Summary
This study reveals an association of NNMT with microRNA-1291-altered pancreatic cell metabolome and tumorigenesis in xenograft tumor mouse models.
Abstract
The cell metabolome comprises abundant information that may be predictive of cell functions in response to epigenetic or genetic changes at different stages of cell proliferation and metastasis. An unbiased ultra-performance liquid chromatography–mass spectrometry-based metabolomics study revealed a significantly altered metabolome for human pancreatic carcinoma PANC-1 cells with gain-of-function non-coding microRNA-1291 (miR-1291), which led to a lower migration and invasion capacity as well as suppressed tumorigenesis in a xenograft tumor mouse model. A number of metabolites, including N-methylnicotinamide, involved in nicotinamide metabolism, and l-carnitine, isobutyryl-carnitine and isovaleryl-carnitine, involved in fatty acid metabolism, were elevated in miR-1291-expressing PANC-1. Notably, N-methylnicotinamide was elevated to the greatest extent, and this was associated with a sharp increase in nicotinamide N-methyltransferase (NNMT) mRNA level in miR-1291-expressing PANC-1 cells. In addition, expression of NNMT mRNA was inversely correlated with pancreatic tumor size in the xenograft mouse model. These results indicate that miR-1291-altered PANC-1 cell function is associated with the increase in N-methylnicotinamide level and NNMT expression, and in turn NNMT may be indicative of the extent of pancreatic carcinogenesis.
Introduction
Pancreatic cancer remains a highly lethal cancer disease with an extremely poor prognosis and the lowest survival rate among all types of malignancies in the USA and worldwide (1,2). Thus, there has been increasing efforts to improve the understanding of pancreatic cancer biology, define more effective druggable targets and identify early detection biomarkers (3–7). Non-coding microRNAs (miRs or miRNAs) are master regulators in the control of cancer cellular processes via modulating target gene expression (8–10). Some miRNAs are aberrantly expressed in pancreatic ductal adenocarcinoma (PDAC) patients (11–13) and a few miRNAs can modulate pancreatic cancer proliferation and tumor progression (14–23), which may serve as novel diagnostic/prognostic biomarkers and/or therapeutic targets.
Recently, we found that miR-1291 is significantly downregulated in human PDAC tissues, and restoration of miR-1291 function represses the tumorigenesis of pancreatic carcinoma cells in a xenograft tumor mouse model (24). Other studies (25,26) also showed that miR-1291 reduces the growth of renal cell carcinoma cells. To gain insight into true endpoints and biomarkers of miR-1291-triggered suppression of pancreatic carcinogenesis, an ultra-performance liquid chromatography-electrospray ionization-quadrupole time-of-flight-mass spectrometry (UPLC-ESI-QTOF-MS)-based metabolomics approach was employed to define the role of miR-1291 in human pancreatic carcinoma cell metabolism. Unbiased and targeted analysis of cellular metabolites led to the identification and validation of N-methylnicotinamide (NMN) as a potential biomarker within PANC-1 cells with lower migration and invasion capacity as a result of gain of miR-1291 function. This was associated with a sharp increase in nicotinamide N-methyltransferase (NNMT) mRNA levels. In addition, an inverse relationship was uncovered between xenograft pancreatic tumor size and cancer cell NNMT mRNA levels, suggesting an important role for NNMT in miR-1291-altered PANC-1 cell metabolome and carcinogenesis.
Materials and methods
Materials
Dulbecco’s modified Eagle’s medium, penicillin sodium and streptomycin sulfate solution were purchased from Mediatech (Manassas, VA). Fetal bovine serum was bought from Lonza (Walkersville, MD), and Trizol was purchased from Life Technologies (Carlsbad, CA). BCA Protein Assay Kit was bought from Thermo Scientific (Rockford, IL). N-methylnicotinamide iodide, taurine, l-carnitine, chlorpropamide and aminopimelic acid were purchased from Sigma-Aldrich (St Louis, MO). Isobutyryl-carnitine and isovaleryl-carnitine were bought from the Metabolic Laboratory, Vrije Universiteit Medical Center (Amsterdam, The Netherlands). All other chemicals and solvents were of the highest grade commercially available.
Cell lines and cell culture
Human pancreatic carcinoma PANC-1 cells were purchased from American Type Culture Collection (Manassas, VA). MiR-1291 and empty vector (control) stably transfected PANC-1 cells were established, as we described recently (27), and maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 100U/ml penicillin sodium, and 100 µg/ml streptomycin sulfate at 37°C in a humidified atmosphere of 5% CO2. MiR-1291-expressing and control PANC-1 cells were subcultured in 10cm polystyrene dishes at a density of 5 × 104 cells per dish and grown to ~90% confluence for metabolomics analysis. The cell lines were re-authenticated at the Animal Health Diagnostic Laboratory (NCI-Frederick) as pathogen free and they were passaged for <6 months after receipt or resuscitation for the metabolomics and animal studies.
Cell migration and invasion
Transwell migration and Matrigel invasion assays were employed to evaluate cell migration and invasion capacities, respectively. In particular, a suspension of 3 × 104 cells were added to cell culture inserts containing a polycarbonate filter with 8 µm diameter pores (Corning, Tewksbury, MA) blocked with 2.5% bovine serum albumin or coated with 1:5 dilution Matrigel (100 µl per well). Cells were incubated for 17 h under standard culture conditions. Cells remaining on the topside of the membrane or gel were removed, and cells migrated to the underside were fixed and stained with Diff-Quik (PolyScience, Warrington, PA). Five fields per insert were photographed and the number of cells was counted under microscope.
Cell sample collection for metabolomics
Cells were rapidly rinsed twice by gently dispensing 5.0 ml isotonic saline (37°C) to the cell surface. With the addition of 1 ml ice cold water, the dish was flash frozen in liquid nitrogen. The cells were detached using a cell scraper, and the cell suspensions were transferred into 1.5 ml tubes. Cell samples were lysed by two freeze-thaw cycles (fresh frozen in liquid nitrogen and thawed at 37°C for 10min), followed by sonication on ice for 30 s. Protein concentrations were measured using the BCA Protein Assay Kit. The cell suspensions were stored at −80°C until analysis.
Sample preparation for metabolomics
To 300 µl of cell suspension, 900 µl of chilled extraction solvent 75% methanol:chloroform (90:10 vol/vol) containing internal standard (5 µM chlorpropamide for reversed-phase liquid chromatography (RPLC) mode; 25 µM aminopimelic acid for hydrophilic interaction liquid chromatography (HILIC) mode) was added. The mixture was vigorously vortexed for 30 s and centrifuged at 14 000g for 15min at 4°C to remove proteins and particles. The supernatants were transferred to fresh glass tubes and dried under nitrogen. The residue was resuspended in 200 µl of 70% acetonitrile (for HILIC mode) or 200 µl of 35% acetonitrile (for RPLC mode). The mixture was centrifuged at 14 000g for 5min at 4°C, and 5 µl of the sample was injected for UPLC-ESI-QTOF-MS analysis. Pooled samples were also made as quality controls for all the extractions, which comprised 5 µl of individual samples.
RPLC and HILIC-UPLC-QTOF-MS analysis
Two complementary chromatographic approaches were used, i.e. RP chromatography for non-polar analytes and HILIC chromatography for polar analytes. For the RPLC metabolomics profiling, samples were separated on a RP 50 × 2.1mm, 1.7 µm ACQUITY BEH C18 column (Waters Corp., Milford, MA) using an ACQUITY UPLC system (Waters Corp). A gradient elution with 0.1% aqueous formic acid (Solution A) and acetonitrile containing 0.1% formic acid (Solution B) was conducted, specifically 2% Solution B for 0.5min and gradually increased to 20% at 4.0min then 95% at 8min. The flow rate was 0.5 ml/min, and the column was washed with 98% Solution B for 1min then equilibrated with 98% Solution A before the next injection.
For the HILIC metabolomics profiling, samples were separated on a 50 × 2.1mm, 1.7 µm ACQUITY BEH Amide column using an ACQUITY UPLC H-class system (Waters Corp.). A gradient elution with 10mM ammonium acetate in 10% acetonitrile (Solution C) and 10mM ammonium acetate in 90% acetonitrile pH 9.0 (Solution D) was carried out at a flow rate of 0.4 ml/min during a 12 min run. In particular, 99% Solution D was held for 0.5 min and decreased to 60% at 6.0min and to 20% at 8min. The gradient was held for 1min and then returned to 99% Solution D for 2min for column equilibration.
RPLC-MS analysis was performed on a Waters Synapt Q-TOF MS system operated in both ESI positive (ESI+) and negative (ESI−) modes. The capillary voltage and cone voltage were set to 3000 and 20V, respectively. Source and desolvation temperature were 120 and 350°C, respectively. Nitrogen was used as both cone gas (50 l/h) and desolvation gas (650 l/h), and argon was used as collision gas. HILIC-MS analysis was conducted on a Waters Xevo G2 Q-TOF MS system operated in ESI+ and ESI− modes. The capillary voltage and cone voltage were set to 3000 and 30V, respectively. Source and desolvation temperatures were 150 and 550°C, respectively. Nitrogen was used as both cone gas (50 l/h) and desolvation gas (850 l/h), and argon was used as collision gas. Mass accuracy was maintained on both systems using Waters LockSpray technology, and sulfadimethoxine was used as the lock mass. For MS scanning, data were acquired in centroid mode from 50 to 850 m/z and for MS/MS fragmentation of target ions, the collision energy was ramped from 5 to 40eV. To avoid artifacts based on sample injection order, the samples were randomized, including pooled, blank and standard samples. Mass chromatograms and mass spectral data were acquired using MassLynx software (Waters) in centroid format.
Multivariate data analysis
The MS data were centroided, integrated and deconvoluted to generate a multivariate data matrix using MarkerLynx. Peak picking, alignment, deisotoping and integration were performed automatically by the software with optimized values of parameters such as mass tolerance, peak width, peak-to-peak baseline noise, intensity threshold, mass window, retention time window and noise elimination level. The raw data were transformed into a multivariate matrix containing aligned peak areas with matched mass-to-charge ratios (m/z) and retention times. The data were normalized to both protein concentration and peak area of the internal standards (chlorpropamide for RPLC mode which exhibited a retention time of 5.3min, m/z 275.026 [M–H]− and 277.041 [M+H]+; aminopimelic acid for HILIC mode which showed a retention time of 4.9min, m/z 174.076 [M–H]− and 176.092 [M+H]+). The multivariate data were thus analyzed with SIMCA-P+12 software (Umetrics, Kinnelon, NJ). The unsupervised segregation of control and miR-1291-expressing PANC-1 cells was done by principal components analysis (PCA) using Pareto-scaled data. The supervised orthogonal projection to latent structures discriminant analysis (OPLS-DA) was also constructed to compare the control and miR-1291-expressing PANC-1 cell metabolomics. A list of ions showing considerable group discriminating power (−0.8 > P(corr.)[1] or P(corr.)[1] > 0.8) was generated from the loading S-plot.
Metabolite identification
Ions that were significantly and consistently altered in the group of miR-1291-expressing cells were used for further identification. Metabolomics databases including HMDB (http://www.hmdb.ca), MMCD (http://mmcd.nmrfam.wisc.edu) and METLIN (http://metlin.scripps.edu) were searched to discover possible candidates for these ions. A mass error of 10 ppm in the respective ionization modes was taken into account. Further confirmation of the identities of individual ions was carried out by comparing the retention times and fragmentation patterns with authentic standards.
Quantitation of intracellular metabolites
Target metabolites were quantitated by multiple reaction monitoring on an ACQUITY UPLC coupled to a XEVO TQ triple-quadrupole tandem MS system (Waters). Multiple reaction monitoring transitions were carried out for 1-methylnicotinamide (137.1 → 94.1 m/z ESI+), l-carnitine (162.2 → 102.8 m/z ESI−), isobutyryl-carnitine (232.2 → 173.1 m/z ESI+), isovaleryl-carnitine (246.2 → 187.2 m/z ESI+) and taurine (125.9 → 43.9 m/z ESI−). UPLC (HILIC) conditions were the same as described above. Cell samples were extracted using 75% methanol:chloroform (90:10, vol/vol) as described above, resuspended in 200 µl acetonitrile:water:methanol (65:35:5, vol/vol/vol) and then diluted 1:10 with acetonitrile:water (80:20, vol/vol). Aminopimelic acid (internal standard, 176.2 → 112.2 m/z ESI+) was added to each sample with a final concentration of 3 µM. Standard calibration curves of 1nM to 10 µM were generated for 1-methylnicotinamide (r 2 = 0.9984), l-carnitine (r 2 = 0.9913), isobutyryl-carnitine (r 2 = 0.9976), isovaleryl-carnitine (r 2 = 0.9951) and taurine (r 2 = 0.9956) using authentic standards. Samples were quantitated using TargetLynx software (Waters), and concentration of each metabolite was normalized to the protein concentration of cells.
Human pancreatic cancer xenograft mouse model
Animal protocol was approved by the Institutional Animal Care and Use Committee at National Cancer Institute. Athymic male nude (CD-1 nu/nu) mice (8 weeks old) were purchased from Charles River Laboratories (Wilmington, MA) and were maintained under standard 12h light/12h dark cycle with water and chow provided ad libitum. Exponentially growing miR-1291-expressing PANC-1 cells and control PANC-1 cells were harvested and injected subcutaneously into the mice (5 × 106 cells in 0.2 ml of 50% Matrigel per animal, and six mice per group). Body weight and tumor size were monitored once a week. Tumor volumes were calculated as V = 0.5 × D × d 2, where D and d represent the larger and the smaller tumor diameter, respectively. Mice were killed when the tumor reached 2cm in diameter, and xenograft tumors harvested and flash frozen in liquid nitrogen.
Pancreatic cancer patient samples and demographic characteristics of donors
The tumor tissues from PDAC patients were collected and validated by clinical pathologists according to a standardized quality control procedure at Roswell Park Cancer Institute (RPCI) in Buffalo, NY. Demographic and clinical information of individual donors were collected as well. A total of 22 tissue specimens from non-Spanish, White patients were used for this study. Among them 7 donors are females and 15 donors are males. The ages of patients were between 31 and 84 years at enrollments, with a mean value of 63.3 and SD of 13.6. Use of the patient samples was reviewed and approved by the Office of Research Subject Protection (ORSP) at RPCI.
Analysis of gene expression in the cell, xenograft and patient samples
Total RNA was isolated from freshly harvested cells and frozen tumor samples with Trizol reagent and quantified with NanoDrop (Thermo Scientific). cDNA was synthesized from 1 µg total RNA using Superscript II reverse transcriptase kit (Invitrogen, Carlsbad, CA). Quantitative real-time PCR (qPCR) analysis was conducted with gene specific primers (Supplementary Table S1, available at Carcinogenesis Online), and β-actin (xenograft tumors) and GAPDH (patient samples) were used as internal control. The stem-loop reverse transcription and qPCR analysis of mature hsa-miR-1291, as well as the U74 control RNA, were conducted as reported (27). All qPCR reactions were performed in triplicate on an Applied Biosystems Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). The relative level of each analyte over internal standard (β-actin, GAPDH or U74) was calculated using the formula 2( −Δ Δ CT), where ΔΔC T = ΔC T (analyte) − ΔC T (internal standard), and then compared between different groups.
Statistical analysis
All values were expressed as mean ± SD. Student’s t-test or two-way analysis of variance was used to analyze the difference in gene expression or other parameters between distinct groups using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Spearman correlation analysis and Kaplan–Meier survival analysis were performed using GraphPad Prism 5. Difference was considered as statistically significant if the probability (P value) was <0.05.
Results
Morphology, migration and invasion of miR-1291-expressing and control PANC-1 cells
MiR-1291-expressing PANC-1 cells showed similar morphology as the control cells (Figure 1A and B), indicating a minimal impact of miR-1291 on overall cell morphology. Nevertheless, the 16-fold increase in miR-1291 expression (Figure 1C) resulted in a significant reduction of cell proliferation, which was mainly due to the difference at late stages (day 6 and 7; Figure 1D). Given the fact that invasive migration is a critical function for tumor progression and metastasis, the effect of miR-1291 on migration and invasion of human pancreatic cancer cells was measured. The data revealed that migration (Figure 2A) and invasion (Figure 2B) capacities of the miR-1291-expressing PANC-1 cells decreased 60 and 80%, respectively, compared to control cells. The results suggest that miR-1291 reduces the migration and invasion ability of PANC-1 cells that are critical for tumor progression and metastasis.
Fig. 1.
The morphology of control (A) and miR-1291-expressing PANC-1 (B) cells showed no difference, whereas an increased miR-1291 expression (C; *P < 0.05, Student’s t-test) led to a significant suppression (*P < 0.05, two-way analysis of variance) of cell proliferation (D). Values are mean ± SD (N = 6 in each group).
Fig. 2.
Transwell migration (A) and Matrigel invasion (B) capacity of the PANC-1-expressing cells with gained miR-1291 function were sharply reduced. Values are mean ± SD (N = 6 in each group). *P < 0.05, as compared to the control.
Metabolomic profiling segregated miR-1291-expressing PANC-1 cells from control cells
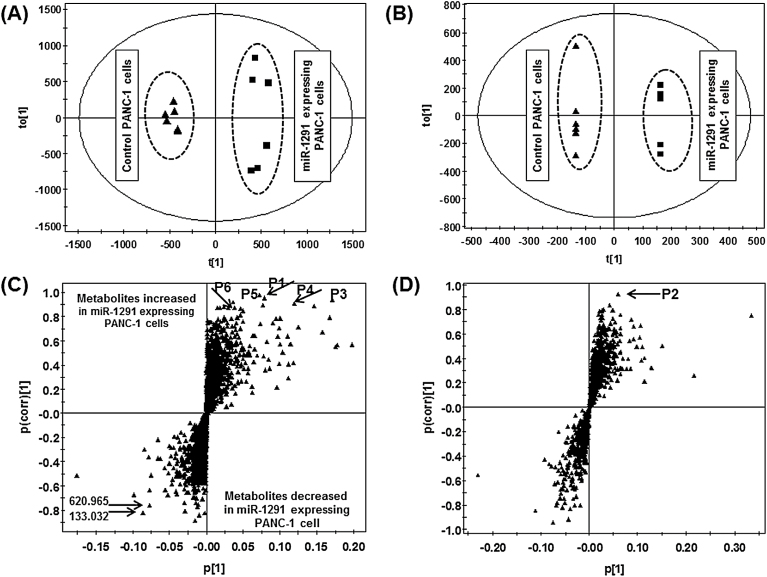
MiR-1291-expressing and control PANC-1 cells were subjected to metabolomics analyses. Although unsupervised PCA of the metabolomic profiles obtained from RPLC-ESI+/ESI−-MS did not yield a separation of the miR-1291-expressing PANC-1 cells from control cells (Supplementary Figure S1A and S1B, available at Carcinogenesis Online), PCA of the data obtained from HILIC-ESI+/ESI−-MS showed an improved segregation of the two groups (Supplementary Figure S1C and S1D, available at Carcinogenesis Online). This is likely due to the difference in analyzing positively versus negatively charged cellular metabolites after RP versus HILIC chromatographic separation. Supervised OPLS-DA successfully determined the differences between control and miR-1291-expressing cells (R 2 > 0.9, Q 2 > 0.7) using HILIC-ESI+-MS (Figure 3A) and RPLC-ESI−-MS (Figure 3B) or HILIC-ESI−-MS and RPLC-ESI+-MS (Supplementary Figure S2A and S2B, available at Carcinogenesis Online) data, which indicates an underlying difference between their cellular metabolomes. The loadings S-plots for the OPLS-DA models (Figure 3C and D and Supplementary Figure S2C and S2D, available at Carcinogenesis Online) were thus used to identify ions (Table I) that were highly altered in the cells (P(corr.)[1] > 0.8). Interestingly, most of the ions were elevated in miR-1291-expressing cells, e.g. m/z 137.072 [M+H]+, 124.007 [M−H]−, 162.113 [M+H]+, 232.156 [M+H]+, 246.171 [M+H]+ and 248.147 [M+H] + (P1, P2, P3, P4, P5 and P6, respectively), which were further investigated in the present study.
Fig. 3.
Scores scatter plot and loading S-plot from the supervised OPLS analyses distinguished miR-1291-expressing PANC-1 cells from the control cells. The top six ions (P1–P6) showing significant differences in two groups are marked in the S-plot. (A) Score plot under ESI+-HILIC mode; (B) score plot under ESI−-RPLC mode; (C) S-plot under ESI+-HILIC mode; (D) S-plot under ESI−-RPLC mode.
Table I.
Ions that were significantly altered in miR-1291-expressing PANC-1 cells, as revealed by HILIC and RPLC-MS metabolomics
| Retention time (min) |
Experimental ion mass (m/z) | Mass error (ppm) |
Trend | Identity |
|---|---|---|---|---|
| HILIC+ | ||||
| 2.5634 | 162.1113 | −6.8 | Increased | l-Carnitine |
| 0.9599 | 232.1541 | −3.4 | Increased | Isobutyryl-carnitine |
| 0.8125 | 246.1697 | −3.2 | Increased | Isovaleryl-carnitine |
| 1.7656 | 248.1502 | −1.4 | Increased | Hydroxybutyryl-carnitine |
| 2.1291 | 137.0718 | 2.2 | Increased | 1-Methylnicotinamide |
| 1.2139 | 133.0324 | — | Decreased | Unknown |
| 2.0519 | 620.9654 | — | Decreased | Unknown |
| 1.1988 | 144.1026 | — | Increased | Unknown |
| 0.9572 | 173.0792 | — | Increased | Unknown |
| 0.3951 | 313.2698 | — | Increased | Unknown |
| RPLC+ | ||||
| 0.4015 | 162.1119 | −8.6 | Increased | l-Carnitine |
| 0.4223 | 258.0986 | — | Increased | Unknown |
| 0.4281 | 160.1319 | — | Increased | Unknown |
| RPLC− | ||||
| 0.3592 | 124.007 | 1.6 | Increased | Taurine |
| HILIC− | ||||
No potential markers were found from HILIC data set.
Identification of metabolomic markers highly altered in miR-1291-expressing cells
The identities of individual ions (Table I) exhibiting consistent and remarkable changes in miR-1291-expressing PANC-1 cells were determined after conducting online database searches [Human Metabolome Database (HMDB) and Madison Metabolomics Consortium Database (MMCD)] and further validated by comparing the retention times and MS fragmentations with those of authentic standards (Supplementary Figures S3 and S4, available at Carcinogenesis Online). The ion P1, m/z 137.072 [M+H]+, was identified as NMN. With the same retention time of 2.11min under the HILIC condition, the authentic standard and cellular NMN both showed characteristic ESI+-TOP MS fragments 94.066+, 78.035+ and 67.054+ (Supplementary Figure S3, available at Carcinogenesis Online). Similarly, ions P2–P5 were defined as taurine, l-carnitine, isobutyryl-carnitine and isovaleryl-carnitine, respectively (Supplementary Figure S4, available at Carcinogenesis Online). Specifically, the authentic and cellular taurine, m/z 124.007 [M–H]−, were fragmented to 81.960−, 79.957− and 17.214−. Tandem MS of the authentic standard and cellular l-carnitine, m/z 162.111 [M+H]+, exhibited the same fragment ions including 120.045+, 103.039+, 85.029+ and 60.081+. Standard and cellular isobutyryl-carnitine, m/z 232.156 [M+H]+, shared the same fragmentation pattern 173.082+, 144.103+, 85.029+ and 60.081+. Cellular isovaleryl-carnitine and the standard, m/z 246.171 [M+H]+, had the same fragments 187.097+, 85.029+ and 60.080+, and the putative cellular hydroxybutyryl-carnitine, m/z 248.147 [M+H]+, exhibited MS/MS fragments of 189.075+, 149.021+, 85.028+ and 77.038+.
Quantitative analyses revealed 1-methylnicotinamide as a potential biomarker for the altered metabolism of miR-1291-expressing PANC-1 cells
The concentrations of cellular metabolites P1–P5 in individual cell samples (Figure 4) were further quantified using specific multiple reaction monitoring methods after establishing the corresponding calibration curves. Intracellular levels of NMN (P1), the most important biomarker revealed by both PCA and OPLS-DA, were elevated to the highest extent in miR-1291-expressing PANC-1 cells compared to the control cells (162±16 versus 0.62±0.60nM/mg protein or 270-fold increase; Figure 4A). Taurine (P2) was abundant in PANC-1 cells and its concentrations were significantly higher in the miR-1291-expressing cells (1200±154 versus 535±130nM/mg protein; Figure 4B). l-Carnitine (P3) was increased 1.8-fold in miR-1291-expressing cells (70.6±13.7 versus 39.3±3.6nM/mg protein; Figure 4C). Low abundant isobutyryl-carnitine (P4) and isovaleryl-carnitine (P5) were ~1- and 5-fold, respectively, higher in miR-1291-expressing cells (Figure 4D and 4E). Last, relative levels of hydroxybutyryl-carnitine were determined due to the lack of authentic standard, which were elevated 2-fold in the miR-1291-expressing cells (Figure 4F).
Fig. 4.
Cellular concentrations of NMN (A), taurine (B), l-carnitine (C), isobutyryl-carnitine (D) and isovaleryl-carnitine (E) as well as the relative levels of hydroxybutyryl-carnitine (F) in the control and miR-1291-expressing PANC-1 cells. Data were normalized to protein concentration of individual samples. Values are mean ± SD (N = 6 in each group). *P < 0.05, compared to the control.
A remarkably upregulated NNMT gene expression was linked to the increased levels of cellular 1-methylnicotinamide
To understand the mechanisms underlying the alteration of these metabolites, the expression of genes (Figure 5) related to their biosynthesis or biodegradation in the cells were determined by qPCR analyses. NNMT [EC 2.1.1.1] is the only enzyme known to produce metabolite NMN, the top candidate biomarker, from nicotinamide in the pathway of nicotinate and nicotinamide metabolism [Kyoto Encyclopedia of Genes and Genomes (KEGG) database, http://www.genome.jp/kegg]. Carnitine and short-chain acylcarnitines such as isobutyryl-carnitine, isovaleryl-carnitine and hydroxybutyryl-carnitine were identified as other possible biomarkers. Two enzymes involved in the carnitine pathway are carnitine O-acetyltransferase (CRAT) and carnitine palmitoyltransferases (CPTs). The relative mRNA levels of NNMT were found to be elevated ~40-fold in miR-1291-expressing PANC-1 cells compared to the control cells (Figure 5A). CPT1A and CPT1C mRNA levels were also significantly higher in miR-1291-expressing PANC-1 cells (Figure 5B and D) that are associated with the increase of carnitine and acylcarnitines, whereas the CPT1B, CPT2 and CRAT mRNA levels remained unchanged (Figure 5C, E and F). The result suggests that CPT1A and CPT1C involved in the carnitine metabolic pathway might play an important role in energy metabolism of PANC-1 cells, and the increase in intracellular NMN may be attributed to the dramatic upregulation of NNMT.
Fig. 5.
Cellular mRNA levels of genes associated with the metabolomics markers including (A) NNMT, (B) CPT1A, (C) CPT1B, (D) CPT1C, (E) CPT2 and (F) CRAT. β-Actin was used as an internal control. Values are mean ± SD (N = 6 per group). *P < 0.05, compared to the control.
NNMT mRNA level was inversely related to the size of xenograft tumor derived from miR-1291-expressing and control PANC-1 cells
Expression of possible biomarker genes in xenograft tumors derived from PANC-1 cells was next determined. Nude mice (N = 6 per group and repeated twice) were inoculated with miR-1291-expressing and control PANC-1 cells, respectively. No mortality occurred during the experiments before tissue collection. The body weights of mice inoculated with miR-1291-expressing PANC-1 cells increased at a higher rate than the control mice (Supplementary Figure S5, available at Carcinogenesis Online). The rate of tumor formation of control PANC-1 cells was 100%, whereas there was minimal or no tumors formed from miR-1291-expressing cells (Figure 6A–D). The mRNAs encoding NNMT, CRAT, CPT1A, CPT1B, CPT1C and CPT2 in the xenograft tumors were thus measured and their possible correlations with tumor sizes were evaluated. The results revealed that the sizes of xenograft tumors derived from miR-1291-expressing and control PANC-1 cells were inversely corrected with NNMT mRNA levels (Spearman R = −0.7626; 95% confidence interval; P = 0.0015; Figure 6E) rather than any other genes investigated, suggesting that NNMT may be a potential biomarker for pancreatic tumor progression.
Fig. 6.
NNMT mRNA level was inversely related to the size of xenograft tumor. (A) Representative mice bearing xenograft tumors derived from miR-1291-expressing or control PANC-1 cells. (B) Growth of xenograft tumors was significantly different (*P < 0.05, two-way analysis of variance) between miR-1291-expressing and control PANC-1 cells. (C) Comparison of tumors excised from xenograft mice at week 7 after inoculation. (D) Weight of xenograft tumors harvested from mice. (E) Inverse relationship between the xenograft tumor size and cancerous NNMT mRNA level. Data mean ± SD (N = 6). *P < 0.05, compared to the control.
Use of NNMT expression to predict overall survival of PDAC patients
To evaluate the effectiveness of NNMT as a possible prognostic biomarker for PDAC, we quantitated NNMT mRNA levels in the tissue specimens collected from a small set of PDAC patients (N = 22). The result showed that there was no different in overall survival rate among PDAC patients with low and high expression of NNMT (P = 0.531, Kaplan–Meier log-rank test), despite that survival rate tended to be higher for patients at later time points (e.g. over 12 months after diagnosis) (Supplementary Figures S6, available at Carcinogenesis Online). Follow-up studies with larger patient sample sizes would permit an improved understanding of NNMT as a possible biomarker for PDAC patients.
Discussion
Metabolism is a critical cellular process, and cell metabolome provides abundant information on the changes of particular metabolic pathways and biochemical reactions in response to genetic, epigenetic, developmental and environmental factors as well as disease progression. Metabolomic profiling would be valuable for disease diagnosis and prognosis (28) and provide insights into mechanisms underlying cell functions. Through UPLC-MS-based untargeted metabolomics and LC-MS/MS-based targeted metabolite analysis, the present study reveals a significant alteration of PANC-1 cell metabolome after gain of miR-1291 function, which results in the suppression of pancreatic cell migration, invasion and xenograft tumorigenesis. Increase of NMN cellular level in miR-1291-expressing cells can be explained by the sharp elevation of NNMT mRNA level. Interestingly, NNMT mRNA levels are inversely correlated with the volumes of pancreatic xenograft tumors. The results indicate that NMN and NNMT are potential biomarkers in miR-1291-altered pancreatic cell metabolism and tumorigenesis.
Cellular NMN levels are elevated to the highest extent among a number of significantly changed metabolites in the miR-1291-expressing PANC-1 cells. NMN is the N-methylation product from nicotinamide catalyzed by NNMT within nicotinate and nicotinamide metabolism pathway (KEGG; http://www.genome.jp/kegg). Nicotinamide is the precursor of coenzyme nicotinamide adenine dinucleotide (NAD) that participates in many critical redox reactions in glycolysis and Krebs cycle of cell respiration. Biotransformation of nicotinamide to NMN requires the use of S-adenosyl-l-methionine (SAM) as methyl group donor that is consequently converted to S-adenosyl-l-homocysteine (29). It is tempting to speculate that the dramatic increase in NMN might be accompanied by a reduction of nicotinamide or SAM and/or increase of S-adenosyl-l-homocysteine. However, no significant changes of nicotinamide, nicotinate, SAM or S-adenosyl-l-homocysteine were found after analysis of the metabolomics data (data not shown). This is consistent with recent findings that addition of NMN does not affect NAD levels in human glioma cells (30) and that overexpression of NNMT does not decrease SAM levels in the Y20A cells (31). These findings indicate that an alteration of NMN and NNMT may not necessarily change cellular nicotinamide, NAD or SAM.
Recent studies also suggest that NMN may exhibit a variety of bioactivities (32–34) despite that NMN has been regarded as biologically inactive (35). A single bout of endurance exercise is shown to induce hepatic NNMT expression and plasma NMN level in animal models (36), and physical excise promotes mean lifespan in humans (37). NMN itself is shown to extend Caenorhabditis elegans lifespan at physiologically relevant concentrations through the induction of a transient reactive oxygen species signal, even in the absence of NAD-dependent protein deacetylase sir-2.1 (38). Interestingly, high doses of NMN exert the opposite effect, i.e. shortening the lifespan, presumably by producing excessive reactive oxygen species load (38). Therefore, in addition to the indication of accumulative impact of miR-1291 on the cell metabolome, the high level of NMN in the miR-1291-expressing PANC-1 cells revealed in the present study is presumably a cause of disrupted cell metabolism such as fatty acid oxidation processes through the production of excessive reactive oxygen species load, as well as the suppressed invasion and tumorigenesis of PANC-1 cells in xenograft mouse models. Nevertheless, our study is limited to the analysis of PANC-1 cell lines and the specificity of miR-1291-triggered alteration in pancreatic cell metabolism such as the change of nicotinamide metabolism and fatty acid oxidations warrants further investigation.
NNMT is the only methyltransferase known to convert nicotinamide to NMN. The dramatic upregulation of NNMT supports its pivotal role in production of stable metabolite NMN in PANC-1 cells. Indeed, the native PANC-1 cells have similar levels of NNMT mRNA as the control PANC-1 cells (data not shown), indicating that the sharp increase in NNMT level is indeed triggered by miR-1291 rather than the transfection process, despite that the precise mechanisms remain elusive. Since NNMT mRNA does not contain any miR-1291 response element, the change of NNMT is likely a result of ‘indirect’ effects such as an influence of this miRNA on a transcription factor regulating the NNMT gene and/or a factor involved in controlling NNMT mRNA stability. For example, hepatocyte nuclear factor 1beta (HIF1β) (39,40) and signal transducer and activator of transcription 3 (STAT3) (41) have been revealed to activate NNMT transcription in human cells. Furthermore, pancreatic carcinoma cell metabolism is modulated by the interactions of HIF1α and Mucin 1 (MUC1) (42), among which the MUC1 is likely a direct target for miR-1291 (43). Whether the miR-1291-triggered dysregulation of NNMT is attributable to the HIF1β, STAT3 or HIF1α-MUC1 pathways needs further investigation. In addition, the alteration of cell membrane transporters such as ABCC1/MRP1 by miR-1291 (27) might contribute to the change of PANC-1 cell metabolites such as NMN.
NNMT promotes epigenetic remodeling via the induction of protein hypomethylation (31), and it is able to modulate cancer cell migration (44) and invasion (45). In agreement with these findings, the present study demonstrated an association between the dysregulation of NNMT and the reduction of proliferation, migration and invasion as well as tumorigenesis of miR-1291-expression PANC-1 cells. While NNMT expression was shown to be positively associated with the degree of malignancy of various cancers including lung, liver, kidney, bladder, colon and pancreas (44,46–50), NNMT seems to be downregulated in human insulinoma (51) and upregulated in malignant pancreatic ductal carcinoma or pancreatic juice (52,53). Interestingly, NNMT expression does not correlate with the stage and grade of renal clear cell carcinoma but tumor size (47). Consistently, there is an inverse correlation between NNMT expression levels and human oral squamous cell carcinomas (50) as well as pancreatic xenograft tumors (this study). Kaplan–Meier survival analysis also reveals a higher overall survival rate for patients bearing oral squamous cell tumors with higher NNMT expression levels, whereas it is not statistically significant (54). Consistently, the present study shows that NNMT does not predict the overall survival for PDAC patients. Therefore, in contrast to other pancreatic cancer biomarkers defined using patient samples and/or data-mining platforms (3,55–58) (e.g. serum carbohydrate antigen 19-9 or CA-19-9, the only biomarker for pancreatic tumor approved by the US Food and Drug Administration), the importance of NNMT may be limited to pancreatic cellular metabolism as it is identified from miR-1291-expressing PANC-1 cells. The potential application of NNMT in PDAC patients warrants more extensive examinations.
In summary, this study employs a UPLC-QTOF-MS-based metabolomics technology to reveal the significant alteration of cell metabolism associated with miR-1291-reduced PANC-1 cell migration, invasion and tumorigenesis. This is manifested by a sharp elevation of NMN in nicotinate and nicotinamide metabolism, as well as other cellular metabolites in fatty acid oxidations. The increase of NMN is attributed to a dramatic upregulation of NNMT expression. In addition, the NNMT mRNA levels are inversely correlated with the sizes of PANC-1-derived xenograft tumors. These results suggest an important role for NNMT in pancreatic cell metabolism and its possible utility as a biomarker for pancreatic carcinogenesis.
Supplementary material
Supplementary Table S1 and Figures S1–S6 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute Intramural Research Program, Center for Cancer Research (1ZIABC005708 and 1ZIABC005562 to F.J.G.); National Institute on Drug Abuse and National Cancer Institute, National Institutes of Health (Award R01DA021172 and U01CA175315 to A.-M.Y.); China Scholarship Council for the State Scholarship Fund (Award number 2010844039 to H.-C.B.).
Supplementary Material
Acknowledgements
We thank Linda Byrd and John Buckley for technical assistance.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- CPT
carnitine palmitoyltransferase
- CRAT
carnitine O-acetyltransferase
- ESI
electrospray ionization
- HILIC
hydrophilic interaction liquid chromatography
- MS
mass spectrometry
- NMN
N-methylnicotinamide
- NNMT
nicotinamide N-methyltransferase
- OPLS-DA
orthogonal projection to latent structures discriminant analysis
- PCA
principal components analysis
- PDAC
pancreatic ductal adenocarcinoma
- qPCR
quantitative real-time PCR
- QTOF
quadrupole time-of-flight
- RPLC
reversed-phase liquid chromatography
- SAM
S-adenosyl-l-methionine
- UPLC
ultra-performance liquid chromatography.
References
- 1. Bayraktar S., et al. (2010). Advanced or metastatic pancreatic cancer: molecular targeted therapies. Mt. Sinai J. Med., 77, 606–619 [DOI] [PubMed] [Google Scholar]
- 2. Nentwich M.F., et al. (2012). Surgery for advanced and metastatic pancreatic cancer–current state and trends. Anticancer Res., 32, 1999–2002 [PubMed] [Google Scholar]
- 3. Schultz N.A., et al. (2014). MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA, 311, 392–404 [DOI] [PubMed] [Google Scholar]
- 4. Jones S., et al. (2008). Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science, 321, 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenfeldt M.T., et al. (2013). p53 status determines the role of autophagy in pancreatic tumour development. Nature, 504, 296–300 [DOI] [PubMed] [Google Scholar]
- 6. Cantoria M.J., et al. (2014). Contextual inhibition of fatty acid synthesis by metformin involves glucose-derived acetyl-CoA and cholesterol in pancreatic tumor cells. Metabolomics, 10, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yabushita S., et al. (2013). Metabolomic and transcriptomic profiling of human K-ras oncogene transgenic rats with pancreatic ductal adenocarcinomas. Carcinogenesis, 34, 1251–1259 [DOI] [PubMed] [Google Scholar]
- 8. Ambros V. (2004). The functions of animal microRNAs. Nature, 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 9. Kasinski A.L., et al. (2011). Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer, 11, 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iorio M.V., et al. (2012). microRNA involvement in human cancer. Carcinogenesis, 33, 1126–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bloomston M., et al. (2007). MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA, 297, 1901–1908 [DOI] [PubMed] [Google Scholar]
- 12. Szafranska A.E., et al. (2007). MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene, 26, 4442–4452 [DOI] [PubMed] [Google Scholar]
- 13. Kent O.A., et al. (2009). A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biol. Ther., 8, 2013–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gironella M., et al. (2007). Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl Acad. Sci. USA, 104, 16170–16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li A., et al. (2010). Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res., 70, 5226–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu S., et al. (2010). miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res., 70, 6015–6025 [DOI] [PubMed] [Google Scholar]
- 17. Lu Z., et al. (2011). miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J., 30, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohuchida K., et al. (2012). MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann. Surg. Oncol., 19, 2394–2402 [DOI] [PubMed] [Google Scholar]
- 19. Wang P., et al. (2014). Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene, 33, 514–524 [DOI] [PubMed] [Google Scholar]
- 20. Nakata K., et al. (2011). MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery, 150, 916–922 [DOI] [PubMed] [Google Scholar]
- 21. Srivastava S.K., et al. (2011). MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis, 32, 1832–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang S., et al. (2011). Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis, 32, 1183–1189 [DOI] [PubMed] [Google Scholar]
- 23. Zhao W.G., et al. (2010). The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis, 31, 1726–1733 [DOI] [PubMed] [Google Scholar]
- 24. Yu A.M. et al. (2013). Therapeutic indications of miR-1291. International patent PCT/US2013/021307.
- 25. Hidaka H., et al. (2012). Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget, 3, 44–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamasaki T., et al. (2013). Tumor-suppressive microRNA-1291 directly regulates glucose transporter 1 in renal cell carcinoma. Cancer Sci., 104, 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan Y.Z., et al. (2013). Small nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug disposition through direct targeting of ABC transporter ABCC1. Drug Metab. Dispos., 41, 1744–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson C.H., et al. (2012). Challenges and opportunities of metabolomics. J. Cell. Physiol., 227, 2975–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aksoy S., et al. (1994). Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J. Biol. Chem., 269, 14835–14840 [PubMed] [Google Scholar]
- 30. Yamada K., et al. (2010). Interferon-gamma elevates nicotinamide N-methyltransferase activity and nicotinamide level in human glioma cells. J. Nutr. Sci. Vitaminol. (Tokyo)., 56, 83–86 [DOI] [PubMed] [Google Scholar]
- 31. Ulanovskaya O.A., et al. (2013). NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol., 9, 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shlomi T., et al. (2013). Metabolism: Cancer mistunes methylation. Nat. Chem. Biol., 9, 293–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bryniarski K., et al. (2008). Anti-inflammatory effect of 1-methylnicotinamide in contact hypersensitivity to oxazolone in mice; involvement of prostacyclin. Eur. J. Pharmacol., 578, 332–338 [DOI] [PubMed] [Google Scholar]
- 34. Chlopicki S., et al. (2007). 1-Methylnicotinamide (MNA), a primary metabolite of nicotinamide, exerts anti-thrombotic activity mediated by a cyclooxygenase-2/prostacyclin pathway. Br. J. Pharmacol., 152, 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fukushima T., et al. (2002). Possible role of 1-methylnicotinamide in the pathogenesis of Parkinson’s disease. Exp. Toxicol. Pathol., 53, 469–473 [DOI] [PubMed] [Google Scholar]
- 36. Chłopicki S., et al. (2012). Single bout of endurance exercise increases NNMT activity in the liver and MNA concentration in plasma; the role of IL-6. Pharmacol. Rep., 64, 369–376 [DOI] [PubMed] [Google Scholar]
- 37. Warburton D.E., et al. (2006). Health benefits of physical activity: the evidence. CMAJ, 174, 801–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmeisser K., et al. (2013). Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol., 9, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu J., et al. (2005). Activation of nicotinamide N-methyltransferase gene promoter by hepatocyte nuclear factor-1beta in human papillary thyroid cancer cells. Mol. Endocrinol., 19, 527–539 [DOI] [PubMed] [Google Scholar]
- 40. Xu J., et al. (2006). Histone deacetylase inhibitor depsipeptide represses nicotinamide N-methyltransferase and hepatocyte nuclear factor-1beta gene expression in human papillary thyroid cancer cells. Thyroid, 16, 151–160 [DOI] [PubMed] [Google Scholar]
- 41. Tomida M., et al. (2008). Stat3 up-regulates expression of nicotinamide N-methyltransferase in human cancer cells. J. Cancer Res. Clin. Oncol., 134, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chaika N.V., et al. (2012). MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc. Natl Acad. Sci. USA, 109, 13787–13792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Friedman R.C., et al. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res., 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu Y., et al. (2008). Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene, 27, 6679–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tang S.W., et al. (2011). Nicotinamide N-methyltransferase induces cellular invasion through activating matrix metalloproteinase-2 expression in clear cell renal cell carcinoma cells. Carcinogenesis, 32, 138–145 [DOI] [PubMed] [Google Scholar]
- 46. Roessler M., et al. (2005). Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin. Cancer Res., 11, 6550–6557 [DOI] [PubMed] [Google Scholar]
- 47. Sartini D., et al. (2006). Identification of nicotinamide N-methyltransferase as a novel tumor marker for renal clear cell carcinoma. J. Urol., 176, 2248–2254 [DOI] [PubMed] [Google Scholar]
- 48. Kim J. et al. (2009). Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J. Exp. Clin. Cancer Res., 28, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomida M., et al. (2009). Serum levels of nicotinamide N-methyltransferase in patients with lung cancer. J. Cancer Res. Clin. Oncol., 135, 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sartini D., et al. (2007). Nicotinamide N-methyltransferase upregulation inversely correlates with lymph node metastasis in oral squamous cell carcinoma. Mol. Med., 13, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nabokikh A., et al. (2007). Reduced TGF-beta1 expression and its target genes in human insulinomas. Exp. Clin. Endocrinol. Diabetes, 115, 674–682 [DOI] [PubMed] [Google Scholar]
- 52. Rogers C.D., et al. (2006). Differentiating pancreatic lesions by microarray and QPCR analysis of pancreatic juice RNAs. Cancer Biol. Ther., 5, 1383–1389 [DOI] [PubMed] [Google Scholar]
- 53. Chen R., et al. (2005). Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology, 129, 1187–1197 [DOI] [PubMed] [Google Scholar]
- 54. Emanuelli M., et al. (2010). Nicotinamide N-methyltransferase upregulation correlates with tumour differentiation in oral squamous cell carcinoma. Histol. Histopathol., 25, 15–20 [DOI] [PubMed] [Google Scholar]
- 55. Fong Z.V., et al. (2012). Biomarkers in pancreatic cancer: diagnostic, prognostic, and predictive. Cancer J., 18, 530–538 [DOI] [PubMed] [Google Scholar]
- 56. Ansari D., et al. (2014). The role of quantitative mass spectrometry in the discovery of pancreatic cancer biomarkers for translational science. J. Transl. Med., 12, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chang P., et al. (2011). Micronuclei levels in peripheral blood lymphocytes as a potential biomarker for pancreatic cancer risk. Carcinogenesis, 32, 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rhodes D.R., et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia, 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.