Summary
The Brahma (SMARCA2) gene product, a cofactor of retinoblastoma (RB), is consistently upregulated by flavonoids and is required for flavonoid-mediated growth arrest and inhibition of tumor development in mice. Flavonoids convert RB into its hypophosporylated state, thereby activating it as well as inducing BRM, and together they function to inhibit growth.
Abstract
Flavonoids have been extensively studied and are well documented to have anticancer effects, but it is not entirely known how they impact cellular mechanisms to elicit these effects. In the course of this study, we found that a variety of different flavonoids readily restored Brahma (BRM) in BRM-deficient cancer cell lines. Flavonoids from each of the six different structural groups were effective at inducing BRM expression as well as inhibiting growth in these BRM-deficient cancer cells. By blocking the induction of BRM with shRNA, we found that flavonoid-induced growth inhibition was BRM dependent. We also found that flavonoids can restore BRM functionality by reversing BRM acetylation. In addition, we observed that an array of natural flavonoid-containing products both induced BRM expression as well as deacetylated the BRM protein. We also tested two of the BRM-inducing flavonoids (Rutin and Diosmin) at both a low and a high dose on the development of tumors in an established murine lung cancer model. We found that these flavonoids effectively blocked development of adenomas in the lungs of wild-type mice but not in that of BRMnull mice. These data demonstrate that BRM expression and function are regulated by flavonoids and that functional BRM appears to be a prerequisite for the anticancer effects of flavonoids both in vitro and in vivo.
Introduction
The Brahma (BRM) protein is a key catalytic subunit of the SWI/SNF complex, and together they function to mediate gene expression. This complex is recruited by a variety of transcription factors and key cellular proteins, such as retinoblastoma (RB) and p53, to specific promoters and DNA regions where SWI/SNF complexes function to shift the position of histones and facilitate gene expression (1,2). Thus, the SWI/SNF complex does not act in only a single signal transduction pathway, but rather it has a broad scope of action that impacts the function of diverse pathways, many of which have anticancer effects (1,2). It is not surprising, then, that the SWI/SNF complex is targeted and inactivated during cancer progression. This complex contains 8–10 subunits, and loss of any of these subunits could alter or negatively impact the function of the complex (3,4). Indeed, BAF180 and BAF250 have recently been found to be targeted and lost in renal cell, breast and cervical cancers (5–10). Likewise, over the last decade, BRM and its homolog Brahma-related gene 1 (BRG1), the two catalytic, complex-nucleating subunits of SWI/SNF, were found to be silenced in a variety of tumor types (1,2). While BRG1 is frequently mutated in cancer cell lines (11,12), in primary tumors BRG1 appears to be infrequently (<3%) silenced by mutations based on the mutational Atlas database (13). Similarly, mutational data from the Atlas database show that BRM is silenced by mutations in <1% of human cancers (13). Non-silencing mutations (e.g. missense mutations) for BRG1 and BRM are observed to occur in <3.3 and in <2.3%, respectively, according to the Atlas database (13). In comparison, immunohistochemistry data for BRG1 and BRM also show that these genes are silenced at rates that range from 15 to 40% in most human tumors (14–20); therefore, these data suggest that other mechanisms, such as epigenetic silencing, may silence BRG1 and BRM expression (1,2) in human cancers. In support of this observation, we have found that BRM can be pharmacologically restored by a variety of compounds (20,21), but the mechanism of action remains unclear.
SWI/SNF has been functionally linked to differentiation, development, cell adhesion, DNA repair and growth control (1,2). As the integrity of these cellular mechanisms is necessary to thwart cancer development, inactivation of this complex impairs or blocks these cellular functions, thereby potentiating the development of cancer. Murine models have shown that the loss of certain SWI/SNF subunits can impact skin, vascular, cardiac and neurologic development (1,2). The determination of how the inactivation of this complex significantly impacts cancer development is a challenge, given its various possible cellular relationships. Perhaps the best-studied link to date between cancer and SWI/SNF has been the interdependence of this complex and growth control (22,23). For example, cancer cell growth is blocked when the BAF47, BRG1 or BRM subunits are reexpressed in cell lines with complexes that lack the expression of these genes (20,23–25). Clearly, this complex is needed for RB-mediated growth inhibition, and by extrapolation of these data, the RB homologs p130 and p107 (26,27) are also likely dependent on the function of SWI/SNF. Similarly, p53 has been functionally linked to SWI/SNF by a number of in vitro studies (1,2). A variety of other anticancer proteins with various functions, such as E-cadherin, BRCA1, CD44, KLF4, Jun and GADD45, have also been linked to SWI/SNF (1,2). Given these associations, the impairment or total inactivation of the SWI/SNF complex would have a significant effect on many distinct cellular functions, thereby effectively knocking out multiple anticancer mechanisms at once.
The dietary compounds known as flavonoids have been extensively studied over the last three decades and have been shown to have both in vitro and in vivo anticancer effects in a variety of tumor types and model systems (28–30). This work has defined six major structural groups derived mainly from different food sources. The mechanism of how flavonoids inhibit cancer has also been extensively studied. Flavonoid compounds are thought to be ATP analogs capable of blocking the function of various kinases, and a subset of flavonoids, including semisynthetic compounds such as flavopiridol, are actually known to inhibit CDK2 and CDK4, both of which control the function of the anticancer protein RB (31,32). Other data link flavonoids to the inhibition of certain chromatin remodeling proteins, such as HDACs (33–37). However, despite two decades of molecular studies, a detailed mechanistic profile of exactly how these compounds function has yet to be elucidated.
Our previous work has shown that BRM is reversibly suppressed in cancer cells and that the restoration of BRM expression inhibits cellular growth (20,38). As such, restoring BRM expression to thwart cancer growth is a potential novel pursuit for cancer therapy. Although HDAC inhibitors were the first agents known to restore BRM (20,39), they were soon found to inactivate BRM via acetylation of the protein’s C-terminus (40). Thus, these compounds cannot be used clinically for restoring BRM (20). However, the induction of BRM by treatment with HDAC inhibitors has shown that BRM is epigenetically suppressed and that the silencing of BRM can be pharmacologically reversed. This appears to be clinically advantageous because BRM is silenced in 15–20% of most solid tumor types (20). As a first step toward the development of BRM reactivation as a therapeutic strategy, we sought to identify agents that could effectively restore BRM expression and its function. From a high-throughput screening assay, we discovered that many flavonoids readily restore BRM expression. We now show that flavonoids act as small-molecule inhibitors by inducing and restoring the function of BRM. Furthermore, we show that the induction of BRM is required in part for the observed anticancer effects of flavonoids. These findings pull together two pieces of the puzzle: namely, how flavonoids act to inhibit cancer by activating both RB and BRM.
Materials and methods
Compounds
Flavonoids and flavonoid-containing supplements were purchased from Indofine (Hillsborough, NJ), Selleck Chemical (Houston, TX), Puritan’s Pride (Oakdale, NY) and Swanson Health Products (Fargo, ND). All compounds were resuspended at a concentration of 10 mM in dimethyl sulfoxide (DMSO). These tablets were pulverized into fine powder. We admixed and incubated each capsule/tablet powder in DMSO to dissolve the flavonoid(s). After several hours of incubation with DMSO, we then removed the minor insoluble ingredients by centrifugation. To derive a relative concentration, we used the difference in the weight of the pill material before and after the DMSO extraction. This net weight was then divided by the volume of the DMSO that was used to dissolve the powder. We have listed the composition of the natural compound sources in Supplementary Table 4, available at Carcinogenesis Online. As shown in Supplementary Table 4, available at Carcinogenesis Online, the vast majority of the weight of each natural extract is a single flavonoid or a combination of flavonoids. A stock concentration was calculated as ‘grams of extract’ divided by the ‘volume of DMSO’ used to dissolve the extract. The final concentration in µg/ml was achieved by multiplying the concentration of the stock (mg/ml) by the volume added and then dividing by the total final volume. The minimum effective concentration for each extract was determined by BRM induction as observed by quantitative PCR (qPCR) (>5-fold induction). Diosmin and Rutin were purchased from Alfa Aesar (Ward Hill, MA) and incorporated into AIN-93G base diet at levels of no flavonoids, 0.05% (low dose), or 0.2% (high dose).
Cell culture/daughter cell line generation and growth inhibition assay
All cell lines were grown and maintained as described in ref. 41. shRNA daughter cell lines were generated, and growth assays were conducted as described previously (41). BRM-negative SW13, C33A and H522 cell lines were obtained from ATCC and are derived from an adrenal, cervical carcinoma and lung adenocarcinoma, respectively. BRM-positive H460 and H441 cell lines were obtained from ATCC and are derived from a lung carcinoma and a lung adenocarcinoma, respectively.
BRM-dependent (MG213) luciferase assay
Cells were treated with 3 μM, or for dose-curve studies, with various doses of flavonoids, for 72 h before luciferase levels were measured. Cells were lysed and each well was evaluated for luciferase signal for 5 s using the OneGlo reagent (Promega, Madison, WI) and the FLx800 microplate reader (Biotek, Winooski, VT) (21).
Drug discovery and high-throughput screen
For this assay, we stably integrated the MMTV promoter linked to the luciferase gene, along with the rat glucocorticoid receptor, into the BRG1/BRM-deficient cell line SW13 as described (20,21,42). The assay was conducted at the Life Science Institute (http://www.msdiscovery.com/) at the University of Michigan by screening the ‘MS2000’ library that contains ~4500 compounds (see Supplementary Table 1, available at Carcinogenesis Online, for a full list of the compounds and Supplementary Table 2, available at Carcinogenesis Online, for the hits including the respective pAC50 values). A total of 5000 cells were plated per 96-well plate with 10 µM of each individual compound or DMSO with or without 0.1 µM dexamethasone for 72 h. The luciferase activity was read by the one-step One-Glo assay (Promega) after 72 h, and the data are reported as a percentage by assay, where 0% is the negative control and 100% is the positive control (Indoprofen). The data are reported as fold induction (or percentage of positive control) of luciferase activity demonstrated by the positive control, Indoprofen.
Quantitative PCR
mRNA levels were examined at 48 h, as our previous time-course experiments showed that maximal mRNA induction by various compounds occurs after 24 h (20). Sequences of the primers specific for BRM, BRM-inducible genes, or RNA polymerase 2A (POLR2A) are listed in the Supplementary Table 5, available at Carcinogenesis Online. All reactions were performed using volumes of 25 μl. EvaGreen was used as the indicator of amplification, and ROX was used for plate/well normalization. Fold change of mRNA expression was computed by comparing the normalized Ct value of the treated cell lines with the untreated parental cell lines.
Western blotting
Following 72-h treatment times with 3 μM of flavonoids, cells were harvested, and total protein was isolated using a urea-based lysis buffer as described previously (20,41). This time point is based on our previous work where maximal protein induction by various compounds is seen after 72 h (20). Antibodies that identify BRM (rabbit polyclonal anti-BRM), acetylated BRM (rabbit anti-acetylated-BRM, a gift from Christian Muchardt) and RB (mouse anti-RB, BD Biosciences, San Jose, CA) were all used at a dilution of 1:500. Appropriate secondary antibodies (GE Healthcare, UK) were used at a 1:2000 dilution. A glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (GeneTex, Irvine, CA) was used as the loading control.
Immunohistochemistry and cell counts
Mouse lungs were fixed in 50% ethanol/50% methanol and paraffin embedded. Antigen retrieval was performed by using either 10 mM Tris (pH 10) or 10 mM sodium citrate buffer (pH 6) and by heating the slides for 15 min on the ‘high’ setting in a standard microwave. Immunohistochemical staining for proliferating cell nuclear antigen (PCNA) (Thermo Scientific, RB-9055-P0, Pittsburgh, PA) and phosphorylated RB (Cell Signaling Technology, 9307, Danvers, MA) was performed with a 2-h incubation at room temperature at dilutions of 1:500 and 1:250, respectively. Sections were incubated for 1 h with a goat anti-rabbit biotinylated secondary antibody at a 1:200 dilution (Vector Labs, Burlingame, CA). We used an ABC staining kit with a DAB/nickel detection reagent (BD Pharmingen, San Diego, CA) and counterstained with Harris hematoxylin. Sections were imaged using a Zeiss Axioplan light microscope. The total number of positive cells was counted in a 60× field in each adenoma present on the slide. The lungs from at least five mice were scored for each genotype and treatment group. The number of positive cells in each adenoma (those that were present on the slide) for each mouse within a group was then divided by the number of total cells per 60× field to calculate the percentage of PCNA- and pRB-positive cells. The average percentage of immunoreactive cells and the standard of the mean were then calculated. More than 5 PCNA counts per mouse and >25 total counts for each group were used for statistical analysis using Student’s t-test.
Mouse breeding/dosing and statistical analysis
Mice experiments followed an experimental design reviewed and approved by the Institutional Animal Care and Use Committee at the University of Florida. This mouse model is described below and has been previously established (20). Both BRMnull (BRM−/−) (SV/129) mice and wild-type BRM (BRM+/+) (SV/129) mice were divided into three experimental treatment groups (six groups in total). Each mouse was then intraperitoneally injected with 1g/kg urethane (ethyl carbamate) weekly for 2 weeks to initiate the development of lung adenomas. At 6 weeks of age, and 2 weeks prior to the first injection of urethane, each mouse group was provided ad libitum with one of the following food variants: 0.05% (low dose), 0.2% (high dose), or food that was not supplemented. The diet was designed and made by Tina Harfel, PhD, at Harlan Laboratories. Mice were fed these diets for an additional 12 weeks (~3 months) at which time they were killed and the number of lung adenomas analyzed. The number of tumors was counted independently by two blinded investigators, and then an average of the tumor counts was obtained for each set of mouse lungs scored. We compared tumors from untreated mice with tumors derived from mice in the two treatment groups and calculated the statistical significance of the difference in tumor counts between each treatment group, using Student’s t-test with alpha 0.05 significance level.
Statistical analysis
Student’s t-test was used to compare the statistical significance of the data. The graphs represent the average of experiments performed in at least triplicate. The error bars represent the standard error of the mean (SEM) of experiments performed in at least triplicate.
Results
Flavonoid structure impacts BRM induction
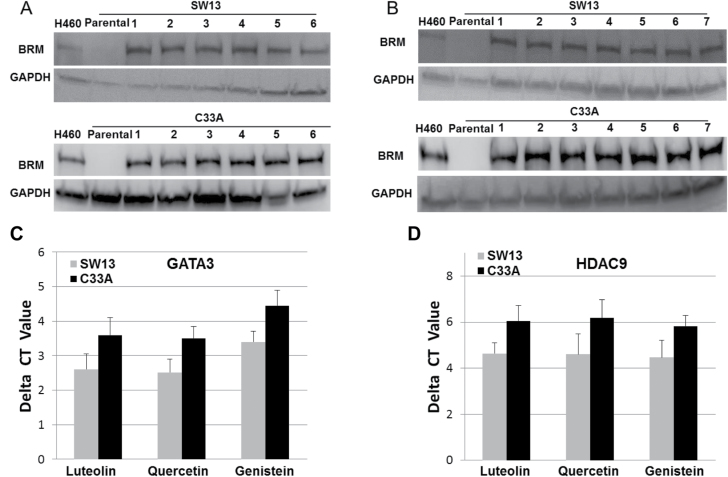
To identify agents that functionally induce Brahma (BRM) or otherwise known as SMARCA2, we conducted high-throughput screening using a BRM functional-dependent assay (21). Using this assay, we screened the ‘MS2000’ chemical library, which contains 384 FDA-approved drugs and naturally occurring compounds. We observed that the most potent, as well as the most frequent hits, were from the flavonoid family of compounds and found that 31 out of 112 hits (~28%) were flavonoids (Supplementary Table 2, available at Carcinogenesis Online) with a pAC50 that ranged from 4 to 5. As flavonoids fall into one of six structural groups, we next tested two compounds from each of the six structural groups to determine if certain types of flavonoids would or would not activate BRM (see Supplementary Tables 3 and 4, available at Carcinogenesis Online, for a list of groups and common corresponding natural product sources). These compounds were selected on the basis that they are frequently used in anticancer studies and cited in the flavonoid literature. We treated two BRG1/BRM-deficient cell lines, SW13 and C33A, with flavonoids from each of these structural groups (listed in Supplementary Table 3, available at Carcinogenesis Online) at a concentration of 3 μM, and we observed that each compound readily induced BRM protein expression (Figure 1A and Supplementary Figure 1, available at Carcinogenesis Online). Many natural products contain high amounts of either a single flavonoid or a combination of flavonoids, and these flavonoids are believed to underlie the health benefits attributed to these products. As such, we further investigated the ability of a number of natural product supplements (Supplementary Table 4, available at Carcinogenesis Online) to restore BRM. We dissolved these supplements (naturally occurring mixture of flavonoids) in DMSO, and we treated SW13 and C33A cell lines with 0.9 µg of extract/ml (stock was 0.9 mg/ml in DMSO; see Materials and methods). We observed that these natural products (Figure 1B) also readily induced BRM in both cell lines, similar to that of the pure flavonoids represented in Figure 1A. Although there are nearly 2000 different flavonoids, testing this subset of 12 flavonoids (listed in Supplementary Table 3, available at Carcinogenesis Online) selected from the six different structural categories suggests that certain flavonoids may, in general, induce BRM.
Fig. 1.
(A) demonstrates the induction of BRM protein in the BRM/BRG1-deficient cell lines, SW13 and C33A, following 72-h treatment with 3 µM of flavonoids selected from each of the known six flavonoid structural groups [1: Luteolin (flavone), 2: Quercetin (flavonol), 3: Genistein (isoflavone), 4: Hespiridin (flavanone), 5: EGCG (flavanol/catechins) and 6: Delphinidin (anthocyanins)]. H460 cell line was used as the positive control and GAPDH was used as the loading control. (B) illustrates the induction of the BRM protein in the SW13 and C33A cell lines after 72-h treatment with ~0.9 μg of extract/ml of each naturally occurring food extract (1: Broccoli, 2: Hibiscus, 3: Celery, 4: Cinnamon, 5: Turmeric, 6: Green tea and 7: Soybean). H460 cell line was used as the positive control and GAPDH was used as the loading control. (C) shows the decrease in the level of GATA3 mRNA following treatment with Luteolin, Quercetin or Genistein in the BRM/BRG1-deficient cell lines, SW13 and C33A for 48h, as measured by qPCR. (D) shows the decrease in the level of HDAC9 mRNA following 48-h treatment with Luteolin, Quercetin or Genistein in the BRM/BRG1-deficient cell lines, SW13 and C33A, as measured by qPCR.
As we have previously demonstrated that BRM silencing is driven by the over expression of GATA3 and HDAC9 (41), we investigated whether certain flavonoids such as Luteolin, Quercetin and Genistein might have an impact upstream of GATA3 and HDAC9 and thereby effect the expression of GATA3, HDAC9 or both as part of the mechanism described above by which flavonoids induce BRM.
As we have also demonstrated that high levels of GATA3 and HDAC9 mRNA and protein inversely mirror changes in BRM expression, we used qPCR to quantitatively measure if GATA3 and HDAC9 mRNA expression levels in BRM-deficient cell lines change after treatment with flavonoids. As such, we treated SW13 and C33A cells with the three most potent of the naturally occurring flavonoids that we tested (Luteolin, Genistein and Quercetin) and then conducted qPCR to measure the GATA3 and HDAC9 mRNA levels as a function of flavonoid treatment. We found that the GATA3 mRNA levels after treatment with each of these three flavonoids differed from the pretreatment levels of GATA3 mRNA, and the net decrease (change) was ~2.7 delta CT values (6.5-fold) and ~3.5 delta CT values (11-fold) for the SW13 and C33A cell lines, respectively (Figure 1C); these differences were statistically significant (P < 0.05). Similarly, the application of the same three flavonoids decreased the mRNA level of HDAC9 (P < 0.05); after treatment with any of these flavonoids, we observed that the levels of HDAC9 mRNA decreased by ~4.3 delta CT values (20-fold) and ~6 delta CT values (32-fold) for the SW13 and C33A cell lines, respectively (Figure 1D). HDAC3 and MEF2D also regulate BRM, but unlike GATA3 and HDAC9, HDAC3 and MEF2D are not overexpressed in BRM-deficient cell lines. As such, we did not observe any noticeable effects of any of these three flavonoids on HDAC3 or MEF2D mRNA levels as measured by qPCR (P > 0.05) in the BRM-deficient cell lines, SW13 and C33A (data not shown). We observed that not only did certain flavonoids, as listed in Supplementary Table 3, available at Carcinogenesis Online, induce BRM, but at least a subset of them also downregulated two proteins that contribute to BRM silencing: GATA3 and HDAC9. This suggests that certain flavonoids can target a BRM-regulating pathway further upstream of HDAC9 and GATA3. For example, the MAP kinase pathway has been reported to be targeted by flavonoids (43,44) and is known to induce BRM (41) when inhibited.
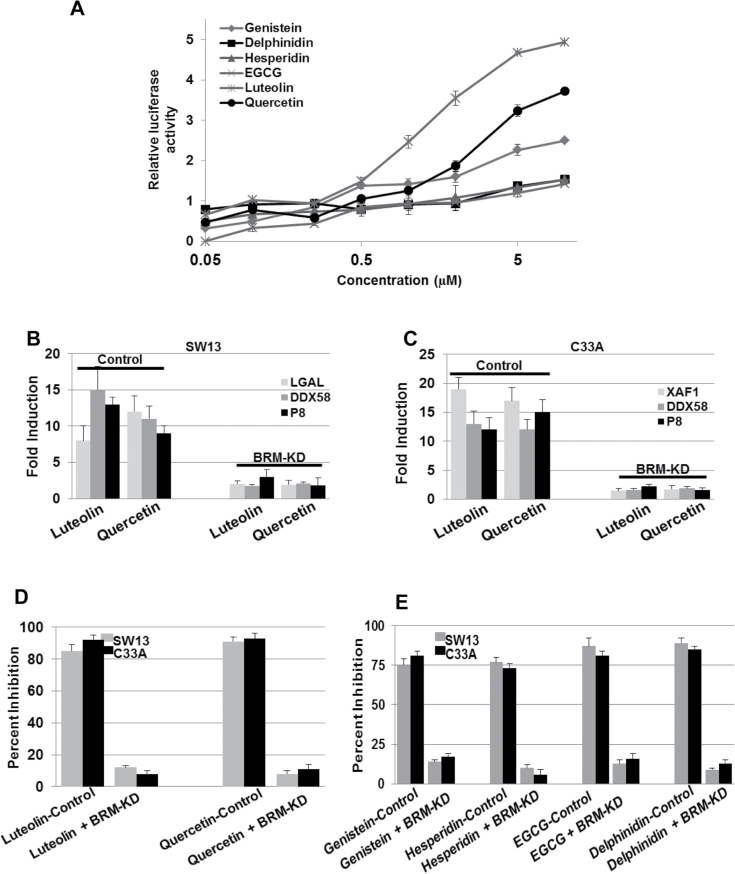
Flavonoids have a common skeleton backbone consisting of three phenolic rings, and each unique flavonoid differs in the number and positions of hydroxyl and methyl groups attached to this conserved ring structure. We conducted dose–response curves again using our luciferase reporter assay to assess which of the six structural flavonoid groups were most potent in inducing BRM. We observed that each tested flavonoid (one from each structural group) progressively induced luciferase (Figure 2A) over a range of concentrations from 0.05 μM to 5 μM. Maximal induction of luciferase activity was observed at 10 μM for each flavonoid tested and was noted to be statistically significant compared with vehicle at concentrations greater than 250 nM for each. Comparing the tested flavonoids, we observed that the most robust luciferase activity (indirect BRM induction) occurred with Luteolin and Quercetin, respectively, at concentrations of 2–5 μM.
Fig. 2.
(A) shows the use of the MMTV-luciferase assay to indirectly measure the potency of flavonoids from the six structural groups at various concentrations. Quercetin and Luteolin show the highest induction of luciferase activity at >5 µM, nearly 11-fold and 15-fold higher, respectively, compared with their respective baseline luciferase levels (~1). (B and C) SW13 and C33A show the induction of BRM-dependent genes in the daughter SW13 and C33A cell lines harboring the scrambled shRNA (control) after treatment with either 3 µM Luteolin or Quercetin for 72h (P < 0.05). In comparison, the induction of these BRM-dependent genes was blunted in daughter SW13 and C33A cell lines that harbored the anti-BRM shRNA (BRM-KD). (D) C33A and SW13 cells were infected with either scrambled shRNA (control) or anti-BRM shRNA (BRM-KD). These cell lines were then treated with 3 µM of either Luteolin or Quercetin for 72h. Compared with C33A and SW13 cell lines transduced with anti-BRM shRNA, the cell lines transduced with the scrambled shRNA elicited significant growth inhibition after Luteolin or Quercetin treatment (P < 0.05). (E) C33A and SW13 cells were transduced with either scrambled shRNA (control) or anti-BRM shRNA (BRM-KD), followed by 72-h treatment with 3 µM of flavonoids from each of the four other structural groups (Genistein, Hesperidin, EGCG and Delphinidin). Compared with C33A and SW13 cell lines transduced with anti-BRM shRNA, the cell lines transduced with scrambled shRNA elicited growth inhibition after treatment with each flavonoid (P < 0.05).
To understand which hydroxyl positions are related to BRM induction, we tested 30 synthetic flavonoids (Supplementary Figure 2, available at Carcinogenesis Online), which had different locations and numbers of hydroxyl groups positioned around the flavonoid ring structure. The lack of specific flavonoids prevented a complete and thorough analysis and a comparison of the potency of each hydroxyl substitution. Nevertheless, we found that hydroxyl groups at the 3′ and 4′ positions as well as at the 5′, 6′and 7′ positions both individually, and in combination, increased their ability to induce BRM (Supplementary Figure 2, available at Carcinogenesis Online).
Flavonoids induce BRM-dependent genes
Although these three compounds induced BRM, we wanted to determine if they might also have preferential side effects that might block a subset of SWI/SNF functions. SWI/SNF complexes facilitate the function of a variety of different transcription factors to foster gene expression (1,45); thus, we further measured BRM functionality by measuring the expression of a variety of BRM-dependent genes. We previously identified a cadre of genes whose expression is dependent on BRM and the activity of this complex. We examined the two most potent of the tested flavonoids in the BRM/BRG1-deficient cell lines, C33A and SW13, for their ability to induce three BRM-dependent genes (SW13: LGAL, DDX58, P8 and C33A: DDX58, P8, XAF1), which were previously established as being BRM dependent by Gramling et al. (38,46). To show that these genes are specifically induced by the reexpression and functional restoration of BRM, the C33A and SW13 cell lines were transduced with either scrambled shRNA (control) or anti-BRM shRNA, capable of blocking the induction of BRM. We observed that cells transduced with scrambled shRNA robustly induced each of the three BRM-dependent genes when treated with either Luteolin or Quercetin, the two most potent inducers of BRM observed in Figure 2A. In comparison, those cells that were transduced with anti-BRM shRNA showed little or no induction of these genes after treatment with Luteolin and Quercetin (Figure 2B and C). Hence, by using the glucocorticoid-based luciferase assay and the expression analysis of BRM-dependent genes, we have shown that a subset of flavonoids is capable of not only inducing BRM expression, but also act to specifically restore its function.
Flavonoids require BRM to inhibit cellular growth
Since the restoration of BRM can result in growth arrest (20), we also sought to determine if treatment with certain pure flavonoids or natural flavonoid-containing extracts could induce growth arrest in BRM-deficient cell lines. Such data could potentially help us to elucidate how certain flavonoids might thwart cancer by inhibiting cell growth. To accomplish this, we treated SW13 and C33A cells with either Luteolin or Quercetin. For these experiments, we generated daughter cell lines from these two parental cell lines by transducing them with scrambled shRNA (control) or anti-BRM shRNA (test), which suppresses the induction of BRM. We conducted growth studies on these SW13 and C33A cells using a 3 µM dose for either Luteolin or Quercetin. At this dose, for each of the compounds tested in both cell lines that harbored the scrambled shRNA, we observed >85% growth inhibition after a 72-h incubation (Figure 2D). However, there was an absence of growth inhibition with either compound when tested in the cell lines that harbored the anti-BRM shRNA. These data indicate that the induction of BRM is a prerequisite for growth inhibition induced by certain flavonoids in these cell lines. To determine if structurally different flavonoids also demonstrate this effect, we repeated this experiment using representative flavonoids from the four remaining flavonoid structural groups (Genistein, Hesperidin, EGCG and Delphinidin). The results of these experiments paralleled the results from the Luteolin and Quercetin experiments and show that each of these four flavonoids required BRM induction in order to inhibit cellular growth (Figure 2E).
Flavonoids reverse BRM acetylation
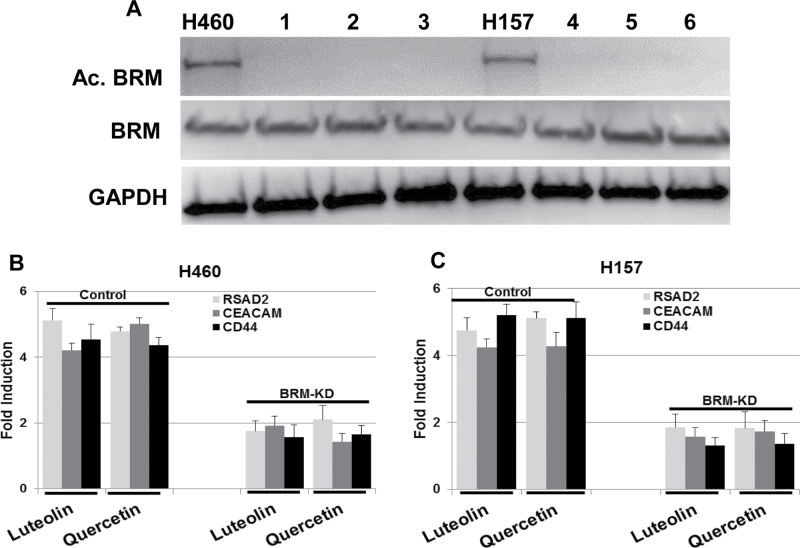
Previous work has demonstrated that BRM can be functionally inactivated by acetylation of lysine residues within its C-terminus (40). It was also shown that the replacement of these lysine residues with arginine residues does not change the function of BRM, but rather prevents BRM inactivation caused by these acetylations (40). Although HDAC inhibitors were one of the first groups of compounds shown to induce BRM (1,2), these compounds cannot be used clinically because they also induce BRM acetylation (40), which inactivates the protein. We have shown that BRM acetylation is controlled by the balance of HDAC2 activity and KAT2B/KAT8 activity (41), whereas certain flavonoids are known to promote HDAC2 activity and inhibit KAT2B (47,48). To determine if certain flavonoids in general could reverse BRM acetylation and thus reactivate BRM, we treated two BRM acetylated cell lines, H460 and H157, with either Luteolin or Quercetin, and then determined if these flavonoids could reverse BRM acetylation. As with our previous results for BRM induction, we observed that the Luteolin or Quercetin that avidly induce BRM also reversed BRM acetylation (Figure 3A). To test BRM functionality, we assayed for the induction for BRM-dependent genes in these cell lines. After a 72-h treatment with Luteolin or Quercetin, we observed the induction of BRM-dependent genes (specifically, RSAD2, CEACAM, CD44) at levels of >4-fold in each of the daughter cell lines that we tested and that harbored scrambled shRNA (Figure 3B and C). However, for those daughter cell lines that harbored anti-BRM shRNA, little to no induction was observed for the BRM-dependent genes tested (Figure 3B and C). These data demonstrate that certain flavonoids restore BRM expression and/or function by both reversing BRM silencing as well as through the reversal of BRM acetylation.
Fig. 3.
(A) illustrates the reversal of BRM acetylation following 3 µM treatment for 72h with (1) Luteolin, (2) Quercetin or (3) Genistein in the H460 or H157 cell lines. (B and C) H460 and H157 demonstrate the induction of BRM-dependent genes by at least 4- to 5-fold in both the H460 and H157 cells that were transduced with scrambled shRNA (control), while these genes were observed to be induced in these same cell lines (P < 0.05) when they were transduced with anti-BRM shRNA (BRM-KD).
Flavonoids fail to inhibit tumor development in BRMnull mice
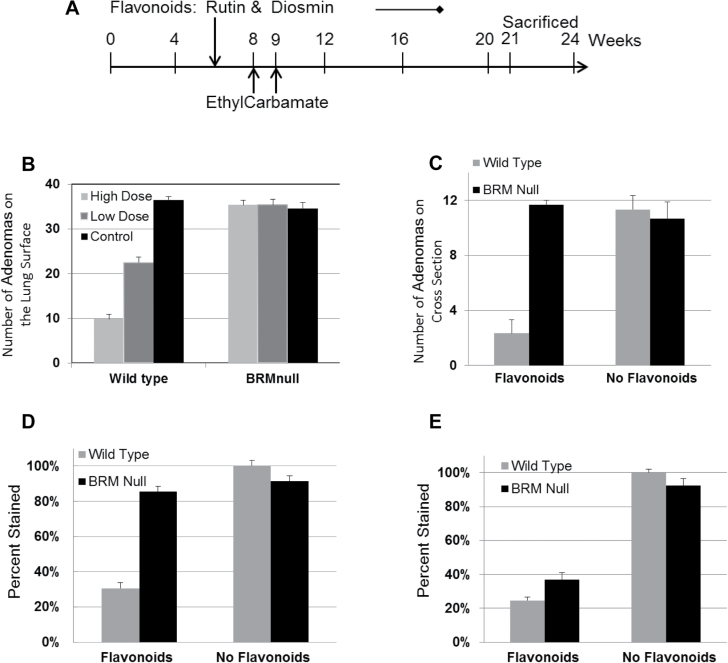
Since abundant data show the anticancer effects of flavonoids in both the prevention and treatment of cancer, we wanted to determine if BRM might play a key role in the in vivo effects of flavonoids. Based on our in vitro experiments, which indicate that BRM contributes to the anticancer effects of flavonoids, we next tested whether there was an interdependence of the anticancer effects of certain flavonoids and BRM expression in vivo. To accomplish this, we used a carcinogen-induced murine lung cancer model described previously (49) to determine if tumors that arise in BRMnull (BRM-deficient) mice are impacted in a similar way by exposure to certain flavonoids as those tumors that arise in wild-type mice (that is, tumors that express BRM). As Quercetin and Luteolin were observed to be the more potent flavonoids for inducing BRM in our in vitro experiments (Figure 2A), these flavonoids were thus used in this in vivo experiment. However, to improve the absorption of these flavonoids, we instead used the respective glycosidic forms of Quercetin and Luteolin, which are Rutin and Diosmin (50,51). Specifically, the glycosides Rutin and Diosmin are more readily bioavailable in that their dietary absorption is significantly favored, and once absorbed, they are metabolized into their respective aglycone forms, Quercetin and Luteolin. Hence, giving the glycosidic forms, Rutin and Diosmin, is essentially the same as giving the aglycone forms, Quercetin and Luteolin. The concentrations of flavonoids used in this experiment were based on a review of a variety of animal studies, in which the concentrations ranged from 0.1 to 1%, that were successfully used to test the in vivo effects of flavonoids (52–58). As such, this experiment was designed to examine the impact of a combination of two flavonoids, Rutin and Diosmin, either at a low (0.05%) or a high dose (0.2%), given to mice that were exposed to a lung-specific carcinogen compared with mice given the same carcinogen, but who were fed a normal diet devoid of significant levels of Rutin and Diosmin. We elected to use a dose approximating the upper range of dietary flavonoid consumption (59,60) but also within a range of concentration previously used in other mouse experiments (52). Most diets, however, are not high in specific flavonoids but rather are composed of a combination of flavonoids at more modest, lower levels. As such, we also chose to feed a second group of mice a diet containing a lower dose of Rutin and Diosmin. Since food sources of flavonoids rarely contain solely one flavonoid, we elected to treat the mice in this experiment with a combination of Rutin and Diosmin in which the combined concentration was also within the range of published flavonoid concentrations (52). We surmised that this combination of flavonoids might show efficacy, though each individual compound is at a lower dose than that used to show effects in most other single flavonoid studies.
To generate the mice for these experiments, we crossbred heterogeneous, BRMnull mice to generate mice containing either wild-type BRM or the BRMnull genotype. To initiate the development of lung adenomas, intraperitoneal injections were performed on these mice using the lung-specific carcinogen ethyl carbamate at 8 weeks and 9 weeks of age as described (61). Starting at 6 weeks of age, before the injections, BRMnull and wild-type mice were fed a regular diet or a combination of Rutin and Diosmin supplemented into the regular diet at a low dose (0.05%) or a high dose (0.2%). All mice were maintained on their specified diets until the completion of the experiment, ~3 months (Figure 4A). We have found from our work and other publications that the adenomas generated in the lungs reach ~90% of their total numbers at 3 months and then slowly increase in number in the next 3 months by another 10% (49,62). After these 6 months, a small percentage of adenomas (1–2%) convert to adenocarcinomas that develop in the 6–12 months post injection (63). As such, in this experiment we focused on the susceptibility of the mice to develop lung adenomas in response to carcinogen exposure as a function of Rutin and Diosmin treatment and BRM expression as the primary endpoints. After the 3-month treatment period, we killed all mice and counted the number of visible adenomas on the surface of the lungs to determine the relative number of tumors that arose in each mouse. We observed that the wild-type mice demonstrated 83 and 55% fewer adenomas, from the high-dose and low-dose Rutin and Diosmin groups, respectively, compared with the untreated ‘control’ wild-type mice (Figure 4B and C). Flavonoid inhibition of carcinogen-induced adenoma formation in murine lungs has been previously demonstrated (64,65). However, in the BRMnull mice, we observed equivalent numbers of adenomas arising from each of the three diet groups, indicating that the lack of BRM expression imparts a ‘resistance’ to the anticancer effects of these flavonoids (Figure 4B and C).
Fig. 4.
(A) shows the time-course outline of the murine experiment with the age of the mice in weeks on the horizontal axis. All mice were given two intraperitoneal injections of ethyl carbamate at a dose of 1g/kg at 8 weeks of age and then again at 9 weeks of age. Mice were given no flavonoids (non-supplemented diet), a combination of low-dose (0.05%) Rutin and Diosmin or a combination of high-dose (0.2%) Rutin and Diosmin in their food starting at 6 weeks of age until 12 weeks after the administration of ethyl carbamate (or ~21 weeks of age). At 21 weeks of age, all mice were killed, and their lungs were subsequently examined to determine the number of visible adenomas on the surface. (B) shows the relative number of tumors visible on the surface of the lungs from either BRM wild-type or BRMnull mice that were fed no flavonoids, a low-dose combination of Rutin and Diosmin (each 0.05%) or a high-dose combination of Rutin and Diosmin (each 0.2%). (C) shows the number of tumors seen in cross-sections of the lungs from wild-type or BRMnull mice that were fed either no flavonoids or a high-dose combination of Rutin and Diosmin. In the wild-type mice, the observed difference in tumor number between control diet group and low-dose flavonoid diet group or high-dose flavonoid group was found to be significant at P = 0.025 and P = 3E−15, respectively, whereas observed difference in tumor number between low and high flavonoid diet was also statistically significant at P = 3E−8. In contrast in the BRMnull mice, we found no statistically significant difference in the number of tumors observed in the control group versus the low flavonoid diet group (P = 0.98) or the high flavonoid diet group (P = 0.59); similarly, there was no statistically significant difference in the number of tumors in low flavonoid diet group versus the high flavonoid diet group (P = 0.35).
(D) Lungs from either wild-type BRM or BRMnull mice were stained for PCNA expression and then scored for the percentage of tumor cells (nuclei) that expressed PCNA per high power field: 60×. PCNA immunoreactivity was ~3- to 4-fold higher in the tumors derived from BRMnull mice regardless of treatment (no flavonoid diet: P = 5.5E−3; high-dose flavonoid diet: P = 0.013) as well as those from untreated wild-type mice (P = 7.0E−5) compared with the tumors derived from wild-type mice treated with a high dose of flavonoids. (E) Lungs from wild-type BRM or BRMnull mice were stained with an antibody to phosphoRB that specifically detects phosphorylation at RB Ser780. The percentage of phosphoRB staining for each of the four groups was scored. Immunopositivity was higher in the tumors derived from untreated mice compared with that in the tumors derived from high-dose flavonoid-treated mice regardless of the genotype.
Flavonoids induce growth inhibition by inducing BRM and the dephosphorylation of retinoblastoma (Rb) protein
As BRM is a cofactor for Rb function, in order for growth inhibition to occur, not only must BRM be functionally active, but Rb also has to be activated. Specifically, Rb must become hypophosphorylated, thus transforming into its active form. This functional interdependence of BRM and Rb to mediate the cellular growth inhibition has been previously demonstrated (22,23). Certain flavonoids are surmised to function as ATP analogs (66), which in turn can act as CDK inhibitors to change the phosphorylation status of Rb and possibly as MEK/MAP kinase inhibitors, which have been shown, in turn, to induce BRM (41). Hence, we next stained the tumors with antibodies against PCNA (a marker of cell proliferation) and phosphoRb (a marker of Rb inactivation). For PCNA staining in these tumors, we observed that the number of tumor cells that maintain their proliferative capability (PCNA positive) is qualitatively ~3-fold lower in tumor cells from the Rutin- and Diosmin-treated wild-type mice compared with the tumor cells derived from either BRMnull mice (Rutin- and Diosmin-treated or untreated) or untreated wild-type mice (Figure 4D and Supplementary Figure 3, available at Carcinogenesis Online). For Rb phosphorylation, we observed a higher percentage of cells stained in the tumors from untreated wild-type mice and untreated BRMnull mice (4- and 2.5-fold higher, respectively), compared with tumors from Rutin- and Diosmin-treated wild-type and flavonoid-treated BRMnull mice (Figure 4E and Supplementary Figure 4, available at Carcinogenesis Online). Together, the PCNA and phosphoRB staining data indicate the decrease in observed growth rate (decreased PCNA immunoreactivity) is not caused by a lack of Rb activation qualitatively, as Rutin and Diosmin treatment caused Rb phosphorylation to decrease similarly in BRMnull and wild-type tumors. Rather, the decrease in observed growth rate (decreased PCNA immunoreactivity) occurs when both BRM is present (wild-type) and Rb becomes activated via dephosphorylation by Rutin and Diosmin treatment.
To further illustrate these results, we conducted western blot analysis with BRG1/BRM-deficient cell lines SW13 and H522 harboring either scrambled shRNA or anti-BRM shRNA and then treated with or without 3 µM Quercetin or Luteolin. After 72 h of Quercetin or Luteolin treatment, we then probed for BRM, phosphoRb and PCNA protein expression under these experimental conditions. Similar to our staining data with lung adenomas, we observed that Quercetin or Luteolin treatment of BRM/BRG1-deficient cancer cell lines SW13 and H522 readily induced BRM and decreased the level of phosphoRb (causing the hypophosphorylated form of Rb to accumulate) in these cell lines (Figure 5A and B). Moreover, proliferation as measured by PCNA only decreases significantly when BRM can be induced (no anti-BRM shRNA) (Figure 5A and B). Certain flavonoids require Rb function for their growth inhibitory effects, while BRM is known to be a necessary cofactor for Rb-mediated growth inhibition (26,27), as illustrated in Figure 6. As such, it is not surprising that these data show that BRM is required for growth inhibition that is induced by certain flavonoids.
Fig. 5.
(A and B) SW13 and H522: SW13 and H522 cell lines were transduced with either scrambled shRNA (control) or anti-BRM shRNA (shBRM) and then treated with 3 µM of Luteolin (Diosmin) and Quercetin (Rutin) for 72h followed by western blotting to detect BRM, phosphoRB and PCNA expression. BRM expression was blunted by the introduction of anti-BRM shRNA, and the levels of phosphoRB expression were reduced in both cell lines that were treated with flavonoids. PCNA expression was only decreased in the SW13 and H522 cells that harbored scrambled shRNA and that were treated with either Quercetin or Luteolin. H460 cell line was used as the positive control and GAPDH was used as the loading control.
Fig. 6.
Illustrates a proposed model underlying the mechanisms of growth inhibition by flavonoids. Based on both our current and published data, we hypothesize that flavonoids activate RB by inhibiting CDK2/4 and by inducing BRM expression via the inhibition of the MAP kinase pathway. Consequently, following treatment with flavonoids, hypophosphorylated RB and BRM bind together to facilitate growth inhibition.
Summary
Reactivation of BRM by flavonoids occurs either by induction or by fostering the deacetylation of BRM. The induction of BRM by flavonoids does not appear to be restricted or limited to a particular subset of flavonoids as we observed that 42 out of 42 tested flavonoids, each structurally different and selected from the six different structural subgroups, induced BRM. Flavonoids cause growth inhibition to occur by both reactivating BRM and by decreasing RB phosphorylation.
Discussion
Flavonoids have long been known to block cancer initiation as well as thwart cancer growth. Detailed studies during the past 30 years have demonstrated the ability of these compounds to inhibit a variety of targets, but recent data mainly focus on their ability to inhibit HDACs and kinases, such as CDK2. Despite this insight into their function, concrete models of how they mechanistically support their anticancer effects have not been clearly elucidated. Nevertheless, a key step in the progress of growth inhibition depends on the ability of flavonoids to activate Rb. To this end, it is not surprising that flavonoids additionally induce BRM and activate the SWI/SNF complex because SWI/SNF is a necessary cofactor for Rb function. A number of papers have shown that in BRM/BRG1-deficient cancer cell lines, the lack of a functional SWI/SNF complex blocks growth inhibition induced by the constitutively active form of Rb (22,23). However, growth inhibition is restored when either BRM or BRG1 is coexpressed with Rb (22,23). This interdependence of BRM/BRG1 with Rb occurs because Rb can effectively bind to both BRM and BRG1 proteins via their LXCXE domains (27,67), which are situated in the C-terminus of each protein following the BRM and BRG1 helicase domain (1,2). Mutation of this LXCXE domain blocks the ability of BRM/BRG1 to bind to Rb and inhibits Rb-mediated growth inhibition (67).
Given their evolutionary and chemical diversity, it is intriguing that flavonoids from the six structural families and the diverse cadre of natural products tested all revealed similar effects on BRM. Flavonoids modulate a large number of specific biological processes and therefore must have a significant number of molecular targets. Understanding how flavonoids interact with biological targets should help provide insight into how they uniformly target the BRM gene and its reexpression, which could be important in the understanding of the anticancer effects of diets rich in flavonoids (68). It is known that flavonoids are nanomolar inhibitors of human CDK2, a cyclin-dependent kinase (69). Interestingly, studies have shown that flavonoids are competitive inhibitors of this enzyme although their structures bear little similarity to ATP. This is supported by an X-ray crystal structure of CDK2 with the inhibitor L868276 (a flavonoid) bound (69), as illustrated in Supplementary Figure 5, available at Carcinogenesis Online.
As there are ~2000 structurally different flavonoids, it is impractical to test each flavonoid to show that there is general or common effect that flavonoids demonstrate. To begin to address this question, we conducted a subset analysis where we tested two flavonoids from each of the six structural groups to determine the potential spectrum of flavonoids that might restore BRM. The tested naturally occurring flavonoids as well as another 30 synthetic flavonoids were each found to restore BRM. Hence, we tested a total 42 different flavonoids as well as 8 flavonoid-containing extracts, all of which demonstrated the same effect: the restoration of BRM expression and activation. There are nine carbon positions within the flavonoid basic ring structure that can be hydroxylated; thus, there are roughly 29 or 512 possible hydroxylation permutations. As such, we tested roughly 8% of the possible hydroxylation combinations. Additional flavonoids can be generated, as theoretically, each hydroxyl group can be methylated. Nevertheless, that none of the compounds we tested failed to induce BRM argues that the regulation of BRM expression is not restricted or limited to a particular subgroup of flavonoids and that flavonoids in general might be able to induce BRM. In this manuscript, we suggest that certain flavonoids induce BRM by inhibiting kinases such as MAPK. However, more recent papers have shown that certain flavonoids can target a diverse array of pathways, which may reflect the structural diversity among different flavonoids, although the mechanisms are just beginning to be explored (70,71). Hence, flavonoids may also induce BRM by targets and mechanisms yet to be defined.
In this paper, we demonstrate that a level of flavonoids that could be feasibly obtained from diets low and high in flavonoids, and the combination of flavonoids used, is effective in inducing BRM and promoting cellular growth inhibition. The work presented here, particularly in the combination of flavonoids used, more closely reflects the human diet, since few, if any, people consume only a single flavonoid for extended periods of time. Of interest would be to broaden the combination of flavonoids given but provide them each at even lower doses to better mimic the normal dietary exposure to flavonoids. Since the flavonoids appear to have similar or the same target proteins regardless of the structural group from which they are derived, the total combined dose would seem to be more important than individual doses of any one flavonoid. This might explain the discrepancy between the higher levels necessary for a single flavonoid to be effective in vitro compared with the lower but effective levels achieved from a regular diet rich in flavonoids. Thus, dose accumulation, which occurs as a result of regular consumption of a variety of flavonoids, may also contribute to the greater benefit provided by lower levels of flavonoids found in foods such as fruits, vegetables and natural supplements.
Moreover, while the yield of tumors is diminished in flavonoid-treated mice, the transformation process is not completely blocked. Incomplete or partial absorption of the flavonoids may have contributed to this observed effect. Specifically, although we chose to use the glycosidic forms of Luteolin and Quercetin based on previously established literature that indicates a higher absorption of these forms, recent work has shown that the aglycone forms may be better absorbed than their glycosidic counterparts (72). Although the benefits of using either the aglycone or the glycosidic forms are not clear, we clearly observed a biological effect in this experiment. As such, these precancerous lesions, adenomas, may still evolve into malignant tumors. The question that remains is whether the reduction in these lesions completely underlies the overall anticancer effects of these compounds. An alternative consideration is whether flavonoids slow or impede the evolution of adenomas into adenocarcinomas. To experimentally examine this question using the model system presented, flavonoids would be provided to wild-type mice after the establishment of the adenomas, beginning at the 3-month time point, and the rate of appearance of adenocarcinomas monitored as a function of flavonoid dose. Since adenomas are precancerous, it would be much more informative to observe the development rate of malignant adenocarcinomas. To this end, the flavonoids found in tea (EGCG) have been shown to inhibit adenocarcinoma development in murine lungs when carcinogenesis is initiated by the tobacco carcinogen NKK (64). This occurs in part because these particular flavonoids lower the phosphorylation rate of Erk1/2 kinase (28) and therefore inhibit Erk1/2 kinase (73), which would presumably induce BRM, as BRM is regulated by the MAP kinase pathway (41). In another study, it was shown that various tea polyphenols inhibited the growth of H661 (BRG1-negative) and H1299 (BRG1- and BRM-negative) lung cancer cell lines (74). The results from these two cell lines support the role of BRM in flavonoid-induced growth inhibition, as the lack of either BRG1 or BRM would typically abrogate growth inhibition. This study also showed that application of tea can continuously inhibit NNK-induced lung tumorigenesis at both the initiation and promotion stages (75). Based on these data, it would be informative to investigate the rate of malignant tumor development in wild-type compared with BRMnull mice as a function of combination low-dose flavonoids versus high-dose flavonoids.
Supplementary material
Supplementary Tables 1–5 and Figures 1–5 can be found at http://carcin.oxfordjournals.org/.
Conflict of Interest Statement: Dr Reisman is the major shareholder of Zenagene, Inc.
Funding
National Cancer Institute (1R01CA136683-01A1 to D.R.); University of Florida: Research Seed Funding.
Supplementary Material
Glossary
Abbreviations:
- BRG1
Brahma-related gene 1
- BRM
Brahma
- CDK2
cyclin-dependent kinase 2
- CDK4
cyclin-dependent kinase 4
- CEACAM1
carcinoembryonic antigen-related cell adhesion molecule 1
- DDX58
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58
- DMSO
dimethyl sulfoxide
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GATA3
GATA-binding protein 3
- HDAC9
histone deacetylase 9
- HDAC3
histone deacetylase 3
- KAT2B
K(lysine) acetyltranferase 2B
- KAT8
K(lysine) acetyltranferase 8
- LGAL
lectin, galactoside-binding protein
- M2K1
mitogen-activated protein kinase kinase 1
- MEF2D
myocyte enhancer factor 2D
- PCNA
proliferating cell nuclear antigen
- qPCR
quantitative PCR
- RSAD2
radical S-adenosyl methionine domain containing 2
- RB
retinoblastoma
- Rb
retinoblastoma protein
- XAF1
XIAP associated factor 1.
References
- 1. Gordon V., et al. (2010). Alteration to the SWI/SNF complex in human cancers. Oncol. Rev., 4, 89–99 [Google Scholar]
- 2. Reisman D., et al. (2009). The SWI/SNF complex and cancer. Oncogene, 28, 1653–1668 [DOI] [PubMed] [Google Scholar]
- 3. Wang W., et al. (1996). Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev., 10, 2117–2130 [DOI] [PubMed] [Google Scholar]
- 4. Wang W., et al. (1996). Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J., 15, 5370–5382 [PMC free article] [PubMed] [Google Scholar]
- 5. Xia W., et al. (2008). BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res., 68, 1667–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Decristofaro M.F., et al. (2001). Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell. Physiol., 186, 136–145 [DOI] [PubMed] [Google Scholar]
- 7. Wiegand K.C., et al. (2010). ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med., 363, 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lichner Z., et al. (2013). The chromatin remodeling gene ARID1A is a new prognostic marker in clear cell renal cell carcinoma. Am. J. Pathol., 182, 1163–1170 [DOI] [PubMed] [Google Scholar]
- 9. Guan B., et al. (2011). ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res., 71, 6718–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cho H., et al. (2013). Loss of ARID1A/BAF250a expression is linked to tumor progression and adverse prognosis in cervical cancer. Hum. Pathol, 44, 1365–1374 [DOI] [PubMed] [Google Scholar]
- 11. Wong A.K., et al. (2000). BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res., 60, 6171–6177 [PubMed] [Google Scholar]
- 12. Medina P.P., et al. (2008). Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum. Mutat., 29, 617–622 [DOI] [PubMed] [Google Scholar]
- 13. The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/
- 14. Oike T., et al. (2013). A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res., 73, 5508–5518 [DOI] [PubMed] [Google Scholar]
- 15. Fukuoka J., et al. (2004). Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin. Cancer Res., 10, 4314–4324 [DOI] [PubMed] [Google Scholar]
- 16. Watanabe T., et al. (2011). Regulation of PTEN expression by the SWI/SNF chromatin-remodelling protein BRG1 in human colorectal carcinoma cells. Br. J. Cancer, 104, 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bai J., et al. (2013). BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS One, 8, e59772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bock V.L., et al. (2011). BRM and BRG1 subunits of the SWI/SNF chromatin remodelling complex are downregulated upon progression of benign skin lesions into invasive tumours. Br. J. Dermatol., 164, 1221–1227 [DOI] [PubMed] [Google Scholar]
- 19. Sun A., et al. (2007). Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate, 67, 203–213 [DOI] [PubMed] [Google Scholar]
- 20. Glaros S., et al. (2007). The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene, 26, 7058–7066 [DOI] [PubMed] [Google Scholar]
- 21. Gramling S., et al. (2011). Discovery of BRM targeted therapies: novel reactivation of an anti-cancer gene. Lett. Drug Des. Discov., 8, 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reisman D.N., et al. (2002). Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene, 21, 1196–1207 [DOI] [PubMed] [Google Scholar]
- 23. Strobeck M.W., et al. (2002). Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem., 277, 4782–4789 [DOI] [PubMed] [Google Scholar]
- 24. Betz B.L., et al. (2002). Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene, 21, 5193–5203 [DOI] [PubMed] [Google Scholar]
- 25. DelBove J., et al. (2011). Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics, 6, 1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dunaief J.L., et al. (1994). The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell, 79, 119–130 [DOI] [PubMed] [Google Scholar]
- 27. Strober B.E., et al. (1996). Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol., 16, 1576–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao H., et al. (2011). Dietary flavonoids as cancer prevention agents. J. Environ. Sci. Health. C. Environ. Carcinog. Ecotoxicol. Rev., 29, 1–31 [DOI] [PubMed] [Google Scholar]
- 29. Nishiumi S., et al. (2011). Dietary flavonoids as cancer-preventive and therapeutic biofactors. Front. Biosci. (Schol. Ed)., 3, 1332–1362 [DOI] [PubMed] [Google Scholar]
- 30. Clere N., et al. (2011). Anticancer properties of flavonoids: roles in various stages of carcinogenesis. Cardiovasc. Hematol. Agents Med. Chem., 9, 62–77 [DOI] [PubMed] [Google Scholar]
- 31. Sedlacek H., et al. (1996). Flavopiridol (L86 8275; NSC 649890), a new kinase inhibitor for tumor therapy. Int. J. Oncol., 9, 1143–1168 [DOI] [PubMed] [Google Scholar]
- 32. Carlson B.A., et al. (1996). Flavopiridol induces G1 arrest with inhibition of cyclin-dependent kinase (CDK) 2 and CDK4 in human breast carcinoma cells. Cancer Res., 56, 2973–2978 [PubMed] [Google Scholar]
- 33. Lee S.J., et al. (2011). Curcumin-induced HDAC inhibition and attenuation of medulloblastoma growth in vitro and in vivo . BMC Cancer, 11, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Attoub S., et al. (2011). Inhibition of cell survival, invasion, tumor growth and histone deacetylase activity by the dietary flavonoid luteolin in human epithelioid cancer cells. Eur. J. Pharmacol., 651, 18–25 [DOI] [PubMed] [Google Scholar]
- 35. Wheatley N.C., et al. (2010). Antimalarial histone deacetylase inhibitors containing cinnamate or NSAID components. Bioorg. Med. Chem. Lett., 20, 7080–7084 [DOI] [PubMed] [Google Scholar]
- 36. Son I.H., et al. (2007). Pomiferin, histone deacetylase inhibitor isolated from the fruits of Maclura pomifera. Bioorg. Med. Chem. Lett., 17, 4753–4755 [DOI] [PubMed] [Google Scholar]
- 37. Bontempo P., et al. (2007). Feijoa sellowiana derived natural Flavone exerts anti-cancer action displaying HDAC inhibitory activities. Int. J. Biochem. Cell Biol., 39, 1902–1914 [DOI] [PubMed] [Google Scholar]
- 38. Gramling S., et al. (2011). Pharmacologic reversal of epigenetic silencing of the anticancer protein BRM: a novel targeted treatment strategy. Oncogene, 30, 3289–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizutani T., et al. (2002). Maintenance of integrated proviral gene expression requires Brm, a catalytic subunit of SWI/SNF complex. J. Biol. Chem., 277, 15859–15864 [DOI] [PubMed] [Google Scholar]
- 40. Bourachot B., et al. (2003). Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation. EMBO J., 22, 6505–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kahali B., et al. (2014). Identifying targets for the restoration and reactivation of BRM. Oncogene, 33, 653–664 [DOI] [PubMed] [Google Scholar]
- 42. Trotter K.W., et al. (2004). Reconstitution of glucocorticoid receptor-dependent transcription in vivo . Mol. Cell. Biol., 24, 3347–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hou D.X., et al. (2010). Flavonoids as protein kinase inhibitors for cancer chemoprevention: direct binding and molecular modeling. Antioxid. Redox Signal., 13, 691–719 [DOI] [PubMed] [Google Scholar]
- 44. Lee S.O., et al. (2007). Silibinin suppresses PMA-induced MMP-9 expression by blocking the AP-1 activation via MAPK signaling pathways in MCF-7 human breast carcinoma cells. Biochem. Biophys. Res. Commun., 354, 165–171 [DOI] [PubMed] [Google Scholar]
- 45. Reisman D., et al. (1995). Glucocorticoid regulation of cyclin D3 gene transcription and mRNA stability in lymphoid cells. Mol. Endocrinol., 9, 1500–1509 [DOI] [PubMed] [Google Scholar]
- 46. Liu G., et al. (2011). Two novel BRM insertion promoter sequence variants are associated with loss of BRM expression and lung cancer risk. Oncogene, 30, 3295–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li L., et al. (2012). Baicalin is anti-inflammatory in cigarette smoke-induced inflammatory models in vivo and in vitro: a possible role for HDAC2 activity. Int. Immunopharmacol., 13, 15–22 [DOI] [PubMed] [Google Scholar]
- 48. Gilbert E.R., et al. (2010). Flavonoids influence epigenetic-modifying enzyme activity: structure—function relationships and the therapeutic potential for cancer. Curr. Med. Chem., 17, 1756–1768 [DOI] [PubMed] [Google Scholar]
- 49. Wang Y., et al. (2006). Prevention of lung cancer progression by bexarotene in mouse models. Oncogene, 25, 1320–1329 [DOI] [PubMed] [Google Scholar]
- 50. Hollman P.C., et al. (1995). Absorption of dietary quercetin glycosides and quercetin in healthy ileostomy volunteers. Am. J. Clin. Nutr., 62, 1276–1282 [DOI] [PubMed] [Google Scholar]
- 51. Hollman P.C., et al. (1998). Bioavailability and health effects of dietary flavonols in man. Arch. Toxicol. Suppl., 20, 237–248 [DOI] [PubMed] [Google Scholar]
- 52. Yang C.S., et al. (2001). Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu. Rev. Nutr., 21, 381–406 [DOI] [PubMed] [Google Scholar]
- 53. Yang K., et al. (2000). Chemoprevention studies of the flavonoids quercetin and rutin in normal and azoxymethane-treated mouse colon. Carcinogenesis, 21, 1655–1660 [DOI] [PubMed] [Google Scholar]
- 54. Singh R.P., et al. (2002). Dietary feeding of silibinin inhibits advance human prostate carcinoma growth in athymic nude mice and increases plasma insulin-like growth factor-binding protein-3 levels. Cancer Res., 62, 3063–3069 [PubMed] [Google Scholar]
- 55. Singh R.P., et al. (2006). Effect of silibinin on the growth and progression of primary lung tumors in mice. J. Natl. Cancer Inst., 98, 846–855 [DOI] [PubMed] [Google Scholar]
- 56. Lambert J.D., et al. (2008). Effect of genistein on the bioavailability and intestinal cancer chemopreventive activity of (-)-epigallocatechin-3-gallate. Carcinogenesis, 29, 2019–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Angst E., et al. (2013). The flavonoid quercetin inhibits pancreatic cancer growth in vitro and in vivo . Pancreas, 42, 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ni F., et al. (2012). Flavonoid ampelopsin inhibits the growth and metastasis of prostate cancer in vitro and in mice. PLoS One, 7, e38802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manach C., et al. (2004). Pharmacokinetics and metabolism of dietary flavonoids in humans. Free Radic. Res., 38, 771–785 [DOI] [PubMed] [Google Scholar]
- 60. Birt D.F., et al. (2001). Dietary agents in cancer prevention: flavonoids and isoflavonoids. Pharmacol. Ther., 90, 157–177 [DOI] [PubMed] [Google Scholar]
- 61. Miller Y.E., et al. (2003). Induction of a high incidence of lung tumors in C57BL/6 mice with multiple ethyl carbamate injections. Cancer Lett., 198, 139–144 [DOI] [PubMed] [Google Scholar]
- 62. Thaete L.G., et al. (1987). Cellular derivation of lung tumors in sensitive and resistant strains of mice: results at 28 and 56 weeks after urethan treatment. J. Natl. Cancer Inst., 78, 743–749 [PubMed] [Google Scholar]
- 63. Meuwissen R., et al. (2005). Mouse models for human lung cancer. Genes Dev., 19, 643–664 [DOI] [PubMed] [Google Scholar]
- 64. Lu G., et al. (2006). Inhibition of adenoma progression to adenocarcinoma in a 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorigenesis model in A/J mice by tea polyphenols and caffeine. Cancer Res., 66, 11494–11501 [DOI] [PubMed] [Google Scholar]
- 65. Dagne A., et al. (2011). Enhanced inhibition of lung adenocarcinoma by combinatorial treatment with indole-3-carbinol and silibinin in A/J mice. Carcinogenesis, 32, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. de Wet H., et al. (2001). Sequence requirements of the ATP-binding site within the C-terminal nucleotide-binding domain of mouse P-glycoprotein: structure-activity relationships for flavonoid binding. Biochemistry, 40, 10382–10391 [DOI] [PubMed] [Google Scholar]
- 67. Dahiya A., et al. (2000). Role of the LXCXE binding site in Rb function. Mol. Cell. Biol., 20, 6799–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ramos S. (2008). Cancer chemoprevention and chemotherapy: dietary polyphenols and signalling pathways. Mol. Nutr. Food Res., 52, 507–526 [DOI] [PubMed] [Google Scholar]
- 69. De Azevedo W.F., Jr, et al. (1996). Structural basis for specificity and potency of a flavonoid inhibitor of human CDK2, a cell cycle kinase. Proc. Natl. Acad. Sci. USA, 93, 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Y., et al. (2010). Interactions of dietary phytochemicals with ABC transporters: possible implications for drug disposition and multidrug resistance in cancer. Drug Metab. Rev., 42, 590–611 [DOI] [PubMed] [Google Scholar]
- 71. Arango D., et al. (2013). Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc. Natl. Acad. Sci. USA, 110, E2153–E2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hostetler G., et al. (2012). Flavone deglycosylation increases their anti-inflammatory activity and absorption. Mol. Nutr. Food Res., 56, 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chung J.Y., et al. (2001). Mechanisms of inhibition of the Ras-MAP kinase signaling pathway in 30.7b Ras 12 cells by tea polyphenols (-)-epigallocatechin-3-gallate and theaflavin-3,3’-digallate. FASEB J., 15, 2022–2024 [DOI] [PubMed] [Google Scholar]
- 74. Yang C.S., et al. (1998). Tea and tea polyphenols inhibit cell hyperproliferation, lung tumorigenesis, and tumor progression. Exp. Lung Res., 24, 629–639 [DOI] [PubMed] [Google Scholar]
- 75. Yang G., et al. (1997). Characterization of early pulmonary hyperproliferation and tumor progression and their inhibition by black tea in a 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorigenesis model with A/J mice. Cancer Res., 57, 1889–1894 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.