Abstract
Molecules that play a role in Plasmodium merozoite invasion of host red blood cells represent attractive targets for blood-stage vaccine development against malaria. In Plasmodium vivax, merozoite invasion of reticulocytes is mediated by the Duffy binding protein (DBP), which interacts with its cognate receptor, the Duffy antigen receptor for chemokines, on the surface of reticulocytes. The DBP ligand domain, known as region II (DBPII), contains the critical residues for receptor recognition, making it a prime target for vaccine development against blood-stage vivax malaria. In natural infections, DBP is weakly immunogenic and DBPII allelic variation is associated with strain-specific immunity, which may compromise vaccine efficacy. In a previous study, a synthetic vaccine termed DEKnull that lacked an immunodominant variant epitope in DBPII induced functional antibodies to shared neutralizing epitopes on the native Sal1 allele. Anti-DEKnull antibody titers were lower than anti-Sal1 titers but produced more consistent, strain-transcending anti-DBPII inhibitory responses. In this study, we further characterized the immunogenicity of DEKnull, finding that immunization with recombinant DEKnull produced an immune response comparable to that obtained with native recombinant DBP alleles. Further investigation of DEKnull is necessary to enhance its immunogenicity and broaden its specificity.
INTRODUCTION
The global control of malaria is threatened with the spread of drug-resistant parasites and insecticide-resistant mosquitoes. The impact of this on the public health infrastructure and economic stability of the countries most affected is a cause for concern. Vaccines are an attractive mode of control since they are cost-effective and easily administered. Despite years of research, effective malaria vaccines have remained elusive and none has been introduced as a commercial product. However, efforts to develop a vaccine remain optimistic for a number of reasons. First, individuals in regions where malaria is endemic develop naturally acquired immunity to clinical manifestation of the disease (1, 2). Second, passive transfer of IgG from immune individuals conveyed protection to naive individuals (3). Third, vaccination with attenuated sporozoites induced partial protection (4). Finally, naturally and artificially acquired antibodies to different Plasmodium antigens have been shown to inhibit parasite invasion of erythrocytes, as well as parasite growth and development, in vitro (5–8).
Successful host red blood cell invasion by Plasmodium merozoites depends on specific ligand-receptor interactions (9, 10). In Plasmodium vivax, a critical step in the invasion of reticulocytes during asexual blood-stage infection is the interaction between the merozoite Duffy binding protein (DBP) and its cognate receptor, the Duffy antigen receptor for chemokines (DARC), on the reticulocyte surface. It is believed that DBP plays an essential role during the irreversible process of junction formation just before invasion (11, 12). This is evident in the virtual absence of P. vivax malaria in populations lacking DARC expression on their erythrocyte surface (13, 14). This dependence of DBP on DARC for erythrocyte invasion makes DBP a promising candidate as a vaccine against the asexual stages of the parasite. Naturally occurring anti-DBP antibodies are prevalent in people living in regions where malaria is endemic (15, 16). There is evidence that these anti-DBP antibodies can block DBP-erythrocyte binding, as well as inhibit parasite invasion in short-term in vitro cultures (5, 17–21). These data further strengthen the case for a vaccine based on DBP. In P. vivax DBP, a cysteine-rich domain, termed region II, consisting of 330 amino acid residues is considered the ligand domain for adherence to DARC during invasion (22, 23). Structural studies have revealed that DBPII can be divided into three subdomains (24, 25), and other studies have demonstrated that critical residues for receptor recognition are located within subdomain 2 (22, 26–28). Interestingly, this region is highly polymorphic, a pattern consistent with high selection pressure on DBPII (26, 29–32). This poses a great challenge to the development of a DBP-based vaccine that will be effective against diverse P. vivax strains.
We have identified B-cell epitopes within the ligand domain of DBP that are associated with protection (17). The immunodominant B-cell epitopes identified are polymorphic, surface-exposed motifs that a previous study determined are not important for receptor recognition but flank residues critical for receptor recognition (22, 25). As most naturally acquired infections with P. vivax tend to elicit weakly reactive and strain-specific antibodies (2, 17, 33), we hypothesize that the polymorphic dominant B-cell epitopes represent an evasion mechanism that misdirects the immune response away from the functional, more conserved Duffy recognition epitopes that are potential targets for broadly neutralizing immunity. Similar to what occurs in other microbial agents (34, 35), these variant immunodominant epitopes in DBPII tend to create an inherent bias toward the induction of a nonprotective, strain-specific humoral immune response. Recently, we designed a novel synthetic DBPII immunogen, termed DEKnull, that lacks a strain-specific immunodominant variant epitope normally present on DBPII (36). We demonstrated that removal of this dominant variant epitope lowered DBP immunogenicity, but importantly, inhibitory anti-DBPII antibodies were elicited against conserved neutralizing epitopes on the native Sal1 strain, which was used as the template and shared with other DBP allelic variants. Therefore, recombinant DEKnull (rDEKnull) was able to produce inhibitory anti-DBP antibodies against diverse DBPII alleles (37). Previous studies have demonstrated that naturally acquired immunity to the erythrocytic stages of malaria parasites is strongly dependent on antibodies (38–40). In areas where malaria is endemic, immunity to vivax malaria is gradually acquired with age as a result of a boosting effect due to repeated exposure to infection (2, 19, 41, 42). Acquired immunity, in addition to being biased toward strain specificity, is relatively slow to develop, never sterile, weak, short-lived (43), and usually unstable (17). There is no long-lasting protective immunologic memory in the absence of continued exposure to infection (17, 43–46) and a failure to consistently boost upon reinfection (47). The present study was designed to further our understanding of the immunogenicity of synthetic DEKnull as a vaccine candidate and to determine whether immunization with rDEKnull can induce an immune response that is relevant to diverse naturally occurring DBPII alleles of P. vivax.
MATERIALS AND METHODS
Production of recombinant proteins.
Recombinant DBPII antigens from three naturally occurring alleles (Sal1, 7.18, and P), the synthetic DBPII allele (DEKnull) (36), and rPvMSP1-19 were expressed and purified as described previously (37, 48). Briefly, the genes coding for the various antigens were synthesized, codon optimized for expression in Escherichia coli, and cloned into expression vector pET21a+ (Novagen) with a C-terminal 6×His tag to facilitate purification by affinity chromatography. The resulting plasmid was transformed into E. coli BL21(DE3) LysE (Invitrogen). Recombinant proteins were expressed, purified under denaturing conditions, and refolded by rapid dilution as previously described (20, 49, 50). The refolded antigens were evaluated for native conformation and further analyzed for function by a standard erythrocyte-binding assay (36, 48). Endotoxins were removed from the antigens with the GenScript ToxinEraser endotoxin removal kit, and endotoxin levels in the final products were determined with the ToxinSensor Chromogenic LAL Endotoxin Assay kit. All antigens had endotoxin levels of ≤40 EU/ml.
Immunization schedule.
Female BALB/c mice (6 to 8 weeks old) were purchased from Harlan Animal Research Laboratories. Immunizations were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of South Florida. Mice were randomly assigned to six groups of 15 mice each, and preimmune serum was collected from each mouse. The first five groups were primed twice 3 weeks apart with rDEKnull at 25 μg/dose subcutaneously, and the sixth group (control) was primed with rSal1 at 25 μg/dose. All antigens were emulsified in TiterMax Gold as an adjuvant according to the manufacturer's specifications. Three weeks after the second dose (day 42), test sera were collected and total serum anti-DBPII IgG titers (baseline titers) were quantified by enzyme-linked immunosorbent assay (ELISA) against the priming antigen. Serum IgG titers were then monitored until an approximately 50% decline in titer was observed (day 217). On day 224 (32 weeks after the first immunization), mice in each group received a booster immunization (anamnestic boost) of 25 μg of rDBPII each from one of three naturally occurring alleles (Sal1, 7.18, and P), rDEKnull, or rPvMSP1-19 and the control group received rSal1 (Table 1). Three weeks following the anamnestic boost (day 245), the mice were bled for serum and individual mouse serum was stored at −20°C in barcoded tubes until needed. The final IgG titers (day 245) for each group were determined against the booster antigen, with the exception of the group boosted with rMSP1-19, whose IgG titer was determined against rDEKnull.
TABLE 1.
Immunization schedule
| Immunization group | Priming antigen (days 0a and 21b) | Anamnestic boost antigen (day 224b) |
|---|---|---|
| 1 | DEKnull | DEKnull |
| 2 | DEKnull | Sal1 |
| 3 | DEKnull | 7.18 |
| 4 | DEKnull | P |
| 5 | DEKnull | MSP1-19 |
| 6 | Sal1 | Sal1 |
Antigen emulsified 1:1 (vol/vol).
Antigen emulsified 1:0.5 (vol/vol) in adjuvant.
Quantitation of anti-DBPII antibody titers.
Immunized mice were evaluated for allele-specific anti-DBPII antibody titers by ELISA as previously reported (37, 48). Briefly, ELISA plates (Nunc) were precoated with 0.2 μg of rDEKnull, Sal1, P, 7.18, or PvMSP1-19 per well, and unbound surfaces were blocked with 5% (wt/vol) skim milk in phosphate-buffered saline–0.05% Tween 20 for 2 h at room temperature. Serial tripling dilutions of individual mouse sera (starting at 1:4,000) in blocking solution was added to triplicate wells and again incubated for 2 h on a shaker at room temperature. After washing, wells were incubated with a goat anti-mouse alkaline phosphatase-conjugated secondary antibody (KPL Inc.) for 90 min as described above, and bound antibody was detected with alkaline phosphatase conjugate (BluPhos Microwell Substrate kit; KPL Inc.). Absorbance at 630 nm was determined on a microplate reader (BioTek Instruments Inc.). DBPII-specific monoclonal antibody 3D10 (48) was used as the standard calibrator on each plate. All optical density (OD) values were normalized at a point on the standard curve where the OD at 630 nm (OD630) was 1.0, and antibody values were expressed in ELISA units (EU), which were determined as the ratio of the OD630 generated by the test antibody to the OD630 of the standard (37). Antibody titers were determined as the serum dilution required to achieve 1.5 EU (51). Anti-MSP1-19 antibody served as a negative control, while preimmune serum was used as a background control.
Lymphocyte proliferative assay.
Splenocytes from immunized mice were prepared as previously reported (52, 53), with some modifications. Briefly, spleens were harvested and washed twice in Iscove's modified Dulbecco's medium (IMDM; Invitrogen). Single-cell suspensions were prepared by teasing spleen tissues into tiny pieces in a petri dish containing IMDM. Red blood cells were lysed with ACK lysis solution (Invitrogen), and membrane debris was removed by passing the cell suspension through a cell strainer (Fisher Scientific). The cells were washed with IMDM, resuspended in IMDM–10% fetal bovine serum (FBS), and layered over Ficoll Paque Plus. The splenocytes were recovered by centrifugation and washed in IMDM–10% FBS. Cell viability was determined by trypan blue staining. The cells were resuspended in IMDM–10% FBS–0.5 mM 2-mercaptoethanol–1× streptomycin-penicillin (Invitrogen). Splenocytes were seeded into 96-well flat-bottom plates at a concentration of 2 × 105/ml in 100 μl of complete medium and stimulated with 1 μg/ml rDEKnull, Sal1, 7.18, P, or PvMSP1-19 in triplicate wells. Additional wells stimulated with concanavalin A (ConA; Sigma-Aldrich) at a final concentration of 1 μg/ml or left unstimulated (with culture medium alone) served as positive and negative controls, respectively. Plates were incubated for 72 h at 37°C in a 5% CO2 humidified incubator. A 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay was used to determine the number of proliferative cells with the CellTiter 96 AQueous One Solution Cell Proliferation Assay reagent (Promega). A 20-μl volume of the reagent was added directly to 100 μl of culture in the wells and incubated for 4 h at 37°C, and absorbance at 490 nm was recorded with a 96-well Synergy 2 ELISA plate reader (BioTek Instruments). Results were expressed as a stimulation index (SI), which is the ratio of the absorbance of the stimulated culture to that of the unstimulated culture.
Inhibition of DBP-erythrocyte binding by COS-7 assay.
Expression plasmid constructs were engineered to target different variants of DBPII alleles on the surface of transiently transfected COS-7 cells as fusion proteins to the N terminus of enhanced green fluorescent protein (EGFP). DBPII-erythrocyte binding inhibition assays were performed as previously reported (21, 54, 55). Briefly, wells of transfected COS-7 cells were preincubated with different concentrations of purified total serum IgG from pooled mouse serum from the different immunization groups prior to the addition of human erythrocytes. Purified total serum IgG from mice immunized with rMSP1-19 served as a negative-control antibody. Binding inhibition was determined by assessing the percentage of rosettes in wells of transfected COS-7 cells in the presence of test IgG relative to that in wells of transfected cells in the presence of purified IgG from preimmune sera.
Statistical analyses.
The distributions of antibody titers and inhibition concentrations for each antiserum were compared between the different alleles tested for any statistically significant differences in antibody reactivity and inhibitory responses by one-way analysis of variance and multiple-comparison analysis by the Bonferroni test with SAS software.
RESULTS
Anamnestic response to rDEKnull.
To determine if rDEKnull vaccine could induce secondary immune responses against native alleles of DBPII, BALB/c mice were primed with two consecutive doses of rDEKnull 3 weeks apart. ELISA was used to determine baseline serum anti-DEKnull IgG antibody titers 3 weeks after the second immunization (day 42). The serum anti-DEKnull IgG titers were monitored until a 50% decline in the IgG titer was observed (day 217), and then a third injection of one of three naturally occurring rDBPII alleles, Sal1, 7.18, or P; rDEKnull; or a control antigen, rPvMSP1-19, was administered (Table 1). Three weeks after the final immunization (day 245), serum anti-DBPII IgG antibody titers were again determined against the boosting antigen. Mice in all of the immunization groups showed strong immune responses to the anamnestic boost (Fig. 1). This was evident by a significant increase in antibody titers between days 217 and 245 (P = 0.0007) in all of the groups, with the exception of the DEKnull/MSP1-19 prime-boost group. No significant differences in antibody titers were observed between days 42 and 245 (P = 0.4).
FIG 1.
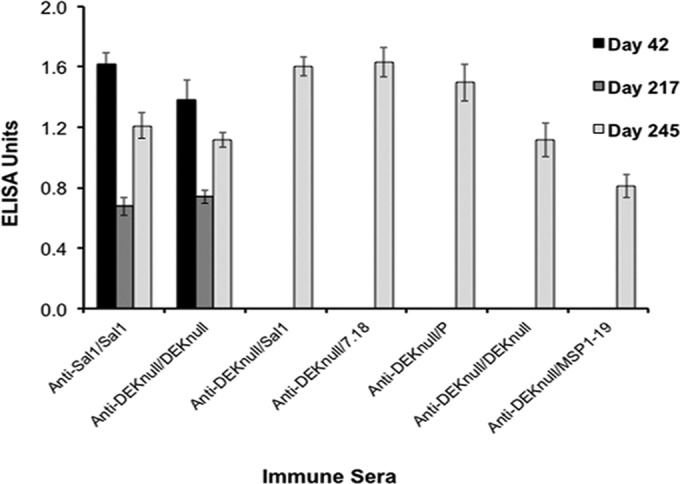
Anamnestic response to rDEKnull. Mice were primed twice 3 weeks apart with either rDEKnull or rSal1 (control), and their serum anti-DEKnull IgG titers were determined (day 42). Antibody titers were allowed to decline by ∼50% (day 217). Mice from each group were boosted on day 224 with a naturally occurring rDBPII allele (Sal1, 7.18, or P), rDEKnull, or rMSP1-19, and their serum anti-DBPII IgG titers were again determined on day 245 (final blood collection). Bars represent the mean antibody titers (EU) for reactivity of immune sera from each group at a 1 × 105 dilution against the boosting antigen for that group. Error bars represent standard errors.
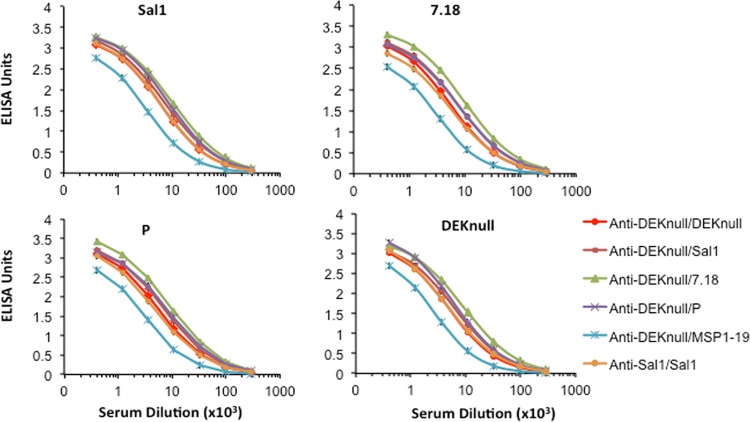
Anti-DBPII reactivity profiles.
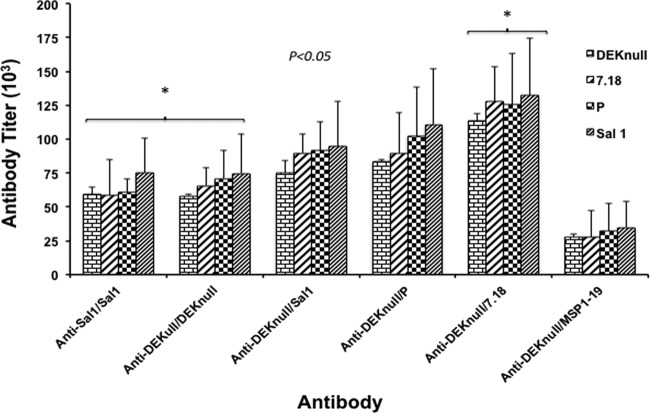
Sera from the anamnestic boosts were evaluated for cross-reactivity with heterologous rDBPII alleles, as well as rPvMSP1-19. All the immunization groups elicited high levels of cross-reactive anti-DBPII antibody responses against heterologous DBPII alleles (Fig. 2). With the exception of the anti-DEKnull/MSP1-19 group, there was no response to rPvMSP1-19 (not shown). The DEKnull-primed and MSP1-19-boost mice showed lower levels of reactivity to rDBPII. We used 1.5 EU as a basis to determine potential differences in antigen-specific anti-DBPII antibody responses. Similar anti-DBPII reactivity profiles were observed for each immune serum against the respective rDBPII alleles (Fig. 3). Only one prime-boost combination (DEKnull/7.18) produced an anamnestic response with antibody titers greater than the homologous prime-boost vaccinations of Sal1/Sal1 and DEKnull/DEKnull (Bonferroni multiple-comparison adjustment, P < 0.05). Interestingly, the DEKnull-primed–heterologous-DBPII-boosted mice showed anamnestic responses to all of the native alleles relatively higher than those of the Sal1/Sal1 homologous prime-boost mice. No anti-DBPII boosting response was observed with the rDEKnull-primed rPvMSP1-19 heterologous-boost mice; however, this group did produce a high anti-MSP1-19 response (not shown).
FIG 2.
Anti-DBPII reactivity profiles. Antisera from the different immunization groups were evaluated in an ELISA by endpoint dilution for cross-reactivity with variant recombinant DBPII alleles. Antigen preparations (2 μg/ml) were allowed to adsorb to the wells of microtiter plates and then allowed to react with different dilutions of antiserum from individual mice. Each curve is a four-parameter logistic regression curve for antisera from each group (n = 14) against the different alleles, and error bars represent standard deviations.
FIG 3.
Quantitative analysis of anti-DBPII antiserum binding specificity. The binding specificity of antiserum from each immunization group was compared against that of recombinant Sal1, 7.18, P, and DEKnull by ELISA. Antibody titers were calculated as the serum dilution required to achieve 1.5 EU. Each bar represents the titer for each antiserum against a specific recombinant DBPII allele, and error bars indicate standard deviations. Asterisks indicate significant differences in antibody titers between the two groups.
Lymphocyte proliferative response.
The immunogenicity of the different antigens was further assessed for a T-cell response. Cultured splenocytes from mice primed with rDEKnull and boosted with the different native rDBPII alleles were stimulated with the same antigens used in the boosting immunization, and antigen-specific T-cell proliferation was determined. The number of proliferating cells was determined as the ratio of cells stimulated with antigens to cells stimulated with culture medium alone. Irrespective of the stimulating antigen, all the splenocytes from the DEKnull-primed mice showed a 3- to 4-fold increase in proliferation (SI, 2.9 to 3.9) compared to that of adjuvant controls (SI, 0.9), but this was significantly smaller than that of the Sal1/Sal1 homologous prime-boost group (SI, 5.3) (P = 0.04). Splenocytes from the Sal1 homologous prime-boost mice stimulated with ConA (control) produced the highest level of proliferation, as expected (Fig. 4).
FIG 4.
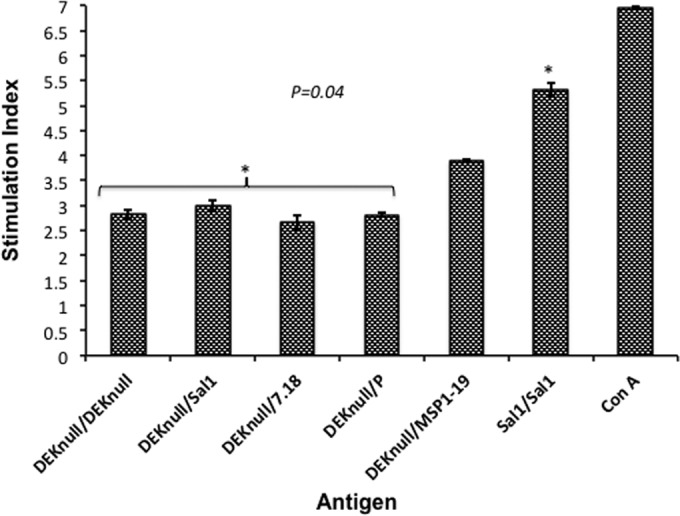
Lymphocyte proliferation. Groups of BALB/c mice were primed with rDEKnull and boosted with recombinant DEKnull, Sal1, 7.18, P, or MSP1-19. An rSal1 prime-boost group served as a control. Cultured splenocytes harvested 3 weeks after boosting were stimulated with the booster antigen. The T-cell proliferative response was quantified by an MTS assay, and the SI was determined as the ratio of the absorbance at 490 nm of stimulated cells to that of unstimulated cells. Bars represent mean SI values ± standard deviations of triplicate wells. The asterisks indicate significant SI differences between the control group (Sal1-Sal1 prime-boost) and the DEKnull prime–heterologous-boost groups.
Assessment of anti-DBPII functional activity.
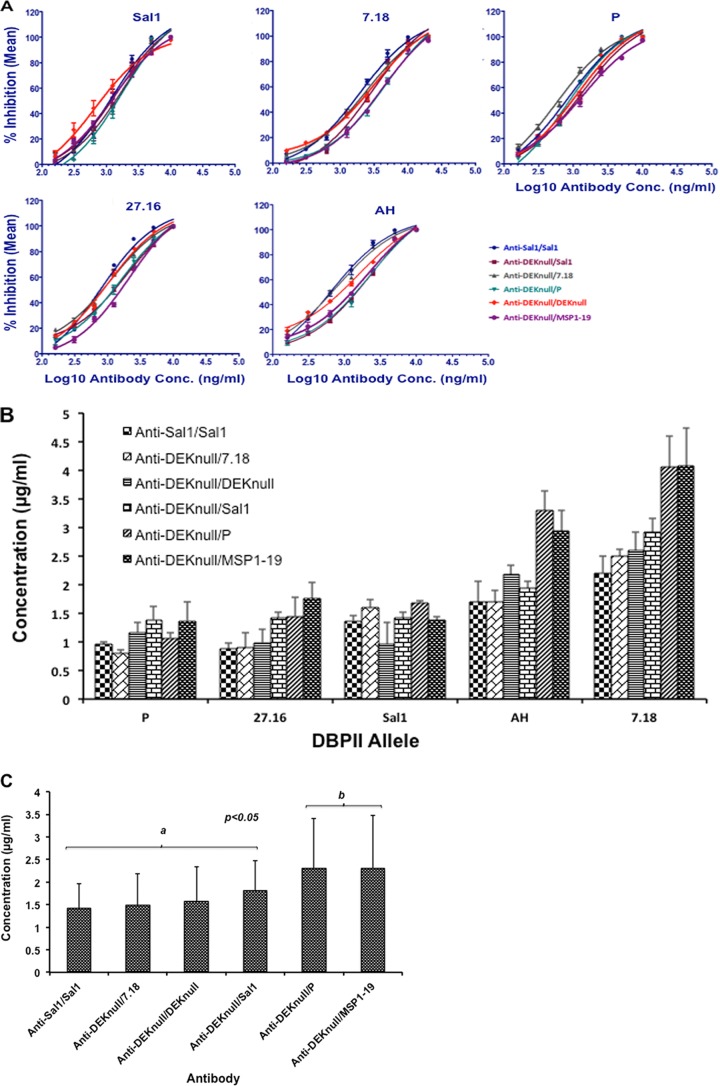
The purified total IgG fraction of pooled sera from final bleeds (day 245 sera) of each immunization group was evaluated for inhibition of COS-7 cell surface-expressed DBPII alleles from binding to human erythrocytes in a standard COS-7 cell assay. Inhibition of DBPII-erythrocyte binding was determined by assessing the number of COS-7 cells with adherent erythrocytes (rosettes) in cultures of transfected COS-7 cells incubated in the presence of different concentrations of purified total serum IgG from the immunized mouse groups relative to IgG from preimmune serum. A dose-dependent anti-DBP binding-inhibitory response was observed with antibodies from each immunization group (Fig. 5A). No inhibition was observed with total IgG from control mice immunized with MSP1-19 alone (not shown). The 50% inhibitory concentration (IC50) was used as a basis for determining the anti-DBPII functional ability of each antibody group to inhibit DBPII-erythrocyte binding (Fig. 5B). Each antibody group showed variations in anti-DBPII inhibitory responses against the different alleles, with the 7.18 allele being the most resistant to inhibition, followed by the AH allele. Statistical analysis with the Bonferroni multiple-comparison adjustment grouped the different antibodies into two distinct groups (a and b; P < 0.05). The anti-DEKnull-primed recombinant P-boost antibodies showed no boosting effect since the inhibitory effect was similar to that of the control anti-DEKnull/MSP1-19 antibodies (Fig. 5C).
FIG 5.
Inhibition of erythrocyte binding to rDBPII expressed on COS-7 cells. Purified total serum IgG from the different prime-boost groups was tested for inhibition of DBPII-erythrocyte binding against a panel of COS-7 cell-expressed DBPII alleles by endpoint dilution. A monolayer of transfected COS-7 cells expressing rDBPII from five different alleles was incubated with the purified serum IgG at different concentrations prior to the addition of human erythrocytes. Binding was scored by counting rosettes in 30 microscopic fields at a magnification of ×200. Percent binding inhibition was determined relative to that of the purified IgG from preimmune sera used as a control. (A) Charts show nonlinear regression curves for the inhibitory activities of the different antibodies against each DBPII allele. Each antibody concentration was tested in triplicate for two independent experiments. (B) Quantitative analysis of anti-DBPII binding inhibition. Bars represent the IC50s of each antibody against individual alleles. (C) Multiple comparisons of anti-DBPII binding-inhibitory responses. The overall inhibitory responses of serum anti-DBPII IgG from each group against all five COS-7-expressed alleles were compared by Bonferroni multiple-comparison adjustment. Bars represent the mean IC50s of each antibody against all the natural alleles. Antibodies were classified into two inhibitory groups (a and b), with a statistically significant difference in inhibitory responses between the groups. Error bars represent standard deviations.
DISCUSSION
It has been established that immunodominant variant epitopes tend to create an inherent bias toward a strain-specific immune response (34, 35) and limit the induction of an immune response toward more conserved protective epitopes. Correspondingly, immunization with a single P. vivax DBPII allele results in strain-specific protective immunity (56). This is also true for the P. falciparum vaccine candidate PfAMA1 (57, 58). On the other hand, it has been demonstrated that antibodies to epitopes common to different DBPII alleles (37) or PfAMA1 alleles are relevant for the induction of broader parasite strain inhibition (59, 60). An effective DBP vaccine therefore needs to focus the immune response towards conserved epitopes that are targets of neutralizing inhibitory antibodies. Recently, we demonstrated that a synthetic DBPII vaccine, DEKnull, lacking a dominant variant B-cell epitope induced inhibitory antibodies to shared neutralizing epitopes on native Sal1 from which it was derived (36). The results demonstrated that removal of the dominant variant epitope from the DEKnull vaccine lowered the immunogenicity of DBPII, but DEKnull immunization produced a more consistent anti-DBPII response against diverse DBPII alleles in an in vitro erythrocyte binding inhibition assay than three single native alleles, confirming that anti-DBP antibodies target shared neutralizing epitopes on DBPII (37).
The development of immunological memory even after a long period of nonexposure forms the basis of a successful vaccine in inducing long-term protective immunity against a given pathogen (61). Therefore, for an immunogen to serve as an effective vaccine, it should be able to produce an enhanced memory response to all of the native variants of the pathogen that either completely prevents reinfection or greatly reduces the severity of disease. In regions where malaria is endemic, individuals develop anti-DBP inhibitory antibodies following natural exposure (17, 21); however, there is no long-lasting protective immunologic memory in the absence of continued exposure to infection (17, 43–46) and sometimes there is failure to consistently boost upon reinfection (47). To further characterize the immunogenicity and vaccine potential of synthetic DEKnull, we conducted a prime-boost immunization study to evaluate the potential of rDEKnull vaccine to induce a B-cell memory response, as well as a T-cell response to naturally occurring DBPII alleles following primary DEKnull vaccination. Mice were immunized twice with rDEKnull, and serum IgG antibody titers were allowed to decline to ∼50% before a boost with a natural DBPII allele. Our data demonstrate that DEKnull prime–heterologous-boost mice generally produced an anamnestic response to native DBPII alleles better than that of native Sal1/Sal1 homologous prime-boost mice (Fig. 3). However, this was not reflected in the anti-DBPII inhibitory responses, as multiple-comparison analysis showed no differences in inhibitory responses between antibodies from the different immunization groups, with the exception of the DEKnull/P prime-boost group (Fig. 5C). This observation supports previous data that showed no correlation between the antibody response to DBPII and inhibition of DBPII-erythrocyte binding (17, 48). Antibodies from all of the vaccination groups showed similar anti-DBPII-specific responses to each native allele, suggesting that the antibodies bind to similar epitopes on DBPII. Even though the DEKnull prime–heterologous-boost groups induced a relatively stronger antibody response to all DBPII alleles than the Sal1/Sal1 homologous prime-boost control group, only the DEKnull/7.18 prime-boost immunization induced an anamnestic response with antibody titers significantly higher (P < 0.05) than those of the Sal1/Sal1 and DEKnull/DEKnull homologous prime-boost groups (Fig. 3).
Previous studies have demonstrated that DBPII contains a cluster of T-cell epitopes that are recognized by individuals in areas where malaria is endemic, with a correlation between cellular immunity and protection and the prevalence and intensity of P. vivax infection (30, 62). This implies that cellular immune responses to DBPII play a role in the development of acquired immunity. To examine the cellular immune response to DEKnull, T-cell responses were determined by lymphocyte proliferation assays following restimulation of splenocytes from each immunization group with the different naturally occurring rDBPII antigens. The Sal1/Sal1 homologous prime-boost group induced a significantly stronger proliferative response than the DEKnull homologous and heterologous prime-boost groups, suggesting that the “DEK” epitope in DBPII represents an important T-cell epitope on DBPII.
We also evaluated the potential biological and functional anti-DBPII activity of the antibodies from the various groups in an in vitro erythrocyte binding inhibition assay. Differences were observed in the anti-DBPII binding-inhibitory responses consistent with anti-DBPII antibodies binding different functional epitopes within the different alleles. No significant difference in inhibition was observed for any antibody group against the Sal1, 27.16, and P alleles. The 7.18 allele was the most refractory to inhibition, followed by the AH allele. These two alleles differ by just one amino acid and so cluster more closely to each other than the other natural alleles (Table 2). These data are in line with other studies that demonstrate that variation in DBPII plays a critical role in altering its antigenic properties and confers significant differences in sensitivity to inhibition by serum antibodies, resulting in a bias toward strain-specific immunity (56). Pairwise comparisons with a Bonferroni multiple-comparison adjustment classified the different antibodies into two statistically significantly different groups (Fig. 5C), with group a antibodies showing stronger inhibitory activity than group b antibodies (P < 0.05). Together, the data are consistent with our hypothetical framework that variation is an evasion mechanism responsible for strain-specific immunity and that stable broadly neutralizing immunity is achieved when antibodies target functionally conserved epitopes, thereby blocking erythrocyte binding and invasion.
TABLE 2.
Panel of DBPII alleles used for protein expression and COS 7 assay
| DBPII allele | Accession no. | Residuea at position: |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 308 | 333 | 371 | 375 | 384 | 385 | 386 | 387 | 388 | 389 | 390 | 391 | 392 | 393 | 417 | 424 | 437 | 447 | 992 | 503 | ||
| DBPII-Sal1 | P22290.2 | R | L | K | N | D | E | K | A | Q | Q | R | R | K | Q | N | L | W | S | K | I |
| DEKnull | NAb | R | . | . | . | A | A | T | A | A | T | S | R | T | S | . | . | . | . | . | . |
| DBPII-27.16c | AAL79076 | S | . | . | . | G | . | . | . | . | . | H | . | . | . | . | I | . | K | E | . |
| DBPII-7.18 | AAL79051.1 | S | . | . | . | G | . | Q | . | . | . | . | . | . | . | K | I | R | . | . | K |
| DBPII-AHc | AAY34130.1 | S | . | E | . | G | . | Q | . | . | . | . | . | . | . | K | I | R | . | . | K |
| DBPII-P | AAL79073.1 | R | F | . | D | G | K | N | . | . | . | H | . | . | . | K | I | . | . | . | K |
Differences in amino acid residues between the different alleles are shown. A dot indicates that the residue is the same as that in DBPII-Sal1.
NA, accession number is not available.
Allele used for COS-7 assay only.
In summary, we have demonstrated that synthetic DEKnull is able to elicit immunological memory responses similar to those elicited by naturally occurring DBPII alleles. While the anti-DBPII inhibitory response induced by rDEKnull is lower than that to DBPII Sal1, the functional activity of these antibodies demonstrated a broader coverage of diverse DBPII alleles. Further investigation is necessary to enhance the immunogenicity of DEKnull and broaden its specificity.
ACKNOWLEDGMENTS
This work was funded by NIAID/DMID contract N01-AI-054210 to Science Application International Cooperation under subcontract PO10035958 and grant R01AI064478 (to J.H.A.).
J. Adams, F. Ntumngia, S. Barnes, A. McHenry, P. Chootong, and J. Schloegel are inventors listed in U.S. provisional patent application 61/525,412, titled “Design and immunogenicity of a novel synthetic antigen based on the ligand domain of the Plasmodium vivax Duffy binding protein.” We have no commercial or other association that poses a conflict of interest.
Footnotes
Published ahead of print 25 June 2014
REFERENCES
- 1.Branch OH, Udhayakumar V, Hightower AW, Oloo AJ, Hawley WA, Nahlen BL, Bloland PB, Kaslow DC, Lal AA. 1998. A longitudinal investigation of IgG and IgM antibody responses to the merozoite surface protein-1 19-kiloDalton domain of Plasmodium falciparum in pregnant women and infants: associations with febrile illness, parasitemia, and anemia. Am. J. Trop. Med. Hyg. 58:211–219 [DOI] [PubMed] [Google Scholar]
- 2.King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, Cole-Tobian JL, Mueller I, Chitnis CE. 2008. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc. Natl. Acad. Sci. U. S. A. 105:8363–8368. 10.1073/pnas.0800371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen S, McGregor IA, Carrington S. 1961. Gamma-globulin and acquired immunity to human malaria. Nature 192:733–737. 10.1038/192733a0 [DOI] [PubMed] [Google Scholar]
- 4.Clyde DF, Most H, McCarthy VC, Vanderberg JP. 1973. Immunization of man against sporozoite-induced falciparum malaria. Am. J. Med. Sci. 266:169–177. 10.1097/00000441-197309000-00002 [DOI] [PubMed] [Google Scholar]
- 5.Grimberg BT, Udomsangpetch R, Xainli J, McHenry A, Panichakul T, Sattabongkot J, Cui L, Bockarie M, Chitnis C, Adams J, Zimmerman PA, King CL. 2007. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 4(12):e337. 10.1371/journal.pmed.0040337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh AP, Puri SK, Chitnis CE. 2002. Antibodies raised against receptor-binding domain of Plasmodium knowlesi Duffy binding protein inhibit erythrocyte invasion. Mol. Biochem. Parasitol. 121:21–31. 10.1016/S0166-6851(02)00017-8 [DOI] [PubMed] [Google Scholar]
- 7.Pattnaik P, Shakri AR, Singh S, Goel S, Mukherjee P, Chitnis CE. 2007. Immunogenicity of a recombinant malaria vaccine based on receptor binding domain of Plasmodium falciparum EBA-175. Vaccine 25:806–813. 10.1016/j.vaccine.2006.09.048 [DOI] [PubMed] [Google Scholar]
- 8.Woehlbier U, Epp C, Hackett F, Blackman MJ, Bujard H. 2010. Antibodies against multiple merozoite surface antigens of the human malaria parasite Plasmodium falciparum inhibit parasite maturation and red blood cell invasion. Malar. J. 9:77. 10.1186/1475-2875-9-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. U. S. A. 89:7085–7089. 10.1073/pnas.89.15.7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnwell JW, Wertheimer SP. 1989. Plasmodium vivax: merozoite antigens, the Duffy blood group, and erythrocyte invasion. Prog. Clin. Biol. Res. 313:1–11 [PubMed] [Google Scholar]
- 11.Dvorak JA, Miller LH, Whitehouse WC, Shiroishi T. 1975. Invasion of erythrocytes by malaria merozoites. Science 187:748–750. 10.1126/science.803712 [DOI] [PubMed] [Google Scholar]
- 12.Aikawa M, Miller LH, Johnson J, Rabbege J. 1978. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J. Cell Biol. 77:72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller LH, Mason SJ, Clyde DF, McGinniss MH. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295:302–304 [DOI] [PubMed] [Google Scholar]
- 14.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. 1975. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189:561–563. 10.1126/science.1145213 [DOI] [PubMed] [Google Scholar]
- 15.Xainli J, Cole-Tobian JL, Baisor M, Kastens W, Bockarie M, Yazdani SS, Chitnis CE, Adams JH, King CL. 2003. Epitope-specific humoral immunity to Plasmodium vivax Duffy binding protein. Infect. Immun. 71:2508–2515. 10.1128/IAI.71.5.2508-2515.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerávolo IP, Bruna-Romero O, Braga EM, Fontes CJ, Brito CF, Souza JM, Krettli AU, Adams JH, Carvalho LH. 2005. Anti-Plasmodium vivax Duffy binding protein antibodies measure exposure to malaria in the Brazilian Amazon. Am. J. Trop. Med. Hyg. 72:675–681 [PubMed] [Google Scholar]
- 17.Chootong P, Ntumngia FB, VanBuskirk KM, Xainli J, Cole-Tobian JL, Campbell CO, Fraser TS, King CL, Adams JH. 2010. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect. Immun. 78:1089–1095. 10.1128/IAI.01036-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutta S, Daugherty JR, Ware LA, Lanar DE, Ockenhouse CF. 2000. Expression, purification and characterization of a functional region of the Plasmodium vivax Duffy binding protein. Mol. Biochem. Parasitol. 109:179–184. 10.1016/S0166-6851(00)00244-9 [DOI] [PubMed] [Google Scholar]
- 19.Michon PA, Arevalo-Herrera M, Fraser T, Herrera S, Adams JH. 1998. Serologic responses to recombinant Plasmodium vivax Duffy binding protein in a Colombian village. Am. J. Trop. Med. Hyg. 59:597–599 [DOI] [PubMed] [Google Scholar]
- 20.Yazdani SS, Shakri AR, Chitnis CE. 2004. A high cell density fermentation strategy to produce recombinant malarial antigen in E. coli. Biotechnol. Lett. 26:1891–1895. 10.1007/s10529-004-6040-4 [DOI] [PubMed] [Google Scholar]
- 21.Michon P, Fraser T, Adams JH. 2000. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect. Immun. 68:3164–3171. 10.1128/IAI.68.6.3164-3171.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanBuskirk KM, Sevova E, Adams JH. 2004. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc. Natl. Acad. Sci. U. S. A. 101:15754–15759. 10.1073/pnas.0405421101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. 1996. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J. Exp. Med. 184:1531–1536. 10.1084/jem.184.4.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. 2006. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature 439:741–744. 10.1038/nature04443 [DOI] [PubMed] [Google Scholar]
- 25.Batchelor JD, Zahm JA, Tolia NH. 2011. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat. Struct. Mol. Biol. 18:908–914. 10.1038/nsmb.2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. 1996. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol. Biochem. Parasitol. 78:269–272. 10.1016/S0166-6851(96)02611-4 [DOI] [PubMed] [Google Scholar]
- 27.Hans D, Pattnaik P, Bhattacharyya A, Shakri AR, Yazdani SS, Sharma M, Choe H, Farzan M, Chitnis CE. 2005. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol. Microbiol. 55:1423–1434. 10.1111/j.1365-2958.2005.04484.x [DOI] [PubMed] [Google Scholar]
- 28.Ranjan A, Chitnis CE. 1999. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 96:14067–14072. 10.1073/pnas.96.24.14067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuboi T, Kappe SH, al-Yaman F, Prickett MD, Alpers M, Adams JH. 1994. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding protein. Infect. Immun. 62:5581–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole-Tobian J, King CL. 2003. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol. Biochem. Parasitol. 127:121–132. 10.1016/S0166-6851(02)00327-4 [DOI] [PubMed] [Google Scholar]
- 31.Ntumngia FB, McHenry AM, Barnwell JW, Cole-Tobian J, King CL, Adams JH. 2009. Genetic variation among Plasmodium vivax isolates adapted to non-human primates and the implication for vaccine development. Am. J. Trop. Med. Hyg. 80:218–227 [PMC free article] [PubMed] [Google Scholar]
- 32.Xainli J, Adams JH, King CL. 2000. The erythrocyte binding motif of Plasmodium vivax Duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol. Biochem. Parasitol. 111:253–260. 10.1016/S0166-6851(00)00315-7 [DOI] [PubMed] [Google Scholar]
- 33.Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, Mueller I, King CL. 2009. Strain-specific Duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect. Immun. 77:4009–4017. 10.1128/IAI.00158-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welsh RM, Fujinami RS. 2007. Pathogenic epitopes, heterologous immunity and vaccine design. Nat. Rev. Microbiol. 5:555–563. 10.1038/nrmicro1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobin GJ, Trujillo JD, Bushnell RV, Lin G, Chaudhuri AR, Long J, Barrera J, Pena L, Grubman MJ, Nara PL. 2008. Deceptive imprinting and immune refocusing in vaccine design. Vaccine 26:6189–6199. 10.1016/j.vaccine.2008.09.080 [DOI] [PubMed] [Google Scholar]
- 36.Ntumngia FB, Adams JH. 2012. Design and immunogenicity of a novel synthetic antigen based on the ligand domain of the Plasmodium vivax Duffy binding protein. Clin. Vaccine Immunol. 19:30–36. 10.1128/CVI.05466-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ntumngia FB, Schloegel J, McHenry AM, Barnes SJ, George MT, Kennedy S, Adams JH. 2013. Immunogenicity of single versus mixed allele vaccines of Plasmodium vivax Duffy binding protein region II. Vaccine 31:4382–4388. 10.1016/j.vaccine.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavanagh DR, Dodoo D, Hviid L, Kurtzhals JA, Theander TG, Akanmori BD, Polley S, Conway DJ, Koram K, McBride JS. 2004. Antibodies to the N-terminal block 2 of Plasmodium falciparum merozoite surface protein 1 are associated with protection against clinical malaria. Infect. Immun. 72:6492–6502. 10.1128/IAI.72.11.6492-6502.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conway DJ, Cavanagh DR, Tanabe K, Roper C, Mikes ZS, Sakihama N, Bojang KA, Oduola AM, Kremsner PG, Arnot DE, Greenwood BM, McBride JS. 2000. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat. Med. 6:689–692. 10.1038/76272 [DOI] [PubMed] [Google Scholar]
- 40.Osier FH, Fegan G, Polley SD, Murungi L, Verra F, Tetteh KK, Lowe B, Mwangi T, Bull Thomas PCAW, Cavanagh DR, McBride JS, Lanar DE, Mackinnon MJ, Conway DJ, Marsh K. 2008. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect. Immun. 76:2240–2248. 10.1128/IAI.01585-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cole-Tobian JL, Cortes A, Baisor M, Kastens W, Xainli J, Bockarie M, Adams JH, King CL. 2002. Age-acquired immunity to a Plasmodium vivax invasion ligand, the Duffy binding protein. J. Infect. Dis. 186:531–539. 10.1086/341776 [DOI] [PubMed] [Google Scholar]
- 42.Fraser T, Michon P, Barnwell JW, Noe AR, Al-Yaman F, Kaslow DC, Adams JH. 1997. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect. Immun. 65:2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerávolo IP, Sanchez BA, Sousa TN, Guerra BM, Soares IS, Braga EM, McHenry AM, Adams JH, Brito CF, Carvalho LH. 2009. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin. Exp. Immunol. 156:502–510. 10.1111/j.1365-2249.2009.03931.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deloron P, Chougnet C. 1992. Is immunity to malaria really short-lived? Parasitol. Today 8:375–378. 10.1016/0169-4758(92)90174-Z [DOI] [PubMed] [Google Scholar]
- 45.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. 2008. Immunity to malaria: more questions than answers. Nat. Immunol. 9:725–732. 10.1038/ni.f.205 [DOI] [PubMed] [Google Scholar]
- 46.Snow RW, Marsh K. 2002. The consequences of reducing transmission of Plasmodium falciparum in Africa. Adv. Parasitol. 52:235–264. 10.1016/S0065-308X(02)52013-3 [DOI] [PubMed] [Google Scholar]
- 47.Bejon P, Mwacharo J, Kai O, Mwangi T, Milligan P, Todryk S, Keating S, Lang T, Lowe B, Gikonyo C, Molyneux C, Fegan G, Gilbert SC, Peshu N, Marsh K, Hill AV. 2006. A phase 2b randomised trial of the candidate malaria vaccines FP9 ME-TRAP and MVA ME-TRAP among children in Kenya. PLoS Clin. Trials 1(6):e29. 10.1371/journal.pctr.0010029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ntumngia FB, Schloegel J, Barnes SJ, McHenry AM, Singh S, King CL, Adams JH. 2012. Conserved and variant epitopes of Plasmodium vivax Duffy binding protein as targets of inhibitory monoclonal antibodies. Infect. Immun. 80:1203–1208. 10.1128/IAI.05924-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran TM, Moreno A, Yazdani SS, Chitnis CE, Barnwell JW, Galinski MR. 2005. Detection of a Plasmodium vivax erythrocyte binding protein by flow cytometry. Cytometry A 63:59–66. 10.1002/cyto.a.20098 [DOI] [PubMed] [Google Scholar]
- 50.Singh S, Pandey K, Chattopadhayay R, Yazdani SS, Lynn A, Bharadwaj A, Ranjan A, Chitnis C. 2001. Biochemical, biophysical, and functional characterization of bacterially expressed and refolded receptor binding domain of Plasmodium vivax Duffy-binding protein. J. Biol. Chem. 276:17111–17116. 10.1074/jbc.M101531200 [DOI] [PubMed] [Google Scholar]
- 51.Mullen GE, Giersing BK, Ajose-Popoola O, Davis HL, Kothe C, Zhou H, Aebig J, Dobrescu G, Saul A, Long CA. 2006. Enhancement of functional antibody responses to AMA1-C1/Alhydrogel, a Plasmodium falciparum malaria vaccine, with CpG oligodeoxynucleotide. Vaccine 24:2497–2505. 10.1016/j.vaccine.2005.12.034 [DOI] [PubMed] [Google Scholar]
- 52.Alaro JR, Lynch MM, Burns JM., Jr 2010. Protective immune responses elicited by immunization with a chimeric blood-stage malaria vaccine persist but are not boosted by Plasmodium yoelii challenge infection. Vaccine 28:6876–6884. 10.1016/j.vaccine.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng YH, Sun Z, Yang XL, Bao L. 2010. Improved immunogenicity of recombinant Mycobacterium bovis bacillus Calmette-Guerin strains expressing fusion protein Ag85A-ESAT-6 of Mycobacterium tuberculosis. Scand. J. Immunol. 72:332–338. 10.1111/j.1365-3083.2010.02444.x [DOI] [PubMed] [Google Scholar]
- 54.Michon P, Woolley I, Wood EM, Kastens W, Zimmerman PA, Adams JH. 2001. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for blood-stage infection. FEBS Lett. 495:111–114. 10.1016/S0014-5793(01)02370-5 [DOI] [PubMed] [Google Scholar]
- 55.Chitnis CE, Miller LH. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497–506. 10.1084/jem.180.2.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, King CL, Adams JH. 2004. Antigenic drift in the ligand domain of Plasmodium vivax Duffy binding protein confers resistance to inhibitory antibodies. J. Infect. Dis. 190:1556–1562. 10.1086/424852 [DOI] [PubMed] [Google Scholar]
- 57.Remarque EJ, Roestenberg M, Younis S, Walraven V, van der Werff N, Faber BW, Leroy O, Sauerwein R, Kocken CH, Thomas AW. 2012. Humoral immune responses to a single allele PfAMA1 vaccine in healthy malaria-naive adults. PLoS One 7(6):e38898. 10.1371/journal.pone.0038898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hodder AN, Crewther PE, Anders RF. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286–3294. 10.1128/IAI.69.5.3286-3294.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kusi KA, Faber BW, Thomas AW, Remarque EJ. 2009. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One 4(12):e8110. 10.1371/journal.pone.0008110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kusi KA, Faber BW, Riasat V, Thomas AW, Kocken CH, Remarque EJ. 2010. Generation of humoral immune responses to multi-allele PfAMA1 vaccines; effect of adjuvant and number of component alleles on the breadth of response. PLoS One 5(11):e15391. 10.1371/journal.pone.0015391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaech SM, Wherry EJ, Ahmed R. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2:251–262. 10.1038/nri778 [DOI] [PubMed] [Google Scholar]
- 62.Xainli J, Baisor M, Kastens W, Bockarie M, Adams JH, King CL. 2002. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J. Immunol. 169:3200–3207. 10.4049/jimmunol.169.6.3200 [DOI] [PubMed] [Google Scholar]