A. fumigatus cyp51A–mediated resistance to azole drugs is rare in the United States.
Keywords: antifungal resistance, azole, azole resistance, fungi, Aspergillus fumigatus, CYP51A, TR34/L98H, susceptibility testing, passive surveillance, United States
Abstract
Emergence of Aspergillus fumigatus strains containing mutations that lead to azole resistance has become a serious public health threat in many countries. Nucleotide polymorphisms leading to amino acid substitutions in the lanosterol demethylase gene (cyp51A) are associated with reduced susceptibility to azole drugs. The most widely recognized mutation is a lysine to histidine substitution at aa 98 (L98H) and a duplication of the untranscribed promoter region, together known as TR34/L98H. This mechanism of resistance has been reported in Europe, Asia, and the Middle East, and is associated with resistance to all azole drugs and subsequent treatment failures. To determine whether isolates with this mutation are spreading into the United States, we conducted a passive surveillance–based study of 1,026 clinical isolates of A. fumigatus from 22 US states during 2011–2013. No isolates harboring the TR34/L98H mutation were detected, and MICs of itraconazole were generally low.
Azole antifungal drugs are the first line of therapy against Aspergillus fumigatus, a common etiologic agent of aspergillosis. These drugs are used as empirical prophylaxis and as targeted therapy for invasive aspergillosis. Resistance to azole drugs has been associated with treatment failure and deaths in patients with aspergillosis. In the past 2 decades, azole resistance in A. fumigatus has been documented in many regions (1–7). The high prevalence of azole resistance in A. fumigatus has prompted the European Centre for Disease Prevention and Control to increase the risk level of this organism to a public health risk (8).
A common mechanism that confers resistance to azole drugs is a mutation in the lanosterol 14 α-demethylase gene that encodes the CYP51A protein. This protein is the primary target of azole drugs and sterol demethylation inhibitor (DMI) fungicides (9–11). However, not all cyp51A mutations contribute to azole resistance (some are benign), and not all azole resistance is caused by mutations in the cyp51A gene (12).
In the past decade, novel cyp51A promoter duplication mutations, especially the cooperative pair of cyp51A mutations known as TR34/L98H, have emerged as a predominant azole-resistance mechanism in A. fumigatus (1,4,6,10,11,13–22). Isolates harboring the TR34/L98H mutation are cross-resistant to multiple azoles (4,15,17). A second cyp51A-promoter duplication genotype, the recently discovered TR46/Y121F+T289A, also displays high tolerance for voriconazole (13,14). Both genotypes are associated with in vitro MICs that exceed by multiple dilutions the established epidemiologic cutoff value (ECV) of ≤1 μg/mL published by the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute members for medical azole agents (9,13). Moreover, these mutations often contribute to failure of azole therapy in aspergillosis patients (13–23). Their presence rules out the use of voriconazole, the treatment recommended by the Infectious Diseases Society of America for aspergillosis, and leaves amphotericin B, an antifungal drug with toxic side effects, as the primary therapy choice.
Since the discovery of the TR34/L98H mutation in the Netherlands by Verweij et al. in 2007 (15), other countries in Europe, including Austria, Belgium, Germany, Spain, France, the United Kingdom, Denmark, and Norway, have also reported isolates harboring TR34/L98H (5,7,16–20,24). A. fumigatus isolates with the TR34/L98H mutation have also been reported in China, India, and Iran (1,4,6,21). Because of rapid increase in detection of the TR34/L98H mutation in many regions and its adverse effect on patient management, isolates harboring this mutation pose a serious public health threat. Isolates bearing the TR34/L98H mutation have not been documented in United States, but given the number of aspergillosis cases in this country, its presence could pose a serious public health threat, as it does in Europe.
The TR34/L98H mutation is not associated with use of long-term medical azole therapy in patients. Instead, prolonged exposure of the fungus to sterol DMI fungicides in the environment, in conjunction with sexual reproduction, has probably led to dissemination of this mutation (19,25). As in many countries in which this genotype has been reported, DMI fungicides are used in various agricultural practices across the United States. According to the US Department of Agriculture data for 2010–2013, DMI fungicides, such as tebuconazole, propiconazole, prothioconazole, tetraconazole, metconazole, and epoxiconazole, have been used by several agricultural producers (26). This shared agriculture practice indicates that the TR34/L98H mutation could potentially arise in the United States.
Thus, the Centers for Disease Control and Prevention initiated passive surveillance for A. fumigatus isolates in 2011 to identify resistance to the representative azole itraconazole. More than 1,000 clinical isolates from 22 states were screened by using an itraconazole antifungal plate assay and the Etest to measure itraconazole MICs. For isolates that require an MIC greater than the ECV, DNA sequence analysis of the cyp51A gene did not yield any isolates with the TR34/L98H genotype.
Materials and Methods
Fungal Isolates
A. fumigatus isolates were obtained through the American Society for Microbiology listservs (DivC) and (ClinMicroNet) (http://www.asm.org/index.php/online-community-groups/listservs) and through the US Association of Public Health Laboratories. The request for isolates read as follows: “To determine the US level of A. fumigatus azole resistance, we are requesting submission of: 1) All new or stored isolates of Aspergillus fumigatus from US collections; 2) Any new isolates of A. fumigatus regardless of clinical relevance.” Isolates were sent to the Centers for Disease Control and Prevention where the species was confirmed by colony color and morphologic features observed by microscopy. Cryptic species that required increased MICs were subsequently ruled out by DNA sequencing of the cyp51A gene as described below.
Itraconazole Susceptibility Assay
All isolates were screened by using 2 methods: antifungal plate culture and itraconazole Etest (bioMérieux, Marcy l’Etoile, France). For plate culture, a 24-well culture plate containing 1 mL of Sabouraud agar supplemented with 4 μg/mL of itraconazole was used. Twenty microliters of a suspension containing ≈ 2 × 104 conidia were transferred to each well and allowed to dry at ambient temperature for 30 min before incubation. Etest determination of itraconazole MICs was performed as described by Pfaller et al. (27) with minor modifications. Isolates of A. fumigatus were allowed to sporulate on potato dextrose agar slants for 5–7 days. Conidial suspensions were prepared in sterile distilled water containing 1% Tween-20. The concentration was adjusted to an optical density of 0.09–0.13 or ≈106 conidia/mL equivalent at 530 nm. An RPMI 1640 agar plate was inoculated by streaking a conidia-laden cotton swab bidirectionally across the surface of the plate. An itraconazole Etest strip was placed across the central surface of the freshly inoculated RPMI plate. An azole-resistant and an azole-susceptible isolate for which broth microdilution azole MICs were known were used as controls (1). The Etest and itraconazole plates were incubated at 37°C, and results were recorded 48 hours postinoculation.
Genetic Profiling of cyp51A
DNA sequence analysis of the cyp51A gene and its promoter region was performed for all isolates that required an MIC above the ECV (1 μg/mL) by using primers described (1). A subset of the first 561 isolates was also examined by using a mixed-format real-time PCR to detect nucleotide polymorphisms that cause single amino acid substitutions at Gly54, Leu98, Gly138, and Met220, and the recently discovered TR46/Y121F/T289A in the CYP51A protein as described by Klaassen et al. (28).
A. fumigatus genomic DNA was purified by using the DNeasy blood and tissue kit (QIAGEN, Valencia, CA, USA) with some modifications. Hyphae were transferred to a 1.5-mL microcentrifuge tube containing 360 μL of Z-buffer (29) (60 mmol/L Na2HPO4, 40 mmol/L NaH2PO4, 10 mmol/L KCl, 1 mmol/L MgSO4, 38 mmol/L β-mercaptoethanol, pH 7) and 40 μL of proteinase K (QIAGEN). A total of 250 mg of 0.5-mm zirconia beads (BioSpec, Bartlesville, OK, USA) was added to the cell suspension, and cells were homogenized for 1 min at maximum speed by using a mini-beadbeater (BioSpec). Four hundred microliters of AL buffer (QIAGEN) was added to the cell lysate and mixed by vortexing for 10 s. The lysate was then centrifuged at 13,200 rpm for 5 min. The supernatant was transferred to a clean 1.5-mL microcentrifuge tube, and an equal volume of absolute ethanol was added to the supernatant. After briefly undergoing vortexing, the DNA suspension was transferred to a spin column (QIAGEN). DNA binding, washes, and elution steps were followed as described by the manufacturer.
Results
Characteristics of A. fumigatus Isolates
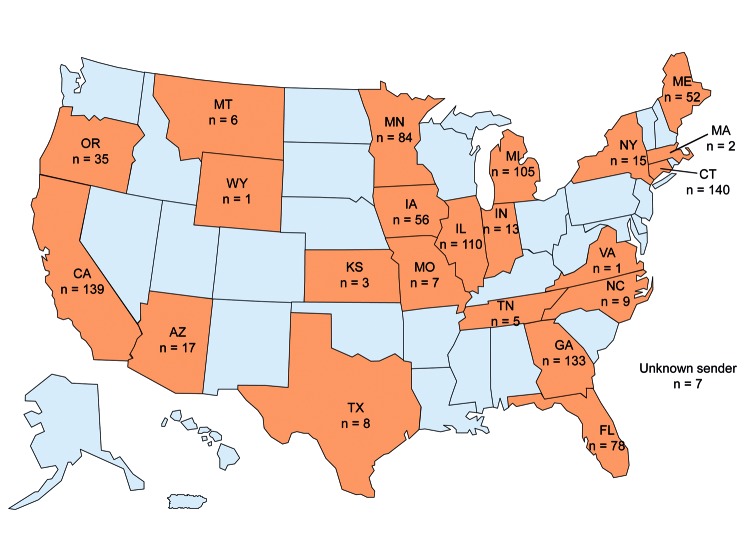
A request for A. fumigatus clinical isolates collected beginning in 2011 was initiated in June 2011. Because the TR34/L98H mutation was believed to be environmentally induced, all A. fumigatus clinical isolates were accepted, not just those causing infection. A total of 1,026 clinical isolates were obtained during September 2011–September 2013 from hospitals, clinics, and state public health laboratories across 22 states (Figure 1), including Arizona (n = 17), California (n = 139), Connecticut (n = 140), Florida (n = 78), Georgia (n = 133), Iowa (n = 56), Illinois (n = 111), Indiana (n = 13), Kansas (n = 3), Massachusetts (n = 2), Maine (n = 52), Michigan (n = 105), Minnesota (n = 84), Missouri (n = 7), Montana (n = 6), North Carolina (n = 9), New York (n = 15), Oregon (n = 35), Tennessee (n = 5), Texas (n = 8), Virginia (n = 1), Wyoming (n = 1), and unknown (n = 7). The isolates were form respiratory tracts (≈57%), ears (≈4%), other tissues (≈5%), and unknown sources (≈34%). Of respiratory tract isolates, 65% were collected from sputum and 25% from bronchoalveolar lavage specimens.
Figure 1.
Distribution of Aspergillus fumigatus isolates, United States, 2011–2013. A total of 1,026 clinical isolates were received from 22 states during October 2011–October 2013.
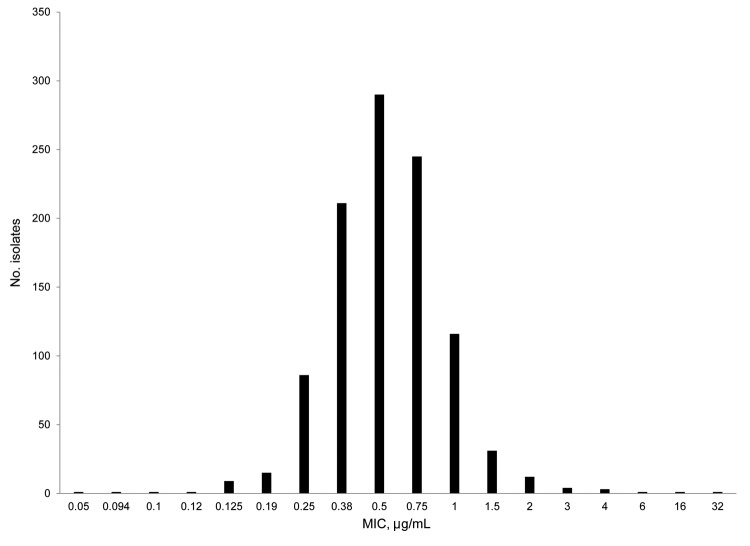
Most A. fumigatus isolates were susceptible to itraconazole. Historically, the TR34/L98H mutation has been detected by itraconazole plate assay. We also used the Etest validate its utility for detecting high MICs. The same 3 isolates were identified by their growth on the itraconazole plate and an itraconazole Etest MIC >4 μg/mL, which indicated congruence for the 2 tests. Overall MICs ranged from 0.05 μg/mL to 32 μg/mL, and 95% of the isolates required an MIC ≤1 μg/mL) (Figure 2). The 50% MIC (MIC50,), MIC90, and MIC mode for isolates were 0.5 μg/mL, 1.0 μg/mL, and 0.5 μg/mL, respectively. Of 51 isolates that required an increased MIC for itraconazole, >94% (n = 48) required an MIC <4 μg/mL and were negative by agar screening, and 3 isolates required MICs of 6 μg/mL (n = 1), 16 μg/mL (n = 1), or ≥32 μg/mL (n = 1), the maximum value on the Etest strip.
Figure 2.
Itraconazole susceptibility profile for Aspergillus fumigatus isolates, United States, 2011–2013. The MIC (μg/mL) required by each isolate was determined by using the Etest method. Approximately 5% of the isolates require an MIC higher than the established epidemiologic cutoff value of 1 μg/mL.
Genetic Analysis of the cyp51A Gene
DNA sequence analysis of the cyp51A gene was performed for 51 isolates that required an itraconazole MIC >1 μg/mL and for 36 isolates that required an itraconazole MIC ≤1 μg/mL. The duplication mutations (TR34 and TR46) in the promoter region of cyp51A and the L98H or Y121F/T289A substitutions in cyp51A were not detected. Eighteen (35%) of the 51 isolates that required an increased MIC had a cyp51A mutation. The most common mutation in this group was a novel valine→isoleucine substitution at residue 242 (I242V), which was found in 13 of the isolates that required increased MICs and 2 of the control isolates; all required MICs ≤4 μg/mL. There was also a set of 4 linked mutations in 2 of the control isolates that are commonly seen in susceptible isolates (1,3). The only mutation found that has been associated with azole treatment failure was an M220I mutation in the isolate that required an MIC ≥32 μg/mL. A mixed-format real-time PCR was also used to screen the first 561 isolates collected for other known resistance-inducing cyp51A single nucleotide polymorphisms. Except for the M220I mutation, no other polymorphisms were identified by using this method.
Discussion
Azoles, specifically voriconazole and (in some instances) itraconazole, are the recommended treatment for aspergillosis (30). These drugs have a high potency against the most common causes of aspergillosis (i.e., A. fumigatus, A. niger, A. flavus, and A. terreus). However, reduced susceptibility to these drugs has been reported (3,7,15,18). Many triazole-resistant isolates from aspergillosis patients who showed treatment failure with triazole therapy have mutations within the CYP51A protein (31–35), but this is not the only mechanism of resistance. The ECV was established to define the limits of the wild-type distribution of MICs. It does not define a breakpoint but it may help identify isolates harboring mutations. In A. fumigatus, 2%–20% of isolates, depending on the country, require MICs above the established ECVs for azoles (2–4,31,32,36). In this study, ≈5% of the isolates required an MIC greater than the established ECV for itraconazole. Isolates with no cyp51A mutations but which require high azole MICs are not uncommon. The mechanisms of resistance in these isolates are not precisely known but are assumed to be changes in drug transporter pumps (37,38).
Azole resistance is typically associated with cyp51A mutations, especially the TR34/L98H and TR46/Y121F/T289A dual mutations. A total of 26%–94% of azole-resistant isolates, especially in the Netherlands, have these mutations (1,15,20,31). However, we found no isolates harboring the TR34/L98H mutation in this study. One isolate was found to have an amino acid substitution at M220 and an itraconazole MIC ≥32 μg/mL. Amino acid substitutions at the M220 residue in A. fumigatus have been shown to confer resistance to all 3 mold-active azoles and are known to be induced by prolonged use of azoles in individual patients (39). Another common mutation that was discovered in isolates that required increased MICs for itraconazole is I242V. The MIC required for isolates with this mutation ranges from 1 μg/mL to 4 μg/mL. There have been no reports linking this mutation to azole resistance and a relationship between this mutation and azole resistance has not been demonstrated experimentally. The mechanism causing increased MICs for other isolates is not known, a common problem for researchers working on drug resistance in Aspergillus spp. (3,32).
Genetic mutations in A. fumigatus cyp51A are believed to be associated with exposure to azole compounds (19,34,37,39). The TR34/L98H and TR46/Y121F/T289A mutations specifically are believed to have originated from exposure to DMI fungicides in the environment (10,13,14,25). According to the US Department of Agriculture, agricultural producers in the United States use a lower tonnage of DMI fungicides than their counterparts in Europe and Asia (26). During 2010–2012, combined DMI fungicide use from states that provided isolates for this study was 381,018 kg. Of these states, California was the highest user (84,051 kg). According to the European Centre for Disease Prevention and Control, the United Kingdom used 271,124 kg of DMI fungicides during 2006–2009 (8). Therefore, the observed low rate of cyp51A mutation–dependent resistance, specifically the absence of TR34/L98H and TR46/Y121F/T289A mutations in this study than in studies from countries in Europe, Asia, and the Middle East, might be caused by differences in the extent of A. fumigatus exposure to DMI fungicides. The low rate of resistance in A. fumigatus shown is supported by results of other smaller studies in the United States (2,40).
There were several limitations to our study. First, isolates were collected passively. Some areas of the United States were overrepresented and others, such as the Midwest, were underrepresented. If the TR34/L98H mutation is found only in isolates from specific localities in the United States, it might have been missed during this surveillance. Second, all isolates were accepted. If surveillance was aimed specifically at isolates from patients who showed therapy failure, there may have been few isolates but those received might have demonstrated a greater prevalence of cyp51A mutations. Third, because isolates were received without personal identifiers and little meta data, there was no way of knowing whether multiple isolates came from 1 person.
In conclusion, this surveillance study indicates that the TR34/L98H mutant A. fumigatus that is now found throughout Europe has not yet emerged in the United States. Approximately 5% of A. fumigatus isolates from the United States required increased MICs for itraconazole, but most isolates did not have a detectable genetic mutation. In light of increasing reports of TR34/L98H-mediated and other cyp51A-mediated drug resistance in Europe and Asia, further surveillance is warranted.
Acknowledgments
We thank Mary Brandt for critically reviewing the manuscript; Shirley McClinton for maintaining the collection of isolates; Joyce Peterson for helping with identification of isolates; and Holly Alexander, Richard Alexander, Beth Albaugh, Sonia Allen, Cydni Boyd, Adrienne Brudie, Suc Boolagangoor, Scott Matushelr, Eileen Burd, Pat Cernoch, Vishnu Chaturvedi, Mary Beth Cook, Michael Costello, Chari Delatore, Rolinda Eddings, Jasmine Estrada, Monica Ferraro, Diane Getsinger, Minoo Ghajar, Paula Gibbs, Joy Henderson, Si Intravichit, Robert Jerris, Inna Kamlet, Peggy Mahlmeister, Paulette Medina, Susan Novak-Weekley, Wanda Petty, Joslyn Pribble, John Radosevic, Douglas Landau, Edward Zollars, Donna Hanah, Eldon Wecler, David Schrock, Irene Ratkiewicz, Linda Roller, Audrey Schuetz, Brenda Sellers, Beth Shade, Susan Sharp, Yolanda Awalwell, Charise Tanner, Natalie Thompson, Richard Thompson, Robert Thompson, Dana Towle, Mark Lewis, Susan Vergara, and Jason Wickstrum for providing isolates.
This study was supported by the Office of Antimicrobial Resistance at the Centers for Disease Control and Prevention.
Biography
Dr Pham is an associate research fellow at the Mycotic Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Georgia. His research interests include molecular genotyping and antifungal resistance mechanisms.
Footnotes
Suggested citation for this article: Pham CD, Reiss E, Hagen F, Meis JR, Lockhart SR. Passive surveillance for azole-resistant Aspergillus fumigatus, United States, 2011–2013. Emerg Infect Dis [Internet]. 2014 Sep [date cited]. http://dx.doi.org/10.3201/eid2009.140142
References
- 1.Lockhart SR, Frade JP, Etienne KA, Pfaller MA, Diekema DJ, Balajee SA. Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance study is primarily due to the TR/L98H mutation in the cyp51A gene. Antimicrob Agents Chemother. 2011;55:4465–8. 10.1128/AAC.00185-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baddley JW, Marr KA, Andes DR, Walsh TJ, Kauffman CA, Kontoyiannis DP, et al. Patterns of susceptibility of Aspergillus isolates recovered from patients enrolled in the Transplant-Associated Infection Surveillance Network. J Clin Microbiol. 2009;47:3271–5. 10.1128/JCM.00854-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bueid A, Howard SJ, Moore CB, Richardson MD, Harrison E, Bowyer P, et al. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J Antimicrob Chemother. 2010;65:2116–8. 10.1093/jac/dkq279 [DOI] [PubMed] [Google Scholar]
- 4.Chowdhary A, Kathuria S, Randhawa HS, Gaur SN, Klaassen CH, Meis JF. Isolation of multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR/L98H mutations in the cyp51A gene in India. J Antimicrob Chemother. 2012;67:362–6. 10.1093/jac/dkr443 [DOI] [PubMed] [Google Scholar]
- 5.Mortensen KL, Mellado E, Lass-Flörl C, Rodriguez-Tudela JL, Johansen SK, Arendrup MC. Environmental study of azole-resistant Aspergillus fumigatus and other aspergilli in Austria, Denmark, and Spain. Antimicrob Agents Chemother. 2010;54:4545–9. 10.1128/AAC.00692-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seyedmousavi S, Hashemi SJ, Zibafar E, Hedayati MT, Mouton JW, Melchers WJ, et al. Azole-resistant Aspergillus fumigatus, Iran. Emerg Infect Dis. 2013;19:832–4. 10.3201/eid1905.130075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgel PR, Baixench MT, Amsellem M, Audureau E, Chapron J, Kanaan R, et al. High prevalence of azole-resistant Aspergillus fumigatus in adults with cystic fibrosis exposed to itraconazole. Antimicrob Agents Chemother. 2012;56:869–74. 10.1128/AAC.05077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control. Risk assessment on the impact of environmental usage of triazoles on the development and spread of resistance to medical triazoles in Aspergillus species [cited 2013 Dec 1]. http://www.ecdc.europa.eu/en/publications/_layouts/forms/Publication_DispForm.aspx?ID=823&List=4f55ad51%2D4aed%2D4d32%2Db960%2Daf70113dbb90
- 9.Pfaller MA, Diekema DJ, Ghannoum MA, Rex JH, Alexander BD, Andes D, et al. Wild-type MIC distribution and epidemiological cutoff values for Aspergillus fumigatus and three triazoles as determined by the Clinical and Laboratory Standards Institute broth microdilution methods. J Clin Microbiol. 2009;47:3142–6. 10.1128/JCM.00940-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhary A, Kathuria S, Xu J, Meis JF. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013;9:e1003633. 10.1371/journal.ppat.1003633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pham CD, Lockhart SR. An invisible threat: mutation-mediated resistance to triazole drugs in Aspergillus. Current Fungal Infection Reports. 2012;6:288–95. [DOI] [PMC free article] [PubMed]
- 12.Escribano P, Recio S, Pelaez T, Bouza E, Guinea J. Aspergillus fumigatus strains with mutations in the cyp51a gene do not always show phenotypic resistance to itraconazole, voriconazole, or posaconazole. Antimicrob Agents Chemother. 2011;55:2460–2. 10.1128/AAC.01358-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Linden JW, Camps SM, Kampinga GA, Arends JP, Debets-Ossenkopp Y, Haas PJ, et al. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin Infect Dis. 2013;57:513–20. 10.1093/cid/cit320 [DOI] [PubMed] [Google Scholar]
- 14.Chowdhary A, Sharma C, Kathuria S, Hagen F, Meis JF. Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India. [Epub ahead of print]. J Antimicrob Chemother. 2014;69:555–7. [DOI] [PubMed]
- 15.Verweij PE, Mellado E, Melchers WJ. Multiple-triazole-resistant aspergillosis. N Engl J Med. 2007;356:1481–3. 10.1056/NEJMc061720 [DOI] [PubMed] [Google Scholar]
- 16.Denning DW, Park S, Lass-Flörl C, Fraczek MG, Kirwan M, Gore R, et al. High-frequency triazole resistance found in non-culturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis. 2011;52:1123–9. 10.1093/cid/cir179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morio F, Aubin GG, Danner-Boucher I, Haloun A, Sacchetto E, Garcia-Hermoso D, et al. High prevalence of triazole resistance in Aspergillus fumigatus, especially mediated by TR/L98H, in a French cohort of patients with cystic fibrosis. J Antimicrob Chemother. 2012;67:1870–3. 10.1093/jac/dks160 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Tudela JL, Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Monzon A, Cuenca-Estrella M, et al. Epidemiological cutoffs and cross-resistance to azole drugs in Aspergillus fumigatus. Antimicrob Agents Chemother. 2008;52:2468–72. 10.1128/AAC.00156-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camps SM, Rijs AJ, Klaassen CH, Meis JF, O'Gorman CM, Dyer PS, et al. Molecular epidemiology of Aspergillus fumigatus isolates harboring the TR34/L98H azole resistance mechanism. J Clin Microbiol. 2012;50:2674–80. 10.1128/JCM.00335-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortensen KL, Jensen RH, Johansen HK, Skov M, Pressler T, Howard SJ, et al. Aspergillus species and other molds in respiratory samples from patients with cystic fibrosis: a laboratory-based study with focus on Aspergillus fumigatus azole resistance. J Clin Microbiol. 2011;49:2243–51. 10.1128/JCM.00213-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Badali H, Vaezi A, Haghani I, Yazdanparast SA, Hedayati MT, Mousavi B, et al. Environmental study of azole-resistant Aspergillus fumigatus with TR34/L98H mutations in the cyp51A gene in Iran. Mycoses. 2013;56:659–63. 10.1111/myc.12089 [DOI] [PubMed] [Google Scholar]
- 22.Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, Samson RA, et al. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008;5:e219. 10.1371/journal.pmed.0050219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denning DW, Venkateswarlu K, Oakley KL, Anderson MJ, Manning NJ, Stevens DA, et al. Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 1997;41:1364–8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader O, Weig M, Reichard U, Lugert R, Kuhns M, Christner M, et al. cyp51A-Based mechanisms of Aspergillus fumigatus azole drug resistance present in clinical samples from Germany. Antimicrob Agents Chemother. 2013;57:3513–7. 10.1128/AAC.00167-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snelders E, Camps SM, Karawajczyk A, Schaftenaar G, Kema GH, van der Lee HA, et al. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS ONE. 2012;7:e31801. 10.1371/journal.pone.0031801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.United States Department of Agriculture. Agricultural chemical usage [cited 2013 Nov 1]. http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do;jsessionid=39589A84B623FFB18D319BF97277F79C?documentID=1560
- 27.Pfaller JB, Messer SA, Hollis RJ, Diekema DJ, Pfaller MA. In vitro susceptibility testing of Aspergillus spp.: comparison of Etest and reference microdilution methods for determining voriconazole and itraconazole MICs. J Clin Microbiol. 2003;41:1126–9. 10.1128/JCM.41.3.1126-1129.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klaassen CH, de Valk HA, Curfs-Breuker IM, Meis JF. Novel mixed-format real-time PCR assay to detect mutations conferring resistance to triazoles in Aspergillus fumigatus and prevalence of multi-triazole resistance among clinical isolates in the Netherlands. J Antimicrob Chemother. 2010;65:901–5. 10.1093/jac/dkq041 [DOI] [PubMed] [Google Scholar]
- 29.Pedley KF, Walton JD. Regulation of cyclic peptide biosynthesis in a plant pathogenic fungus by a novel transcription factor. Proc Natl Acad Sci U S A. 2001;98:14174–9. 10.1073/pnas.231491298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, et al. Treatment of aspergillosis: clinical practice guidelines of the Infectious Disease Society of America. Clin Infect Dis. 2008;46:327–60. 10.1086/525258 [DOI] [PubMed] [Google Scholar]
- 31.van der Linden JW, Snelders E, Kampinga GA, Rijnders BJ, Mattsson E, Debets-Ossenkopp YJ, et al. Clinical implications of azole resistance in Aspergillus fumigatus, the Netherlands, 2007–2009. Emerg Infect Dis. 2011;17:1846–54. 10.3201/eid1710.110226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard SJ, Cerar D, Anderson MJ, Albarrag A, Fisher MC, Pasqualotto AC, et al. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis. 2009;15:1068–76. 10.3201/eid1507.090043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snelders E, Karawajczyk A, Schaftenaar G, Verweij PE, Melchers WJ. Azole resistance profile of amino acid changes in Aspergillus fumigatus cyp51A based on protein homology modeling. Antimicrob Agents Chemother. 2010;54:2425–30. 10.1128/AAC.01599-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Li H, Li R, Bu D, Wan Z. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J Antimicrob Chemother. 2005;55:31–7. 10.1093/jac/dkh507 [DOI] [PubMed] [Google Scholar]
- 35.Mellado E, Garcia-Effron G, Alcazer-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. Substitutions at methionine 220 in the 14alpha-sterol demethylase (cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob Agents Chemother. 2004;48:2747–50. 10.1128/AAC.48.7.2747-2750.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tashiro M, Izumikawa K, Minematsu A, Hirano K, Iwanaga N, Ide S, et al. Antifungal susceptibilities of Aspergillus fumigatus clinical isolates obtained in Nagasaki, Japan. Antimicrob Agents Chemother. 2012;56:584–7. 10.1128/AAC.05394-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slaven JW, Anderson MJ, Sanglard D, Dixon GK, Bille J, Roberts IS, et al. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet Biol. 2002;36:199–206 . 10.1016/S1087-1845(02)00016-6 [DOI] [PubMed] [Google Scholar]
- 38.Bowyer P, Mosquera J, Anderson M, Birch M, Bromley M, Denning DW. Identification of novel genes conferring altered azole susceptibility in Aspergillus fumigatus. FEMS Microbiol Lett. 2012;332:10–9. 10.1111/j.1574-6968.2012.02575.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.da Silva Ferreira ME, Capellaro JL, dos Res Marques E, Malavazi I, Perlin D, Park S, et al. In vitro evolution of itraconazole resistance in Aspergillus fumigatus involves multiple mechanisms of resistance. Antimicrob Agents Chemother. 2004;48:4405–13. 10.1128/AAC.48.11.4405-4413.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaller MA, Messer SA, Woosley LN, Jones RN, Castanheira M. Echinocandin and triazole antifungal susceptibility profiles for clinical opportunistic yeast and mold isolates collected from 2010 to 2011: application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic and temporal trends of antifungal resistance. J Clin Microbiol. 2013;51:2571–81 . 10.1128/JCM.00308-13 [DOI] [PMC free article] [PubMed] [Google Scholar]