Abstract
Papillary thyroid carcinoma (PTC) is a common endocrine malignancy with low death rate but increased incidence and recurrence in recent years. MicroRNAs (miRNAs) are small non-coding RNAs with diverse regulatory capacities in eukaryotes and have been frequently implied in human cancer. Despite current progress, however, a panoramic overview concerning miRNA regulatory networks in PTC is still lacking. Here, we analyzed the expression datasets of PTC from The Cancer Genome Atlas (TCGA) Data Portal and demonstrate for the first time that immune responses are significantly enriched and under specific regulation in the direct miRNA−target network among distinctive PTC variants to different extents. Additionally, considering the unconventional properties of miRNAs, we explore the protein-coding competing endogenous RNA (ceRNA) and the modulatory networks in PTC and unexpectedly disclose concerted regulation of immune responses from these networks. Interestingly, miRNAs from these conventional and unconventional networks share general similarities and differences but tend to be disparate as regulatory activities increase, coordinately tuning the immune responses that in part account for PTC tumor biology. Together, our systematic results uncover the intensive regulation of immune responses underlain by miRNA-mediated networks in PTC, opening up new avenues in the management of thyroid cancer.
Thyroid carcinoma (THCA) is a common type of endocrine malignancies, the majority of which are derived from the thyroid follicular cell origin. Despite the relatively lower death rate compared with other tumors, the incidence rate of thyroid cancer has increasingly arisen in recent years, accompanied by its still high recurrence1,2,3. Among all thyroid malignancies, papillary thyroid carcinoma (PTC) is the predominant, well-differentiated neoplasm intimately associated with radiation exposure4,5 and is characteristic of aberrant activation of mitogen-activated protein kinase (MAPK) signaling pathway1,2,3. Importantly, some genetic alterations have been identified to drive PTC pathogenesis, such as BRAF mutations (especially BRAFV600E)6, RAS mutations7, RET rearrangements (the most common chimeric forms including RET−PTC1 and RET−PTC3)8, or neurotrophic receptor tyrosine kinase 1 (NTRK1) rearrangements9. Until recently, the copy-number gain of IQ-motif-containing GTPase-activating protein 1 (IQGAP1)10 and the genome-wide hypermethylation and hypomethylation associated with BRAF-V600E11 have also been implied in PTC. Conjointly, these genetic or epigenetic events could amplify MAPK cascade activation, accounting for tumorigenesis or progression of PTC.
MicroRNAs (miRNAs) are small non-coding RNA molecules (19–24 nucleotides long) initially identified in Caenorhabditis elegans with a temporal expression pattern during the larval development12,13, but to date accumulating knowledge has been expanding their regulatory spectra with diverse functional implication in all eukaryotes14,15. Through the sequence complementarity of seed pairing and the site context, miRNAs could counteract the effects of their cognate mRNAs and thus protein output via mRNA degradation or translational repression14. Notably, miRNA dysregulation was ubiquitously detectable in all types of tumors and therefore has incurred broad attention as a targeting choice in cancer therapies16 or as the diagnostic or prognostic markers17. Regarding their cell type-specific action, some miRNAs were involved consistently across multiple cancer types, while some could be either oncogenic or tumor-suppressive depending on the malignant context. For example, the miR-34 family was tumor-suppressive and consistently down-regulated in several solid tumors18, whereas the let-7 family was oncogenic and overexpressed in acute myeloid leukemia (AML), but, generally, tumor-suppressive and down-regulated in other malignancies than AML19.
Beyond post-transcriptional repression, the regulatory complexity of miRNAs has been manifested by some current discoveries about the versatility of miRNAs. For instance, miRNAs could not only down-regulate but up-regulate protein translation, as evidenced by the capability of miR-369-3 to interact with AU-rich elements in the tumor necrosis factor (TNF) mRNA, ultimately leading to increased protein translation during cell cycle arrest20. In addition, miRNAs could communicate with various RNA species in the competing endogenous RNA (ceRNA) network with significant implications in human diseases21, as exemplified by the fact that the pseudogene PTENP1 was tumor-suppressive via acting as a decoy for phosphatase and tensin homolog (PTEN)-targeting miRNAs, whose loss concomitantly amplified PTEN loss and was frequently detected in human cancers22. Moreover, an unconventional role for miRNAs has been highlighted in their ability to directly bind as agonists to Toll-like receptors (TLRs) and trigger the downstream signaling in diseased states23,24. Similarly, a recent study has also implied an indirect, modulatory contribution for miRNAs other than miRNA-mRNA targeting in breast cancer25.
Concerning our molecular understandings in PTC tumor, it has been reported that miR-221, miR-222, and miR-146b were significantly overexpressed, accompanied with their target KIT down-regulation26. Similarly, other lines of evidence have also identified a role of miR-221, miR-222 and miR-181b overexpression in thyroid cell transformation and carcinogenesis27 and the contribution of miR-146b to tumor aggressiveness in PTC28. Besides, several studies based on miRNA expression profiling have presented some sets of miRNAs as diagnostic markers to help distinguish malignancies from indeterminate thyroid samples29. Despite the current progress, however, how miRNAs might contribute to PTC pathogenesis still remains incompletely understood and a panoramic study in terms of miRNA networks in PTC is required.
In this study, we utilized publicly available miRNA and mRNA expression and SNP array datasets of PTC from The Cancer Genome Atlas (TCGA) and computationally performed a series of miRNA network analyses in different aspects. Specifically, we evaluated the contribution of PTC miRNAs in the direct miRNA−target network, the protein-coding competing endogenous RNA (ceRNA) network, and the indirect, modulatory network25. We first demonstrated that miRNAs among these networks, sharing general similarities and differences, cooperatively regulate the immune responses of PTC. Moreover, the clear discrepancies among PTC histological variants were also noticed in terms of differentially expressed miRNAs and mRNAs and associated functional enrichments. Interestingly, by incorporating the SNP array data, we manifested several genes with somatic copy number alterations (SCNAs) underlying PTC biology. Collectively, our findings provide a comprehensive miRNA-associated network landscape with an unprecedented implication of miRNAs in PTC immunity and present several high-confidence molecular candidates for further research, which could potently serve as new therapeutic targets for this common thyroid neoplasia.
Results
Identification of differentially expressed miRNAs and mRNAs in PTC
We accessed TCGA Data Portal to retrieve miRNA and mRNA expression datasets of thyroid carcinoma, which comprised approximately 500 samples belonging to three PTC histological variants, i.e., classical PTC (CPTC), follicular PTC (FPTC), and tall-cell PTC (TCPTC). By performing the significance analysis of microarrays (SAM) on these datasets, we demonstrated that histological variants are good features in terms of differentially expressed (DE) miRNAs and mRNAs (Table 1) but not gender, age at initial pathologic diagnosis, and T and N scores (stand for tumor and node, respectively) in the TNM staging system (a classification tool for physicians to stage types of cancers) (Supplementary Table 1). Notably, the differential expression was manifested to some extent between N0 (cancer, not spread to regional lymph nodes) and N1 (cancer, spread to regional lymph nodes) categories, and we reasoned that this was greatly due to the confounding effect of histological variants within these categories, where CPTC accounted for a large portion of N1 (all N1 = 208; CPTC N1 = 166; FPTC N1 = 13; TCPTC N1 = 22) while FPTC also contributed a lot to N0 (all N0 = 216; CPTC N0 = 138; FPTC N0 = 61; TCPTC N0 = 13), which could be further confirmed by SAM results between N0 and N1 but restricted within each PTC variant (Supplementary Table 1).
Table 1. Identification of DE miRNAs and mRNAs regarding histological variants in PTC.
| SAM group | miRNA | mRNA | |||
|---|---|---|---|---|---|
| Source | Reference | Up | Down | Up | Down |
| Histological variants | |||||
| CPTC | Normal | 63 | 118 | 2719 | 2850 |
| FPTC | Normal | 74 | 100 | 1867 | 2224 |
| TCPTC | Normal | 70 | 84 | 3067 | 2174 |
| FPTC | CPTC | 74 | 95 | 2352 | 2394 |
| TCPTC | FPTC | 93 | 80 | 2216 | 2234 |
| CPTC | TCPTC | 12 | 24 | 238 | 846 |
CPTC, classical PTC; FPTC, follicular PTC; TCPTC, tall-cell PTC.
Concerning differential expression with respect to the normal (Table 1), we found that CPTC and TCPTC were relatively alike while dissimilar to FPTC (Fig. 1a, b). Utilizing the comparative results in Table 1, we highlighted miRNAs with differential expression to some extent among CPTC, FPTC, and TCPTC (Supplementary Table 2); for instance, there were 22 miRNAs with the relative DE intensity “TCPTC > CPTC > FPTC” (i.e., TCPTC > CPTC, CPTC > FPTC, and TCPTC > FPTC), 54 miRNAs with “CPTC ≈ TCPTC > FPTC” (i.e., CPTC > FPTC, TCPTC > FPTC, but CPTC ≈ TCPTC), and 18 miRNAs with “FPTC > TCPTC” (i.e., FPTC > TCPTC, but FPTC ≈ CPTC and TCPTC ≈ CPTC). Additionally, we also wondered whether driver genes30, which were characterized by somatic copy number alterations (SCNAs) and of top 50% expressed genes in each selected sample, should play a role in PTC biology. Using the TCGA copy number data processed by GISTIC31, we found no gene with significant amplification (GISTIC Q-value < 0.25) as the potential driver genes in PTC. However, we were still interested in evaluating SCNAs using a tumor−normal paired approach to show genes with concurrent differential expression and copy number changes in PTC (Supplementary Fig. 1, 2, Supplementary Table 3). From Supplementary Fig. 1, the extent of SCNA gains was higher than losses in all PTC variants, while a greater loss of genomic contents covering chromosome 22 was detectable in FPTC than in CPTC or TCPTC. Notably, some genes with SCNA gains of the highest scores were present in all PTC variants, including signal-regulatory protein beta 1 (SIRPB1), muscular LMNA-interacting protein (MLIP), WW domain containing oxidoreductase (WWOX), heat shock transcription factor 2 binding protein (HSF2BP), and polypeptide N-acetylgalactosaminyltransferase-like 6 (GALNTL6) (Supplementary Table 3). Several DE up-regulated genes with SCNA gains, such as cadherin 13 (CDH13), CUB and Sushi multiple domains 1 (CSMD1), deleted in malignant brain tumors 1 (DMBT1), GALNTL6, and the protocadherin alpha (PCDHA) gene cluster, were further manifested commonly in PTC (Supplementary Fig. 2).
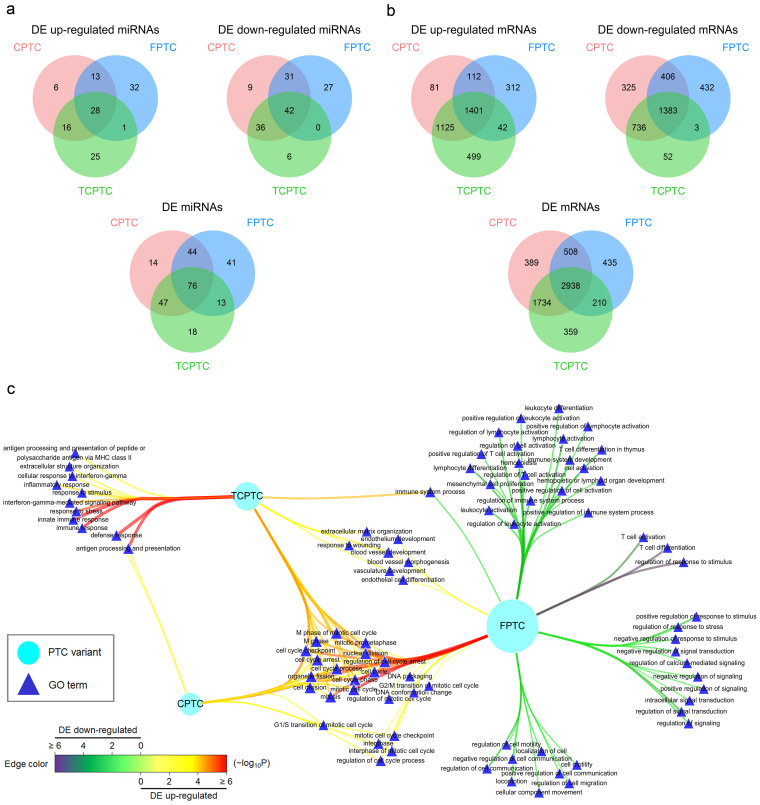
Figure 1. Relationships of DE miRNAs and mRNAs and functional enrichments spanning DE mRNAs among PTC variants.
The differentially expressed (DE) miRNAs (a) and mRNAs (b) were obtained from significant analysis of microarrays (SAM) results (Mann−Whitney U test with 1000 permutations and 90th percentile false discovery rate (FDR) < 0.01) among CPTC (red), FPTC (blue), and TCPTC (green) variants compared to the normal (see Table 1). (c) DE up-regulated or down-regulated mRNAs in each PTC variant were subject to Gene Ontology (GO) association tests using the R package “GOstats” and the results were visualized using Cytoscape (version 3.0.2) (see Methods). Nodes contain PTC variants (ellipse) and GO terms (triangle). Node size is proportional to the degree of node itself. Edge color and width reflect the significance of enrichment (−log10 transformed Q-values) for a GO term to a PTC variant (from green to purple for DE down-regulated genes, while from yellow to red for DE up-regulated genes). Clearly, cell cycle-relevant GO terms were consistently enriched for DE up-regulated mRNAs in all PTC variants, while other terms showed distinctive enrichment in certain variants.
Diverse functional enrichments spanning DE mRNAs among PTC variants
To gain a functional perspective on DE mRNAs of three PTC variants, we conducted the Gene Ontology (GO) enrichment analysis. Intriguingly, cell cycle-related GO terms were consistently overrepresented for DE up-regulated mRNAs among all PTC variants (Fig. 1c), suggesting an anomalous cell cycle event inherently in this cancer. TCPTC displayed several terms associated with immune responses, while CPTC showed a little, for DE up-regulated mRNAs. Additionally, TCPTC and FPTC shared some terms involved in the endothelium or vasculature development (Fig. 1c). For DE down-regulated mRNAs, however, no significant functional annotation was observed in CPTC and TCPTC, but an enrichment of the immune system process, especially “leukocyte activation” and many of its descendants, as well as of the motility and signal transduction, was underscored in FPTC (Fig. 1c). Furthermore, we also manifested overrepresented GO terms enriched for the comparative DE mRNAs between PTC variants, demonstrating 18 GO terms with “TCPTC > CPTC > FPTC” (i.e., enriched for DE mRNAs satisfying TCPTC > CPTC, CPTC > FPTC, and TCPTC > FPTC) and 136 GO terms with “CPTC ≈ TCPTC > FPTC”, almost all of which were related to activation or regulation of the immune or inflammatory responses (Supplementary Table 4). Collectively, our results indicate discernible functional enrichments with a common cell cycle dysregulation spanning DE mRNAs in PTC variants.
Direct miRNA−target network links immune activities to PTC
We investigated the network spanning direct miRNA−mRNA targeting in PTC. With the results in Table 1 and the aid of miRNA target site prediction databases TargetScan32 and miRanda33, and Spearman correlation coefficient (SCC), we generated several candidate DE miRNA−mRNA pairs. By incorporating SCNAs from Supplementary Table 3, we also revealed DE genes with paired miRNAs and concurrent SCNAs, in which miR-138-5p:GALNTL6 was identified consistently in all PTC variants (Supplementary Fig. 3). Through GO association tests on those candidate DE paired mRNAs in each variant, we disclosed an exclusive enrichment of “immune system process” and many of its descendants in this direct miRNA−target network (Fig. 2a), which was devoid of cell cycle-related GO terms as observed for simply DE mRNAs in Fig. 1c. Obviously, CPTC and TCPTC shared several GO terms specifically concerning “antigen processing and presentation” for candidate DE up-regulated mRNAs, whereas TCPTC and FPTC shared some significant terms like “immune response” and “leukocyte activation” but for candidate DE up-regulated and down-regulated mRNAs, respectively (Fig. 2a). Moreover, TCPTC had some singular terms dealing with responses to stimuli and cytokine signaling pathway (e.g., “defense response”, “interferon-γ-mediated signaling pathway”) for candidate DE up-regulated mRNAs, while FPTC possessed many individual terms linking to activation of T and natural killer (NK) lymphocytes and cell killing (Fig. 2a).
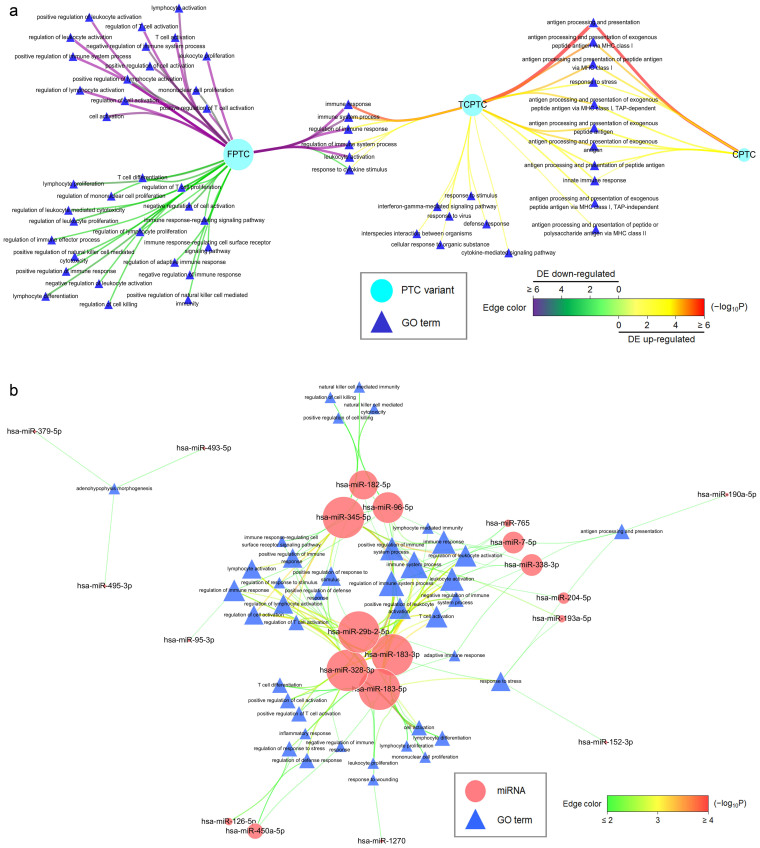
Figure 2. MicroRNA−target network specifically directs immune activity in PTC.
(a) Candidate DE up-regulated or down-regulated mRNAs (paired with oppositely DE miRNAs) in each PTC variant were subject to GO association tests and the results were visualized using Cytoscape. Nodes contain PTC variants (ellipse) and GO terms (triangle). Node size is proportional to the degree of node itself. Edge color and width reflect the significance of enrichment (−log10 transformed Q-values) for a GO term to a PTC variant (from green to purple for DE down-regulated genes, while from yellow to red for DE up-regulated genes). Immune response-associated GO terms were specifically enriched in the miRNA−target network of PTC. (b) Regulatory spectrum analyses of the miRNA−target network were conducted by collecting miRNA's cognate targets per miRNA for individual GO association tests and the results were visualized using Cytoscape. Nodes contain miRNAs (ellipse) and GO terms (triangle). Node size is proportional to the degree of node itself. Edge color and width reflect the significance of enrichment (−log10 transformed Q-values) for a GO term to a miRNA (from green then yellow to red). Some miRNAs showed immunity-regulatory spectra in the direct miRNA−target network of PTC.
To evaluate whether miRNAs repress their cognate mRNAs with a specific biological function in PTC, we performed the regulatory spectrum analysis for each candidate DE miRNA. In line with our findings in Fig. 2a, we revealed that some miRNAs repressed their targets with a propensity toward immune response activities in PTC, such as miR-345-5p, miR-328-3p, miR-183-3p and -5p, and miR-29b-2-5p (Fig. 2b). In general, however, we noted that most of candidate DE miRNAs with stronger expression in PTC, such as miR-221-3p, miR-222-3p, and miR-146b-5p frequently reported in thyroid cancer26,27,28,34, were not statistically enriched for their cognate targets with a specific function, consistent with the observation that broadly conserved miRNAs tend to repress genes with a wide range of biological processes or molecular functions14.
With regard to exclusive immune-regulatory activities in the PTC direct miRNA−target network (Fig. 2a, b), we illustrated the miRNA−target pairs of paramount importance for mRNAs annotated with at least one GO term at or downstream of “immune system process” with statistical significance by assigning the combinatorial scores for each miRNA−target pair (Fig. 3, Supplementary Table 5).
Figure 3. MicroRNA−target−GO term network underlying immune system process in PTC.
MicroRNA−target pairs in which mRNA targets having at least one GO annotation descendant of “immune system process” defined by the bimap object “org.Hs.egGO2ALLEGS” in the R package “org.Hs.eg.db” (version 2.8.0) were visualized using Cytoscape. Nodes contain miRNAs (ellipse), mRNA targets (triangle), and GO terms (rounded rectangle). Node size is proportional to the degree of node itself. For miRNA−target edges, the edge color indicates up-regulation (dark) o down-regulation (light) of that miRNA in CPTC (red), FPTC (blue), and TCPTC (green), and the edge width proportionally reflects the combinatorial score defined in the text for that miRNA−target pair. For mRNA target−GO term edges, an edge (gray) only indicates an annotation.
In Fig. 3, we perceived that several potential miRNA−target pairs were shared between CPTC and TCPTC with miRNA down-regulation (light red and light green edges, respectively) whereas some pairs were uniquely present in FPTC with miRNA up-regulation (dark blue edge). In agreement with the previous study26, we remarked that miR-221-3p and miR-222-3p were consistently overexpressed among three PTC variants targeting KIT mRNA with the highest scores, suggesting a potential role of these miRNA−target pairs in immune responses as part of PTC pathogenesis. Furthermore, miR-146b-5p was also highlighted in this network targeting other immunity-related mRNAs like recombination activating gene 2 (RAG2), cytochrome P450, family 27, subfamily B (CYP27B1), and chemokine binding protein 2 (CCBP2) (Fig. 3). Interestingly, miR-34a-5p was DE up-regulated accordantly in all PTC variants with several high-score targets linking to immune response activities, such as protein kinase C theta (PRKCQ) or epsilon (PRKCE), engulfment and cell motility 1 (ELMO1), as well as KIT (Fig. 3). Evidently, for those pairs in Fig. 3 with miRNA up-regulation in FPTC (dark blue edge), several miRNAs with multiple top-score targets were indeed those miRNAs with a specific regulatory spectrum for their cognate targets toward immune response activities in Fig. 2b.
On the other hand, from the score-ranked miRNA−target pair list in Supplementary Table 5, we could clearly discern high-confidence miRNA−target pairs with a possible immune function in PTC, as exemplified by miR-450a-5p:DUSP10 (dual specificity phosphatase 10) with miRNA up-regulation in CPTC and miR-363-3p:LYST (lysosomal trafficking regulator) with miRNA down-regulation in both CPTC and TCPTC. We further manifested critical miRNAs (e.g., miR-204-5p, miR-183-5p, miR-182-5p) and mRNAs (e.g., KIT, LYST, PRKCQ) underlying “immune system process” in the PTC direct miRNA−target network by extracting information with combinatorial scores greater than 70 (Supplementary Table 6, 7). Finally, rather than this general GO term “immune system process”, we could also use other specific GO terms of statistical significance to highlight potential miRNA−target pairs of interest. For example, miR-193a-5p:HLA-F (human leukocyte antigen F) and miR-30c-5p:STAT1 (signal transducer and activator of transcription 1) were the only two miRNA−target pairs with scores greater than 70 underlying “interferon-γ-mediated signaling pathway” (Supplementary Table 8). Taken together, these data suggest that miRNAs in the direct miRNA−target network potently regulate immune responses in PTC.
Protein-coding ceRNA network manifests immune responses in PTC
Considering the versatility of miRNAs in the competing endogenous RNA (ceRNA) crosstalk21, we conducted the protein-coding ceRNA network analysis on PTC and inferred several putative miRNA:ceRNA tuples (Supplementary Fig. 4). To acquire a global sense of biological function, we performed a GO association test on all identified ceRNAs, revealing unexpectedly that immune activities were also enriched in PTC (Supplementary Table 9).
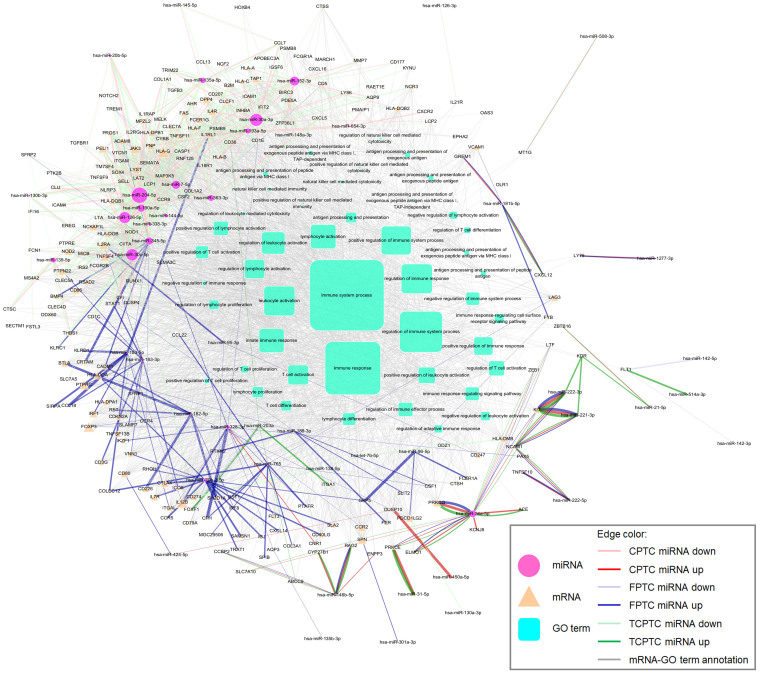
By the same token, we elucidated all the ceRNA interactions in PTC by assigning the combinatorial score for each miRNA:ceRNA tuple (Fig. 4, Supplementary Table 10). Distinctly, a large highly connected cluster was manifested in the ceRNA network in PTC (Fig. 4), with several ceRNAs having at least one GO term being or downstream of the “immune system process” (red color node). To better underline promising miRNA:ceRNA tuples, we unveiled crucial miRNAs (e.g., miR-98-5p, miR-148b-3p, miR-103a-3p) and ceRNAs (e.g., protein tyrosine phosphatase, non-receptor type 22 (PTPN22), protein tyrosine phosphatase, receptor type, C (PTPRC), Z-DNA binding protein 1 (ZBP1), cytotoxic and regulatory T cell molecule (CRTAM), LYST, STAT1) relating to “immune system process” by distilling information with combinatorial scores greater than 60 (Supplementary Table 11, 12). Notably, some ceRNAs with experimental validation in other reports were also contained in PTC, such as versican (VCAN) and fibronectin 1 (FN1)35,36,37 (Supplementary Table 12).
Figure 4. Panoramic overview of protein-coding ceRNA interaction in PTC.
The potential miRNA:ceRNA tuples identified in our screen (context+ < −0.4 or miRSVR < −1.0) were visualized using Cytoscape. Nodes represent ceRNAs (rounded rectangle). Node color indicates whether a ceRNA has at least one GO annotation being or descendent of “immune system process” (red for yes; cyan for no). Node size is proportional to the degree of node itself. An edge indicates a miRNA mediating correspondent ceRNAs. Edge size proportionally reflects the combinatorial score defined in the text for that miRNA:ceRNA tuple. Multiple edges between two nodes are allowed, which means multiple miRNAs mediated two ceRNAs given. Obviously, there was a large, highly connected cluster with several ceRNAs linking to the GO term “immune system process”.
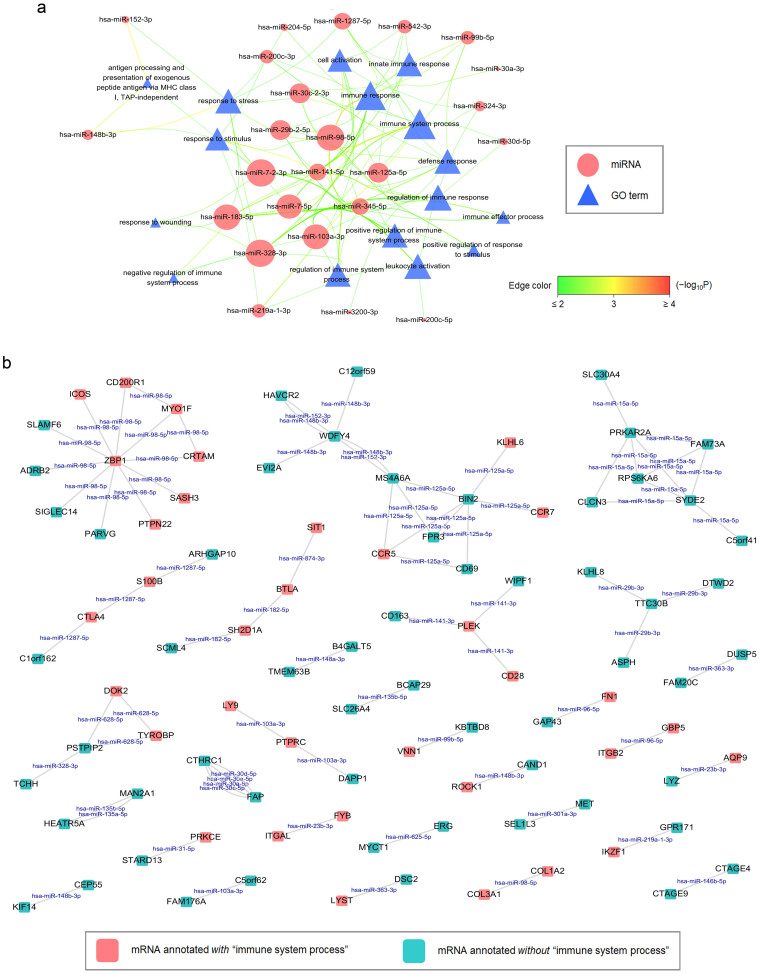
To examine whether miRNAs regulate their corresponding ceRNAs with a specific functional spectrum in PTC, we performed the regulatory spectrum analysis in a similar fashion. Remarkably, immune response-associated terms were principally enriched for ceRNAs underlying specific miRNA regulation in PTC (Fig. 5a). To further reflect an essential role of immune responses in PTC ceRNA crosstalk, we presented a high-confidence ceRNA interaction network by gathering information from Supplementary Table 10, confirming that many ceRNAs with the highest scores were indeed tied to “immune system process” in PTC (Fig. 5b). Conjointly, these data indicate that protein-coding ceRNA regulatory network underlies immune activities in PTC.
Figure 5. Protein-coding ceRNA network intensively regulates immunity in PTC.
(a) Regulatory spectrum analyses of the ceRNA network were performed by gathering correspondent ceRNAs per miRNA for individual GO association tests and the results were visualized using Cytoscape. Nodes contain miRNAs (ellipse) and GO terms (triangle). Node size is proportional to the degree of node itself. Edge color and width reflect the significance of enrichment (−log10 transformed Q-values) for a GO term to a miRNA (from green then yellow to red). Some ceRNA-associated miRNAs revealed immunity-regulatory spectra in PTC. (b) A high-confidence ceRNA interaction network by extracting the miRNA:ceRNA tuples with combinatorial scores > 80 defined in the text. Node color indicates whether a ceRNA has at least one GO annotation being or descendent of “immune system process” (red for yes; cyan for no). Node size is proportional to the degree of node itself. An edge indicates a miRNA (shown on the edge) mediating correspondent ceRNAs.
MicroRNA modulatory network contributes to immune regulation in PTC
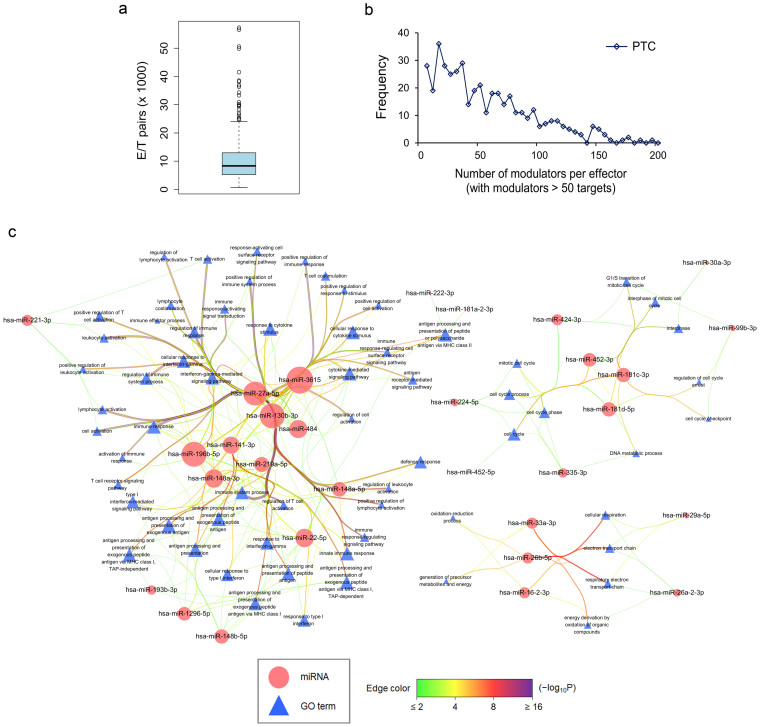
With an unconventional property of miRNAs documented in a recent study25, we also explored the possibilities that miRNAs (modulators) could behave as co-activators or co-repressors through modulating the relationship between certain mRNAs (effectors and targets) independently of miRNA target recognition in PTC (Supplementary Fig. 5). To this end, we demonstrated the miRNA modulatory activity of PTC in terms of the effector−target pairs per modulator (Fig. 6a) and the distribution showing connectivity between modulators and effectors (Fig. 6b). Next, we devised the combinatorial scores for each target and therefore a step-by-step procedure to extract critical targets from the ranked score distribution. Conspicuously, immune response-relevant GO terms were unequivocally enriched for significant targets with top scores across all modulators in PTC modulatory network (Supplementary Table 13). Similarly, we also applied this procedure within each modulator to analyze the regulatory spectrum in the modulatory network. Intriguingly, we found that three functional clusters were enriched for specific miRNA modulation in PTC, with the largest one dealing with immune activities (e.g., miR-27a-5p, miR-3615, miR-148a-3p) and the remaining two concerning cell cycle regulation (e.g., miR-181c-3p, miR-181d-5p, miR-452-3p) and electron transport chain (e.g., miR-26b-5p, miR-33a-3p, miR-16-2-3p) (Fig. 6c). Noticeably, miR-27a-5p was especially prominent in the immune cluster with several most significant P-values (Fig. 6c). Finally, we further revealed strong modulators ranked by effector counts with variable target number restrictions (Supplementary Table 14) and important targets ranked by the combinatorial scores across all modulators with signs indicative of “immune system process” GO annotation (for example, interferon regulatory factor 5 (IRF5), selectin L (SELL), fms-related tyrosine kinase 3 ligand (FLT3LG), and PYD and CARD domain containing (PYCARD) with positive signs) (Supplementary Table 15). Altogether, our data imply that miRNA modulatory network closely engages in immune regulation in PTC.
Figure 6. MicroRNA modulatory network contributes to immune responses in PTC.
Modulatory activities in PTC were shown in terms of the number of total effector/target pairs per modulator (a) or the number of bona fide modulators per effector (b) (a bona fide relationship was defined for a given modulator−effector pair with > 50 targets). (c) Regulatory spectrum analyses of the modulatory network were conducted by gathering critical targets per modulator for individual GO association tests and the results were visualized using Cytoscape. Nodes contain miRNAs (ellipse) and GO terms (triangle). Node size is proportional to the degree of node itself. Edge color and width reflect the significance of enrichment (−log10 transformed Q-values) for a GO term to a miRNA (from green, yellow, red, to purple). Some modulators revealed immunity-regulatory spectra as a major functional cluster in the modulatory network of PTC.
MicroRNAs share similarities and differences from these networks and collaboratively regulate immune responses in PTC
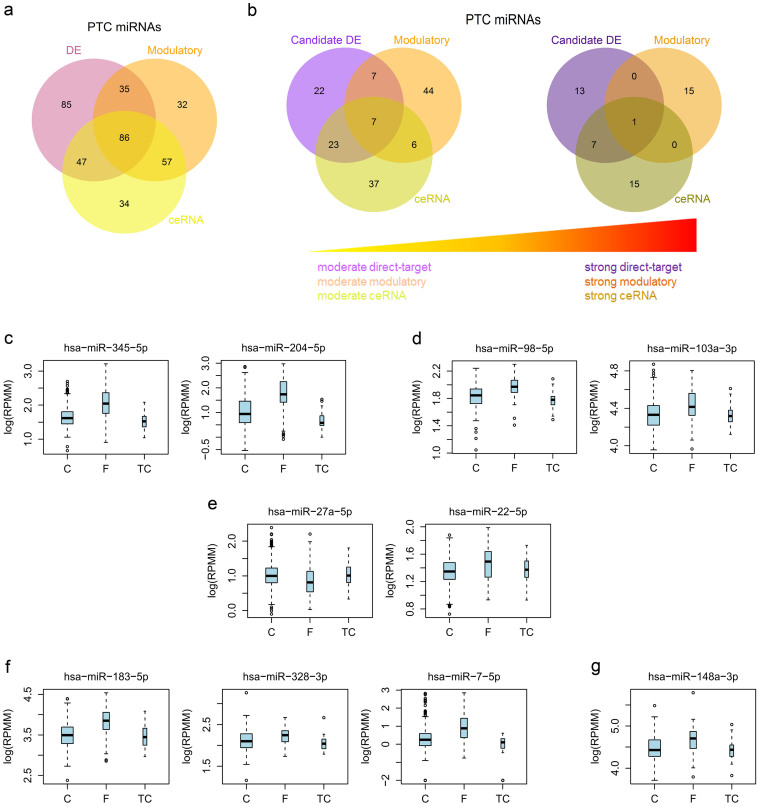
Motivated by our network results from three perspectives with commensurate contribution to immune responses, we wondered the interrelationships of miRNAs by tuning respective intensities from these networks in PTC. Under minimal restriction on the selection criterion, we found that miRNAs shared general similarities and differences among the DE, the ceRNA, and the modulatory networks in PTC (Fig. 7a). By tuning the restriction to moderate and strong stringencies, however, we observed that miRNAs with strong regulatory activities tended to be exclusive in any of the candidate DE, the ceRNA, and the modulatory networks in PTC (Fig. 7b).
Figure 7. MicroRNAs from different networks show cooperative regulation of immune responses in PTC.
Venn diagrams demonstrating the logical relations among DE, ceRNA-associated, and modulatory miRNAs (a) or for different miRNA-mediated networks with increasing regulatory activities (b) were shown. From miRNAs with strong regulatory activity defined in the right panel in (b), we displayed the expression of representative miRNAs associated with set regions in PTC variants in terms of boxplots as follows: miRNAs with strong activities only in candidate DE (c), only in ceRNA-associated (d), only in modulatory (e), in both candidate DE and ceRNA-associated but not modulatory (f), and in all networks (g). For boxplots, expression values of miRNAs were log10-transformed (a zero value was assigned −2) and box width was proportional to the square root of sample size in each variant.
For a check on each set region of the Venn diagram in Fig. 7b displaying correspondent miRNAs with strong activities (right panel), we first illustrated two miRNAs, miR-345-5p and miR-204-5p, with strong activity only in the direct miRNA−target network with respect to their expression among three PTC variants (Fig. 7c). Noteworthily, both miR-345-5p and miR-204-5p repressed their cognate targets with a regulatory spectrum of immune responses (Fig. 2b) and, in addition, miR-345-5p had significant differential expression among PTC variants with “FPTC > CPTC > TCPTC” while miR-204-5p with “FPTC > CPTC ≈ TCPTC” (Supplementary Table 2). Next, we exemplified two miRNAs, miR-98-5p and miR-103a-3p, with strong activity only in the protein-coding ceRNA network (Fig. 7d). Both miR-98-5p and 103a-3p regulated their corresponding ceRNAs in PTC with a specific functional spectrum toward immune responses (Fig. 5a) but were not differentially expressed among PTC variants (Supplementary Table 2). Third, we marked two miRNAs, miR-27a-5p and miR-22-5p, with strong activity only in the modulatory network (Fig. 7e). Impressively, miR-27a-5p, as well as miR-22-5p, modulated their targets with a biological preference to immune regulation in PTC (Fig. 6c), while miR-27a-5p showed differential expression with “CPTC ≈ TCPTC > FPTC” but miR-22-5p did not (Supplementary Table 2). Fourth, we examined three miRNAs, miR-183-5p, miR-328-3p and miR-7-5p, with strong activity in both the direct miRNA−target and ceRNA networks (Fig. 7f). Outstandingly, all miR-183-5p, miR-328-3p and miR-7-5p had strong propensities to immune regulation for both their cognate targets and the corresponding ceRNAs in PTC (Fig. 2b, 5a) and, furthermore, they all revealed differential expression among PTC variants with “FPTC > CPTC ≈ TCPTC” (Supplementary Table 2). Last, we highlighted miR-148a-3p (a miR-148/152 family member) with strong activity in all the direct miRNA−target, ceRNA, and modulatory networks (Fig. 7g). As generally tumor-suppressive in malignancies, miR-148a-3p in our screen was also DE down-regulated in CPTC and TCPTC but not in FPTC, while its differential expression among PTC variants was relatively indeterminate with “FPTC > CPTC” (Supplementary Table 2). Concerning the regulatory spectrum, however, miR-148a-3p possessed a strong preference to immune regulation only in the modulatory network (Fig. 6c), without specific propensity in the direct miRNA−target and ceRNA networks. As a whole, our comprehensive analyses indicate that miRNAs have multifaceted contribution at different network levels but cooperatively regulate immune responses in PTC.
Discussion
Our data revealed unprecedentedly that miRNAs in PTC concertedly regulate immune response processes at different levels. We explored the conventional (i.e., the miRNA−target relationships) and unconventional properties (i.e., the protein-coding ceRNA network and the indirect, modulatory contribution) of PTC miRNAs, providing valuable information about critical miRNAs and mRNAs from respective networks linking to the tumor biology. Consistent with our findings, a subset of Hashimoto's thyroiditis harbored some precursor lesions with molecular characteristics of PTC38,39, suggesting frequent lymphocytic infiltration in this malignancy, and a higher incidence rate of TCPTC in patients with Graves' disease was also reported40. In this study, we demonstrated that TCPTC and, secondly, CPTC possess more immune activities under miRNA regulation than FPTC, indicating the involvement of immunological factors to different extents in PTC variants.
From the conventional miRNA−target network perspective, we identified miR-34a-5p up-regulation among CPTC, FPTC, and TCPTC targeting PRKCQ (or PKC-θ) and, to a lesser extent, KIT with highest scores underlying “immune system process”, inconsistent with a general tumor-suppressive role of miR-34a-5p in p53-mediated effects in almost solid tumors18, suggesting a unique regulatory dependency for miR-34a-5p on this malignant context. Furthermore, considering the ability of PKC-θ to regulate KIT expression in gastrointestinal stromal tumors (GIST)41, miR-34a-5p targeting to PRKCQ and KIT could augment the effects of miR-221-3p: and miR-222-3p:KIT interactions26, which may accordantly lead to PTC pathogenesis.
Notably, several descendants of “antigen processing and presentation” were enriched for candidate mRNA targets coupled with miRNA down-regulation in TCPTC and CPTC. Some of these targets contained major histocompatibility complex (MHC) molecules (or HLAs), where MHC class II proteins should be normally expressed by such professional antigen-presenting cells (APCs) as macrophages, B lymphocytes, and especially dendritic cells. In contrast with this notion, however, thyroid epithelial cells have been reported to express HLA-DRA, HLA-DMB, class II transactivator (CIITA), and invariant chain (Ii) in an IFNγ-responsive manner, indicating an antigen processing and presenting capability for thyrocytes as non-professional APCs42. Moreover, similar HLA-DRA and CIITA expression was detectable in thyroid carcinoma cells43 and antigen presentation coactivators B7-1 and B7-2 were positively correlated with lymphocytic infiltration and poor prognosis for differentiated thyroid carcinomas44. Interestingly, STAT1 is required for enhanced activation of IFNγ-inducible HLA-DR expression following thyroid hormone stimulation in responsive cells45. As a support, miR-193a-5p:HLA-F and miR-30c-5p:STAT1 interactions with highest scores underlying “interferon-γ-mediated signaling pathway” were marked in TCPTC or CPTC, indicating antigen processing and presentation events to stronger extents in these variants than FPTC.
Concerning the involvement of miR-146b-3p in KIT targeting26 and tumor aggressiveness28, we also noted other putative miR-146b-3p targets with highest scores linking immune responses in PTC such as RAG2, whose product is involved in the V(D)J recombination for immunoglobulin (Ig) heavy chains and T cell receptors. Intriguingly, along with mature B lymphocytes and plasma cells, papillary thyroid cancer cells are able to express immunoglobulin G (IgG) as well as RAG1, RAG2, and activation-induced cytidine deaminase (AID)46, leaving the putative miR-146b-3p:RAG2 interaction in our screen linking to PTC cancer biology worthy to be determined.
Our protein-coding ceRNA analysis within PTC revealed an exceptional role of miR-98-5p with a frequently coupled ceRNA ZBP1 in immune responses. Analogous to membrane-associated TLRs, ZBP1 is a cytosolic DNA sensor which potently induces type I IFNs and activates other innate immune responses47. Noticeably, miR-98-5p is a let-7 family member and has been implied in several neoplastic events as a tumor suppressor48,49. Although miR-98-5p and ZBP1 did not show apparent differential expression in any PTC variant, several immunity-related ceRNAs coupled with them did show, such as MYO1F up-regulated in TCPTC and CPTC, PTPN22 up-regulated in TCPTC but down-regulated in FPTC, and SASH3 up-regulated in TCPTC, underpinning a potential contribution of ZBP1 ceRNA network to PTC pathogenesis. Additionally, miR-98-5p regulated correspondent ceRNAs with a functional tendency to immune responses, further underscoring the importance of miR-98-5p in the PTC ceRNA network.
Other than ZBP1, we noticed several high-confidence ceRNAs which have been implied in a repertoire of thyroid diseases, e.g., PTPN22 and cytotoxic T-lymphocyte-associated protein 4 (CTLA4) as susceptibility genes to thyroid autoimmunity50, PTPRC as a biomarker for thyroid cancer lymph node metastasis51, and STAT1, runt-related transcription factor 1 (RUNX1), and FN1 responsible for RET−PTC3 oncogene-induced gene expression in early modulation of the immune response in thyroid follicular cells52. Interestingly, albeit LYST is well-known for mediating intracellular protein trafficking whose mutation is associated with a lysosomal storage disorder Chediak-Higashi syndrome53, it has been also evidenced for a requirement of this gene to sort MHC class II molecules, in accordance with severe immunodeficiencies in patients with this disease54. Higher LYST expression in TCPTC and CPTC and its active participation in the ceRNA network were observed in this study, suggesting intensive adaptive immunity under miRNA regulation in these PTC variants.
We also identified miRNAs with strong regulatory activity in both direct miRNA−target and ceRNA networks, among which some also possessed a specific immunity-regulatory spectrum. As exemplified by miR-7-5p that was down-regulated in TCPTC and CPTC, its cognate mRNA targets cardiotrophin-like cytokine factor 1 (CLCF1) and insulin receptor substrate 2 (IRS2) were noted linking immune responses in the PTC direct miRNA−target network with highest scores, of which each mRNA was also entangled in the PTC ceRNA network mediated by miR-7-5p, consistent with a recent finding that miR-7-5p:IRS2 interaction is responsible for migration and invasion in melanoma cells55. Alternatively, despite the oncogenic potential in several neoplastic contexts56,57,58, miR-182-5p, miR-183-5p, and miR-328-5p, which showed significant up-regulation in FPTC, were remarked for their capability of tuning immune responses from these networks in thyroid cancer.
As suggested by an unconventional, modulatory properties of miRNAs in a subset of breast tumors25, we also demonstrated that this indirect modulation also contributed in large part to immune responses in PTC, albeit the modulatory activity was similar to the estrogen receptor (ER)+ tumors while relatively weaker than the genomically quiescent integrative cluster 4 subtype identified in that study. Several miRNAs functionally dealt with immune responses in the PTC modulatory network, whereas some specifically engaged in cell cycle regulation and electron transport chain as minority groups. Outstandingly, miR-27a-5p linked several immunity-associated GO terms with the most significant P-values, whose expression was significantly down-regulated in FPTC, suggesting a potential contribution for this miRNA to immune responses in PTC from the modulatory aspect. Remarkably, this modulatory property should be reminiscent of some recent discoveries that miRNAs act as the RNA ligands for TLRs directly triggering downstream signaling pathways in different settings23,24, indicating that our findings regarding the PTC modulatory network could be re-checked as a consequence of putative TLR or other pattern recognition receptor (PRR)-mediated signaling events. In line with this concept, the most significant GO enrichment for globally top-scored targets was “innate immune response” in the PTC modulatory network and, indeed, several top-scored targets are engaged in PRR-mediated signaling contexts, such as IRF5, TRAF5, PYCARD, and TRIM14.
Our results particularly highlighted miR-148a-3p, which belongs to the miR-148/152 family and showed significant down-regulation in TCPTC and CPTC, for strong regulatory activities in all direct miRNA−target, ceRNA, and modulatory networks but with a specific immunity-regulatory spectrum only in the modulatory network. Noticeably, innate immune responses and antigen presentation could be dampened by miR-148a-3p and miR-152-3p in TLR-triggered dendritic cells via direct targeting59, but here we suggest that miR-148a-3p may play a potential role in PTC immunity via an indirect modulatory effect. Intriguingly, miR-152-3p, whose expression was significantly down-regulated in all PTC variants, possessed strong regulatory activities and immunity-specific functional spectra in both the direct miRNA−target and ceRNA networks, together with miR-148a-3p tuning PTC immunity in a reciprocal manner from respective networks.
Interestingly, although driver genes30 were not apparent in PTC, the importance of several SCNA genes identified from our tumor-normal paired scheme was confirmed by their consistent changes in all PTC variants. Noteworthily, SIRPB1, which was of the most significant SCNA gain in each variant, also belongs to the immunoglobulin superfamily with a role in the immune signaling and, concerning our results, should deserve more investigative efforts in PTC. While there is no clear association for GALNTL6 with immune system process, miR-138-5p:GALNTL6 was prominent as a direct miRNA−target pair for their consistent appearances with GALNTL6 SCNA gains among all PTC variants, possibly suggesting an unknown function to the cancer biology.
In summary, we have unveiled that immune responses are under specific and intensive regulation by miRNAs in PTC, the extent of which differs among three histological variants, complementing our knowledge about the biology of this endocrine neoplasia. Apart from miR-221-3p, miR-222-3p, and miR-146b-5p for an established link to thyroid carcinogenesis, we further point out other miRNAs of putative yet great importance regarding conventional and unconventional properties that in concert tune the immune responses of PTC. Additionally, we have identified several SCNAs previously uncharacterized that might suggest their great importance in the pathogenesis of PTC. As a whole, our comprehensive studies not only prioritize several miRNA and gene candidates from corresponding networks for detailed investigation but also provide a new vision for seeking more effective therapeutics of papillary thyroid cancer.
Methods
Data acquisition, preprocessing, and DE analysis
The miRNA and mRNA expression datasets of PTC were retrieved from TCGA Data Portal. For a given miRNA in a sample, we summed up normalized expression values with the same MIMAT accession number and referenced miRBase release 20 for nomenclature60. Expression datasets from primary tumor and solid tissue normal groups were merged, excluding any expression vector (reads per million miRNA mapped for miRNA; RPKM for mRNA) with third quartile (Q3) < 2 times of first quartile (Q1) or with the maximum expression value of any miRNA or mRNA < 0.5. The expression data were quantile-normalized using R package “preprocessCore” (version 1.20.0) and subject to SAM (two class unpaired, Mann−Whitney U test with 1000 permutations, and 90th percentile FDR < 0.01) using R package “samr” (version 2.0). At this step, the number of selected miRNAs = 490 and mRNAs = 8235. Venn diagrams showing DE results for histological variants were plotted using R package “VennDiagram” (version 1.6.5).
Analysis of SCNAs
To demonstrate somatic copy number alterations (SCNAs) using a tumor−normal paired approach in PTC, we accessed the SNP array data (level 3) from TCGA. Instead of “nocnv_hg18.seg.txt” files, where a fixed set of probes frequently associated with germline copy number variations (CNVs) were excluded before segmentation, we made use of “hg18.seg.txt” files to uncover SCNAs by the steps as follows. In each PTC variant, for each patient available, we took the difference of segment means between the primary tumor and the paired blood derived normal (i.e., tumor − normal) across entire chromosomal regions as the representative SCNA of that patient, and summed up all the SCNAs divided by number of selected patients as the representative SCNA of that PTC variant. To allow computation of the difference of region-unaligned segment data, we resorted to R package “CNTools” (version 1.20.0) to obtain a reduced segment matrix (in a region-aligned format) for this purpose, where the “geneInfo” object (made from human genes based on build 36) was used for computation by genes and the median would be taken for regions with multiple segment values.
Functional enrichments for histological DE mRNAs
Gene Ontology (GO) association tests were conducted using R package “GOstats” within ontology class “Biological Process” referencing to R package “org.Hs.eg.db” (version 2.8.0) for gene annotations. Hits of GO terms were considered independent hypergeometric distributions and P-values were corrected by Benjamini−Hochberg method (as Q-values). Enrichment results were presented for DE mRNAs in each variant and visualized using Cytoscape (version 3.0.2).
Discovery of miRNA−target pairs
DE miRNAs were bipartitely paired with oppositely DE mRNAs, with incorporation of TargetScan context+32 and miRanda miRSVR33 scores and Spearman correlation coefficient (SCC) across tumor samples. For TargetScan prediction, the minimum score for a given miRNA−target pair from all conserved and non-conserved sites was representative of the context+ score throughout this study.
Scoring design and characterization for miRNA−target pairs
We devised a combinatorial score for each miRNA−target pair (as CMBdirect; context+ < −0.2 or miRSVR < −0.5, and SCC < −0.3) by partitioning the contribution of context+, miRSVR, and SCC using linear transformation without ceiling: [0, −0.4] was mapped to [0, 40] for context+; [0, −1.0] to [0, 40] for miRSVR; and [0, −0.7] to [0, 20] for SCC. To characterize miRNA−target pairs, we first derived descendants of a selected GO BP term defined by the bimap object “GOBPOFFSPRING” in R package “GO.db” (version 2.8.0). We then extracted pairs with presence in at least one histological variant with significantly enriched GO terms (Q-value < 0.001) underlying that selected term via checking Entrez IDs of associated mRNAs using the bimap object “org.Hs.egGO2ALLEGS” in R package “org.Hs.eg.db” (version 2.8.0) and visualized results using Cytoscape. For “immune system process”, we summarized critical miRNAs and mRNAs from pairs satisfying CMBdirect > 70.
Functional enrichments for direct miRNA−target network
Global functional enrichments were performed for candidate paired DE mRNAs in each variant (context+ < −0.2 or miRSVR < −0.5, and SCC < −0.3) and visualized using Cytoscape (Q-values < 0.01; only top 40 significant GO terms for FPTC). To demonstrate regulatory spectra, for each miRNA, we took the union of all its cognate targets across variants (context+ < −0.1 or miRSVR < −0.2, and SCC < −0.3) followed by a GO association test. Results were visualized using Cytoscape for GO terms (Q-values < 0.05) by at least three miRNAs.
Discovery of miRNA:ceRNA tuples
First, we filtered out miRNAs (Q3 > 1.5 × Q1 and 20th percentile > 0 and 80th percentile > 1) and mRNAs (Q3 > 1.5 × Q1 and 20th percentile > 1) across PTC samples. At this step, the number of miRNAs = 442 and mRNAs = 9312. Next, for each miRNA, we extracted uppermost 20% samples by its expression values, imposed context+ < −0.2 or miRSVR < −0.5, and SCC < −0.3, computed SCCs between predicted mRNAs (SCCuppermost), and eventually produced mRNAs with SCCuppermost > 0.4 as ceRNAs, in which SCCs between predicted mRNAs within lowermost 20% samples (SCClowermost) were also recorded for the scoring design (Supplementary Fig. 4). Note that we used SCC rather than Pearson's correlation coefficient (PCC) to allow nonlinearity in miRNA−mRNA relationships.
Scoring design and characterization for miRNA:ceRNA tuples
We devised a combinatorial score for each ceRNA1:miRNA:ceRNA2 tuple (as CMBceRNA; context+ < −0.2 or miRSVR < −0.5) by partitioning the contribution of two context+miRNA:ceRNA, two miRSVRmiRNA:ceRNA, SCCuppermost, and SCClowermost, using linear transformation: [0, −0.4] was mapped to [0, 15] for context+ without ceiling; [0, −1.0] to [0, 15] for miRSVR without ceiling; [0.4, 1.0] to [0, 30] for SCCuppermost; and [0.4, 1.0] to [0, 10], [−0.4, −1.0] to [0, −20] for SCClowermost with ceiling. To characterize miRNA:ceRNA tuples, we determined whether a ceRNA has at least one GO annotation underlying “immune system process” using the bimap object “org.Hs.egGO2ALLEGS” in R package “org.Hs.eg.db” (version 2.8.0) and plotted a panoramic (context+ < −0.4 or miRSVR < −1.0) and a high-confidence (CMBceRNA > 80) ceRNA interaction networks using Cytoscape. Similarly, we also summarized critical miRNAs and ceRNAs from tuples satisfying CMBceRNA > 60.
Functional enrichments for ceRNA network
A global functional enrichment was conducted for ceRNAs identified from all miRNA:ceRNA tuples. To display regulatory spectra, for each miRNA, we gathered all of its corresponding ceRNAs (context+ < −0.2 or miRSVR < −0.5) followed by a GO test, collected information for GO terms with Q-values < 0.05, took a geometric sum of Q-values for each single term, and visualized final results using Cytoscape for each GO term (average Q-value < 0.01) by at least three miRNAs.
Discovery of effector-modulator-target tuples
Following the analytic framework by Dvinge et al.25, we filtered out effectors and targets (Q3 > 1.5 × Q1 and 25th percentile > 1) and modulators (Q3 > 1.5 × Q1 and 25th percentile > 0 and 75th percentile > 1) across PTC samples. We confined effectors to at least one GO annotation underlying five selected terms concerning transcription factor binding or activity (GO:0001071, GO:0000988, GO:0008134, GO:0005667, GO:0051090) using the “is a” ontology structure constructed from Gene Ontology website (http://www.geneontology.org/). At this step, the number of effectors = 436, targets = 9476, and modulators = 435. Next, for each modulator (miRNA), we defined uppermost 25% and lowermost 25% samples by its expression values, imposed |SCC| < 0.3 on modulator−effector and modulator−target relationships, confirmed no differential expression for effectors (and targets) between the uppermost 25% and lowermost 25% samples (using simple t-tests, excluding pairs with P < 0.01 and fold change > 1.5), calculated SCCs between effector and target within uppermost (as SCCHIGH) and lowermost (as SCCLOW), and finally derived effector−modulator−target tuples with both |SCCHIGH| or |SCCLOW| > 0.4 and |SCCHIGH − SCCLOW| > 0.45 (Supplementary Fig. 5).
Scoring design, modulatory activity, and target extraction
We devised a combinatorial score for each target within a given modulator or across all modulator (as CMBmod). Specifically, for each target under a modulator−effector pair, we defined the weighted score as products of max{|SCCHIGH|, |SCCLOW|} × 2, |SCCHIGH − SCCLOW|, and Ntarget−1 (where Ntarget is the number of targets under that modulator−effector). For computation within a modulator, we only included information in which Ntarget > 50 and a given target had ≥ 5 modulator−effector pairs (NEM pair | target > 5) and summed up all weighted scores for a given target. Similarly, for computation across all modulators, we instead imposed higher stringencies (Ntarget > 100 and NEM pair | target > 10). To exemplify modulatory activities, we summarized the number of total effector/target pairs per modulator and of bona fide modulators per effector (Ntarget > 50).
To manifest critical targets from the elbow-like target score distribution (Supplementary Fig. 6), we devised an algorithm that approximates this transition point as follows:
Find the maximum and minimum values (head and tail).
Define and calculate the expected increment c as follows: c = (head − tail) × gene count−1.
Define a variable penalty p and a penalty threshold pt (set pt = 20).
Starting from the head, calculate the difference d between the current value and next value.
If d < c, then p = p + 1; else, p = p – 1 if p > 0 (non-negative).
If p < pt, repeat procedures d−e until reaching the tail; else, report the current gene index i and extract (i – pt) genes from the head.
If the tail has been achieved but p < pt, extract all the genes.
Functional enrichments for modulatory network
GO tests were performed for critical targets across all modulators (global enrichment) or within each modulator (regulatory spectrum). In case of regulatory spectra, we collected information for GO terms with Q-values < 0.05, took a geometric sum of Q-values for each single term, and visualized results using Cytoscape for each GO term (average Q-value < 0.001) by at least three modulators.
Logical relations of miRNAs from different networks
To demonstrate logical relations, we plotted Venn diagrams using the following selection criteria. Minimum restriction case: select DE miRNAs from all variants, ceRNA-associated miRNAs with total CMBceRNA > 100, and modulatory miRNAs with > 50 effectors each of which corresponded to > 50 targets (Neffector | target > 50 > 50). Moderate restriction case: select candidate DE miRNAs with ≥ 3 paired DE mRNAs satisfying CMBdirect > 20, ceRNA-associated miRNAs with total CMBceRNA > 300 (under tuples with CMBceRNA > 60), and modulatory miRNAs satisfying Neffector | target > 100 > 50. Strong restriction case: select candidate DE miRNAs with ≥ 3 paired DE mRNAs satisfying CMBdirect > 70, ceRNA-associated miRNAs with total CMBceRNA > 3000 (under tuples with CMBceRNA > 60), and modulatory miRNAs satisfying Neffector | target > 200 > 50.
Author Contributions
C.-T.H., H.-C.H. and H.-F.J. designed the research methods, interpreted the results, and wrote the paper. C.-T.H. analyzed the data. Y.-J.O. helped the data analysis. H.-C.H. and H.-F.J. defined the research theme and supervised the work.
Supplementary Material
Supplementary Information
Acknowledgments
We acknowledged Mrs. Chia-Hsien Lee and Andy Ho for their initial efforts in the design and analysis for microRNA−target network. This work was supported by the National Science Council of Taiwan [NSC 102-2628-B-002-041-MY3, 102-2627-B-002-002, and 102-2311-B-010-004] and the National Taiwan University Cutting-Edge Steering Research Project [103R7602C3].
References
- Kondo T., Ezzat S. & Asa S. L. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat. Rev. Cancer 6, 292–306, 10.1038/nrc1836 (2006). [DOI] [PubMed] [Google Scholar]
- Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat. Rev. Cancer 13, 184–199, 10.1038/nrc3431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D. F. & Chen H. New developments in the diagnosis and treatment of thyroid cancer. CA Cancer J. Clin. 63, 374–394, 10.3322/caac.21195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov V. S., Demidchik E. P. & Astakhova L. N. Thyroid-Cancer after Chernobyl. Nature 359, 21–21, Doi 10.1038/359021a0 (1992). [DOI] [PubMed] [Google Scholar]
- Williams D. Cancer after nuclear fallout: lessons from the Chernobyl accident. Nat. Rev. Cancer 2, 543–549, 10.1038/nrc845 (2002). [DOI] [PubMed] [Google Scholar]
- Xing M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer 12, 245–262, Doi 10.1677/Erc.1.0978 (2005). [DOI] [PubMed] [Google Scholar]
- Lemoine N. R. et al. High-Frequency of Ras Oncogene Activation in All Stages of Human Thyroid Tumorigenesis. Oncogene 4, 159–164 (1989). [PubMed] [Google Scholar]
- Santoro M., Melillo R. M. & Fusco A. RET/PTC activation in papillary thyroid carcinoma: European Journal of Endocrinology Prize Lecture. Eur. J. Endocrinol. 155, 645–653, 10.1530/eje.1.02289 (2006). [DOI] [PubMed] [Google Scholar]
- Rabes H. M. et al. Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-Chernobyl papillary thyroid carcinomas: Biological, phenotypic, and clinical implications. Clin. Cancer. Res. 6, 1093–1103 (2000). [PubMed] [Google Scholar]
- Liu Z. et al. IQGAP1 Plays an Important Role in the Invasiveness of Thyroid Cancer. Clin. Cancer. Res. 16, 6009–6018, Doi 10.1158/1078-0432.Ccr-10-1627 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P., Liu D. X. & Xing M. Z. Genome-wide alterations in gene methylation by the BRAF V600E mutation in papillary thyroid cancer cells. Endocr. Relat. Cancer 18, 687–697, Doi 10.1530/Erc-11-0212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L. & Ambros V. The C-Elegans Heterochronic Gene Lin-4 Encodes Small Rnas with Antisense Complementarity to Lin-14. Cell 75, 843–854, Doi 10.1016/0092-8674(93)90529-Y (1993). [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I. & Ruvkun G. Posttranscriptional Regulation of the Heterochronic Gene Lin-14 by Lin-4 Mediates Temporal Pattern-Formation in C-Elegans. Cell 75, 855–862, Doi 10.1016/0092-8674(93)90530-4 (1993). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 136, 215–233, DOI 10.1016/j.cell.2009.01.002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres S. L. & Zamore P. D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488, 10.1038/nrm3611 (2013). [DOI] [PubMed] [Google Scholar]
- Ling H., Fabbri M. & Calin G. A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 12, 847–865, 10.1038/nrd4140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A. & Croce C. M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866, 10.1038/nrc1997 (2006). [DOI] [PubMed] [Google Scholar]
- He L. et al. A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134, 10.1038/nature05939 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. M. et al. RAS is regulated by the let-7 MicroRNA family. Cell 120, 635–647, DOI 10.1016/j.cell.2005.01.014 (2005). [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y. C. & Steitz J. A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 318, 1931–1934, DOI 10.1126/science.1149460 (2007). [DOI] [PubMed] [Google Scholar]
- Tay Y., Rinn J. & Pandolfi P. P. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352, 10.1038/nature12986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038, 10.1038/nature09144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M. et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 109, E2110–2116, 10.1073/pnas.1209414109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S. M. et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835, 10.1038/nn.3113 (2012). [DOI] [PubMed] [Google Scholar]
- Dvinge H. et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497, 378–382, 10.1038/nature12108 (2013). [DOI] [PubMed] [Google Scholar]
- He H. et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. U. S. A. 102, 19075–19080, 10.1073/pnas.0509603102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P. et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer 13, 497–508, 10.1677/erc.1.01209 (2006). [DOI] [PubMed] [Google Scholar]
- Chou C. K. et al. Prognostic Implications of miR-146b Expression and Its Functional Role in Papillary Thyroid Carcinoma. J. Clin. Endocrinol. Metab. 98, E196–E205, 10.1210/jc.2012-2666 (2013). [DOI] [PubMed] [Google Scholar]
- Menon M. P. & Khan A. Micro-RNAs in thyroid neoplasms: molecular, diagnostic and therapeutic implications. J. Clin. Pathol. 62, 978–985, 10.1136/jcp.2008.063909 (2009). [DOI] [PubMed] [Google Scholar]
- Zaman N. et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell Rep 5, 216–223, 10.1016/j.celrep.2013.08.028 (2013). [DOI] [PubMed] [Google Scholar]
- Broad Institute TCGA Genome Data Analysis Center (2014): SNP6 Copy number analysis (GISTIC2). Broad Institute of MIT and Harvard. doi:10.7908/C1M61HZV.
- Grimson A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105, 10.1016/j.molcel.2007.06.017 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Koppal A., Agius P., Sander C. & Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90, 10.1186/gb-2010-11-8-r90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo M. V., Yamashita A. S. & Kimura E. T. MicroRNA miR-146b-5p regulates signal transduction of TGF-beta by repressing SMAD4 in thyroid cancer. Oncogene 31, 1910–1922, 10.1038/onc.2011.381 (2012). [DOI] [PubMed] [Google Scholar]
- Lee D. Y. et al. Expression of versican 3′-untranslated region modulates endogenous microRNA functions. PLoS One 5, e13599, 10.1371/journal.pone.0013599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutnam Z. J. & Yang B. B. The non-coding 3′ UTR of CD44 induces metastasis by regulating extracellular matrix functions. J. Cell Sci. 125, 2075–2085, 10.1242/jcs100818 (2012). [DOI] [PubMed] [Google Scholar]
- Fang L. et al. Versican 3′-untranslated region (3′-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 27, 907–919, 10.1096/fj.12-220905 (2013). [DOI] [PubMed] [Google Scholar]
- Gasbarri A. et al. Detection and molecular characterisation of thyroid cancer precursor lesions in a specific subset of Hashimoto's thyroiditis. Br. J. Cancer 91, 1096–1104, 10.1038/sj.bjc.6602097 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M. L., Huang Y., Pellegata N. S., de la Chapelle A. & Kloos R. T. Hashimoto's thyroiditis with papillary thyroid carcinoma (PTC)-like nuclear alterations express molecular markers of PTC. Histopathology 45, 39–46, 10.1111/j.1365-2559.2004.01876.x (2004). [DOI] [PubMed] [Google Scholar]
- Boutzios G. et al. Higher incidence of tall cell variant of papillary thyroid carcinoma in graves' disease. Thyroid 24, 347–354, 10.1089/thy.2013.0133 (2014). [DOI] [PubMed] [Google Scholar]
- Ou W. B., Zhu M. J., Demetri G. D., Fletcher C. D. & Fletcher J. A. Protein kinase C-theta regulates KIT expression and proliferation in gastrointestinal stromal tumors. Oncogene 27, 5624–5634, 10.1038/onc.2008.177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. et al. HLA-DMB expression by thyrocytes: indication of the antigen-processing and possible presenting capability of thyroid cells. Clin. Exp. Immunol. 116, 62–69 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahat M. A., Chernichovski I. & Lahat N. Increased binding of IFN regulating factor 1 mediates the synergistic induction of CIITA by IFN-gamma and tumor necrosis factor-alpha in human thyroid carcinoma cells. Int. Immunol. 13, 1423–1432 (2001). [DOI] [PubMed] [Google Scholar]
- Shah R. et al. Intense expression of the b7-2 antigen presentation coactivator is an unfavorable prognostic indicator for differentiated thyroid carcinoma of children and adolescents. J. Clin. Endocrinol. Metab. 87, 4391–4397, 10.1210/jc.2002-011262 (2002). [DOI] [PubMed] [Google Scholar]
- Lin H. Y. et al. Potentiation by thyroid hormone of human IFN-gamma-induced HLA-DR expression. J. Immunol. 161, 843–849 (1998). [PubMed] [Google Scholar]
- Qiu Y. et al. Immunoglobulin G expression and its colocalization with complement proteins in papillary thyroid cancer. Mod. Pathol. 25, 36–45, 10.1038/modpathol.2011.139 (2012). [DOI] [PubMed] [Google Scholar]
- Takaoka A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505, 10.1038/nature06013 (2007). [DOI] [PubMed] [Google Scholar]
- Hebert C., Norris K., Scheper M. A., Nikitakis N. & Sauk J. J. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer 6, 5, 10.1186/1476-4598-6-5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alajez N. M. et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 1, e85, 10.1038/cddis.2010.64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson E. M. & Tomer Y. The genetic basis of thyroid autoimmunity. Thyroid 17, 949–961, 10.1089/thy.2007.0153 (2007). [DOI] [PubMed] [Google Scholar]
- Cerutti J. M. et al. Molecular profiling of matched samples identifies biomarkers of papillary thyroid carcinoma lymph node metastasis. Cancer Res. 67, 7885–7892, 10.1158/0008-5472.CAN-06-4771 (2007). [DOI] [PubMed] [Google Scholar]
- Puxeddu E. et al. RET/PTC-induced gene expression in thyroid PCCL3 cells reveals early activation of genes involved in regulation of the immune response. Endocr. Relat. Cancer 12, 319–334, 10.1677/erc.1.00947 (2005). [DOI] [PubMed] [Google Scholar]
- Barbosa M. D. et al. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 382, 262–265, 10.1038/382262a0 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faigle W. et al. Deficient peptide loading and MHC class II endosomal sorting in a human genetic immunodeficiency disease: the Chediak-Higashi syndrome. J. Cell Biol. 141, 1121–1134 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K. M., Brown R. A., Epis M. R., Kalinowski F. C. & Leedman P. J. miRNA-7-5p inhibits melanoma cell migration and invasion. Biochem. Biophys. Res. Commun. 430, 706–710, 10.1016/j.bbrc.2012.11.086 (2013). [DOI] [PubMed] [Google Scholar]
- Sarver A. L., Li L. & Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 70, 9570–9580, 10.1158/0008-5472.CAN-10-2074 (2010). [DOI] [PubMed] [Google Scholar]
- Arora S. et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int. J. Cancer 129, 2621–2631, 10.1002/ijc.25939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskwa P. et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 41, 210–220, 10.1016/j.molcel.2010.12.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. et al. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J. Immunol. 185, 7244–7251, 10.4049/jimmunol.1001573 (2010). [DOI] [PubMed] [Google Scholar]
- Kozomara A. & Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–157, 10.1093/nar/gkq1027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information