Abstract
Purpose
The circadian rhythm of endogenous corticosterone (CS) may produce fluctuations of downstream gene expression in normal rats. This study examined changes in glucocorticoid receptor (GR) and glutamine synthetase (GS) expression in rat skeletal muscle in relation to plasma CS over a 24-h period.
Methods
Fifty-four normal male Wistar rats were sacrificed at 18 time points (n = 3) over 24 h. Plasma CS concentrations and gastrocnemius muscle GR and GS mRNA and GS activity were measured.
Results
The circadian rhythm of plasma CS was captured by a two-harmonic function. The expression of GR and GS mRNA and GS activity follow a circadian rhythm in normal rat skeletal muscle. GR mRNA reaches a trough at 4 h after the peak of plasma CS and it fluctuates between 0.55 and 0.9 fmol g tissue−1. GS mRNA and activity reach peaks at 6 and 12 h after the endogenous CS peak. GS mRNA oscillates between 3 and 6 fmol g tissue−1, whereas GS activity fluctuates between 17 and 23 µmol min−1 g protein−1. Mechanistic receptor/gene-mediated pharmacodynamic models were applied to describe the temporal patterns of GR mRNA, GS mRNA, and GS activity within the circadian cycle.
Conclusions
The integrated models were able to capture the circadian expression patterns of plasma CS, and GR and GS in normal rat skeletal muscle showing a dependence of tissue gene expression on plasma CS.
Keywords: biological rhythm, mathematical model, pharmacodynamics, pharmacokinetics
INTRODUCTION
Plasma levels of endogenous adrenal steroids display a circadian rhythm that is under the control of the hypothalamus–pituitary–adrenal axis (HPA). In humans, plasma cortisol is high in the early morning and progressively declines toward the evening (1). In nocturnal animals such as rats, the pattern is reversed, with the highest corticosterone concentrations occurring near the onset of darkness and the lowest levels in the early morning.
It is expected that the circadian rhythm of endogenous steroids may lead to daily fluctuations of downstream gene expression in peripheral tissues. It has been shown that expression of tyrosine aminotransferase (TAT) and cholesterol-7a-hydroxylase in rat liver follow circadian rhythms (2,3). Both are prototype glucocorticoid-regulated genes. The rhythmic expression of both genes is controlled by endogenous corticosterone (CS). Adrenalectomy completely abolishes the circadian oscillation of cholesterol-7a-hydroxylase in rats (3). A recent report supports the hypothesis that glucocorticoids act as a potential linking signal between the central nervous system pacemaker suprachiasmatic nucleus and peripheral oscillator genes in various tissues (4).
Steroids are known to cause down-regulation of their own receptors (5). Previous studies from our laboratory and others have demonstrated that glucocorticoid receptor (GR) and glutamine synthetase (GS) are regulated by glucocorticoids through a receptor/gene-mediated pathway (6–8). Both GS mRNA and activity were up-regulated in adrenalectomized (ADX) rat skeletal muscle following 50 mg kg−1 methylprednisolone treatment, whereas receptor mRNA was down-regulated (8). The GS expression vs. time profile in muscle was similar to that of TAT in liver following the same drug treatment (9,10). Thus, we hypothesized that expression of physiological GR and GS in skeletal muscle may follow a circadian rhythm in normal rats. Our aim is to characterize the 24-h circadian rhythm of those genes in untreated rats.
Development of pharmacokinetic/pharmacodynamic (PK/PD) models is necessary for the quantitative understanding of physiological and pharmacological mechanisms underlying steroid regulatory effects. Mechanistic PD models have been developed in our laboratory to describe the receptor/gene-mediated effects of steroids (8,10). Our current fifth-generation model is an integrated model describing multiple events including receptor dynamics, receptor mRNA suppression, and target gene induction (10).
In this report, we determine whether GS and GR receptor follow a circadian rhythm under the control of plasma CS. Integrated pharmacodynamic models were used to describe steroid effects on target gene transcription. To solve the problem of parameter correlation, data from single bolus dose studies with dexamethasone and hydrocortisone treatment were incorporated in the modeling.
MATERIALS AND METHODS
Animals
Fifty-four normal (150–175 g) male Wistar rats were purchased from Harlan Sprague–Dawley Inc. (Indianapolis, IN, USA) and experiments were initiated at body weights between 225 and 275 g. Animals were housed and allowed to acclimatize in a constant-temperature environment (22°C) equipped with a 12-h light–dark cycle. Rats were randomly divided into two groups. Twenty-seven rats (group I) were acclimatized for 2 weeks prior to study to a normal light–dark cycle, where lights went on at 8 a.m. and off at 8 p.m. The onset of light cycle was considered as time zero. The other 27 rats (group II) were acclimatized for 2 weeks prior to study to a reversed light–dark cycle, where lights went on at 8 p.m. and off at 8 a.m. All rats had free access to rat chow and 0.9% NaCl drinking water. Our research protocol adheres to the “Principles of Laboratory Animal Care” (NIH publication 85-23, revised in 1985) and was approved by the University at Buffalo Institutional Animal Care and Use Committee.
Experimental
Circadian Rhythm Study
Rats in group I were killed at 0.25, 1, 2, 4, 6, 8, 10, 11, and 11.75 h after lights on to capture the light cycle. Rats in group II were killed at 12.25, 13, 14, 16, 18, 20, 22, 23, and 23.75 h after lights on to capture the dark cycle. Three animals were sacrificed at each time point. Because normal rats were used in the study, minimal animal handling with least possible environmental disturbances was employed to minimize stress. Night vision goggles were used to carry out animal procedures conducted in the dark period. At sacrifice, rats were weighed, anesthetized by ketamine/xylazine, and sacrificed by aortic exsanguination. Blood was drawn from the abdominal aortic artery into syringes using ethylenediaminetetraacetic acid as anticoagulant. Plasma was harvested from blood and frozen at −20°C until analyzed for CS. Gastrocnemius muscles were excised from both legs and were frozen in liquid nitrogen immediately after excision and stored in −80°C until RNA preparation or enzymatic assay.
Single Bolus Study
Gastrocnemius muscle tissues were obtained from a previous animal study performed in our laboratory (11). In brief, 36 ADX rats were randomly divided into two groups. One group received 0.1 mg kg−1 of dexamethasone sodium phosphate (American Regent Laboratories, Inc., Shirley, NY, USA). Rats in the other group were given 50 mg kg−1 of hydrocortisone sodium succinate (Solu-Cortef, Pharmacia & Upjohn Company, Kalamazoo, MI, USA). Three untreated rats served as control animals. Drugs were administered as single penile vein injections under ketamine/xylazine anesthesia. Rats were sacrificed at various time points between 0.5 and 12 h by exsanguination under ketamine/xylazine anesthesia. Blood was drawn and plasma was harvested as described before. Gastrocnemius muscles were excised and stored at −80°C until analysis.
Plasma Steroid Assays
Plasma CS concentrations in normal rats or dexamethasone and hydrocortisone concentrations in ADX rats were determined by a sensitive normal-phase high-performance liquid chromatography (HPLC) method as previously described (12). The limit of quantitation was 10 ng ml−1. The interday and intraday coefficients of variation (CV) were less than 10%.
RNA Preparation
Skeletal muscle tissues were ground into powder in liquid nitrogen. Muscle total RNA was extracted in Trizol (Invitrogen Corp, Carlsbad, CA, USA) according to the manufacturer’s instructions. Before extraction, an external “pseudomessage” glucose repressible gene (GRG) cRNA was added to each muscle sample to correct for variable extraction yields. The integrity of extracted RNA was confirmed by agarose–formaldehyde gel electrophoresis. The quantity of extracted RNA was determined by its optical density taken at 260 nm. The total RNA was diluted to desired concentrations in nuclease-free water (Ambion, Austin, TX, USA) and stored at −80°C until use.
Quantitative Real-Time Reverse Transcription–Polymerase Chain Reaction
The yield of RNA extraction was determined for each muscle sample by comparing the quantity of exogenous GRG cRNA added into muscle tissue with that recovered after extraction as previously described for Northern hybridization (13). The target mRNA expression per gram tissue was calculated as the measured mRNA expression level after extraction normalized by extraction yield. This procedure corrects for experimental variations in RNA yields between multiple tissue samples and allows conversion of mRNA expression levels to moles of message per gram wet weight of tissue. GRG was originally cloned from Neorospora and does not share homology with any mammalian gene (14).
The quantity of GRG cRNA following extraction and muscle GR and GS mRNA were determined by quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR) using TaqMan probes and gene-specific in vitro transcribed cRNA standards for quantification. Primer and probe sequences were designed using PrimerExpress software (Applied Biosystems, Foster City, CA, USA). Sequences sharing homology with other genes were excluded. The probe was labeled with FAM (or HEX) reporter dye at the 5′ end and a BHQ quencher at the 3′ end and custom synthesized by Biosearch Technologies, Inc. (Novato, CA, USA). The RT-PCR was performed using Brilliant QRT-PCR Core Reagent Kit, 1-Step (Stratagene, La Jolla, CA, USA) according to instructions of the manufacturer. An absolute standard curve for each individual real-time RT-PCR assay was generated using an in vitro transcribed sense cRNA. The cRNA was constructed using conventional cloning methods and in vitro transcribed using T7 MEGAscript™ In Vitro Transcription Kits (Ambion, Austin, TX, USA).
For quantitation of GR mRNA, a 76-bp segment of PCR product was amplified using a forward primer (5′-AACATGTTAGGTGGGCGTCAA- 3′), a reverse primer (5′-GGTGTAAGTTTCTCAAGCCTAGTATCG-3′), and a FAM-labeled probe (5′-TGATTGCAGCAGTGAAATGGGCAAAG-3′).
For quantitation of GRG cRNA, the following primers and probe were used (forward primer: 5′-CGGTTCTGGTGTAATGCTAAAGCT-3′, reverse primer: 5′-AGTTCGCCAAGGGCTTCTC-3′, and a HEX labeled probe: 5′-CCCTTCGAAATTCCAAGCCAAGTATGTCAT-3′). The produced amplicon was a 76-bp sequence.
GS mRNA expression was determined by amplification of a 110-bp sequence using a forward primer (5′-CGCCCGCCGTCTGA-3′), a reverse primer (5′-TCTCCTGGCCGACAATCC-3′), and a FAM labeled probe (5′-TCCACGAAACCTCCAACATCAACGACTTT-3′).
As a time-, labor-, and reagent-saving technique, multiplex GR/GRG real-time RT-PCR was performed to amplify both GR and GRG cDNA in a single tube without apparent reduction in sensitivity compared to individual reactions. The average intraday and interday CVs were 6.4 and 16.1% for GR, 10.6 and 14.0% for GRG, and 18.0 and 10.0% for GS.
GS Activity Assay
GS activity was determined by a colorimetric assay monitoring its γ-glutamyltransferase activity (15). Frozen muscle powder was homogenized in ice-cold buffer (0.05 M imidazole, pH 6.8) and centrifuged at 4500 × g at 4°C for 20 min to produce tissue cytosol. A reaction mix including 0.05 M imidazole, 0.05 M imidazole-L-glutamine, 25 mM sodium arsenate, 2 mM manganese chloride, 0.16 mM ADP, and 25 mM hydroxylamine was prepared and prewarmed to 37°C. Muscle tissue cytosol was added into the reaction mix and incubated at 37°C for 30 min. The reaction was stopped by addition of stop solution (2.42% ferric chloride, 1.45% trichloroacetate acid, 1.82% HCl). The reaction product γ-glutamylhydroxamate forms a purplish, brown complex in the presence of trivalent ion. Blanks were made for each muscle sample by adding stop solution before the reaction mix. Both assay tubes and blanks were centrifuged at 3400 rpm for 30 min. The supernatant was collected and the optical density was taken at 540 nm. The standard curve was generated by mixing reaction mix, stop solution with various concentrations of purchased γ-glutamyl-hydroxamate (Sigma, St. Louis, MO, USA). The absorbances obtained from blanks were deducted from those obtained from corresponding assay tubes and GS activities were calculated from standard curves. GS activity was expressed as micromoles of γ-glutamylhydroxamate formed per gram of protein added per minute. The interday and intraday CVs were less than 15%.
THEORY
Pharmacokinetics
The plasma concentrations of CS (CCS) in the 24-h circadian rhythm study were modeled by harmonic functions (16,17).
| (1) |
where ai and bi are Fourier coefficients and i represents the frequency of the harmonic functions. The circadian rhythm period T equals 24 h. Plasma CS data were fitted to this equation using the FOURPHARM software (17).
The plasma dexamethasone or hydrocortisone concentrations (C) vs. time (t) following a single intravenous injection was described by (11):
| (2) |
where Ci and λi are the coefficients for the intercepts and slopes. The pharmacokinetics of both steroids was analyzed previously by Mager et al. (11). Such parameters were used for simulations of the steroid kinetics (Table I).
Table I.
Corticosterone Circadian Rhythm in Normal Rats and Pharmacokinetics of Hydrocortisone/Dexamethasone in ADX Rats
| Parameter | Definition | Value | ||
|---|---|---|---|---|
| CS circadian rhythm | ||||
| T (h) | Biorhythmic period | 24 | ||
| n | Number of harmonics | 0 | 1 | 2 |
| an | Fourier coefficient | 116 | −121 | 38.8 |
| bn | Fourier coefficient | – | −41.1 | 1.58 |
| Hydrocortisone/dexamethasone pharmacokinetics | ||||
| C1 (µg ml−1) | Intercept coefficient | 85.1a/0.0582b | ||
| C2 (µg ml−1) | Intercept coefficient | 2.22a/0.0753b | ||
| λ1 (h−1) | Slope coefficient | 5.04a/1.66b | ||
| λ2 (h−1) | Slope coefficient | 0.542a/0.125b | ||
Hydrocortisone 50 mg kg−1 i.v. injection.
Dexamethasone 0.1 mg kg−1 i.v. injection.
Mechanistic Basis for Pharmacodynamics
Glucocorticoids exert their functions mainly through a receptor/gene-mediated pathway. Unbound steroid molecules diffuse through cell membranes and bind with cytosolic- free receptors. The receptor is activated upon binding and is released from heat shock protein and then undergoes conformational changes. Drug–receptor complex (DR) is translocated into the nucleus where it dimerizes and binds to glucocorticoid response elements (GREs) in the target gene promoter region. The binding of DR and GRE enhances or inhibits the transcription of target genes.
GS Circadian Rhythm
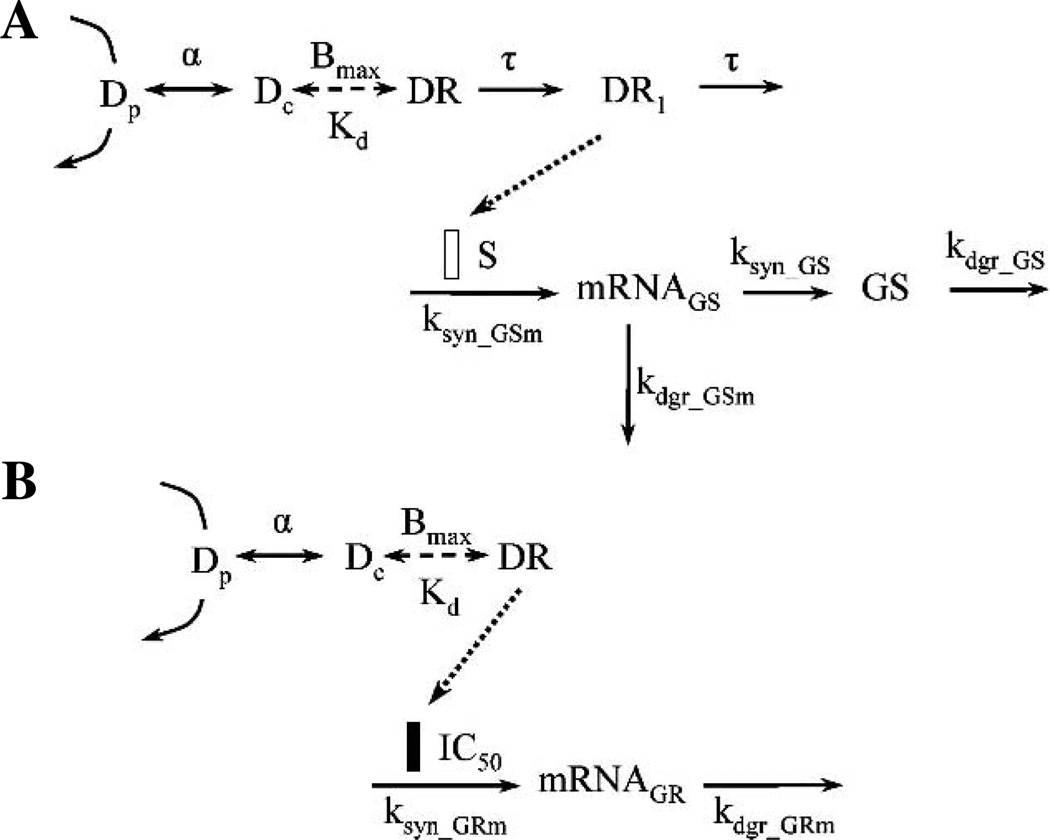
Based on the cellular mechanisms of steroid action, the integrated pharmacodynamic model shown in Fig. 1A was used to describe GS mRNA and activity profiles under the control of steroid. GS activity here represents the abundance of GS protein. It is assumed that the GS activity is proportional to the amount of GS protein in skeletal muscle. Endogenous CS serves as the driving force in the circadian rhythm study, whereas the drug concentrations (Dp) serve as the driving force in the single bolus dose studies. The equations for the various components of the model controlling receptor dynamics and GS induction are:
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where Dp and Dc are the total plasma and free intracellular steroid concentrations. It is assumed that the free intracellular steroid concentration is a constant fraction of the total plasma steroid concentration. The Bmax and Kd are parameters characterizing the binding of steroid to its receptor, representing the total binding capacity (total number of GRs) and the equilibrium dissociation constant. The Kd values were calculated from a quantitative structure–property relationship (QSPR) model (18) and were fixed in the fitting. The calculated Kd values for dexamethasone, hydrocortisone, and CS are 1.32, 13.5, and 5.13 nM, respectively. A transduction process with τ as the mean transit time was incorporated into the mechanistic PD model representing the signaling steps following steroid and receptor binding, which lead to GS gene induction and account for the induction time delay. GS mRNA (GSm) is synthesized through a zero-order process (ksyn_GSm) and degraded via a first-order process (kdgr_GSm) in the absence of steroid. GS protein is synthesized from its mRNA. First-order rate constants represent the synthesis efficiency (ksyn_GS) and the degradation rate (kdgr_GS) of GS protein. DR1 serves as the direct driving force for the induction of GS mRNA transcription and S is a linear coefficient representing the stimulatory efficiency.
Fig. 1.
Pharmacodynamic models of glucocorticoid effects in rat skeletal muscle. Pharmacodynamic model of (A) GS induction by glucocorticoids and (B) GR mRNA suppression by glucocorticoids. Symbols are defined in the text.
The total GR density in ADX rat skeletal muscle is much higher than that in normal rat skeletal muscle (19). Thus, different Bmax values were assumed for normal and ADX rats. The value of Bmax in ADX rats was fixed to 66.3 fmol mg protein−1 (8), whereas Bmax in normal rats was estimated from the fitting. It was also assumed that the synthesis rates for GS mRNA and GS activity were different between ADX and normal rats, whereas all other parameters were the same.
Stationary baselines were assumed for ADX rats in the single bolus studies since no endogenous steroid was present. The following initial conditions were derived from Eqs. (6) and (7):
| (8) |
| (9) |
where and GS0 are the mean observed GS mRNA and activity in the control ADX rat skeletal muscle. Because no stationarity was assumed for normal rats, their ksyn_GSm and ksyn_GS values were estimated with other parameters.
GR Circadian Rhythm
An integrated pharmacodynamic model shown in Fig. 1B was used to describe the GR mRNA profiles under the regulation of steroid based on the cellular mechanisms of steroid action. It is assumed that the total receptor number in muscle does not change during the period of drug administration and the 24-h period in normal rats. In the model, DR itself instead of DR1 serves as the direct driving force for gene repression. The differential equation for the inhibition of GR mRNA transcription is:
| (10) |
where GRm is the receptor mRNA. It is synthesized through a zero-order process (ksyn_GRm) and degraded by a first-order process (kdgr_GRm) in the absence of steroid. The IC50 is the concentration of DR at which the synthesis rate of receptor mRNA is reduced to 50% of its baseline value.
The Bmax for normal rats and α values were fixed from GS induction modeling. It was assumed that the synthesis rate for GR mRNA was different between ADX and normal rats, whereas all other parameters were same.
Stationarity was assumed for ADX rats without drug treatment so ksyn_GRm was defined as
| (11) |
where is the mean observed GR mRNA in the control ADX rat skeletal muscle. Because no stationarity was assumed for normal rats, their ksyn_GRm value was estimated with other parameters.
Data Analysis
The giant rat study, a typical animal study design in our laboratory, was utilized for both single-dose and circadianrhythm studies (8–10). Pharmacodynamic data obtained from multiple animals were pooled. The GS and GR data from both hydrocortisone and dexamethasone treatments and the circadian study were fitted simultaneously in the modeling. Assuming that the errors from the observed and predicted responses were normally distributed, the ADAPT II program (20) with the maximum likelihood method was applied for all fittings. The following variance model was used in all fitting processes:
| (12) |
where Vi is the variance of the response at the ith time point, ti is the actual time at the ith time point, θ represents the systemic parameter vector for the respective PD model, σ is defined as the vector of variance parameters for the model, and Y(θ, ti) is the predicted response value at time ti from the model. Variance parameters σ1 and σ2 were estimated along with systemic parameters during fitting and were incorporated in objective function-minimization procedures. Individual data from all animals were used in fitting the models. The goodness-of-fit criteria include visual inspection of the fitted curves, estimator criterion value, sum of squared residuals, Akaike information criterion, Schwartz criterion, and CV of the estimated parameters.
Statistical analysis was performed to test if the GS activities at peak and trough were significantly different. The GS activity data around time zero (t = 0.25 and 23.75 h) were pooled together to represent the GS activities at peak. The GS activity data around time 12 h (t = 11.75 and 12.25 h) were pooled together to represent the GS activities at trough. There were six animals in each group. Unpaired Student t test was applied and a p value of less than 0.05 was assumed to be significantly different.
RESULTS
CS Circadian Rhythm
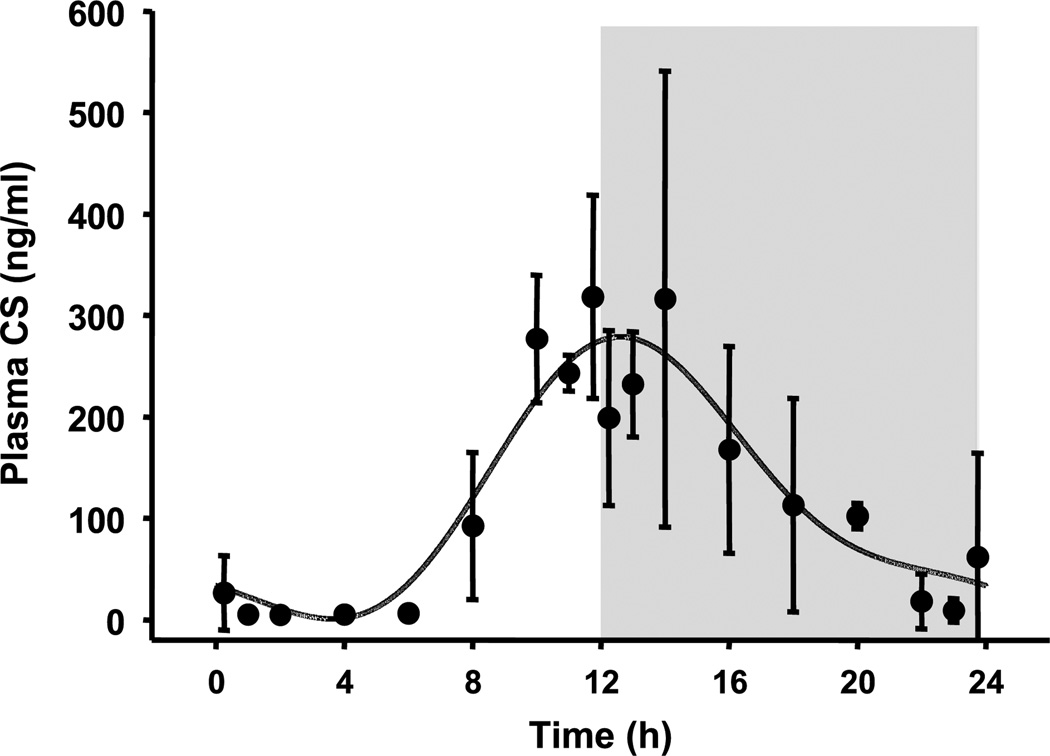
The plasma CS concentration vs. time profile obtained from normal rats and the fitted curve are shown in Fig. 2. The harmonic functions that contributed more than 1% to the overall response were reported. A two-harmonic function was able to capture the most apparent circadian fluctuations of plasma CS in normal rats during a 24-h period (16). The estimated Fourier coefficients are listed in Table I. Plasma CS reaches a peak of about 300 ng ml−1 around the onset of darkness. There is little endogenous steroid in rat plasma during the early morning with the trough occurring at about 4 h following lights on.
Fig. 2.
Plasma corticosterone concentration vs. time profile in normal male Wistar rats. Symbols are mean data (±SD) and the lines are model fitted profiles. The onset of light period was t = 0 h, whereas the onset of dark period was 12 h. The shaded area represents the dark period.
Dexamethasone and Hydrocortisone PK
The actual and simulated plasma steroid concentrations vs. time profiles are shown in Fig. 3. Plasma concentrations of both dexamethasone and hydrocortisone can be described by biexponential kinetics. Dexamethasone has a t1/2 of 5.56 h, which is much longer than the t1/2 of hydrocortisone, which is 1.28 h.
Fig. 3.
Plasma dexamethasone (●) or hydrocortisone (○) concentration vs. time profiles following i.v. injection of 0.1 mg kg−1 dexamethasone or 50 mg kg−1 hydrocortisone to ADX rats. Symbols are mean data (±SD) and the lines are model-fitted profiles. Data were taken from Mager et al. (11).
Gene-Mediated GS Induction
Figure 4 shows the fittings of the GS dynamic model to the data from both the circadian and single-dose drug studies. The estimated parameters are listed in Table II. The model seemed to capture the data from both studies well despite limited information from the single bolus dose study.
Fig. 4.
Time course of muscle GS mRNA and activity profiles in normal rats (circadian rhythm study) or following i.v. injection of 0.1 mg kg−1 dexamethasone or 50 mg kg−1 hydrocortisone in ADX rats (single bolus dose study). Symbols are mean data (± SD) and the lines are model-fitted profiles. The dashed line represents simulated plasma CS concentrations over the 24-h period. The shaded areas represent the dark period. Note differences in scales.
Table II.
Estimated and Fixed Pharmacodynamic Parameters for GS Modeling
| Parameter | Definition | Value | CV (%) |
|---|---|---|---|
| α | Intracellular/plasma ratio | 0.0175 | 67 |
| Bmax_normal (fmol mg protein−1) | Cytosolic GR density in normal rats | 44.0 | 90 |
| τ (h) | Mean transit time | 1.13 | 65 |
| kdgr_GSm (h−1) | GS mRNA degradation rate | 0.203 | 41 |
| S [(fmol/mg protein)−1] | Stimulatory coefficient | 0.142 | 28 |
| kdgr_GS (h−1) | GS Degradation Rate | 0.0512 | 19 |
| ksyn_GSm_normal (fmol g tissue−1 h−1) | GS mRNA synthesis rate in normal rats | 0.267 | 109 |
| ksyn_GS_normal (µmol min−1 g protein−1 h−1) | GS synthesis rate in normal rats | 0.210 | 20 |
| ksyn_GSm_ADX (fmol g tissue−1 h−1) | GS mRNA synthesis rate in ADX rats | 0.487 | –a |
| ksyn_GS_ADX (µmol min−1 g protein−1 h−1) | GS synthesis rate in ADX rats | 0.370 | –a |
| Kd_CS (nM) | Corticosterone dissociation constant | 5.13 | Fixed |
| Kd_HYD (nM) | Hydrocortisone dissociation constant | 13.5 | Fixed |
| Kd_DEX (nM) | Dexamethasone dissociation constant | 1.32 | Fixed |
| Bmax_ADX (fmol mg protein−1) | Cytosolic GR density in ADX rats | 66.3 | Fixed |
| (fmol g tissue−1) | GS mRNA baseline in ADX rats | 2.43 | Fixed |
| (µmol min−1 g protein−1) | GS activity baseline in ADX rats | 17.6 | Fixed |
Secondary parameter calculated from degradation rate and baseline.
The circadian oscillation of GS mRNA in normal rat skeletal muscle is apparent despite the interindividual variability. Its peak occurs at about 18 h, 6 h following the peak of plasma CS concentrations. It reaches its trough at 8 h during the light period, 4 h following the plasma CS trough. Comparison of GS mRNA and plasma CS circadian profiles reveals a 4- to 6-h time delay in GS mRNA expression. The GS mRNA expression level changes from 3 to 6 fmol g tissue−1 in a 24-h period. The lowest expression level of GS mRNA in normal rats is comparable to that expressed in ADX rats without drug treatment.
The statistical analysis shows that the GS activity around time zero is significantly higher than that around 12 h (p < 0.01). However, examination of the GS activity profile during a 24-h period exhibits a very flat fluctuation around 20 µmol min−1 g protein−1. The GS activity reaches the peak during the onset of the light period and a trough near the beginning of the dark period. Comparison of GS activity and plasma CS circadian profiles reveals a 12-h time delay in the GS activity expression. As for GS mRNA, the lowest GS activity level (17.4 µmol min−1 g protein−1) is comparable to that in ADX animals without drug treatment. The highest expression level at time zero is about 23.2 µmol min−1 g protein−1.
In the single bolus dose studies, expression of both GS mRNA and activity show a slow rise following drug treatment. GS mRNA reaches a peak at 6 h following hydrocortisone treatment and then declines. Expression of GS mRNA in dexamethasone-treated ADX rats keeps increasing until the end of the study at 12 h. This difference might be due to the longer t1/2 of dexamethasone. Additionally, dexamethasone has a higher receptor-binding affinity than hydrocortisone, which may lead to prolonged effects. Administration of both steroids induces a 9-fold increase of GS mRNA expression at peak time in ADX rat skeletal muscles. The enhanced GS activity occurs much later and increases with a much slower pace than GS mRNA in both groups, which is expected because increased GS protein is due to enhanced mRNA transcription by steroids. At 12 h, steroid treatment only induces about a 2-fold enhancement in GS activity in rat skeletal muscle in both groups. However, the single bolus study only provided limited information about GS induction before 12 h and we do not know if GS activities would increase further or return to baseline after 12 h.
The estimated α in skeletal muscle was 0.0175, which was similar to the corresponding parameter estimate (0.02) in liver (11). It indicates that the intracellular concentration of free steroid is 1.75% of total plasma concentration. Those steroid molecules are a fraction of total steroids that are capable of binding directly with receptor in cytosol. The estimated total intracellular GR (Bmax) was 44.0 fmol mg protein−1 in normal rat skeletal muscle. Fitting the proposed model yielded reasonable estimations for GS mRNA and protein degradation rates. The estimated kdgr_GSm (0.203 h−1) and kdgr_GS (0.0512 h−1) indicate the degradation half-lives of GS mRNA and GS activity are 3.41 and 13.5 h, respectively. The synthesis rates of GS mRNA and GS activity were estimated as 0.267 fmol g tissue−1 h−1 and 0.210 µmol min−1 g protein−1 h−1 in normal rats. The same two parameters were calculated as secondary parameters in ADX rats, yielding 0.487 fmol g tissue−1 h−1 and 0.370 µmol min−1 g protein−1 h−1. A one-step transduction process was able to describe the time delay between drug–receptor binding and GS mRNA induction. The mean transit time τ was estimated to be 1.19 h. The linear coefficient S was estimated at 0.142 (fmol/mg protein)−1, representing a moderate stimulatory efficiency of DR1 on the transcriptional induction of GS mRNA. A nonlinear Hill function affecting ksyn_GSm was tried in the modeling. However, only one dose level was studied and the model was overparameterized and could not resolve all parameters reliably.
Gene-Mediated GR Suppression
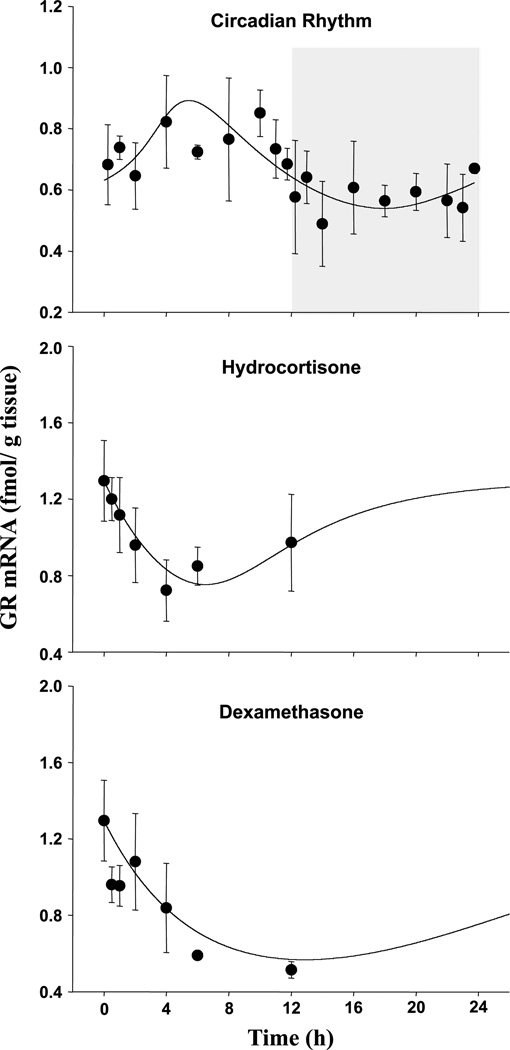
Fittings of the PD model to the GR mRNA data from both circadian and single-dose drug studies are shown in Fig. 5. The estimated parameters are listed in Table III. The proposed model was able to capture the receptor mRNA transcription profiles in normal rat muscle or following treatments of both steroids in ADX rat muscle.
Fig. 5.
Time course of muscle GR mRNA profiles in normal rats (circadian rhythm study) or following i.v. injection of 0.1 mg kg−1 dexamethasone or 50 mg kg−1 hydrocortisone in ADX rats (single bolus dose study). Symbols and lines are defined as in Fig. 4. Note differences in scales.
Table III.
Estimated and Fixed Pharmacodynamic Parameters for GR mRNA Modeling
| Parameter | Definition | Value | CV (%) |
|---|---|---|---|
| kdgr_GRm (h−1) | GR mRNA degradation rate | 0.170 | 12 |
| IC50 (fmol mg protein−1) | Inhibitory coefficient | 17.4 | 17 |
| ksyn_GRm_normal (fmol−1 g tissue−1 h−1) | GR mRNA synthesis rate in normal rats | 0.218 | 9 |
| ksyn_GRm_ADX (fmol−1 g tissue−1 h−1) | GR mRNA synthesis rate in ADX rats | 0.220 | –a |
| α | Intracellular/plasma ratio | 0.0175 | Fixed |
| Kd_CS (nM) | Corticosterone dissociation constant | 5.13 | Fixed |
| Kd_HYD (nM) | Hydrocortisone dissociation constant | 13.5 | Fixed |
| Kd_DEX (nM) | Dexamethasone dissociation constant | 1.32 | Fixed |
| Bmax_normal (fmol mg protein−1) | Cytosolic GR density in normal rats | 44.0 | Fixed |
| Bmax_ADX (fmol mg protein−1) | Cytosolic GR density in ADX Rats | 66.3 | Fixed |
| (fmol g tissue−1) | GR mRNA baseline in ADX rats | 1.30 | Fixed |
Secondary parameter calculated from degradation rate and baseline.
In normal rat skeletal muscles, the expression of receptor mRNA also follows a circadian pattern. Because receptor transcription is inhibited by its own ligand, its expression profile exhibits an opposite trend compared to GS mRNA expression. Generally, GR mRNA is expressed at a higher level in the light period than in the dark period. The trough of receptor mRNA occurs at the middle of the dark period, about 4 h after the peak of plasma CS. The time delay between plasma CS and mRNA expression is reasonable and comparable to the time delay for GS mRNA expression. The peak occurs at 6 h after the beginning of the light period. Skeletal muscle receptor RNA level fluctuates between 0.55 and 0.9 fmol g tissue−1 in normal rats under the control of endogenous CS.
In the single bolus dose studies, GR mRNA expression immediately declines following both dexamethasone and hydrocortisone administration. It reaches a trough about 6 h following hydrocortisone treatment, where it decreases from 1.30 ± 0.21 to 0.72 ± 0.16 fmol g tissue−1. Receptor mRNA expression exhibits a slight increasing trend around 12 h. In dexamethasone-treated animals, it reaches the trough at a much later time. At 12 h following dexamethasone treatment, GR mRNA decreases from baseline (1.30 ± 0.21) to 0.59 ± 0.02 fmol g tissue−1. Again, only limited information was obtained from the single bolus studies.
It was assumed that receptor as well as GS transcription is directly under the control of intracellular free GR. Thus, in the analysis, the α value was fixed to the estimate obtained from fittings of GS data. The Bmax for normal rats was also fixed according to GS mRNA modeling because both receptor and GS data were taken from the same animals. The degradation rate of receptor mRNA was estimated at 0.170 h−1, yielding a t1/2 of 4.08 h. The synthesis rate of GR mRNA in normal rat was 0.218 fmol g tissue−1 h−1. In ADX rat skeletal muscle, the synthesis rate was calculated from the degradation rate and baseline, yielding 0.220 fmol g tissue−1 h−1. The IC50 was estimated at 17.4 fmol mg protein−1. Reasonable CVs, between 9 and 17%, were obtained for all fitted parameters.
DISCUSSION
It is quite apparent that biological clocks have an impact on most aspects of physiology. About 10% of genes that are actively transcribed tend to oscillate in a circadian fashion, most of which are involved in carbohydrate and lipid metabolism (21). Disruption of the normal temporal profiles of those genes may lead to physiological dysfunction or even disease. For example, it has been suggested that dysregulated clock functions are associated with metabolic syndrome, a condition affecting about 47% of the US population (21,22). Understanding circadian rhythms is prerequisite in the course of studying and managing those diseases. Mathematical modeling provides a useful tool to assess the temporal profiling of genes and their perturbations by drugs or environmental factors.
Our overall aim is to evaluate the circadian rhythm of endogenous CS and its effects on its own receptor and downstream gene transcription in normal rat skeletal muscle. The plasma cortisol concentrations in a 24-h period in humans were modeled by several mathematical functions (23) and the two-harmonic function worked well. In the present report, the circadian episodic profile of CS in rat plasma was also described by a two-harmonic function, which explained 67.3% of the total variance (r = 0.820). In the present report, plasma CS concentrations were modeled by FOURPHARM program (17). FOURPHARM does not do traditional fitting. It first generates a baseline, which connects all mean data at each time point using a Fourier transform. The harmonics with significant contributions to the overall baseline were selected. The relative contributions of the mean, 24- and 12-h harmonics were 55.8, 38.4, and 3.4%, respectively. The harmonic functions contributing less than 1% of overall baseline concentrations were not used.
Our giant rat study demonstrates that both GS mRNA and activity exhibit daily circadian changes. Their expression profiles follow the similar pattern as plasma CS with a right shift of 6 and 12 h, suggesting the dependence of muscle GS expression on plasma CS. The dynamics of receptor mRNA follows an inverse pattern as plasma CS with a time shift of 4 h, which is consistent with the inhibiting effect of steroids on their own receptor. In spite of the rich literature studying the regulation of GS and receptor gene expression by glucocorticoids, variable baselines were not considered. This report shows that the peak/trough variations of GS and receptor mRNA are 2- and 1.6-fold in rat muscle, respectively. Despite the relatively flat GS activity profile, the highest GS activity at the onset of the light period is 30% higher than that at the beginning of the dark period. The abundances of GS and receptor depend on the study time and the light–dark cycle in addition to drug treatment. Conclusions made from normal rat studies are distorted without consideration of the daily oscillations of gene expression. With this complexity, future studies with GR and GS gene expression in muscle and possibly other tissues need careful design, or variable baselines have to be incorporated in the studies.
Circadian rhythms elicit changes in sensitivity of response, depending on the timing of drug administration. Studies of the biological rhythm of human plasma cortisol suggest that cortisol suppression by inhaled corticosteroids is at maximum when administered at 3 a.m. and minimized when injected in the afternoon (24). Dexamethasone injected prior to the zenith results in prolonged and/or stronger expression of its downstream gene in mice, whereas the same treatment induces a new surge of gene expression if injected after the peak time (4). The circadian rhythm of GS expression in skeletal muscle leads to the expectation that managing corticosteroid dosing time may lead to augmented or reduced drug response.
Integrated PD models were used to quantitatively characterize the rhythmic expression of receptor and downstream genes under the control of steroids. Our previously published work has demonstrated that post-receptor-binding events seem largely drug independent and the principle components explaining the pharmacological differences between various corticosteroids are the drug kinetics and relative receptor affinity (11). Our present PD model is based on this premise. Data from studies of three steroids including both endogenous and exogenous glucocorticoids were pooled. Different equilibrium dissociation constant (Kd) values for each steroid and one set of parameters for the postreceptor events were applied to the PD model. The Kd values were calculated from a QSPR model (18) previously used in the integrated modeling of liver TAT expression (11). In our circadian rhythm study, the time delay for each measured component following plasma CS is similar to that observed following single-dose hydrocortisone, supporting our assumption that the steps following ligand–receptor binding are probably common for all glucocorticoids. However, the time delay is longer for all measured components following dexamethasone treatment, probably due to its longer half-life or greater receptor affinity.
One of our assumptions was that only intracellular free glucocorticoid molecules are able to bind with the cytosolic receptor and exert its function. The α parameter was included in the model, representing the fraction of intracellular free/plasma total steroid concentration. It was assumed that the intracellular concentration was proportional to the total drug concentration in plasma. Glucocorticoids have moderate protein binding. Both CS and hydrocortisone bind to corticosteroid binding globulin (CBG) and albumin. The CBG has a high affinity and a low capacity for both steroids, whereas albumin has a low affinity but a high capacity. Nonlinearity has been observed for CS in rat plasma (25) as well as for cortisol in human plasma (26). Dexamethasone has no affinity to CBG and binds only to albumin in a linear manner. Therefore, different α values for each steroid with or without nonlinearity were tried in the modeling. However, a common linear α value was adequate to capture all profiles without consideration of nonlinear protein binding. This result is consistent with our previous study in liver (11). Although protein binding may be the major factor restricting glucocorticoid molecules from entering cells, other processes may also be involved.
A transduction model was used to describe the signaling steps linking receptor binding and downstream gene transcription for GS expression. Following receptor binding, a series of events including receptor conformational change, dissociation of multiprotein complex, translocation of receptor to nucleus, receptor dimerization, binding with GRE, and activation or repression occur prior to gene transcription. The mean transit time τ was able to describe part of the time delay between CS and GS mRNA induction. However, a transduction step was not required for GR mRNA modeling. This is consistent with the phenomenon that inhibition of GR mRNA is faster than stimulation of GS mRNA transcription. It may be due to different GRE binding affinities or efficiencies of activation or repression. It also suggests that the signaling process is different for each gene and different transduction models or parameters may be needed.
There is an increase in the number of receptor sites in the cytosol of skeletal muscle following adrenalectomy which is consistent with glucocorticoid inhibition of its own receptor transcription. The ratio of total GR density in ADX to normal rat muscle is between 1.39 and 1.88 depending on age (19). In the present report, Bmax_normal was estimated at 44.0 fmol mg protein−1, yielding an ADX to normal rat ratio of 1.51.
Because both ADX and normal rats were studied in the present report, different baselines were observed. Our assumption was that the synthesis rates of various components differed between these rats, whereas the degradation rates were the same. However, it is possible that both production and degradation processes are altered when endogenous steroid is removed. This possibility will be difficult to test, as the parameters will not be accurately estimated. A similar approach was applied to pharmacodynamic data generated from different studies using ADX rats when significantly distinct baseline levels were observed (11). Our modeling revealed different estimates of synthesis rates for GS mRNA and GS activity between normal and ADX rats. The synthesis rates of GR mRNA are similar for both ADX and normal rat skeletal muscle. The half-lives of mRNA were estimated around 4 h, whereas that of protein was estimated at 13 h.
In normal rat skeletal muscle, the circadian variation in GS mRNA is more apparent than that of GS activity. The difference in the level of induction in GS mRNA and activity levels was also observed in a previous study, which showed that following dexamethasone treatment, GS mRNA increased to a much greater extent (20-fold) than the enzyme activity (3-fold) (7). Other translational and/or posttranslational events might be involved in GS enzyme activity regulation. The translation or activity of GS protein might be regulated by other hormones or growth factors. Another explanation is that the relatively high animal interindividual variability may disguise the true circadian pattern of the protein.
The necessity of estimating mRNA synthesis rate with other parameters in the circadian study results in correlation between the linear coefficient S and the mRNA synthesis rate. This problem was solved by including our single bolus dose studies, as S can then be estimated, yielding reasonable parameter estimates and CV values. Three steroids were studied, indicating that these effects are not restricted to a single steroid. One set of parameters was adequate to capture the pharmacodynamic data profiles for both exogenous and endogenous steroid-induced effects from two different studies except for certain parameters such as Bmax and synthesis rates.
In summary, the studies demonstrate that GR and GS expression in rat muscle follows a circadian rhythm as does TAT in liver. Our integrated PD models well captured the 24-h rhythmic oscillations of GS and receptor under the control of plasma CS and facilitated understanding of the underlying molecular and physiological mechanisms. It suggests that the rhythmic baselines of GR and GS in skeletal muscle have to be considered in future studies of gene expression when normal rats are used as the animal model.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Donald E. Mager and Ms. Nancy A. Pyszczynski for performance of animal studies and Ms. Suzette M. Mis for HPLC analysis. We would also like to thank Dr. Lori Badura from Pfizer Inc. for her advice in the experimental design. This study was supported by grant GM 24211 from the National Institutes of Health and by a research grant from NASA.
ABBREVIATIONS
- ADX
adrenalectomized
- CBG
corticosteroid binding globulin
- CS
corticosterone
- DR
drug–receptor complex
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- GRG
glucose repressible gene
- GS
glutamine synthetase
- HPA
hypothalamus–pituitary–adrenal axis
- PK/PD
pharmacokinetic/pharmacodynamic
- QSPR
quantitative structure–property relationship
- RT-PCR
reverse transcription–polymerase chain reaction
- TAT
tyrosine aminotransferase.
REFERENCES
- 1.Retana-Marquez S, Bonilla-Jaime H, Vazquez-Palacios G, Dominguez-Salazar E, Martinez-Garcia R, Velazquez-Moctezuma J. Body weight gain and diurnal differences of corticosterone changes in response to acute and chronic stress in rats. Psychoneuroendocrinology. 2003;28:207–227. doi: 10.1016/s0306-4530(02)00017-3. [DOI] [PubMed] [Google Scholar]
- 2.Angelova KC, Angelov CG. Cosinor analysis of circadian oscillations of amino acid catabolizing enzymes in temporal pattern of nutrient input. Z. Ernahrungswiss. 1998;37(Suppl. 1):98–102. [PubMed] [Google Scholar]
- 3.Van Cantfort J, Gielen JE. Comparison of rat and mouse circadian rhythm of cholesterol-7 alpha-hydroxylase activity. J. Steroid Biochem. 1979;10:647–651. doi: 10.1016/0022-4731(79)90518-1. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 5.Oakley RH, Cidlowski JA. Homologous down-regulation of the glucocorticoid receptor: the molecular machinery. Crit. Rev. Eukaryot. Gene Expr. 1993;3:63–88. [PubMed] [Google Scholar]
- 6.Abcouwer SF, Bode BP, Souba WW. Glucocorticoids regulate rat glutamine synthetase expression in a tissue-specific manner. J. Surg. Res. 1995;59:59–65. doi: 10.1006/jsre.1995.1132. [DOI] [PubMed] [Google Scholar]
- 7.Max SR, Mill J, Mearow K, Konagaya M, Konagaya Y, Thomas JW, Banner C, Vitkovic L. Dexamethasone regulates glutamine synthetase expression in rat skeletal muscles. Am. J. Physiol. 1988;255:E397–E402. doi: 10.1152/ajpendo.1988.255.3.E397. [DOI] [PubMed] [Google Scholar]
- 8.Sun YN, McKay LI, DuBois DC, Jusko WJ, Almon RR. Pharmacokinetic/Pharmacodynamic models for corticosteroid receptor down-regulation and glutamine synthetase induction in rat skeletal muscle by a receptor/gene-mediated mechanism. J. Pharmacol. Exp. Ther. 1999;288:720–728. [PubMed] [Google Scholar]
- 9.Sun YN, DuBois DC, Almon RR, Jusko WJ. Fourth-generation model for corticosteroid pharmacodynamics: a model for methylprednisolone effects on receptor/gene-mediated glucocorticoid receptor down-regulation and tyrosine aminotransferase induction in rat liver. J. Pharmacokinet. Biopharm. 1998;26:289–317. doi: 10.1023/a:1023233409550. [DOI] [PubMed] [Google Scholar]
- 10.Ramakrishnan R, DuBois DC, Almon RR, Pyszczynski NA, Jusko WJ. Fifth-generation model for corticosteroid pharmacodynamics: application to steady-state receptor down-regulation and enzyme induction patterns during seven-day continuous infusion of methylprednisolone in rats. J. Pharmacokinet. Pharmacodyn. 2002;29:1–24. doi: 10.1023/a:1015765201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mager DE, Pyszczynski NA, Jusko WJ. Integrated QSPR-pharmacodynamic model of genomic effects of several corticosteroids. J. Pharm. Sci. 2003;92:881–889. doi: 10.1002/jps.10343. [DOI] [PubMed] [Google Scholar]
- 12.Haughey DB, Jusko WJ. Analysis of methylprednisolone, methylprednisone and corticosterone for assessment of methylprednisolone disposition in the rat. J. Chromatogr. 1988;430:241–248. doi: 10.1016/s0378-4347(00)83159-x. [DOI] [PubMed] [Google Scholar]
- 13.DuBois DC, Almon RR, Jusko WJ. Molar quantification of specific messenger ribonucleic acid expression in northern hybridization using cRNA standards. Anal. Biochem. 1993;210:140–144. doi: 10.1006/abio.1993.1164. [DOI] [PubMed] [Google Scholar]
- 14.McNally MT, Free SJ. Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 1988;14:545–551. doi: 10.1007/BF00434079. [DOI] [PubMed] [Google Scholar]
- 15.Minet R, Villie F, Marcollet M, Meynial-Denis D, Cynober L. Measurement of glutamine synthetase activity in rat muscle by a colorimetric assay. Clin. Chim. Acta. 1997;268:121–132. doi: 10.1016/s0009-8981(97)00173-3. [DOI] [PubMed] [Google Scholar]
- 16.Hazra A, Jusko WJ, Almon RA, Dubois DC. Pharmacodynamics of circadian rhythm of corticosterone effects on tyrosine aminotransferase in normal rats. AAPS J. 2004;6 Abstract T3355. [Google Scholar]
- 17.Krzyzanski W. FOURPHARM User’s Guide: A Computer Program Applying Fourier Analysis to Biorhythmic Data. Buffalo, NY: 2000. [Google Scholar]
- 18.Wolff ME, Baxter JD, Kollman PA, Lee DL, Kuntz ID, Bloom E, Matulich DT, Morris J. Nature of steroid–glucocorticoid receptor interactions: thermodynamic analysis of the binding reaction. Biochemistry. 1978;17:3201–3208. doi: 10.1021/bi00609a005. [DOI] [PubMed] [Google Scholar]
- 19.DuBois DC, Almon RR. Glucocorticoid sites in skeletal muscle: adrenalectomy, maturation, fiber type, and sex. Am. J. Physiol. 1984;247:E118–E125. doi: 10.1152/ajpendo.1984.247.1.E118. [DOI] [PubMed] [Google Scholar]
- 20.D’Argenio DZ, Schumitzky A. ADAPT II User’s Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software. Los Angeles: Biomedical Simulation Resource; 1997. [Google Scholar]
- 21.Johnson N. Beating the clock for a better understanding of metabolism. Scientist. 2005;19:16. [Google Scholar]
- 22.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty A, Krzyzanski W, Jusko WJ. Mathematical modeling of circadian cortisol concentrations using indirect response models: comparison of several methods. J. Pharmacokinet. Biopharm. 1999;27:23–43. doi: 10.1023/a:1020678628317. [DOI] [PubMed] [Google Scholar]
- 24.Meibohm B, Hochhaus G, Rohatagi S, Mollmann H, Barth J, Wagner M, Krieg M, Stockmann R, Derendorf H. Dependency of cortisol suppression on the administration time of inhaled corticosteroids. J. Clin. Pharmacol. 1997;37:704–710. doi: 10.1002/j.1552-4604.1997.tb04357.x. [DOI] [PubMed] [Google Scholar]
- 25.Jobin M, Perrin F. Evaluation of three constants involved in the binding of corticosterone to plasma proteins in the rat. Can. J. Biochem. 1974;52:101–105. doi: 10.1139/o74-016. [DOI] [PubMed] [Google Scholar]
- 26.Rocci ML, Jr, D’Ambrosio R, Johnson NF, Jusko WJ. Prednisolone binding to albumin and transcortin in the presence of cortisol. Biochem. Pharmacol. 1982;31:289–292. doi: 10.1016/0006-2952(82)90172-1. [DOI] [PubMed] [Google Scholar]