Abstract
Objective
To determine the roles of complement C4A and C4B gene CNVs and their plasma protein concentrations in residual insulin secretion and loss of pancreatic beta-cell function in new-onset type 1 diabetes patients.
Methods
We studied 34 patients of European ancestry with new-onset type 1 diabetes, aged between 3 and 17 years (10.7±3.45), at Nationwide Children's Hospital in Columbus, Ohio. Gene copy-number and size variations of complement C4A and C4B were determined by genomic Southern blot analyses. C4A and C4B protein phenotypes were elucidated by immunofixation and radial immunodiffusion. Two-digit HLA-DRB1 genotypes were determined by sequence-specific PCR. At 1 month and 9-month post diagnosis, stimulated C-peptide levels were measured after a standardized mixed-meal tolerance test.
Results
The diploid gene copy-numbers of C4A varied from 0 to 4, and those of C4B from 0 to 3. Patients with higher copy-number of C4A or higher C4A plasma protein concentrations at diagnosis had higher C-peptide levels at 1 month post diagnosis (p=0.008; p=0.008). When controlled by the Z-score of body-mass index, C4A copy-numbers, C4A protein concentrations, the age of disease-onset, the number of HLA-DR3 but not DR4 alleles were significant parameters in determining C-peptide levels. At 9-month post diagnosis, 42.3% of patients remained in partial remission, and these patients were characterized by lower total C4B copy-numbers or lower C4B protein concentrations (p=0.02, p=0.0004).
Conclusions
C4A appears to associate with the protection of residual beta-cell function in new-onset type 1 diabetes; C4B is correlated with the end of disease remission at 9-month post diagnosis.
Keywords: Type 1 Diabetes, Partial disease remission, C-Peptide, Complement C4, CNV, HLA-DRB1
Introduction
Type 1 diabetes (T1D) is an autoimmune disease characterized by the destruction of β-cells in the pancreas, leading to inability to synthesize endogenous insulin and poor glycemic controls. Multiple genetic and environmental factors have been implicated in the etiology of T1D (1, 2). The strongest genetic factor associated with human T1D lies in the major histocompatibility complex (MHC), also known as the HLA, on the short arm of chromosome 6. In particular, specific polymorphic variants of HLA class II genes are thought to contribute to over 50% of the genetic risk for T1D (2, 3), with the highest risk conferred by haplotypes with DRB1*0301-DQB1*0201-DQA1*0501 (also known as HLA-DR3 haplotype), DRB1*0405-DQB1*0302-DQA1*0301, or DRB1*0401-DQB1*0302-DQA1*0301 (HLA-DR4 haplotypes) (4, 5). While T1D is considered an organ-specific disease that mainly involves T-cell mediated autoimmunity, B-cells and other immune effector proteins may also be involved in disease pathogenesis. Autoantibodies against antigens of pancreatic islet cells are detectable years before the disease onset or development of hyperglycemia (6).
At disease presentation, most T1D patients have already lost 80–90% of their original beta-cell mass (7, 8). After diagnosis and medical treatment, many patients experience a partial remission period or “honeymoon” characterized by residual endogenous insulin secretion, requiring less frequent or lower doses of exogenous insulin injections, which may last between months and years after disease onset. T1D patients who produce more endogenous insulin for a longer duration have better prognosis: a lower total daily insulin requirement, better glucose control, fewer episodes of severe hypoglycemia and relatively less nephropathy and retinopathy (9–13). While variants of HLA class II genes are important genetic factors in T1D disease onset or susceptibility, they do not appear to play a major role in modulating the partial disease remission. Understanding the etiologic factors that differentiate preservation of beta-cell function and development of full-blown T1D disease may yield new insights into pathogenesis and effective intervention and therapies.
Complement C4 gene maps to the class III region of human MHC, which is 544 kb telomeric from HLA-DRB1 (14). Recent advance in the molecular genetic studies of the C4 locus lead to the establishment of a novel concept in human genetics, which features frequent, multi-allelic copy-number variations (CNV) of C4 genes. One to four copies of C4 gene in an HLA haplotype, or two to eight copies of total C4 genes in a diploid genome, are present among different individuals (15–18). The CNV of C4 genes is a result of segmental duplications involving four contiguous genes encoding for serine/threonine kinase RP, complement C4, steroid 21-hydroxylase CYP21, and tenascin TNX, a phenomenon termed as the RCCX modular duplications (15, 19). C4-CNV is coupled with complex secondary variations in (a) the physical size of C4 genes with a long form (L) of 20.6 kb and a short form (S) of 14.2 kb, and (b) DNA sequences leading to two classes of polymorphic proteins, the acidic C4A and the basic C4B with a large range of plasma protein concentrations (17, 20, 21). Dysregulation of complement activation can lead to complement-mediated cellular destruction or tissue injuries and thus affect autoimmunity. Although extremely rare in prevalence, human subjects with complete deficiencies of both C4A and C4B are almost always afflicted with systemic lupus erythematosus (SLE) or a lupus-like syndrome (22, 23). Furthermore, low gene copy-number of C4A is a risk factor for SLE disease susceptibility, while high copy-number of C4A is a protective factor against SLE (17). Thus, complement C4, and C4A in particular, is important in the protection against systemic autoimmunity (23). However, the roles of C4A and C4B multi-allelic CNVs in organ-specific autoimmunity such as T1D have not been investigated vigorously.
Here we determine whether complement C4 is associated with changes in pancreatic β-cell function at T1D disease onset and during the partial remission period. Pancreatic beta-cell function was examined through measurements of serum C-peptide levels before and after mixed-meal stimulation. Plasma C4 protein levels, C4A and C4B protein polymorphisms, gene CNVs of C4A and C4B, plus HLA-DRB1 genotypes were determined. Our data reveals a novel phenomenon with significant associations between C4A gene copy-numbers or C4A protein concentrations and C-peptide levels at 1-month post T1D diagnosis. By contrast, patients who remained in partial disease remission at 9-month post T1D onset are characterized by low copy-number of C4B genes and low C4B protein levels.
Methods
Study population
We recruited 35 pediatric subjects of European ancestry who were recently diagnosed with T1D at Nationwide Children's Hospital (NCH; Columbus, OH) from 2009–2010. These new onset T1D patients aged between 3 and 17 years old, among them were 28 males and 6 females. The prevalence of T1D in males and females is roughly similar (24). The higher participation of male patients in this serial clinical study reflected voluntary participations in this study, and young boys were more willing and amendable to several venipunctures during the mixed meal tolerance tests. The diagnosis of T1D was based on a fasting blood sugar >126 mg/dL (7 mmol/L) or a random blood sugar >200 mg/dL (11.1 mmol/L) with symptoms of hyperglycemia, including polydipsia and polyuria. Autoimmune diabetes was confirmed by the presence of at least one of the diabetes associated autoantibodies, including GAD65, ICA512 and insulin autoantibody. All diagnosed patients used insulin therapy with multiple daily insulin injections of rapid-acting insulin and a single daily injection of long-acting insulin, or insulin pump therapy. Informed consent and assent (for subjects 8–17 years of age) were obtained in accordance with Institutional Review Board approved protocols for human subject research.
Mixed-meal tolerance test (MMTT)
MMTT was performed in the study subjects 1-month and 9-month post diabetes diagnosis in the NCH Clinical Research Services Center. Subjects were fasted overnight, and the MMTT was performed in the next morning. The usual morning dose of rapid acting insulin was withheld, though subjects still continued basal insulin. Baseline blood samples were obtained to determine fasting glucose and C-peptide levels (25–27). The subjects ingested 6ml/kg (maximum of 360 ml) of the mixed-meal, Boost (8 fluid ounces with 240 kcal; 33g carbohydrate, 15g protein, and 6g fat) in a period less than 10 minutes. Ninety minutes after ingestion, a blood sample was obtained for determination of stimulated C-peptide and glucose levels. Glucose levels were determined in the clinical laboratory at NCH. Plasma C-peptide levels were measured by Esoterix Laboratories (Calabassas Hills, CA).
Determination of T1D-associated autoantibodies
Serum samples were processed and kept frozen at −86°C until the evaluation of the diabetes-associated autoantibodies against GAD-65, ICA-512 and insulin were performed (Esoterix Laboratories, Calabassas Hills, CA). Briefly, the measurements of anti-GAD-65 and anti-ICA-512 were performed by immunoprecipitation using radioactive recombinant proteins for GAD-65 and ICA-512, respectively, and protein-A as a carrier for immune complexes. The sensitivity of assays and intra-assay precision were 0.5 units and >97%, respectively, for anti-GAD-65; and 0.75 units and >90%, respectively, for anti-ICA512. As provided by Esoterix, Inc., for anti-GAD assays, the intra-assay variance for quadruplicate assays of same samples were between 4.8 and 10%. The inter-assay variance among 246 assays was between 12–15%. For the assays of ICA-512, the intra-assay variance (quadruplicates) was between 1.3 and 2.8%. The inter-assay variance among 153 assays was between 6 and 7%. For the measurement of total insulin antibodies (all Ig classes), insulin complexed with antibodies were dissociated from immune complexes, and free insulin in serum were removed by binding to acidified-charcoal and centrifugation. Serum autoantibodies against insulin were then measured through interaction with radioactive insulin and precipitation with polyethylene glycol (total binding). Nonspecific binding (NSB) was determined by adding unlabelled insulin to replicate samples to compete with radiolabeled insulin for binding to insulin autoantibodies. Specific binding counts were calculated by subtracting NSB from total counts and converted to μU of insulin based on specific activity of labeled insulin used in the assay, and data were shown as total insulin antibody binding capacity in μU of insulin/ml of serum tested (28). The intra-assay and inter-assay variations for insulin autoantibodies (20 assays) were between 2.5 and 6.5%, and between 8.2 and 11%, respectively.
HLA-DRB1 genotyping
Two-digit genotyping of HLA-DRB1 alleles was determined using genomic DNA for sequence-specific primer (SSP) PCR. Using customized oligos for PCR, the DRB1 alleles were determined with 20 independent reactions for each sample, followed by agarose gel electrophoresis (29). Specific patterns of PCR products after electrophoresis were interpreted based on the presence or absence of PCR products.
Complement C4 genotyping and phenotyping
Detailed procedures for genotyping and phenotyping of complement C4 have been described (18, 30). Briefly, genomic DNA samples were isolated from peripheral blood leukocytes by Gentra Kits (Valencia, CA). CNVs and genotypes of complement C4 and RCCX modules were determined by genomic Southern blot analyses (Fig. 1A). TaqI restriction fragment length polymorphisms (RFLP) were determined using specific probes corresponding to 3' region of RP and 5' region of C4, CYP21, and TNX (RCCX). Results of TaqI RFLP yielded information on the C4 CNV, C4 gene size and CNVs of other three constituent genes in RCCX modules. The relative copy-numbers of C4A and C4B genes were determined by PshAI-PvuII RFLP, using a C4d-specific probe for hybridization (Fig. 1B).
Fig. 1.
Genotypic and phenotypic characterizations of complement C4 in the MHC class III region. A. Genomic TaqI RFLP to determine RP-C4-CYP21-TNX (RCCX) modules and copy-number variations of long and short C4 genes. B.PshAI-PvuII RFLP to elucidate the relative gene copy-numbers of C4A and C4B. C. High voltage agarose gel electrophoresis (HVAGE) and immunofixation of plasma protein to demonstrate polymorphisms of native C4A and C4B allotypes based on gross differences in electric charges. D. Interpretations of C4 genotypes and phenotypes in eight T1D patients shown in panels A to C.
To determine the polymorphism and relative levels of C4A and C4B proteins, EDTA-plasma samples were digested with neuraminidase and carboxyl peptidase B to eliminate irregularities in glycosylations, and at the carboxyl termini of the beta and alpha chains, and resolved through their gross difference in electric charges by high voltage agarose gel electrophoresis (Fig. 1C). The C4 proteins were immunofixed with goat antisera against human C4, washed and stained. The C4A and C4B protein allotypes were scanned and their relative intensities quantified by ImageQuant Software. The C4A and C4B protein concentrations were calculated from their total C4. Plasma protein levels of C4 and C3 were determined by single radial immunodiffusion using EDTA-plasma and commercial RID-kits (The Binding Site, UK).
Statistical analyses
All statistical analyses were performed using JMP software (SAS Institute, Cary). Linear regressions or bivariate fits were used to evaluate the correlation between two parameters such as between C4 protein levels and C-peptide levels. Student's t-test was used to compare means of two groups. Logistic regression models were built to evaluate independent contribution of multiple parameters for C-peptides at 1-month or 9-month, or for the presence and absence of disease remission at 9-month post T1D diagnosis.
Results
Patient demographics and summary statistics
Of the 35 subjects enrolled in the study, one subject was negative for all three autoantibodies (GAD65, ICA-512 and insulin autoantibody) and was therefore excluded for data analyses. The mean age of T1D disease onset was 10.66±3.45 years (mean±SD). The average body mass index (BMI) at disease onset was 18.35±3.60 (Tables 1 and 2). Thirty-two subjects completed the MMTT at 1-month post diabetes diagnosis, and twenty-six subjects completed the MMTT at both 1-month and 9-month post diabetes diagnosis. Six patients had a first-degree relative with T1D (frequency: 0.176), and two patients had a family history of other autoimmune diseases.
TABLE 1.
Demographics, complement and HLA-DRB1 in new onset T1D
| Patient | Sex | Age (yrs) | BMI (kg/m2; | [C3] (mg/L) | [C4] (mg/L) | [C4]-9 mon. (mg/L) | Total C4 CNV | C4A CNV | C4B CNV | RCCX H1 | RCCX H2 | DRB1 H1 | DRB1 H2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 9 | 15.7 | 1532 | 158 | 188 | 3 | 0 | 3 | LL | S | 17(3)* | 13.1 |
| 2 | F | 11 | 16.3 | 1137 | 187 | 138 | 4 | 3 | 1 | LL | LL | 17(3) | 4 |
| 3 | M | 10 | 15.6 | 1592 | 456 | 406 | 4 | 2 | 2 | LS | LS | 1 | 4 |
| 4 | M | 12 | 23.0 | 1414 | 254 | 236 | 4 | 3 | 1 | LL | LL | 4 | 13 |
| 6 | M | 9 | 23.9 | 2681 | 504 | 556 | 5 | 2 | 3 | LL | LLS | 7 | 9 |
| 7 | M | 13 | 17.2 | 1157 | 242 | 434 | 3 | 1 | 2 | LS | S | 17(3)* | 4 |
| 8 | M | 11 | 17.3 | 1288 | 179 | 362 | 4 | 4 | 0 | LL | LL | 1 | 4 |
| 9 | M | 4 | 14.4 | 2244 | 202 | 266 | 3 | 1 | 2 | LL | S | 17(3)* | 4 |
| 10 | M | 9 | 15.5 | 1222 | 209 | 271 | 3 | 1 | 2 | LL | S | 17(3)* | 11 |
| 11 | M | 10 | 17.5 | 1855 | 341 | n.d. | 3 | 1 | 2 | LL | S | 17(3)* | 4 |
| 12 | M | 14 | 15.4 | 849 | 181 | 128 | 4 | 3 | 1 | LL | LS | 1 | 4 |
| 13 | M | 14 | 17.0 | 1355 | 241 | 195 | 4 | 2 | 2 | LS | LS | 7 | 8 |
| 14 | M | 13 | 12.5 | 849 | 273 | n.d. | 3 | 1 | 2 | LL | S | 17(3)* | 4 |
| 15 | M | 10 | 21.8 | 1493 | 371 | 484 | 5 | 3 | 2 | LS | LSS | 12 | 13.1 |
| 16 | F | 12 | 17.0 | 1031 | 163 | 102 | 2 | 1 | 1 | L | S | 1 | 17(3) |
| 17 | M | 14 | 15.5 | 1493 | 469 | 222 | 5 | 2 | 3 | LS | LSS | 1 | 4 |
| 18 | M | 9 | 17.3 | 1306 | 278 | 261 | 4 | 3 | 1 | LL | LL | 4 | 4 |
| 19 | M | 12 | 16.3 | 1183 | 289 | 276 | 4 | 2 | 2 | LS | LS | 7 | 8 |
| 20 | M | 16 | 29.2 | 732 | 245 | 279 | 4 | 3 | 1 | LL | LS | 4 | 7 |
| 21 | M | 8 | 23.9 | 1984 | 298 | 399 | 4 | 2 | 2 | LS | LS | 17(3) | 17(3) |
| 22 | M | 4 | 19.4 | 1006 | 297 | 397 | 6 | 3 | 3 | LSS | LLS | 7 | 13.1 |
| 23 | M | 10 | 25.4 | 1306 | 335 | n.d. | 3 | 1 | 2 | LL | S | 4 | 4 |
| 24 | M | 16 | 17.6 | 1006 | 197 | 237 | 4 | 3 | 1 | LL | LL | 4 | 12 |
| 25 | F | 11 | 21.3 | 1123 | 236 | 196 | 3 | 1 | 2 | LS | S | 4 | 15 |
| 26 | F | 8 | 17.1 | 2441 | 404 | 321 | 4 | 2 | 2 | LS | LS | 4 | 4 |
| 27 | M | 11 | 17.5 | 485 | 158 | 246 | 3 | 2 | 1 | LS | L | 13.1 | 1 |
| 28 | M | 6 | 19.7 | 1006 | 153 | 323 | 3 | 2 | 1 | LL | L | 4 | 7 |
| 30 | M | 12 | 15.4 | 1386 | 341 | 340 | 5 | 3 | 2 | LSS | LL | 1 | 7 |
| 31 | M | 7 | 15.2 | 912 | 198 | n.d. | 2 | 0 | 2 | S | S | 17(3)* | 17(3)* |
| 32 | M | 13 | 19.8 | 1386 | 150 | 175 | 2 | 1 | 1 | L | S | 17(3)* | 17(3) |
| 33 | M | 17 | 20.7 | 1761 | 244 | n.d. | 4 | 3 | 1 | LL | LS | 13.1 | 12 |
| 34 | F | 3 | 14.9 | 727 | <30 | n.d. | 3 | 2 | 1 | LS | L | 13.1 | 4 |
| 35 | M | 16 | 17.0 | 735 | 135 | n.d. | 3 | 1 | 2 | LL | S | 17(3)* | 14.1 |
an asterisk signifies an extended haplotype with HLA-DR3 and C4A deficiency; [C4], C4 protein concentration; [C3], C3 protein concentration; H1, haplotype 1; H2, haplotype 2; n.d., not determined.
TABLE 2.
Summary statistics of demographic and laboratory data for new onset T1D patients
| at diagnosis or 1-month | at 9 months | |
|---|---|---|
| mean ± s. d. | mean ± s. d. | |
| Age (yrs) | 10.73 ± 3.45 | |
| Body Weight (kg) | 43.21 ± 15.92 | |
| Height (m) | 1.51 ± 0.22 | |
| Body mass index | 18.35 ± 3.6 | |
| Z-score, BMI | −0.04 ± 3.6 | |
| anti-GAD-65 (U/ml) | 10.78 ± 27.98 | |
| anti-ICA512 (U/ml) | 11.39 ± 14.36 | |
| anti-insulin (U/ml) | 16.84 ± 30.78 | |
| Total C4-CNV | 3.65 ± 0.95 | |
| C4A-CNV | 1.97 ± 0.10 | |
| C4B-CNY | 1.71 ± 0.72 | |
| frequency | ||
| HLA-DR3 (0/1/2) | 0.647/ 0.265/ 0.088 | |
| HLA-DR4 (0/1/2) | 0.441/ 0.471/ 0.088 | |
| DR3/DR4 (0/1) | 0.853/ 0.157 | |
| mean ± s. d. | mean ± s. d. | |
| Hb-A1c (%) | 11.37 ± 2.43 | 7.45 ± 1.42 |
| Fasting glucose (mg/dL) | 108.7 ± 38.6 | 151.9 ± 73.8 |
| Stimulated glucose (mg/dL) | 189.7 ± 54.3 | 277.1 ± 101.9 |
| Fasting C-peptide (ng/ml) | 0.774 ± 0.484 | 0.695 ± 0.418 |
| Stimulated C-peptide (ng/ml) | 2.10 ± 1.02 | 1.63 ± 1.01 |
| Total C4 cone. (mg/L) | 245.1 ± 113.0 | 288.0 ± 110.7 |
| C4A cone. (mg/L) | 130.5 ± 66.8 | 161.6 ± 74.7 |
| C4B cone. (mg/L) | 114.7 ± 71.2 | 121.9 ± 77.4 |
| C3 cone., mg/L | 1325.5 ± 490.5 | 1752.9 ± 562.6 |
Complement C3 and C4 protein levels
Total C4 protein level in each patient at diagnosis was measured three times and averaged. C4 protein ranged from <30 to 505 mg/L with a mean of 245 mg/L. C4A protein levels ranged from 0 (C4A deficiency) to 248 mg/L with a mean of 131 mg/L. C4B protein levels ranged from 0 (C4B deficiency) to 303 mg/L with a mean of 115 mg/L. Total C3 protein levels ranged from 485 to 2,681 mg/L with a mean of 1,326 mg/L (Tables 1 and 2).
Gene CNVs of total C4, C4A, C4B, long C4, short C4 and RCCX modules
The total C4 CNV in a diploid genome ranged from 2 to 6 copies among the subjects (Figure 1, Table 1). The median C4 CNV was 4 and had a frequency of 0.38. C4A CNV ranged from 0 to 4 and C4B ranged from 0 to 3 copies. Approximately one-third of T1D subjects had 0 or 1 copies of C4A. Two subjects were found to have homozygous deficiency (0 copy) of C4A. Most subjects have 2 copies of C4B (frequency=0.50); one subject was homozygously deficient of C4B. Overall, 65% of the C4 genes were in the long form, and 35% were in the short form.
RCCX and HLA-DRB1
RCCX haplotypes with monomodular-L had a frequency of 0.07, while monomodular-S had a frequency of 0.19. Bimodular-LL was the most common haplotype with a frequency of 0.35, followed by bimodular-LS with a frequency of 0.28. Trimodular haplotypes of LLS and LSS were far less common in our patients with a frequency of 0.03 and 0.07 respectively.
The two most common HLA-DRB1 genotypes among our subjects were DRB1*04 (also known as DR4) and DRB1*03(17) (or DR3) with a frequency of 0.32 and 0.22, respectively. Five subjects were heterozygous for DRB1*03/*04 (frequency: 0.15). Only one patient had a DRB1*15 (or DR2) haplotype. Detailed results for patient demographics, CNVs of total C4, C4A, C4B and RCCX modules, C3 and C4 protein levels, and HLA-DRB1 are shown in Tables 1 and 2.
C-peptide levels
Stimulated C-peptide levels at 1-month post diabetes diagnosis ranged from 0.5 to 3.9 ng/mL with a mean (±SD) of 2.1±1.0 ng/mL. Nine-month stimulated C-peptide levels ranged from 0.0 to 3.8 ng/mL with a mean of 1.6±1.0 ng/mL. (SI conversion: C-peptide nmol/L= 0.333 × C-peptide ng/mL)
Complement C4A gene CNV and C4A plasma protein concentrations strongly correlate with stimulated C-peptide levels at 1-month post T1D diagnosis
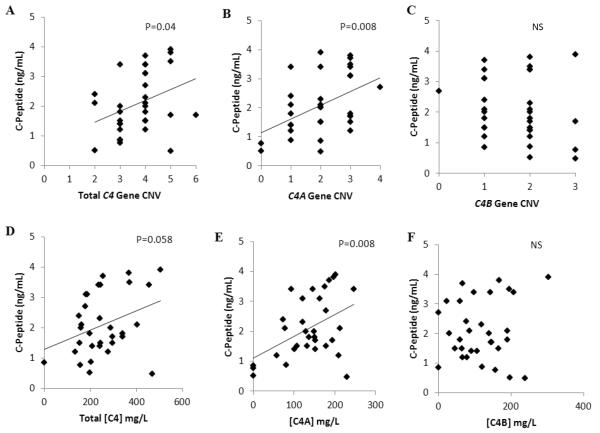
To investigate if variations of C4 genes and proteins are associated with the outcome for the loss of beta-cell function in the new onset T1D patients, we determined the relationship between stimulated C-peptide levels at 1-month post diagnosis and the C4 gene copy-numbers. Total C4 CNV was significantly correlated with 1-month C-peptide levels (R2=0.13; p=0.04), such that subjects with higher C4 CNV had higher C-peptide levels (Fig. 2A). The correlation with C-peptide was even greater for C4A CNV, which had an R2 of 0.21 and a p-value of 0.008 (Fig. 2B). By contrast, there was no apparent relationship between C4B CNV and 1-month C-peptide values (Fig. 2C).
Fig. 2.
Correlation of total C4 gene CNV (A), C4A gene CNV (B), C4B gene CNV (C), total C4 protein (D), C4A protein (E) and C4B protein (F) concentrations with stimulated C-peptide levels at 1-month after disease onset. Note that higher total C4 CNV or C4A gene is positively correlated with C-Peptide levels (p=0.04, p=0.008 respectively), and higher total C4 protein concentration or C4A protein concentration is positively correlated with C-peptide levels (p=0.058, p=0.008 respectively).
The correlation of C4 CNVs with stimulated C-peptide levels was further manifested in the positive correlation between protein levels of plasma C4 and C-peptides. Total C4 protein concentrations were marginally correlated with C-peptide levels (R2=0.11; p=0.058) at 1-month post T1D diagnosis (Fig. 2D). When the C4 proteins were separated into C4A and C4B, it became clear that C4A protein at disease diagnosis was significantly correlated with 1-month C-peptide levels (R2=0.21; p=0.008) (Fig. 2E). Similar to the case for the CNV of C4B genes, there was no clear relationship between 1-month C-peptide levels and C4B protein at diagnosis (Fig. 2F).
There were no significant relationships between diabetes-associated autoantibodies, anti-GAD-65, anti-ICA-512 or insulin autoantibody with C-peptide levels at 1 month or 9 months post diagnosis.
C4B CNV was inversely correlated with C-peptide levels at 9-month post T1D diagnosis
At 9-month post diabetes diagnosis, C-peptide levels were no longer significantly correlated with total C4 CNV or C4A CNV. On the contrary, a negative correlation between 9-month C-peptide levels and C4B gene CNV emerged (R2=0.15; p=0.048). Higher C4B copy-number correlated with lower C-peptide levels. C4A and C4B protein levels at diagnosis also did not correlate with 9- month C-peptide levels.
C4A, BMI or BMI z-score are independent parameters for stimulated C-peptide levels at 1-month
C4A CNV (R2=0.215, p=0.0075), C4A protein concentration (R2=0.209; p=0.0085), dosage of HLA-DR3 allele (R2=0.138; p=0.036), BMI (R2=0.310; p=0.0009) or BMI Z-score (R2=0.184; p=0.014) were each significantly correlated with 1-month (stimulated) C-peptide levels (Table 3). We further investigated if the correlation of C4A with stimulated C-peptide was independent of other parameters such as age of disease onset, BMI, and Z-score of BMI by performing multiple logistic regression analyses. BMI and C4A gene CNVs co-existed as positive and independent parameters for variations of C-peptides (overall R2=0.40, p=0.0005; BMI, p=0.005; C4A CNV, p=0.04). The same phenomenon held for BMI and C4A protein levels at diagnosis (overall R2=0.423, p=0.0003; BMI, p=0.003; C4A conc., p=0.024). Similar but smaller effects on C-peptide levels were observed for BMI Z-score plus C4A CNV or C4A protein concentrations. Age of onset, however, could not coexist in multiple logistic regression models with BMI, and C4A CNV or C4A protein concentration as an independent parameter of C-peptide levels.
Table 3.
Effects of individual contributing factors and multiple logistic regression models for risk factors associated with C-peptide levels at 1-month (A), and the end of partial remission at 9-month (B) post T1D diagnosis.
| A. Stimulated C-peptides at 1-month post diagnosis | |||
|---|---|---|---|
| R2 | t-ratio | P | |
| Individual parameters | |||
| C4A CNV | 0.215 | 2.87 | 0.0075 |
| C4A cone. | 0.209 | 2.82 | 0.0085 |
| HLA-DR3 * | 0.138 | −2.20 | 0.036 |
| BMI | 0.310 | 3.67 | 0.0009 |
| Z-BMI | 0.184 | 2.61 | 0.014 |
| age of onset | 0.06 | 1.43 | 0.162 |
| Regression Model 1: C4A CNV and BMI | |||
| model | 0.405 | 9.85* | 0.0005 |
| C4A CNV | 2.15 | 0.040 | |
| BMI | 3.04 | 0.005 | |
| Regression Model 2: C4A protein cone, and BMI | |||
| model | 0.423 | 10.6¶ | 0.0003 |
| C4A cone. | 2.39 | 0.024 | |
| BMI | 3.28 | 0.0027 | |
| Regression Model 3: HLA-DR3 and BMI | |||
| model | 0.405 | 9.87¶ | 0.0005 |
| HLA-DR3 | −2.16 | 0.040 | |
| BMI | 3.61 | 0.0012 | |
| B. End of partial disease remission at 9-month | |||
|---|---|---|---|
| R2 | χ 2 | P | |
| Individual parameters | |||
| C4B CNV | 0.162 | 5.74 | 0.0166 |
| C4B cone. | 0.355 | 12.6 | 0.0004 |
| DR3/DR4 * | 0.161 | 5.71 | 0.0169 |
| Regression Model 4: C4B-CNV and DR3/DR4 | |||
| model | 0.355 | 12.6 | 0.0018 |
| C4B-CHV | 6.88 | 0.0087 | |
| DR3/DR4 * | 6.85 | 0.0089 | |
| Regression Model 4: C4B cone. andDR3/DR4 | |||
| model | 0.471 | 16.8 | 0.0002 |
| C4B cone. | 11.0 | 0.0009 | |
| DR3/DR4 * | 4.09 | 0.043 | |
inverse association
F-ratio value is presented.
Partial disease remission at 9-month post diabetes diagnosis had significantly lower copy-number of C4B genes, lower plasma C4B proteins, and the presence of HLA-DR3/DR4 heterozygotes
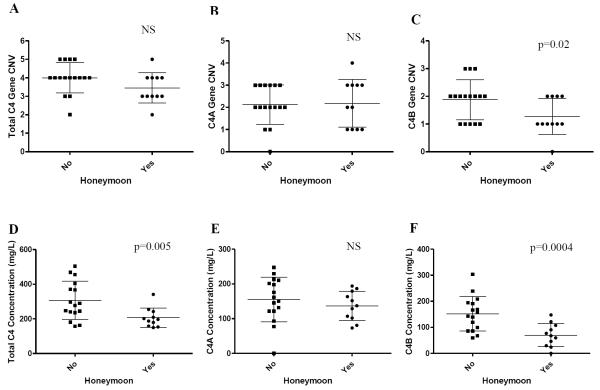
Twenty-six subjects were followed through 9 months. All had been previously studied at 1-month. Mean hemoglobin (Hgb) A1c for all subjects at 9-month was 7.5%. These subjects were divided into two groups based on insulin dose adjusted hemoglobin A1c (IDAA1c): HbA1c(%) + 4×insulin dose (units/kg/day). A patient with an IDAA1c < 9 is defined as being in partial remission (31). Eleven of 26 subjects (42.3%) at 9-month remained in partial remission. Of the subjects who had ended the partial remission period, they had higher C4B CNV (mean copy-number±SD: 1.88±0.7 vs 1.27±0.62; p=0.02), higher C4B protein concentrations at diagnosis (mean±SD: 156.2±66.2 mg/L vs 70.0±44 mg/L; p=0.0004; t-test) (Fig. 3C, 3F), and higher C4B protein concentration at 9-month post-diagnosis (148.5±79.3 mg/L vs 90.3±56.7 mg/L; p=0.02; t-test). While there was no relationship between C4A CNV or C4A protein concentration and subjects with a defined honeymoon period (Fig. 3B, 3E), the patient group who had ended partial remission at 9-month had significantly higher total C4 protein concentrations at diagnosis (310±112 mg/L vs. 207±56 mg/L; p=0.005; t-test) (Fig. 3D) and marginal difference in total C4 gene copy-number (p=0.056) (Fig. 3A).
Fig. 3.
Comparison of C4 gene CNV (A), C4A gene CNV (B), C4B CNV (C), total C4 protein (D), C4A protein (E), and C4B protein (F) concentrations in subjects with and without disease partial remission (honeymoon) at 9-month post disease onset. Lower total C4 protein concentration, C4B CNV or C4B protein concentration are correlated with the presence of partial disease remission (p=0.005, p=0.02, p=0.0004, respectively).
The dosages of HLA-DR3 alleles (p=0.26, χ2 analysis) or HLA-DR4 alleles (p= 0.90, χ2 analysis) did not associate with partial disease remission at 9 months. On the other hand, T1D subjects who had HLA DR3/DR4 heterozygotes all remained in partial disease remission at 9 months (p=0.017, χ2 analysis).
To determine if DR3/DR4 heterozygotes and C4B CNV or C4B protein concentrations are independent risk factors for partial disease remission at 9 months, multiple logistic regression analyses were performed (Table 3, panel B). Lower C4B CNV (χ2=6.88, p=0.0087) and the presence of DR3/DR4 heterozygotes (χ2=6.85, p=0.0089) independently contributed to disease remissions at 9-month (model p-value=0.0018; R2=0.355). In an alternative model, DR3/DR4 heterozygotes (χ2=4.09, p=0.043) and lower C4B protein concentration (χ2=11.0, p=0.0009) were both significant and independent contributing factor for partial disease remission (model p-value=0.0002, R2=0.471).
As a negative control, no apparent correlations were observed between plasma C3 concentration and stimulated C-peptide levels at 1 or 9-month post diabetes diagnosis (p=0.51; p=0.36), or partial disease remission (p=0.21).
Discussion
T1D has been extensively characterized as a T-cell mediated autoimmune disease but the roles for other immune effector proteins including those of the complement system in disease pathogenesis and progression are unclear. We determined the gene CNVs and their corresponding plasma protein levels for the two isotypes of complement C4, C4A and C4B in new-onset T1D patients. We showed that T1D subjects with higher gene copy-numbers of C4A, or with higher plasma C4A protein concentrations had higher levels of stimulated C-peptide at 1-month post diabetes diagnosis. On the other hand, T1D patients with higher C4B gene copy-number or higher C4B protein concentrations were less likely to retain the capability to secrete endogenous insulin or C-peptide at 9-month post diagnosis and therefore ended partial disease remission. The gene CNVs of C4A and C4B and their protein concentrations at T1D disease onset can be novel biomarkers relevant for the prediction and intervention of T1D disease remission.
From a physiologic standpoint, activated C4A proteins bind effectively to antigens with amino groups such as proteins or immune complexes. Through covalent binding to target molecules and serving as a ligand to complement receptor CR1 on erythrocytes, activated C4, particularly those from C4A, is engaged in the clearance of immune complexes and waste materials in the circulation (32, 33). Activated C4B proteins bind vehemently to antigens with hydroxyl groups such as carbohydrates or glycoproteins (34, 35), and is highly efficient in the propagation of complement activation pathways that cumulate to the formation of the membrane attack complex. Such differential chemical reactivity between C4A and C4B offers an explanation for the “Yin-Yang” phenomenon of C4A and C4B in the pathogenesis and progression of the new-onset T1D.
At least two sets of parameters appeared to be confounding factors. First, there is a strong and inverse correlation between the number of HLA-DR3 alleles and the copy-number of C4A genes (R2=0.388, p=0.000084), or C4A protein concentration (R2=0.248, p=0.0027). Second, serum C4 levels among healthy subjects modestly correlated with BMI (20), probably because adipose tissues also synthesize complement proteins.
To resolve the confounding factors in determining the residual beta-cell activities 1-month post disease onset, we performed multiple logistic regression analyses. C4A gene CNV or dosage of HLA-DR3 alleles each can co-exist with BMI Z-score as significant and independent parameters on determining the stimulated C-peptide levels in regression models. When controlled by BMI Z-scores, the strengths of association with C-peptide for C4A CNV (t-ratio=2.68, p=0.012) and C4A protein concentration (t-ratio=2.58, p=0.015) appeared to be slightly stronger than that for HLA-DR3 allele dosage (t-ratio=2.35, p=0.026).
At 9-month post diagnosis, BMI Z-scores, HLA-DR3 alleles, C4A CNV or C4A protein concentrations at diagnosis were no longer significant determining factors for stimulated C-peptides. Instead, the copy-number of C4B genes (p=0.048) and the age of disease onset (p=0.022) become significant determining factors. Among those subjects who had ended partial remission, they had significantly higher copy number of C4B genes and higher C4B protein concentration levels. Under hyperglycemic conditions, high C4B gene copy-number and/or high levels of activated C4B protein could potentiate complement-mediated tissue-injuries during an inflammatory response and aggravate pancreatic beta-cell destructions, leading to shorter honeymoon period in new-onset T1D. An alternative explanation would be that hyperglycemia associated with the end of remissions might have altered C4 secretion. Against the latter are earlier findings that C4 levels were not significantly altered in poorly controlled diabetes patients without infection (36), or among patients with intensive insulin therapy during coronary bypass surgery (37).
Previous studies have identified several risk factors for a non-existence or shortened honeymoon period including younger age of disease onset, severity of disease at presentation (DKA), female gender, and higher HbA1c at T1D onset (11, 12, 38–42). One study investigated the association of HLA-DRB1 alleles DR3 and DR4 with stimulated C-peptides after disease onset and found no significant correlation (11). Our data revealed that HLA-DR4 has no role on stimulated C-peptide levels during the honeymoon period. Therefore, lumping HLA-DR3 and DR4 alleles together in an analysis would have masked the effect of DR3. In our study population, 83.3% of T1D subjects who had an HLA-DR3 allele also had a specific RCCX haplotype characterized by the presence of a short C4B gene (coding for C4B1 protein) and the absence of C4A. This haplotype is similar or probably identical to the described “extended” or “ancient” haplotype with HLA A1 B8 DR3 and complotype SC01 (which stands for complement factor B-slow, C2C, C4AQ0 and C4B1) (43, 44).
This study enrolled a limited number of new-onset T1D patients in a serial clinical study to identify risk factors of large effect-size (i.e., prominent risk factors) associated with residual C-peptide levels at one month after disease onsets, and the existence of partial remissions 9 months after diagnosis. Results of this study favorably justify an in-depth prospective study with greater number of new-onset patients for quarterly mixed meal tolerance tests, assays of complement C4A and C4B gene CNVs, and serial variations of C4A and C4B protein levels over a two-year period. Such extended study will allow us to identify medium and low effect-size risk factors associated with T1D pathogenesis, disease remission and progression. If the Yin-Yang phenomenon for C4A and C4B in T1D “honeymoon period” is replicated in larger and more extensive studies, then enhancing the expression of C4A and reducing the levels of functional C4B would be a reasonable goal to intervene the disease progression and to achieve prolonged disease remission of T1D. Future investigations of the remission period following diagnosis with reagents that modulate C4A and C4B biosynthesis or turnover can lead to new exciting discoveries for treatment and therapies of T1D.
Acknowledgements
We are indebted to the new onset T1D patients and their parents for their support. We thank Dr. Amanda S. Dye and Dr. Stacy L. Meyer (Division of Endocrinology, Nationwide Children's Hospital, Columbus, OH) for their generous contributions.
This work was supported by National Institutes of Health grant 1R01 AR054459 from the NIAMS (C.Y.Y.), and Nationwide Children's Hospital institutional clinical research pilot grant 242309 (S.E.K.).
Abbreviations
- (Hgb)
Hemoglobin
- (MMTT)
mixed meal tolerance test
- (T1D)
Type 1 Diabetes
Footnotes
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
References
- 1.VAN BELLE TL, COPPIETERS KT, VON HERRATH MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev. 2011;91:79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 2.BLUESTONE JA, HEROLD K, EISENBARTH G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NERUP J, PLATZ P, ANDERSEN OO, et al. HL-A antigens and diabetes mellitus. Lancet. 1974;2:864–866. doi: 10.1016/s0140-6736(74)91201-x. [DOI] [PubMed] [Google Scholar]
- 4.LAMBERT AP, GILLESPIE KM, THOMSON G, et al. Absolute risk of childhood-onset type 1 diabetes defined by human leukocyte antigen class II genotype: a population-based study in the United Kingdom. J Clin Endocrinol Metab. 2004;89:4037–4043. doi: 10.1210/jc.2003-032084. [DOI] [PubMed] [Google Scholar]
- 5.ERLICH H, VALDES AM, NOBLE J, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.TAPLIN CE, BARKER JM. Autoantibodies in type 1 diabetes. Autoimmunity. 2008;41:11–18. doi: 10.1080/08916930701619169. [DOI] [PubMed] [Google Scholar]
- 7.AKIRAV E, KUSHNER JA, HEROLD KC. Beta-cell mass and type 1 diabetes: going, going, gone? Diabetes. 2008;57:2883–2888. doi: 10.2337/db07-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.SHERRY NA, TSAI EB, HEROLD KC. Natural history of beta-cell function in type 1 diabetes. Diabetes. 2005;54(Suppl 2):S32–S39. doi: 10.2337/diabetes.54.suppl_2.s32. [DOI] [PubMed] [Google Scholar]
- 9.VON HERRATH M, SANDA S, HEROLD K. Type 1 diabetes as a relapsing-remitting disease? Nat Rev Immunol. 2007;7:988–994. doi: 10.1038/nri2192. [DOI] [PubMed] [Google Scholar]
- 10.ALY H, GOTTLIEB P. The honeymoon phase: intersection of metabolism and immunology. Curr Opin Endocrinol Diabetes Obes. 2009;16:286–292. doi: 10.1097/MED.0b013e32832e0693. [DOI] [PubMed] [Google Scholar]
- 11.MORTENSEN HB, SWIFT PG, HOLL RW, et al. Multinational study in children and adolescents with newly diagnosed type 1 diabetes: association of age, ketoacidosis, HLA status, and autoantibodies on residual beta-cell function and glycemic control 12 months after diagnosis. Pediatr Diabetes. 2010;11:218–226. doi: 10.1111/j.1399-5448.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- 12.BOWDEN SA, DUCK MM, HOFFMAN RP. Young children (<5 yr) and adolescents (>12 yr) with type 1 diabetes mellitus have low rate of partial remission: diabetic ketoacidosis is an important risk factor. Pediatr Diabetes. 2008;9:197–201. doi: 10.1111/j.1399-5448.2008.00376.x. [DOI] [PubMed] [Google Scholar]
- 13.GREENBAUM CJ. Type 1 diabetes intervention trials: what have we learned? A critical review of selected intervention trials. Clin Immunol. 2002;104:97–104. doi: 10.1006/clim.2002.5234. [DOI] [PubMed] [Google Scholar]
- 14.HORTON R, WILMING L, RAND V, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 15.YANG Z, MENDOZA AR, WELCH TR, ZIPF WB, YU CY. Modular variations of HLA class III genes for serine/threonine kinase RP, complement C4, steroid 21-hydroxylase CYP21 and tenascin TNX (RCCX): a mechanism for gene deletions and disease associations. J Biol Chem. 1999;274:12147–12156. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- 16.BLANCHONG CA, ZHOU B, RUPERT KL, et al. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in Caucasians: the load of RCCX genetic diversity on MHC-associated disease. J Exp Med. 2000;191:2183–2196. doi: 10.1084/jem.191.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.YANG Y, CHUNG EK, WU YL, et al. Gene copy number variation and associated polymorphisms of complement component C4 in human systemic erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against European American SLE disease susceptibility. Am J Hum Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WU YL, SAVELLI SL, YANG Y, et al. Sensitive and specific real-time PCR Assays to accurately determine copy-number variations (CNVs) of human complement C4A, C4B, C4-Long, C4-Short and RCCX modules: Elucidation of C4 CNVs in 50 consanguineous subjects with defined HLA genotypes. J Immunol. 2007;179:3012–3025. doi: 10.4049/jimmunol.179.5.3012. [DOI] [PubMed] [Google Scholar]
- 19.SHEN LM, WU LC, SANLIOGLU S, et al. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and C4B genes in the HLA class III region: Molecular cloning, exon-intron structure, composite retroposon and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- 20.YANG Y, CHUNG EK, ZHOU B, et al. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities and body mass index. J Immunol. 2003;171:2734–2745. doi: 10.4049/jimmunol.171.5.2734. [DOI] [PubMed] [Google Scholar]
- 21.SAXENA K, KITZMILLER KJ, WU YL, et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: A comparison of Asian-Indian and European American populations. Mol Immunol. 2009;46:1289–1303. doi: 10.1016/j.molimm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WU YL, HAUPTMANN G, VIGUIER M, YU CY. Molecular basis of complete complement C4 deficiency in two North-African families with systemic lupus erythematosus. Genes Immun. 2009;10:433–445. doi: 10.1038/gene.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ATKINSON JP, YU CY. Genetic susceptibility and class III complement genes. In: Lahita RG, Buyon JP, Koike T, Tsokos GC, editors. Systemic Lupus Erythematosus. Elsevier Academic Press; Amsterdam: 2011. pp. 21–45. [Google Scholar]
- 24.WHITACRE CC. Sex differences in autoimmune disease. Nature Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 25.GREENBAUM CJ, MANDRUP-POULSEN T, MCGEE PF, et al. Mixed-meal tolerance test versus glucagon stimulation test for the assessment of beta-cell function in therapeutic trials in type 1 diabetes. Diabetes Care. 2008;31:1966–1971. doi: 10.2337/dc07-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.THE DCCT GROUP Effects of age, duration and treatment of insulin-dependent diabetes mellitus on residual beta-cell function: observations during eligibility testing for the Diabetes Control and Complications Trial (DCCT) J Clin Endocrinol Metab. 1987;65:30–36. doi: 10.1210/jcem-65-1-30. [DOI] [PubMed] [Google Scholar]
- 27.PALMER JP, FLEMING GA, GREENBAUM CJ, et al. C-peptide is the appropriate outcome measure for type 1 diabetes clinical trials to preserve beta-cell function: report of an ADA workshop, 21–22 October 2001. Diabetes. 2004;53:250–264. doi: 10.2337/diabetes.53.1.250. [DOI] [PubMed] [Google Scholar]
- 28.MOXNESS M, FOLEY J, STENE M, et al. Development and validation of radioligand binding assays to measure total, IgA, IgE, IgG, and IgM insulin antibodies in human serum. Ann N Y Acad Sci. 2003;1005:265–268. doi: 10.1196/annals.1288.040. [DOI] [PubMed] [Google Scholar]
- 29.HUI KM, BIDWELL JL. Handbook of HLA Typing Techniques. CRC Press; Boca Raton: 1998. [Google Scholar]
- 30.CHUNG EK, WU YL, YANG Y, ZHOU B, YU CY. Human complement components C4A and C4B genetic diversities: complex genotypes and phenotypes. Curr Protoc Immunol. 2005:13.8.1–13.8.35. doi: 10.1002/0471142735.im1308s68. [DOI] [PubMed] [Google Scholar]
- 31.MORTENSEN HB, HOUGAARD P, SWIFT P, et al. New definition for the partial remission period in children and adolescents with type 1 diabetes. Diabetes Care. 2009;32:1384–1390. doi: 10.2337/dc08-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.CORNACOFF JB, HEBERT LA, SMEAD WL, VANAMAN ME, BIRMINGHAM DJ, WAXMAN FJ. Primate erythrocyte-immune complex-clearing mechanism. J Clin Invest. 1983;71:236–247. doi: 10.1172/JCI110764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SCHIFFERLI JA, NG YC, PETERS DK. The role of complement and its receptor in the elimination of immune complexes. N Engl J Med. 1986;315:488–495. doi: 10.1056/NEJM198608213150805. [DOI] [PubMed] [Google Scholar]
- 34.CARROLL MC, FATHALLAH DM, BERGAMASCHINI L, ALICOT EM, ISENMAN DE. Substitution of a single amino acid (aspartic acid for histidine) converts the functional activity of human complement C4B to C4A. Proc Natl Acad Sci U S A. 1990;87:6868–6872. doi: 10.1073/pnas.87.17.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DODDS AW, REN X-D, WILLIS AC, LAW SKA. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379:177–179. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- 36.VAN EEDEN SF, STRACHAN AF, HOUGH SF. Circulating acute phase reactive proteins as indicators of infection in poorly controlled diabetes mellitus. Diabetes Res Clin Pract. 1988;5:99–105. doi: 10.1016/s0168-8227(88)80048-2. [DOI] [PubMed] [Google Scholar]
- 37.ALBACKER T, CARVALHO G, SCHRICKER T, LACHAPELLE K. High-dose insulin therapy attenuates systemic inflammatory response in coronary artery bypass grafting patients. Ann Thorac Surg. 2008;86:20–27. doi: 10.1016/j.athoracsur.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 38.GREENBAUM CJ, HARRISON LC. Guidelines for intervention trials in subjects with newly diagnosed type 1 diabetes. Diabetes. 2003;52:1059–1065. doi: 10.2337/diabetes.52.5.1059. [DOI] [PubMed] [Google Scholar]
- 39.ORTQVIST E, FALORNI A, SCHEYNIUS A, PERSSON B, LERNMARK A. Age governs gender-dependent islet cell autoreactivity and predicts the clinical course in childhood IDDM. Acta Paediatr. 1997;86:1166–1171. doi: 10.1111/j.1651-2227.1997.tb14837.x. [DOI] [PubMed] [Google Scholar]
- 40.KOMULAINEN J, LOUNAMAA R, KNIP M, KAPRIO EA, AKERBLOM HK. Ketoacidosis at the diagnosis of type 1 (insulin dependent) diabetes mellitus is related to poor residual beta cell function. Childhood Diabetes in Finland Study Group. Arch Dis Child. 1996;75:410–415. doi: 10.1136/adc.75.5.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DOST A, HERBST A, KINTZEL K, et al. Shorter remission period in young versus older children with diabetes mellitus type 1. Exp Clin Endocrinol Diabetes. 2007;115:33–37. doi: 10.1055/s-2007-948214. [DOI] [PubMed] [Google Scholar]
- 42.ABDUL-RASOUL M, HABIB H, AL-KHOULY M. 'The honeymoon phase' in children with type 1 diabetes mellitus: frequency, duration, and influential factors. Pediatr Diabetes. 2006;7:101–107. doi: 10.1111/j.1399-543X.2006.00155.x. [DOI] [PubMed] [Google Scholar]
- 43.DAWKINS R, LEELAYUWAT C, GAUDIERI S, et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 44.AWDEH ZL, RAUM D, YUNIS EJ, ALPER CA. Extended HLA/complement allele haplotypes: Evidence for T/t-like complex in man. Proc Natl Acad Sci USA. 1983;80:259–263. doi: 10.1073/pnas.80.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]