL3), pellino homolog 2 (PELI2), platelet/endothelial cell adhesion molecule (PECAM1), interleukin 1 family, member 6 (epsilon)-like (IL1F6), transmembrane 4 L six family member 19 (TM4SF19).
Discussion
This study provided further evidence that B. abortus is capable of actively modulating the host innate immune response during the early stages of infection in target cells that are highly relevant for disease transmission, i.e. bovine trophoblasts. A previous study from our group demonstrated that B. abortus is able to modulate the innate immune response of bovine trophoblastic cells by suppressing the expression of proinflammatory cytokines and chemokines at early stages of infection [13]. In this study we expanded this notion by demonstrating that the absence of a functional T4SS due to deletion of virB2 as well as deletion of the btpB gene impairs the ability of B. abortus to suppress transcription of proinflammatory genes at early stages of infection in bovine trophoblasts. Although suppression of a proinflammatory response by trophoblastic cells may conflict with the fact that B. abortus causes acute placentitis in pregnant cows [7], our previous study [13] also demonstrated expression of proinflammatory chemokines at later stages of infection (i.e., 12 h after inoculation) in B. abortus-infected CAM explants, with a pattern that is similar to that observed in vivo in the placentomes of experimentally infected pregnant cows [13].
BtpB is known to interfere with innate immunity, since it inhibits TLR signaling in dendritic cells [19]. Our results suggest that BtpB could play a similar role in trophoblastic cells, suppressing an innate immune response. The virB operon- encoded T4SS is required for Brucella spp. to interfere with intracellular trafficking, which mediates exclusion of lysosomal markers from the Brucella-containing vacuole, and ultimately allows the pathogen to reach its intracellular replication niche [26]–[28]. Therefore, we hypothesize that the marked differences in host transcriptional profile when trophoblastic cells are infected with a Brucella strain lacking a functional T4SS is likely to be a result of differences in intracellular trafficking. Since virB mutants are killed and degraded, liberation of TLR ligands from degraded virB mutant bacteria couldcontribute to the increased proinflammatory transcriptional profile observed in our study [29]. Alternatively, since the VirB T4SS was recently shown to promote translocation of BtpB into host cells, the increased inflammatory signature in cells infected with the virB2 mutant could result from reduced translocation of BtpB to the cytosol of infected trophoblasts within the CAM explants, and consequently, reduced suppression of TLR signaling.
Confirming previous results from our group that demonstrated that B. abortus inhibits a proinflammatory responses in infected bovine CAM at early stages of infection [13], microarray analysis revealed decreased transcription of genes related to immune response and cellular stress such as chemokine (CXC motif) ligand 12 (CXCL12), Pellino homolog 2 (PELI2), TRAF2 and NCK interacting kinase (TNIK), N-myc downstream regulated 1 (NDRG1) and serine peptidase inhibitor, Kazal type 5 (SPINK5). CXCL12, or stromal-derived factor 1 (SDF-1), is the only ligand for CXCR4 and its decrease may affect various biological processes, including hematopoiesis, cardiogenesis, vascular and neuronal development (processes that may be relevant for fetal development), and traffic of immune cells [30]. In trophoblastic cells infected with B. abortus, the reduction of CXCL12 can also cause decreased interaction with CXCR4, which can be deleterious for the developing fetus and affect immune responses. PELI2 is involved in signaling pathways by TLR1, TLR2, TLR4 and IL-1 by interaction with the complex containing IRAK kinases and TRAF6. It mediates polyubiquination of IRAK1 and can activate MAP (mitogen activated protein) kinase pathways. TNIK, in turn, is regulated by TRAF2, and it is induced by stress, external stimuli and by signal transducers like JNK and NF-κB after stimulation by TNF-α. It also acts protecting cells from apoptosis [31]. NDRG1 is involved in regulation of cellular proliferation and differentiation, as well as cellular response to stress. In human trophoblastic cells, this gene promotes cell viability and protection against injury due to hypoxia, a condition that is commonly associated with placental injury and impaired fetal development [32]. SPINK5 is a serine protease inhibitor, probably related to an anti-inflammatory response. In humans, it is important for protection against pathogens, and it plays a role in formation and physiological renewal of the epidermal barrier [33]–[35]. Decreased transcription of these genes confirms the notion of a negative modulation of the immune response at early stages of infection of bovine trophoblastic cells with B. abortus. Conversely, two genes within this category had increased transcription in bovine trophoblastic cells infected with B. abortus: TLR6 and HSPA1L. Recently, a study showed the importance of TLR6 in triggering the innate immune response against B. abortus in vivo and activation of dendritic cells and production of proinflammatory cytokine [36]. HSPA1L (70 kDa heat shock protein 1-like - Hsp70) is a chaperone that may play a role in the internalization of Brucella sp. Tropism of B. abortus for placental tissues has important implications for the occurrence of B. abortus-induced abortion, although the molecular mechanism for this tropism is unknown. Watanabe et al. [37] demonstrated that heat shock cognate protein (Hsc70) plays a role in Brucella sp. internalization in trophoblastic cells. The administration of anti-Hsc 70 to pregnant mice prevents abortion [37]. In this study there was increased transcription of this gene in trophoblastic cells infected with the ΔvirB strain, supporting the notion that Hsp70 may be involved at early stages of Brucella infection (4 hpi).
The virB operon encodes structural components of the T4SS, and therefore it is required for secretion of effector molecules. The T4SS is required for persistence of Brucella spp. in vivo, and for intracellular survival in macrophages, which are considered one of the primary target cells for Brucella infection. Induction of the T4SS expression occurs after the initial acidification of the Brucella-containing vacuole (BCV). Moreover, the absence of markers of phagolysosomes in BCVs as well as the maturation of compartments derived from the endoplasmic reticulum are mediated by the T4SS of Brucella [15], [26]–[28]. Thus, the virB operon is related to survival and multiplication of Brucella in host cells, since virB mutant strains fail to multiply and localize to a lysosomal compartment [38], [39]. This study demonstrated that bovine trophoblastic cells infected with the ΔvirB mutant strain had significant increases in transcription of several proinflammatory genes at early stages of infection, when compared to trophoblastic cells infected with the wild type strain, although the elucidation of the mechanism of this phenotype is beyond the scope of this study, our data support the notion that suppression of proinflammatory responses by trophoblastic cells that is induced by B. abortus apparently requires a functional T4SS.
Transcripts of several proinflammatory cytokines and chemokines were significantly increased in trophoblastic cells infected with B. abortus ΔvirB compared to uninfected cells. These transcripts included interleukin 15 (IL15), interleukin 1 family, member 6 (epsilon)-like (IL1F6), interleukin 2 receptor alpha (IL2RA), tumor necrosis factor receptor superfamily, member 9 (TNFRSF9) and chemokines such as chemokine (CC motif) receptor-like 2 (CCRL2), chemokine (CC motif) ligand 5 (CCL5), chemokine binding protein 2 epsilon (CCBP2), chemokine (CXC motif) receptor 5 (CXCR5). CXC chemokines act primarily on neutrophil chemotaxis, whereas CC chemokines are chemoattractants for monocytes, lymphocytes, and eosinophils [40]. There was also an increased transcription of the gene encoding Complement component 3a receptor 1 (C3AR1), which is the 3a peptide receptor, one of the proteins of the complement cascade that opsonizes pathogens, and induces a series of inflammatory responses that help fight infection [41]. Opsonization of B. abortus influences the internalization of this pathogen by phagocytic cells [42], [43]. Opsonized bacteria are internalized via receptors for complement and Fc and are more susceptible to the bactericidal action of the macrophages than non-opsonized bacteria which, in turn, are internalized via fibronectin receptor [42], [43]. In the first case, most of the internalized bacteria are destroyed within phagolysosomes before reaching the sites of intracellular multiplication [42], [43]. Therefore, the route of internalization interferes with the intracellular traffic in professional phagocytic cells [43], although opsonized B. abortus is capable of surviving intracellularly in human macrophages [44]. Apolipoprotein L, 3 (APOL3) transcripts were also increased in ΔvirB B. abortus -infected trophoblasts. APOL3 is part of the apolipoproteins family. Increase in APOL proteins is related to signaling by different pro-inflammatory molecules including IFN-α [45], IFN-β [46], IFN-γ [47], and TNF-α [48], which suggests that APOL proteins are involved in immune response.
To a lesser extent when compared to ΔvirB B. abortus infection, bovine trophoblastic cells infected with ΔbtpB B. abortus also had increased mRNA levels of genes related to inflammation. When compared to non-infected controls, there was increased transcription of genes related to complement and also of the TNF family. Interestingly, these results contrast with the transcription profile of the spleen from mice infected with B. abortus or B. melitensis at 3 days post infection, in which there is a T4SS-dependent proinflammatory response [49]. The btpB gene is present in Brucella and encodes a protein with a TIR-domain, and it inhibits innate immune response probably by binding to MyD88, restricting the TLR signaling and therefore, it contributes to the control of inflammation and establishment of infection [19]. Salcedo et al. [19] reported that a virB mutant translocated less of BtpA and BtpB into mouse macrophages. Therefore, the increased expression of inflammation-related transcripts could be related to reduced translocation of the Btp proteins into infected trophoblasts. Furthermore, since virB mutants are killed more efficiently after uptake by cells, this could release more microbe associated molecular patterns (MAMPs) into the phagolysosome where they can be detected by TLRs. Two genes belonging to the TNF family had significant increases in transcription: tumor necrosis factor receptor superfamily, member 13C (TNFRSF13C), which is associated with increased survival of B cells in vitro and regulating the population of peripheral B cells, and tumor necrosis factor (ligand) superfamily, member 10-like (TNFSF10), is a member of TNF family of cytokines and primarily related to apoptosis. TNFSF10 playing important roles in regulating cell death, immune response, and inflammation. TNFSF10 binding to its receptors promotes activation of MAPK8/JNK, caspase 8, and caspase 3 [50]–[52]. There was also an increased transcription of the complement factor H (CFH), which is a member of the Regulator of Complement Activation (RCA) cluster. It plays an essential role in the regulation of complement activation, restricting the activation of the complement cascade innate immune response against pathogens and CCAAT/enhancer binding protein (C/EBP) epsilon (CEBPE). It is also a critical mediator of myelopoiesis and it is related to the functional maturation of neutrophils and monocytes/macrophages [53]–[55].
In conclusion, our study demonstrated that infection with B. abortus induces a downregulation of the bovine trophoblastic innate immune response during the first hours of infection. The virB-encoded T4SS, and to a lesser extent the btpB gene, play a direct or indirect role in this mechanism.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The microarray data set has been submitted to the Gene Expression Omnibus database at NCBI (http://www.ncbi.nlm.nih.gov/geo/) and assigned accession number GSE58216.
Funding Statement
Work in RLS' lab is supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), PRONEX grant from FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Brazil), and a PNPD fellowship for JPSM from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Boschiroli ML, Foulongne V, O'Callaghan D (2001) Brucellosis: a worldwide zoonosis. Curr Opin Microbiol 4: 58–64. [DOI] [PubMed] [Google Scholar]
- 2. Cutler SJ, Whatmore AM, Commander NJ (2005) Brucellosis - new aspects of an old disease. J Appl Microbiol 98: 1270–1281. [DOI] [PubMed] [Google Scholar]
- 3. Santos RL, Martins TM, Borges ÁM, Paixão TA (2013) Economic losses due to bovine brucellosis in Brazil. Pesq Vet Bras 33: 759–764. [Google Scholar]
- 4. Samartino LE, Enright FM (1993) Pathogenesis of abortion of bovine brucellosis. Comp Immunol Microbiol Infect Dis 16: 95–101. [DOI] [PubMed] [Google Scholar]
- 5.Acha PN, Szyfres B (2003) Zoonoses and communicable diseases common to man and animals. 3rd ed. Washington: Pan American Health Organization, 3v. (Scientific and Technical Publication, 580). [Google Scholar]
- 6.Nicoletti P (1990) Bovine abortion caused by Brucella sp. In: Kirkbride CA (Ed.) Laboratory Diagnosis of Livestock Abortion 3rd Ed. Ames: Iowa State University Press. p. 22–26. [Google Scholar]
- 7. Xavier MN, Paixão TA, Poester FP, Lage AP, Santos RL (2009) Pathology, immunohistochemistry and bacteriology of tissues and milk of cows and fetuses experimentally infected with Brucella abortus . J Comp Pathol 140: 149–157. [DOI] [PubMed] [Google Scholar]
- 8. Anderson T, Cheville NF (1986) Ultrastructural morphometric analysis of Brucella abortus-infected trophoblasts in experimental placentitis. Bacterial replication occurs in rough endoplasmic reticulum. Am J Pathol 124: 226–237. [PMC free article] [PubMed] [Google Scholar]
- 9. Meador VP, Deyoe BL, Cheville NF (1989) Pathogenesis of Brucella abortus infection of the mammary gland and supramammary lymph node of the goat. Vet Pathol 26: 357–368. [DOI] [PubMed] [Google Scholar]
- 10. Samartino LE, Enright FM (1996) Brucella abortus differs in the multiplication within bovine chorioallantoic membrane explants from early and late gestation. Comp Immunol Microbiol Infect Dis 19: 55–63. [DOI] [PubMed] [Google Scholar]
- 11. Carvalho Neta AV, Mol JPS, Xavier MN, Paixão TA, Lage AP, et al. (2010) Pathogenesis of bovine brucellosis. Vet J 184: 146–155. [DOI] [PubMed] [Google Scholar]
- 12. Samartino LE, Enright FM (1992) Interaction of bovine chorioallantoic membrane explants with three strains of Brucella abortus . Am J Vet Res 53: 359–363. [PubMed] [Google Scholar]
- 13. Carvalho Neta AV, Steynen APR, Paixão TA, Miranda KL, Silva FL, et al. (2008) Modulation of bovine trophoblastic innate immune response by Brucella abortus . Infect Immun 76: 1897–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G (2002) The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A 99: 1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, et al. (2002) Type IV secretion and Brucella virulence. Vet Microbiol 90: 341–348. [DOI] [PubMed] [Google Scholar]
- 16. De Jong MF, Tsolis RM (2012) Brucellosis and type IV secretion. Future Microbiol 7: 47–58. [DOI] [PubMed] [Google Scholar]
- 17. Kahl-McDonagh MM, Elzer PH, Hagius SD, Walker JV, Perry QL, et al. (2006) Evaluation of novel Brucella melitensis unmarked deletion mutants for safety and efficacy in the goat model of brucellosis. Vaccine 24: 5169–5177. [DOI] [PubMed] [Google Scholar]
- 18. Cirl C, Wieser A, Yadav M, Duerr S, Schubert SR, et al. (2008) Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain–containing proteins. Nature 14: 399–406. [DOI] [PubMed] [Google Scholar]
- 19. Salcedo SP, Marchesini MI, Degos C, Terwagne M, Von Bargen K, et al. (2013) BtpB a novel Brucella TIR-containing effector protein with immune modulatory functions. Front Cell Infect Microbiol 8: 3–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salcedo SP, Marchesini M, Lelouard H, Fugier E, Jolly G, et al. (2008) Brucella control of dendritic cell maturation is dependent on the TIR-Containing Protein Btp1. PLoS Pathog 4: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Den Hartigh AB, Sun YH, Sondervan D, Heuvelmans N, Reinders MO, et al. (2004) Differential requirements for virB1 and virB2 during Brucella abortus infection. Infect Immun 72: 5143–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun YH, Rolan HG, den Hartigh AB, Sondervan D, Tsolis RM (2005) Brucella abortus virB12 is expressed during infection but is not an essential component of the type IV secretion system. Infect Immun 73: 6048–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Silva TM, Paixão TA, Costa EA, Xavier MN, Sá JC, et al. (2011) Putative ATP-binding cassette transporter is essential for Brucella ovis pathogenesis in mice. Infect Immun 79: 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans HE, Sack WO (1973) Prenatal development of domestic and laboratory mammals: growth curves, external features and selected references. Zentralbl Veterinarmed 2: 11–45. [DOI] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 26. Celli J, De Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, et al. (2003) Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med 198: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Celli J, Salcedo SP, Gorvel JP (2005) Brucella coopts the small GTPase Sar1 for intracellular replication. Proc Natl Acad Sci U S A 102: 1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J (2008) Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic 9: 678–694. [DOI] [PubMed] [Google Scholar]
- 29. Wolf AJ, Arruda A, Reyes CN, Kaplan AT, Shimada T, et al. (2011) Phagosomal degradation increases TLR access to bacterial ligands and enhances macrophage sensitivity to bacteria. J Immunol 187: 6002–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hansen TR, Smirnova NP, Van Campen H, Shoemaker ML, Ptitsyn AA, et al. (2010) Maternal and fetal response to fetal persistent infection with bovine viral diarrhea virus. Am J Reprod Immunol 64: 295–306. [DOI] [PubMed] [Google Scholar]
- 31. Gui J, Li Z, Zhou X (2013) Dynamic change of TNIK in response to tumor necrosis factor alpha in a TRAF2-dependent manner. Hum Cell 26: 67–72. [DOI] [PubMed] [Google Scholar]
- 32. Shi XH, Larkin JC, Chen B, Sadovsky Y (2013) The expression and localization of N-myc downstream regulated gene 1 in human trophoblasts. PLoS One 8: e75473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M (2000) Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet 25: 141–142. [DOI] [PubMed] [Google Scholar]
- 34. Bitoun E, Micheloni A, Lamant L, Bonnart C, Tartaglia-Polcini A, et al. (2003) LEKTI proteolytic processing in human primary keratinocytes, tissue distribution and defective expression in Netherton syndrome. Hum Mol Genet 12: 2417–2430. [DOI] [PubMed] [Google Scholar]
- 35. Descargues P, Deraison C, Prost C, Fraitag S, Mazereeuw-Hautier J, et al. (2006) Corneodesmosomal cadherins are preferential targets of stratum corneum trypsin-like and chymotrypsin-like hyperactivity in Netherton syndrome. J Invest Dermatol 126: 1622–1632. [DOI] [PubMed] [Google Scholar]
- 36. Almeida LA, Macedo GC, Marinho FA, Gomes MT, Corsetti PP, et al. (2013) Toll-like receptor 6 plays an important role in host innate resistance to Brucella abortus infection in mice. Infect Immun 81: 1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe K, Tachibana M, Tanaka S, Furuoka H, Horiuchi M, et al. (2008) Heat shock cognate protein 70 contributes to Brucella invasion into trophoblast giant cells that cause infectious abortion. BMC Microbiol 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seleem MN, Stephen BM, Sriranganathan N (2007) Brucella: A pathogen without classic virulence genes. Vet Microbiol 129: 1–14. [DOI] [PubMed] [Google Scholar]
- 39. Celli J, Gorvel JP (2004) Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr Opin Microbiol 7: 93–97. [DOI] [PubMed] [Google Scholar]
- 40.Mcgee DW (1999) Inflammation and mucosal cytokine production. In: OGRA PL. Mucosal immunology 2nd ed. San Diego: Academic Press, p. 559–573. [Google Scholar]
- 41. Wende E, Laudeley R, Bleich A, Bleich E, Wetsel RA, et al. (2013) The complement anaphylatoxin C3a receptor (C3aR) contributes to the inflammatory response in Dextran Sulfate Sodium (DSS)-induced colitis in mice. PLoS One 8: e62257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Campbell GA, Adams LG, Sowa BA (1994) Mechanism of binding of Brucella abortus to mononuclear phagocytes from cows naturally resistant or susceptible to brucellosis. Vet Immunol Immunopathol 41: 295–306. [DOI] [PubMed] [Google Scholar]
- 43. Gorvel JP, Moreno E (2002) Brucella intracellular life: from invasion to intracellular replication. Vet Microbiol 90: 281–297. [DOI] [PubMed] [Google Scholar]
- 44. Bellaire BH, Roop RM 2nd, Cardelli JA (2005) Opsonized virulent Brucella abortus replicates within nonacidic, endoplasmic reticulum-negative, LAMP-1-positive phagosomes in human monocytes. Infect Immun 73: 3702–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hayashi J, Stoyanova R, Seeger C (2005) The transcriptome of HCV replicon expressing cell lines in the presence of alpha interferon. Virology 335: 264–275. [DOI] [PubMed] [Google Scholar]
- 46. Stojdl DF, Lichty BD, Tenoever BR, Paterson JM, Power AT, et al. (2003) Vsv strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4: 263–275. [DOI] [PubMed] [Google Scholar]
- 47. Sana TR, Janatpour MJ, Sathe M, Mcevoy LM, McClanahan TK (2005) Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine 29: 256–269. [DOI] [PubMed] [Google Scholar]
- 48. Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ (2002) The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics 79: 539–546. [DOI] [PubMed] [Google Scholar]
- 49. Roux CM, Rolán HG, Santos RL, Beremand PD, Thomas TL, et al. (2007) Brucella requires a functional Type IV secretion system to elicit innate immune responses in mice. Cell Microbiol 9: 1851–1869. [DOI] [PubMed] [Google Scholar]
- 50. Baker SJ, Reddy EP (1996) Transducers of life and death: TNF receptor superfamily and associated proteins. Oncogene 12: 1–9. [PubMed] [Google Scholar]
- 51. Bossen C, Schneider P (2006) BAFF, APRIL and their receptors: structure, function and signaling. Semin Immunol 18: 263–275. [DOI] [PubMed] [Google Scholar]
- 52. Nomura S, Ishii K, Inami N, Uoshima N, Ishida H, et al. (2007) Role of soluble tumor necrosis factor-related apoptosis-inducing ligand concentrations after stem cell transplantation. Transpl Immunol 18: 115–121. [DOI] [PubMed] [Google Scholar]
- 53. Diaz-Guillen MA, De Cordoba SR, Heine-Suner D (1999) A radiation hybrid map of complement factor H and factor H-related genes. Immunogenetics 49: 549–552. [DOI] [PubMed] [Google Scholar]
- 54. Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL (2000) Complement factor H: sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Mol Immunol 37: 41–52. [DOI] [PubMed] [Google Scholar]
- 55. Kyme P, Thoennissen NH, Tseng CW, Thoennissen GB, Wolf AJ, et al. (2012) C/EBPε mediates nicotinamide enhanced clearance of Staphylococcus aureus in mice. J Clin Invest 122: 3316–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The microarray data set has been submitted to the Gene Expression Omnibus database at NCBI (http://www.ncbi.nlm.nih.gov/geo/) and assigned accession number GSE58216.